ABSTRACT
Acetic acid bacteria catalyze the two-step oxidation of ethanol to acetic acid using the membrane-bound enzymes pyrroloquinoline quinone-dependent alcohol dehydrogenase and molybdopterin-dependent aldehyde dehydrogenase (ALDH). Although the reducing equivalents from the substrate are transferred to ubiquinone in the membrane, intramolecular electron transport in ALDH is not understood. Here, we purified the AldFGH complex, the membrane-bound ALDH that is physiologically relevant to acetic acid fermentation in Gluconacetobacter diazotrophicus strain PAL5. The purified AldFGH complex showed acetaldehyde:ubiquinone (Q2) oxidoreductase activity. c-type cytochromes of the AldFGH complex (in the AldF subunit) were reduced by acetaldehyde. Next, we genetically dissected the AldFGH complex into AldGH and AldF units and reconstituted them. The AldGH subcomplex showed acetaldehyde:ferricyanide oxidoreductase activity but not Q2 reductase activity. The ALDH activity of AldGH was not found in membranes but was found in the soluble fraction of the recombinant strain, suggesting that the AldF subunit is responsible for membrane binding of the AldFGH complex. The absorption spectra of the purified AldGH subcomplex suggested the presence of an [Fe-S] cluster, which can be reduced by acetaldehyde. The AldFGH complex reconstituted from the AldGH subcomplex and AldF showed Q2 reductase activity. We propose a model in which electrons from the substrate are abstracted by a molybdopterin in the AldH subunit and transferred to the [Fe-S] cluster(s) in the AldG subunit, followed by electron transport to c-type cytochrome centers in the AldF subunit, which is the site of ubiquinone reduction in the membrane.
IMPORTANCE Two membrane-bound enzymes of acetic acid bacteria, pyrroloquinoline quinone-dependent alcohol dehydrogenase and molybdopterin-dependent aldehyde dehydrogenase (ALDH), are responsible for vinegar production. Upon the oxidation of acetaldehyde, ALDH reduces ubiquinone in the cytoplasmic membrane. ALDH is an enzyme complex of three subunits. Here, we tried to understand how ALDH works by using a classical biochemical approach and genetic engineering to dissect the enzyme complex into soluble and membrane-bound parts. The soluble part had limited activity in vitro and did not reduce ubiquinone. However, the enzyme complex reconstituted from the soluble and membrane-bound parts showed ubiquinone reduction activity. The proposed working model of ALDH provides a better understanding of how the enzyme works in the vinegar fermentation process.
KEYWORDS: acetic acid fermentation, Gluconacetobacter diazotrophicus, membrane-bound aldehyde dehydrogenase, [Fe-S] cluster, cytochrome c
INTRODUCTION
Acetic acid fermentation is the oxidation of ethanol by acetic acid bacteria. This is a process involving successive oxidation reactions catalyzed by membrane-bound enzymes: alcohol dehydrogenase (ADH) and aldehyde dehydrogenase (ALDH) (1). ADH catalyzes the oxidization of ethanol to acetaldehyde, coupled with the reduction of ubiquinone in the cytoplasmic membrane (2). ADHs of different species of acetic acid bacteria differ in subunit composition. ADHs of Acetobacter spp. and Gluconobacter spp. are composed of three subunits, AdhA, AdhB, and AdhS, whereas those of Komagataeibacter spp. and Gluconacetobacter spp. are composed of two subunits, AdhA and AdhB (2). The AdhA and AdhB subunits are a quinohemoprotein subunit containing pyrroloquinoline quinone (PQQ) and one c-type cytochrome center as the prosthetic groups and a c-type cytochrome subunit containing three hemes, respectively (2, 3). AdhB is the site of ubiquinone reduction (3). AdhS has no characteristic cofactors, but several reports suggest a function as a molecular chaperone that assists in the maturation of the AdhA subunit (4, 5).
The functions of the membrane-bound ALDH of acetic acid bacteria, which catalyzes the oxidation of acetaldehyde to produce acetic acid, have been studied for a long time (6–8). In both Acetobacter pasteurianus strain SKU1108 (9) and Gluconacetobacter diazotrophicus strain PAL5 (10), there are two molecular species of ALDH, AldFGH and AldSLC. Amino acid identities between AldH and AldL, AldF and AldC, and AldG and AldS of the PAL5 strain are 24%, 32%, and 59%, respectively. AldFGH is physiologically important for acetic acid fermentation (9, 10). However, the presence of multiple enzymes with ALDH activity complicates the purification of AldFGH. According to the predicted amino acid sequences, the AldH protein is an 80-kDa molybdopterin-containing subunit that is the site of the oxidation of the substrate (11), AldF is a 45-kDa c-type cytochrome containing three hemes, and AldG is a 17-kDa protein possessing two binding motifs for [2Fe-2S] clusters (7).
The ALDH activity of PQQ-dependent ADH might perturb the purification of ALDH (12, 13), which is presumably one of the reasons why the prosthetic group of ALDH was a matter of debate for a long time: either PQQ or molybdopterin (7, 8, 14, 15). Recently, we confirmed that PQQ is not involved in ALDH, but a form of molybdopterin is required by this enzyme (10). This work used a triple-deletion strain, G. diazotrophicus PAL5 ΔaldSLC ΔadhAB ΔPQQ, called strain MR17, that lacks the genes encoding AldSLC and ADH and has AldFGH as the sole ALDH (10); in this strain, the genes for the biosynthesis of PQQ (pqqABCDE) were also deleted to enable the identification of the prosthetic group of AldFGH.
In the present study, we purified the AldFGH complex from G. diazotrophicus strain MR17 while maintaining the important functions, that is, the ubiquinone reduction ability upon acetaldehyde oxidation and the reduction of ALDH with acetaldehyde. Ubiquinone reductase activity is relevant to the in vivo function of the enzyme, but only a few studies have considered this activity (8), and the purification of ALDH having ubiquinone reductase activity has not yet been reported. The reduction of the purified ALDH with the substrate is also functionally critical (8) because AldFGH is predicted to possess c-type cytochrome centers and an [Fe-S] cluster. We genetically dissected the AldFGH complex into an AldGH subcomplex and the AldF subunit because the presence of cytochromes in AldF would mask the absorption of the [Fe-S] cluster of the AldG subunit and inhibit spectral analysis. By the dissection and reconstitution of the AldFGH complex, we propose a model for intramolecular electron transport upon acetaldehyde oxidation coupled with ubiquinone reduction.
RESULTS
Stabilizing agent for ALDH of G. diazotrophicus.
Before starting the purification of the AldFGH complex, we experimented with the purification of the AldGH subcomplex (see “The AldF subunit is responsible for membrane binding of the AldFGH complex,” below). The AldGH subcomplex rapidly lost its ferricyanide reductase activity. We determined whether the substrate could repress the inactivation of the AldGH subcomplex. Because aldehydes are volatile, we tried aldehydes with relatively high boiling points, namely, propionaldehyde and butyraldehyde. The KM values for the ferricyanide reductase activities of the AldGH subcomplex for acetaldehyde, propionaldehyde, and butyraldehyde were determined to be 0.48, 0.68, and 3.2 mM, respectively (see Fig. S1 in the supplemental material). We examined 2 mM butyraldehyde as a stabilizing agent for the AldGH subcomplex over 4 days at 4°C, as well as 50 mM benzaldehyde, which we previously used as a stabilizing agent for the membrane-bound ALDH of Gluconobacter sp. (6). Butyraldehyde maintained the activity of the AldGH subcomplex, but benzaldehyde did not (Fig. S2). Thus, 2 mM butyraldehyde was included in all buffers used in the purification procedures in this study.
Properties of the AldFGH complex.
We purified the AldFGH complex from G. diazotrophicus strain MR17 (PAL5 ΔadhAB ΔaldSLC ΔPQQ) (see Materials and Methods), which lacks the genes encoding ADH, AldSLC, and PQQ biosynthesis. Strain MR17 produces AldFGH as the sole molecular species that exhibits ALDH activity in the membrane (10). A summary of the purification is shown in Table 1. The final recovery of activity was approximately 20% with specific acetaldehyde:ferricyanide (FC3−) oxidoreductase activity of 350 U (mg protein)−1 and acetaldehyde:ubiquinone-2 (Q2) oxidoreductase activity of 20 U (mg protein)−1. The Q2 reductase activity was approximately 5% of the FC3− reductase activity in each purification step (Table 1), indicating that the purification procedures in this study maintained physiologically relevant ubiquinone reductase activity.
TABLE 1.
Summary of purification of the Gluconacetobacter diazotrophicus AldFGH complex from the triple-deletion strain MR17 (PAL5 ΔaldSLC ΔadhAB ΔPQQ)a
| Purification step | Total protein concn (mg) | Sp act (U/mg) |
Total activity (U) |
Yield (%) |
|||
|---|---|---|---|---|---|---|---|
| Ferricyanide reductase | Q2 reductase | Ferricyanide reductase | Q2 reductase | Ferricyanide reductase | Q2 reductase | ||
| Membranes | 4,900 | 2.5 | 0.12 | 12,000 | 580 | 100 | 100 |
| Solubilized membranes | 860 | 12 | 0.55 | 9,900 | 460 | 83 | 82 |
| CM-Toyopearl | 250 | 36 | 1.6 | 8,300 | 400 | 69 | 68 |
| Hydroxyapatite | 8.9 | 310 | 15 | 3,800 | 170 | 31 | 30 |
| DEAE-Toyopearl | 5.7 | 350 | 20 | 2,500 | 120 | 21 | 21 |
Activity was assayed under standard conditions as described in Materials and Methods.
In sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels under denaturing conditions, our final enzyme preparation showed the presence of three main bands (Fig. 1A). Large amounts of protein (33 μg) were loaded onto the SDS-PAGE gel consisting of high concentrations of acrylamide to see the small subunit AldG. However, the large subunit AldH migrated as a larger molecular species than the expected size (80 kDa) in the gel. We anticipated that the migration of the large protein in the SDS-PAGE gel was perturbed by the large amount of the detergent because the detergent in the enzyme preparation may have been concentrated by ultrafiltration. Therefore, smaller amounts of the ALDH (2 μg) were loaded onto an SDS-PAGE gel consisting of lower acrylamide concentrations (Fig. 1B). The large subunit AldH migrated at the expected molecular size. Taken together, we concluded that the enzyme preparation contains three subunits of 80, 45, and 17 kDa, which correspond to the expected molecular masses of AldH, AldF, and AldG, respectively. Moreover, the N-terminal amino acid sequence of the 17-kDa band was TTFRL, which corresponded to the expected sequence for AldG (16). Heme staining based on heme-dependent peroxidase activity showed that the 45-kDa band contained heme covalently associated with the protein, namely, heme c (Fig. 1C).
FIG 1.
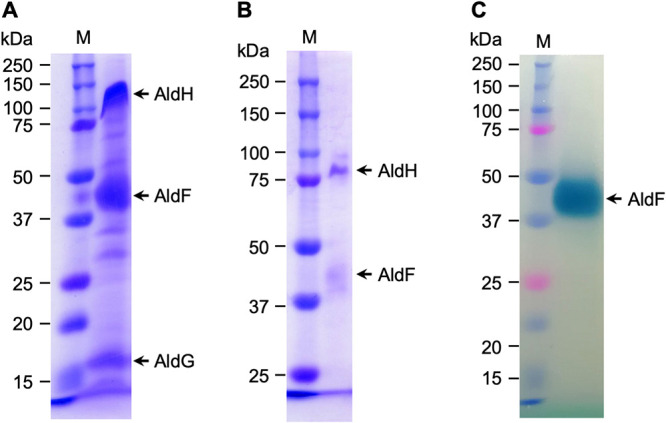
SDS-PAGE of the purified AldFGH complex of Gluconacetobacter diazotrophicus strain MR17 (PAL5 ΔaldSLC ΔadhAB ΔPQQ). (A) Thirty-three micrograms of purified protein separated by SDS-PAGE (10% [wt/vol] acrylamide) followed by staining with Coomassie brilliant blue R-250. (B) Similar to panel A but with 2 μg of purified protein and 7.0% (wt/vol) acrylamide. (C) Twenty-two micrograms of purified protein and staining for heme-dependent peroxidase activity using tetramethylbenzidine. Precision Plus Protein Dual Color Standards (Bio-Rad Laboratories, Hercules, CA) were loaded in the lanes M as molecular mass markers.
Visible spectroscopy of the as-purified AldFGH complex showed an absorption peak at 410 nm but no obvious peaks at around 520 or 550 nm (black line in Fig. 2). However, when the AldFGH complex was incubated with the substrate acetaldehyde at a final concentration of 10 mM, absorption peaks were observed at 417, 522, and 550 nm, which correspond to the γ, β, and α absorption bands of a typical c-type cytochrome, respectively (red line in Fig. 2). We suggest that the as-purified AldFGH complex is mostly in the oxidized form, even though we added butyraldehyde to all the buffers used for purification. The addition of acetaldehyde reduced the AldFGH complex. Importantly, the addition of sodium dithionite after the addition of acetaldehyde did not change the absorption spectrum much (dashed blue line in Fig. 2, compared with the red line), suggesting that almost all cytochromes in the purified AldFGH complex were reduced in the presence of the substrate. The absorption spectrum also showed shoulders at around 530 and 556 nm (inset in Fig. 2), similar to previous reports that suggested the presence of a b-type cytochrome (7, 8).
FIG 2.
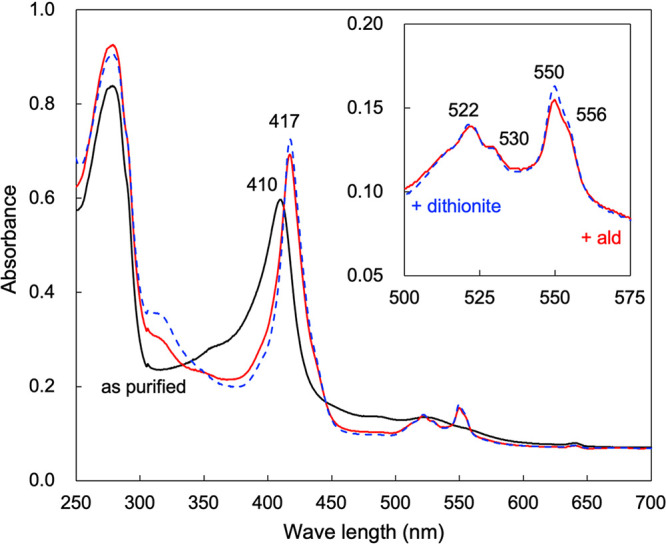
Absorption spectra of the purified AldFGH complex from G. diazotrophicus strain MR17. Shown are absorption spectra from 250 to 700 nm with an enzyme concentration of 0.7 mg protein mL−1 in 10 mM K+-phosphate buffer (pH 6.0). The solid black line is the absorption spectrum of the “as-purified” AldFGH complex. The AldFGH complex was reduced with 10 mM acetaldehyde (solid red line), and a few grains of sodium dithionite were then added to the protein solution to chemically reduce the AldFGH complex (dashed blue line). The inset shows enlarged absorption spectra of the acetaldehyde-reduced and dithionite-reduced AldFGH complex from 500 to 575 nm.
Similar to previous reports by Thurner et al. (7) and Gómez-Manzo et al. (8), the absorption spectrum of the purified AldFGH complex had a shoulder at 556 nm (inset in Fig. 2). Because previous reports suggested the presence of heme b, we attempted to detect it in our purified AldFGH complex. The reduced-minus-oxidized difference spectrum of the pyridine hemochromogen failed to detect heme b, showing only one peak (absorption maximum at 550 nm), consistent with heme c (Fig. S3). Moreover, we attempted to extract hemes by acid-acetone treatment (17). Heme c cannot be extracted from protein by this method because the heme is covalently bound to the polypeptide chain, but noncovalently bound hemes such as hemes a and b are extracted. However, we did not detect any heme in the acetone-soluble fraction extracted from AldFGH. For comparison, hemes a and b were extracted by acid-acetone treatment from partially purified cytochrome ba3 ubiquinol oxidase (18), which was derived from subfractions of the AldFGH purification (Fig. S3). We conclude that the AldFGH complex does not contain heme b.
For the determination of heme c, a reduced-minus-oxidized difference spectrum was recorded for the pyridine hemochromogen of the AldFGH complex (Fig. S3). The heme c content in the AldFGH complex was calculated to be 4.4 μM with 0.30 mg protein mL−1. Assuming that the molecular mass of the AldFGH complex is 142 kDa, the heme c content would be 2.1 mol per mol of AldFGH complex. However, even though the nearest integer was 2, we suggest that the AldFGH complex possesses three hemes c because impurities in the enzyme preparation may decrease the experimentally observed value. The value of 3 is also consistent with that reported in previous work (8) and with the number of heme c binding motifs (Cys-Xxx-Xxx-Cys-His) in AldF (7).
The AldF subunit is responsible for membrane binding of the AldFGH complex.
We examined the membrane association of the AldFGH complex. The ALDH activity of G. diazotrophicus strain MR17 was mostly detected in the membranes: 77% of the total activity was found in the membrane fractions (Fig. 3). The remaining ALDH activity (in the crude soluble fraction) was not a soluble form of the enzyme: we tried to isolate ALDH from the crude soluble fraction by column chromatography, but the elution profile of the ALDH activity was different from that of the solubilized AldFGH complex or the AldGH subcomplex. ALDH activity in the crude soluble fraction is presumably derived from small membrane fragments that were not sedimented by ultracentrifugation.
FIG 3.
