Abstract
Background
The mayhem COVID-19 that was ushered by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV2) was declared pandemic by the World Health Organization in March 2020. Since its initial outbreak in late 2019, the virus has affected hundreds of million adults in the world and killing millions in the process. After the approval of newly developed vaccines, severe challenges remain to manufacture and administer them to the adult population globally in quick time. However, we have witnessed several mutations of the virus leading to ‘waves’ of viral spread and mortality. WHO has categorized these mutations as variants of concern (VOCs) and variants of interest (VOIs). The mortality due to COVID-19 has also been associated with various comorbidities and improper immune response. This has created further complications in understanding the nature of the SARS-CoV2–host interaction that has fuelled doubts in the efficacy of the approved vaccines. Whether there is requirement of booster dose and whether the impending wave could affect the children are some of the hotly debated topics.
Materials and Methods
A systematic literature review of PubMed, Medline, Scopus, Google Scholar was utilized to understand the nature of Delta variant and how it alters our T-cell responses and cytokine production and neutralizes vaccine-generated antibodies.
Conclusion
In this review, we discuss the variants of SARS-CoV2 with specific focus on the Delta variant. We also specifically review the T-cell response against the virus and bring a narrative of various factors that may hold the key to fight against this marauding virus.
Keywords: COVID-19, Delta variant, T cells, Vaccines, Variant of concern, Variant of interest
Introduction
Pandemics have befallen human civilization multiple times in the past. From the Spanish Flu in 1918 to the Ebola outbreak of 2013, the deadly effect of these microscopic infective agents like viruses has been witnessed by mankind several times. In December 2019, the world took notice of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV2) leading to the novel coronavirus disease (COVID-19). The disease was declared a global pandemic by the World Health Organization (WHO) in March 2020. We are still reeling from the devastation of the pandemic that has caused immense damage to the health and the economy globally. The virus has infected more than 200 million people claiming more than 4 million lives worldwide [https://www.worldometers.info/coronavirus/]. Despite the rapid development and approval of vaccines, there are several challenges that might have limited the success stories of the vaccines: (a) failure to adhere to COVID-19 appropriate behavior, (b) lack of quick production of vaccines in large numbers, (c) variants of concerns emerging from the parent strain, (d) comorbidities associated with a large number of COVID-19-positive patients, (e) fatalities despite taking the necessary doses of approved vaccines, and (f) concerns about the duration of the memory responses mediated by the vaccines and whether there is any requirement of booster doses.
On May 31, 2021, the WHO designated different variants of COVID-19 using Greek alphabets like alpha, beta, gamma, and delta among others. India witnessed a surge of COVID-19 cases during the second wave with more than 400,000 cases and over 4000 deaths reported in early May 2021 [1]. The B.1.617.2 variant, otherwise known as the delta variant, was first detected in October 2020 in the country, which became the most obvious variant in April 2021, leading to a huge uptick in the number of positive cases [2]. This variant had been detected in more than 40 countries across six continents. According to CDC, the delta variant is twice as fast in terms of spreading infection that might lead to ‘hyperlocal outbreaks’. The adaptive immune response, especially the T-cell response against COVID-19 is critical to mount resistance against the virus. However, the rapid induction and the amount of humoral responses in fighting the virus is equally important. Reports have suggested the role of CD4 + and CD8 + T cells in the resolution of SARS-CoV2 infection, including modulating disease severity in humans [3]. The coordinated action of both CD4 + and CD8 + T cells leads to a milder form of the disease aided with faster viral clearance [4]. The incubation time to the onset of the disease and its subsequent progression to the severe form takes approximately 4–10 days according to studies assessing the clinical features of patients infected with the virus [5]. Patients suffering from the severe form of COVID-19 display an impaired function of CD4 + T cells, associated with diminished IFN-γ production [6]. In contrast, other studies report the over-activation of these T cells. Patients who have recovered from COVID-19 demonstrated an increased frequency of CD4 + T-cell responses against the virus spike protein [3, 6]. This has an uncanny resemblance to what has been observed during influenza virus infections. The function of cytotoxic CD8 + T cells against COVID-19 has also remained unclear. While a few studies suggested that CD8 + T cells from severe COVID-19 patients had attenuated cytokine production following in vitro stimulation, some studies strongly suggest the potential exhaustion of these T cells [7]. However, other reports seem to highlight an over-exuberant CD8 + T-cell response associated with augmented cytotoxic response in COVID-19 patients [6]. Strong CD4 + and CD8 + T-cell memory responses are reported to be induced after COVID-19; accordingly, several COVID-19 vaccines have been demonstrated to elicit CD4 + and CD8 + T-cell responses [8, 9].
Needless to say, that vaccines hold the key to unlock the deadly mayhem unleashed by COVID-19. These include RNA and DNA vaccines, adenovirus vector vaccines, inactivated virus vaccines, and subunit vaccines. While there has been debate about the efficacy of such vaccines against the variants of concern, it goes without saying that additional interventional, non-randomized studies are required. In this review, we highlight the COVID-19 variants of concern and variants of interest. Furthermore, we discuss the immune responses against such variants with a specific focus on the T-cell responses that ideally would play a pivotal role to fend off the viruses. Additionally, the mechanisms of the approved vaccines in regulating T-cell responses in hosts will be highlighted. To conclude, we will discuss the key factors that play a crucial role in the pathogenicity of SARS-CoV2. With additional waves knocking on the door of different countries, our timely review provides the much-needed insights into the role of the new VOCs in keeping the pandemic alive, and how we can combat the situation to end the pandemic.
SARS-CoV2 variants
From its first detection in the later half of 2019, a substantial amount of research has investigated the principal causative agent of the ongoing pandemic. The SARS-CoV2 viral genome has been extensively studied and has been phylogenetically mapped to the betacoronavirus genus under the coronaviridae family. It shows about 79.5% and 50% sequence similarity to the previously identified SARS-CoV (Severe Acute Respiratory Syndrome Coronavirus) and MERS-CoV (Middle Eastern Respiratory Syndrome Coronavirus), respectively [10–12]. Apart from being a single-stranded enveloped RNA virus with spherical morphology, it has shown remarkable ability to infect multiple mammalian species with humans being their principal host. The RNA-dependent RNA polymerase (RdRP) of this novel member of the SARS family has proof-reading capabilities, making it fairly stable to changes in its genome unlike some of the other RNA viruses like Influenza A [13]. However, due to antigenic drift, and the natural need for adaptation in the host, mutations beneficial for the virus can be predicted to occur over time. Agreeably, a period of genomic steadiness was observed in the virus after its initial emergence from Wuhan in 2019 [14]. From the middle of 2020, changes in the genome of the SARS-CoV2 virus have been detected that resulted in the inception of new variants with significantly altered transmissibility, disease severity, and immune escape abilities.
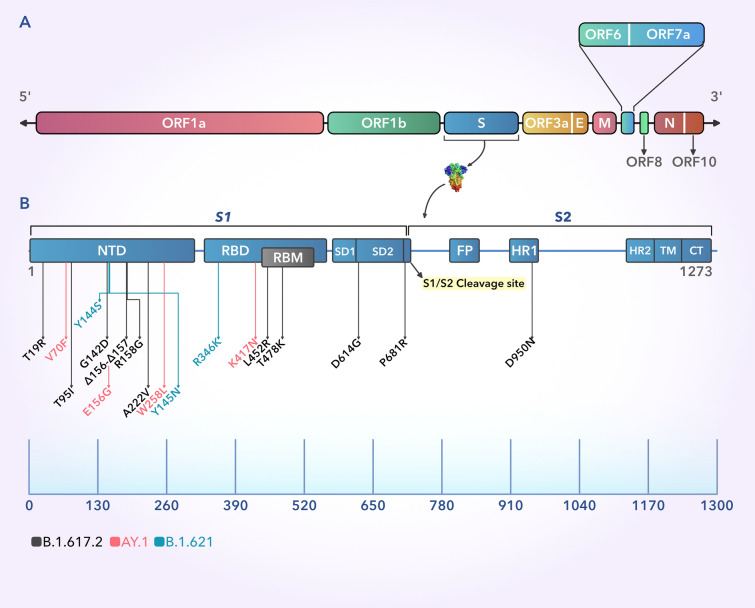
The SARS-CoV2 genome is 29.9 kbp (kilo base pair) in length and consists of several ORFs [15]. The ORFs1a/b in the 5’ part is responsible for directing the synthesis of 16 non-structural proteins (Nsp1-16), while the 3’ ORFs constitute the codons that give rise to 4 structural proteins, namely the Spike (S), Envelope (E), Matrix (M), and Nucleocapsid (N) along with 6 other accessory proteins [16]. The homotrimeric glycosylated S-protein of SARS-CoV-2 is the key for entering into host cells [17]. This bipartite, 180–200 KDa protein, primarily comprises of an exposed extracellular NTD, a transmembrane section TM, and a short C-terminal region CT [18]. The S-protein has 2 major subunits, named S1 and S2. The S1 unit is responsible for receptor recognition and binding, while the S2 unit assists in host cell membrane fusion and virus entry. Evidently, the S1 unit contains the NTD, RBD and RBM, SD1 and SD2, while the S2 unit encompasses the FP, HR1, HR2, TM, and CT regions. On binding to the host cell receptor, the trimeric S-protein undergoes substantial alteration in its structural conformation which results in splitting of the S1 and S2 subunits at their cleavage site in S1/S2 junction [18] (Fig. 2).
Fig. 2.
A Schematic representation of the 29.9 kbp-long SARS-CoV-2 genome where the different regions have been highlighted. B The 1273 amino acid long S protein of the SARS-CoV-2 showing different domains with mutations belonging to the delta, delta plus, and mu variants. The black arrows point to the major mutations across the S protein for the B.1.617.2 (Delta) variant, while the red and cyan arrows indicate the exclusive mutations present in the AY.1 (delta plus) and B.1.621 (Mu) variants, respectively. The scale shows the relative position of the amino acids in the S protein. Index: S1 and S2: S protein subunits 1 and 2, NTD: N-terminal domain, RBD: Receptor-binding domain, RBM: Receptor-binding motif, SD1 and SD2: Sub-domains 1 and 2, FP: Fusion peptide, HR1 and HR2: Heptad repeats 1 and 2, TM: Transmembrane domain, CT: Cytoplasmic domain (Color figure online)
The occurrences of mutations in the non-coding regions of 5’ and 3’ UTRs were found to be quite common. One study identified 105 and 158 nucleotide-level mutations in the 5’ and 3’ UTRs, respectively, after analyzing 2492 genome samples, while another study reported 260 and 224 mutations in the same non-coding regions in a sample size of 10,022 genomes [16, 19]. However, all of these mutations have been predicted to have little impact on the evolution of the pandemic. However, changes in the coding region of the viral genome present a different challenge as they can modify the host–pathogen interactions vastly. ORF1a/b being the largest portion of the genome has also been found to harbor the majority of the mutations and it is closely followed by the S protein in the second position [20]. In the previously mentioned study by Koyama et. al. consisting of 10,022 genomes, a total number of 1905 missense mutations in ORF1a/b could be detected along with 394 in the S protein [19]. All these data readily support the phenomenon of viral evolution and the subsequent rise of newer variants. These viral strains with impactful mutations in the genome results in enhanced viral characteristics that could pose a serious threat to our health in the upcoming waves.
Various classification systems, including Nexstrain system (https://nextstrain.org/) and PANGOLIN classification system (https://cov-lineages.org/resources/pangolin.html/), have been developed to keep track of all the new emerging variants as the pandemic has progressed. The extensive amount of information available from all these organizations along with a constant effort from scientists around the world has helped the WHO understand the impact of these variants on human health on a global scale. For ease of study and monitoring, the variants have been divided into various subtypes each consisting of several lineages, including Variants of Concern (VOCs), and Variants of Interest (VOIs). Table 1 summarizes all the important variants that have been detected to date, their defining substitutions, and their impact on viral characteristics.
Table 1.
