ABSTRACT
Azoles, the most commonly used antifungal drugs, specifically inhibit the fungal lanosterol α-14 demethylase enzyme, which is referred to as Erg11. Inhibition of Erg11 ultimately leads to a reduction in ergosterol production, an essential fungal membrane sterol. Many Candida species, such as Candida albicans, develop mutations in this enzyme which reduces the azole binding affinity and results in increased resistance. Candida glabrata is also a pathogenic yeast that has low intrinsic susceptibility to azole drugs and easily develops elevated resistance. In C. glabrata, these azole resistant mutations typically cause hyperactivity of the Pdr1 transcription factor and rarely lie within the ERG11 gene. Here, we generated C. glabrata ERG11 mutations that were analogous to azole resistance alleles from C. albicans ERG11. Three different Erg11 forms (Y141H, S410F, and the corresponding double mutant (DM)) conferred azole resistance in C. glabrata with the DM Erg11 form causing the strongest phenotype. The DM Erg11 also induced cross-resistance to amphotericin B and caspofungin. Resistance caused by the DM allele of ERG11 imposed a fitness cost that was not observed with hyperactive PDR1 alleles. Crucially, the presence of the DM ERG11 allele was sufficient to activate the Pdr1 transcription factor in the absence of azole drugs. Our data indicate that azole resistance linked to changes in ERG11 activity can involve cellular effects beyond an alteration in this key azole target enzyme. Understanding the physiology linking ergosterol biosynthesis with Pdr1-mediated regulation of azole resistance is crucial for ensuring the continued efficacy of azole drugs against C. glabrata.
KEYWORDS: azole drugs, Candida glabrata, Cdr1, Erg11, Pdr1, ergosterol, transcriptional regulation
INTRODUCTION
Ergosterol is essential for fungal plasma membrane homeostasis (1, 2). It maintains membrane integrity and fluidity, as well as facilitates membrane-bound enzymatic reactions (1). The azole class of antifungal therapy targets the fungal ability to synthesize ergosterol through inhibition of the function of lanosterol 14-alpha-demethylase, which is encoded by the ERG11 gene (3, 4). The Erg11 enzyme catalyzes a three-step reaction that results in the demethylation of lanosterol. In each step, one oxygen molecule is oxidized, leading to the production of one NADPH molecule (4, 5). The nucleophilic N-4 atom of the azole ring binds to heme, while the N-1 group interacts with the enzyme active site, directly competing with the normal substrate (lanosterol) binding (6, 7). While azoles bind selectively to the Erg11 enzyme (8) the affinity of this enzyme for different azoles varies; with fluconazole binding with lower affinity than either itraconazole or voriconazole (4).
In Candida albicans (Ca), mutations in the Erg11 enzyme have been shown to alter azole binding capacity and significantly reduce the drug inhibitory effect resulting in an enhanced azole resistant phenotype (9–14). The majority of these mutations cluster into three hot spots, located within residues 105–165, 266– 287, and 405–488 (15). The ability to confer azole resistance by some of these mutations has also been confirmed by introducing these mutations on plasmids into a heterologous Saccharomyces cerevisiae system and then assessing azole susceptibility. With this approach, single amino acid mutations, such as Y132H, S405F, G464S, R467K, and double mutations, such as Y132H with S405F, Y132H with G464S, R467K with G464S, have been shown to confer azole resistance in S. cerevisiae (9). In addition, several CaERG11 constructs of the enzyme catalytic domain containing amino acid substitutions, including Y132H, F145L, I471T, S279F, and G464S have been expressed in Escherichia coli, purified, and showed to exhibit a lower binding capacity to azoles (15). Finally, many of these mutant alleles have also been introduced back into an azole-susceptible C. albicans strain, at the ERG11 gene native locus, to confirm the relative contributions of individual and combined mutation to azole resistance (10).
Unlike in C. albicans, mutations in the ERG11 gene have been rarely reported among Candida glabrata azole resistant clinical isolates. The most common amino acid substitution mutations in C. glabrata are in the gene encoding a Zn2Cys6 zinc cluster-containing transcription factor called Pdr1 (16). These mutations yield a gain-of-function (GOF) phenotype and lead to the elevated transcription of downstream target genes, such as the ATP-binding cassette (ABC) transporter-encoding CDR1 gene. The Cdr1 transporter is required for the azole resistant mechanism in both wild-type and GOF PDR1 C. glabrata isolates (17).
We recently reported that there is biological cross talk between the ergosterol biosynthesis pathway and the drug efflux pump system in C. glabrata (18). Alteration in ergosterol synthesis directly induced expression of both Pdr1 and Cdr1. The Zn2Cys6 zinc cluster-containing factor Upc2A is essential for such cross-interaction, through its functions as a master transcription factor regulating the expression levels of both ergosterol synthesis genes (ERG) and PDR1-CDR1 genes (19, 20). Since our earlier experiments indicated that activity of the Pdr1/CDR1 system was responsive to ergosterol biosynthesis, we further wanted to determine if mutations in ERG11 that reduced azole susceptibility might act via induction of this separate azole resistance pathway. Considering the conserved inhibitory effects of azoles on the Erg11 protein in both C. albicans and C. glabrata, we first generated isogenic C. glabrata isolates carrying mutant ERG11 alleles, analogs of the CaERG11 Y132H, S405F or Y132H-S405F mutations. Then we examined the effects of these mutant alleles on ergosterol synthesis, antifungal drug resistance, and abilities to cross-regulate the drug efflux pump Pdr1-Cdr1 system. Our data indicate that azole-resistant ERG11 mutations can activate function of Pdr1 with attendant induction of genes (like CDR1) downstream of this factor, even in the absence of drug. These findings illustrate the importance of considering secondary effects of ERG11 mutations on activation of other resistance pathways when evaluating the azole resistance caused by these lesions. Our data provide further evidence directly tying expression of the Pdr1-dependent resistance pathway to ergosterol biosynthesis.
RESULTS
Generation of azole-resistant forms of C. glabrata Erg11.
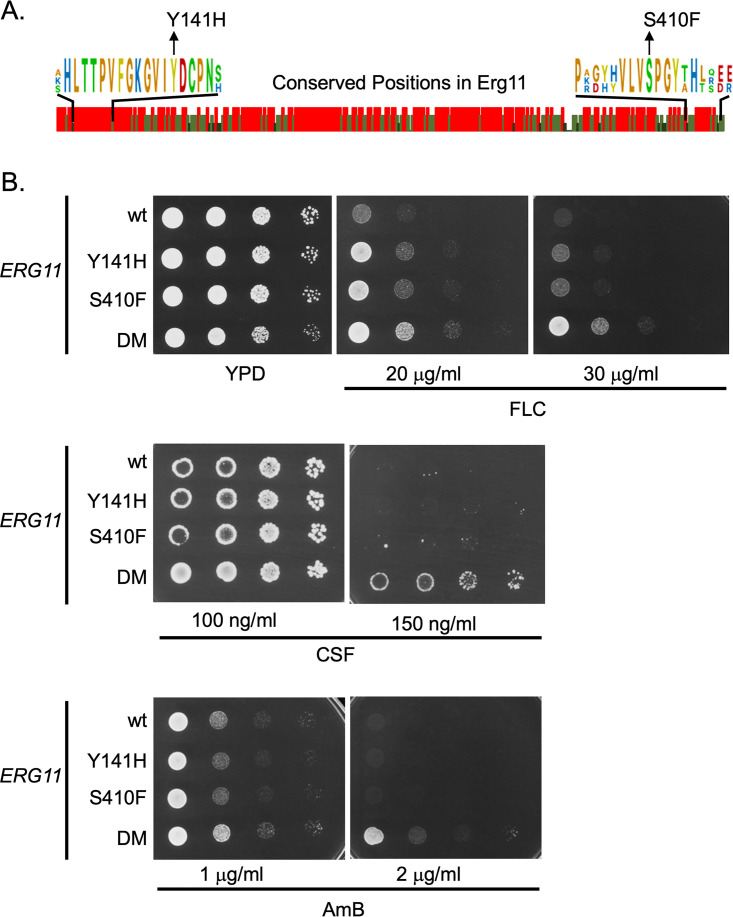
Detailed structural information is available for the Erg11 enzymes from these two pathogenic Candida species as their three-dimensional structures have recently been solved by X-ray crystallography, along with the previously determined structure of S. cerevisiae Erg11 (6, 21). Figure 1A shows an alignment of the amino acid sequences of these three closely related proteins.
FIG 1.
Construction of mutant ERG11 alleles in Candida glabrata. A. Amino acid sequence comparison between Erg11 proteins from Saccharomyces cerevisiae, Candida glabrata and Candida albicans. Graphic indication of amino acid identity between these three yeast enzymes is shown with a red bar indicating 100%, while light and dark green denotes 66% or no conservation. Expanded regions with conserved residues are shown to illustrate the positions that were mutated in C. glabrata to match changes associated with fluconazole resistance originally found in C. albicans. B. Isogenic C. glabrata strains containing the indicated ERG11 alleles were grown to mid-log phase and plated as serial dilutions on rich medium (YPD) or the same medium containing the indicated concentrations of antifungal drugs. The strain containing both the Y141H- and S410F-encoding alleles was designated double mutant (DM). Plates were allowed to develop at 30°C and then photographed. Antifungal drugs are abbreviated as fluconazole (FLC), caspofungin (CSF) and amphotericin B (AmB).
Previous analyses of azole resistant clinical isolates of C. albicans identified changes in residue tyrosine 132 to histidine (Y132H), serine 405 to phenylalanine (S405F), and a double mutant containing both of these lesions (Y132H S405F, referred to as double mutant or DM here) to trigger a decrease in fluconazole susceptibility when present in the C. albicans Erg11 enzyme (9). Inspection of the aligned sequences shown in Fig. 1A indicated that both Y141 and S410 (in C. glabrata) were conserved residues across the enzymes from S. cerevisiae and the two Candida species. To determine if these same substitution mutations would cause similar fluconazole resistance phenotypes when introduced into the C. glabrata Erg11, we used site-directed mutagenesis to generate the appropriate mutant enzymes. These were returned to C. glabrata as the sole source of Erg11 using a plasmid shuffling approach (see Materials and Methods). Appropriate transformants were tested for their resistance to fluconazole, caspofungin and amphotericin B (Fig. 1B).
The presence of either single mutant form of Erg11 (Y141H or S410F) led to a slight increase in fluconazole resistance compared to the wild-type enzyme while the double mutant (Y141H and S410F: DM) caused the strongest elevation in fluconazole resistance. The enhanced effect on azole resistance caused by combining these two mutations into a single enzyme has been seen before (9). Only the DM Erg11 was able to enhance resistance to caspofungin or amphotericin B. These data indicated that these mutant forms of C. glabrata Erg11 were able to increase fluconazole resistance like their C. albicans counterparts and, when combined in the DM Erg11, increased resistance to two other antifungal drugs. We also tested these strains for their susceptibility to voriconazole (Fig. S1) and found that the mutants caused voriconazole phenotypes similar to those seen for fluconazole. This differential response of a given ERG11 mutant to different azole drugs has been observed previously (9, 22).
