Abstract
Pacific Ocean rockfishes (genus Sebastes) exhibit extreme variation in lifespan with some species amongst the most long-lived extant vertebrates. We de-novo assembled the genomes of 88 rockfish species and from this identified repeated signatures of positive selection in DNA repair pathways in long-lived taxa and 137 longevity-associated genes with effects on lifespan through insulin signaling, and pleiotropic effects through size and environmental adaptations. A genome-wide screen of structural variation reveals copy number expansions in the immune modulatory butyrophilin gene family in long-lived species. The evolution of different rockfish life-histories is coupled to genetic diversity and reshapes the mutational spectrum driving segregating CpG->TpG variants in long-lived species. These analyses highlight the genetic innovations that underlie life-history trait adaptations and, in turn, how they shape genomic diversity.
One Sentence Summary:
Genomes of 88 rockfish species identify the genetic drivers of longevity and how life-history adaptations can reshape patterns of genetic diversity.
Decades of theoretical work on the evolution of aging have demonstrated that senescence is a “theoretically inevitable” consequence of the increased selective impact of genes that influence early life survival and fecundity compared to genes that act late in life (1). Nonetheless, across vertebrates alone lifespan varies by more than 3 orders of magnitude ranging from the 5-week lifecycle of the pygmy goby (2) to the ~400-year lifespan of the Greenland shark (3) highlighting exceptional diversity of this trait across species. Understanding evolutionary transitions in lifespan can highlight diverse genetic mechanisms of lifespan control and the subsequent impacts of life-history shifts on patterns of genetic diversity.
Across organisms aging is accompanied by several molecular hallmarks including genome instability, loss of protein homeostasis, and mitochondrial dysfunction among others (4). Vertebrates in particular also exhibit a number of distinct hallmarks of aging which are directly linked to human disease and healthspan such as immuno-senescence, inflammation, and stem-cell exhaustion (5). Thus, understanding the underpinnings of lifespan-variation across vertebrate taxa can provide critical insights into the maintenance of human health and vigor in old age.
Rockfishes (genus Sebastes) of the Pacific Ocean exhibit lifespans ranging from 11 years (Sebastes minor) to more than 200 years for the Rougheye Rockfish (Sebastes aleutianus) (6) (Fig. 1A). This phenotype exhibits a relatively uniform distribution across the more than 75 different species of this clade for which detailed longevity information is available (Fig. S1). Rockfishes are thus unique in that while some species are among the longest-lived vertebrates known to exist, lifespan can widely range even among closely related taxa. More than 120 different species of rockfish are found throughout the Northeast and Northwest Pacific Ocean (7) (Fig. 1B). This abundance of species with vastly differing life histories represents a unique example of repeated, recent adaptations that have reshaped longevity phenotypes.
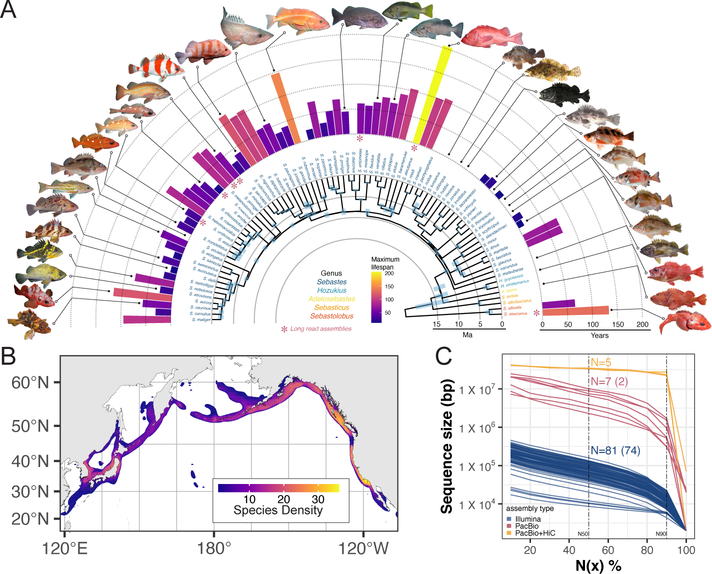
Figure 1 -. Genome assemblies and relationships among rockfish species:
A) Ultrametric tree of the rockfish species sequenced in this study and their associated maximum lifespans along with representative images (node timing confidence intervals in light blue) created using IQ-tree, ASTRAL and BPPR. Asterisks indicate individuals for which long-read sequencing-based genomes were assembled. B) The density of rockfish species (heatmap colors) throughout the Pacific Ocean. C) Genome assembly statistics for 81 species, blue and pink represent N(x)-contig lengths while orange indicate N(x) scaffold lengths.
