ABSTRACT
Klebsiella oxytoca complex comprises nine closely related species causing human infections. We curated genomes labeled Klebsiella (n = 14,256) in GenBank and identified 588 belonging to the complex, which were examined for precise species, sequence types, K- and O-antigen types, and virulence and antimicrobial resistance genes. The complex and Klebsiella pneumoniae share many K- and O-antigen types. Of the complex, K. oxytoca and Klebsiella michiganensis appear to carry more virulence genes and be more commonly associated with human infections.
KEYWORDS: resistance, Klebsiella oxytoca, virulence, taxonomy, Klebsiella, antimicrobial resistance
INTRODUCTION
Klebsiella oxytoca and eight closely related species—Klebsiella grimontii, Klebsiella huaxiensis, Klebsiella michiganensis, Klebsiella pasteurii, Klebsiella spallanzanii, and three new unnamed ones (taxons 1, 2, and 3)—comprise a complex, that are difficult, if not impossible, to be reliably differentiated by phenotypic characteristics (1). K. oxytoca complex is a member of the normal gut microflora (2, 3) and also an important human pathogen causing antibiotic-associated hemorrhagic colitis (AAHC) and a variety of other infections (4–7). Although several studies have addressed antimicrobial resistance and virulence (8, 9), there are large knowledge gaps in our understanding clinically relevant aspects of the complex, such as the prevalence or the proportion of each species of K. oxytoca complex in clinical samples, the virulence factors other than cytotoxins causing AAHC, and the strain clonal background (1). Genome sequences available in the public domain may provide useful information to complement literature for understanding these aspects. We therefore examined all available genomes of K. oxytoca complex in GenBank and generated a curated database for reference.
Precise species identification for all Klebsiella genomes identified 588 genomes of K. oxytoca complex.
We used txid570 [Organism:exp] AND “latest” [filter] to search NCBI and found 14,256 genome sequences labeled as Klebsiella (accessed by 1 January 2021). We discarded 855 genomes for any of the following reasons: (i) assemblies are of low quality as defined by the NCBI, such as being labeled as “many frameshifted protein,” “fragmented assembly,” “genome length too small/large,” etc. (n = 472); (ii) assemblies are duplicated, as they share the same BioSample (n = 225); (iii) assemblies are labeled as “contaminated” (n = 118); (iv) assemblies are labeled as “derived from metagenome” (n = 36); (v) assemblies belong to genetically modified organisms (GMO; n = 3); or (vi) assemblies are derived from an uncultured source (n = 1). Therefore, 13,401 genome sequences labeled as Klebsiella (see Data Set S1 in the supplemental material) were included. We therefore determined their precise species assignations using the average nucleotide identity (ANI) based on BLAST to compare with type strains of Klebsiella and Raoultella species (see Table S1), as described previously with a 95 to 96% ANI as the species cutoff (10). For genomes with a <97% ANI comparing with any type strains of the Enterobacteriaceae, in silico DNA-DNA hybridization (isDDH) was further determined between the genome and that of the closest (sharing the highest ANI value) type strain to confirm or to determine the species status with a 70.0% isDDH as the species cutoff (11). Among the 13,401 genomes, 18 do not belong to the genus Klebsiella, and 55 are Klebsiella spp. but could not be assigned to a known species (see Table S2 and Data Set S1). Instead, the 55 strains could be assigned to two novel Klebsiella species named here taxon 4 (n = 16) and taxon 5 (n = 39) (see Table S2). Taxons 4 and 5 do not belong to K. oxytoca complex but are most closely related to K. aerogenes (95.92% ANI and 66.7% isDDH) and K. pneumoniae (95.87% ANI and 66.8% isDDH), respectively.
Among the 13,383 remaining Klebsiella genomes, there were only 588 (4.39%) belonging to K. oxytoca complex. The numbers of each species are as follows: K. oxytoca (n = 183), K. michiganensis (n = 233), K. grimontii (n = 128), K. huaxiensis (n = 3), K. pasteurii (n = 31), K. spallanzanii (n = 4), taxon 1 (n = 1), taxon 2 (n = 1), and taxon 3 (n = 4) (see Data Set S2). Species of K. oxytoca complex have similar genomic characteristics (genome size, GC content, and numbers of coding sequences and tRNA) with other Klebsiella species and Raoultella spp. (see Table S3).