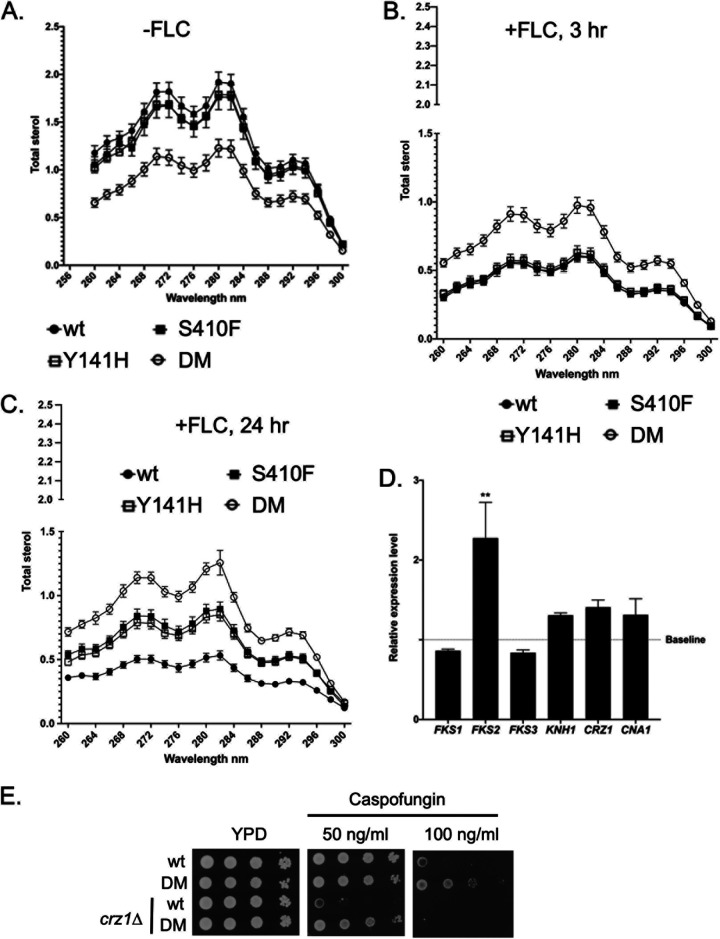
The multidrug resistant nature of the DM ERG11 mutant strain was not expected. This finding prompted us to evaluate the levels of ergosterol supported by each of these mutant strains as well as expression of genes that could explain the increased resistance to caspofungin. The three mutant strains along with their wild-type ERG11 counterpart were grown to mid-log phase and allowed to grow for 3 or 24 h with or without fluconazole challenge. Total sterols were extracted from these cultures and assayed for the level of ergosterol produced in each strain. Total RNA was also prepared from isogenic wild-type and DM ERG11 strains without fluconazole challenge and assayed by RT-qPCR for the mRNA levels of the genes indicated.
Ergosterol levels were the lowest in the DM Erg11-expressing strains in the absence of fluconazole stress (Fig. 2A). The two single mutants produced ergosterol levels that were very similar to those seen in the wild-type strain. A 3-h treatment with fluconazole failed to effect the ergosterol levels in the DM Erg11 strain, while the ergosterol levels produced in the other 3 strains were markedly lowered (Fig. 2B). No differences were observed between the two mutant forms of Erg11 compared to the wild-type protein until the fluconazole treatment was extended to 24 h (Fig. 2C). At this time, the DM Erg11 still produced the highest level of ergosterol but now both the Y141H and S410F single mutant strains had levels of ergosterol that were higher than the wild-type but still lower than the DM Erg11.
FIG 2.
Ergosterol levels and caspofungin resistance-related mRNA production in ERG11 mutant strains. Mid-log phase C. glabrata strains containing the indicated ERG11 alleles were either not treated (A) with fluconazole (20 μg/ml) (-FLC) or exposed to this drug for 3 (B) or 24 (C) hours. Total lipid extracts were prepared and analyzed for the levels of ergosterol in each sample (45). D. Total RNA was extracted from isogenic wild-type and DM ERG11 strains without fluconazole treatment. Samples were then assayed for levels of the indicated mRNAs using RT-qPCR. Relative expression level refers to the ratio of mRNA produced in DM ERG11 strains/wild-type strain. Baseline indicates equivalent expression in the wildtype strain. E. Loss of CRZ1 prevents increased caspofungin resistance of a DM ERG11 strain. The CRZ1 gene was disrupted in wild-type and DM ERG11 strains. These crz1Δ derivatives and their isogenic wild-type strains were grown to mid-log phase and then analyzed by serial dilution on YPD plates without (YPD) or containing the indicated levels of caspofungin. Plates were incubated at 30°C for 2 days and photographed.
Analysis of mRNA levels from genes directly involved in production of β-glucan synthase (FKS1, FKS2, FKS3, KNH1) as well as regulators of these genes (CRZ1, CNA1) indicated that only the mRNA for the FKS2 gene was elevated in the DM ERG11 mutant compared to its cognate wild-type strain (Fig. 2D). This transcriptional induction of FKS2 could explain the increase in caspofungin resistance seen in the DM Erg11-expressing strain.
We analyzed the contribution of the Crz1 transcription factor to the DM Erg11-triggered increase in caspofungin resistance by disrupting this gene in both wild-type and the isogenic DM ERG11 strains (Fig. 2E). At low concentrations of caspofungin, DM Erg11 was still able to increase drug resistance in a crz1Δ strain compared to the wild-type ERG11 (see 50 ng/ml plate). However, loss of Crz1 eliminated high level (100 ng/ml) caspofungin resistance. We interpret these data to suggest there are at least two effects of the DM ERG11 allele on caspofungin resistance: a minor one that is Crz1 independent and a major one that requires the presence of Crz1.
Communication between ERG11 and Pdr1/CDR1 pathway.
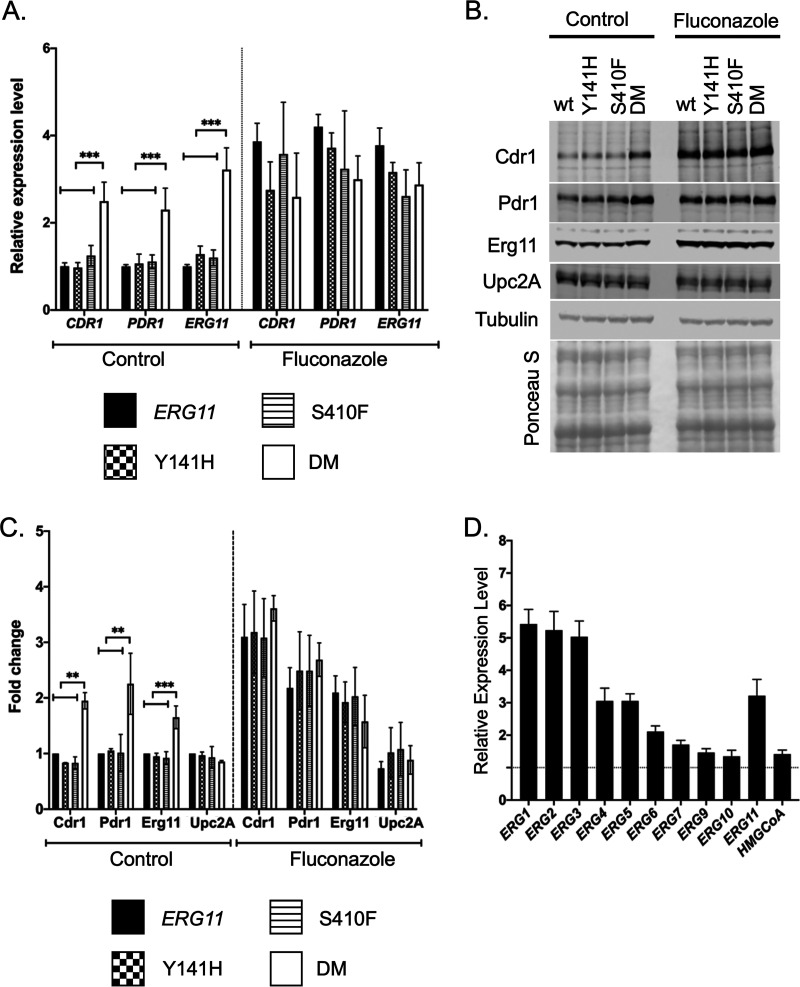
Our previous work demonstrated that reduction in Erg11 expression led to an induction of the Pdr1 transcription factor and its target genes such as CDR1 (19). To determine if these mutant forms of Erg11 triggered a similar response, we measured expression of Pdr1 and Cdr1 at both the protein and mRNA levels. Transformants containing the various ERG11 alleles were grown to mid-log phase and either treated with fluconazole for 3 h or left untreated. Total RNA was prepared from these cultures and mRNA for CDR1, PDR1 and ERG11 measured by RT-qPCR (Fig. 3A).
FIG 3.
Expression changes in response to ERG11 alleles. A. Levels of mRNA for PDR1, CDR1, and ERG11 were assayed by RT-qPCR in the indicated ERG11 mutant strains during growth in the absence (Control) or presence of fluconazole stress (Fluconazole). Fluconazole treatment was 20 μg/ml for 2 h. B. Whole cell protein extracts were prepared from the same strains in A. These extracts were analyzed by Western blotting using the indicated antisera against Cdr1, Pdr1, Erg11, Upc2A or Tubulin. Equal loading and transfer of these extracts was ensured by staining the nitrocellulose membrane with Ponceau S prior to antisera incubation. C. Quantitation of 4 replicates of this Western blotting is shown with the data presented as the ratio of each strain relative to the wild-type cells. D. RT-qPCR analysis of ERG gene expression in the DM ERG11 mutant background relative to the wild-type level. HMGCoA (HMG-CoA reductase) refers to the HMG1 or CAGL0L11506g locus.
In the absence of fluconazole stress, only the DM ERG11 allele led to induction of all three of these mRNAs. Upon challenge with fluconazole, all of these genes were induced to similar relative levels. The enhanced fluconazole resistance seen in the DM ERG11 strain may be explained by elevated PDR1 and CDR1 transcription, at least in part.
The levels of these proteins were also evaluated by Western blots using appropriate rabbit polyclonal antibodies for each C. glabrata protein (Fig. 3B) and quantitated (Fig. 3C). Both Pdr1 and Cdr1 were elevated in untreated cells along with Erg11 in the DM ERG11 strain. Treatment with fluconazole led to similar levels of induction of all these proteins. Upc2A protein levels were not changed under any of these conditions or genetic backgrounds. This is consistent with Upc2A regulation occurring at the post-translational step as has been suggested (23–25).