Results
Sequencing and assembly of 102 rockfish genomes
To dissect the genetic underpinnings of lifespan variation and adaptation we sequenced and de novo assembled the genomes of 102 rockfish individuals encompassing 88 different species, including 79 members of the Sebastes clade and 9 closely related outgroup taxa (Fig. S1, Fig. S2). Long-read Pacific Biosciences genome assemblies were generated for 6 Sebastes species and the outgroup Sebastolobus alascanus (6.1Mbp mean contig N50, Fig. 1C). Five of these Sebastes genomes were scaffolded with long-range HiC data resulting in chromosome-scale assemblies (34.1Mbp mean scaffold N50, Fig. 1C, Table S1, Fig. S3). High-coverage Illumina based assemblies were generated for 71 additional Sebastes species (73.8Kbp mean contig N50). Genome completeness (assessed by BUSCO scores (8)) ranged from an average of 97.9% for long-read based genomes to 91.2% for Illumina based assemblies and the majority (5 of 7) of the long-read based genomes reached QV40 base quality (average QV39 for all 7) (Fig. S4). Transcriptomes were generated from brain and eye tissue for 8 species including 6 individuals for which we generated long-read based genome assemblies to assist in gene annotation. In total, we identified ~25,000 protein-coding genes on average across the seven long-read genome assemblies (Table S3). Together, these genome assemblies encompass the majority of species represented in the Sebastes genus and almost all representatives for which detailed lifespan information has been cataloged, including a chromosome level assembly of the ultra-long-lived S. aleutianus.
We established the phylogenetic relationships and speciation times among rockfish species by constructing an ultrametric species phylogeny (See supplementary methods (9)) (Fig. 1A). The topology of this tree is consistent with previous phylogenetic analyses of rockfishes (7, 10). The varied placement of long-lived taxa in the tree suggests repeated life-span related evolution (Fig. 1A), which is supported by character mapping analysis which shows multiple independent gains of long lifespan (Fig. S7).
Repeated signatures of positive selection in DNA repair pathways in long-lived taxa
We next sought to determine the genetic underpinnings of lifespan in Pacific rockfish. We used a branch-site model test of codon evolution to identify candidates of positive selection in the longest-lived (≥105 years) and shortest-lived (≤20 years) species (lifespan decile tails, n=7) identifying 772 and 873 genes, respectively. Most of these positively selected genes (PSGs) were lineage-specific, although ~12–15% of genes (127 and 118 respectively) were present in two or more species representing either selection on the ancestral branch or convergent evolution. Additionally, 180 PSGs were found in both long- and short-lived individuals, highlighting that many of these genes may be under selection for other traits. While no pathways were enriched in PSGs in short-lived species after multiple testing correction, PSGs in long-lived species were enriched for DNA double stranded break repair pathways (Fig. 2A, Table S5).
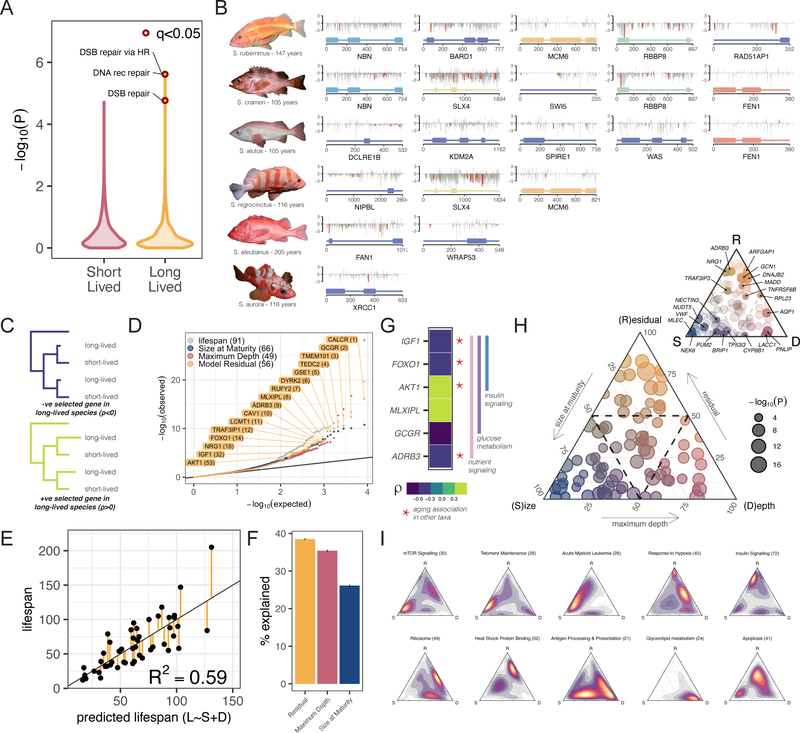
Figure 2 – Genetic underpinnings of lifespan adaptations:
A) Distribution of pathway enrichment P-values for genes under positive selection in short- and long-lived species. B) Schematics of the 16 DNA replication/repair/maintenance genes exhibiting positive selection in long-lived rockfish. Domain structure (bottom), grey lines indicate putative functional consequence of rockfish amino acid compared to human (PROVEAN score). Red and green bars indicate mutations distinct from all non-long-lived taxa colored by SIFT score - Tolerated (green) / Damaging (red). C) Schematic representation of relative evolutionary rate test and D) P-value distribution of genes from relative evolutionary rate test (RERconverge) of