To identify genes unique to the complex, a subset of genomes (n = 1,410), including 191 of K. oxytoca complex was retrieved from all 13,383 Klebsiella assemblies to adapt local computational resources using Assembly Dereplicator v0.1.0 (https://github.com/rrwick/Assembly-Dereplicator). Coding sequences annotated using Prokka v1.14.6 (12) were clustered using PIRATE v1.0.4 (13) to identify genes that present in 99% K. oxytoca complex genomes but absent from all other Klebsiella genomes. These candidate genes were then challenged with the entire data set of 13,383 genomes using BLASTP algorithm, and hits with coverage or identity of <80% were discarded. As such, 35 genes unique to K. oxytoca complex were identified. Among these, 19 genes, including the intrinsic blaOXY (see Data Set S3), could be assigned with a known KEGG orthology number of 11 KEGG pathways using KofamKOALA v2021-11-01 (14), KEGG mapper v5.0 (15), and KOBAS v3.0 (16). Notably, a type II secretion system to secrete pullulanase, a lipoprotein allowing growth on branched maltodextrin polymers (17), was the solely over-represented protein among all pathways (see Data Set S3, corrected P value of <0.001 [as determined by the Fisher exact test and Benjamini-Hochberg P value adjustment]). The contribution of these unique genes to the evolution of K. oxytoca complex warrants further studies.
K. oxytoca and K. michiganensis are the two main species associated with human infections.
The prevalence of each species of K. oxytoca complex in human colonization and infection is largely unknown since strains called K. oxytoca in most clinical studies were not subjected to precise species identification (1). Of the 588 genomes, 154 were recovered from human samples other than feces and rectal swabs as we focused on extraintestinal infections. The 154 genomes belong to K. oxytoca (n = 96), K. michiganensis (n = 46), K. grimontii (n = 5), K. pasteurii (n = 4), K. spallanzanii (n = 2), and K. huaxiensis (n = 1) (see Data Set S2). K. oxytoca and K. michiganensis appear to be the two main species of the complex associated with human extraintestinal infections. However, genome sequencing is usually highly biased, and well-designed surveillance studies are therefore required to demonstrate the true distribution of each species within K. oxytoca complex.
Several sequence types were relatively common.
The 588 genomes can be assigned to 227 sequence types (STs), including 121 known STs and 116 new STs (assigned as N1 to N116 here) by query the multilocus sequence typing database (https://pubmlst.org/organisms/klebsiella-oxytoca) (see Data Set S2 and Table S4 for the number of genomes belonging to each ST). This is consistent with previous reports (18, 19), suggesting that very diverse clonal background of the complex. Only six STs have at least 10 genomes and have been seen in at least three countries, including ST2 (gapA-infB-mdh-pgi-phoE-rpoB-tonB allele profile, 1-2-2-1-2-1-2; n = 25, in Denmark, Spain, the UK, and the USA) and ST176 (1-7-2-1-65-1-2; n = 12, in Australia, Canada, the UK, and the USA) of K. oxytoca, ST29 (3-4-15-8-18-6-11; n = 11, in Germany, South Africa, and the UK), ST85 (3-5-21-13-24-6-19; n = 16, in China, the UK, and the USA), and ST88 (3-8-24-33-20-6-23; n = 10, in Australia, the UK, and the USA) of K. michiganensis, and ST215 (21-6-14-10-46-11-6; n = 10, in China, the UK, and the USA) of K. grimontii. Single nucleotide polymorphisms (SNPs) between genomes of the same ST were called using Snippy v4.6.0 (https://github.com/tseemann/snippy) and were filtered to remove recombination using Gubbins v2.4.1 (20). No isolates of the aforementioned six STs in different countries belonged to the same clone based on SNP calling (see Data Set S4 for SNP numbers of these STs). This suggests that no international transmission of the same clone was identified at present, but these STs may need to be monitored to reveal their potential to become high-risk clones.
The K- and O-antigen types of K. oxytoca complex could overlap K. pneumoniae.
The major surface antigens of Klebsiella are capsular polysaccharide (CPS; K antigen) and lipopolysaccharide (LPS; containing O antigen), which are common virulence factors of K. pneumoniae (21–24) but are less studied in K. oxytoca complex. We performed the serotyping of K and O antigens for the 588 genomes using Kleborate (25). Of 588 genomes, known K-antigen types can only be assigned to 194 (33.0%) genomes, which belong to K. grimontii, K. michiganensis, K. oxytoca, or K. pasteurii, while the remaining 394 genomes contain K-antigen loci of novel K types. A total of 14 known K types (K26, K29, K41, K43, K66, K70, K74, K102, K109, K145, K152, K157, K164, and K169) were identified, among which K74 (9.2%, 54/588), K29 (3.4%, 20/588), and K43 (2.9%, 17/588) were the most common (see Data Set S2). K102, K109, K145, K152, K157, K164, and K169 have not yet been reported. All of the K types identified in K. oxytoca complex other than K157 and K164 have also been seen in K. pneumoniae (25–31). Currently, there are no published reports of the O antigen in K. oxytoca complex (1). Known O-antigen types could be assigned to the vast majority (96.1%, 565/588) of the genomes across all species of K. oxytoca complex, and 7 O antigen types (O1, O5, OL104, O2, O3/O3a, O3b, and O4) were identified, with O1 (52%, 306/588), O5 (14.3%, 86/588), and OL104 (11.9%, 70/588) as the most common types. All of the O-antigen types have also been reported in K. pneumoniae (24, 32, 33).