The levels of mRNA of a range of genes in the ergosterol biosynthetic pathway were also determined in both the DM and wild-type Erg11 strains (Fig. 3D). These data are presented as a ratio of mRNA level in the DM Erg11 strain over the wild-type level for each transcript. Most of these ERG genes were induced at least 2-fold in the DM Erg11-expressing cells, consistent with the ergosterol limitation in this strain although several ERG genes including ERG9, ERG7, ERG10, and HMG1 showed little to no increase. Clearly, the presence of the DM Erg11 enzyme induced a wide range of transcriptional activation across the ergosterol pathway in addition to its effects on PDR1 and CDR1 expression. We also suggest that the DM ERG11 strain may activate Crz1 leading to the previously observed increase in FKS2 expression and caspofungin resistance. These effects on Pdr1 and Crz1 would explain the multidrug resistance seen in DM ERG11 mutant strains.
Upc2A function is increased in the ERG11 mutants.
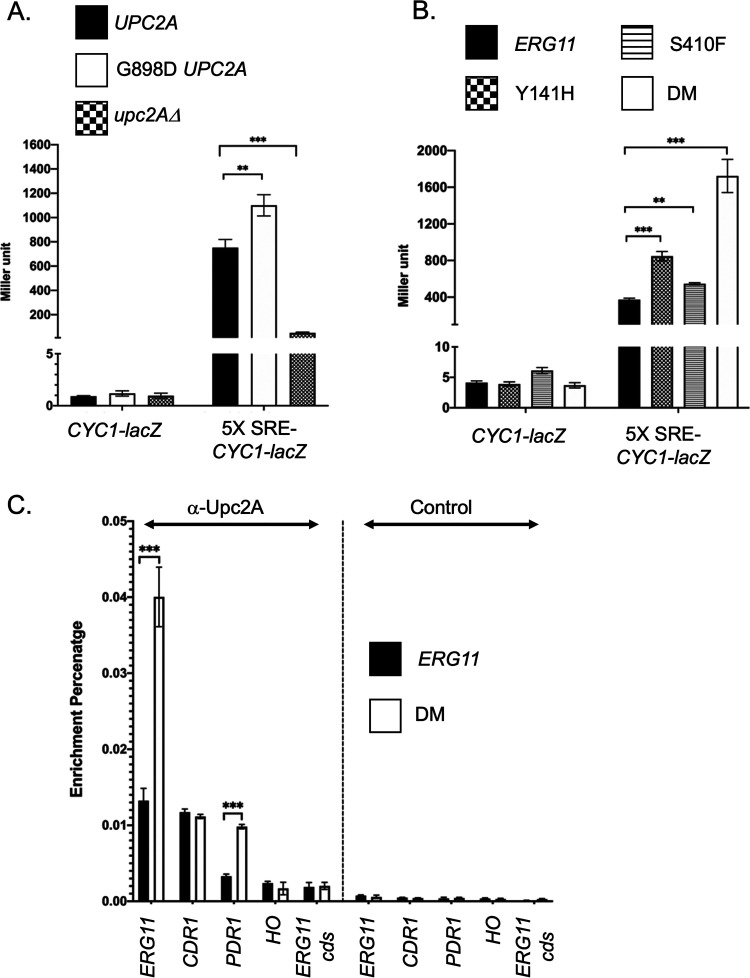
Our previous data showed that genetic inhibition of ERG11 gene expression could induce the expression of the drug efflux pump system Pdr1-Cdr1 (19). Upc2A was essential for this interaction in which it directly induced the expression levels of genes in both the ERG and Pdr1/CDR1 pathways (19, 20). Because these two pathways were also induced in the DM ERG11 mutant, we wanted to determine if Upc2A function was activated in response to these changes in the ERG11 gene. To test this, we generated an Upc2A reporter construct that contains five copies of the Upc2A binding site (Sterol Response Element: SRE) from the ERG1 promoter. These concatemerized SREs were placed upstream from the S. cerevisiae CYC1 promoter that was fused to E. coli lacZ. To confirm that this reporter system was sensitive to the UPC2A allele, we introduced this plasmid into isogenic C. glabrata strains containing the 3X HA-UPC2A, 3X HA-G898D UPC2A gain-of-function mutation (20) or a null allele of UPC2A. A plasmid containing only the CYC1-lacZ fusion gene with no upstream activation sequence was also used as negative control (Fig. 4A).
FIG 4.
Regulation of Upc2A in response to ERG11 alleles. A. Two different reporter constructs carried on low-copy-number C. glabrata vectors that contained either the S. cerevisiae CYC1 promoter region with a translational fusion to E. coli lacZ (CYC1-lacZ) or the same reporter construct with five copies of the C. glabrata ERG1 sterol response element (SRE) cloned upstream of the CYC1 promoter (5X SRE-CYC1-lacZ) were introduced into the three different C. glabrata strains indicated at the top. These strains varied at their UPC2A allele corresponding to a wild-type strain (UPC2A), gain-of-function form of UPC2A (G898D UPC2A) or a null mutation (upc2AΔ). Transformants were grown to mid-log phase and then assayed for the level of β-galactosidase produced in each strain. B. The two different reporter plasmids from A were introduced into the 4 isogenic ERG11 mutant strains indicated. Transformants were grown to mid-log phase and assayed for β-galactosidase levels. C. Chromatin immunoprecipitation (ChIP) of Upc2A DNA-binding to genomic target sites. Total sheared and cross-linked chromatin was prepared from either wild-type or DM ERG11 cells. ChIP reactions were carried out with either anti-Upc2A or a nonspecific control antisera. Immunopurified DNA was quantitated with qPCR using primer pairs that detected the promoters of ERG11 (ERG11), CDR1 (CDR1), PDR1 (PDR1) or HO (HO) along with a primer pair that detected a segment of the ERG11 coding sequence (ERG11 cds). Data are plotted as the percentage of DNA recovered in the immunopurified sample/total input DNA.
The introduction of the gain-of-function form of Upc2A (G898D UPC2A) led to production of the highest level of β-galactosidase seen from the 5X SRE-CYC1-lacZ plasmid with nearly 1200 Miller units of activity compared to approximately 800 Miller units when the wild-type UPC2A gene was present. Loss of UPC2A lowered expression to approximately 80 Miller units. All C. glabrata isolates carrying the CYC1-lacZ control plasmid lacking an upstream activation sequence showed minimal levels of β-galactosidase activity. These data are consistent with this reporter system faithfully detecting the status of the UPC2A gene.
Next, we transformed these two plasmids into strains containing the various ERG11 alleles. Appropriate transformants were grown to mid-log phase and enzyme levels determined (Fig. 4B). The highest level of β-galactosidase activity was produced in the presence of the DM ERG11 allele, consistent with this strain triggering the highest level of Upc2A activity. Both single mutant ERG11 alleles induced smaller but significant increases in expression of the 5X SRE-CYC1-lacZ plasmid compared to the wild-type ERG11-containing strain. Only minimal levels of β-galactosidase activity were produced in strains containing the CYC1-lacZ reporter plasmid lacking any SREs as upstream control elements. These data support the view that the DM ERG11 allele caused the largest increase in Upc2A function, likely through production of the most compromised form of the Erg11 enzyme, but each single mutant had a less dramatic but still detectable impact on Upc2A. To directly assess Upc2A function, we carried out single gene chromatin immunoprecipitation analysis of Upc2A binding to target promoters.
Previous work from several labs including ours (20, 25) has provided evidence that ergosterol limitation increased Upc2A DNA-binding to nuclear target genes. We also found that these Upc2A target genes included the PDR1 and CDR1 genes (19), linking control of ergosterol biosynthesis with expression of genes involved in drug resistance. To determine if the presence of the fluconazole-resistant, DM allele of ERG11 caused increased levels of Upc2A DNA binding, chromatin immunoprecipitation was used to detect binding of this factor to SREs present in the ERG11, CDR1 and PDR1 promoters. Nonspecific binding was also evaluated by examining Upc2A enrichment on the HO promoter and ERG11 coding sequence. Isogenic wild-type and DM ERG11 strains were grown to mid-log phase and Upc2A DNA-binding analyzed by ChIP using anti-Upc2A or a nonspecific control polyclonal antiserum (Fig. 4C).
Upc2A binding to the ERG11 SRE was strongly induced when cells contained the DM ERG11 allele compared to the wild-type gene. Upc2A association with the PDR1 SRE was also induced under these conditions although binding to CDR1 did not change. Control ChIP reactions confirmed that the Upc2A binding was antibody-specific and showed no enrichment with either the HO gene or the ERG11 coding sequence. The increased Upc2A-dependent transcriptional activation and DNA-binding indicate that Upc2A function was increased in strains containing the DM ERG11 allele compared to the wild-type gene.
Phenotypic comparison of PDR1 GOF and DM ERG11 mutations indicate mutant Pdr1 factors drive higher level azole resistance.
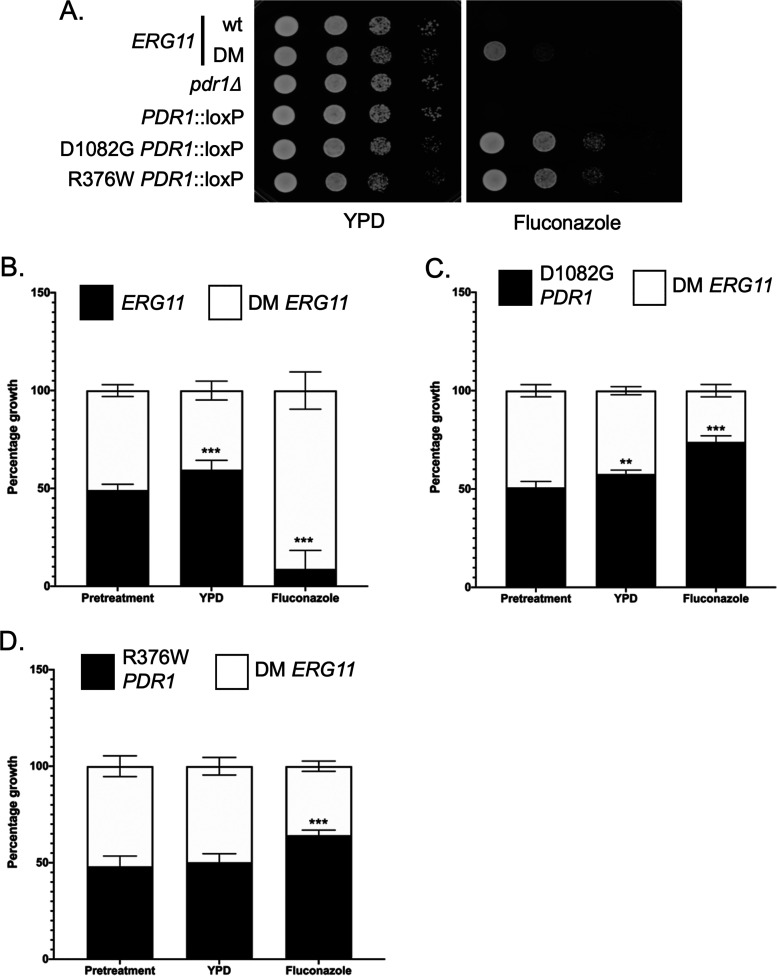
Since the DM ERG11 mutant was capable of inducing strong fluconazole resistance, we wanted to compare its behavior to that seen in the presence of PDR1 GOF alleles. These GOF mutants of PDR1 represent the vast majority of azole resistant mutants found in C. glabrata isolates (26–28). An isogenic series of strains was produced that produced wild-type or DM mutant forms of Erg11 containing the following alleles of PDR1: pdr1Δ, wild-type, D1082G or R376W. These strains were grown to mid-log phase and tested for their relative susceptibility to 30 μg/ml fluconazole in a serial dilution plate assay (Fig. 5A).