correlation between evolutionary rate and individual traits. Lifespan and the individual components of a predictive linear model of lifespan (E) are used as traits. Gene names are highlighted for the top 12 genes associated with the linear model residual as a trait in addition to four other significant gene candidates previously associated with aging (full gene list in Table S14). E) Linear regression model between maximum lifespan and predicted lifespan based on body size and depth for different species. F) Proportion of variation in lifespan explained by size, depth, and the residual from the linear model. G) Relative evolutionary rates of six genes involved in glucose, insulin, and nutrient signaling four of which have been associated with aging in other taxa (Table S14). H) Ternary plot of 91 lifespan associated genes plotted as a function of the relative importance along linear model axes with dotted line at 50% (inset with gene labels) and heatmaps of pathway enrichment along axes (I).
The enrichment in DNA replication, repair, and maintenance is driven by 16 genes (Fig. 2B), five of which exhibit selective signatures in more than one species. These include genes in pathways associated with lifespan across organisms, such as telomere maintenance (e.g. WRAP53, DCLRE1B) and base excision repair (e.g. FEN1). Intriguingly adaptive signatures in DCLRE1B and FEN1 have been identified in the Bowhead whale and giant tortoise respectively (11, 12). MCM6, a core member of the DNA replication helicase machinery, exhibits signatures of positive selection in S. ruberrimus and S. nigrocinctus. These signatures of selection suggest that repeated transitions in lifespan in rockfish are likely enabled by parallel evolution of different genes in shared critical DNA maintenance pathways.
Genes associated with lifespan adaptations through direct and pleiotropic effects
We next examined the role of evolution in convergent genes and pathways on rockfish lifespan by comparing the relative evolutionary rates of genes across the phylogeny (13). This approach allows us to identify genes with evolutionary rates that are correlated, either positively or negatively, with lifespan (Fig. 2C, Table S14). While these correlations do not prove causation, they can guide insights in experimentally intractable systems, such as rockfish. At a q-value cutoff of 0.05 we discovered 91 genes significantly associated with lifespan including candidates with roles in cell growth and proliferation (e.g. NRG1), DNA repair (e.g. BRIP1) and suppression of apoptosis (e.g. TNFRSF6B) (Fig. 2D, Fig. S9). However, lifespan in rockfish is correlated with body size and environmental factors, such as depth (14). Thus, some of these genes associated with lifespan may act by influencing growth and size or may facilitate adaptations to environments that promote longevity.
To identify genes associated with rockfish longevity, independent of other factors, we constructed a linear model using the two most predictive variables for lifespan, size at maturity and maximum depth (Fig. 2E). This model described 59% of the variation in lifespan (Fig. 2F), comparable to simple models of lifespan and body size in mammals (15). Using the residuals of this model as phenotypes we identified 56 genes associated with lifespan, independent of size at maturity or depth (Fig. 2D, Table S14). These were overrepresented for insulin and glucose signaling (Hypergeometric test, P=1.8e-6, Fig. 2G, Fig. S10) including genes with roles in lifespan extension across many organisms (19, 20). The rate of evolution for most of these genes was negatively correlated with lifespan emphasizing the importance of nutrient sensing maintenance in long-lived species. We also identified genes associated with reproductive aging in mice (16), the antiviral innate immunity factor TRAF3IP3 (17), and the tumor suppressor DYRK2 (18).