Summarized list of all the VOCs and VOIs that have been identified to date along with their characteristic features
| Sl. No. | Lineage (PANGO) |
Assignment | Variant identity | COO | Key S-protein mutations | Effect(s) of mutation | |
|---|---|---|---|---|---|---|---|
| Nextstrain clade | WHO nomenclature | ||||||
| 1 | B.1.1.7 | 20I (V1) | Alpha | VOC | UK | N501Y, P681H, ΔH69/ΔV70, ΔY144 | Total 8 mutations are present in S protein apart from D614G. ΔY144 in the NTD restricts neutralization by NTD supersite directed antibodies. N501Y is predicted to confer superior binding to ACE2 [114] |
| 2 | B.1.351 | 20H (V2) | Beta | VOC | RSA | L18F, K417N, E484K, N501Y, A701V, R246I, D215G | E484K reduces monoclonal antibody binding to the RBD of S protein. R246I in the NTD results in reduced effectiveness of the NTD supersite directed antibodies as seen in the Alpha variant [114] |
| 3 | P.1 | 20 J (V3) | Gamma | VOC | BRA | L18F, R190S, D138Y, K417T, E484K, N501Y, H655Y | Similar to the previous variant, E484K and K417T facilitate immune escape and N501Y enhances attachment to ACE2 [114]–[116] |
| 4 | B.1.617.1 | 21B | Kappa | VOI | IND | P681R, L452R, E484Q, Q1071H | Although these mutations may result in a reduction of viral entry into the host, they primarily assist the virus to evade vaccine-elicited antibody targeting. P681R enhances syncitium formation and efficient cleavage for faster transmission and aggravates disease pathogenesis [117, 118] |
| 5 | B.1.617.2 | 21A | Delta | VOC | IND | L452R, T478K, D950N, P681R, T19R, G142D, E156G, T95I | L452R and T478K facilitate antibody escape against the RBD. T19R maps to the NTD supersite, further decreasing antibody interaction in the NTD [2]. The P681R mutation is also present in this variant, functioning similarly as mentioned before |
| 6 | B.1.617.3 | - | IND | L452R, D950N, P681R, T19R, E156G, E484Q | Similar to the above-mentioned variants | ||
| 7 |
AY.1 /B.1.617.2.1 |
Delta Plus | VOI | IND | K417N, V70F, W258L, E156G, including mutations similar to the Delta variant | K417N reduces antibody binding to the RBD of S protein. Mutations which can also be found in the Delta variant are present in this variant at the higher prevalence and act similarly [22] | |
| 8 | B.1.525 | 21D/20A | Eta | VOI | MUL | Q52R, A67V, E484K, Q677H, F888L | Q677H and E484K result in reduced neutralization by various antibody treatments [119, 120] |
| 9 | B.1.526 | 21F/20C | Iota | VOI | USA | L5F, T95I, D253G, E484K | Mutations in common with the Beta, Gamma and Delta variants perform similarly |
| 10 | C.37 | 21G/20D | Lambda | VOI | PER | G75V, T76I, L452Q, F490S, T859N | L452Q and F490S are on the RBD of the S protein. The former is exclusive to C.37 and is responsible for enhancing the affinity for the ACE2 receptor, while the latter is responsible for decreased neutralization by antibodies [121, 122]). T76I is also involved in increasing viral infectivity [123] |
| 11 | B.1.621 | 21H | Mu | VOI | COL | T95I, Y144S, Y145N, R346K, E484K, N501Y, P681H | Mutations that are also found in the previously mentioned Alpha, Beta, Gamma and Delta variants show similar effects as described previously |
| 12 | B.1.427 and B.1.429 | 21C | Epsilon | VOI (currently De-escalated) | USA | S13I, W152C, L452R | L452R mutation decreases the activity of multiple RBD specific monoclonal antibodies, while W152C and L452R abrogate the action of the NTD supersite directed antibodies [124] |
| 13 | B.1.1.519 | 20B/S:732A | - | - | MUL | T478K, P681H, T732A | T478K and P681H mutations present in the Delta, Alpha and Gamma variants show similar effects as mentioned before [125] |
| 14 | B.1.619 | 20A/S:126A | - | - | MUL | I210T, N440K, E484K, D936N, S939F, T1027I | Further studies are needed |
| 15 | P.3 | 20B/S:265C | Theta | Currently De-escalated | PHL | E484K, N501Y, P681H, E1092K, H1101Y, V1176F | One study reported this variant had a high potential to spread among the population owing to its increased immune evasion abilities [126] |
R.1, B.1.466.2, B.1.1.318, B.1.1.319, C.36.3, B.1.214.2, B.1.1.523, B.1.620, C.1.2, B.1.618- are some of the important variants that have also been detected. Some of these variants may not be prevalent anymore; however, further investigations are required to shed some light on their roles and characteristics. All the data have been curated from Outbreak.info, Co-Variants.org, Nexstrain.org, Cov-lineages.org, https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/, and the above-mentioned references
INDEX: COO country of origin, MUL multiple countries, PER Peru, PHL Philippines, COL Colombia, BRA Brazil, RSA Republic of South Africa, IND India, USA United States of America, UK United Kingdom
Delta and delta plus variants
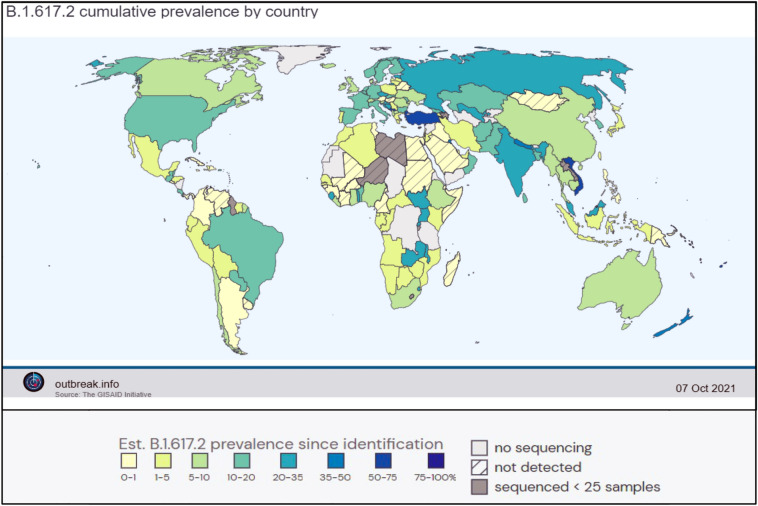
The first traces of the B.1.617 family were detected in India in October of 2020. In the second wave of the pandemic, the B.1.617 lineage and B.1.618.1 (Bengal variant) were primarily responsible for the disastrous situation in India which resulted in over 400,000 new infections per day during its peak in May 2021 [21]. Now, as the signs of a third wave are being observed in some countries, the delta variant or B.1.617.2 from the same lineage is shaping up to be an even deadlier enemy. Currently, the delta variant is watchlisted as a VOC by the WHO and has been detected in 149 countries according to cov-lineages.org (https://cov-lineages.org/global_report_B.1.617.2.html), while outbreak.info shows that at least 160 countries have been affected (https://outbreak.info/situation-reports?pango=B.1.617.2), making it the predominant strain across the globe (Fig. 1). The B.1.617 lineage consists of three sub-lineages, B.1.617.1 (Kappa), B.1.617.2 (Delta), and B.1.617.3. This parental B.1.617 family has originated from the B.1 lineage with the original D614G mutation in the S protein [2]. Reports have identified multiple non-uniform substitutions prevalent in one sub-lineage of the B.1.617 than the others, indicating the influence of selection pressure and the need for adaptation in the host [2]. Hence, it can be concurred that majority of these mutations are non-synonymous and have been found modify viral attributes (Table 1). The B.1.617.2 or delta sub-lineage contains the exclusive T478K (in the receptor-binding motif of the RBD) and G142D (in the NTD) of the S protein, while the E154K mutation is only observed in the B.1.617.1 subvariant [2]. Another study has observed a high frequency of T95I mutation in the delta variant adding to the list of changes (Fig. 2) [22]. These unique mutations make the delta variant more resistant to neutralization by monoclonal antibodies and also facilitate immune escape.
Fig. 1.
Worldwide incidence of the B.1.617.2 (Delta) variant as of 7th October 2021 courtesy of Outbreak.info. It has become the most widespread strain of SARS-CoV 2 after its initial detection in India in 2020 (Color figure online)
The delta plus or AY.1 or B.1.617.2.1 variant is further characterized by the addition of K417N change in the parental delta variant which causes the loss of proper binding of neutralizing antibodies with the S protein and thus making it even more prone to immune evasion [22]. Kannan et. al. reported 269 unique mutations in the delta plus variant including V70F and W258L (Fig. 2) [22]. Unique mutations in other non-structural proteins (A328T in Nsp3) were also found to be present in the delta plus variant, indicating that AY.1 is not a subvariant of the delta strain, but a distinct evolutionary jump consisting of unique mutational features that demand attention [22]. Additionally, those mutations already found in the S protein of the delta variant (like T95I and G142D) were present at a greater prevalence in the delta plus variant (37% and 69% more, respectively) [22]. Apart from India, this variant has been detected in the United States, United Kingdom, and Japan (cov-lineage.org), and as time progresses, it is predicted to spread across other countries owing to its more enhanced ability to escape the immune system and lesser neutralization by various antibodies courtesy of its distinctive mutations. On August 30, 2021, the mu variant (B.1.621) from Colombia has been added as a VOI by the WHO. It is currently being tracked as it has the potential to become the next major strain. One study reported that B.1.621 S protein is the most resistant version of all previously reported variants to be neutralized by convalescent sera and vaccines (BNT162b2) [23]. Thus, it is of paramount importance to continue our research and understanding of these rapidly evolving variants to face the oncoming days of the pandemic with little consequence.
Immune response and evasion strategies of SARS-CoV2
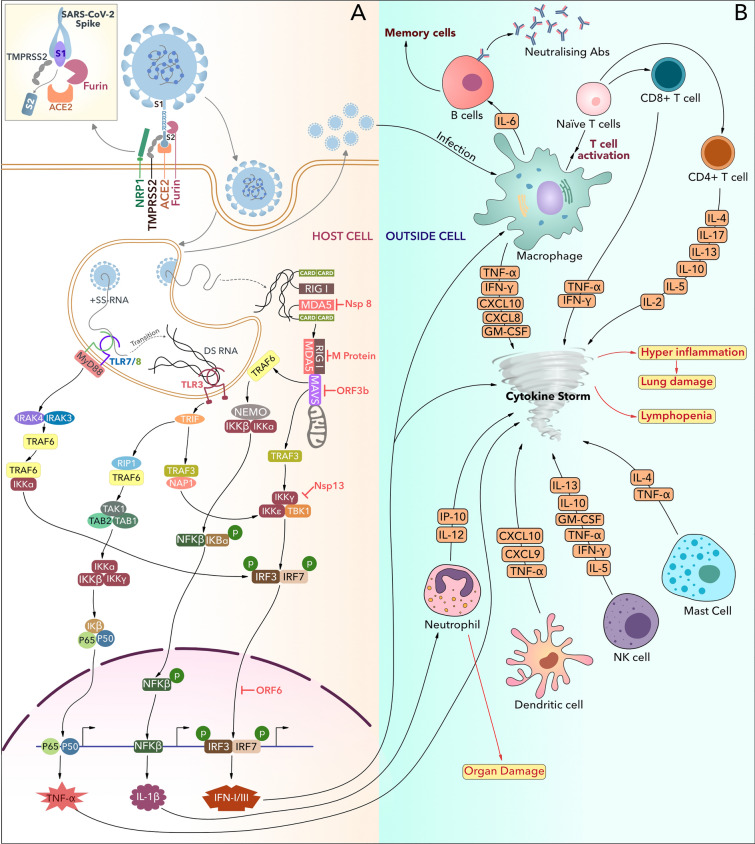
Distinct immune evasion strategies and dysregulated cellular responses are the prime contributors to SARS-CoV2-mediated pathogenesis. Being airborne, this virus enters the host body through respiratory tract and eventually entire respiratory system [trachea, bronchi, bronchioles, and lungs] get infected. The viral S protein targets the angiotensin-converting enzyme 2 [ACE2] receptor present on the surface of various host cells, including epithelial cells, enterocytes, and goblet cells [24]. This tropism provokes the virus to attack other organs such as kidney, brain, eyes, and gastrointestinal tract [25]. Recent studies suggest that SARS-CoV2 could also access the host cell by interacting with CD147, Neuropilin1 [NRP1], and Dipeptidyl-peptidase-4 [DPP-4] through S protein, which makes the lymphocytes also susceptible to the SARS-CoV2 virus [26–28]. Upon infection, the viral recognition is mediated by tissue-resident innate cells, which later activate other immune components to remove the pathogen (Fig. 3). However, SARS-CoV2 has evolved intricate evasion strategies to dodge the surveillance of the innate immune system and replicate inside the host [29]. To avoid identification by intracellular receptor Mda5 and IFIT family of proteins, SARS-CoV-2 Nsp16 modifies the 5’ capping of viral mRNA by 2’O methylation [30, 31]. Further research has concluded that SARS-CoV-2 not only avoids detection inside the host but also the Nsp1 and Nsp14 shuts down antiviral protein synthesis at the level of translation [32–34]. SARS-CoV-2-specific Nsp8 and Nsp9 seize Sec61 mediated trafficking pathway of the host cell to enter the ER and halts the host cell protein fabrication machinery [35]. NK cells are known to play a pivotal role in fine-tuning the interaction between innate and adaptive immune systems. Therefore, SARS-CoV2 suppresses the production of IFN-I to inhibit this interaction [36]. To suppress the IFN-I and II synthesis, SARS-CoV-2 additionally uses M, ORF3b, ORF6, and Nsp13. The virus-specific M protein suppresses immune component PRR and RIG-I from forming protein complexes with Mda5 and MAVs and hampers the production of IFN I [37]. Virus-specific ORF6 blocks the nuclear translocation of STAT1 to suppress IFN I production [38], while ORF3b hinders the nuclear translocation of IRF3 to block IFN I-mediated signaling [39]. Several reports have proclaimed that Nsp13 along with Nsp8 also plays an inhibitory role in the host innate immunity, specifically in MDA5 derived IFN I signaling pathway [40, 41]. As a unique evasion mechanism, ORF8 protein of SARS-CoV-2 has shown the ability to re-route the developing MHC-I molecule from ER to autophagosome where they get degraded and effect adaptive immunity [42]. COVID-19-mediated infection is also characterized by cytokine storm, leading to an enhanced production of pro-inflammatory cytokines. After the virus enters the host, various immune cells, such as T cells, B cells, DCs, neutrophils, NK cells, macrophages, start to release pro-inflammatory cytokines to fight against the offending pathogen. In contrast to healthy individuals, COVID-19 patients display elevated levels of the cytokines IL-1β, IL-2, IL-6, IL-7, IL-10, TNF-α, and IFN-γ (Table 2) [43]. In the coming section, we will highlight the dynamism of adaptive immune responses against COVID-19, especially the T cells.