FIG 5.
Growth phenotypic comparison of DM ERG11 with gain-of-function (GOF) alleles of PDR1. A. Isogenic mid-log phase C. glabrata strains containing ERG11 wild-type or DM alleles, PDR1 null (pdr1Δ), wild-type PDR1 (PDR1::loxP), or two GOF alleles of PDR1 D1082G and R376W were serially diluted and spotted on YPD agar plates supplemented with or without fluconazole (20 μg/ml). Plates were incubated at 30°C and photographed. B. Fitness comparisons between the DM ERG11 strain compared with wild-type (B), the GOF D1082G (C) and R376W (D) PDR1 mutant strains. Equal number of mid-log phase cells from each strain were mixed together (Pretreatment). Cultures were then allowed to grow in the absence (YPD) or presence of azole drug (Fluconazole) (20 μg/ml). At the end of incubation, samples of each culture were plated and final distribution of each strain determined by plating. Deviations from the starting 50:50 mix of strains would indicate a relative fitness advantage for one strain versus the other.
Both GOF forms of PDR1 lowered fluconazole susceptibility more than the DM ERG11 strain. To probe the relative effects of mutations in either PDR1 or ERG11, we carried out a competitive growth assay in which equal numbers of cells containing mutations of interest were mixed together. These mixed cultures were then allowed to grow in the presence or absence of fluconazole challenge and the relative contribution of each mutant strain to the final population determined. This competitive growth assay allows subtle differences to be more readily detected than on a plate-based assay.
We prepared mixed cultures of isogenic wild-type and DM ERG11 cells (Fig. 5B) and allowed these cultures to grow under untreated conditions (YPD) or in YPD medium containing fluconazole to late log phase. Growth in rich medium led to an increase in the population of the wild-type (60% final) compared to the DM ERG11 strain (40% final). This analysis indicated that the DM ERG11 had a growth defect compared to wild-type cells in the absence of fluconazole but this mutant allele conferred a significant growth advantage in the presence of this drug.
The relative growth of the DM ERG11 strain was then compared to each of the PDR1 GOF mutant strains (Fig. 5C and D). In both cases, the GOF PDR1 mutant outcompeted the DM ERG11 strain in the presence of fluconazole (D1082G-74%: DM-26%; R376W-65%: DM-35%) while the only difference in growth in the absence of drug was seen in comparison of the D1082G PDR1 strain compared to the DM ERG11: 58% to 42%, respectively. Both GOF PDR1 mutants were more effective in conferring fluconazole resistance while growing at least as well (R376W) if not better (D1082G) than the DM ERG11 strain. While we are only testing a single mutant form of ERG11 here, growth differences associated with azole-resistant forms of Erg11 may help explain the predominance of GOF mutants of PDR1 in clinical azole-resistant isolates of C. glabrata.
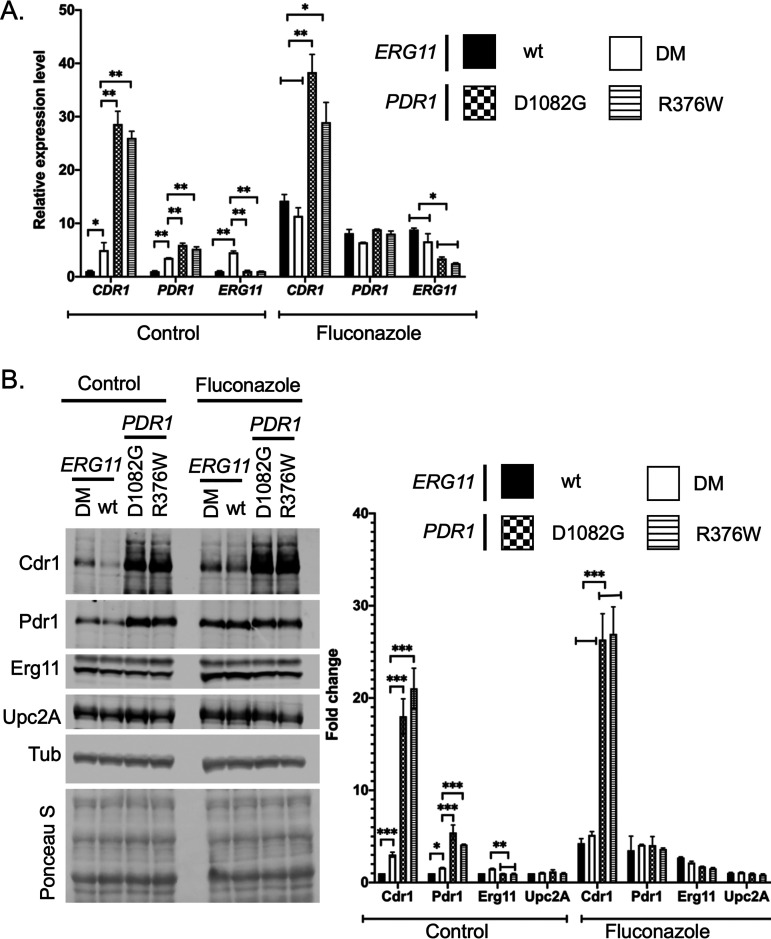
Increased activation of PDR1 and CDR1 transcription seen in the presence of GOF PDR1 mutants compared to DM ERG11.
The phenotypic data above indicated that both GOF PDR1 strains produced a stronger effect on fluconazole resistance than the presence of the DM ERG11 allele. To probe the basis of this phenotypic difference, we examined transcription of CDR1, PDR1 and ERG11 by RT-qPCR. Strains containing wild-type, D1082G or R376W forms of PDR1 or the DM ERG11 were grown to mid-log phase in the absence or presence of fluconazole. Levels of mRNA from these three genes were assayed as before.
CDR1 mRNA levels were strongly elevated in the presence of both GOF PDR1 mutations, well above those induced by the presence of the DM ERG11 mutation (Fig. 6A). This increase was seen both in the absence as well as the presence of fluconazole. PDR1 mRNA levels were significantly higher in the presence of the two GOF alleles than in the DM ERG11 but this difference was restricted to the absence of fluconazole. Drug treatment elevated both wild-type and DM ERG11 strains to levels similar to those of the GOF mutants. Finally, ERG11 mRNA levels were higher in the DM ERG11 strain than in the other three strains in the absence of fluconazole. Strikingly, fluconazole exposure induced ERG11 mRNA to higher levels in the strains containing wild-type PDR1 but was less effective in the two GOF PDR1-containing strains. These data suggest that the increased PDR1/CDR1 expression seen in the GOF PDR1 strains may reduce the level of fluconazole stress (and associated ERG11 induction) seen. The changes seen in mRNA levels were well-correlated with Western blot analyses for each protein (Fig. 6B and C). Upc2A protein levels were constant under all conditions tested, consistent with changes due to this factor being caused by alterations in Upc2A function rather than its expression.
FIG 6.
Relative gene expression responses to the DM ERG11 or two different GOF PDR1 strains. A. Strains varying at the ERG11 or PDR1 loci as indicated were grown in the presence or absence of fluconazole as in Fig. 3 and processed for RT-qPCR analyses of the indicated mRNAs. Fluconazole was included at 20 μg/ml. B. The strains above were analyzed by Western blotting as in Fig. 3 using antisera for Cdr1, Pdr1, Erg11, Upc2A or Tubulin (Tub). The membrane was stained after transfer with Ponceau S as above. Quantitation of 4 western experiments is provided on the right hand side of the figure.
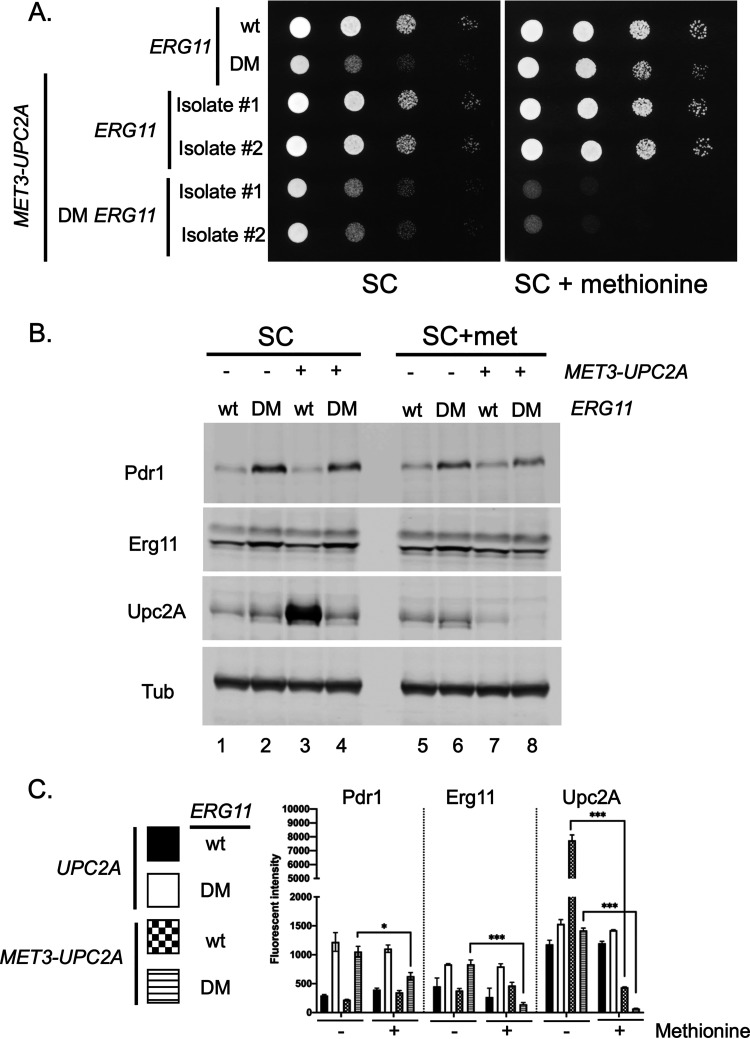
Reduced levels of Upc2A exacerbate the growth defect of the DM ERG11 strain.