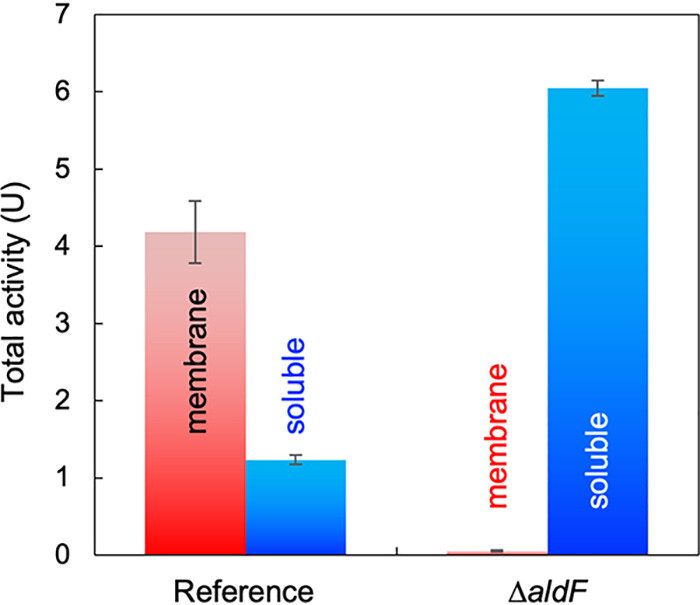
The AldF subunit is responsible for membrane binding of the AldFGH complex. G. diazotrophicus strain MR17, referred to as the reference strain, and strain MR25 (MR17 ΔaldF), referred to as the ΔaldF strain, were cultivated in 100 mL YPG medium. The membranes (red) and the crude soluble fraction (blue) were separated by ultracentrifugation. Acetaldehyde:ferricyanide oxidoreductase activity was determined at pH 6.0. Mean values and standard deviations from triplicate assays are shown.
In the quadruple-deletion strain G. diazotrophicus MR25 (ΔadhAB ΔaldSLC ΔPQQ ΔaldF), almost all of the ALDH activity was detected in the crude soluble fraction; only 1% of the total activity was detected in the membrane fraction. These results suggest that AldF is responsible for membrane binding of the AldFGH complex.
Heterologous expression of G. diazotrophicus AldFGH.
We genetically dissected the AldFGH complex using heterologous expression in A. pasteurianus strain mNS4 (SKU1108 ΔadhAB ΔaldSLC ΔaldFGH) (see Materials and Methods), from which the genes encoding AldFGH, AldSLC, and ADH have been deleted. The membranes of A. pasteurianus strain mNS4 show negligible levels of acetaldehyde:FC3− oxidoreductase activity at pH 4: <0.01 U (mg protein)−1 (S. Nina, K. Matsushita, and T. Yakushi, unpublished data). As described in Materials and Methods, we constructed plasmids carrying several combinations of the aldFGH genes of G. diazotrophicus strain PAL5 and expressed them in A. pasteurianus mNS4.
The membranes of the recombinant Acetobacter strain harboring the plasmid carrying G. diazotrophicus aldFGH showed acetaldehyde:FC3− oxidoreductase reductase (see “Reconstitution of the AldFGH complex,” below). The crude soluble fraction of the recombinant Acetobacter strain harboring the plasmid carrying aldGH also showed this activity (Fig. S4), suggesting that the AldGH subcomplex can function as a dehydrogenase even without the AldF subunit. However, cells harboring the aldH-carrying plasmid did not show any activity (Fig. S4), suggesting a functionally critical role of AldG.
The membranes of recombinant Acetobacter expressing the G. diazotrophicus AldFGH complex reduced ubiquinone-1 [45 mU (mg protein)−1 at pH 4.0] and consumed dissolved oxygen [56 mU (mg protein)−1 at pH 5.0], while the cell extract of recombinant Acetobacter expressing AldGH failed to reduce ubiquinone or molecular oxygen.
Properties of the AldGH subcomplex.
To obtain clues about the function of the [Fe-S] cluster in the AldG subunit, we attempted to purify the G. diazotrophicus AldGH subcomplex from recombinant A. pasteurianus mNS4 as described in Materials and Methods. We finally obtained 1.8 mg of the AldGH subcomplex with specific FC3− reductase activity of 100 U (mg protein)−1 (Table 2). In SDS-PAGE gels, the final preparation of the AldGH subcomplex showed two main bands with molecular masses of approximately 80 and 17 kDa (Fig. S5). We attempted to purify the AldGH subcomplex further by Superdex 200 gel filtration chromatography. The elution profile of the two bands corresponded to that of FC3− reductase activity. The data suggest that the 80-kDa and 17-kDa bands are AldH and AldG, respectively (Fig. S5). Furthermore, the 5 amino acids at the N terminus of the 17-kDa band were as expected for AldG.
TABLE 2.
Summary of purification of the AldGH subcomplex from recombinant Acetobacter pasteurianus mNS4a
| Purification step | Total protein concn (mg) | Sp act (U/mg) | Total activity (U) | Yield (%) |
|---|---|---|---|---|
| Crude soluble fraction | 1,000 | 0.64 | 660 | 100 |
| DEAE-Toyopearl | 95 | 6.1 | 580 | 88 |
| Phenyl-Toyopearl | —b | —b | 280 | 42 |
| Hydroxyapatite | 1.8 | 100 | 180 | 28 |
Acetaldehyde:ferricyanide oxidoreductase activity was measured at pH 5.0.
—, accurate protein content was not determined because of the presence of ammonium sulfate that may affect the Lowry method.
The absorption spectra of the purified AldGH subcomplex suggested the presence of an [Fe-S] cluster (Fig. 4). We performed a spectrophotometric analysis of the AldGH subcomplex in fraction 19 from gel filtration column chromatography because the fewest impurities were observed in this fraction in the SDS-PAGE analysis (Fig. 4 and Fig. S5). The overall absorption spectrum of the as-purified AldGH subcomplex was that of a typical protein with an absorption peak at around 280 nm (Fig. 4B), but a small shoulder was also observed at around 480 nm (inset of Fig. 4B). We anticipated that the [Fe-S] cluster of the purified AldGH subcomplex would be present in a reduced form because our purification system contained butyraldehyde. Therefore, we added potassium ferricyanide (final concentration of 0.5 mM) to oxidize the subcomplex. The shoulder at around 480 nm showed an increased absorbance, and a new shoulder appeared at around 580 nm (blue line in the inset of Fig. 4B). Finally, we added acetaldehyde (final concentration of 1 mM) to reduce the AldGH subcomplex. The two shoulders were now close to those in the original “as-purified” spectrum (compare the red and black lines in the inset of Fig. 4B). These results suggest that the AldGH subcomplex contains an [Fe-S] cluster that is involved in intramolecular electron transport for the catalytic reaction of the enzyme. They also suggest that the AldGH subcomplex purified in this study was in a reduced form.
FIG 4.
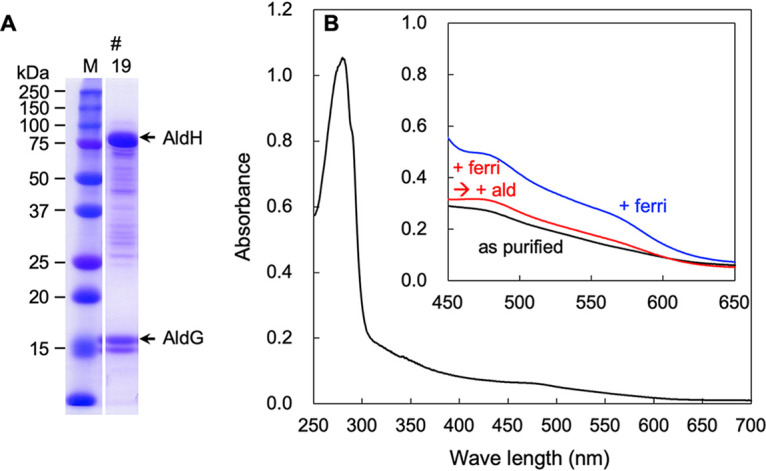
The [Fe-S] cluster of the AldGH subcomplex is reduced by acetaldehyde. (A) SDS-PAGE of fraction 19 from Superdex 200 gel filtration chromatography of the AldGH subcomplex. Thirty micrograms of protein was separated by SDS-PAGE, followed by staining with Coomassie brilliant blue R-250. (B) Absorption spectrum from 250 to 700 nm of as-purified fraction 19 (0.55 mg protein mL−1). The inset shows the absorption spectrum from 450 to 650 nm of as-purified fraction 19 (2.2 mg protein mL−1) (black line). Potassium ferricyanide was added to a final concentration of 0.5 mM (blue line). Next, acetaldehyde was added to a final concentration of 1 mM (red line).
Reconstitution of the AldFGH complex.
We constructed a recombinant A. pasteurianus mNS4 strain harboring the plasmid carrying the G. diazotrophicus aldF gene. Because we suggest (see above) that the AldF subunit is responsible for the membrane binding of the AldFGH complex, we prepared the membranes of the recombinant strain (referred to as “AldF membrane” here) for experiments to try to reconstitute the AldFGH complex using the AldGH subcomplex. The AldF membranes showed low ALDH activity of <0.01 U (mg protein)−1 at pH 4. When the crude soluble fraction containing G. diazotrophicus AldGH was mixed with AldF membranes, the acetaldehyde:FC3− oxidoreductase activity at pH 4.0 was significantly elevated in an AldF-dependent manner (Fig. S6). These results suggest that the AldF membrane contains active molecules that enhance the activity of AldGH and that genetically dissected Ald subunits can be reconstituted to form the AldFGH complex.