Fig. 3.
A The initial target of SARS CoV2 spike protein is the ACE2 receptor, present on the cell surface. After binding to ACE2, Furin cleaves the S protein at R–R–R–R stretch of S1/S2 junction and TMPRSS2 at S2 subunit. This protease cleavage helps the virus to enter the host cell via the endocytic pathway. Subsequently, the viral RNA is recognized by cytosolic RNA sensors such as TLR 7/8, TLR 3, and RIG-I/MDA5 leading to the activation of downstream kinases of the NFκβ signaling pathway. This pathway elicits the production of various pro-inflammatory cytokines including TNFα, IL1β, and IFN-I/III, but gets suppressed by SARS CoV2 components like Nsp8, M protein, ORF3b, and Nsp13. B All the pro-inflammatory cytokines depicted in Fig. 2A induce the activation and cytokine production from other innate immune cells. This uncontrolled cytokine production initiates cytokine storm followed by hyperinflammation and organ damage. Increased cytokine levels also trigger activation and proliferation of adaptive immune cells which further contributes in cytokine storm (Color figure online)
Table 2.
Cytokine profile of COVID-19
| Cytokines | Disease onset | Source | Role in COVID-19 | Refs | |
|---|---|---|---|---|---|
| Week 1–2 | Week 3–4 | ||||
| IL-1β | √ | Macrophages, endothelial cells, activated monocytes | Induces activation of pro-inflammatory cytokines IL-6 and TNF-α; recruits neutrophil and T cells | [127, 128] | |
| IL-2 | √ | Th1 cells | Triggers differentiation and survival of Treg cells; takes part in cytokine storm | [129] | |
| IL-4 | √ | Mast cells, Th2 cells | Activates macrophages and increases the secretion of TGF-β | [130] | |
| IL-5 | √ | NK cells, mast cells, airway smooth muscle cells, epithelial cells, Th2 cells | Eosinophil expansion and activation in severe patients; triggers cytokine storm | [131] | |
| IL-6 | √ | Fibroblasts, macrophages, Th2 cells | Accelerates the lung inflammation, fever, and fibrosis; cytokine storm inducer; induces B-cell differentiation | [132] | |
| IL-7 | √ | Epithelial cells, dendritic cells | Initiates rapid elevation of lymphocyte count by modulating the effect of IL-6, IL-10, IFN-γ, TNF-α | [133] | |
| IL-8 | √ | Endothelial cells, epithelial cells, macrophages, fibroblasts | Promotes neutrophil chemotaxis and angiogenesis | [134] | |
| IL-10 | √ | Endothelial cells, macrophages, fibroblasts, Th2 cells | Increased expression with the disease severity which leads to immune suppression | [135] | |
| IL-12 | √ | Dendritic cells, macrophages | Regulates maintenance of NK cell numbers; inducer of Th1 cell differentiation | [136] | |
| IL-13 | √ | Mast cells, granulocytes, Th2 cells, NK-T cells | Accelerates cytokine storm via macrophage activation | [130] | |
| IL-17 | √ | Th17 cells | Downregulates Treg cell response; induces pulmonary inflammation | [137] | |
| IL-21 | √ | Tfh, Th2, NK-T cells | Triggers IFN-γ production by dendritic and NK cells; inhibits differentiation to Th1 cell | [138] | |
| IL-23 | √ | Macrophages, dendritic cells | Promotes cytokine storm via proliferation of Th17 cells | [139] | |
| IL-33 | √ | Macrophages, dendritic cells, pulmonary epithelial cells | Suppressing IFN-γ production by NK cells leads to impaired NK cell responses; inhibits innate antiviral immune response by suppressing TLR7 | [140] | |
| IFN-γ | √ | Endothelial cells, macrophages | Downregulates viral replication; triggers cytokine production from T cell; prevents Th2 response; can induce cytokine storm in high levels which leads to organ damage | [141] | |
| TNF-α | √ | Endothelial cells, macrophages | Promotes organ damage by accelerating cell death | [142] | |
Antibody responses in COVID-19
During the first wave of SARS-CoV-2, little data were available regarding the antibody response against the virus. In infected individuals, it is important to understand the nature of the neutralizing antibody response against SARS-CoV-2 for evaluating the efficacy of the developed vaccines. The presence of neutralizing antibodies against SARS-CoV-2 can act as an indicator for protection against reinfection for previously infected individuals. The antibodies that target the S1 domain of the SARS-CoV-2 spike protein, including the RBD, severely impair virus cell entry. In a study from Italy, antibody response to distinct antigens of SARS-CoV-2 was elucidated in patients who were admitted to hospital during the first wave in March and April 2020. The study evaluated neutralizing antibodies, IgG against NP and IgG, and IgM and IgA against S1 [44]. There was increased S1 IgM seroconversion rate for patients who succumbed compared to patients who survived. However, no difference in S1 antibody titers was observed in patients who survived compared to patients who did not survive. The peak antibody response was observed between days 6 and day 18–20, which was different from other studies [44]. This difference could be attributed to time recruitment of such patients to the hospital and the severity of the disease. Another study from Germany that recruited patients in April and May 2020, observed a very small fraction of the patients’ serum samples, could be classified as high neutralizers [45]. In contrast, some symptomatic patients failed to develop antibodies against SARS-CoV-2, suggesting the varied antibody response against the virus.
The neutralization capacity of polyclonal antibodies in serum has a positive correlation with IgG against spike protein or against RBD. A detailed systematic review of 150 studies observed an early detection of virus-specific IgM over IgG with a peak in weeks 2 and 5 and a decline in the subsequent 3–5-week time point after symptoms [46]. The peak of IgG could last for at least 8 weeks. The neutralizing antibodies can be detectable within 7–15 days of disease onset, increase until days 14–22, before decreasing [46]. During convalescence, the decrease in serum antibody level could indicate a downtick of the immune response as the presence of virus-specific B cells in the bone marrow could be detected [47]. One study has found that high level of IgG antibodies could be detected 11 months post-symptom onset [48]. The same study observed that the development of neutralizing antibody is associated with the activation of T cells.
One study noted that across 4 variants of SARS-CoV-2 (A.1, B.1.1.7, B.1, N501Y), infection and vaccine-mediated immunity could be retained against the B.1.1.7 variant [49]. However, the study had several limitations including small sample size and lack of definitive clinical outcome. The epitope-based (from the S and N proteins) antibody responses could differentiate the outcome of COVID-19 against different SARS-CoV-2 variants. The mutations including S235F (in the N-protein) and P681H (in the S-Protein) associated with B.1.1.7 could alter the specificity of the corresponding epitopes [50]. The efficacy of various vaccines in inducing prime-boost immunity has been evaluated against different SARS-CoV-2 variants. The BioNTech/Pfizer’s BNT162b2 vaccine demonstrated relatively high titers of neutralizing antibodies against the B.1.351, B.1.1.7 and P.1 variants [51]. While the Pfizer and the AstraZeneca vaccines were found to be efficient against the Delta variant, compared to the Alpha variant, the neutralizing ability of the vaccines was about three-to-fivefold less potent [2]. Importantly, serum samples from single-dose recipient of either of the two above-mentioned vaccines failed to elicit a proper inhibitory effect against the Delta variant. Such studies are pointers toward understanding the importance of the impact of different variants of the virus in regulating the antibody response.
T cells regulate COVID-19 disease progression
Similar to innate immune and humoral immune cells, the cell-mediated immune system plays as a prime regulator in viral disease pathophysiology. The cytotoxic CD8 + T cells function by attacking the virus-infected cells to reduce the viral load. SARS-CoV2-specific S, M, N, and ORF3a are the highly prevalent antigenic regions that trigger the activation of CD8 + T cells [52]. Cytotoxic CD8 + T cell usually takes few days to become activated after the onset of disease. However, COVID-19-specific CD8 + T-cell responses get further delayed due to the prevalent inflammatory environment and viral load [53]. Besides the killing ability of infected cells, virus-specific memory CD8 + T cells formation is also reported in the COVID-19 patients. In a study with patient recovered from COVID-19, functional SARS-CoV2-specific memory CD8 + T cells was detected even after 6 months following infection, revealing the impact of T cells on the clinical outcome of the disease [54]. Another cohort study with a hospitalized patient group following their recovery reported the percentage of circulating SARS-CoV2 memory CD8 + T cells to be 70% and 50%, respectively, at 1 month post-symptom onset [PSO] and 6 month PSO [55].
Virus-specific CD4 + T cells differentiate into heterogeneous populations with their diverse effects on disease pathogenesis. To eliminate the invading pathogens, Th1 cells induce inflammatory responses, while Th2 cells trigger the activation of B cells. In COVID-19, the increased response of CD4 + T cells relative to CD8 + T cells has been reported during primary infection [52, 56, 57]. SARS-CoV2-specific S, M, and N proteins trigger the activation of CD4 + T cells, although substantial responses against ORF3a and Nsp3 have also been detected [52]. Recent studies have reported the presence of SARS-CoV2-specific CD4 + T cell as early as 2–4 days PSO [58]. During COVID-19, antiviral Th1 cell induces cytokine storm by producing IFN-γ, while Th2 secretes IL-4 and IL-10 [59, 60]. Although a higher level of IL-10 is a potent inhibitor of Th1 cells and pro-inflammatory cytokines, a report has suggested that Th1 suppression is not beneficial for COVID-19 disease prognosis [28]. In addition, the increased concentrations of IL-4 and IL-10 induce the accumulation of a high proportion of basophils and degranulated eosinophils in the lungs, leading to cytokine storm and organ damage [60–62]. Overactive Th2 cells also trigger acute respiratory distress syndrome [ARDS] by increasing the level of IL-6 and IL-15 [28]. IL-6 is a typical innate immune cell-induced cytokine; however, but the abnormality of COVID-19 triggers the secretion of this pro-inflammatory cytokine by Th2 cells [28]. IL-21, produced by Th2 cells, significantly helps in Th17 cell differentiation. With increased disease severity, enhanced expression of the chemokine receptor CCR6 + on Th17 cells explains the increased number of these cells on mucosal tissue and the subsequent effect in COVID-19 disease pathophysiology [52, 63]. Th17 cells trigger aggressive inflammatory response during COVID-19 by producing the pro-inflammatory cytokine IL-17, aiding in the cytokine storm [64]. In severe COVID-19 cases, Th17 cells induce acute lung injury [ALI] and ARDS by promoting neutrophil migration to the lungs via suppressing Treg cell response [60–62].