Our analysis of the cellular response to the presence of the DM ERG11 mutation demonstrated the involvement of Upc2A as activity of this factor was induced in this genetic background compared to wild-type cells. We introduced the highly repressible MET3 promoter (29) in place of the chromosomal UPC2A cognate region to test the effect of changing Upc2A production via repression in the presence of exogenous methionine. This MET3-UPC2A fusion gene was generated in both wild-type and DM ERG11 strains. Appropriate transformants were placed on media lacking (SC) or containing excess methionine (SC+methionine) and allowed to grow. Two independent isolates of each MET3-UPC2A transformant were tested to evaluate strain variability in these assays.
Addition of methionine to strains containing the MET3-UPC2A fusion gene in addition to the DM ERG11 allele caused the growth of this strain to be further reduced when compared to the presence of the wild-type UPC2A gene (Fig. 7A, right hand panel). This result suggests that reduction of Upc2A levels (by repression of the MET3 promoter with methionine) caused a growth defect in the presence of the DM ERG11 mutation. When these UPC2A and MET3-UPC2A strains containing the DM ERG11 mutation were grown in the absence of exogenous methionine, their growth was similar (Fig. 7A, left hand panel).
FIG 7.
Depletion of Upc2A enhances growth defect of a DM ERG11 strain. A. The ERG11 promoter was replaced with the methionine-repressible MET3 promoter in wild-type and DM ERG11 cells. Two different isolates of each MET3 promoter replacement were grown to mid-log phase, along with the isogenic wild-type and DM ERG11 strains. These cells were serially diluted onto plates containing either synthetic complete (SC) medium or the same medium containing an excess of methionine (SC + methionine). Transformants were grown at 30°C and the plates photographed. B. The indicated strains were grown to mid-log phase in the presence or absence of methionine and whole cell protein extracts prepared. These extracts were analyzed as before using the indicated antisera. C. Quantitation of protein levels in multiple Western blots is shown. Significant changes are indicated with asterisks.
To determine the levels of Upc2A protein in these different strains, we prepared whole cell protein extracts from these strains. These extracts were analyzed by Western blotting using the anti-Upc2A antiserum. The MET3 promoter activity is highest in SC medium (29) and we will consider the data from these media conditions first (Fig. 7B, left hand panel). The MET3 promoter drove much higher levels of Upc2A protein than were produced from the native UPC2A promoter in the presence of wild-type ERG11 (compare lanes 1 and 3). The elevated expression of Upc2A seen in wild-type cells containing MET3-UPC2A did not lead to a corresponding increase in Erg11 expression (lanes 1 and 3). Interestingly, Upc2A levels were dramatically reduced when the MET3-UPC2A fusion gene was present in the DM ERG11 strain, albeit to levels similar to those seen when Upc2A was produced by the native UPC2A gene (compare lanes 2 and 4). The presence of the MET3-UPC2A allele did not affect the observed induction of Erg11 in the presence of the DM ERG11 allele (compare lanes 2 and 4), potentially due to the surprisingly low levels of Upc2A driven by the MET3-UPC2A allele when the DM ERG11 mutant was present. The lack of any detectable Upc2A expression differences in the two DM ERG11 strains (varying due to their different UPC2A promoters) also correlated well with the equivalent ability of these strains to grow on SC medium (Fig. 7A above).
When methionine was added to repress MET3-UPC2A transcription, the levels of the corresponding Upc2A protein were strongly reduced as expected (Fig. 7B, right hand panel). However, the reduction of Upc2A upon methionine repression led to different levels of this transcription factor in a manner dependent on the ERG11 allele present in the cell. Methionine repression of the MET3-UPC2A in the DM ERG11 strain blocked the production of a detectable level of Upc2A while this same treatment of cells carrying wild-type ERG11 still produced readily detectable Upc2A (compare lanes 7 and 8). The extremely low level Upc2A production seen in the methionine-repressed MET3-UPC2A with DM ERG11 may explain the methionine-dependent growth defect seen in this strain (see Fig. 7A). These low levels of Upc2A also blocked the normal increase in Erg11 and Pdr1 caused by the DM ERG11 allele (compare lanes 6 and 8), suggesting that both Erg11 and Pdr1 expression were, in part, driven by Upc2A function in this mutant background.
We carried out this same analysis but using a MET3-PDR1 gene (Fig. S2). In the absence of fluconazole, there was no significant effect of the MET3-PDR1 fusion gene on growth of either the wild-type or DM ERG11 strain irrespective of the presence of methionine (Fig. S2A). The addition of fluconazole revealed the strong increase in resistance shown by the DM ERG11 strains in the presence of either the wild-type or MET3-driven PDR1 gene, in the absence of methionine (Fig. S2A, top panels). The addition of methionine to these media to repress MET3-PDR1 (Fig. S2A, bottom panels) caused a loss of fluconazole resistance at the highest concentration tested. These data illustrate the critical contribution made by Pdr1 to the fluconazole resistance phenotype of the DM ERG11 strain.
Western blot analyses were carried out to determine the effect of the MET3-PDR1 gene on expression of Pdr1 itself, Cdr1 and Erg11 (Fig. S2B). Steady-state levels of Pdr1 were much higher in the presence of the wild-type ERG11 gene than in the DM ERG11-containing strain (compare lanes 1 and 3). The expression of Pdr1 in these two strains was inversely correlated with expression of the Pdr1 target gene CDR1 as evidenced by the high Cdr1 levels in the DM ERG11 strain compared to the wild-type strain (compare lanes 1 and 3). This finding suggests that Pdr1 is activated in the presence of the DM ERG11 allele as its steady-state expression is lower yet Cdr1 expression is higher. Previously, we used the MET3 promoter to produce either wild-type or GOF Pdr1 proteins (30). The GOF proteins were found to be less stable than the corresponding wild-type factor. Production of this hypomorphic form of Erg11, in the absence of any azole drug, was sufficient to strongly induce Pdr1 activity and Cdr1 synthesis.
DISCUSSION
One of the nearly inevitable outcomes from the extensive use of fluconazole as an antifungal drug has been the development of resistance. Mutant forms of ERG11 have been found in every Candida species with the exception of C. glabrata and C. krusei (31). Detailed analyses of the C. albicans Erg11 enzyme have clearly implicated structural changes within azole-resistant mutant forms of this enzyme leading to reduced azole susceptibility (reviewed in Ref. 32). Azole resistance caused by mutational alteration within Erg11 is most parsimoniously interpreted in this manner.
Our data provide a second means of ERG11 mutants impacting azole resistance via a secondary effect on expression of Pdr1/CDR1 in C. glabrata. While it is clear that the mutants we have characterized here have effects in addition to activation of Pdr1, genetic depletion by methionine repression of MET3-PDR1 caused an approximate 5-fold reduction in fluconazole resistance in the DM ERG11 strain (Fig. S2). This finding indicates that the presence of the DM ERG11 allele is not sufficient to confer the normal elevation in fluconazole resistance seen and requires the function of Pdr1. This type of a secondary effect can have major implications in the observed azole resistance of a given ERG11 mutant background and is an important consideration in evaluation of this phenotype.
Fluconazole resistant clinical isolates of C. albicans are readily recovered with lesions in the CaERG11 gene but azole resistant isolates with mutations in this locus have rarely been reported in C. glabrata (33). We constructed C. glabrata ERG11 mutations based on cognate changes in the C. albicans gene. We selected positions Y141 and S410 in the C. glabrata enzyme as the analogous positions in the C. albicans Erg11 could be found mutated either alone or together in azole-resistant clinical isolates (9). The C. glabrata equivalent mutations caused azole resistance as expected but the DM allele also exhibited a growth defect, even on rich YPD medium, which was not reported for the C. albicans mutant. This growth defect, at least for the DM ERG11 mutant, may be part of the reason that mutations in ERG11 are not found in C. glabrata isolates. Confirmation of this suggestion will require more detailed study of the corresponding mutations in C. albicans as well as analyses of additional ERG11 mutants in C. glabrata.
The DM ERG11 allele was also found to cause several other drug phenotypes. The increased resistance to caspofungin was not expected and may involve induction of the FKS2 gene (Fig. 2D). We have preliminary evidence that Upc2A may act to control expression of β-glucan synthase (20) but have not yet linked this to FKS2. Crz1 is a well-established transcriptional activator of FKS gene expression (34, 35) and we provide evidence that the CRZ1 gene must be intact for the DM ERG11 allele to induce caspofungin resistance (Fig. 2E).
The DM ERG11 seems to be acting as a reduced function or hypomorphic allele of this gene. This was evidenced by the reduced level of ergosterol accumulation in the DM mutant background and the increased amphotericin B resistance. We suggest that this reduction in function is critical for the observed induction of the ERG pathway genes as well as the Pdr1/CDR1 regulatory circuit. Previous work from our lab showed that genetically depleting Erg11 by repression of two different artificial promoters driving ERG11 caused the same changes in gene regulation (19). The behavior of the DM ERG11 mutant strain provides further support for our hypothesis that a physiological link exists between the ergosterol biosynthetic pathway and Pdr1 (18).
Comparison of the DM ERG11 allele with each of the two single mutant alleles suggests that there is a synthetic interaction between the Y141H and S410F mutations. In neither of the two single mutants was evidence obtained that endogenous downstream gene expression was being induced in the presence of these altered enzymes. However, when both of these lesions were present in the same protein, a range of downstream responses were triggered. These include FKS2 induction and Pdr1 activation along with Upc2A. This was not observed with the single mutants. We suggest that the single mutants seem to cause increased fluconazole resistance primarily through their action on activity of the altered enzyme. The very sensitive 5X SRE-CYC1-lacZ reporter plasmid was modestly induced in the presence of each single mutant Erg11 (Fig. 4B), but their effect was small compared to the induction caused by the DM Erg11 protein. The combination of the two single mutations led to production of a more defective Erg11 protein that in turn caused intracellular responses such as activation of Upc2A and Pdr1.
