ABSTRACT
Pyridoxal 5′-phosphate (PLP) is an essential cofactor for organisms in all three domains of life. Despite the central role of PLP, many aspects of vitamin B6 metabolism, including its integration with other biological pathways, are not fully understood. In this study, we examined the metabolic perturbations caused by the vitamin B6 antagonist 4-deoxypyridoxine (dPN) in a ptsJ mutant of Salmonella enterica serovar Typhimurium LT2. Our data suggest that PdxK (pyridoxal/pyridoxine/pyridoxamine kinase [EC 2.7.1.35]) phosphorylates dPN to 4-deoxypyridoxine 5′-phosphate (dPNP), which in turn can compromise the de novo biosynthesis of PLP. The data are consistent with the hypothesis that accumulated dPNP inhibits GlyA (serine hydroxymethyltransferase [EC 2.1.2.1]) and/or GcvP (glycine decarboxylase [EC 1.4.4.2]), two PLP-dependent enzymes involved in the generation of one-carbon units. Our data suggest that this inhibition leads to reduced flux to coenzyme A (CoA) precursors and subsequently decreased synthesis of CoA and thiamine. This study uncovers a link between vitamin B6 metabolism and the biosynthesis of CoA and thiamine, highlighting the integration of biochemical pathways in microbes.
IMPORTANCE PLP is a ubiquitous cofactor required by enzymes in diverse metabolic networks. The data presented here expand our understanding of the toxic effects of dPN, a vitamin B6 antagonist that is often used to mimic vitamin B6 deficiency and to study PLP-dependent enzyme kinetics. In addition to de novo PLP biosynthesis, we define a metabolic connection between vitamin B6 metabolism and synthesis of thiamine and CoA. This work provides a foundation for the use of dPN to study vitamin B6 metabolism in other organisms.
KEYWORDS: pyridoxal 5′-phosphate, vitamin B6, 4-deoxypyridoxine, PtsJ, thiamine, CoA, coenzyme A, PLP, ptsJ
INTRODUCTION
Pyridoxal 5′-phosphate (PLP) is the biologically active form of vitamin B6 and a required cofactor in the metabolism of all organisms. Due to its unique chemical properties, PLP can facilitate diverse enzymatic reactions, including transamination, elimination, decarboxylation, and racemization (1). PLP-dependent enzymes, which are classified into seven fold types based on structural similarities (2), account for ∼4% of all activities described by the Enzyme Commission (3). Most PLP-dependent enzymes, with the exception of glycogen phosphorylases, are associated with metabolic reactions that involve amino substrates (3).
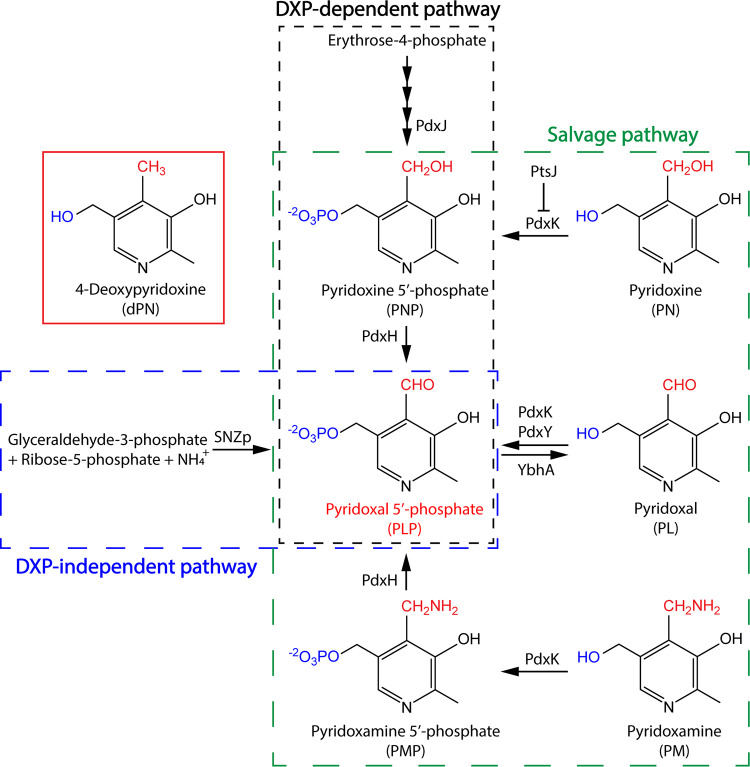
In nature, PLP is synthesized de novo via one of two biosynthetic routes (Fig. 1). The deoxyxylulose 5-phosphate (DXP)-dependent pathway is found primarily in gammaproteobacteria such as Salmonella enterica and Escherichia coli, while the DXP-independent pathway is widespread among other bacteria, fungi, and archaea (4, 5). Alternatively, PLP can be acquired from other B6 vitamers using the salvage pathway, as found in humans and animals (4, 5) (Fig. 1). Due to the reactivity of this cofactor and the prevalence of PLP-dependent enzymes in metabolic networks, perturbations in PLP pools can inhibit both PLP-independent and PLP-dependent enzymes, causing adverse metabolic consequences (6–10). Despite the importance of this cofactor, the mechanism (or mechanisms) used by cells to control PLP homeostasis is not fully understood.
FIG 1.
PLP biosynthesis and salvage. In S. enterica, PLP can be synthesized de novo via the DXP-dependent pathway (black box) or salvaged from other B6 vitamers (green box). Other organisms, such as Saccharomyces cerevisiae, use the DXP-independent pathway (blue box) to synthesize PLP in addition to salvage. Animals, including humans, lack the biosynthetic pathway and rely exclusively on salvage to acquire this cofactor. Relevant enzymes involved in biosynthesis and salvage of PLP are shown next to the arrows, each of which represents a biochemical reaction catalyzed by the corresponding enzyme.
One approach to study vitamin B6 metabolism involves the use of analogs, i.e., compounds that are structurally similar to B6 vitamers but lack the biochemical properties to aid enzyme catalysis. In particular, 4-deoxypyridoxine (dPN) (Fig. 1) and its phosphorylated derivative 4-deoxypyridoxine 5′-phosphate (dPNP) have been used as potent vitamin B6 antagonists in research for decades. Early studies in bacteria, fungi, plants, and animals focused on the inhibitory effects of dPN on growth, morphology, and the metabolism of other vitamins (11). Later work showed that dPN and dPNP could competitively inhibit the in vitro activity of the human pyridoxal (PL)/pyridoxine (PN)/pyridoxamine (PM) kinase (PdxK) (EC 2.7.1.35) (12) and pyridoxine 5′-phosphate (PNP)/pyridoxamine 5′-phosphate (PMP) oxidase (PdxH) (EC 1.4.3.5) (13), which are conserved enzymes essential for the salvage of B6 vitamers (Fig. 1). This study was initiated to understand the mechanism of dPN toxicity in S. enterica serovar Typhimurium LT2 (referred to here as S. enterica) using phenotypic and metabolite analyses. Our results uncover a link between vitamin B6 metabolism and the biosynthesis of thiamine and coenzyme A (CoA), highlighting the complexity of metabolic networks and the primary consequences of perturbed pools of B6 vitamers.
RESULTS AND DISCUSSION
Sensitivity of a ptsJ mutant to dPN is dependent on PdxK.