K. oxytoca complex carries multiple virulence factors seen in K. pneumoniae.
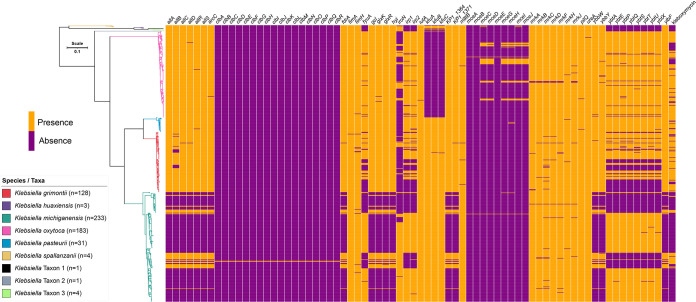
K. oxytoca is well known for causing AAHC, which is due to the production of cytotoxins tilimycin and tilivalline (a derivative of tilimycin) (34, 35). Among the 9 species of K. oxytoca complex, the tilimycin gene cluster is absent from K. huaxiensis, K. spallanzanii, and taxons 1, 2, and 3 (Table 1), suggesting that these species may not cause AAHC. Other than the AAHC-associated cytotoxins, virulence factors of K. oxytoca complex remain largely unknown. As such, in addition to examining the presence of the tilimycin gene cluster, we also screened the 588 genomes for known virulence factors of K. pneumoniae (n = 80; https://bigsdb.pasteur.fr/cgi-bin/bigsdb/bigsdb.pl?db=pubmlst_klebsiella_seqdef&page=downloadAlleles&scheme_id=4&render=1) (36) using BLASTn with a threshold of 90% for both coverage and identity. A total of 72 virulence genes were detected (see Table 1 and Data Set S2). These genes encode adhesins or biofilm formation (fimA, fimH, KP1, mrkABCDFHJ, and pilQ), allantoin utilization (allABCDRS, fdrA, gcl, glxKR, hyi, ybbWY, and ylbF), capsules (matB), microcin (mceABCDEHGIJ), iron acquisition (clbA-R, fyuA, iroN, irp, iutA, kfuABC, and ybtAEPQTUX), and urease (ureA). Among these genes, fimA, fimH, iutA, matB, mrkABCDFHJ, pilQ, and ureA were present in almost all of the 588 genomes (Table 1). Of note, rmpA and rmpA2 (regulators of the hypermucoviscous phenotype) were not found in K. oxytoca complex. Among the 9 species, there appeared to be two patterns of the distribution of virulence genes. The four species most often seen in clinical samples in the present data set (see Data Set S2)—K. michiganensis, K. oxytoca, K. grimontii, and K. pasteurii—carried ≥45 virulence genes, with K. michiganensis carrying all 72 genes, followed by K. oxytoca (carrying 51 genes). In contrast, the other five species had 30 to 35 genes (Table 1). Iron acquisition genes fyuA, iroN, irp, and ybtAEPQTUX were enriched in the genomes of K. michiganensis, K. oxytoca, K. grimontii, and K. pasteurii but absent from those of the other five species (Fig. 1 and Table 1). Another iron acquisition gene, iroN, was present in most (75.9%, 177/233) K. michiganensis samples but was only seen in only a minority of K. oxytoca (21.3%, 39/183) and K. grimontii (11.7%, 15/128) and was absent from K. pasteurii (Table 1). Although the distribution of virulence genes may provide clues to understand the clinical significance of individual species of K. oxytoca complex, the association of these genes with pathogenesis in the complex remains to be determined.
TABLE 1.