The activation of Upc2A-dependent transcription is a central feature of the cellular response to the presence of the DM ERG11 allele. We produced a conditional MET3-UPC2A strain that contained the DM ERG11 mutation to avoid any problematic interactions caused by introduction of a upc2AΔ null allele into this strain. The essential contribution of Upc2A to the maintenance of the DM ERG11-containing strain could be shown by the severe growth defect caused by methionine repression of this UPC2A allele (Fig. 7A). We suggest that the presence of the DM ERG11 allele led to induction of Upc2A function which in turn is linked to an increased level of degradation of this active transcription factor. This suggestion is supported by the inhibition of MET3-UPC2A transcription by the addition of methionine leading to a drop in Upc2A levels below the limit of detection in the DM ERG11 background (Fig. 7B). This is roughly the equivalent of a direct protein stability assay as we block synthesis at the level of transcription while allowing degradation to proceed without intervention. The apparent instability of the wild-type Upc2A after activation by the presence of the DM Erg11 protein is very similar to the instability found when GOF forms of Pdr1 are compared to wild-type Pdr1 (17). These factors are evidently deleterious for the cell to maintain in their activated states unless their presence is crucial for a given stress response. We have previously observed this as a hyperactive mutant form of Pdr1 can be lethal (17). These data suggest that an important control point for Upc2A may lie at the level of protein stability.
One reason for constructing these different fluconazole resistant forms of Erg11 in C. glabrata was to see if their presence would influence fluconazole induction of Pdr1. It is possible that fluconazole resistant forms of Erg11 would no longer respond to challenge by this azole drug. This was precisely what was observed in the case of the DM ERG11 allele in which the elevated CDR1, PDR1 and ERG11 expression seen in the absence of fluconazole was unaffected by the addition of this azole drug (Fig. 3). This readily interpreted as the DM Erg11 enzyme is no longer subject to fluconazole inhibition of function. In addition, the striking elevation of PDR1 and CDR1 transcription in the DM ERG11-containing strain in the absence of any drug argues that these genes and the pathways they define are normally co-regulated, irrespective of the presence of drug. The corresponding single mutant forms of Erg11 were still seen to trigger gene induction in the presence of fluconazole, indicating that these mutant forms of the enzyme were still sensitive to the presence of this drug.
The simplest interpretation for azole resistant ERG11 mutations in multiple organisms has been that the altered enzymes may be less able to interact with azole drugs and this can explain their resistance phenotype. While this is certainly true for some mutants (such as the two single mutant alleles of ERG11 analyzed here), there are likely to be more complex physiological responses to these mutant enzymes that can alter the resistance phenotype in ERG11 mutant strains. Ergosterol is an essential constituent of the fungal cell membrane and limiting its biosynthesis triggers transcriptional correction in most fungi examined (5, 36, 37). In C. glabrata, there appears to be a threshold of ergosterol limitation that is not crossed by the single mutant forms of Erg11 but is exceeded by the DM Erg11 enzyme. Exceeding this threshold leads to activation of downstream pathways to deal with the detected ergosterol limitation.
Our finding of these deeper regulatory connections between ergosterol biosynthesis and transcription circuitry including but not restricted to factors directly involved in ERG gene control suggests that additional linkages of this type may be participating in azole resistance in other fungi. Azole-resistant forms of Aspergillus fumigatus are commonly associated with substitution mutations within the cyp51A gene (ERG11 equivalent in Aspergilli) (37). Some of the most common azole-resistant alleles found in A. fumigatus are the result of linked changes in which a residue in the coding sequence is changed and a region of the promoter is duplicated (reviewed in Ref. 38). These linked changes act together to enhance azole resistance more than either alone (39), suggesting that alleles of this sort engage more than one mechanism to affect azole resistance. We have previously shown that the L98H form of Cyp51A in A. fumigatus produces lower levels of this enzyme than the wild-type yet is still resistant to voriconazole (40). In addition, mutations in the HMGCoA reductase gene (hmg1) from A. fumigatus also causes a strong decrease in voriconazole susceptibility although cyp51A gene expression appeared to be unaffected (41). These observations suggest that other fungi may also employ additional transcriptional circuitry to impact azole resistance in response to alterations in ergosterol biosynthesis. Identifying and characterizing these other transcriptional contributors to azole resistance are important goals in the dissection of the molecular basis of azole resistance.
MATERIALS AND METHODS
Strains and growth conditions.
C. glabrata was grown in rich YPD medium (1% yeast extract, 2% peptone, 2% glucose) or under amino acid-selective conditions in complete supplemental medium (CSM) (Difco yeast nitrogen extract without amino acids, amino acid powder from Sunrise Science Products, 2% glucose). YPD media supplemented with 50 μg/ml nourseothricin (Jena Bioscience, Jena, Germany) and 2 mM methionine was used to select strains containing pBV65 vector derivatives (42). CSM media (2% glucose, 1 mM estradiol) without methionine was used to recycle the selection cassette on pBV65. YPD solid agar supplemented with 1 mg/ml 5-fluoro-orotic acid was used to cure the pBV43 plasmid. All strains used in this study are listed in Table 1.
TABLE 1.
Strains used in this work
| Strain name | Parent | Genotype |
|---|---|---|
| SPG96 | KKY2001 | his3Δ::FRT leu2Δ::FRT trp1Δ::FRT ura3Δ(-85 + 932)::Tn903NeoR |
| BVGC14 | SPG96 | his3Δ::FRT trp1Δ::FRT CEN-ScERG11/URA3 erg11Δ::LEU2 |
| BVGC344 | BVGC14 | his3Δ::FRT leu2Δ::FRT trp1Δ::FRT ura3Δ(-85 + 932)::Tn903NeoR ERG11::HIS3 |
| BVGC346 | BVGC14 | his3Δ::FRT leu2Δ::FRT trp1Δ::FRT ura3Δ(-85 + 932)::Tn903NeoR Y141H ERG11::HIS3 |
| BVGC336 | BVGC14 | his3Δ::FRT leu2Δ::FRT trp1Δ::FRT ura3Δ(-85 + 932)::Tn903NeoR S410F ERG11::HIS3 |
| BVGC340 | BVGC14 | his3Δ::FRT leu2Δ::FRT trp1Δ::FRT ura3Δ(-85 + 932)::Tn903NeoR Y141H S410F ERG11::HIS3 |
| BVGC367 | BVGC344 | his3Δ::FRT leu2Δ::FRT trp1Δ::FRT ura3Δ(-85 + 932)::Tn903NeoR ERG11::HIS3 pdr1Δ::loxP |
| BVGC383 | BVGC367 | his3Δ::FRT leu2Δ::FRT trp1Δ::FRT ura3Δ(-85 + 932)::Tn903NeoR ERG11::HIS3 PDR1::loxP |
| BVGC387 | BVGC367 | his3Δ::FRT leu2Δ::FRT trp1Δ::FRT ura3Δ(-85 + 932)::Tn903NeoR ERG11::HIS3 D1082G PDR1::loxP |
| BVGC391 | BVGC367 | his3Δ::FRT leu2Δ::FRT trp1Δ::FRT ura3Δ(-85 + 932)::Tn903NeoR ERG11::HIS3 R376W PDR1::loxP |
| BVGC361 | BVGC340 | his3Δ::FRT leu2Δ::FRT trp1Δ::FRT ura3Δ(-85 + 932)::Tn903NeoR Y141H S410F ERG11::HIS3 PDR1::loxP |
| BVGC365 | BVGC361 | his3Δ::FRT leu2Δ::LEU2 trp1Δ::FRT ura3Δ(-85 + 932)::Tn903NeoR Y141H S410F ERG11::HIS3 PDR1-LOXP |
| BVGC374 | BVGC344 | his3Δ::FRT leu2Δ::FRT trp1Δ::FRT ura3Δ(-85 + 932)::Tn903NeoR ERG11::HIS3 URA3::MET3-PDR1 |
| BVGC378 | BVGC344 | his3Δ::FRT leu2Δ::FRT trp1Δ::FRT ura3Δ(-85 + 932)::Tn903NeoR ERG11::HIS3 URA3::MET3-UPC2A |
| BVGC395 | BVGC340 | his3Δ::FRT leu2Δ::FRT trp1Δ::FRT ura3Δ(-85 + 932)::Tn903NeoR Y141H S410F ERG11::HIS3 URA3::MET3-PDR1 |
| BVGC404 | BVGC340 | his3Δ::FRT leu2Δ::FRT trp1Δ::FRT ura3Δ(-85 + 932)::Tn903NeoR Y141H S410F ERG11::HIS3 URA3::MET3-UPC2A |
| BVGC511 | BVGC344 | his3Δ::FRT leu2Δ::FRT trp1Δ::FRT ura3Δ(-85 + 932)::Tn903NeoR ERG11::HIS3 crz1Δ::loxP |
| BVGC514 | BVGC340 | his3Δ::FRT leu2Δ::FRT trp1Δ::FRT ura3Δ(-85 + 932)::Tn903NeoR Y141H S410F ERG11::HIS3 crz1Δ::loxP |
Plasmid construction and promoter mutagenesis.
All constructs used for homologous recombination into the chromosome were constructed in the pUC19 vector (New England Biolabs, Ipswich, MA). PCR was used to amplify DNA fragments. PCR products were run on 1.5% agarose gel (RPI, Troy, NY), excised and purified with Purelink quick gel extraction and PCR cleaning combo kit (Invitrogen, Carlsbad, CA). Gibson assembly cloning (New England Biolabs) was routinely used to assemble fragments together into appropriate plasmid backbones. All isogenic deletion constructs were made by assembling the recyclable cassette from pBV65 and fragments from the immediate upstream and downstream regions of the target genes. Fragments of the target genes were amplified from CBS138 background. Eviction of the recyclable cassette left a single copy of loxP in place of the excised target gene coding region.
PDR1 constructs were made by using Gibson assembly (43) to produce fragments corresponding to the wild-type, R376W or D1082G alleles of PDR1 with a recyclable cassette located downstream from the natural stop codon. Eviction of the recyclable cassette in the integrated constructs left a single copy of loxP 250–300 base-pairs downstream of the target gene stop codons. R376W and D1082G forms of PDR1 were amplified from pSK70 and pSK71 backgrounds (17).
ERG11 wild-type and mutant integration constructs were made as above with the resulting fragments corresponding to the wild-type, Y141H, S410F, or the Y141H S410F alleles of ERG11 with the his3MX6 (19) marker located 270 bp from the natural stop codon.
Conversion of strains to LEU2 was done by PCR amplifying the LEU2 coding region along with 500 bp immediately upstream and downstream from the CBS138 background. Linear DNA was then transformed into KKY2001 and the colonies were selected on CSM agar without leucine.
To generate the S. cerevisiae ERG11 complementing plasmid (pBV43), the ScERG11 promoter (800 bp upstream of AUG), ScERG11 coding sequence, and ScERG11 terminator (350 bp downstream of the stop codon) were PCR amplified from the S288c wild-type strain (SEY6210). This fragment was then cloned into the pCU-MET3 vector (29), which was pre-digested with SacI and XhoI.