Of the 91 genes associated with lifespan, prior to correcting for size and depth, only 10 overlapped with those identified as associated with the lifespan residual suggesting that the majority (n=81) of the genes we identified in Sebastes act indirectly by influencing size or facilitating adaptations to depth. To parse apart the axes along which these lifespan-associated genes act, we correlated their relative evolutionary rates with either the growth-associated component of lifespan in our linear model (S, size); the depth associated component of lifespan (D); or the residual of the model (R) (Fig. 2H). The size axis was associated with the most genes (n=33, S>0.5) in comparison to depth or the residual (n=18, D>0.5; n=17, R>0.5) in agreement with the importance of growth and size related pathways underlying differences in lifespan among different organisms (19). Pathways enriched along this axis included mTOR signaling, DNA and telomere maintenance, and cancer (Fig. 2I). Genes and pathways along the depth axis reflect environmental adaptations including lipid metabolism and synthesis. The R-axis was enriched for insulin signaling, and protein homeostasis with ribosome assembly pathways along the DR axis. Apoptosis genes were more closely clustered with the R axis intermediate to S and D and antigen processing and presentation genes were also clustered along the SD axis. Together, these results suggest that the genetic basis of longevity in rockfish species may be due to the combined effect of genes acting directly on lifespan alongside genes impacting ecological and growth phenotypes which pleiotropically influence lifespan.
Expansion of the immune modulatory butyrophilin gene family in long-lived species
Gene duplications and structural rearrangements can drive evolutionary innovations (20, 21). To identify copy number changes associated with variation in lifespan, we performed a genome-wide screen using windowed read-depth based copy estimates across chromosomes. Controlling for phylogenetic signal, we found an enrichment for positive associations between lifespan and copy number, whether we used a short- or long-lived species reference (χ-squared, P < 1e-15). The greatest cluster of positive associations was on chromosome 1 (Fig. 3A); it extended for almost 250kb and overlapped 9 genes (Fig. 3B) ranging from 0–7 copies among different species (Fig. 3C). These 9 genes included 6 members of the butyrophilin gene family (BTN and BTNL genes, Fig. 3D), a family of immune regulators associated with human inflammatory diseases (22). Using our linear model we find that this signature is primarily driven by the depth-associated component of lifespan (Fig. 2H, Fig. S15). Mutations in BTNL2 cause sarcoidosis in humans (23) and variants in BTN3A2 have been associated with human lifespan (24).
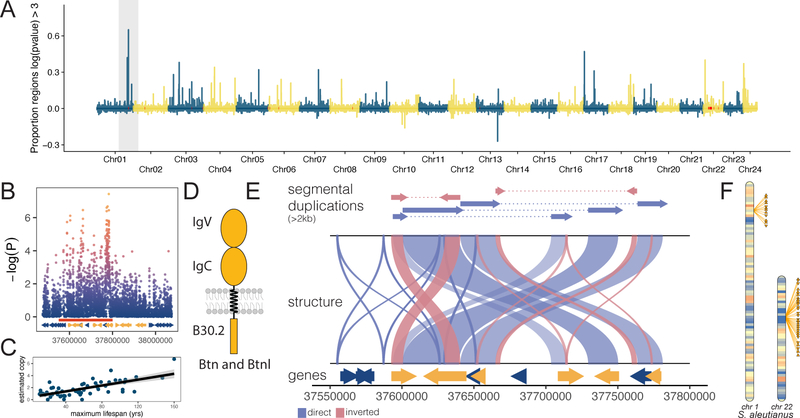
Figure 3 – Lifespan associated butyrophilin gene duplications:
A) Miami plot of proportion of 100bp regions with significant phylogenetic least squares linear model (PGLS) correlations between read-depth inferred copy number and lifespan (10kb windows) across the S. aleutianus reference genome. B) Manhattan plot of ~250kb chromosome 1 butyrophilin locus and copy number association in C. Butyrophilin genes shown in orange. D) Protein structural organization of BTN and BTNL gene family members. E) Genome structure of the butyrophilin locus in the S. aleutianus genome assembly. Segmental duplications are highlighted above a self-alignment of the locus to show the underlying duplication architecture with genes below. F) Karyotypes of chromosomes 1 and 22 in S. aleutianus (color indicates gene-density) highlighting the two butyrophilin gene family clusters in Sebastes species.
Butyrophilins are members of the B7 immunoglobulin superfamily and in response to inflammatory stimuli inhibit cytokine secretion and production in T-cells, thus serving an immuno-suppressive function. We resolved the underlying segmental duplication (SD) architecture of the lifespan-associated butyrophilin duplication in the S. aleutianus genome (Fig. 3E) identifying 22 large (>1kb, >90% identity) SD blocks across the locus. The majority of the locus is contained in three pairs of large adjacent SDs in direct orientation which together encompass 163kb. Two large, high-identity (>95%) inverted duplications add additional complexity to the locus. While the copy number of BTN/BTNL genes at this locus correlates significantly (Fig. 3C) with lifespan in rockfish, we identified non-lifespan associated BTN/BTNL copies as well, largely localized to two clusters on chromosomes 1 and 22 (Fig. 3F). Among our 6 Sebastes reference genome assemblies the total number of BTN/BNTL genes ranges from 18–36 highlighting expansion of this gene family across taxa. Teleost fishes exhibit diverse immune defense strategies and gene expansions of the major histocompatibility complex (MHC) I locus have occurred numerous times (25). Here a distinct class of immuno-regulatory genes in rockfish may have facilitated adaptations to extreme lifespan.