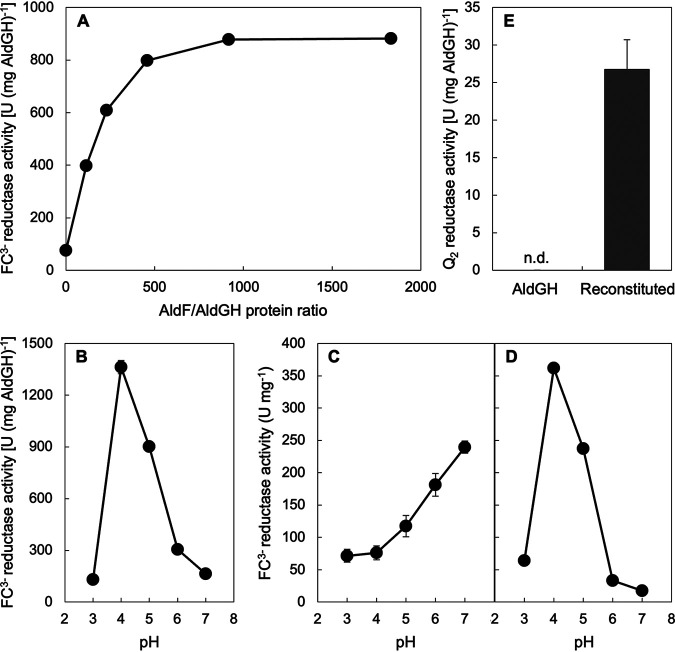
We tried to reconstitute acetaldehyde:ubiquinone-2 oxidoreductase activity with the crude soluble fraction containing AldGH and the AldF membranes. However, because high protein concentrations due to impurities perturb the enzyme assay, we did not detect Q2 reductase activity. Therefore, we partially purified AldF from detergent (Mydol 10 [decyl glucoside])-solubilized membranes by column chromatography to reconstitute the AldFGH complex with the purified AldGH subcomplex. When the purified AldGH with a specific activity of 120 U (mg protein)−1 at pH 5.0 was mixed with various amounts of the partially purified AldF, the reconstituted ALDH showed a saturation curve of FC3− reductase activity at pH 4.0 (Fig. 5A). The FC3− reductase activity of the AldGH subcomplex at pH 4.0 was activated in an AldF-dependent manner, and an AldF/AldGH protein ratio of 920 (mg protein/mg protein) showed saturated activation in this experiment. The pH dependence profile of the FC3− reductase activity of the reconstituted ALDH with an AldF/AldGH protein ratio of 920 (Fig. 5B) was different from that of the purified AldGH subcomplex (Fig. 5C) but similar to that of the purified AldFGH complex (Fig. 5D).
FIG 5.
Dissection of the AldFGH complex and reconstitution of the AldGH subcomplex with AldF. (A) Saturation curve of the ferricyanide (FC3−) reductase activity as a function of a protein ratio in milligrams of partially purified AldF to the purified AldGH subcomplex. The mixture of the AldGH subcomplex and AldF was incubated at 25°C for 10 min to accelerate reconstitution. Next, the acetaldehyde:FC3− oxidoreductase activity of the mixture was assayed at pH 4.0. (B) Specific FC3− reductase activity of the reconstituted AldFGH complex (protein ratio of 920) as a function of pH. (C) Specific FC3− reductase activity of the purified AldFG subcomplex as a function of pH for comparison with the data in panel B. (D) Specific FC3− reductase activity of the purified AldFG subcomplex as a function of pH for comparison with the data in panel B. (E) Q2 reductase activity of the purified AldGH subcomplex (AldGH) and the reconstituted AldFGH complex (protein ratio of 920) (Reconstituted) at pH 4.0. Mean values and standard deviations from triplicate assays are shown as the specific activity to the protein amount of AldGH (A to C and E) or AldFGH (D). n.d., not detected.
We detected acetaldehyde-dependent Q2 reductase activity with 27 U (mg AldGH)−1 by the reconstitution of AldGH with the partially purified AldF with an AldF/AldGH protein ratio of 920 (Fig. 5E). The Q2 reductase activity was detected with neither the purified AldGH subcomplex alone nor the partially purified AldF subunit (Table S1). For the AldFGH complex purified from G. diazotrophicus MR17, the Q2 reductase activity was approximately 5% of the FC3− reductase activity (Table 1). By considering that the FC3− reductase activity of the reconstituted ALDH with an AldF/AldGH protein ratio of 920 is approximately 1,000 U (mg AldGH)−1, Q2 reductase activity with 50 U (mg AldGH)−1 can be expected for the reconstituted ALDH. Taken together, our data show that the AldF subunit enhances catalysis with FC3− as the electron acceptor, presumably pulling the electrons from the AldGH subcomplex and passing them to FC3−. More importantly, the results clearly indicate that AldF is the site of ubiquinone reduction.
DISCUSSION
We purified the AldFGH complex from strain MR17 of G. diazotrophicus, which lacks the genes encoding AldSLC and ADH to avoid contaminating ALDH activity and the genes for PQQ biosynthesis because PQQ is not involved in AldFGH function (10). Gómez-Manzo et al. (8) purified ALDH from G. diazotrophicus strain PAL5. Even though the wild-type strain has the aldFGH, aldSLC, and adhAB gene clusters, the purified ALDH consisted of two subunits, and the large and middle subunits of ALDH were identified as the aldH and aldF gene products, respectively, by determining the amino acid sequences (8). Our enzyme preparation contained a 17-kDa band, the N-terminal amino acid sequence of which was identical to that of AldG, which possesses two [2Fe-2S] cluster binding motifs (7, 16). The previous report did not mention the presence of this small subunit (8). The AldFGH complex purified in the present study showed acetaldehyde:Q2 oxidoreductase activity, whereas this activity was not detected for purified ALDH in the previous report (8). More importantly, the previous report failed to reduce the cytochromes of the enzyme with the substrate. However, almost all cytochromes of the purified AldFGH complex were reduced when using acetaldehyde as the substrate in the present study (Fig. 2). We suggest that the detergent used for purification may be an important factor to maintain the intrinsic properties of the AldFGH complex, although we did not try to purify the AldFGH complex with Triton X-100 as used in the previous report (8). We summarize the comparison between the previous study and our present study in Table 3.
TABLE 3.
Summary of the properties of ALDH complexes purified from Gluconacetobacter diazotrophicus strain PAL5
| Property | Value in study |
|
|---|---|---|
| Gomez-Manzo et al.b | This study | |
| Specific FC3− reductase activity (U mg−1) | 800 | 300 |
| Optimum pH for FC3− reductase activity | 3.5 | 4.0 |
| Presence of ubiquinone reductase | No | Yes |
| Presence of small subunit | No | Yes |
| Size of subunit detected by heme staining | Middle | Middle |
| No. of hemes c | 3 | 3a |
| No. of hemes b | 1 | None |
| Reduction of cytochromes by the substrate | No | Yes |
Determination of heme c showed 2.1 mol per mol of the AldFGH complex. However, we suggest that there is 3 mol of heme c per mol of the AldFGH complex, even though the nearest integer was 2, because impurities in the enzyme preparation may decrease the experimentally observed value.
See reference 8.
We genetically dissected the AldFGH complex of G. diazotrophicus to understand the function of each subunit and the catalytic mechanism of the complex. The results shown in Fig. 3 clearly indicate that even in the absence of the AldF subunit, the AldG and AldH proteins show acetaldehyde:ferricyanide oxidoreductase activity, and Fig. S5 in the supplemental material indicates that the two proteins work as an AldGH subcomplex. As shown in Fig. 3, AldF is responsible for the membrane binding of the complex. The acetaldehyde:ferricyanide oxidoreductase activities of the AldFGH complex (Fig. 5D) and the AldGH subcomplex (Fig. 5C) as a function of pH were completely different from each other. Upon reconstitution with AldF, the pH dependence of the AldGH subcomplex changed to be similar to that of the AldFGH complex (Fig. 5B). Furthermore, the reconstituted ALDH complex showed acetaldehyde:Q2 oxidoreductase activity, although the AldGH subcomplex alone did not (Fig. 5E). Taken together, it is reasonable to conclude that we prepared an active AldF subunit and AldGH subcomplex and functionally reconstituted an AldFGH complex.
Several discrepancies in the properties of purified ALDHs have been reported by different research groups (6, 8, 19). Fukaya et al. (19) reported the purification and characterization of the ALDH of Komagataeibacter polyoxogenes (formerly Acetobacter polyoxogenes) strain NBI1028. They reported that the ALDH possessed no cytochromes, but the absorption spectrum showed an absorption shoulder at around 480 nm, similar to the AldGH subcomplex purified in the present study. However, they did not mention the presence of an [Fe-S] cluster in the purified enzyme. The optimum pH for the ferricyanide reductase activity of K. polyoxogenes ALDH was also similar to that of the AldGH subcomplex in the present work. A summary of the comparison of the ALDHs is shown in Table 4. We suggest that the ALDH purified by Fukaya et al. was the AldGH subcomplex of K. polyoxogenes.
TABLE 4.
Summary of the properties of the ALDH complex from Komagataeibacter polyoxogenes strain NBI1028 and the AldGH subcomplex of Gluconacetobacter diazotrophicus strain PAL5a
| Property | Value in studyd |
|
|---|---|---|
| Fukaya et al.a | This study | |
| Specific FC3− reductase activity (U mg−1)b | 800 | 100 |
| Optimum pH for FC3− reductase activity | 7.0 | 7.0c |
| Presence of ubiquinone reductase | NI | No |
| Sizes of subunits | Large and small | Large and small |
| Presence of cytochromes | No | No |
| Absorption shoulder (nm) | ∼480 | ∼480 |
| Presence of absorption changes upon redox | NI | Yes |
Komagataeibacter polyoxogenes strain NBI1028 was formerly referred to as Acetobacter polyoxogenes (19).