The function of cytokines is not limited to regulate immune cell activation; eventually, they also impact the function of humoral immune cells. Although increased levels of SARS-CoV2-specific cytokines indicate disease severity, disease-specific Tfh cell-secreted IL-21 induces neutralizing antibody titers to help in memory B-cell formation for long-term use humoral immunity. SARS-CoV2-specific circulating Tfh [cTfh] cells are formed during disease severity [57, 65]. Despite this, no association has been found between cTfh cell frequency and antibody titer [55, 57]. At the later stage of the disease, a high level of SARS-CoV2-specific memory cTfh responses has been reported, which may be result from prolonged antigen exposure [66]. One study has found memory cTfh responses up to 6 months PSO in convalescent patients [55]. Analogous to SARS-CoV2-specific CD8 + memory T cells, CD4 + memory T cells were found against virus-specific S, M, N, ORF3a, and Nsp3 proteins [52]. SARS-CoV2 memory CD4 + T cells show quite robust response with a detection rate of 93% after 1 month and 92% after 6 months, respectively [55]. Other cohort studies have reported the presence of SARS-CoV2 [B.1.1.7] N-specific memory CD4 + T cells in unexposed individuals as the consequence of previous exposure to the common cold virus, circulating widely in humans [67]. Apart from N-reactive memory CD4 + T cells, S-reactive CD4 + T cells have also been found in healthy donors, which indicate the possibility of S sequence similarity of SARS-CoV2 with other HCoVs [63]. Thus, the above-mentioned evidence supports the role of immunological memory of T cells in COVID-19 patients. The defining response of distinct classes of T cells can reveal novel opportunities for treatment and disease prevention.
Impact of delta variant in immune system
The highly transmissible new B.1.617.2 variant with escalated replication kinetics evades the immune system with an enhanced mutation at the RBD and NTD site of SARS-CoV2 S protein [68, 69]. The T19R substitution mutation, present on the NTD of S protein, led to the B.1.617.2 and B.1.617.2.1 variants [70]. This mutation leads to the reduced binding affinity of the NTD neutralizing antibodies to the NTD of S protein [70]. Another highly prevalent mutation of the delta and delta plus variant is G142D, which forms a structural alteration at its side chain that clashes with its neighbor R158 side chain. To avoid this collision, R158 undergoes a structural change, which may be one of the reasons for the emergence of new variants [22]. Δ156–157 mutations help the variant escape immune recognition more efficiently due to changes in the configuration of the N-terminal domain. The flexibility of S-specific NTD of these new variants was decreased for the deletion mutation at the Arg158, and Phe157 with substitution mutation at E156G compared to the B.1.351 variant, which may affect the transmissibility [71]. A substitution mutation at P681R, situated at the furin cleavage site of S1/S2 junction, significantly increases the replication kinetics of the delta variant via higher fusogenic activity [72, 73]. The D950N mutation, which resides in the trimer interface, may regulate the spike protein dynamics by influencing the refolding of S2, which increases the efficiency of membrane fusion [74, 75]. The D614G affects viral protein glycosylation and the cleavage of S protein by host serine protease to increase membrane fusion, which can escalate the infectivity of this strain [76]. Molecular dynamic simulation of the RBD has reported that both L452R and T478K mutations exert reduced binding ability to the host ACE2 and antibody binding of B.1.617.2 variant compared to B.1.351 and B.1.1.7 variants. Consequently, the infectivity and transmissibility of the B.1.617.2 variant has enhanced [77].
Vaccines against COVID-19
The emergence and escalation of the current pandemic of SARS-CoV2 has posed an overwhelming burden to our current vaccine technology. Traditional live-attenuated or inactivated pathogen-based vaccines have worked wonders for many pathogens such as hepatitis B, polio, human papilloma virus, and measles [78]. Even though the current vaccine technology and production strategies have proved successful, the exigent nature of the SARS-CoV2 pandemic requires a rapid and mass producibility of vaccines. Nucleic acid (DNA and mRNA)-based vaccines, recombinant protein subunit-based vaccines, and viral vector-based vaccines such as recombinant adenoviruses (rAds) have been a major focus for COVID-19 vaccine development as they can be quickly advanced to phase III clinical trials compared to the traditional vaccines. Nucleic acid-based vaccines with their unique advantage of ease in design and synthesis as well as simplicity in packaging of either DNA or mRNA in lipid nanoparticle capsules have become the most sought out vaccine platform [79, 80]. Especially, the immunostimulatory effect of RNA upon recognition by pattern recognition receptors (PRRs) and robust antigenic protein production is more desired as vaccine candidate over DNA vaccines [80]. During the 1990s, when mRNA vaccines were first introduced, it did not garner much interest due to its inherent instability, unavailability of efficient in vivo delivery system, and high innate immunogenicity [81]. However, mRNA vaccine has become major focus for fight against SARS-CoV2 pandemic because of several advantages such as (i) mRNA is non-infectious, thus possessing no risk of insertional mutagenesis, (ii) anti-vector immunity can be avoided using mRNA vectors as it is a minimal genetic vector, and (iii) it can be produced rapidly and administered repeatedly due to its inexpensive and scalable manufacturing methods [80]. Protein subunit vaccines are known to have strong safety as well as immunogenicity. These vaccines can be made relatively at low cost in large quantities, and can be administered in large heterozygous population regardless of their genetic makeup. The recombinant viral vector vaccines stimulate the host cells to produce pathogenic antigens endogenously and simulate a partial infectious setting inside the host. This results in a greater immunogenicity and greater efficacy. For example, recombinant adenovirus rAd26 and rAd5 have been used in clinical practice and are known to induce both cellular and humoral immunity after single dose of immunization [82]. A total of 22 vaccines against SARS-CoV2 have been approved for use in 192 countries of which 8 are inactivated pathogen-based vaccines and 6 are non-replicating viral vector-based vaccines (https://covid19.trackvaccines.org/). Albeit the ‘doubt’ regarding the effectiveness of current vaccines, there are more than 400 different vaccines which are under clinical trials (https://covid19.trackvaccines.org/). Recent landscape of novel COVID-19 candidate vaccines development by WHO (https://www.who.int/publications/m/item/draft-landscape-of-COVID-19-candidate-vaccines) shows 114 candidates in clinical assessment (including 8 at phase IV) and 185 vaccines under pre-clinical development. The phase IV vaccines consist of a variety of vaccine platforms: vector vaccines (ChAdOx1-S, Ad26.COV2.S), inactivated viral vaccines (CoronaVac, BBIBP-CorV), and mRNA vaccines (mRNA-1273, BNT162b2, mRNA-1273.351).
Efficacy of current vaccines against the delta variant
As a consequence of new mutations, the delta variant decreases the neutralization antibody titers by four-to-sixfold, attenuating the effect of monoclonal antibodies like Bamlanivimab [2]. This new SARS-CoV2 variant also becomes less sensitive to TMPRSS2 inhibitor, Camostat [83]. Due to the alteration of the ACE2 binding site, all the RBD targeting vaccines elicit a threefold lower neutralization response [2]. Another cohort study has been conducted to evaluate the effect of vaccine-elicited neutralizing serum between ancestral B1 variant and B.1.1.7, Q.1-Q.8 variant G614S. This study showed that Pfizer/BioNTech BNT162b2-elicited plasma level was reduced to threefold even after two doses. Moderna mRNA-1273-elicited plasma was reduced twofold after two doses, while Janssen-induced plasma neutralization was reduced fourfold after a single dose. The recent study with the Pfizer vaccine and the AstraZeneca vaccine demonstrated that the level of neutralizing antibody titers was undetectable against D614G in the delta variant after a single dose [2]. Notably, the level of vaccine-induced neutralizing antibody increased after the second dose, which concludes that the single dose of those vaccines is not much reliable against the new strain. Another 12-month cohort study of 21 individuals [9 received AstraZeneca vaccine, 9 Pfizer vaccine, and 3 Moderna vaccine] has displayed increased neutralizing antibody titers against the delta variant compared to the normal convalescent individuals who have not received any vaccine [2]. Another study with collected sera of 12 infected individuals of the first wave has expressed 5.7-fold less sensitive to delta variant [68]. The working vaccines may not be fully effective against the delta variant, and vaccinated individuals will have a higher level of neutralizing antibodies in their blood compared to non-vaccinated individuals. However, the reduced effectiveness of the working vaccine demands further research for a more viable vaccine target.
It is of paramount importance to assess the effectiveness of the currently approved as well as under trials vaccines against new and emerging variants of concerns. It is also required to monitor the preventive duration of the current vaccines against the new delta variant. A recent report has compared the efficacies of ChAdOx1 nCOV-19 (AZD1222) and BNT162b2 vaccines in alpha and delta variants [84]. The first dose of AZD1222 was shown to be less effective in the delta variant (30.7%; 95% confidence interval [CI] 25.2–35.7) as compared to the alpha variant (48.7%; 95% CI 45.5–51.7). Interestingly, the BNT162b2 vaccine was more effective compared to AZD1222 in second doses, but similarly 88.0% effective against the delta variant compared to 93.7% effectiveness of AZD1222 [84]. Recently, the mRNA-1273 vaccine was shown to be 94.1% effective against the alpha variant; however, this study failed to provide any indication regarding its effectiveness against the delta variant [85]. Another cohort study compared and reported the efficacies of mRNA-1273 and BNT162b2 vaccines against the alpha and delta variant in the Mayo clinic health system over a time period from January to July 2021 [86]. Both mRNA-1273 (91.6%, 95% CI 81–97%) and BNT162b2 (85%, 95% CI 73–93%) vaccines were highly effective against SARS-CoV2 infection; however, the effectiveness against infection in the month of July reduced mRNA-1273 to 76%, 95% CI 58–87% and BNT162b2 to 42%, 95% CI 13–62%. Interestingly, the prevalence of the delta variant increased over 70%, while the prevalence of the alpha variant decreased from 85 to 13% in the month of July [86]. One study measured the effectiveness of vaccines against the delta variant using data from large-scale outbreak of COVID-19 in China. They reported that patients with two doses of inactivated vaccines (fully vaccinated) had an 88% reduced risk to develop severe stage symptoms, whereas, this protective effect was missing from the individuals with just one dose of vaccine [87]. Moreover, the effectiveness of vaccines BNT162b2 and ChAdOx1 against B.1.617.2 was notably lower in symptomatic disease cases in UK. Two doses of BNT162b2 vaccine had reduced effectiveness from 93.4% with B.1.1.7 to 87.9% with B.1.617.2 [84]. Similarly, two doses of ChAdOx1 vaccine had reduced effectiveness from 86.8% with B.1.1.7 to 80.6% with B.1.617.2, albeit the reduction was non-significant [84]. Similar findings were reported using the real-world data collected from population in Ontario, Canada [88]. The effectiveness against the delta variant compared to the alpha was notably lower for partial vaccinations with mRNA-1273 (72 vs 83%) and BNt162b2 (56 vs 66%), but with ChAdOx1, it was similar to the alpha (67 vs 64%) [88]. BBV152 vaccine has been proved quite effective against B.1.1.7 variant with the seroconversion rates of neutralizing antibodies being 98.6% in plaque-reduction neutralizing test (PRNT50)-based assay [89]. However, its effectiveness against the new delta variant is observed to be 65.2% in another phase 3 clinical trial study [90]. The recombinant adenovirus rAd26- and rAd5-based vaccine Gam-COVID-vac has been approved in more than 70 countries while being under clinical trial. The safety and efficacy of the Gam-COVID-vac is still under debate, but the interim analysis of phase III trials showed 91.6% efficacy against COVID-19 and was well tolerated in a large cohort [91]. However, further study is needed to assess its efficacy against particular variants of concerns specifically recently emerging B.1.617.2. These reports indicate that the phase IV vaccines such as ChAdOx1 nCOV-19, BNT162b2, and mRNA-1273 have lower efficacy against the delta variants; however, fully vaccinated individual seems to have more protective effect against severe illness in spite of increase in the delta variant infections. The efficacies of various vaccines in individuals either partially or completely vaccinated are summarized in Table 3. Albeit the availability of multiple vaccines as well as vaccine platforms, we need to keep in mind few critical features that make a vaccine effective, successful and desirable against pathogens. First, safety: the vaccine has to be safe even in immunocompromised individuals. Second, efficacy: the vaccine has to be highly efficient and induce optimal ‘sterilizing’ immunity. Vaccines should prevent the subsequent challenge from the said pathogen, which can be achieved by formation of memory immune cells. Third, the economics: the cost of production and administration of vaccines must be kept to minimal and at a level where a mass can be effectively vaccinated.