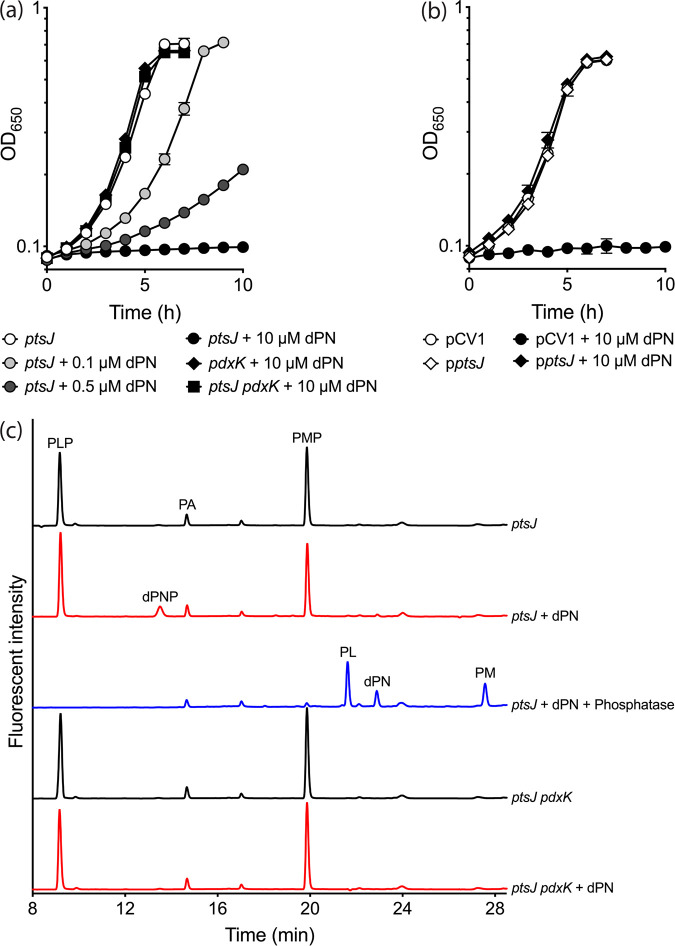
While growth of an S. enterica wild-type strain in minimal no-carbon E (NCE) glycerol was not affected by 10 μM dPN, the presence of this antagonist inhibited growth if pdxK was overexpressed in trans (data not shown). Consistently, a strain lacking PtsJ, a MocR-type transcriptional repressor that regulates expression of pdxK (14), was sensitive to dPN in minimal NCE glucose in a dose-dependent manner (Fig. 2a). A null mutation in pdxK (Fig. 2a) or expression of ptsJ in trans (Fig. 2b) overcame the growth defect caused by dPN of the ptsJ mutant. These data showed the importance of PtsJ and PdxK in modulating dPN sensitivity.
FIG 2.
dPN sensitivity of a ptsJ mutant is PdxK-dependent. (a) S. enterica ptsJ (DM17239), pdxK (DM17238), and ptsJ pdxK (DM17168) were grown in minimal NCE glucose without (white symbols) or with addition of 0.1 μM (light gray symbols), 0.5 μM (dark gray symbols), or 10 μM (black symbols) dPN. (b) ptsJ strains carrying an empty vector control (pCV1) (DM17242) or a vector expressing ptsJ in trans (pptsJ) (DM17243) were grown in minimal NCE glucose supplemented with 0.02% arabinose in the absence (white symbols) or presence (black symbols) of 10 μM dPN. (c) HPLC chromatogram of intracellular vitamin B6 content of ptsJ (DM17239) and ptsJ pdxK (DM17168) strains grown in minimal NCE glucose with or without addition of 0.5 μM dPN. Representative data from at least two independent experiments with three biological replicates are shown. Error bars depict standard deviations from the mean values.
Unphosphorylated B6 vitamers are thought to enter cells via facilitated diffusion and subsequently be trapped by phosphorylation (15–17). In addition, the human homolog of PdxK could use dPN as a substrate in vitro (18). Taken together, the data suggest that, to be toxic, dPN was phosphorylated to dPNP by PdxK. Consistent with this hypothesis, high-performance liquid chromatography (HPLC) analysis showed that a ptsJ strain grown in minimal glucose medium supplemented with dPN (0.5 μM) accumulated intracellular dPNP (∼555 ± 42 pmol/optical density at 650 nm [OD650] unit). No dPNP was detected in cells grown in the absence of dPN (Fig. 2c). When a mutation in pdxK was introduced in the pstJ background, the profiles of intracellular B6 vitamers in the absence and presence of dPN were indistinguishable. In total, these data support the conclusion that dPNP is the molecule that exerts toxic effects on S. enterica and, by extension, other organisms that possess homologs of PdxK.
DXP-dependent biosynthesis of PLP is compromised in the presence of dPNP.
dPNP inhibited both the E. coli and human homologs of PdxH by competing with the native substrates PNP and PMP in vitro (13, 19). If such inhibition were to occur in vivo, then the growth defect of the S. enterica ptsJ mutant in the presence of dPN could be due to PLP limitation resulting from decreased PdxH activity. Growth of the ptsJ mutant in the presence of dPN was restored to different degrees with supplementation with 1 μM PL, PN, or PM (Fig. 3a). However, it was unclear whether these vitamers rescued growth by competing with dPN for transport, or for PdxK as the substrates or by increasing intracellular PLP content.
FIG 3.
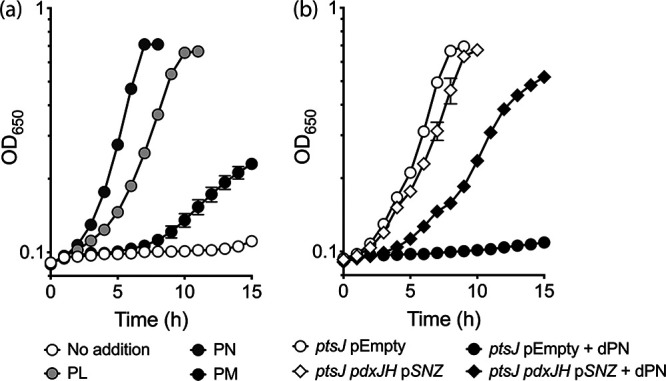
dPNP affects PLP synthesis via the DXP-dependent pathway. (a) The S. enterica ptsJ strain (DM17239) was grown in minimal NCE glucose supplemented with dPN (10 μM) in the absence (white symbols) or presence of PL (1 μM) (light gray symbols), PN (1 μM) (dark gray symbols), or PM (1 μM) (black symbols). (b) S. enterica ptsJ strains with the native DXP-dependent pathway intact (ptsJ pEmpty) (DM17244) or replaced with the S. cerevisiae DXP-independent pathway (ptsJ pdxJH pSNZ) (DM17246) were grown in minimal NCE glycerol supplemented with 0.02% arabinose in the absence (white symbols) or presence (black symbols) of dPN (10 μM). Representative data from at least two independent experiments with three biological replicates are shown. Error bars depict standard deviations from the mean values.
To help distinguish between these possibilities, the DXP-dependent PLP biosynthesis was eliminated in the ptsJ mutant by deleting pdxJ (encoding a PNP synthase [EC 2.6.99.2]) and pdxH. As a replacement for the native synthetic route, Saccharomyces cerevisiae SNZ3 (encoding a PLP synthase [EC 4.3.3.6]) was expressed in trans. The resulting strain synthesizes PLP from endogenous glyceraldehyde-3-phosphate, ribose-5-phosphate, and exogenous ammonium in the medium using Snz3p (20) (Fig. 1). Growth of ptsJ strains carrying different PLP biosynthetic pathways in the presence of dPN was examined in minimal NCE glycerol (Fig. 3b). Strains lacking ptsJ and utilizing either the native (DXP-dependent) or heterologous (Snz3p-dependent) pathway for PLP synthesis grew similarly in the absence of dPN. When dPN was added, growth inhibition of the ptsJ strain synthesizing PLP with Snz3p was significantly less severe than that of the ptsJ strain using the native pathway. These data are consistent with the hypothesis that dPNP compromises the DXP-dependent PLP synthesis, while the remaining growth defect suggests that PdxH is not the only target of dPNP toxicity.
dPNP impacts ThiC-dependent thiamine synthesis.