Virulence factors in genomes of K. oxytoca complex (n = 588)
| Virulence factor(s) |
Klebsiella
b
|
Taxon |
Frequency (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| grimo | huaxi | michi | oxyto | paste | spall | 1 | 2 | 3 | ||
| No. of genomes | 128 | 3 | 233 | 183 | 31 | 4 | 1 | 1 | 4 | |
| No. of genesa | 45 | 33 | 72 | 51 | 45 | 35 | 30 | 33 | 33 | |
| allA, allS | 128 | 3 | 42 | 183 | 31 | 4 | 1 | 1 | 4 | 67.5 |
| allB | 111 | 3 | 42 | 183 | 31 | 4 | 1 | 1 | 4 | 64.6 |
| allC | 127 | 3 | 42 | 183 | 31 | 4 | 1 | 1 | 4 | 67.3 |
| allD | 127 | 3 | 42 | 182 | 31 | 4 | 1 | 1 | 4 | 67.2 |
| allR | 128 | 3 | 42 | 182 | 30 | 4 | 1 | 1 | 4 | 67.2 |
| arcC | 128 | 3 | 42 | 182 | 31 | 4 | 1 | 1 | 4 | 67.3 |
| clb genesc | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0.3 |
| fdrA | 127 | 3 | 42 | 183 | 31 | 4 | 1 | 1 | 4 | 67.3 |
| fimA | 127 | 3 | 233 | 183 | 31 | 4 | 1 | 1 | 4 | 99.8 |
| fimH | 123 | 3 | 229 | 182 | 31 | 4 | 1 | 1 | 4 | 98.3 |
| fyuA | 72 | 0 | 158 | 175 | 31 | 0 | 0 | 0 | 0 | 74.1 |
| gcl, hyi | 128 | 3 | 42 | 183 | 31 | 4 | 1 | 1 | 4 | 67.5 |
| glxK | 128 | 3 | 42 | 181 | 31 | 4 | 1 | 1 | 4 | 67.2 |
| glxR | 128 | 3 | 41 | 183 | 31 | 4 | 1 | 1 | 4 | 67.3 |
| iroN | 15 | 3 | 177 | 39 | 0 | 3 | 1 | 1 | 4 | 41.3 |
| irp1 | 70 | 0 | 158 | 172 | 31 | 0 | 0 | 0 | 0 | 73.3 |
| irp2 | 69 | 0 | 158 | 175 | 29 | 0 | 0 | 0 | 0 | 73.3 |
| iutA | 128 | 3 | 233 | 182 | 31 | 4 | 1 | 1 | 4 | 99.8 |
| kfuA | 128 | 3 | 233 | 0 | 31 | 3 | 0 | 1 | 1 | 68.0 |
| kfuB, kfuC | 127 | 3 | 233 | 0 | 31 | 3 | 0 | 1 | 1 | 67.9 |
| KP1_1364, ylbF | 128 | 3 | 42 | 183 | 31 | 4 | 1 | 1 | 4 | 67.5 |
| KP1_1371 | 127 | 3 | 42 | 183 | 31 | 4 | 1 | 1 | 4 | 67.3 |
| matB | 125 | 3 | 233 | 182 | 31 | 4 | 1 | 1 | 4 | 99.3 |
| mceA, mceB, mceE | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0.3 |
| mceC | 0 | 0 | 1 | 29 | 0 | 2 | 0 | 0 | 0 | 5.4 |
| mceD, mceJ | 0 | 0 | 1 | 28 | 0 | 0 | 0 | 0 | 0 | 4.9 |
| mceG | 0 | 0 | 2 | 29 | 0 | 0 | 0 | 0 | 0 | 5.3 |
| mceH | 0 | 0 | 2 | 29 | 2 | 0 | 0 | 0 | 0 | 5.6 |
| mceI | 0 | 0 | 1 | 28 | 0 | 2 | 0 | 0 | 0 | 5.3 |
| mrkA | 125 | 3 | 232 | 180 | 31 | 4 | 1 | 1 | 4 | 98.8 |
| mrkB | 126 | 3 | 229 | 180 | 31 | 4 | 1 | 1 | 4 | 98.5 |
| mrkC | 119 | 3 | 225 | 180 | 31 | 4 | 1 | 1 | 4 | 96.6 |
| mrkD | 125 | 3 | 231 | 180 | 31 | 4 | 1 | 1 | 4 | 98.6 |
| mrkF | 125 | 3 | 228 | 180 | 31 | 4 | 1 | 1 | 4 | 98.1 |
| mrkH | 127 | 3 | 231 | 182 | 30 | 4 | 1 | 1 | 4 | 99.1 |
| mrkJ | 127 | 3 | 230 | 178 | 31 | 4 | 1 | 1 | 4 | 98.5 |
| pilQ | 128 | 3 | 233 | 182 | 30 | 4 | 1 | 1 | 4 | 99.7 |
| ureA | 128 | 3 | 233 | 182 | 31 | 4 | 1 | 1 | 4 | 99.8 |
| ybbW | 127 | 3 | 42 | 181 | 31 | 4 | 1 | 1 | 4 | 67.0 |
| ybbY | 128 | 3 | 41 | 179 | 31 | 4 | 1 | 1 | 4 | 66.7 |
| ybtA | 70 | 0 | 159 | 175 | 31 | 0 | 0 | 0 | 0 | 74.0 |
| ybtE | 72 | 0 | 158 | 175 | 31 | 0 | 0 | 0 | 0 | 74.1 |
| ybtP | 68 | 0 | 159 | 172 | 31 | 0 | 0 | 0 | 0 | 73.1 |
| ybtQ | 70 | 0 | 159 | 176 | 31 | 0 | 0 | 0 | 0 | 74.1 |
| ybtS | 68 | 0 | 159 | 175 | 31 | 0 | 0 | 0 | 0 | 73.6 |
| ybtT | 72 | 0 | 157 | 175 | 31 | 0 | 0 | 0 | 0 | 74.0 |
| ybtU | 71 | 0 | 157 | 175 | 31 | 0 | 0 | 0 | 0 | 73.8 |
| ybtX | 71 | 0 | 158 | 176 | 30 | 0 | 0 | 0 | 0 | 74.0 |
| Tilimycin gene cluster | 110 | 0 | 34 | 141 | 26 | 0 | 0 | 0 | 0 | 52.9 |
Genes with an identical distribution pattern are clustered together. The number of genes does not contain the tilimycin gene cluster.