Table 3.
Efficacies of phase 4 vaccines against B.1.1.7 (Alpha) and B.1.617.2 (delta) variant reported in various demographic regions
| Sl. No. | Vaccines | Country/region | Dose | Vaccine efficacy B.1.17 (%) | Vaccine efficacy B.1.617.2 (%) | References |
|---|---|---|---|---|---|---|
| 1 | ChAdOx1 | Canada | 1 | 64 | 67 | [88] |
| 2 | BNT162b2 | 1 | 66 | 56 | ||
| BNT162b2 | 2 | 89 | 87 | |||
| 3 | mRNA-1273 | 1 | 83 | 72 | ||
| mRNA-1273 | 2 | 92 | / | |||
| 4 | ChAdOx1 | UK | 1 | 51.1 | 33.5 | [84] |
| 5 | BNT162b2 | 1 | 51.1 | 33.5 | ||
| 6 | ChAdOx1 | 2 | 86.8 | 80.6 | ||
| 7 | BNT162b2 | 2 | 93.4 | 87.9 | ||
| 8 | ChAdOx1 | Scotland | 1 | 37 | 18 | [143] |
| 9 | BNT162b2 | 1 | 38 | 30 | ||
| 10 | ChAdOx1 | 2 | 73 | 60 | ||
| 11 | BNT162b2 | 2 | 92 | 79 | ||
| 12 | BBV152 | India | 2 | 98.6 | 65.2 | [89, 90] |
INDEX:
BNT162b2: Pfizer–BioNTech COVID-19 vaccine
ChAdOx1: Oxford–AstraZeneca COVID-19 vaccine
mRNA-1273: Moderna COVID-19 vaccine
BBV152: Covaxin
Discussion
The SARS-CoV2 is evolving rapidly with time. Presently, new variants are being monitored and reported regularly by the scientists. Despite having a fairly stable genome, SARS-CoV2 has demonstrated a rapid rate of mutations, which led to the emergence of many new variants. Some of these variants have displayed altered receptor binding, increased infectivity, or reduced neutralization by previously generated antibodies (by vaccination or infection) compared to their predecessors. These newly acquired properties might have made the new variants such as B.1.617.2 to be more infective, lethal, and immune to vaccination. B.1.617.2 was first reported in India back in late 2020; it outcompeted pre-existing variants such as kappa (B.1.617.1) and alpha (B.1.1.7) variants, via its spread all over the globe. Two key mutations (L452R and E484Q) in the receptor-binding domain have helped the delta variant evade neutralizing antibodies, and reduced effectiveness of vaccination [92]. In vitro, B.1.617.2 is sixfold less sensitive to the acquired serum neutralizing antibodies, and eightfold less sensitive to vaccine-generated (ChAdOx-1 or BNT162b2) antibodies compared to the original variant [93]. Further research suggests that the effectiveness after one dose of vaccination (both BNT162b2 and ChAdOx1 nCoV-19) is significantly lower in patients with delta variant compared to patients with alpha variant (B.1.1.7). Even after complete vaccination (both BNT162b2 and ChAdOx1 nCoV-19), the effectiveness remains lower in the delta variant [84]. The continuous evolution of the spike protein of SARS-CoV2 will definitely give the virus a long-term evolutionary edge over active immunity. However, developing next-generation multi-epitope vaccine candidates against the virus might solve the very problem [94]. Additionally, four mutated epitopes are identified in the delta variant that significantly reduced CD8 + T-cell responses [95]. Emerging research suggests, the new variants engage our immune system differently. Plethora of factors, including demographic, nutrition status, comorbidities, age, and gender, may play important roles in modulating the disease severity [96].
Diet and nutrition are the essential pillars of a healthy immune system. However, some micronutrients play dominant role in shaping our immunity compared to the rest. Vitamins such as vitamin A, and D play crucial roles in immunomodulation. Vitamin D is a group of fat-soluble seco-steroid hormone, majorly responsible for maintaining calcium homeostasis in the body. Along with facilitating calcium, magnesium, and phosphate absorption in intestine, it also acts as a major player when it comes to the maintaining immune-barrier integrity. Vitamin D has already been reported to have antiviral roles [97, 98]. Low-serum vitamin D level is associated with increased risks and severity of several respiratory diseases such as asthma and allergic-rhinitis [99, 100]. Effect of vitamin D has also been investigated in mitigating influenza with varied effects [101]. Two major clinical trials are ongoing; one with 2,700 participants, and another with 6,200 participants to investigate whether vitamin D supplementation reduces COVID-19 severity in patients (https://clinicaltrials.gov/). Similarly, there are multiple small- and large-scale interventional clinical trials going on under EU Clinical Trials Register on the same. The completion of these studies will shed much-needed light on this matter. The efficacy of vitamin D supplementation to ameliorate virus infection has been extensively debated. However, it has long been clear that the groups that exhibit vitamin D deficiency, including elderly adults, Asians, and minority ethnic groups, are the population that have also been impacted disproportionately by SARS-CoV2. The lockdown has kept a huge population indoors, further decreasing the exposure time to direct sunlight. This reduced physiological level of calcitriol and triggered concerns that this might inversely impacted COVID-19 scenario during the second wave [102]. Similar to vitamin D, vitamin A is also an important immunomodulator, and is responsible for normal differentiation of epithelial tissues [103]. Vitamin A deficiency is a common risk factor for virus induced respiratory infections [103]. Along with vitamins, essential trace elements such as zinc, selenium, and magnesium are also required to maintain a healthy immunity. Deficiency of these trace elements is widely associated with increased inflammatory disorders, upper respiratory tract infection, and viral pneumonia [104–106]. Even though any direct correlation between vitamin or trace-element deficiency and COVID-19 severity is yet to be drawn; it is evident that malnutrition can exacerbate the severity of COVID-19, especially in children and lactating mothers [107]. Hence, supplementation of vitamins and minerals may be incorporated in the treatment regimen of COVID-19 patients, and susceptible individuals as a preventive measure, especially in low- and middle-income countries where a huge percentage of population suffers from malnutrition.
Reports from early 2020 indicated markedly low severity of SARS-CoV2 among children [108]. However, with the spread of the delta variant, the scenario has changed drastically. Children under 10 are more susceptible to delta transmission than the people above 30 [109]. The prevalence of infection in young adults (18–24 years of age) is also significantly higher with B.1.617.2. A huge demography, which was previously exempted from COVID-19, is now at risk. The hospitalization rate and emergency attendance risk also increased for the patients with the delta variant compared to the Alpha variant [110]. We are yet to ascertain whether such parameters depend upon the ethnicity. During the first wave of the pandemic, a clear association between comorbidities and patient mortality was noticed. Elderly patients with diabetes, hypertension, chronic obstructive pulmonary disease, cardiovascular diseases, HIV, and other comorbidities could develop a life-threatening situation [95]. During the second wave, the major players were the B.1.617.1 and B.1.617.2 variants [21]. Incidentally, the onset of these variants correlated with the beginning of vaccination. With the change in dominating variant, the target demographic also changed from elderly population to younger individuals. This shift will definitely alter the association between comorbidities and patient mortality. The SARS-CoV2 genome is still undergoing rapid mutations. The delta plus variant (AY.1 or B.1.617.2.1) has significant number of high-prevalence mutations (≥ 20%) than in B.1.617.2. AY.1 has three new spike protein mutations (W258L, K417N, and V70F), along with five key predominant mutations (T95I, A222V, G142D, R158G, and K417N), already been recognized as a VOC [22]. Another VOI from South Africa reported in May, 2021 was named C.1.2. C.1.2 got evolved from C.1 lineage; one of the lineages that dominated the first wave of COVID-19. C.1.2 contains multiple substitution mutations (R190S, D215G, N484K, N501Y, H655Y, and T859N), and deletion mutations (Y144del and L242-A243del) in the spike protein, which have also been observed in other VOCs [111]. The Mu variant (B.1.621), now considered as a VOI, inherited several key mutations (amino acid changes in T95I, Y144S, Y145N, and insertion of 146 N in the N-terminal domain, R346K, E484K, and N501Y in the RBD and P681H in the S1/S2 cleavage site of the Spike protein) associated with lower antibody neutralization (outbreak.info). The N501Y and K417N mutations on the RBD of the spike protein increase ACE2 receptor-binding affinity, but diminish antibody binding [112]. K417N shows more binding affinity toward ACE2 receptor compared to its British counterpart N501Y [113]. One major concern is the accumulation of additional novel mutations by these new variants, that might increase their transmissibility, further reducing antibody neutralization and increasing disease severity [111]. Till date, information on these new variants is incomplete, but a mosaic of studies hint that they might have an edge over the other variants circulating worldwide; and might cause another surge of cases in the upcoming months. New Zealand already went under level-4 lockdown when it witnessed a spike in COVID-19 cases dominated by the delta variant in August, 2021. Now, New Zealand is in a level-3 lockdown after the number of cases gone relatively low. On the other hand, South Africa imposed travel restriction during the third wave. After 10 weeks of decline, the number of cases is also in rise throughout Europe driven by increased gatherings, social interactions, travels, and easing social restrictions. In this era of globalization, it is hard to contain a variant within its country of origin. Considering highly contagious nature of these new VOIs and VOCs, it is just a matter of time before they spread all over the world. The only hope to fight the pandemic still remains vaccination, strictly adhering to COVID-19 appropriate behavior, and keeping a healthy immune system through diet and healthy habits. With current settings and scientific advancements, it is next to impossible to stop a pandemic midway. However, with better understanding of the new variants, and further rapid development of vaccines, we can fight COVID-19 in a more-efficient manner by lowering mortality and reducing severity until the natural cycle of the pandemic meets its end.
Acknowledgements
BB acknowledges Council for Scientific and Industrial Research [File 09/081(1333)/2019-EMR-I], Government of India for fellowship. SH acknowledges University Grant Commission, Government of India for fellowship assistance. SC and AKH acknowledge Ministry of Education, Government of India for providing fellowships. RG is funded by SERB (File No. CRG/2019/001049), Government of India.
Abbreviations
- COVID-19
Coronavirus disease 2019
- WHO
World Health Organization
- CDC
Centers for Disease Control and Prevention
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
- SARS-CoV
Severe acute respiratory syndrome coronavirus
- IFN-γ
Interferon gamma
- CD
Cluster of differentiation
- VOC
Variant of concern
- VOI
Variant of interest
- MERS-CoV
Middle Eastern Respiratory Syndrome Coronavirus
- ORF
Open-reading frame
- UTR
Untranslated region
- S
Spike protein
- E
Envelope protein
- M
Matrix protein
- N
Nucleocapsid protein
- NTD
N-terminal domain
- RBD
Receptor-binding domain
- RBM
Receptor-binding motif
- SD1 and SD2
Sub-domains 1 and 2
- FP
Fusion peptide
- HR1
Heptad repeat 1
- HR2
Heptad repeat 2
- TM
Transmembrane region
- CT
C-Terminal region
- Nsp
Non-structural protein
- ACE2
Angiotensin converting enzyme 2
- TMPRSS2
Transmembrane serine protease 2
- NRP1
Neuropilin 1
- DPP-4
Dipeptidyl-peptidase-4
- Mda5
Melanoma differentiation-associated protein 5
- IFIT
Interferon-induced protein with tetratricopeptide repeats
- NK cells
Natural killer cells
- RIG-I
Retinoic acid inducible gene 1
- MAVs
Mitochondrial antiviral-signaling proteins
- IL
Interleukins
- PSO
Post symptom onset
- ARDS
Acute respiratory distress syndrome
- ALI
Acute lung injury
- PRR
Pattern recognition receptor
- CI
Confidence interval
Declarations
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Shagnik Chattopadhyay, Sayantee Hazra, and Arman Kunwar Hansda have equally contributed to this work.