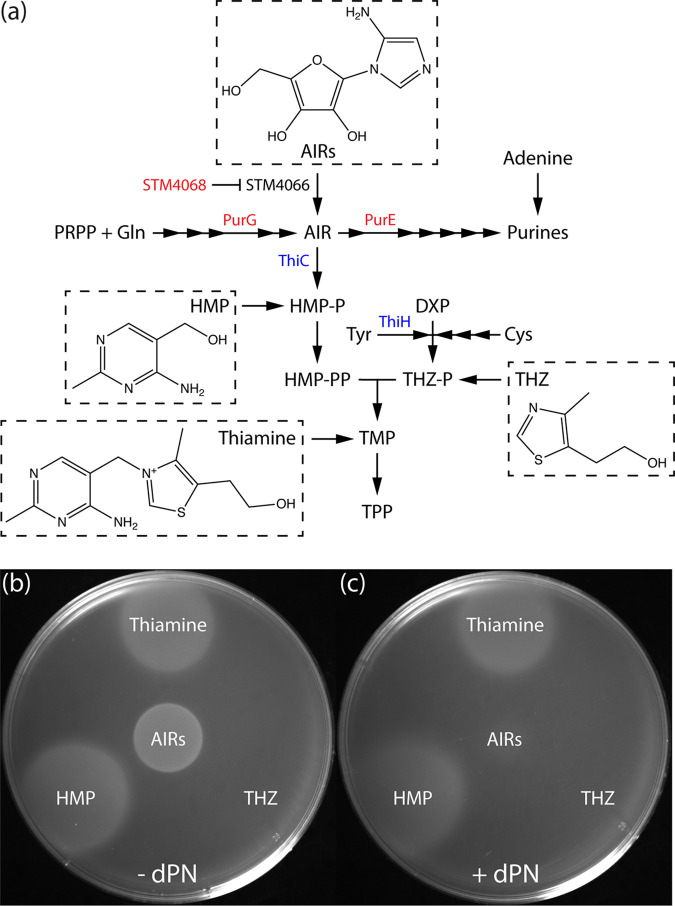
Probing the mechanism of dPNP toxicity beyond the presumed inhibition of PdxH required that the concentration of dPN be reduced to allow growth. Nutritional experiments showed that, in the presence of low levels of dPN (0.5 μM), the ptsJ strain was sensitive to adenosine, which was rescued by thiamine addition (Fig. 4). The ability of thiamine to abolish the synergistic effect of adenosine and dPN was reminiscent of previous studies, in which adenosine sensitivity indicated compromised synthesis of the pyrimidine moiety of thiamine (4-amino-5-hydroxymethyl-2-methylpyrimidine [HMP]). In those cases, adenosine reduced flux toward 5-aminoimidazole ribotide (AIR) formation, which lowered HMP synthesis only if the efficiency of the AIR→4-amino-5-hydroxymethyl-2-methylpyrimidine phosphate (HMP-P) reaction was diminished (21–24) (Fig. 5a). We hypothesized that the AIR→HMP-P conversion, catalyzed by ThiC (25, 26), was decreased in the presence of dPNP. In this scenario, the synergetic effects of adenosine and dPN would be due to reduced flux to AIR and reduced conversion of AIR to HMP-P, respectively. The effect of dPN on the efficiency of the AIR→HMP-P conversion was tested using a biosensor strain (DM7060) carrying a ptsJ mutation. The genotype of this strain allows the efficiency of the ThiC-dependent step to be assessed using 5-aminoimidazole riboside (AIRs) as a proxy (27, 28). This strain carries null alleles of purG and purE, which prevent flux to and from AIR, respectively, in de novo purine synthesis. In addition to purine auxotrophy, these mutations cause a thiamine requirement that can be rescued by supplementation of HMP or AIRs (Fig. 5a). A mutation in stm4068 enables efficient utilization of exogenous AIRs by derepressing the kinase responsible for converting AIRs to AIR (29). Thus, the amount of AIRs that allows growth in this background is a reflection of the efficiency of the AIR→HMP-P conversion by ThiC.
FIG 4.
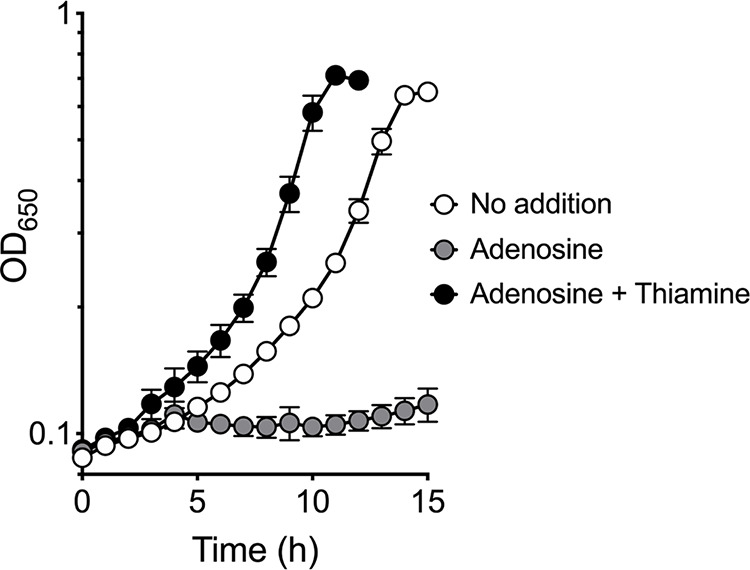
Adenosine exacerbates dPN sensitivity. The S. enterica ptsJ strain (DM17239) was grown in minimal NCE glucose supplemented with dPN (0.5 μM) in the absence (white symbols) or presence of adenosine (1 mM) (gray symbols) and thiamine (0.1 μM) (black symbols). Representative data from two independent experiments with three biological replicates are shown. Error bars depict standard deviations from the means.
FIG 5.
dPN increases the AIRs requirement for ThiC-dependent thiamine synthesis. (a) Thiamine and purine biosynthesis and salvage in S. enterica. Each arrow represents a biochemical reaction, and relevant enzymes are indicated next to the arrows. The DM7060 background carries lesions in genes that encode proteins in red. Blue indicates radical AdoMet enzymes that utilize iron-sulfur clusters. Compounds accompanied by chemical structures were used in subsequent feeding experiments. (b and c) Molten soft agar (0.7% [wt/vol]) inoculated with purGE stm4068 ptsJ culture (DM17274) was overlaid on minimal NCE glucose containing 0.4 mM adenine in the absence (b) or presence (c) of 0.1 μM dPN. Other supplementation was added by spotting 1 μL of AIRs (∼300 mM), thiamine (0.1 mM), HMP (0.1 mM), and THZ (0.1 mM) on top of the solidified soft agar layer. Representative growth from three independent experiments is shown. PRPP, phosphoribosyl pyrophosphate; HMP-PP, 4-amino-5-hydroxymethyl-2-methylpyrimidine diphosphate; THZ-P, 4-methyl-5-(2-hydroxyethyl)-thiazole phosphate; TMP, thiamine monophosphate; TPP, thiamine pyrophosphate; Gln, l-glutamine; Tyr, l-tyrosine; Cys, l-cysteine.
The AIRs requirement of the purGE stm4068 ptsJ strain was assessed with soft agar overlay on minimal NCE glucose medium supplemented with adenine as a source for purines (Fig. 5b and c). In the absence of dPN, the mutant grew with addition of AIRs, HMP, or thiamine (Fig. 5b), as expected. When dPN (0.1 μM) was added to the medium, overall growth was not as robust. Nonetheless, it was still clear that growth was not supported by the amount of AIRs that allowed growth in the absence of dPN (Fig. 5c). These results indicated that the ptsJ mutant required more AIRs to grow in the presence of dPN, supporting the conclusion that the efficiency of AIR→HMP-P conversion by ThiC was compromised in the presence of dPNP.
dPNP inhibits the pantoate branch of CoA biosynthesis.