The Sterol Response Element (SRE) reporter construct (pBV382) was produced by cloning the E. coli lacZ gene as a PCR fragment from pSK80 (17). This fragment was then cloned into the pCL vector (29), which was pre-digested with SaII and XhoI. A Upc2A-responsive promoter cassette, containing the 5 concatemerized copies of the ERG1 promoter SRE (-725 to -775) located downstream from the ADH1 terminator and upstream of the CYC1 minimal promoter, was generated by Genscript (Piscataway, NJ). The Upc2A responsive promoter was PCR amplified and cloned into the pCL-lacZ vector, digested with SacI and SalI. The control vector (pBV378) was constructed by cloning the ADH1 terminator and the CYC1 minimal promoter, PCR amplified from BVGC7 (19), into the pCL-lacZ vector, digested with SacI and SalI.
To generate the MET3-driven UPC2A and PDR1 strains, the MET3 promoter and the URA3 gene were PCR amplified from the pUC-MET3 plasmid. This URA3-MET3 promoter cassette was placed immediately upstream of the target gene AUG codon by recombination into the gene of interest.
C. glabrata transformation.
Cell transformations were performed using a lithium acetate method. After being heat shocked, cells were either directly plated onto selective CSM agar plates (for auxotrophic complementation) or grown at 30°C at 200 rpm overnight (for nourseothricin selection). Overnight cultures were then plated on YPD agar plates supplemented with 50 μg/ml of nourseothricin. Plates were incubated at 30°C for 24 to 48 h. In case of chromosomal insertion, individual colonies were isolated and screened by PCR for correct insertion of the targeted construct.
Construction of mutant ERG11 strains.
Since Erg11 is essential for C. glabrata aerobic growth (44), A URA3 (ScURA3)-containing plasmid shuffling technique was used to construct ERG11 mutant alleles in the SPG96 (ura3Δ) C. glabrata strain (17). An autonomous plasmid pBV43 marked with ScURA3 and containing the wild-type ScERG11 was first transformed into the SPG96 strain. Then ERG11 was deleted in the chromosome by homologous recombination, leaving ScERG11 on pBV43 functioning as the sole copy of the ERG11 gene. Next, the wildtype C. glabrata ERG11 and the three mutant alleles were individually reintroduced to the normal locus by homologous recombination with selection for His+ transformants. Finally, the pBV43 plasmid was cured with 1 mg/ml 5-FOA on a YPD agar plate to construct the C. glabrata strains, BVGC344 (ERG11), BVGC346 (Y141H ERG11), BVGC336 (S410F ERG11), and a strain containing an ERG11 gene with both of these mutations present (BVGC340: DM ERG11).
Total sterol estimation.
Cell total sterol was extracted and measured as previously described (19). In short, cell pellets were lysed in 25% alcoholic potassium hydroxide at 90°C for 2 h. Total sterol was detected by spectrophotometric scanning between the wavelengths of 240 nm and 300 nm. The presence of ergosterol in the extracted sample resulted in a four-peak curve with peaks located at approximately 262, 270, 281, and 290 nm.
Quantification of transcript levels by RT-qPCR.
Total RNA was extracted from cells by extraction using TRIzol (Invitrogen) and chloroform (Fisher Scientific, Hampton, NH) followed by purification with RNeasy minicolumns (Qiagen, Redwood City, CA). 500 ng to 1 μg total RNA was reverse-transcribed using an iScript cDNA synthesis kit (Bio-Rad, Des Plaines, IL). Assay of RNA via quantitative PCR (qPCR) was performed with iTaq universal SYBR green supermix (Bio-Rad). Target gene transcript levels were normalized to transcript levels of 18S rRNA. Expression levels of each gene in the 4 different ERG11 backgrounds are normalized to wild-type levels in the absence of fluconazole.
Spot test assay.
Cells were grown in YPD medium to mid-log-phase. Cultures were then 10-fold serially diluted and spotted onto YPD agar plates containing different concentrations of fluconazole (LKT laboratories, St Paul, MN), caspofungin (Apexbio, Houston, TX) or Amphotericin B (Sigma). All agar plates were incubated at 30°C for 24 to 48 h before imaging was performed.
β-galactosidase assay.
Harvested cells were lysed with glass beads (Scientific Industries Inc) in breaking buffer (100 mM Tris pH 8, 1 mM Dithiothreitol, and 20% Glycerol) at 4°C for 10 min. Lysate was collected and β-galactosidase enzymatic reaction was carried out in Z-buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 50 mM 2-Mercaptoethanol) with 650 μg/ml O-nitrophenyl-β-d-galactopyranoside (ONPG). Miller units were calculated based on the equation: (OD420 × 1.7)/(0.0045 × total protein concentration × used extract volume × time). Bradford assay (Bio-Rad) was used to measure the total protein concentration in the lysate.
Chromatin immunoprecipitation (ChIP).
The detailed ChIP protocol was previously described (19). The sheared chromatin was incubated with rabbit polyclonal anti-Upc2A antibody (1:50 dilution) or rabbit IgG control antibody, NBP2-24891 (Novus Biologicals, Centennial, CO) for 2 h before being incubated together with 30 μl of washed protein G Dynabeads (Life Technologies) overnight on a nutator at 4°C. Washing, reversal of cross-links, and purification of DNA processed by the use of ChIP were performed as described in Ref. 19.
Real-time PCR was performed on ChIP purified DNA, under the following conditions: 1 cycle of 95°C for 30 s followed by 40 cycles of 95°C for 15 s and 57°C for 30 s on a MyiQ2 Bio-Rad machine. A 1 μl volume of the DNA processed by the use of ChIP (diluted 5-fold) or of input (diluted 20-fold) DNA was used in a reaction mixture with a 20 μl total volume using SYBR green master mix (Bio-Rad) and a 0.4 μM concentration of each primer. The percent input method was used to calculate the signal of enrichment of the promoter region for each gene. ERG11, CDR1, and PDR1 promoters were analyzed with primers specifically targeting the ERG11 promoter (−561 to −694 relative to the ATG as +1), CDR1 promoter (−476 to −665), and PDR1 promoter (−551 to −651) regions. A region of ERG11, located within the coding sequence (+939 to +1042) and the HO promoter (-585 to -751) region were used as negative controls.
Competitive growth assay.
The LEU2 coding region along with its immediate 500 bp up- and downstream sequences were amplified from CBS138 genomic DNA. The product was then used to transform the leu2Δ::FRT allele to LEU2 in DM ERG11 background at the LEU2 native locus to generate BVGC365 (DM ERG11/LEU2). A mid-log growth culture [between O.D. of 1 and 2] was diluted to 0.2 O.D. in fresh YPD. Each culture to be tested: wild-type, R376W PDR1 or D1082G PDR1, was mixed at a 1:1 ratio with DM ERG11/LEU2, and the mixed cultures were treated either with fluconazole (20 μg/ml) or ethanol for 24 h at 30°C. Cultures were collected, serially diluted, and plated on YPD and CSM media without leucine for CFU analysis.
Antibodies and western immunoblotting.
Cells were lysed with lysis buffer (1.85 M NaOH, 7.5% 2-Mercaptoethanol). Proteins were precipitated with 50% Trichloroacetic acid and resuspended in Urea buffer (40 mM Tris pH 8, 8.0 M Urea, 5% SDS, 1% 2-Mercaptoethanol). Cdr1, Pdr1, and Upc2A rabbit polyclonal antibodies were previously described (19). Anti-HA monoclonal antibody was purchased from Invitrogen. Secondary antibodies were purchased from LI-COR Biosciences. Imaging was performed with Odyssey CLx Imaging System (LI-COR Biosciences) and analyzed by Image Studio Lite Software (LI-COR Biosciences). Detected target band fluorescence intensity was normalized against tubulin fluorescence intensity and compiled from two biological replicate experiments and two technical replicates in each experiment, giving four replicates in total. Erg11 anti-peptide rabbit polyclonal antibody was produced by GenScript (Piscataway, NJ). The peptide sequence was AKIYWEKRHPEQKY, and it covered the 520th to 533rd amino-acids in the Erg11 protein primary sequence. The peptide antibody was confirmed by Western blotting comparing wild-type and a strain carrying an ERG11-3X HA allele. Anti-peptide antibody detected native Erg11 from wild-type cells and the increased molecular mass of the Erg11-3X HA protein. Dual color LI-COR secondary antibodies were used to overlap the band intensity of Erg11 protein detected with anti-HA mouse monoclonal antibody and Erg11 peptide rabbit polyclonal antibody.
Statistics.
The Student’s t-test was used to assess the statistical significance of results of comparisons of samples. Paired conditions were used for comparisons of results from the same isolate obtained under different treatment conditions, while unpaired conditions were used for comparisons of results from isolates obtained under the same treatment conditions (*, P < 0.5; **, P < 0.01; ***, P < 0.001).