References
- 1.Ranjan R, Sharma A, Verma MK. Characterization of the second wave of COVID-19 in India. medRxiv. 2021 doi: 10.1101/2021.04.17.21255665. [DOI] [Google Scholar]
- 2.Planas D, Veyer D, Baidaliuk A, Staropoli I, Guivel-benhassine F, Rajah MM, et al. Reduced sensitivity of SARS-CoV-2 variant delta to antibody neutralization. Nature. 2021;596(7871):276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 3.Le Bert N, Tan AT, Kunasegaran K, Tham CYL, Hafezi M, Chia A, et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584(7821):457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 4.Vabret N, Britton GJ, Gruber C, Hegde S, Kim J, Kuksin M, et al. Immunology of COVID-19: current state of the science. Immunity. 2020;52(6):910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walsh KA, Jordan K, Clyne B, Rohde D, Drummond L, Byrne P, et al. SARS-CoV-2 detection, viral load and infectivity over the course of an infection. J Infect. 2020;81(3):357–371. doi: 10.1016/j.jinf.2020.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Z, John WE. T cell responses in patients with COVID-19. Nat Rev Immunol. 2020;20(9):529–536. doi: 10.1038/s41577-020-0402-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang Y, Liu J, Zhang D, Xu Z, Ji J, Wen C. Cytokine storm in COVID-19: the current evidence and treatment strategies. Front Immunol. 2020;11:1708. doi: 10.3389/fimmu.2020.01708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sahin U, Muik A, Derhovanessian E, Vogler I, Kranz LM, Vormehr M, et al. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature. 2020;586(7830):594–599. doi: 10.1038/s41586-020-2814-7. [DOI] [PubMed] [Google Scholar]
- 9.Ewer KJ, Barrett JR, Belij-Rammerstorfer S, Sharpe H, Makinson R, Morter R, et al. T cell and antibody responses induced by a single dose of ChAdOx1 nCoV-19 (AZD1222) vaccine in a phase 1/2 clinical trial. Nat Med. 2021;27(2):270–278. doi: 10.1038/s41591-020-01194-5. [DOI] [PubMed] [Google Scholar]
- 10.Petrosillo N, Viceconte G, Ergonul O, Ippolito G, Petersen E. COVID-19, SARS and MERS: are they closely related? Clin Microbiol Infect. 2020;26(6):729–734. doi: 10.1016/j.cmi.2020.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maldonado LL, Bertelli AM, Kamenetzky L. Molecular features similarities between SARS-CoV-2, SARS, MERS and key human genes could favour the viral infections and trigger collateral effects. Sci Rep. 2021;11(1):4108. doi: 10.1038/s41598-021-83595-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Groves DC, Rowland-Jones SL, Angyal A. The D614G mutations in the SARS-CoV-2 spike protein: implications for viral infectivity, disease severity and vaccine design. Biochem Biophys Res Commun. 2021;538:104–107. doi: 10.1016/j.bbrc.2020.10.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harvey WT, Carabelli AM, Jackson B, Gupta RK, Thomson EC, Harrison EM, et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021;19(7):409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khailany RA, Safdar M, Ozaslan M. Genomic characterization of a novel SARS-CoV-2. Gene Rep. 2020;19:100682. doi: 10.1016/j.genrep.2020.100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Islam MR, Hoque MN, Rahman MS, Alam ASMRU, Akther M, Puspo JA, et al. Genome-wide analysis of SARS-CoV-2 virus strains circulating worldwide implicates heterogeneity. Sci Rep. 2020;10(1):14004. doi: 10.1038/s41598-020-70812-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shang J, Wan Y, Luo C, Ye G, Geng Q, Auerbach A, et al. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci USA. 2020;117(21):11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang Y, Yang C, Xu XF, Xu W, Liu SW. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol Sin. 2020;41(9):1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koyama T, Platt D, Parida L. Variant analysis of SARS-cov-2 genomes. Bull World Health Organ. 2020;98(7):495–504. doi: 10.2471/BLT.20.253591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohammadi E, Shafiee F, Shahzamani K, Ranjbar MM, Alibakhshi A, Ahangarzadeh S, et al. Novel and emerging mutations of SARS-CoV-2: biomedical implications. Biomed Pharmacother. 2021;139:111599. doi: 10.1016/j.biopha.2021.111599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vaidyanathan G. Coronavirus variants are spreading in India—what scientists know so far. Nature. 2021;593(7859):321–322. doi: 10.1038/d41586-021-01274-7. [DOI] [PubMed] [Google Scholar]
- 22.Kannan SR, Spratt AN, Cohen AR, Naqvi SH, Chand HS, Quinn TP, et al. Evolutionary analysis of the delta and delta plus variants of the SARS-CoV-2 viruses. J Autoimmun. 2021;124:102715. doi: 10.1016/j.jaut.2021.102715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uriu K, Kimura I, Shirakawa K, et al. Neutralization of the SARS-CoV-2 Mu variant by convalescent and vaccine serum. N Engl J Med. 2021;385(25):2397–2399. doi: 10.1056/NEJMc2114706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perlman S, Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol. 2009;7(6):439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jain U. Effect of COVID-19 on the organs. Cureus. 2020;12(8):e9540. doi: 10.7759/cureus.9540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang K, Chen W, Zhang Z, Deng Y, Lian JQ, Du P, et al. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct Target Ther. 2020;5(1):283. doi: 10.1038/s41392-020-00426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abebe EC, Ayele TM, Muche ZT, Dejenie TA. Neuropilin 1: a novel entry factor for sars-cov-2 infection and a potential therapeutic target. Biol Targets Ther. 2021;15:143–152. doi: 10.2147/BTT.S307352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gil-Etayo FJ, Suàrez-Fernández P, Cabrera-Marante O, Arroyo D, Garcinuño S, Naranjo L, et al. T-helper cell subset response is a determining factor in COVID-19 progression. Front Cell Infect Microbiol. 2021;11:624483. doi: 10.3389/fcimb.2021.624483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schultze JL, Aschenbrenner AC. COVID-19 and the human innate immune system. Cell. 2021;184(7):1671–1692. doi: 10.1016/j.cell.2021.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Y, Su C, Ke M, Jin X, Xu L, Zhang Z, et al. Biochemical and structural insights into the mechanisms of sars coronavirus RNA ribose 2’-O-methylation by nsp16/nsp10 protein complex. PLoS Pathog. 2011;7(10):e1002294. doi: 10.1371/journal.ppat.1002294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krafcikova P, Silhan J, Nencka R, Boura E. Structural analysis of the SARS-CoV-2 methyltransferase complex involved in RNA cap creation bound to sinefungin. Nat Commun. 2020;11(1):3717. doi: 10.1038/s41467-020-17495-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thoms M, Buschauer R, Ameismeier M, Koepke L, Denk T, Hirschenberger M, et al. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science. 2020;369(6508):1249–1256. doi: 10.1126/science.abc8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schubert K, Karousis ED, Jomaa A, Scaiola A, Echeverria B, Gurzeler LA, et al. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat Struct Mol Biol. 2020;27(10):959–966. doi: 10.1038/s41594-020-0511-8. [DOI] [PubMed] [Google Scholar]
- 34.Hsu JCC, Laurent-Rolle M, Pawlak JB, Wilen CB, Cresswell P. Translational shutdown and evasion of the innate immune response by SARS-CoV-2 NSP14 protein. Proc Natl Acad Sci USA. 2021;118(24):e2101161118. doi: 10.1073/pnas.2101161118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM, et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583(7816):459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Acharya D, Liu GQ, Gack MU. Dysregulation of type I interferon responses in COVID-19. Nat Rev Immunol. 2020;20(7):397–398. doi: 10.1038/s41577-020-0346-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng Y, Zhuang MW, Han L, Zhang J, Nan ML, Zhan P, et al. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) membrane (M) protein inhibits type I and III interferon production by targeting RIG-I/MDA-5 signaling. Signal Transduct Target Ther. 2020;5(1):299. doi: 10.1038/s41392-020-00438-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miorin L, Kehrer T, Sanchez-Aparicio MT, Zhang K, Cohen P, Patel RS, et al. SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc Natl Acad Sci USA. 2020;117(45):28344–28354. doi: 10.1073/pnas.2016650117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Konno Y, Kimura I, Uriu K, Fukushi M, Irie T, Koyanagi Y, et al. SARS-CoV-2 ORF3b is a potent interferon antagonist whose activity is increased by a naturally occurring elongation variant. Cell Rep. 2020;32(12):108185. doi: 10.1016/j.celrep.2020.108185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kasuga Y, Zhu B, Jang KJ, Yoo JS. Innate immune sensing of coronavirus and viral evasion strategies. Exp Mol Med. 2021;53(5):723–736. doi: 10.1038/s12276-021-00602-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuen CK, Lam JY, Wong WM, Mak LF, Wang X, Chu H, et al. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg Microbes Infect. 2020;9(1):1418–1428. doi: 10.1080/22221751.2020.1780953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, Chen Y, Li Y, Huang F, Luo B, Yuan Y, et al. The ORF8 protein of SARS-CoV-2 mediates immune evasion through down-regulating MHC-Ι. Proc Natl Acad Sci USA. 2021;118(23):e2024202118. doi: 10.1073/pnas.2024202118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ricci D, Etna MP, Rizzo F, Sandini S, Severa M, Coccia EM. Innate immune response to sars-cov-2 infection: from cells to soluble mediators. Int J Mol Sci. 2021;22(13):7017. doi: 10.3390/ijms22137017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marchi S, Viviani S, Remarque EJ, Ruello A, Bombardieri E, Bollati V, et al. Characterization of antibody response in asymptomatic and symptomatic SARS-CoV-2 infection. PLoS ONE. 2021;16(7):e0253977. doi: 10.1371/journal.pone.0253977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruetalo N, Businger R, Althaus K, Fink S, Ruoff F, Pogoda M, et al. Antibody response against SARS-CoV-2 and seasonal coronaviruses in nonhospitalized COVID-19 patients. mSphere. 2021;6(1):01145–1220. doi: 10.1128/mSphere.01145-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Post N, Eddy D, Huntley C, van Schalkwyk MCI, Shrotri M, Leeman D, et al. Antibody response to SARS-CoV-2 infection in humans: a systematic review. PLoS ONE. 2020;15(12):e0244126. doi: 10.1371/journal.pone.0244126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hartley GE, Edwards ESJ, Aui PM, Varese N, Stojanovic S, McMahon J, et al. Rapid generation of durable B cell memory to SARS-CoV-2 spike and nucleocapsid proteins in COVID-19 and convalescence. Sci Immunol. 2020;5(54):eabf8891. doi: 10.1126/sciimmunol.abf8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pan Y, Jiang X, Yang L, Chen L, Zeng X, Liu G, et al. SARS-CoV-2-specific immune response in COVID-19 convalescent individuals. Signal Transduct Target Ther. 2021;6(1):256. doi: 10.1038/s41392-021-00686-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Edara VV, Hudson WH, Xie X, Ahmed R, Suthar MS. Neutralizing antibodies against SARS-CoV-2 variants after infection and vaccination. JAMA. 2021;325(18):1896–1898. doi: 10.1001/jama.2021.4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Voss C, Esmail S, Liu X, Knauer MJ, Ackloo S, Kaneko T, et al. Epitope-specific antibody responses differentiate COVID-19 outcomes and variants of concern. JCI Insight. 