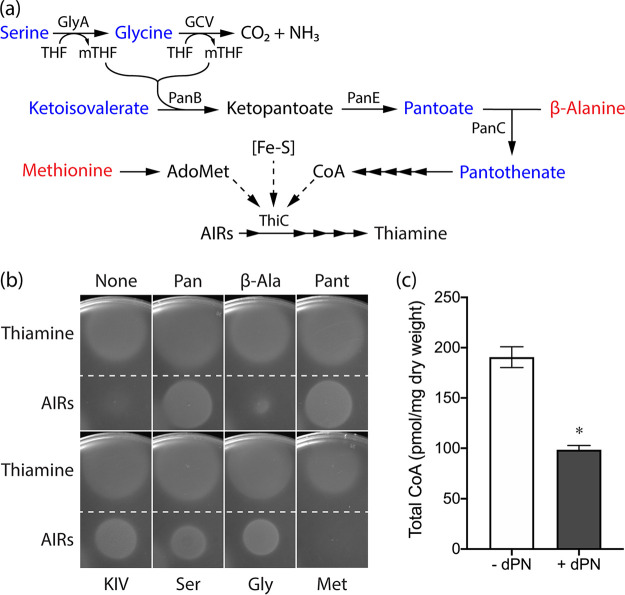
Defects in several metabolic processes affect ThiC activity in vivo, including iron-sulfur cluster metabolism (28), synthesis of S-adenosylmethionine (AdoMet) from methionine (30), and intracellular CoA levels (31) (Fig. 6a). If dPNP caused a defect in iron-sulfur cluster synthesis/repair, both the HMP and thiazole (THZ) branches of thiamine synthesis would be expected to be compromised (28, 32–35) (Fig. 5a). However, only HMP-P synthesis was affected by dPNP (Fig. 5c). If the synthesis of AdoMet and/or CoA was compromised by dPNP, then methionine or pantothenate, respectively, should alter growth. The impact of these additions on the AIRs requirement of the purGE stm4068 ptsJ strain was determined in the presence of dPN (Fig. 6b). Methionine had no effect on the AIRs requirement when dPN was present, making it unlikely that AdoMet synthesis was compromised. In contrast, growth in the presence of AIRs was significantly enhanced when either pantothenate or pantoate was added. These data implicated CoA biosynthesis, specifically the pantoate branch, as a target for the inhibitory effect of dPNP. Consistent with this hypothesis, a ptsJ mutant had a significant ∼2-fold decrease in the total CoA pool when grown in medium supplemented with dPN (0.5 μM) (Fig. 6c). The weak stimulation by β-alanine was considered to be due to increased flux through PanC when the pantoate substrate level was decreased.
FIG 6.
Precursors for CoA synthesis eliminate dPN sensitivity of a ptsJ mutant. (a) The integrated serine-glycine metabolic node, CoA, and thiamine synthesis in S. enterica. Blue indicates metabolites that mitigate the inhibition of dPN on ThiC-dependent thiamine synthesis, while those in red have little to no effect. Solid arrows indicate biochemical reactions in the pathways. Dashed arrows depict metabolic processes that affect ThiC activity. (b) The purGE stm4068 ptsJ strain (DM17274) was mixed with soft agar and overlaid onto minimal NCE glucose containing adenine (0.4 mM) and dPN (0.1 μM) without or with one of the following supplementations: pantothenate (0.1 mM), pantoate (0.1 mM), β-alanine (0.1 mM), KIV (0.1 mM), serine (2.5 mM), glycine (0.67 mM), or methionine (0.3 mM). The AIRs requirement was assessed by spotting 1 μL of AIRs (∼300 mM) and thiamine (0.1 mM) as a control. (c) Total intracellular CoA level of a ptsJ mutant (DM17239) grown in minimal NCE glucose in the absence or presence of dPN (0.5 μM). *, P < 0.0001, as determined by two-tailed unpaired Student's t test. Representative data from two experiments, each with three biological replicates, are shown. Error bars depict standard deviations from the means. THF, tetrahydrofolate; Pan, pantothenate; β-Ala, β-alanine; Pant, pantoate; Ser, l-serine; Gly, l-glycine; Met, l-methionine.
Addition of α-ketoisovalerate (KIV), serine, or glycine also decreased the AIRs requirement for growth in dPN (Fig. 6b). Serine and glycine are substrates in the generation of one-carbon units by GlyA (serine hydroxymethyltransferase [EC 2.1.2.1]) and the glycine cleavage (GCV) system, respectively (Fig. 6a). Both reactions produce 5,10-methylenetetrahydrofolate (mTHF) (36, 37), which is a cosubstrate for PanB (ketopantoate hydroxymethyltransferase [EC 2.1.2.11]) to convert KIV to ketopantoate (Fig. 6a). The PanB-dependent reaction was suggested to be the rate-limiting step for CoA synthesis (38). Notably, GlyA and GcvP (glycine decarboxylase [EC 1.4.4.2]) of the GCV system are PLP-dependent enzymes, which could be targeted by dPNP. There is precedent for damaged GlyA causing a decrease in CoA levels in S. enterica (39). In addition, mitogen-induced GlyA activity measured in crude extracts was decreased when human leukocytes and T cells were cultured with dPN supplementation, and the activity was rescued by the addition of vitamin B6 (40). Although the effect of dPNP on GcvP has not been reported, PNP, a B6 vitamer with a structure similar to that of dPNP (Fig. 1), could inhibit the in vitro activity of E. coli and Bacillus subtilis GCV systems (41). In total, the data are consistent with a model in which dPNP inhibits GlyA and/or the GCV system by competing with PLP. Such inhibition would lead to a reduction in mTHF levels and decrease flux to CoA synthesis.
Working model for growth limitation by dPN.
The cumulative effects of dPN toxicity, exacerbated by a ptsJ mutation, are depicted in Fig. 7. Our working model suggests that exogenous dPN is phosphorylated to dPNP by PdxK (Fig. 2). Intracellular dPNP then competes with PNP for binding to PdxH and thus compromises the DXP-dependent synthesis of PLP (Fig. 3). Additionally, dPNP inhibits the activity of the GlyA and/or GcvP PLP-dependent enzymes, which reduces the levels of mTHF. Finally, lower-than-optimal mTHF levels compromise the activity of PanB, reducing formation of pantothenate and decreasing flux to CoA (Fig. 6). Reduced CoA levels then indirectly affect the synthesis of the pyrimidine moiety of thiamine, resulting in overall lower thiamine formation (Fig. 4 and 5).
FIG 7.
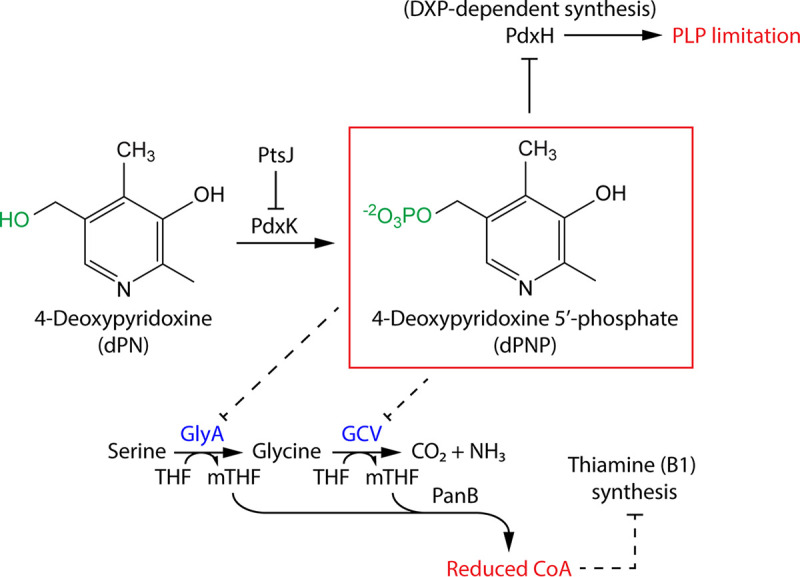
Working model regarding how dPN affects S. enterica metabolism. PdxK converts exogenous dPN to dPNP, which inhibits PLP biosynthesis via PdxH, leading to PLP limitation. In addition, dPNP inhibits the PLP-dependent enzyme GlyA and/or GcvP, decreasing flux through the pantoate branch of CoA synthesis. Reduced CoA levels indirectly dampen ThiC-dependent thiamine synthesis. THF, tetrahydrofolate.