Species: grimo, grimontii; huaxi, huaxiensis; michi, michiganensis; oxyto, oxytoca; paste, pasteurii; spall, spallanzanii.
The clb cluster contains 18 genes (clbA, clbB, clbC, clbD, clbE, clbF, clbG, clbH, clbI, clbJ, clbK, clbL, clbM, clbN, clbO, clbP, clbQ, and clbR). All 18 genes have an identical distribution pattern.
FIG 1.
Phylogenomic tree of 588 genomes of K. oxytoca complex and distribution of virulence factors. The core genes of the K. oxytoca complex (n = 588) were identified and concatenated using PIRATE v.1.0.4 (13) with default settings, followed by the phylogenetic tree inference using IQ-TREE v2.1.3 (39) under GTR+G+ASC model with a 1,000-bootstrap test. Tree was then visualized and annotated using web-based tools Phandango v1.3.0 (40) and iTOL v6.4.2 (41).
K. oxytoca complex has a large number of acquired antimicrobial resistance genes.
K. oxytoca complex carries intrinsic β-lactamase-encoding blaOXY and fosfomycin resistance gene fosA (1, 37). Species other than K. huaxiensis, K. spallanzanii, taxon 1, and taxon 3 also have intrinsic low-level quinolone resistance genes oqxA-oqxB (1). A variety of acquired genes mediating resistance to β-lactams (e.g., penicillins, cephalosporins, and carbapenems), aminoglycosides, colistin, chloramphenicol, macrolides, quinolones, rifampicin, sulfonamides, tetracyclines, and trimethoprim have been reported in literature (38) and have been summarized previously (1). By analyzing the 588 genomes, many antimicrobial resistance genes that have not been reported in literature were identified using ResFinder (http://genomicepidemiology.org/) (Table 2), highlighting the large defense arsenal in the complex. Among the 588 genomes, 109 contain one or more carbapenemase-encoding genes, including blaGES-5, blaIMP-4, -13, -29, -38, blaKPC-2, -3, blaNDM-1, -5, blaOXA-48, -181, and blaVIM-1 (Table 2), which are commonly carried by plasmids (1). The most common one was blaKPC-2, present in 63 genomes. However, there was no single ST that carried a carbapenemase gene and was present in at least three countries (see Data Set S2).
TABLE 2.
Acquired antimicrobial resistance genes in genomes of K. oxytoca complex (n = 588)
| Antimicrobial type | Acquired antimicrobial resistance gene(s)a |
|---|---|
| Carbapenems | blaGES-5, blaIMP-4, -13, -29, -38, blaKPC-2, -3, blaNDM-1, -4, -5, blaOXA-48, -181, blaVIM-1 |
| Other β-lactamsb | blaACC-1, blaCARB-2, -12, blaCMY-6, blaCTX-M-1, -3, -8, -14, -15, -62, blaDHA-1, blaFOX-5, blaLAP-2, blaOXA-1, -2, -9, -10, -101, blaSCO-1, blaSFO-1, blaSHV-2, -5, -7, -12, -14, -30, blaTEM-1, -3, -26, -116, blaTLA-3 |
| Colistin | mcr-9 |
| Aminoglycosides | aac(2′)-IIa, aac(3)-Ia, -IId, -IIe, -IIg, -IVa, aac(6′)-30, -Ib, -Ib′, -Ib3, -Ib4, -Ib-cr, -Ib-cr5, -IIa, -IIc, aadA1, aadA2, aadA4, aadA5, aadA10, aadA11, aadA13, aadA16, ant(2″)-Ia, aph(3′)-Ia, -IIa, -VI, -XV, aph(3″)-Ib, aph(4)-Ia, aph(6)-Id, aphA16, armA, rmtC, sat2 |
| Quinolones | qnrA1, qnrB1, qnrB2, qnrB4, qnrB6, qnrB19, qnrS1 |
| Sulfonamides | sul1, sul2, sul3 |
| Trimethoprim | dfrA1, dfrA3b, dfrA10, dfrA12, dfrA14, dfrA15, dfrA17, dfrA19, dfrA21, dfrA27, dfrA32, dfrB1 |
| Chloramphenicol | catA1, catA2, catB2, catB3, catB8, catB11, cmlA1, cmlA4, cmlA5, cmlA10, cmx, floR |
| Rifampicin | arr-2, arr-3 |
| Tetracyclines | tet(A), tet(B), tet(C), tet(D), tet(M), tet(34), tet(39) |
| Macrolides | ere(A), mph(A), mph(E), msr(E) |
Resistance genes that have not been reported in the literature (1) but are identified in genomes available in the NCBI database are underlined (see Data Set S2).