ACKNOWLEDGMENTS
This work was supported by NIH AI152494 to W.S.M.-R. We thank Damian Krysan for useful suggestions and Tom Conway and Lucia Simonicova for critically reading this manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Rodrigues ML. 2018. The multifunctional fungal ergosterol. mBio 9: e01755-18. 10.1128/mBio.01755-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sokolov SS, Trushina NI, Severin FF, Knorre DA. 2019. Ergosterol turnover in yeast: An interplay between biosynthesis and transport. Biochemistry (Mosc) 84:346–357. 10.1134/S0006297919040023. [DOI] [PubMed] [Google Scholar]
- 3.Zhang J, Li L, Lv Q, Yan L, Wang Y, Jiang Y. 2019. The fungal CYP51s: Their functions, structures, related drug resistance, and inhibitors. Front Microbiol 10:691. 10.3389/fmicb.2019.00691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Warrilow AG, Martel CM, Parker JE, Melo N, Lamb DC, Nes WD, Kelly DE, Kelly SL. 2010. Azole binding properties of Candida albicans sterol 14-alpha demethylase (CaCYP51). Antimicrob Agents Chemother 54:4235–4245. 10.1128/AAC.00587-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jorda T, Puig S. 2020. Regulation of ergosterol biosynthesis in Saccharomyces cerevisiae. Genes (Basel) 11:795. 10.3390/genes11070795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keniya MV, Sabherwal M, Wilson RK, Woods MA, Sagatova AA, Tyndall JDA, Monk BC. 2018. Crystal structures of full-length lanosterol 14alpha-demethylases of prominent fungal pathogens Candida albicans and Candida glabrata provide tools for antifungal discovery. Antimicrob Agents Chemother 62(11). 10.1128/AAC.01134-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sagatova AA, Keniya MV, Wilson RK, Monk BC, Tyndall JD. 2015. Structural insights into binding of the antifungal drug fluconazole to Saccharomyces cerevisiae lanosterol 14alpha-demethylase. Antimicrob Agents Chemother 59:4982–4989. 10.1128/AAC.00925-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warrilow AG, Parker JE, Kelly DE, Kelly SL. 2013. Azole affinity of sterol 14alpha-demethylase (CYP51) enzymes from Candida albicans and Homo sapiens. Antimicrob Agents Chemother 57:1352–1360. 10.1128/AAC.02067-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanglard D, Ischer F, Koymans L, Bille J. 1998. Amino acid substitutions in the cytochrome P-450 lanosterol 14alpha-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents. Antimicrob Agents Chemother 42:241–253. 10.1128/AAC.42.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flowers SA, Colon B, Whaley SG, Schuler MA, Rogers PD. 2015. Contribution of clinically derived mutations in ERG11 to azole resistance in Candida albicans. Antimicrob Agents Chemother 59:450–460. 10.1128/AAC.03470-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelly SL, Lamb DC, Kelly DE. 1999. Y132H substitution in Candida albicans sterol 14alpha-demethylase confers fluconazole resistance by preventing binding to haem. FEMS Microbiol Lett 180:171–175. 10.1111/j.1574-6968.1999.tb08792.x. [DOI] [PubMed] [Google Scholar]
- 12.Kelly SL, Lamb DC, Loeffler J, Einsele H, Kelly DE. 1999. The G464S amino acid substitution in Candida albicans sterol 14alpha-demethylase causes fluconazole resistance in the clinic through reduced affinity. Biochem Biophys Res Commun 262:174–179. 10.1006/bbrc.1999.1136. [DOI] [PubMed] [Google Scholar]
- 13.Warrilow AG, Mullins JG, Hull CM, Parker JE, Lamb DC, Kelly DE, Kelly SL. 2012. S279 point mutations in Candida albicans Sterol 14-alpha demethylase (CYP51) reduce in vitro inhibition by fluconazole. Antimicrob Agents Chemother 56:2099–2107. 10.1128/AAC.05389-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marichal P, Koymans L, Willemsens S, Bellens D, Verhasselt P, Luyten W, Borgers M, Ramaekers FCS, Odds FC, Vanden Bossche H. 1999. Contribution of mutations in the cytochrome P450 14alpha-demethylase (Erg11p, Cyp51p) to azole resistance in Candida albicans. Microbiology (Reading) 145:2701–2713. 10.1099/00221287-145-10-2701. [DOI] [PubMed] [Google Scholar]
- 15.Warrilow AG, Nishimoto AT, Parker JE, Price CL, Flowers SA, Kelly DE, Rogers PD, Kelly SL. 2019. The evolution of azole resistance in Candida albicans sterol 14alpha-demethylase (CYP51) through incremental amino acid substitutions. Antimicrob Agents Chemother 63(5): e02586-18. 10.1128/AAC.02586-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moye-Rowley WS. 2019. Multiple interfaces control activity of the Candida glabrata Pdr1 transcription factor mediating azole drug resistance. Curr Genet 65:103–108. 10.1007/s00294-018-0870-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khakhina S, Simonicova L, Moye-Rowley WS. 2018. Positive autoregulation and repression of transactivation are key regulatory features of the Candida glabrata Pdr1 transcription factor. Mol Microbiol 107:747–764. 10.1111/mmi.13913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moye-Rowley WS. 2020. Linkage between genes involved in azole resistance and ergosterol biosynthesis. PLoS Pathog 16:e1008819. 10.1371/journal.ppat.1008819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vu BG, Thomas GH, Moye-Rowley WS. 2019. Evidence that ergosterol biosynthesis modulates activity of the Pdr1 transcription factor in Candida glabrata. mBio 10(3). 10.1128/mBio.00934-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vu BG, Stamnes MA, Li Y, Rogers PD, Moye-Rowley WS. 2021. Azole resistance is mediated by integration of sterol gene regulation and membrane transporter production by the zinc cluster-containing transcription factor Upc2A in Candida glabrata. bioRxiv
- 21.Monk BC, Tomasiak TM, Keniya MV, Huschmann FU, Tyndall JD, O'Connell JD, 3rd, Cannon RD, McDonald JG, Rodriguez A, Finer-Moore JS, Stroud RM. 2014. Architecture of a single membrane spanning cytochrome P450 suggests constraints that orient the catalytic domain relative to a bilayer. Proc Natl Acad Sci USA 111:3865–3870. 10.1073/pnas.1324245111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiang MJ, Liu JY, Ni PH, Wang S, Shi C, Wei B, Ni YX, Ge HL. 2013. Erg11 mutations associated with azole resistance in clinical isolates of Candida albicans. FEMS Yeast Res 13:386–393. 10.1111/1567-1364.12042. [DOI] [PubMed] [Google Scholar]
- 23.Davies BS, Wang HS, Rine J. 2005. Dual activators of the sterol biosynthetic pathway of Saccharomyces cerevisiae: similar activation/regulatory domains but different response mechanisms. Mol Cell Biol 25:7375–7385. 10.1128/MCB.25.16.7375-7385.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marie C, Leyde S, White TC. 2008. Cytoplasmic localization of sterol transcription factors Upc2p and Ecm22p in S. cerevisiae. Fungal Genet Biol 45:1430–1438. 10.1016/j.fgb.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang H, Tong J, Lee CW, Ha S, Eom SH, Im YJ. 2015. Structural mechanism of ergosterol regulation by fungal sterol transcription factor Upc2. Nat Commun 6:6129. 10.1038/ncomms7129. [DOI] [PubMed] [Google Scholar]
- 26.Vermitsky JP, Edlind TD. 2004. Azole resistance in Candida glabrata: coordinate upregulation of multidrug transporters and evidence for a Pdr1-like transcription factor. Antimicrob Agents Chemother 48:3773–3781. 10.1128/AAC.48.10.3773-3781.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsai HF, Krol AA, Sarti KE, Bennett JE. 2006. Candida glabrata PDR1, a transcriptional regulator of a pleiotropic drug resistance network, mediates azole resistance in clinical isolates and petite mutants. Antimicrob Agents Chemother 50:1384–1392. 10.1128/AAC.50.4.1384-1392.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferrari S, Ischer F, Calabrese D, Posteraro B, Sanguinetti M, Fadda G, Rohde B, Bauser C, Bader O, Sanglard D. 2009. Gain of function mutations in CgPDR1 of Candida glabrata not only mediate antifungal resistance but also enhance virulence. PLoS Pathog 5:e1000268. 10.1371/journal.ppat.1000268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zordan RE, Ren Y, Pan SJ, Rotondo G, De Las Penas A, Iluore J, Cormack BP. 2013. Expression plasmids for use in Candida glabrata. G3 3:1675–1686. 10.1534/g3.113.006908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simonicova L, Moye-Rowley WS. 2020. Functional information from clinically-derived drug resistant forms of the Candida glabrata Pdr1 transcription factor. PLoS Genet 16:e1009005. 10.1371/journal.pgen.1009005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berkow EL, Lockhart SR. 2017. Fluconazole resistance in Candida species: a current perspective. Infect Drug Resist 10:237–245. 10.2147/IDR.S118892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parker JE, Warrilow AG, Price CL, Mullins JG, Kelly DE, Kelly SL. 2014. Resistance to antifungals that target CYP51. J Chem Biol 7:143–161. 10.1007/s12154-014-0121-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hull CM, Parker JE, Bader O, Weig M, Gross U, Warrilow AG, Kelly DE, Kelly SL. 2012. Facultative sterol uptake in an ergosterol-deficient clinical isolate of Candida glabrata harboring a missense mutation in ERG11 and exhibiting cross-resistance to azoles and amphotericin B. Antimicrob Agents Chemother 56:4223–4232. 10.1128/AAC.06253-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matheos DP, Kingsbury TJ, Ahsan US, Cunningham KW. 1997. Tcn1p/Crz1p, a calcineurin-dependent transcription factor that differentially regulates gene expression in Saccharomyces cerevisiae. Genes Dev 11:3445–3458. 10.1101/gad.11.24.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stathopoulos AM, Cyert MS. 1997. Calcineurin acts through the CRZ1/TCN1-encoded transcription factor to regulate gene expression in yeast. Genes Dev 11:3432–3444. 10.1101/gad.11.24.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lv QZ, Yan L, Jiang YY. 2016. The synthesis, regulation, and functions of sterols in Candida albicans: Well-known but still lots to learn. Virulence 7:649–659. 10.1080/21505594.2016.1188236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dhingra S, Cramer RA. 2017. Regulation of sterol biosynthesis in the human fungal pathogen Aspergillus fumigatus: Opportunities for therapeutic development. Front Microbiol 8:92. 10.3389/fmicb.2017.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chowdhary A, Kathuria S, Xu J, Meis JF. 2013. Emergence of azole-resistant aspergillus fumigatus strains due to agricultural azole use creates an increasing threat to human health. PLoS Pathog 9:e1003633. 10.1371/journal.ppat.1003633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mellado E, Garcia-Effron G, Alcazar-Fuoli L, Melchers WJ, Verweij PE, Cuenca-Estrella M, Rodriguez-Tudela JL. 2007. A new Aspergillus fumigatus resistance mechanism conferring in vitro cross-resistance to azole antifungals involves a combination of cyp51A alterations. Antimicrob Agents Chemother 51:1897–1904. 10.1128/AAC.01092-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paul S, Diekema D, Moye-Rowley WS. 2017. Contributions of both ATP-binding cassette transporter and Cyp51A proteins are essential for azole resistance in Aspergillus fumigatus. Antimicrob Agents Chemother 61: e02748-16. 10.1128/AAC.02748-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rybak JM, Ge W, Wiederhold NP, Parker JE, Kelly SL, Rogers PD, Fortwendel JR. 2019. Mutations in hmg1, challenging the paradigm of clinical triazole resistance in Aspergillus fumigatus. mBio 10:e00437-19. 10.1128/mBio.00437-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vu BG, Moye-Rowley WS. 2018. Construction and use of a recyclable marker to examine the role of major facilitator superfamily protein members in Candida glabrata drug resistance phenotypes. mSphere 3:e00099-18. 10.1128/mSphere.00099-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, 3rd, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 44.Geber A, Hitchcock CA, Swartz JE, Pullen FS, Marsden KE, Kwon-Chung KJ, Bennett JE. 1995. Deletion of the Candida glabrata ERG3 and ERG11 genes: effect on cell viability, cell growth, sterol composition, and antifungal susceptibility. Antimicrob Agents Chemother 39:2708–2717. 10.1128/AAC.39.12.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arthington-Skaggs BA, Jradi H, Desai T, Morrison CJ. 1999. Quantitation of ergosterol content: novel method for determination of fluconazole susceptibility of Candida albicans. J Clin Microbiol 37:3332–3337. 10.1128/JCM.37.10.3332-3337.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 and S2. Download aac.02098-21-s0001.pdf, PDF file, 1.8 MB (1.8MB, pdf)