Life history transitions are tightly coupled to patterns of genetic diversity
To assess genetic diversity across the Sebastes clade we performed variant calling on samples mapped to each of their respective genome assemblies. We identified a more than 13-fold range in heterozygosity among Sebastes species (π = 2.9×10−4–3.8×10−3 per bp, Fig. 4A) and a 20-fold range when the outgroup taxa were also considered. These levels of diversity correspond to a range in effective population size (Ne) of 7.2×104–9.6×105 in the Sebastes clade assuming a mutation rate of 1e-9.
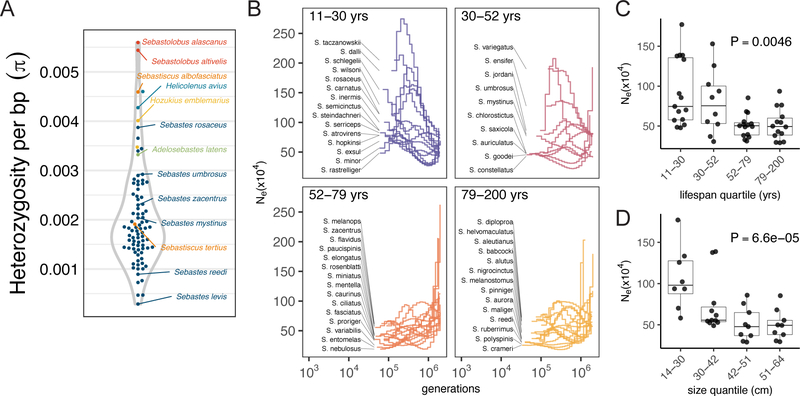
Figure 4 -. Life history transitions are associated with patterns of diversity:
A) Nucleotide diversity across 88 different species. B) MSMC based estimates of effective population sizes grouped by lifespan quartile over the last ~106 generations and averaged Ne grouped by lifespan quartile in C or grouped by size quartile in D. Box and whisker plots indicate the median and interquartile range (IQR) with tails at the min and maximum values within 1.5*IQR. P-values from PGLS linear model.
We applied multiple sequentially Markovian coalescent (26) (MSMC) analysis to dissect the demographic histories underlying these differences in diversity. We grouped MSMC trajectories by lifespan quartile and compared the mean Ne over the last 106 generations (Fig. 4B). While we observed a wide range of different demographic trajectories, short-lived species exhibited increases in Ne over the past ~106 generations. Note that the short-lived African killifish exhibits reduced lifespan linked to severe population bottlenecks (27). However, rockfish lifespan was significantly negatively correlated with Ne as a function of maximum lifespan (PGLS P=0.005, Fig. 4C).
Body mass is strongly positively correlated with both census population sizes and lifespans across terrestrial vertebrates (28, 29). In rockfish too, size is correlated with lifespan (Fig. 2E) and indeed size is a better predictor of Ne than lifespan (Fig. 4D, PGLS P=6.6e-5). These results suggest that life-history transitions to shortened life and smaller body size are potentially adaptive, in contrast to the drift-driven transitions observed in killifish. These results also underscore the inherent genetic vulnerability of larger long-lived marine fish species which are often the targets of commercial fisheries (30).