The conclusions made from phenotypic analyses are bolstered by considering the kinetic parameters of PdxH. Based on HPLC data, the intracellular concentration of dPNP was estimated to be ∼4-fold and 350-fold higher than those of PMP and PNP, respectively, under conditions where growth was significantly impaired (data not shown). The binding affinity of dPNP for E. coli PdxH is similar to that of PMP and ∼50-fold lower than that of PNP (19). Together, these considerations support the ability of dPNP to compete with PNP and/or PMP for PdxH and reduce its ability to generate PLP. Similar kinetic parameters with GlyA and GcvP have not been reported, and additional in vitro experiments are required to further support the genetic analyses.
It was somewhat surprising that PdxY was not involved in dPNP toxicity, since it is another PL kinase (EC 2.7.1.35) involved in B6 salvage (42). PdxK and PdxY have promiscuous kinase activity with pyrimidine and pyridine derivatives, including the HMP moiety of thiamine and unphosphorylated B6 vitamers (42–45). A potential explanation for the lack of a role for PdxY is that S. enterica evolved differential regulation of pdxK and pdxY. Strains lacking PtsJ, like those used here, led to an ∼80-fold increase in expression of pdxK, while only a 2- to 3-fold increase was observed with pdxY (14). Alternatively, PdxY may not phosphorylate dPN with the efficiency that would make it relevant under the conditions tested.
This work did not address whether dPN/dPNP could be metabolized or detoxified in S. enterica. Rapid excretion and modification of dPN was proposed to be a detoxifying mechanism in humans and rats, in which dPN appeared to be converted to 4-deoxy-5-pyridoxic acid and deoxypyridoxine-3-(hydrogen sulfate), respectively (46). Isolation and characterization of S. enterica mutants that are more or less sensitive to dPN could address whether such a mechanism exists in bacteria.
In total, this study contributes to our understanding of the mechanism by which exogenous dPN inhibits growth in S. enterica. The data presented here provide a framework for probing the metabolic network structure involving vitamin B6 in other organisms. Furthermore, use of dPN provides an additional means to disrupt PLP metabolism, which can be coupled with mutations that impact PLP-dependent enzymes and PLP homeostasis, such as ridA and yggS, respectively, to better understand additional metabolic aspects of this critical cofactor.
MATERIALS AND METHODS
Strains, plasmids, and primers.
Strains and plasmids are presented in Table 1. Primers used for strain and plasmid construction are listed in Table 2. Strains are derivatives of S. enterica serovar Typhimurium LT2. Escherichia coli DH5α and BL21AI (Invitrogen, Carlsbad, CA) were used to propagate plasmids and purify recombinant protein, respectively. Phage λ Red recombineering (47) was adapted for S. enterica to generate gene deletions. Initial deletions were transduced into appropriate strain backgrounds using phage P22 HT105/1 int-201 (48) following published protocol (49) and confirmed by colony PCR.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| S. enterica strains | ||
| DM17239 | ptsJ617::Km | This study |
| DM17238 | pdxK682::Cm | This study |
| DM17168 | ptsJ617::Km pdxK682::Cm | This study |
| DM17274 | purG3111 purE3043 stm4068-6::Tn10(d)Tc ptsJ617::Km | This study |
| DM16666 | Wild-type/pCV1 | This study |
| DM17210 | Wild-type/pDM1643 | This study |
| DM17242 | ptsJ617::Km/pCV1 | This study |
| DM17243 | ptsJ617::Km/pDM1647 | This study |
| DM17253 | ptsJ617::Km pdxK682::Cm/pCV1 | This study |
| DM17244 | ptsJ617::Km/pBAD33-SD1 | This study |
| DM17246 | pdxJ664 pdxH673 ptsJ617::Km/pDM1594 | This study |
| Plasmids | ||
| pBAD33-SD1 | Modified pBAD33 (Cmr) | 55 |
| pCV1 | Modified pBAD24 (Amr) | 50 |
| pDM1594 | pBAD33-SD1 expressing S. cerevisiae SNZ3 (Cmr) | 55 |
| pDM1643 | pCV1 expressing S. enterica pdxK (Amr) | This study |
| pDM1647 | pCV1 expressing S. enterica ptsJ (Amr) | This study |
| pBsPTA1 | pTEV5 expressing B. subtilis 6×His-tagged PTA (Amr) | Escalante-Semerenaa |
Provided by Jorge C. Escalante-Semerena at the University of Georgia (Athens, GA).
TABLE 2.
Primers used in this study
| Primer | Sequence (5′ to 3′) | Description |
|---|---|---|
| PR1383 | GGAAAATTTTATGGGACAAGAGAGTGATATTCAGTCAGTGGTGTAGGCTGGAGCTGCTTC | Inactivation of pdxK |
| PR1384 | CGATAACTCTTCATCGCGCCTCCCCTGCCGGCGGCAGAATCATATGAATATCCTCCTTAGTTCCTATTCC | Inactivation of pdxK |
| PR1461 | ATTAAAGGCAGTCCGTCTCCCGTAGCGCTGGAAGGCGGCGTGTAGGCTGGAGCTGCTTC | Inactivation of ptsJ |
| PR1462 | TCCCTCACGTACCAGCCAGCCGGATTTTGCCAACGTAAACATATGAATATCCTCCTTAGTTCCTATTCC | Inactivation of ptsJ |
| PR1318 | NNGCTCTTCNTTCATGGGACAAGAGAGTGATATTCAGTCAGTG | Construction of pDM1643 |
| PR1319 | NNGCTCTTCNTTATCATCGCGCCTCCCCTG | Construction of pDM1643 |
| PR1449 | NNGCTCTTCNTTCATGATCGACGGAAAAACCGCTAACGAAATTTTTGACAG | Construction of pDM1647 |
| PR1450 | NNGCTCTTCNTTATTAGCGATTTAGCGCCTGATGGATATCAGCC | Construction of pDM1647 |
Plasmid pDM1643 and pDM1647 were constructed by cloning S. enterica pdxK and ptsJ, respectively, into pCV1 (50) as described (51) and were validated using Sanger sequencing (Eton Bioscience, San Diego, CA). Plasmid pBsPTA1 was generated by cloning pta (encoding a phosphotransacetylase [PTA] [EC 2.3.1.8]) from B. subtilis 168 into pTEV5 (52) and was a gift from Jorge C. Escalante-Semerena at the University of Georgia (Athens, GA). Plasmids were electroporated into S. enterica and E. coli using a standard protocol.
Media and chemicals.
Strains were grown at 37°C in nutrient broth (8 g/L Difco mix, 5 g/L NaCl) or minimal NCE medium supplemented with 1 mM MgSO4, trace metals (53), and 11 mM glucose or 22 mM glycerol as the sole carbon source. Lysogeny broth (10 g/L Bacto tryptone, 5 g/L yeast extract, 5 g/L NaCl) and super broth (32 g/L Bacto tryptone, 20 g/L yeast extract, 5 g/L NaCl, 5 mM NaOH) were used to culture E. coli for protein purification. Agar was added to a final concentration of 1.5% (wt/vol) to obtain solid media. Additional supplements included dPN (0.1 to 10 μM), PL (1 μM), PN (1 μM), PM (1 μM), adenosine (1 mM), adenine (0.4 mM), pantothenate (0.1 mM), pantoate (0.1 mM), β-alanine (0.1 mM), KIV (0.1 mM), serine (2.5 mM), glycine (0.67 mM), methionine (0.3 mM), or thiamine (0.1 μM) as indicated in the text. When needed, antibiotics were added as followed: ampicillin (Am), 150 μg/mL in rich medium and 15 μg/mL in minimal medium; chloramphenicol (Cm), 20 μg/mL in rich medium and 5 μg/mL in minimal medium; kanamycin (Km), 50 μg/mL.
Unless otherwise stated, all chemicals were purchased from MilliporeSigma (formerly Sigma-Aldrich, St. Louis, MO). Crude dPNP was obtained from Cayman Chemical (Ann Arbor, MI). PNP was synthesized from PLP by reduction with NaBH4 (45). PMP was synthesized by dissolving PLP in concentrated NH4OH, which was subsequently reduced by the addition of NaBH4 (54). Restriction enzymes and Taq polymerase were purchased from New England BioLabs (Ipswich, MA). Primers were synthesized by Eton Bioscience (San Diego, CA).