AldGH activity was routinely determined at pH 5.0 in this study but at pH 6.0 in the study by Fukaya et al. The ferricyanide reductase activity at pH 5.0 would be half of that at pH 6.0.
The ferricyanide reductase activity at pH 8.0 was higher than that at pH 7.0, but a weak chemical reduction of ferricyanide by acetaldehyde occurred. We tentatively conclude that pH 7.0 is optimal for the ferricyanide reductase activity of the AldGH subcomplex.
NI, no information.
We observed the reduction of an [Fe-S] cluster by acetaldehyde in the purified AldGH subcomplex (Fig. 4). These results strongly support the idea that the [Fe-S] cluster is involved in catalysis by the AldFGH complex. The experiments in Fig. 5 reconstituted electron transfer from the AldGH subcomplex to the AldF subunit where the Q2 molecule finally accepts the electrons. The intramolecular electron transport pathway in the AldFGH complex is proposed in Fig. 6A. A molybdopterin cofactor in the AldH subunit is the site of oxidation of acetaldehyde. Electrons from molybdopterin are transported to the [Fe-S] (presumably [2Fe-2S]) cluster in the AldG subunit and then to hemes c in the AldF subunit, which is responsible for membrane binding and ubiquinone reduction.
FIG 6.
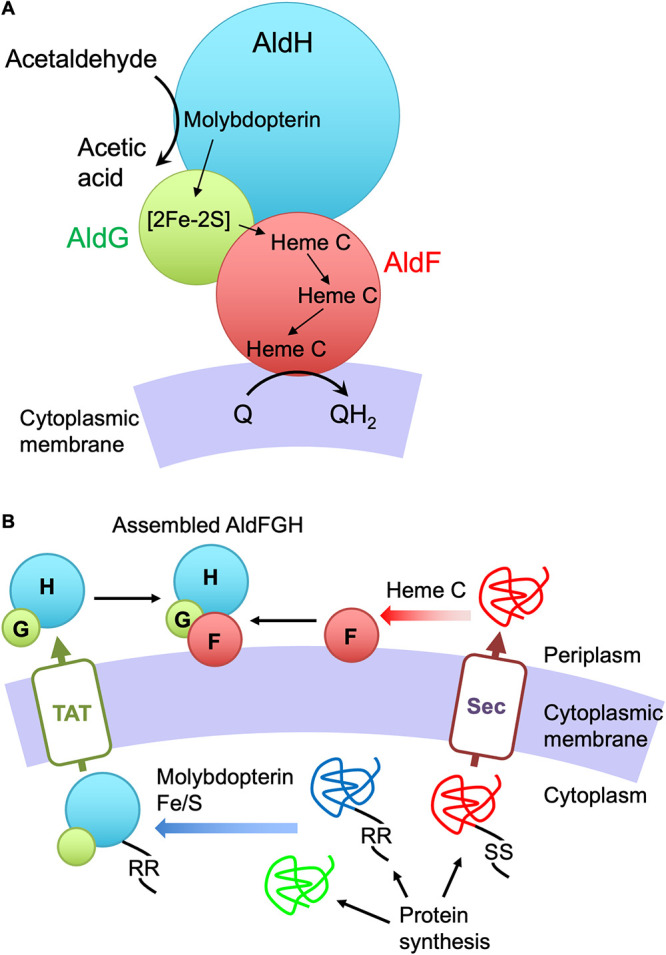
Models of intramolecular electron transport (A) and molecular assembly (B) of the AldFGH complex of G. diazotrophicus. (A) A molybdopterin cofactor in the AldH subunit is the site of acetaldehyde oxidation. Electrons from molybdopterin are transferred to the [Fe-S] cluster in the AldG subunit, presumably the [2Fe-2S] cluster suggested previously (7), and they are then transferred to c-type cytochrome centers in the AldF subunit, which is responsible for the membrane binding of the complex and ubiquinone reduction. (B) All the Ald subunits are synthesized in the cytoplasm. AldH and AldG are folded and assembled with a molybdopterin cofactor and an [Fe-S] cluster, respectively, followed by complex formation of AldG and AldH in the cytoplasm. The folded, assembled, and complexed AldG and AldH precursor is translocated to the periplasm via the TAT (“twin-arginine transport”) protein translocator. The AldF precursor is translocated to the periplasm via the Sec translocator without protein folding or assembly with any cofactors. After translocation, the hemes c are covalently attached to the polypeptide, the AldF subunit is folded, and the protein associates with the cytoplasmic membrane. Finally, the AldGH subcomplex and the AldF subunit meet at the cytoplasmic membrane to form the AldFGH complex. Black strings indicate the signal sequences for the TAT (RR) and Sec (SS) protein translocators. TAT, TAT protein translocator; Sec, Sec protein translocator.
The reconstitution experiment in this study may have reproduced the final step of the assembly of the AldFGH complex in vivo. In this study, we constructed two recombinant Acetobacter strains that respectively produced the AldGH subcomplex and the AldF subunit to reconstitute the AldFGH complex in vitro. All the Ald proteins are synthesized in the cytoplasm (Fig. 6B). The AldG and AldH precursors are expected to be folded and assembled with the cofactors molybdopterin and the [Fe-S] cluster in the cytoplasm. The precursors of these proteins do not possess a signal peptide for the Sec protein translocator system, but the AldH precursor possesses a signal peptide for the twin-arginine protein translocator (TAT) for the secretion of folded, cofactor-bound proteins to the periplasm (20). The folded and assembled AldG would associate with AldH to hitchhike to the periplasm. The AldF precursor would be secreted to the periplasm by the Sec system and assembled with the c hemes in the periplasm (21). The mature AldF would associate with the cytoplasmic membrane, but its mode of binding to the membrane remains unknown because AldF apparently does not have a transmembrane segment for membrane anchoring. Finally, the AldGH subcomplex and the AldF subunit would meet in the cytoplasmic membrane to complete AldFGH complex assembly, as we reconstituted the complex in vitro.
The determination of heme c showed 2.1 mol of heme c per mol of the AldFGH complex. However, we suggest that there is likely to be 3 mol of heme c per mol of the AldFGH complex, even though the nearest integer was 2, because our enzyme preparation contained protein impurities that can decrease the experimentally observed value. The value of 3 corresponds to there being three heme c binding CXXCH motifs in AldF (7) and the experimental results in a previous report (8). Although we failed to detect heme b in the purified AldFGH complex (Fig. S3), the absorption spectrum showed a shoulder at 556 nm (Fig. 2), similar to a previous report (8). The AldF, AldG, and AldH proteins do not contain the amino acid sequence for heme b binding suggested previously by Li et al. (22). Because the absorption peak of the α-band of c-type cytochromes is variable (23), we suggest that one of the three hemes c is responsible for the absorption at 556 nm. Intriguingly, the as-purified AldFGH complex in the present work was oxidized even though 2 mM butyraldehyde was included in the buffers used in the purification steps (Fig. 2), but the AldGH subcomplex was reduced (Fig. 4), hinting that the AldFGH complex can be oxidized by molecular oxygen in an AldF-dependent manner. Such an oxygen reduction ability of the AldFGH complex will be addressed in the future.
MATERIALS AND METHODS
Materials.
Acetaldehyde (99.5%) and n-dodecyl-β-d-maltoside (DDM) were purchased from Fluka (St. Louis, MO, USA) and Dojindo (Kumamoto, Japan), respectively. Yeast extract and Hipolypepton were obtained from Oriental Yeast (Tokyo, Japan) and Nihon Pharmaceutical (Tokyo, Japan), respectively. Mydol 10 (decyl glucoside) was obtained from Kao (Tokyo, Japan). All other materials were purchased from commercial sources and were of analytical grade.
Bacterial strains, plasmids, and growth conditions.
The G. diazotrophicus and A. pasteurianus strains and plasmids used in this study are listed in Table 5. G. diazotrophicus strain PAL5 (ATCC 49037) was obtained from the American Type Culture Collection (Manassas, VA). Suicide vector pK18mobSacB was provided by Alfred Pühler, Universität Bielefeld, Bielefeld, Germany, and was used for markerless gene deletion. A. pasteurianus strain mNS4 (ΔadhAB ΔaldSLC ΔaldFGH) was used as a host for the heterologous expression of the G. diazotrophicus aldFGH genes. The mNS4 strain shows only negligible levels of acetaldehyde:FC3− oxidoreductase activity [<0.01 U (mg membrane protein)−1] at pH 4 but does have activity at pH 6 [0.02 to 0.03 U (mg membrane protein)−1] (Nina et al., unpublished).
TABLE 5.