Shifts in lifespan reshape the mutational spectrum of segregating genetic variation
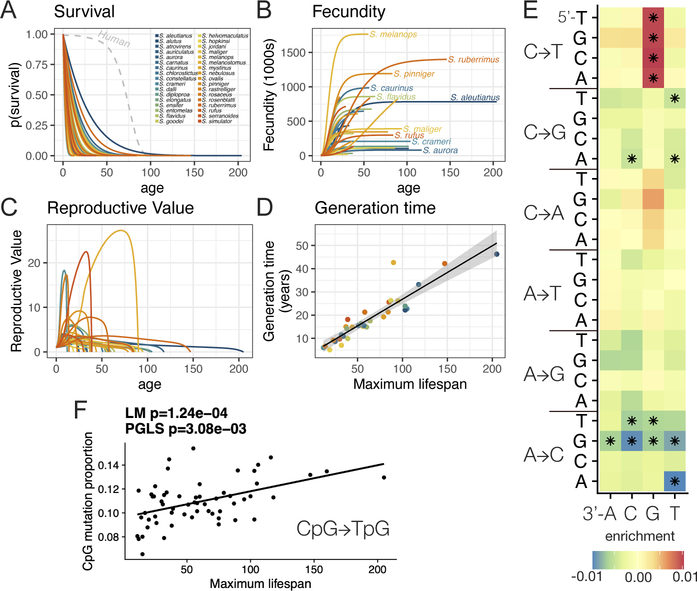
Long-lived, larger, terrestrial vertebrates with smaller Ne often exhibit low rates of fecundity, increased maturation times, and higher survival of offspring (31). Long-lived rockfish species also exhibit extremely long maturation times ranging to over a decade (3). We modelled survival, maturation, and fecundity in 34 different rockfish species for which detailed information could be ascertained to understand the evolutionary tradeoffs driving rockfish life histories (Fig. 5A–D, Tables S15–17). Similar to other marine fishes, rockfish exhibit a “type-III” survivorship curve with high mortality at young ages and very few individuals surviving to old age (32) (Fig. 5A). However, fecundity increases rapidly as a function of age, proportional to size (Fig. 5B) and thus aging imparts minimal declines in their reproductive values (Fig. 5C). In extreme cases such as in yelloweye rockfish (Sebastes ruberrimus), 150 year-old individuals can produce more than 1 million offspring per season. Even compared to other marine fishes, which exhibit non-linear scaling in reproductive output with size (33), older, larger rockfish have significantly higher reproductive output per unit weight (Fig. S20, Wilcoxon P=6.3e-6).
Figure 5 – Shifts in life history traits reshape the mutational spectrum of segregating genetic variation:
A) Survival curves for 34 rockfish species. Humans, shown for comparison, exhibit similar maximum lifespans to many rockfish species but different survival curves (Type I versus Type III survival). B) Rockfish fecundity (births per season) plotted as a function of age. C) Reproductive value plotted as a function of age. D) Generation times estimated from survival and fecundity plotted as a function of lifespan. E) Association of segregating single nucleotide variant mutation types with lifespan across all trinucleotide contexts. Asterisks indicate age-associated mutational profiles significant after multiple testing correction (q<0.05). F) The proportion of segregating CpG->TpG mutations as a function of maximum lifespan.
The disproportionate reproductive output of older fish results in generation times that span from 5–45 years across different species (Fig. 5D, Table S17). We find that these differences in generation time and lifespan are associated with reduced nucleotide substitution rates (resulting in shortened phylogenetic branch lengths) in longer-lived species, as observed in terrestrial mammals (Fig. S21) (34). Generation time can also have an influence on the mutational spectrum with certain classes of mutations more likely to occur in older parents (35, 36). We thus classified species-specific segregating single nucleotide variants (SNVs) by mutation type and tri-nucleotide context (Fig. S22). Correlations between mutation types and lifespan (Fig. 5E) were seen with enrichments of CpG->TpG mutations; longer-lived species exhibited a significantly increased proportion of CpG transitions in all *CG->*TG contexts (Fig. 5F, PGLS P=1×10−4). CpG->TpG transitions are characteristic of spontaneous methylated cytosine deamination, a mutational signature that occurs independent of DNA-replication (37) and is the dominant age-associated mutational profile of human tumors (38).
In humans a reduced proportion of CpG variants in European populations has been hypothesized to have been driven by shortened generation times (39), which, if due to similar processes in fish, may be consistent with our findings. A number of mutational signatures were also at reduced frequency across long-lived Rockfish species, including A->C and C->G transversions (Fig. 5E), however it is unclear what the underlying mutational mechanism of this signature might be. Together, these results indicate that shifts in life histories can in turn reshape patterns of segregating genetic diversity.
Discussion
The vast diversity of life histories in Pacific Ocean rockfishes presents an opportunity to dissect the genetic adaptations that shape life-history transitions in vertebrates. Our results highlight selective signatures in pathways underlying “hallmarks of aging” (4) that are conserved across all eukaryotes (e.g. DNA damage and nutrient sensing pathways) as well as in vertebrate-specific hallmarks such as immunity and inflammation (5). Chronic inflammation (“inflammaging”) in particular has emerged as a key therapeutic target in humans and our results highlight a novel gene family, the butyrophilins, which may play a role in modulating lifespan in rockfish. We also highlight that the genetic adaptations that enable extreme longevity in rockfish do so both directly, by influencing insulin signaling and other key pathways, as well as indirectly, by influencing size and adaptations to depth. Our results further highlight that such life-history transitions themselves reshape patterns of genetic diversity. Long-lived rockfish species exhibit reduced genetic diversity in contrast to short-lived species and the mutational spectrum of segregating genetic variation is also altered by lifespan. Indeed, the disproportionately higher reproductive output in older, larger rockfish, is likely directly linked to their extreme longevity. Our work further highlights the utility of genus-wide genome assembly efforts to answer questions that have thus far been limited to representative species from broad taxonomic groups (e.g. vertebrates). The advent of additional complete genome assemblies from vertebrates will further our understanding of the generality of our findings across evolutionary and physical scales.