Growth analysis.
Strains were precultured in 2 mL nutrient broth at 37°C for 6 to 8 h in an Innova 43 shaker (New Brunswick Scientific, Edison, NJ) at 250 rpm. Cells were washed with an equal volume of 0.85% NaCl and inoculated (2.5% [vol/vol]) into minimal NCE medium containing appropriate carbon sources and supplements. Growth was monitored in a 96-well plate by following the OD650 over time using a Elx808 plate reader (BioTek Instruments, Winooski, VT). Data were visualized using GraphPad Prism v8.4.3 (GraphPad Software, La Jolla, CA).
HPLC analysis of B6 vitamers.
Intracellular vitamin B6 profiles of ptsJ (DM17239) and ptsJ pdxk (DM17168) strains grown in minimal NCE glucose without or with addition of 0.5 μM dPN were determined following a described protocol (55) except that dPN was replaced by 4-pyridoxic acid (PA) as an internal standard. The identity of the dPNP peak was confirmed by coinjection of crude dPNP with the samples and by treatment with wheat germ acid phosphatase (55). The level of internal dPNP accumulation was inferred by quantifying the concentration of dPN in phosphatase-treated samples.
Assessment of AIRs requirement.
S. enterica purGE stm4068 ptsJ (DM17274) was precultured in 2 mL nutrient broth and washed as described for growth analysis. Three milliliters of molten soft agar (0.7% [wt/vol]) seeded with 100 μL washed cells was overlaid onto minimal NCE glucose containing adenine (0.4 mM), without or with the addition of dPN (0.1 μM), pantothenate (0.1 mM), pantoate (0.1 mM), β-alanine (0.1 mM), KIV (0.1 mM), serine (2.5 mM), glycine (0.67 mM), or methionine (0.3 mM). One microliter of ∼300 mM AIRs, 0.1 mM thiamine, 0.1 mM HMP, and 0.1 mM THZ was subsequently spotted on top of the solidified agar layer. After overnight incubation at 37°C, plates were photographed using a FOTO/Analyst FX workstation (Fotodyne, Hartland, WI).
Purification and activity assay of B. subtilis PTA.
The E. coli BL21AI strain carrying pBsPTA1 was precultured in 10 mL lysogeny broth in duplicate at 37°C in an Innova 43 shaker (New Brunswick Scientific) at 250 rpm. Overnight cultures were inoculated into two Fernbach flasks, each of which contained 1.5 L of super broth. Growth was resumed at 37°C in an Innova 44 shaker (New Brunswick Scientific) at 180 rpm until mid-log phase (OD650 of ∼0.6), as determined by a Spectronic 20D+ instrument (Thermo Fisher Scientific, Waltham, MA). After the addition of arabinose (0.02% [wt/vol]) and isopropyl-β-d-thiogalactopyranoside (0.5 mM) to induce expression of pta, the temperature was shifted to 30°C. Cells were harvested (6,000 × g for 15 min at 4°C) after 20 h and stored at −80°C until purification.
Protein purification and dialysis steps were performed at 4°C. The frozen cell pellet was resuspended (3 mL/g wet weight) in buffer A (50 mM HEPES, 500 mM NaCl, 20 mM imidazole [pH 7.5]) containing lysozyme (1 mg/mL), DNase (0.14 mg/mL), and phenylmethylsulfonyl fluoride (PMSF) (1 mM) and was lysed at 18,000 lb/in2 using a Constant Systems Limited One Shot cell disruptor (Northants, UK). The cell lysate was clarified (45,000 × g for 45 min at 4°C), filtered (0.45-μm polyvinylidene difluoride [PVDF]), and loaded onto a preequilibrated 5-mL HisTrap HP Ni-Sepharose column (GE Healthcare, Chicago, IL) using an NGC Quest 10 chromatography system (Bio-Rad, Hercules, CA). The loaded column was washed and eluted with 10 column volumes (CVs) of buffer A, 6 CVs of 4% buffer B (50 mM HEPES, 500 mM NaCl, 500 mM imidazole [pH 7.5]), and a 10-CV gradient of 4 to 100% buffer B at a flow rate of 2 mL/min. Fractions containing PTA were visualized on a 14% SDS-PAGE gel and pooled. Protein was concentrated using an Amicon Ultra-15 30K centrifugal filter unit (MilliporeSigma), dialyzed overnight against buffer C (50 mM HEPES, 150 mM NaCl, 10% glycerol [pH 7.5]), flash-frozen with liquid nitrogen, and stored at −80°C until assay. The protein concentration was determined spectrophotometrically using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific), from the calculated molecular mass and extinction coefficient (ExPASy).
Enzyme activity was assayed using a previously described protocol (56) adapted for 96-well plates. Briefly, 0 to 10 ng of PTA was added to a 300-μL reaction mixture containing 100 mM Tris-HCl (pH 7.4), 1.6 mM glutathione, 0.4 mM CoA, 7.2 mM acetyl phosphate, and 13.3 mM (NH4)2SO4 to initiate the reaction. Acetyl-CoA formation was monitored at 233 nm (ε233 = 4.44 mM−1 cm−1) in a 96-well quartz plate using a SpectraMax 398 Plus plate reader (Molecular Devices, Sunnyvale, CA) and normalized to a 1-cm pathlength. One unit of PTA converts 1 μmol of CoA to acetyl-CoA per minute at 25°C (pH 7.4) using acetyl phosphate as a substrate.
CoA quantification.
Total CoA was determined as described (57, 58) with modifications. The S. enterica ptsJ strain (DM17239) was precultured in 10 mL nutrient broth in triplicate overnight. Cells were washed with an equal volume of 0.85% NaCl and inoculated (2% [vol/vol]) into Klett flasks containing 200 mL minimal NCE glucose without or with 0.5 μM dPN supplementation. After reaching an OD650 of 0.7 to 0.8, cells were harvested (8,000 × g for 15 min at 4°C), washed with 5 mL cold phosphate-buffered saline (PBS), and stored at −80°C until analysis.
Frozen cell pellets were resuspended in cold PBS and lysed by the addition of formic acid to a final concentration of 0.3 N. After a 30-min incubation on ice with occasional agitation, cell debris was pelleted (17,000 × g for 10 min at 4°C), and the lysate was neutralized with NH4OH (pH ∼6.5 to 7.5). Dithiothreitol (0.7% [vol/vol]) was added to aliquots of neutralized lysate to reductively cleave CoA thioesters. The assay mixture (120 μL) contained 250 mM Tris-HCl (pH 7.2), 50 mM KCl, 15 mM malate, 6 mM acetyl phosphate, 1 mM NAD+, 0.4 U porcine citrate synthase, 2 U porcine malate dehydrogenase, 40 μL treated extract, and 0.8 U B. subtilis PTA, which was added last to initiate the reaction. NADH formation was monitored at 340 nm in a 96-well quartz plate and normalized to a 1-cm pathlength using a SpectraMax 398 Plus plate reader (Molecular Devices). The CoA level for each biological sample was measured with four technical replicates and normalized to dry cell weight. A 7-point standard curve was generated with the addition of 0 to 1.2 nmol CoA and was used as reference for CoA quantification.
Data availability.
All relevant data are included in the content of the manuscript.
ACKNOWLEDGMENTS
We thank Jorge C. Escalante-Semerena for access to his laboratory strain collection and Rachel M. Burckhardt for constructing plasmid pBsPTA1.
This work was supported by an award from the competitive grants program at the NIH (grant GM095837) to D.M.D.
Contributor Information
Diana M. Downs, Email: dmdowns@uga.edu.
Anke Becker, Philipps University Marburg.