Bacterial strains and plasmids used in this studya
| Strain or plasmid | Description | Source and/or reference |
|---|---|---|
| Bacterial strains | ||
| Gluconacetobacter diazotrophicus | ||
| ATCC 49037 | Wild type; synonym of PAL5 | ATCC; 37 |
| MR17 | PAL5 ΔaldSLC ΔadhAB ΔpqqABCDE | 10 |
| MR25 | PAL5 ΔaldSLC ΔadhAB ΔpqqABCDE ΔaldF | This study |
| Acetobacter pasteurianus | ||
| SKU1108 | Wild type; synonym of NBRC101655 | 38 |
| mNS4 | SKU1108 ΔadhAB ΔaldSLC ΔaldFGH | Nina et al., unpublished |
| Plasmids | ||
| pT7Blue | Cloning vector; Apr | Novagen |
| pRK2013 | Plasmid-mediated plasmid transfer; Kmr | 30 |
| pK18mobsacB | Suicide vector; mob sacB; Kmr | 28 |
| pCM62 | Broad-host-range plasmid; mob lacZα; Tcr | 29 |
| pMRG | pK18mobsacB with a 2-kb fragment containing the aldF allele | This study |
| pTM8 | pCM62 aldGH | This study |
| pTM10 | pCM62 aldH | This study |
| pTM14 | pCM62 aldF with the putative aldFGH promoter in the orientation opposite that of the lac promoter | This study |
| pTM15 | pT7Blue aldFGH with a putative aldFGH promoter | This study |
| pTM16 | pCM62 aldFGH with a putative aldFGH promoter in the orientation opposite that of the lac promoter | This study |
Apr, ampicillin resistance; Kmr, kanamycin resistance; Tcr, tetracycline resistance.
G. diazotrophicus cells were grown at 30°C in DYGS medium (2.0 g glucose, 1.5 g Hipolypepton, 2.0 g yeast extract, 0.5 g KH2PO4, 0.5 g MgSO4·7H2O, and 1.5 g sodium l-glutamate monohydrate per L [pH 6.0]), YPGD medium (5 g yeast extract, 5 g Hipolypepton, 5 g glycerol, and 5 g glucose per L), or YPG medium (5 g yeast extract, 5 g Hipolypepton, and 10 g glycerol per L). Kanamycin was used at a final concentration of 200 μg mL−1. A. pasteurianus cells were cultivated at 30°C in YPGD medium or YPGDP medium (5 g yeast extract, 5 g Hipolypepton, and 10 g glycerol per L, supplemented with 50 mM K+-phosphate [pH 6.5]). Tetracycline was used at a final concentration of 50 μg mL−1.
Escherichia coli strains DH5α (24) and HB101 (25) were used for strain construction and triparental mating, respectively. E. coli cells were grown in modified Luria-Bertani medium (10 g Hipolypepton, 5 g yeast extract, and 5 g NaCl per L, adjusted to pH 7 with NaOH). Ampicillin, kanamycin, and tetracycline were used at final concentrations of 50 μg mL−1, 50 μg mL−1, and 10 μg mL−1, respectively.
Construction of plasmids.
The outline of the construction of plasmids used in this study is shown in Fig. S7 in the supplemental material. Genomic DNA of G. diazotrophicus strain PAL5 was isolated according to the method described previously by Marmur (26), with some modifications (27). The aldFGH genes (Gdia_3086 to Gdia_3088) were amplified by PCR with Herculase II fusion DNA polymerase (Stratagene, CA, USA), PAL5 genomic DNA, and several pairs of oligonucleotides (Table S2).
For the elimination of the aldF gene from the genome of G. diazotrophicus PAL5, the pair of oligonucleotides Pal5-D-aldF-Hin(+) and Pal5-D-aldF-5-RI(−) was used to amplify the 1.0-kb DNA fragment containing the 5′-flanking region, and the primer pair Pal5-D-aldF-3-RI(+) and Pal5-D-aldF-Xba(−) was used to amplify the 1.0-kb 3′-flanking region. The two resulting DNA fragments were treated with HindIII and EcoRI and with EcoRI and XbaI, respectively, and inserted into the HindIII and XbaI sites of K18mobsacB (28) to yield pMRG (Fig. S7, ΔaldF).
For the construction of the aldFGH expression plasmid pTM16, the 4.4-kb DNA fragment containing aldFGH, including the putative promoter region, was amplified using genomic DNA of G. diazotrophicus PAL5 as the template and oligonucleotide pair Pal5-aldpro-RI(+) and Pal5-ex-aldH-Xba(−) and inserted into plasmid pT7Blue (Novagen) to yield pTM15. The 4.4-kb DNA fragment obtained by XbaI treatment of pTM15 was inserted into the corresponding site of pCM62 (29) to yield pTM16 (Fig. S7, aldFGH). pTM8, pTM14, and pTM10 for the expression of aldGH, aldH, and aldF, respectively, were constructed using similar procedures (outlined in Fig. S7), and the oligonucleotides used are shown in Table S2.
Transformation of acetic acid bacteria.
G. diazotrophicus and A. pasteurianus were transformed with plasmids via a triparental mating method using E. coli HB101 harboring pRK2013 as the helper strain (30), as described previously (9).
Construction of the ΔaldF deletion strain.
G. diazotrophicus MR17, a ΔaldSLC ΔadhAB ΔPQQ derivative of strain PAL5, was transformed with suicide plasmid pMRG (ΔaldF) to construct the quadruple-deletion mutant MR25 (ΔaldSLC ΔadhAB ΔPQQ ΔaldF), as described previously (10).
Preparation of membranes and crude soluble fractions.
G. diazotrophicus strains were precultivated in 100 mL YPGD medium at 30°C for 24 h, and 5 mL of the preculture was transferred to 500 mL YPG medium in a 3-L flask and incubated at 30°C for 24 h. Recombinant A. pasteurianus strains were precultivated in 100 mL YPGDP medium containing 50 μg mL−1 tetracycline at 30°C for 48 h, and 5 mL of the preculture was transferred to 500 mL of the same medium in a 3-L flask and incubated at 30°C for 48 h. The cells were collected by centrifugation at 8,000 × g for 10 min at 4°C, washed with ice-chilled 10 mM K+-MES (2-morpholinoethanesulfonic acid) (pH 6.0), and resuspended in the same buffer supplemented with 0.5 mM phenylmethanesulfonyl fluoride. The cell suspension was passed through a French pressure cell press (American Instrument Company, Silver Spring, MD) at 1,100 kg cm−2, twice. After the removal of intact cells and cell debris by centrifugation at 8,000 × g for 10 min at 4°C, the supernatant was further centrifuged at 100,000 × g for 1 h at 4°C. The supernatant was used as the crude soluble fraction, and the precipitate was suspended in ice-chilled 10 mM K+-MES (pH 6.0) and used as the membrane fraction.
Purification of the AldFGH complex.
The membranes of G. diazotrophicus strain MR17 were solubilized with 1.0% (wt/vol) DDM in 10 mM Na+-acetate (pH 5.0) with a protein concentration of 10 mg mL−1. After incubation at 4°C for 1 h with gentle stirring, the membrane suspension was centrifuged at 100,000 × g for 1 h at 4°C. The supernatant was applied to a CM-Toyopearl column (Tosoh, Tokyo, Japan). After washing the column with 10 mM Na+-acetate (pH 5.0) containing 0.02% (wt/vol) DDM and 2 mM butyraldehyde, ALDH was eluted with a linear NaCl gradient from 0 to 0.5 M. The acetaldehyde:FC3− oxidoreductase activity in the eluate fractions was determined.
Active fractions from CM-Toyopearl column chromatography were pooled; K+-phosphate (pH 6.0) was added to a final concentration of 10 mM, followed by pH adjustment to 6.0 with potassium hydroxide; and the solution was applied to a ceramic hydroxyapatite column (Bio-Rad, Hercules, CA). After washing the column with 10 mM K+-phosphate (pH 6.0) containing 0.02% (wt/vol) DDM and 2 mM butyraldehyde, ALDH was eluted by a stepwise increase in the phosphate concentration: 50, 100, 200, 300, 400, and 500 mM. The acetaldehyde:FC3− oxidoreductase activity in the eluate fractions was determined.
Active fractions from the ceramic hydroxyapatite column eluted with 500 mM K+-phosphate (pH 6.0) containing 0.02% (wt/vol) DDM and 2 mM butyraldehyde were pooled, concentrated by ultrafiltration with Amicon Ultra centrifugal filters (molecular weight of 50,000 [50K]; Merck, Darmstadt, Germany), and dialyzed into 10 mM K+-phosphate (pH 6.0) containing 0.02% (wt/vol) DDM and 2 mM butyraldehyde at 4°C.
The dialysate was applied to a DEAE-Toyopearl column (Tosoh). ALDH was eluted with a linear NaCl gradient from 0 to 0.5 M, after washing the column with 10 mM K+-phosphate (pH 6.0) containing 0.02% (wt/vol) DDM and 2 mM butyraldehyde. The active fractions were pooled, concentrated as described above, and stored at 4°C.
Purification of the AldGH subcomplex.
The crude soluble fraction of the recombinant A. pasteurianus mNS4 strain harboring pTM8 (aldGH+) was applied to a DEAE-Toyopearl column. After washing the column with 10 mM K+-MES (pH 6.0) containing 2 mM butyraldehyde, ALDH was eluted with a linear NaCl gradient from 0 to 0.3 M. The acetaldehyde:FC3− oxidoreductase activity in the eluate fractions was determined.