Supplementary Material
Acknowledgements:
We acknowledge L. Smith and A. Bentley of the University of Kansas Biodiversity Institute and M. Miya at Chiba Natural History Museum and Institute for their assistance in sample acquisition. We thank NOAA’s Alaska Fisheries Science Center, RACE Division, for specimens collected during resource assessment surveys. Fish image credits: Kyoto University, Fish Collection; SWFSC ROV Team; R. R. Lauth, Alaska Fisheries Science Center; D.E. Stevenson, Alaska Fisheries Science Center; J.W. Orr; J. Nichols; C. Grossman; D. Karimoto; P. Ridings; K. Lee; M. Chamberlain; M. Guimaraes; S. Geitler. We would also like to thank M. Slatkin for helpful discussions;
Funding:
R35GM142916 to PHS;
Footnotes
Competing interests: The authors declare no competing interests.
Data and materials availability:
All sequencing data have been depositing in the European Nucleotide Archive under accession PRJEB42258. Complete methods are described in the supplementary materials (9) and all code required to recapitulate analyses, including BUSCO gene alignments and gene trees, has been posted at https://zenodo.org/record/5535007#.YVOk9y1h3wo (40).
References and Notes
- 1.Charlesworth B, Fisher, Medawar, Hamilton and the Evolution of Aging. Genetics. 156, 927–931 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Depczynski M, Bellwood DR, Shortest recorded vertebrate lifespan found in a coral reef fish. Current Biology. 15, R288–R289 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Nielsen J, Hedeholm RB, Heinemeier J, Bushnell PG, Christiansen JS, Olsen J, Ramsey CB, Brill RW, Simon M, Steffensen KF, Steffensen JF, Eye lens radiocarbon reveals centuries of longevity in the Greenland shark (Somniosus microcephalus). Science. 353, 702–704 (2016). [DOI] [PubMed] [Google Scholar]
- 4.López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G, The Hallmarks of Aging. Cell. 153, 1194–1217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh PP, Demmitt BA, Nath RD, Brunet A, The Genetics of Aging: A Vertebrate Perspective. Cell. 177, 200–220 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Love MS, Yoklavich M, Thorsteinson L, The Rockfishes of the Northeast Pacific (University of California Press, Berkeley, First American Edition edition., 2002). [Google Scholar]
- 7.Rabosky DL, Chang J, Title PO, Cowman PF, Sallan L, Friedman M, Kaschner K, Garilao C, Near TJ, Coll M, Alfaro ME, An inverse latitudinal gradient in speciation rate for marine fishes. Nature. 559, 392–395 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Seppey M, Manni M, Zdobnov EM, in Gene Prediction: Methods and Protocols, Kollmar M, Ed. (Springer, New York, NY, 2019; 10.1007/978-1-4939-9173-0_14), Methods in Molecular Biology, pp. 227–245. [DOI] [Google Scholar]
- 9. Materials and methods are available as supplementary materials.