REFERENCES
- 1.Di Salvo ML, Budisa N, Contestabile R. 2012. PLP-dependent enzymes: a powerful tool for metabolic synthesis of non-canonical amino acids, p 27–66. In Proceedings of the Beilstein Bozen Symposium on Molecular Engineering and Control. Beilstein-Istitut, Frankfort, Germany. [Google Scholar]
- 2.Percudani R, Peracchi A. 2009. The B6 database: a tool for the description and classification of vitamin B6-dependent enzymatic activities and of the corresponding protein families. BMC Bioinformatics 10:273. 10.1186/1471-2105-10-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Percudani R, Peracchi A. 2003. A genomic overview of pyridoxal‐phosphate‐dependent enzymes. EMBO Rep 4:850–854. 10.1038/sj.embor.embor914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mittenhuber G. 2001. Phylogenetic analyses and comparative genomics of vitamin B6 (pyridoxine) and pyridoxal phosphate biosynthesis pathways. J Mol Microbiol Biotechnol 3:1–20. [PubMed] [Google Scholar]
- 5.Tanaka T, Tateno Y, Gojobori T. 2005. Evolution of vitamin B6 (pyridoxine) metabolism by gain and loss of genes. Mol Biol Evol 22:243–250. 10.1093/molbev/msi011. [DOI] [PubMed] [Google Scholar]
- 6.Venegas A, Martial J, Valenzuela P. 1973. Active site-directed inhibition of E. coli DNA-dependent RNA polymerase by pyridoxal 5′-phosphate. Biochem Biophys Res Commun 55:1053–1059. 10.1016/s0006-291x(73)80001-4. [DOI] [PubMed] [Google Scholar]
- 7.Bartzatt R, Beckmann JD. 1994. Inhibition of phenol sulfotransferase by pyridoxal phosphate. Biochem Pharmacol 47:2087–2095. 10.1016/0006-2952(94)90085-x. [DOI] [PubMed] [Google Scholar]
- 8.Vermeersch JJ, Christmann-Franck S, Karabashyan LV, Fermandjian S, Mirambeau G, Der Garabedian PA. 2004. Pyridoxal 5′-phosphate inactivates DNA topoisomerase IB by modifying the lysine general acid. Nucleic Acids Res 32:5649–5657. 10.1093/nar/gkh897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whittaker MM, Penmatsa A, Whittaker JW. 2015. The Mtm1p carrier and pyridoxal 5′-phosphate cofactor trafficking in yeast mitochondria. Arch Biochem Biophys 568:64–70. 10.1016/j.abb.2015.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boycheva S, Dominguez A, Rolcik J, Boller T, Fitzpatrick TB. 2015. Consequences of a deficit in vitamin B6 biosynthesis de novo for hormone homeostasis and root development in Arabidopsis. Plant Physiol 167:102–117. 10.1104/pp.114.247767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coburn SP. 2018. The chemistry and metabolism of 4′-deoxypyridoxine. CRC Press, Boca Raton, FL. [Google Scholar]
- 12.Hanna MC, Turner AJ, Kirkness EF. 1997. Human pyridoxal kinase: cDNA cloning, expression, and modulation by ligands of the benzodiazepine receptor. J Biol Chem 272:10756–10760. 10.1074/jbc.272.16.10756. [DOI] [PubMed] [Google Scholar]
- 13.Salamon N, Gurgui C, Leistner E, Drewke C. 2009. Influence of antivitamins ginkgotoxin 5′-phosphate and deoxypyridoxine 5′-phosphate on human pyridoxine 5′-phosphate oxidase. Planta Med 75:563–567. 10.1055/s-0029-1185482. [DOI] [PubMed] [Google Scholar]
- 14.Tramonti A, Milano T, Nardella C, di Salvo ML, Pascarella S, Contestabile R. 2017. Salmonella typhimurium PtsJ is a novel MocR‐like transcriptional repressor involved in regulating the vitamin B6 salvage pathway. FEBS J 284:466–484. 10.1111/febs.13994. [DOI] [PubMed] [Google Scholar]
- 15.Mulligan JH, Snell E. 1976. Transport and metabolism of vitamin B6 in Salmonella typhimurium LT2. J Biol Chem 251:1052–1056. 10.1016/S0021-9258(17)33800-0. [DOI] [PubMed] [Google Scholar]
- 16.Yamada R, Tsuji T, Nose Y. 1977. Uptake and utilization of vitamin B6 and its phosphate esters by Escherichia coli. J Nutr Sci Vitaminol (Tokyo) 23:7–17. 10.3177/jnsv.23.7. [DOI] [PubMed] [Google Scholar]
- 17.Yamada R, Furukawa Y. 1981. Role of pyridoxal kinase in vitamin B6 uptake by Escherichia coli. J Nutr Sci Vitaminol (Tokyo) 27:177–191. 10.3177/jnsv.27.177. [DOI] [PubMed] [Google Scholar]
- 18.Kästner U, Hallmen C, Wiese M, Leistner E, Drewke C. 2007. The human pyridoxal kinase, a plausible target for ginkgotoxin from Ginkgo biloba. FEBS J 274:1036–1045. 10.1111/j.1742-4658.2007.05654.x. [DOI] [PubMed] [Google Scholar]
- 19.Zhao G, Winkler ME. 1995. Kinetic limitation and cellular amount of pyridoxine (pyridoxamine) 5′-phosphate oxidase of Escherichia coli K-12. J Bacteriol 177:883–891. 10.1128/jb.177.4.883-891.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paxhia MD, Downs DM. 2019. SNZ3 encodes a PLP synthase involved in thiamine synthesis in Saccharomyces cerevisiae. G3 (Bethesda) 9:335–344. 10.1534/g3.118.200831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moyed H. 1964. Inhibition of the biosynthesis of the pyrimidine portion of thiamine by adenosine. J Bacteriol 88:1024–1029. 10.1128/jb.88.4.1024-1029.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rolfes RJ, Zalkin H. 1988. Escherichia coli gene purR encoding a repressor protein for purine nucleotide synthesis: cloning, nucleotide sequence, and interaction with the purF operator. J Biol Chem 263:19653–19661. 10.1016/S0021-9258(19)77686-8. [DOI] [PubMed] [Google Scholar]
- 23.Zhou G, Smith J, Zalkin H. 1994. Binding of purine nucleotides to two regulatory sites results in synergistic feedback inhibition of glutamine 5-phosphoribosylpyrophosphate amidotransferase. J Biol Chem 269:6784–6789. 10.1016/S0021-9258(17)37444-6. [DOI] [PubMed] [Google Scholar]
- 24.Petersen L, Enos-Berlage J, Downs DM. 1996. Genetic analysis of metabolic crosstalk and its impact on thiamine synthesis in Salmonella typhimurium. Genetics 143:37–44. 10.1093/genetics/143.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chatterjee A, Li Y, Zhang Y, Grove TL, Lee M, Krebs C, Booker SJ, Begley TP, Ealick SE. 2008. Reconstitution of ThiC in thiamine pyrimidine biosynthesis expands the radical SAM superfamily. Nat Chem Biol 4:758–765. 10.1038/nchembio.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez-Gomez NC, Downs DM. 2008. ThiC is an [Fe-S] cluster protein that requires AdoMet to generate the 4-amino-5-hydroxymethyl-2-methylpyrimidine moiety in thiamin synthesis. Biochemistry 47:9054–9056. 10.1021/bi8010253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dougherty MJ, Downs DM. 2004. A mutant allele of rpoD results in increased conversion of aminoimidazole ribotide to hydroxymethyl pyrimidine in Salmonella enterica. J Bacteriol 186:4034–4037. 10.1128/JB.186.12.4034-4037.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dougherty MJ, Downs DM. 2006. A connection between iron-sulfur cluster metabolism and the biosynthesis of 4-amino-5-hydroxymethyl-2-methylpyrimidine pyrophosphate in Salmonella enterica. Microbiology (Reading) 152:2345–2353. 10.1099/mic.0.28926-0. [DOI] [PubMed] [Google Scholar]