Active fractions from DEAE-Toyopearl column chromatography were pooled, and solid ammonium sulfate was added to 25% saturation, followed by application to a Phenyl-Toyopearl column (Tosoh). After washing the column with 10 mM K+-MES (pH 6.0) containing 2 mM butyraldehyde and ammonium sulfate (25% saturation), ALDH was eluted with a linear ammonium sulfate gradient from 25% to 0% saturation. Active fractions from Phenyl-Toyopearl column chromatography were pooled and dialyzed into 10 mM K+-phosphate (pH 6.0) containing 2 mM butyraldehyde at 4°C.
The dialysate was applied to a ceramic hydroxyapatite column. ALDH was eluted with a stepwise increase in the phosphate concentration: 10, 20, 50, 100, and 1,000 mM. Most activity was recovered in the 100 mM K+-phosphate fraction. Active fractions from the ceramic hydroxyapatite column were pooled, concentrated by ultrafiltration as described above, and stored at 4°C.
Partial purification of the AldF subunit.
The membranes of the recombinant A. pasteurianus mNS4 strain harboring pTM14 (aldF+) were solubilized with 1.0% (wt/vol) Mydol 10 (decyl glucoside) in 10 mM Na+-acetate (pH 5.0) at a protein concentration of 10 mg mL−1. After incubation at 4°C for 1 h with gentle mixing, the membrane suspension was centrifuged at 100,000 × g for 1 h at 4°C. The supernatant was applied to a CM-Toyopearl column. After washing the column with 10 mM Na+-acetate (pH 5.0) containing 0.2% (wt/vol) Mydol 10, AldF was eluted with a linear NaCl gradient from 0 to 0.2 M. AldF in the eluate fractions was detected by the activation ability of the AldGH subcomplex (Fig. 4 and Fig. S5). The acetaldehyde:FC3− oxidoreductase activity of the AldGH subcomplex at pH 4.0 is low, but it is enhanced by AldF. Thus, we detected AldF by examining the ability of the eluate from the chromatography column to activate AldGH. Active fractions were pooled and dialyzed into 10 mM K+-phosphate (pH 6.0) containing 0.2% (wt/vol) Mydol 10 at 4°C.
The dialysate was applied to a ceramic hydroxyapatite column. ALDH was eluted with a stepwise increase in the phosphate concentration: 100, 200, 500, and 1,000 mM. AldGH activation activity was recovered in the 200 and 500 mM K+-phosphate fractions. Active fractions were pooled and dialyzed into 10 mM Na+-acetate (pH 5.0) containing 0.2% (wt/vol) Mydol 10, followed by application to a small CM-Toyopearl column to concentrate the AldF. The activity was eluted with 10 mM Na+-acetate (pH 5.0) containing 0.2% (wt/vol) Mydol 10 and 0.2 M NaCl.
Reconstitution of the AldFGH complex.
The purified AldGH subcomplex and the partially purified AldF subunit were mixed in 10 mM Na+-acetate (pH 5.0) containing 0.2% (wt/vol) Mydol 10 and incubated at 25°C for 10 min.
Enzyme assays.
Acetaldehyde:ferricyanide oxidoreductase activity was determined by the ferric-Dupanol assay at 25°C using 100 mM acetaldehyde, as described previously (6). McIlvaine buffer (a mixture of 100 mM citric acid and 200 mM Na2HPO4) was used at pH 4.0 to 7.0. Acetaldehyde:ubiquinone-1 (Q1) [2,3-dimethoxy-5-methyl-6-(3-methyl-2-butenyl)-1,4-benzoquinone] and acetaldehyde:ubiquinone-2 (Q2) (2,3-dimethoxy-5-geranyl-6-methyl-1,4-benzoquinone) oxidoreductase activities were determined at 25°C by monitoring the decrease in the absorbance at 275 nm in McIlvaine buffer (pH 4.0) containing either 50 μM Q1 or 20 μM Q2, 4 mM NaN3, membranes or enzyme solution, and 100 mM acetaldehyde. The molecular absorption coefficient used was an ε275 of 12.25 mM−1 cm−1 (31). One unit of enzyme was defined as the amount that oxidized 1 μmol of the substrate per min. Oxygen consumption activity was measured by using a Clark-type oxygen electrode (YSI model 5300; Yellow Springs Instrument, Yellow Springs, OH, USA) at 25°C. The electrode was calibrated by using air-saturated McIlvaine buffer (pH 5.0), assuming the concentration of molecular oxygen to be 249 μM (32). Sodium dithionite was used for calibration to reduce molecular oxygen completely. The reaction mixture contained membranes, McIlvaine buffer (pH 5.0), and 100 mM acetaldehyde. One unit was defined as 1 μmol of half a molecular oxygen (equivalent to an oxygen atom) consumed per min. The protein content was determined by the modified Lowry method, and bovine serum albumin was used as the standard (33).
Absorption spectra.
The absorption spectra of the purified AldFGH complex and AldGH subcomplex were recorded at 25°C using a UV-1900i spectrophotometer (Shimadzu, Kyoto, Japan) and 1-cm-light-path cuvettes.
Determination of hemes in the AldFGH complex.
For the quantification of heme, the purified AldFGH complex, pyridine, and NaOH were mixed at final concentrations of 0.30 mg mL−1, 20% (vol/vol), and 0.5 M, respectively. The sample was reduced with a few grains of sodium dithionite or oxidized with a few grains of potassium ferricyanide. Reduced-minus-oxidized differences in the absorption spectra were recorded at 300 to 700 nm. The heme c content was determined using a extinction coefficient 23.97 mM−1 cm−1 for the reduced-minus-oxidized absorbance difference at 550 nm minus 535 nm (34).
Alternatively, hemes in the purified AldFGH complex (1.0 mg protein) or partially purified cytochrome ba3 ubiquinol oxidase (1.0 mg protein), which was obtained by anion-exchange column chromatography of subfractions in the purification of the AldFGH complex, was separated into protein-bound and free forms by treatment with acid-acetone (65% [vol/vol] acetone and 35 mM HCl). The acetone was evaporated, and the extracted materials were dissolved in 50% acetonitrile as described previously by Puustinen and Wikström (17), followed by the application of the above-described procedure for the quantification of hemes.
SDS-PAGE.
SDS-PAGE was performed as described previously (35), followed by staining with Coomassie brilliant blue R-250 or, for heme-dependent peroxidase activity, tetramethylbenzidine (36). Broad-range (250- to 10-kDa) Precision Plus protein dual-color standards (Bio-Rad) were used as molecular size standards.
For the determination of N-terminal amino acid sequences, proteins in a gel were electrophoretically transferred onto a polyvinylidene difluoride membrane (Merck) in 10 mM Na+-CAPS (N-cyclohexyl-3-aminopropanesulfonic acid) (pH 11.0) containing 20% (vol/vol) methanol. The membrane was stained with Coomassie brilliant blue R-250. N-terminal amino acid sequencing was performed by automated Edman degradation (catalog number PPSQ-33A; Shimadzu, Kyoto, Japan).
ACKNOWLEDGMENTS
We thank Oriental Yeast (Tokyo, Japan), Kao (Tokyo, Japan), and Toyobo (Osaka, Japan) for gifting us yeast extract, Mydol 10 (decyl glucoside), and restriction endonucleases, respectively. We thank Rio Izumi for her skillful technical assistance. We are grateful to Tomoyuki Kosaka and Mamoru Yamada for invaluable suggestions. We thank Osao Adachi and Hirohide Toyama for continuously encouraging our work on ALDH. We thank James Allen from Edanz for editing a draft of the manuscript.
We are grateful to the Japanese Government (Monbukagakusho [MEXT]) for scholarships supporting the work of R.M. This work was supported by KAKENHI grant number 23580115.
T.Y. designed the study and wrote the manuscript. R.M. performed most experiments, analyzed the data, prepared the figures, and wrote a draft manuscript. S.N. and T.M. assisted with constructing bacterial strains and edited the manuscript. M.M. performed bioinformatic analyses. T.Y., N.K., Y.A., and K.M. supervised the work and edited the manuscript. All authors read and approved the final manuscript.
Footnotes
Supplemental material is available online only.
Contributor Information
Toshiharu Yakushi, Email: juji@yamaguchi-u.ac.jp.