2021;6(13):e148855. doi: 10.1172/jci.insight.148855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barros-Martins J, Hammerschmidt SI, Cossmann A, Odak I, Stankov MV, Morillas Ramos G, et al. Immune responses against SARS-CoV-2 variants after heterologous and homologous ChAdOx1 nCoV-19/BNT162b2 vaccination. Nat Med. 2021;27(9):1525–1529. doi: 10.1038/s41591-021-01449-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181(7):1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kared H, Redd AD, Bloch EM, Bonny TS, Sumatoh H, Kairi F, et al. SARS-CoV-2-specific CD8+ T cell responses in convalescent COVID-19 individuals. J Clin Invest. 2021;131(5):e145476. doi: 10.1172/JCI145476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zuo J, Dowell AC, Pearce H, Verma K, Long HM, Begum J, et al. Robust SARS-CoV-2-specific T cell immunity is maintained at 6 months following primary infection. Nat Immunol. 2021;22(5):620–626. doi: 10.1038/s41590-021-00902-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dan JM, Mateus J, Kato Y, Hastie KM, Yu ED, Faliti CE, et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021;371(6529):eabf4063. doi: 10.1126/science.abf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sekine T, Perez-Potti A, Rivera-Ballesteros O, Strålin K, Gorin JB, Olsson A, et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020;183(1):158–168.e14. doi: 10.1016/j.cell.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rydyznski Moderbacher C, Ramirez SI, Dan JM, Grifoni A, Hastie KM, Weiskopf D, et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell. 2020;183(4):996–1012.e19. doi: 10.1016/j.cell.2020.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Iwasaki N, Terawaki S, Shimizu K, Oikawa D, Sakamoto H, Sunami K, et al. Th2 cells and macrophages cooperatively induce allergic inflammation through histamine signaling. PLoS ONE. 2021;16(3):e0248158. doi: 10.1371/journal.pone.0248158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cancer Therapy Evaluation Program (CTEP). Common Terminology Criteria for Adverse Events (CTCAE).v.5.0 [5x7]. Cancer Ther Eval Progr. 2017;155. https://ctep.cancer.gov/protocolDevelopment/electronic_applications/ctc.htm#ctc_50. Accessed 27 Nov 2017
- 60.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan. China Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weiskopf D, Schmitz KS, Raadsen MP, Grifoni A, Okba NMA, Endeman H, et al. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci Immunol. 2020;5(48):eabd2071. doi: 10.1126/sciimmunol.abd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roncati L, Nasillo V, Lusenti B, Riva G. Signals of Th2 immune response from COVID-19 patients requiring intensive care. Ann Hematol. 2020;99(6):1419–1420. doi: 10.1007/s00277-020-04066-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Braun J, Loyal L, Frentsch M, Wendisch D, Georg P, Kurth F, et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587(7833):270–274. doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- 64.Xu Z, Shi L, Wang Y, Zhang J, Huang L, Zhang C, et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8(4):420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Meckiff BJ, Ramírez-Suástegui C, Fajardo V, Chee SJ, Kusnadi A, Simon H, et al. Imbalance of regulatory and cytotoxic SARS-CoV-2-reactive CD4+ T cells in COVID-19. Cell. 2020;183(5):1340–1353.e16. doi: 10.1016/j.cell.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Boppana S, Qin K, Files JK, Russell RM, Stoltz R, Bibollet-Ruche F, et al. SARS-CoV-2-specific circulating T follicular helper cells correlate with neutralizing antibodies and increase during early convalescence. PLoS Pathog. 2021;17(7):e1009761. doi: 10.1371/journal.ppat.1009761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sette A, Crotty S. Pre-existing immunity to SARS-CoV-2: the knowns and unknowns. Nat Rev Immunol. 2020;20(8):457–458. doi: 10.1038/s41577-020-0389-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mlcochova P, Kemp S, Dhar MS, Papa G, Meng B, Mishra S, et al. SARS-CoV-2 B.1.617.2 delta variant replication and immune evasion. Nature. 2021;599(7883):114–119. doi: 10.1038/s41586-021-03944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li B, Deng A, Li K, Hu Y, Li Z, Xiong Q, et al. Viral infection and transmission in a large, well-traced outbreak caused by the SARS-CoV-2 delta variant. Nat Commun. 2022;13(1):460. doi: 10.1038/s41467-022-28089-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McCallum M, Walls AC, Sprouse KR, Bowen JE, Rosen LE, Dang HV, et al. Molecular basis of immune evasion by the delta and kappa SARS-CoV-2 variants. Science. 2021;374(6575):1621–1626. doi: 10.1126/science.abl8506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chaudhari AM, Kumar D, Joshi M, Patel A, Joshi C. E156/G and Arg158, Phe-157/del mutation in NTD of spike protein in B.1.167.2 lineage of SARS-CoV-2 leads to immune evasion through antibody escape. bioRxiv. 2021 doi: 10.1101/2021.06.07.447321. [DOI] [Google Scholar]
- 72.Saito A, Irie T, Suzuki R, Maemura T, Nasser H, Uriu K, et al. Enhanced fusogenicity and pathogenicity of SARS-CoV-2 delta P681R mutation. Nature. 2021 doi: 10.1038/s41586-021-04266-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu Y, Liu J, Johnson BA, Xia H, Ku Z, Schindewolf C, et al. Delta spike P681R mutation enhances SARS-CoV-2 fitness over alpha variant. bioRxiv. 2021 doi: 10.1101/202108.12.456173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Plante JA, Mitchell BM, Plante KS, Debbink K, Weaver SC, Menachery VD. The variant gambit: COVID-19’s next move. Cell Host Microbe. 2021;29(4):508–515. doi: 10.1016/j.chom.2021.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang J, Xiao T, Cai Y, Lavine CL, Peng H, Zhu H, et al. Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 delta variant. Science. 2021;374(6573):1353–1360. doi: 10.1126/science.abl9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Daniloski Z, Jordan TX, Ilmain JK, Guo X, Bhabha G, Tenoever BR, et al. The spike d614g mutation increases sars-cov-2 infection of multiple human cell types. Elife. 2021;10:e65365. doi: 10.7554/eLife.65365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Baral P, Bhattarai N, Hossen ML, Stebliankin V, Gerstman BS, Narasimhan G, et al. Mutation-induced changes in the receptor-binding interface of the SARS-CoV-2 Delta Variant B.1.617.2 and implications for immune evasion. Biochem Biophys Res Commun. 2021;574:14–19. doi: 10.1016/j.bbrc.2021.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gerberding JL, Haynes BF. Vaccine innovations—past and future. N Engl J Med. 2021;384(5):393–396. doi: 10.1056/NEJMp2029466. [DOI] [PubMed] [Google Scholar]
- 79.Liu MA. DNA vaccines: a review. J Intern Med. 2003;253(4):402–410. doi: 10.1046/j.1365-2796.2003.01140.x. [DOI] [PubMed] [Google Scholar]
- 80.Pardi N, Hogan MJ, Porter FW, Weissman D. mRNA vaccines—a new era in vaccinology. Nat Rev Drug Discov. 2018;17(4):261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Suschak JJ, Williams JA, Schmaljohn CS. Advancements in DNA vaccine vectors, non-mechanical delivery methods, and molecular adjuvants to increase immunogenicity. Hum Vaccines Immunother. 2017;13(12):2837–2848. doi: 10.1080/21645515.2017.1330236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tatsis N, Ertl HCJ. Adenoviruses as vaccine vectors. Mol Ther. 2004;10(4):616–629. doi: 10.1016/j.ymthe.2004.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gunst JD, Staerke NB, Pahus MH, Kristensen LH, Bodilsen J, Lohse N, et al. Efficacy of the TMPRSS2 inhibitor camostat mesilate in patients hospitalized with Covid-19-a double-blind randomized controlled trial. eClinicalMedicine. 2021;35:100849. doi: 10.1016/j.eclinm.2021.100849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lopez Bernal J, Andrews N, Gower C, Gallagher E, Simmons R, Thelwall S, et al. Effectiveness of covid-19 vaccines against the B.1.617.2 (delta) variant. N Engl J Med. 2021;385(7):585–594. doi: 10.1056/NEJMoa2108891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R, et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med. 2021;384(5):403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Puranik A, Lenehan PJ, Silvert E, Niesen MJM, Corchado-Garcia J, O’Horo JC, et al. Comparison of two highly-effective mRNA vaccines for COVID-19 during periods of alpha and delta variant prevalence. medRxiv. 2021 doi: 10.1101/08.06.21261707. [DOI] [Google Scholar]
- 87.Hu Z, Tao B, Li Z, Song Y, Yi C, Li J, et al. Effectiveness of inactive COVID-19 vaccines against severe illness in B.1.617.2 (delta) variant-infected patients in Jiangsu, China. Int J Infect Dis. 2022;116:204–209. doi: 10.1016/j.ijid.2022.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nasreen S, Chung H, He S, Brown KA, Gubbay JB, Buchan SA, et al. Effectiveness of COVID-19 vaccines against variants of concern in Ontario, Canada. medRxiv. 2021 doi: 10.1101/06.28.21259420. [DOI] [Google Scholar]
- 89.Sapkal GN, Yadav PD, Ella R, Deshpande GR, Sahay RR, Gupta N, et al. Inactivated COVID-19 vaccine BBV152/COVAXIN effectively neutralizes recently emerged B.1.1.7 variant of SARS-CoV-2. J Travel Med. 2021;28(4):taab051. doi: 10.1093/jtm/taab051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ella R, Reddy S, Blackwelder W, Potdar V, Yadav P, Sarangi V, et al. Efficacy, safety, and lot to lot immunogenicity of an inactivated SARS-CoV-2 vaccine (BBV152): a, double-blind, randomised, controlled phase 3 trial. Lancet. 2021;398(10317):2173–2184. doi: 10.1016/S0140-6736(21)02000-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Logunov DY, Dolzhikova IV, Shcheblyakov DV, Tukhvatulin AI, Zubkova OV, Dzharullaeva AS, et al. Safety and efficacy of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine: an interim analysis of a randomised controlled phase 3 trial in Russia. Lancet. 2021;397(10275):671–681. doi: 10.1016/S0140-6736(21)00234-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ferreira I, Kemp S, Datir R, Saito A, Meng B, Rakshit P, et al. SARS-CoV-2 B1617 mutations L452 and E484Q are not synergistic for antibody evasion. J Infect Dis. 2021;224(6):989–994. doi: 10.1093/infdis/jiab368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mlcochova P, Kemp SA, Dhar MS, Papa G, Meng B, Ferreira IA, et al. SARS-CoV-2 B.1.617.2 delta variant replication and immune evasion. Nature. 2021;599(7883):114–119. doi: 10.1038/s41586-021-03944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Saha R, Ghosh P, Burra VLSP. Designing a next generation multi—epitope based peptide vaccine candidate against SARS—CoV-2 using computational approaches. 3 Biotech. 2021;11(2):47. doi: 10.1007/s13205-020-02574-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang H, Deng S, Ren L, Zheng P, Hu X, Jin T, et al. Profiling CD8+ T cell epitopes of COVID-19 convalescents reveals reduced cellular immune responses to SARS-CoV-2 variants. Cell Rep. 2021;36(11):109708. doi: 10.1016/j.celrep.2021.109708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gallo Marin B, Aghagoli G, Lavine K, Yang L, Siff EJ, Chiang SS, et al. Predictors of COVID-19 severity: a literature review. Rev Med Virol. 2021;31(1):1–10. doi: 10.1002/rmv.2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Beard JA, Bearden A, Striker R. Vitamin D and the anti-viral state. J Clin Virol. 2011;50(3):194–200. doi: 10.1016/j.jcv.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Biswas B, Goswami R. Differential gene expression analysis in 1,25(OH)2D3 treated human monocytes establishes link between AIDS progression, neurodegenerative disorders, and aging. Meta Gene. 2021;28:100886. [Google Scholar]