All strains of K. oxytoca complex also have intrinsic blaOXY genes.
In conclusion, the O- and K-antigen types of K. oxytoca complex overlap those of K. pneumoniae. Although several STs appear to be relatively common, no internationally distributed high-risk clones associated with the spread of carbapenem resistance were identified at present. Among the 9 species of K. oxytoca complex, K. oxytoca and K. michiganensis appear to be the major ones seen in human infections, which could be associated with the relatively more virulence factors that they harbor. Although the information generated by analysis of currently available genomes is helpful, due to the bias of sampling for genome sequencing, well-designed surveillance studies are required to provide insights in the prevalence, pathogenesis, antimicrobial resistance, and spread of K. oxytoca complex.
ACKNOWLEDGMENTS
The relevant works of the authors were supported by the National Natural Science Foundation of China (81861138055) and by a grant from the West China Hospital of Sichuan University (1.3.5 project for disciplines of excellence, grant ZYYC08006).
We declare there are no conflicts of interest.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Yang J, Long H, Hu Y, Feng Y, McNally A, Zong Z. 2022. Klebsiella oxytoca complex: update on taxonomy, antimicrobial resistance, and virulence. Clin Microbiol Rev 35:e00006-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Savino F, Cordisco L, Tarasco V, Calabrese R, Palumeri E, Matteuzzi D. 2009. Molecular identification of coliform bacteria from colicky breastfed infants. Acta Paediatr 98:1582–1588. 10.1111/j.1651-2227.2009.01419.x. [DOI] [PubMed] [Google Scholar]
- 3.Podschun R, Ullmann U. 1998. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev 11:589–603. 10.1128/CMR.11.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghasemian A, Mohabati Mobarez A, Najar Peerayeh S, Talebi Bezmin Abadi A, Khodaparast S, Mahmood SS. 2019. Expression of adhesin genes and biofilm formation among Klebsiella oxytoca clinical isolates from patients with antibiotic-associated haemorrhagic colitis. J Med Microbiol 68:978–985. 10.1099/jmm.0.000965. [DOI] [PubMed] [Google Scholar]
- 5.Bouchillon SK, Badal RE, Hoban DJ, Hawser SP. 2013. Antimicrobial susceptibility of inpatient urinary tract isolates of Gram-negative bacilli in the United States: results from the study for monitoring antimicrobial resistance trends (SMART) program: 2009–2011. Clin Ther 35:872–877. 10.1016/j.clinthera.2013.03.022. [DOI] [PubMed] [Google Scholar]
- 6.Mineau S, Kozak R, Kissoon M, Paterson A, Oppedisano A, Douri F, Gogan K, Willey BM, McGeer A, Poutanen SM. 2018. Emerging antimicrobial resistance among Escherichia coli strains in bloodstream infections in Toronto, 2006–2016: a retrospective cohort study. Cmajo 6:E580–E586. 10.9778/cmajo.20180039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoban DJ, Badal R, Bouchillon S, Hackel M, Kazmierczak K, Lascols C, Hawser S. 2014. In vitro susceptibility and distribution of β-lactamases in Enterobacteriaceae causing intra-abdominal infections in North America 2010–2011. Diagn Microbiol Infect Dis 79:367–372. 10.1016/j.diagmicrobio.2014.03.026. [DOI] [PubMed] [Google Scholar]
- 8.Cuenod A, Wuthrich D, Seth-Smith HMB, Ott C, Gehringer C, Foucault F, Mouchet R, Kassim A, Revathi G, Vogt DR, von Felten S, Bassetti S, Tschudin-Sutter S, Hettich T, Schlotterbeck G, Homberger C, Casanova C, Moran-Gilad J, Sagi O, Rodriguez-Sanchez B, Muller F, Aerni M, Gaia V, van Dessel H, Kampinga GA, Muller C, Daubenberger C, Pfluger V, Egli A. 2021. Whole-genome sequence-informed MALDI-TOF MS diagnostics reveal importance of Klebsiella oxytoca group in invasive infections: a retrospective clinical study. Genome Med 13:150. 10.1186/s13073-021-00960-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moradigaravand D, Martin V, Peacock SJ, Parkhill J. 2017. Population structure of multidrug resistant Klebsiella oxytoca within hospitals across the UK and Ireland identifies sharing of virulence and resistance genes with K. pneumoniae. Genome Biol Evol 9:574–587. 10.1093/gbe/evx019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richter M, Rossello-Mora R. 2009. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106:19126–19131. 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meier-Kolthoff JP, Auch AF, Klenk HP, Goker M. 2013. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 13.Bayliss SC, Thorpe HA, Coyle NM, Sheppard SK, Feil EJ. 2019. PIRATE: a fast and scalable pangenomics toolbox for clustering diverged orthologues in bacteria. Gigascience 8. 10.1093/gigascience/giz119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aramaki T, Blanc-Mathieu R, Endo H, Ohkubo K, Kanehisa M, Goto S, Ogata H. 2020. KofamKOALA: KEGG Ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics 36:2251–2252. 10.1093/bioinformatics/btz859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanehisa M, Sato Y, Kawashima M. 2021. KEGG mapping tools for uncovering hidden features in biological data. Protein Sci 31:37–53. 10.1002/pro.4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bu D, Luo H, Huo P, Wang Z, Zhang S, He Z, Wu Y, Zhao L, Liu J, Guo J, Fang S, Cao W, Yi L, Zhao Y, Kong L. 2021. KOBAS-i: intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res 49:W317–W325. 10.1093/nar/gkab447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Francetic O, Pugsley AP. 2005. Towards the identification of type II secretion signals in a nonacylated variant of pullulanase from Klebsiella oxytoca. J Bacteriol 187:7045–7055. 10.1128/JB.187.20.7045-7055.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herzog KA, Schneditz G, Leitner E, Feierl G, Hoffmann KM, Zollner-Schwetz I, Krause R, Gorkiewicz G, Zechner EL, Hogenauer C. 2014. Genotypes of Klebsiella oxytoca isolates from patients with nosocomial pneumonia are distinct from those of isolates from patients with antibiotic-associated hemorrhagic colitis. J Clin Microbiol 52:1607–1616. 10.1128/JCM.03373-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Izdebski R, Fiett J, Urbanowicz P, Baraniak A, Derde LP, Bonten MJ, Carmeli Y, Goossens H, Hryniewicz W, Brun-Buisson C, Brisse S, Gniadkowski M, MOSAR WP2 WP3 and WP5 Study Groups. 2015. Phylogenetic lineages, clones and β-lactamases in an international collection of Klebsiella oxytoca isolates non-susceptible to expanded-spectrum cephalosporins. J Antimicrob Chemother 70:3230–3237. [DOI] [PubMed] [Google Scholar]
- 20.Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, Parkhill J, Harris SR. 2015. Rapid phylogenetic analysis of large samples of recombinant bacterial whole-genome sequences using Gubbins. Nucleic Acids Res 43:e15. 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bengoechea JA, Sa Pessoa J. 2019. Klebsiella pneumoniae infection biology: living to counteract host defences. FEMS Microbiol Rev 43:123–144. 10.1093/femsre/fuy043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wyres KL, Lam MMC, Holt KE. 2020. Population genomics of Klebsiella pneumoniae. Nat Rev Microbiol 18:344–359. 10.1038/s41579-019-0315-1. [DOI] [PubMed] [Google Scholar]
- 23.Paczosa MK, Mecsas J. 2016. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev 80:629–661. 10.1128/MMBR.00078-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi M, Hegerle N, Nkeze J, Sen S, Jamindar S, Nasrin S, Sen S, Permala-Booth J, Sinclair J, Tapia MD, Johnson JK, Mamadou S, Thaden JT, Fowler VG, Jr, Aguilar A, Terán E, Decre D, Morel F, Krogfelt KA, Brauner A, Protonotariou E, Christaki E, Shindo Y, Lin YT, Kwa AL, Shakoor S, Singh-Moodley A, Perovic O, Jacobs J, Lunguya O, Simon R, Cross AS, Tennant SM. 2020. The diversity of lipopolysaccharide (O) and capsular polysaccharide (K) antigens of invasive Klebsiella pneumoniae in a multi-country collection. Front Microbiol 11:1249. 10.3389/fmicb.2020.01249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wyres KL, Wick RR, Gorrie C, Jenney A, Follador R, Thomson NR, Holt KE. 2016. Identification of Klebsiella capsule synthesis loci from whole genome data. Microb Genom 2:e000102. 10.1099/mgen.0.000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hermansen LT, Loft AG, Christiansen AA, Munk HL, Gilbert L, Jurik AG, Arnbak B, Manniche C, Weber U, Ostergaard M, Pedersen SJ, Barington T, Junker P, Horslev-Petersen K, Hendricks O. 2017. No diagnostic utility of antibody patterns against Klebsiella pneumoniae capsular serotypes in patients with axial spondyloarthritis versus patients with non-specific low back pain: a cross-sectional study. Scand J Rheumatol 46:296–302. 10.1080/03009742.2016.1205659. [DOI] [PubMed] [Google Scholar]
- 27.Nordberg V, Jonsson K, Giske CG, Iversen A, Aspevall O, Jonsson B, Camporeale A, Norman M, Naver L. 2018. Neonatal intestinal colonization with extended-spectrum β-actamase-producing Enterobacteriaceae-a 5-year follow-up study. Clin Microbiol Infect 24:1004–1009. 10.1016/j.cmi.2017.12.028. [DOI] [PubMed] [Google Scholar]