Michael Y. Galperin, NCBI, NLM, National Institutes of Health
REFERENCES
- 1.Matsushita K, Toyama H, Adachi O. 1994. Respiratory chains and bioenergetics of acetic acid bacteria. Adv Microb Physiol 36:247–301. 10.1016/s0065-2911(08)60181-2. [DOI] [PubMed] [Google Scholar]
- 2.Yakushi T, Matsushita K. 2010. Alcohol dehydrogenase of acetic acid bacteria: structure, mode of action, and applications in biotechnology. Appl Microbiol Biotechnol 86:1257–1265. 10.1007/s00253-010-2529-z. [DOI] [PubMed] [Google Scholar]
- 3.Matsushita K, Yakushi T, Toyama H, Shinagawa E, Adachi O. 1996. Function of multiple heme c moieties in intramolecular electron transport and ubiquinone reduction in the quinohemoprotein alcohol dehydrogenase-cytochrome c complex of Gluconobacter suboxydans. J Biol Chem 271:4850–4857. 10.1074/jbc.271.9.4850. [DOI] [PubMed] [Google Scholar]
- 4.Kondo K, Beppu T, Horinouchi S. 1995. Cloning, sequencing, and characterization of the gene encoding the smallest subunit of the three-component membrane-bound alcohol dehydrogenase from Acetobacter pasteurianus. J Bacteriol 177:5048–5055. 10.1128/jb.177.17.5048-5055.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Masud U, Matsushita K, Theeragool G. 2010. Cloning and functional analysis of adhS gene encoding quinoprotein alcohol dehydrogenase subunit III from Acetobacter pasteurianus SKU1108. Int J Food Microbiol 138:39–49. 10.1016/j.ijfoodmicro.2009.12.027. [DOI] [PubMed] [Google Scholar]
- 6.Adachi O, Tayama K, Shinagawa E, Matsushita K, Ameyama M. 1980. Purification and characterization of membrane-bound aldehyde dehydrogenase from Gluconobacter suboxydans. Agric Biol Chem 44:503–515. 10.1271/bbb1961.44.503. [DOI] [Google Scholar]
- 7.Thurner C, Vela C, Thony-Meyer L, Meile L, Teuber M. 1997. Biochemical and genetic characterization of the acetaldehyde dehydrogenase complex from Acetobacter europaeus. Arch Microbiol 168:81–91. 10.1007/s002030050473. [DOI] [PubMed] [Google Scholar]
- 8.Gómez-Manzo S, Chavez-Pacheco JL, Contreras-Zentella M, Sosa-Torres ME, Arreguín-Espinosa R, Pérez de la Mora M, Membrillo-Hernández J, Escamilla JE. 2010. Molecular and catalytic properties of the aldehyde dehydrogenase of Gluconacetobacter diazotrophicus, a quinoheme protein containing pyrroloquinoline quinone, cytochrome b, and cytochrome c. J Bacteriol 192:5718–5724. 10.1128/JB.00589-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yakushi T, Fukunari S, Kodama T, Matsutani M, Nina S, Kataoka N, Theeragool G, Matsushita K. 2018. Role of a membrane-bound aldehyde dehydrogenase complex AldFGH in acetic acid fermentation with Acetobacter pasteurianus SKU1108. Appl Microbiol Biotechnol 102:4549–4561. 10.1007/s00253-018-8940-6. [DOI] [PubMed] [Google Scholar]
- 10.Miah R, Nina S, Murate T, Kataoka N, Matsutani M, Matsushita K, Yakushi T. 2021. Major aldehyde dehydrogenase AldFGH of Gluconacetobacter diazotrophicus is independent of pyrroloquinoline quinone but dependent on molybdopterin for acetic acid fermentation. Appl Microbiol Biotechnol 105:2341–2350. 10.1007/s00253-021-11144-x. [DOI] [PubMed] [Google Scholar]
- 11.Huber R, Hof P, Duarte RO, Moura JJ, Moura I, Liu MY, LeGall J, Hille R, Archer M, Romão MJ. 1996. A structure-based catalytic mechanism for the xanthine oxidase family of molybdenum enzymes. Proc Natl Acad Sci USA 93:8846–8851. 10.1073/pnas.93.17.8846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanchanarach W, Theeragool G, Yakushi T, Toyama H, Adachi O, Matsushita K. 2010. Characterization of thermotolerant Acetobacter pasteurianus strains and their quinoprotein alcohol dehydrogenases. Appl Microbiol Biotechnol 85:741–751. 10.1007/s00253-009-2203-5. [DOI] [PubMed] [Google Scholar]
- 13.Gómez-Manzo S, Contreras-Zentella M, González-Valdez A, Sosa-Torres M, Arreguín-Espinoza R, Escamilla-Marván E. 2008. The PQQ-alcohol dehydrogenase of Gluconacetobacter diazotrophicus. Int J Food Microbiol 125:71–78. 10.1016/j.ijfoodmicro.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 14.Ameyama M, Matsushita K, Ohno Y, Shinagawa E, Adachi O. 1981. Existence of a novel prosthetic group, PQQ, in membrane-bound, electron transport chain-linked, primary dehydrogenases of oxidative bacteria. FEBS Lett 130:179–183. 10.1016/0014-5793(81)81114-3. [DOI] [PubMed] [Google Scholar]
- 15.Takemura H, Tsuchida T, Yoshinaga F, Matsushita K, Adachi O. 1994. Prosthetic group of aldehyde dehydrogenase in acetic acid bacteria not pyrroloquinoline quinone. Biosci Biotechnol Biochem 58:2082–2083. 10.1271/bbb.58.2082. [DOI] [Google Scholar]
- 16.Giongo A, Tyler HL, Zipperer UN, Triplett EW. 2010. Two genome sequences of the same bacterial strain, Gluconacetobacter diazotrophicus PAl 5, suggest a new standard in genome sequence submission. Stand Genomic Sci 2:309–317. 10.4056/sigs.972221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Puustinen A, Wikström M. 1991. The heme groups of cytochrome o from Escherichia coli. Proc Natl Acad Sci USA 88:6122–6126. 10.1073/pnas.88.14.6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsushita K, Shinagawa E, Adachi O, Ameyama M. 1990. Cytochrome a1 of Acetobacter aceti is a cytochrome ba functioning as ubiquinol oxidase. Proc Natl Acad Sci USA 87:9863–9867. 10.1073/pnas.87.24.9863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fukaya M, Tayama K, Okumura H, Kawamura Y, Beppu T. 1989. Purification and characterization of membrane-bound aldehyde dehydrogenase from Acetobacter polyoxogenes sp. nov. Appl Microbiol Biotechnol 32:176–180. 10.1007/BF00165884. [DOI] [Google Scholar]
- 20.Palmer T, Stansfeld PJ. 2020. Targeting of proteins to the twin-arginine translocation pathway. Mol Microbiol 113:861–871. 10.1111/mmi.14461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kranz RG, Richard-Fogal C, Taylor JS, Frawley ER. 2009. Cytochrome c biogenesis: mechanisms for covalent modifications and trafficking of heme and for heme-iron redox control. Microbiol Mol Biol Rev 73:510–528. 10.1128/MMBR.00001-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li T, Bonkovsky HL, Guo JT. 2011. Structural analysis of heme proteins: implications for design and prediction. BMC Struct Biol 11:13. 10.1186/1472-6807-11-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamanaka T. 1992. The biochemistry of bacterial cytochromes. Springer-Verlag, Tokyo, Japan. [Google Scholar]
- 24.Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580. 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 25.Boyer HW, Roulland-Dussoix D. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol 41:459–472. 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 26.Marmur J. 1961. A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J Mol Biol 3:208–218. 10.1016/S0022-2836(61)80047-8. [DOI] [Google Scholar]
- 27.Kawai S, Goda-Tsutsumi M, Yakushi T, Kano K, Matsushita K. 2013. Heterologous overexpression and characterization of a flavoprotein-cytochrome c complex fructose dehydrogenase of Gluconobacter japonicus NBRC3260. Appl Environ Microbiol 79:1654–1660. 10.1128/AEM.03152-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73. 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 29.Marx CJ, Lidstrom ME. 2001. Development of improved versatile broad-host-range vectors for use in methylotrophs and other Gram-negative bacteria. Microbiology (Reading) 147:2065–2075. 10.1099/00221287-147-8-2065. [DOI] [PubMed] [Google Scholar]
- 30.Figurski DH, Helinski DR. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA 76:1648–1652. 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Redfearn ER. 1967. Isolation and determination of ubiquinone. Methods Enzymol 10:381–384. 10.1016/0076-6879(67)10071-2. [DOI] [Google Scholar]
- 32.Mitchell P, Moyle J, Mitchell R. 1979. Measurement of translocation of H+/O in mitochondria and submitochondrial vesicles. Methods Enzymol 55:627–640. 10.1016/0076-6879(79)55071-x. [DOI] [PubMed] [Google Scholar]
- 33.Dulley JR, Grieve PA. 1975. A simple technique for eliminating interference by detergents in the Lowry method of protein determination. Anal Biochem 64:136–141. 10.1016/0003-2697(75)90415-7. [DOI] [PubMed] [Google Scholar]
- 34.Berry EA, Trumpower BL. 1987. Simultaneous determination of hemes a, b, and c from pyridine hemochrome spectra. Anal Biochem 161:1–15. 10.1016/0003-2697(87)90643-9. [DOI] [PubMed] [Google Scholar]
- 35.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 36.Thomas PE, Ryan D, Levin W. 1976. An improved staining procedure for the detection of the peroxidase activity of cytochrome P-450 on sodium dodecyl sulfate polyacrylamide gels. Anal Biochem 75:168–176. 10.1016/0003-2697(76)90067-1. [DOI] [PubMed] [Google Scholar]
- 37.Yamada Y, Hoshino K, Ishikawa T. 1997. The phylogeny of acetic acid bacteria based on the partial sequences of 16S ribosomal RNA: the elevation of the subgenus Gluconoacetobacter to the generic level. Biosci Biotechnol Biochem 61:1244–1251. 10.1271/bbb.61.1244. [DOI] [PubMed] [Google Scholar]
- 38.Saeki A, Theeragool G, Matsushita K, Toyama H, Lotong N, Adachi O. 1997. Development of thermotolerant acetic acid bacteria useful for vinegar fermentation at higher temperatures. Biosci Biotechnol Biochem 61:138–145. 10.1271/bbb.61.138. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1-S7; Tables S1 and S2. Download jb.00558-21-s0001.pdf, PDF file, 0.7 MB (709.6KB, pdf)