- 29.Dougherty M, Downs DM. 2003. The stm4066 gene product of Salmonella enterica serovar Typhimurium has aminoimidazole riboside (AIRs) kinase activity and allows AIRs to satisfy the thiamine requirement of pur mutant strains. J Bacteriol 185:332–339. 10.1128/JB.185.1.332-339.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palmer LD, Dougherty MJ, Downs DM. 2012. Analysis of ThiC variants in the context of the metabolic network of Salmonella enterica. J Bacteriol 194:6088–6095. 10.1128/JB.01361-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frodyma M, Rubio A, Downs D. 2000. Reduced flux through the purine biosynthetic pathway results in an increased requirement for coenzyme A in thiamine synthesis in Salmonella enterica serovar Typhimurium. J Bacteriol 182:236–240. 10.1128/JB.182.1.236-240.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skovran E, Downs DM. 2000. Metabolic defects caused by mutations in the isc gene cluster in Salmonella enterica serovar Typhimurium: implications for thiamine synthesis. J Bacteriol 182:3896–3903. 10.1128/JB.182.14.3896-3903.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leonardi R, Fairhurst SA, Kriek M, Lowe DJ, Roach PL. 2003. Thiamine biosynthesis in Escherichia coli: isolation and initial characterisation of the ThiGH complex. FEBS Lett 539:95–99. 10.1016/s0014-5793(03)00204-7. [DOI] [PubMed] [Google Scholar]
- 34.Martinez-Gomez NC, Robers M, Downs DM. 2004. Mutational analysis of ThiH, a member of the radical S-adenosylmethionine (AdoMet) protein superfamily. J Biol Chem 279:40505–40510. 10.1074/jbc.M403985200. [DOI] [PubMed] [Google Scholar]
- 35.Leonardi R, Roach PL. 2004. Thiamine biosynthesis in Escherichia coli: in vitro reconstitution of the thiazole synthase activity. J Biol Chem 279:17054–17062. 10.1074/jbc.M312714200. [DOI] [PubMed] [Google Scholar]
- 36.Florio R, di Salvo ML, Vivoli M, Contestabile R. 2011. Serine hydroxymethyltransferase: a model enzyme for mechanistic, structural, and evolutionary studies. Biochim Biophys Acta 1814:1489–1496. 10.1016/j.bbapap.2010.10.010. [DOI] [PubMed] [Google Scholar]
- 37.Kikuchi G, Motokawa Y, Yoshida T, Hiraga K. 2008. Glycine cleavage system: reaction mechanism, physiological significance, and hyperglycinemia. Proc Jpn Acad Ser B 84:246–263. 10.2183/pjab.84.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rubio A, Downs D. 2002. Elevated levels of ketopantoate hydroxymethyltransferase (PanB) lead to a physiologically significant coenzyme A elevation in Salmonella enterica serovar Typhimurium. J Bacteriol 184:2827–2832. 10.1128/JB.184.10.2827-2832.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Flynn JM, Christopherson MR, Downs DM. 2013. Decreased coenzyme A levels in ridA mutant strains of Salmonella enterica result from inactivated serine hydroxymethyltransferase. Mol Microbiol 89:751–759. 10.1111/mmi.12313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trakatellis A, Dimitriadou A, Exindari M, Scountzou J, Koliakos G, Christodoulou D, Malissiovas N, Antoniadis A, Polyzoni T. 1992. Effect of pyridoxine deficiency on immunological phenomena. Postgrad Med J 68(Suppl 1):S70–S77. [PubMed] [Google Scholar]
- 41.Ito T, Hori R, Hemmi H, Downs DM, Yoshimura T. 2020. Inhibition of glycine cleavage system by pyridoxine 5′‐phosphate causes synthetic lethality in glyA yggS and serA yggS in Escherichia coli. Mol Microbiol 113:270–284. 10.1111/mmi.14415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang Y, Tsui H-CT, Man T-K, Winkler ME. 1998. Identification and function of the pdxY gene, which encodes a novel pyridoxal kinase involved in the salvage pathway of pyridoxal 5′-phosphate biosynthesis in Escherichia coli K-12. J Bacteriol 180:1814–1821. 10.1128/JB.180.7.1814-1821.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reddick JJ, Kinsland C, Nicewonger R, Christian T, Downs DM, Winkler ME, Begley TP. 1998. Overexpression, purification and characterization of two pyrimidine kinases involved in the biosynthesis of thiamin: 4-amino-5-hydroxymethyl-2-methylpyrimidine kinase and 4-amino-5-hydroxymethyl-2-methylpyrimidine phosphate kinase. Tetrahedron 54:15983–15991. 10.1016/S0040-4020(98)01006-0. [DOI] [Google Scholar]
- 44.di Salvo ML, Hunt S, Schirch V. 2004. Expression, purification, and kinetic constants for human and Escherichia coli pyridoxal kinases. Protein Expr Purif 36:300–306. 10.1016/j.pep.2004.04.021. [DOI] [PubMed] [Google Scholar]
- 45.Vu HN, Downs DM. 2021. An unexpected role for the periplasmic phosphatase PhoN in the salvage of B6 vitamers in Salmonella enterica. Appl Environ Microbiol 87:e02300-20. 10.1128/AEM.02300-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Coburn S, Mahuren J. 1976. In vivo metabolism of 4′-deoxypyridoxine in rat and man. J Biol Chem 251:1646–1652. 10.1016/S0021-9258(17)33697-9. [DOI] [PubMed] [Google Scholar]
- 47.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97:6640–6645. 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmieger H. 1971. A method for detection of phage mutants with altered transducing ability. Mol Gen Genet 110:378–381. 10.1007/BF00438281. [DOI] [PubMed] [Google Scholar]
- 49.Chan RK, Botstein D, Watanabe T, Ogata Y. 1972. Specialized transduction of tetracycline resistance by phage P22 in Salmonella typhimurium. II. Properties of a high-frequency-transducing lysate. Virology 50:883–898. 10.1016/0042-6822(72)90442-4. [DOI] [PubMed] [Google Scholar]
- 50.VanDrisse C, Escalante-Semerena J. 2016. New high-cloning-efficiency vectors for complementation studies and recombinant protein overproduction in Escherichia coli and Salmonella enterica. Plasmid 86:1–6. 10.1016/j.plasmid.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Galloway NR, Toutkoushian H, Nune M, Bose N, Momany C. 2013. Rapid cloning for protein crystallography using type IIS restriction enzymes. Crystal Growth Design 13:2833–2839. 10.1021/cg400171z. [DOI] [Google Scholar]
- 52.Rocco C, Dennison K, Klenchin VA, Rayment I, Escalante-Semerena J. 2008. Construction and use of new cloning vectors for the rapid isolation of recombinant proteins from Escherichia coli. Plasmid 59:231–237. 10.1016/j.plasmid.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Davis R, Botstein D, Roth J. 1980. Advanced bacterial genetics. Cold Spring Harbor Laboratory Press. Cold Spring Harbor, NY. [Google Scholar]
- 54.Yang ES, Schirch V. 2000. Tight binding of pyridoxal 5′-phosphate to recombinant Escherichia coli pyridoxine 5′-phosphate oxidase. Arch Biochem Biophys 377:109–114. 10.1006/abbi.2000.1737. [DOI] [PubMed] [Google Scholar]
- 55.Vu HN, Ito T, Downs DM. 2020. The role of YggS in vitamin B6 homeostasis in Salmonella enterica is informed by heterologous expression of yeast SNZ3. J Bacteriol 202:e00383-20. 10.1128/JB.00383-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Klotzsch HR. 1969. Phosphotransacetylase from Clostridium kluyveri. Methods Enzymol 13:381–386. 10.1016/0076-6879(69)13065-7. [DOI] [Google Scholar]
- 57.Allred JB, Guy DG. 1969. Determination of coenzyme A and acetyl CoA in tissue extracts. Anal Biochem 29:293–299. 10.1016/0003-2697(69)90312-1. [DOI] [PubMed] [Google Scholar]
- 58.Ernst DC, Borchert AJ, Downs DM. 2018. Perturbation of the metabolic network in Salmonella enterica reveals cross-talk between coenzyme A and thiamine pathways. PLoS One 13:e0197703. 10.1371/journal.pone.0197703. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are included in the content of the manuscript.