- 28.Zhou W, Liu L, Feng Y, Zong Z. 2018. A p7 phage-like plasmid carrying mcr-1 in an ST15 Klebsiella pneumoniae clinical isolate. Front Microbiol 9:11. 10.3389/fmicb.2018.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hernandez-Alles S, Conejo M, Pascual A, Tomas JM, Benedi VJ, Martinez-Martinez L. 2000. Relationship between outer membrane alterations and susceptibility to antimicrobial agents in isogenic strains of Klebsiella pneumoniae. J Antimicrob Chemother 46:273–277. 10.1093/jac/46.2.273. [DOI] [PubMed] [Google Scholar]
- 30.Mbelle NM, Feldman C, Sekyere JO, Maningi NE, Modipane L, Essack SY. 2020. Pathogenomics and evolutionary epidemiology of multidrug resistant clinical Klebsiella pneumoniae isolated from Pretoria, South Africa. Sci Rep 10:1232. 10.1038/s41598-020-58012-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sapugahawatte DN, Li C, Zhu C, Dharmaratne P, Wong KT, Lo N, Ip M. 2020. Prevalence and characteristics of extended-spectrum β-lactamase-producing and carbapenemase-producing Enterobacteriaceae from freshwater fish and pork in wet markets of Hong Kong. mSphere 5:e00107-20. 10.1128/mSphere.00107-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Follador R, Heinz E, Wyres KL, Ellington MJ, Kowarik M, Holt KE, Thomson NR. 2016. The diversity of Klebsiella pneumoniae surface polysaccharides. Microb Genom 2:e000073. 10.1099/mgen.0.000073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guachalla LM, Stojkovic K, Hartl K, Kaszowska M, Kumar Y, Wahl B, Paprotka T, Nagy E, Lukasiewicz J, Nagy G, Szijártó V. 2017. Discovery of monoclonal antibodies cross-reactive to novel subserotypes of K. pneumoniae O3. Sci Rep 7:6635. 10.1038/s41598-017-06682-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dornisch E, Pletz J, Glabonjat RA, Martin F, Lembacher-Fadum C, Neger M, Högenauer C, Francesconi K, Kroutil W, Zangger K, Breinbauer R, Zechner EL. 2017. Biosynthesis of the enterotoxic pyrrolobenzodiazepine natural product tilivalline. Angew Chem Int Ed Engl 56:14753–14757. 10.1002/anie.201707737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tse H, Gu Q, Sze KH, Chu IK, Kao RY, Lee KC, Lam CW, Yang D, Tai SS, Ke Y, Chan E, Chan WM, Dai J, Leung SP, Leung SY, Yuen KY. 2017. A tricyclic pyrrolobenzodiazepine produced by Klebsiella oxytoca is associated with cytotoxicity in antibiotic-associated hemorrhagic colitis. J Biol Chem 292:19503–19520. 10.1074/jbc.M117.791558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bialek-Davenet S, Criscuolo A, Ailloud F, Passet V, Jones L, Delannoy-Vieillard AS, Garin B, Le Hello S, Arlet G, Nicolas-Chanoine MH, Decre D, Brisse S. 2014. Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerg Infect Dis 20:1812–1820. 10.3201/eid2011.140206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Merla C, Rodrigues C, Passet V, Corbella M, Thorpe HA, Kallonen TVS, Zong Z, Marone P, Bandi C, Sassera D, Corander J, Feil EJ, Brisse S. 2019. Description of Klebsiella spallanzanii sp. nov. and of Klebsiella pasteurii sp. nov. Front Microbiol 10:2360. 10.3389/fmicb.2019.02360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blair JM, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJ. 2015. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol 13:42–51. 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- 39.Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol 37:1530–1534. 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hadfield J, Croucher NJ, Goater RJ, Abudahab K, Aanensen DM, Harris SR. 2018. Phandango: an interactive viewer for bacterial population genomics. Bioinformatics 34:292–293. 10.1093/bioinformatics/btx610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Letunic I, Bork P. 2021. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 49:W293–W296. 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental figure and tables. Download aac.02183-21-s0001.pdf, PDF file, 0.2 MB (250KB, pdf)
Supplemental Data Set S1. Download aac.02183-21-s0002.xlsx, XLSX file, 0.5 MB (475.5KB, xlsx)
Supplemental Data Set S2. Download aac.02183-21-s0003.xlsx, XLSX file, 0.1 MB (111KB, xlsx)
Supplemental Data Set S3. Download aac.02183-21-s0004.xlsx, XLSX file, 0.01 MB (11.5KB, xlsx)
Supplemental Data Set S4. Download aac.02183-21-s0005.xlsx, XLSX file, 0.02 MB (22.5KB, xlsx)