- 99.Bener A, Ehlayel MS, Tulic MK, Hamid Q. Vitamin D deficiency as a strong predictor of asthma in children. Int Arch Allergy Immunol. 2012;157(2):168–175. doi: 10.1159/000323941. [DOI] [PubMed] [Google Scholar]
- 100.Wjst M. Vitamin D serum levels and allergic rhinitis. Allergy. 2007;62(9):1085–1086. doi: 10.1111/j.1398-9995.2007.01437.x. [DOI] [PubMed] [Google Scholar]
- 101.Grant WB, Lahore H, McDonnell SL, Baggerly CA, French CB, Aliano JL, et al. Evidence that vitamin d supplementation could reduce risk of influenza and covid-19 infections and deaths. Nutrients. 2020;12(4):988. doi: 10.3390/nu12040988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.The Lancet Diabetes and Endocrinology Vitamin D and COVID-19: why the controversy? Lancet Diabetes Endocrinol. 2021;9(2):53. doi: 10.1016/S2213-8587(21)00003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gasmi A, Tippairote T, Mujawdiya PK, Peana M, Menzel A, Dadar M, et al. Micronutrients as immunomodulatory tools for COVID-19 management. Clin Immunol. 2020;220:108545. doi: 10.1016/j.clim.2020.108545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Prasad AS. Clinical, immunological, anti-inflammatory and antioxidant roles of zinc. Exp Gerontol. 2008;43(5):370–377. doi: 10.1016/j.exger.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 105.Beck MA. Selenium as an antiviral agent. Selenium: Springer; 2001. [Google Scholar]
- 106.Shaikh MN, Malapati BR, Gokani R, Patel B, Chatriwala M. Serum magnesium and vitamin D levels as indicators of asthma severity. Pulm Med. 2016;2016:1643717. doi: 10.1155/2016/1643717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fore HH, Dongyu Q, Beasley DM, Ghebreyesus TA. Child malnutrition and COVID-19: the time to act is now. Lancet. 2020;396(10250):517–518. doi: 10.1016/S0140-6736(20)31648-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Davies NG, Klepac P, Liu Y, Prem K, Jit M, Pearson CAB, et al. Age-dependent effects in the transmission and control of COVID-19 epidemics. Nat Med. 2020;26(8):1205–1211. doi: 10.1038/s41591-020-0962-9. [DOI] [PubMed] [Google Scholar]
- 109.McLaws ML. COVID-19 in children: time for a new strategy. Med J Aust. 2021;215(5):212–213. doi: 10.5694/mja2.51206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Twohig KA, Nyberg T, Zaidi A, Thelwall S, Sinnathamby MA, Aliabadi S, et al. Hospital admission and emergency care attendance risk for SARS-CoV-2 delta (B.1.617.2) compared with alpha (B.1.1.7) variants of concern: a cohort study. Lancet Infect Dis. 2021;22(1):35–42. doi: 10.1016/S1473-3099(21)00475-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Scheepers C, Everatt J, Amoako DG, Mnguni A, Ismail A, Mahlangu B, et al. The continuous evolution of SARS-CoV-2 in South Africa: a new lineage with rapid accumulation of mutations of concern and global detection. medRxiv. 2021 doi: 10.1101/2021.08.20.21262342. [DOI] [Google Scholar]
- 112.Fratev F. N501Y and K417N mutations in the spike protein of SARS-CoV-2 alter the interactions with both hACE2 and human-derived antibody: a free energy of perturbation retrospective study. J Chem Inf Model. 2021;61(12):6079–6084. doi: 10.1021/acs.jcim.1c01242. [DOI] [PubMed] [Google Scholar]
- 113.Khan A, Zia T, Suleman M, Khan T, Ali SS, Abbasi AA, et al. Higher infectivity of the SARS—CoV-2 new variants is associated with K417N/T, E484K, and N501Y mutants: an insight from structural data. J Cell Physiol. 2021;236(10):7045–7057. doi: 10.1002/jcp.30367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wang P, Nair MS, Liu L, Iketani S, Luo Y, Guo Y, et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593(7857):130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 115.Nonaka CKV, Gräf T, De Lorenzo Barcia CA, Costa VF, De Oliveira JL, Da Hora Passos R, et al. SARS-CoV-2 variant of concern P1 (gamma) infection in young and middle-aged patients admitted to the intensive care units of a single hospital in salvador, Northeast Brazil, February 2021. Int J Infect Dis. 2021;111:47–54. doi: 10.1016/j.ijid.2021.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Laiton-Donato K, Franco-Muñoz C, Álvarez-Díaz DA, Ruiz-Moreno HA, Usme-Ciro JA, Prada DA, et al. Characterization of the emerging B.1.621 variant of interest of SARS-CoV-2. Infect Genet Evol. 2021;95:105038. doi: 10.1016/j.meegid.2021.105038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ferreira I, Datir R, Kemp S, Papa G, Rakshit P, Singh S, et al. SARS-CoV-2 B.1.617 emergence and sensitivity to vaccine-elicited antibodies. bioRxiv. 2021;2021.05.08.443253.
- 118.Callaway E. The mutation that helps delta spread like wildfire. Nature. 2021;596(7873):472–473. doi: 10.1038/d41586-021-02275-2. [DOI] [PubMed] [Google Scholar]
- 119.Annavajhala MK, Mohri H, Wang P, et al. Emergence and expansion of SARS-CoV-2 B1526 after identification in New York. Nature. 2021;597(7878):703–708. doi: 10.1038/s41586-021-03908-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mohammadi M, Shayestehpour M, Mirzaei H. The impact of spike mutated variants of SARS-CoV2 [alpha, beta, gamma, delta, and lambda] on the efficacy of subunit recombinant vaccines. Brazilian J Infect Dis. 2021;25(4):101606. doi: 10.1016/j.bjid.2021.101606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Romero PE, Dávila-Barclay A, Salvatierra G, González L, Cuicapuza D, Solis L, et al. The emergence of Sars-CoV-2 variant lambda (C.37) in South America. Microbiol Spectr. 2021;9(2):e0078921. doi: 10.1128/Spectrum.00789-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Baj A, Novazzi F, Ferrante FD, Genoni A, Cassani G, Prestia M, et al. Introduction of SARS-COV-2 C.37 (WHO VOI lambda) from Peru to Italy. J Med Virol. 2021;93(12):6460–6461. doi: 10.1002/jmv.27235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kimura I, Kosugi Y, Wu J, Zahradnik J, Yamasoba D, Butlertanaka EP, et al. SARS-CoV-2 lambda variant exhibits higher infectivity and immune resistance. Cell Rep. 2022;38(2):110218. doi: 10.1016/j.celrep.2021.110218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.McCallum M, Bassi J, de Marco A, Chen A, Walls AC, Di Iulio J, et al. SARS-CoV-2 immune evasion by the B.1.427/B.1.429 variant of concern. Science. 2021;373(6555):648–654. doi: 10.1126/science.abi7994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cedro-Tanda A, Gómez-Romero L, Alcaraz N, de Anda-Jauregui G, Peñaloza F, Moreno B, et al. The evolutionary landscape of SARS-CoV-2 variant B11519 and its clinical impact in Mexico city. Viruses. 2021;13(11):2182. doi: 10.3390/v13112182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ferraz MVF, Moreira EG, Coêlho DF, Wallau GL, Lins RD. Immune evasion of SARS-CoV-2 variants of concern is driven by low affinity to neutralizing antibodies. Chem Commun. 2021;57(49):6094–6097. doi: 10.1039/d1cc01747k. [DOI] [PubMed] [Google Scholar]
- 127.Nieto-Torres JL, DeDiego ML, Verdiá-Báguena C, Jimenez-Guardeño JM, Regla-Nava JA, Fernandez-Delgado R, et al. Severe acute respiratory syndrome coronavirus envelope protein ion channel activity promotes virus fitness and pathogenesis. PLoS Pathog. 2014;10(5):e1004077. doi: 10.1371/journal.ppat.1004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Siu KL, Yuen KS, Castano-Rodriguez C, Ye ZW, Yeung ML, Fung SY, et al. Severe acute respiratory syndrome coronavirus ORF3a protein activates the NLRP3 inflammasome by promoting TRAF3-dependent ubiquitination of ASC. FASEB J. 2019;33(8):8865–8877. doi: 10.1096/fj.201802418R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yazan A. Interleukin-2 level for normal people and COVID-19 infection: is it our concern is COVID-19 infection or interleukin-2 level before the infection? Eurasian J Med Oncol. 2021;5(1):1–5. [Google Scholar]
- 130.Vaz de Paula CB, de Azevedo MLV, Nagashima S, Martins APC, Malaquias MAS, Miggiolaro AFR dos S, et al. IL-4/IL-13 remodeling pathway of COVID-19 lung injury. Sci Rep. 2020;10(1):18689. [DOI] [PMC free article] [PubMed]
- 131.Pala D, Pistis M. Anti-IL5 drugs in COVID-19 patients: role of eosinophils in SARS-CoV-2-induced immunopathology. Front Pharmacol. 2021;12:622554. doi: 10.3389/fphar.2021.622554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Costela-Ruiz VJ, Illescas-Montes R, Puerta-Puerta JM, Ruiz C, Melguizo-Rodríguez L. SARS-CoV-2 infection: the role of cytokines in COVID-19 disease. Cytokine Growth Factor Rev. 2020;54:62–75. doi: 10.1016/j.cytogfr.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Monneret G, de Marignan D, Coudereau R, Bernet C, Ader F, Frobert E, et al. Immune monitoring of interleukin-7 compassionate use in a critically ill COVID-19 patient. Cell Mol Immunol. 2020;17(9):1001–1003. doi: 10.1038/s41423-020-0516-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Li J, Rong L, Cui R, Feng J, Jin Y, Chen X, et al. Dynamic changes in serum IL-6, IL-8, and IL-10 predict the outcome of ICU patients with severe COVID-19. Ann Palliat Med. 2021;10(4):3706–3714. doi: 10.21037/apm-20-2134. [DOI] [PubMed] [Google Scholar]
- 135.Zhao Y, Qin L, Zhang P, Li K, Liang L, Sun J, et al. Longitudinal COVID-19 profiling associates IL-1RA and IL-10 with disease severity and RANTES with mild disease. JCI Insight. 2020;5(13):e139834. doi: 10.1172/jci.insight.139834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tjan LH, Furukawa K, Nagano T, Kiriu T, Nishimura M, Arii J, et al. Early differences in cytokine production by severity of coronavirus disease 2019. J Infect Dis. 2021;223(7):1145–1149. doi: 10.1093/infdis/jiab005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Martonik D, Parfieniuk-Kowerda A, Rogalska M, Flisiak R. The role of th17 response in COVID-19. Cells. 2021;10(6):1550. doi: 10.3390/cells10061550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Acet Öztürk NA, Ursavaş A, Görek Dilektaşli A, Demirdöğen E, Coşkun NF, Ediger D, et al. Interleukin-21: a potential biomarker for diagnosis and predicting prognosis in covid-19 patients. Turkish J Med Sci. 2021;51(5):2274–2284. doi: 10.3906/sag-2102-24. [DOI] [PubMed] [Google Scholar]
- 139.Wang CJ, Truong AK. COVID-19 infection on IL-23 inhibition. Dermatol Ther. 2020;33(6):e13893. doi: 10.1111/dth.13893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Liang Y, Ge Y, Sun J. IL-33 in COVID-19: friend or foe? Cell Mol Immunol. 2021;18(6):1602–1604. doi: 10.1038/s41423-021-00685-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gadotti AC, de Castro Deus M, Telles JP, Wind R, Goes M, Garcia Charello Ossoski R, et al. IFN-γ is an independent risk factor associated with mortality in patients with moderate and severe COVID-19 infection. Virus Res. 2020; 289:198171. [DOI] [PMC free article] [PubMed]
- 142.Karki R, Sharma BR, Tuladhar S, Williams EP, Zalduondo L, Samir P, et al. Synergism of TNF-α and IFN-γ triggers inflammatory cell death, tissue damage, and mortality in SARS-CoV-2 infection and cytokine shock syndromes. Cell. 2021;184(1):149–168.e17. doi: 10.1016/j.cell.2020.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sheikh A, McMenamin J, Taylor B, Robertson C. SARS-CoV-2 delta VOC in Scotland: demographics, risk of hospital admission, and vaccine effectiveness. Lancet. 2021;397(10293):2461–2462. doi: 10.1016/S0140-6736(21)01358-1. [DOI] [PMC free article] [PubMed] [Google Scholar]