- 10.Hyde JR, Vetter RD, The origin, evolution, and diversification of rockfishes of the genus Sebastes (Cuvier). Molecular Phylogenetics and Evolution. 44, 790–811 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Keane M, Semeiks J, Webb AE, Li YI, Quesada V, Craig T, Madsen LB, van Dam S, Brawand D, Marques PI, Michalak P, Kang L, Bhak J, Yim H-S, Grishin NV, Nielsen NH, Heide-Jørgensen MP, Oziolor EM, Matson CW, Church GM, Stuart GW, Patton JC, George JC, Suydam R, Larsen K, López-Otín C, O’Connell MJ, Bickham JW, Thomsen B, de Magalhães JP, Insights into the Evolution of Longevity from the Bowhead Whale Genome. Cell Reports. 10, 112–122 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quesada V, Freitas-Rodríguez S, Miller J, Pérez-Silva JG, Jiang Z-F, Tapia W, Santiago-Fernández O, Campos-Iglesias D, Kuderna LFK, Quinzin M, Álvarez MG, Carrero D, Beheregaray LB, Gibbs JP, Chiari Y, Glaberman S, Ciofi C, Araujo-Voces M, Mayoral P, Arango JR, Tamargo-Gómez I, Roiz-Valle D, Pascual-Torner M, Evans BR, Edwards DL, Garrick RC, Russello MA, Poulakakis N, Gaughran SJ, Rueda DO, Bretones G, Marquès-Bonet T, White KP, Caccone A, López-Otín C, Giant tortoise genomes provide insights into longevity and age-related disease. Nat Ecol Evol. 3, 87–95 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kowalczyk A, Meyer WK, Partha R, Mao W, Clark NL, Chikina M, RERconverge: an R package for associating evolutionary rates with convergent traits. Bioinformatics. 35, 4815–4817 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mangel M, Kindsvater HK, Bonsall MB, Evolutionary Analysis of Life Span, Competition, and Adaptive Radiation, Motivated by the Pacific Rockfishes (sebastes). Evolution. 61, 1208–1224 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Austad SN, Fischer KE, Mammalian Aging, Metabolism, and Ecology: Evidence From the Bats and Marsupials. Journal of Gerontology. 46, B47–B53 (1991). [DOI] [PubMed] [Google Scholar]
- 16.Umehara T, Kawai T, Kawashima I, Tanaka K, Okuda S, Kitasaka H, Richards JS, Shimada M, The acceleration of reproductive aging in Nrg1flox/flox;Cyp19-Cre female mice. Aging Cell. 16, 1288–1299 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng M, Tam JW, Wang L, Liang K, Li S, Zhang L, Guo H, Luo X, Zhang Y, Petrucelli A, Davis BK, Conti BJ, June Brickey W, Ko C-C, Lei YL, Sun S, Ting JP-Y, TRAF3IP3 negatively regulates cytosolic RNA induced anti-viral signaling by promoting TBK1 K48 ubiquitination. Nature Communications. 11, 2193 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tandon V, de la Vega L, Banerjee S, Emerging roles of DYRK2 in cancer. Journal of Biological Chemistry. 296 (2021) (available at https://www.jbc.org/article/S0021-9258(20)00348-8/abstract). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Promislow DE, On size and survival: progress and pitfalls in the allometry of life span. J Gerontol. 48, B115–123 (1993). [DOI] [PubMed] [Google Scholar]
- 20.Todesco M, Owens GL, Bercovich N, Légaré J-S, Soudi S, Burge DO, Huang K, Ostevik KL, Drummond EBM, Imerovski I, Lande K, Pascual-Robles MA, Nanavati M, Jahani M, Cheung W, Staton SE, Muños S, Nielsen R, Donovan LA, Burke JM, Yeaman S, Rieseberg LH, Massive haplotypes underlie ecotypic differentiation in sunflowers. Nature. 584, 602–607 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Sudmant PH, Mallick S, Nelson BJ, Hormozdiari F, Krumm N, Huddleston J, Coe BP, Baker C, Nordenfelt S, Bamshad M, Jorde LB, Posukh OL, Sahakyan H, Watkins WS, Yepiskoposyan L, Abdullah MS, Bravi CM, Capelli C, Hervig T, Wee JTS, Tyler-Smith C, van Driem G, Romero IG, Jha AR, Karachanak-Yankova S, Toncheva D, Comas D, Henn B, Kivisild T, Ruiz-Linares A, Sajantila A, Metspalu E, Parik J, Villems R, Starikovskaya EB, Ayodo G, Beall CM, Di Rienzo A, Hammer MF, Khusainova R, Khusnutdinova E, Klitz W, Winkler C, Labuda D, Metspalu M, Tishkoff SA, Dryomov S, Sukernik R, Patterson N, Reich D, Eichler EE, Global diversity, population stratification, and selection of human copy-number variation. Science. 349, aab3761 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arnett HA, Viney JL, Immune modulation by butyrophilins. Nature Reviews Immunology. 14, 559–569 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Valentonyte R, Hampe J, Huse K, Rosenstiel P, Albrecht M, Stenzel A, Nagy M, Gaede KI, Franke A, Haesler R, Koch A, Lengauer T, Seegert D, Reiling N, Ehlers S, Schwinger E, Platzer M, Krawczak M, Müller-Quernheim J, Schürmann M, Schreiber S, Sarcoidosis is associated with a truncating splice site mutation in BTNL2. Nature Genetics. 37, 357–364 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Wright KM, Rand KA, Kermany A, Noto K, Curtis D, Garrigan D, Slinkov D, Dorfman I, Granka JM, Byrnes J, Myres N, Ball CA, Ruby JG, A Prospective Analysis of Genetic Variants Associated with Human Lifespan. G3: Genes, Genomes, Genetics. 9, 2863–2878 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malmstrøm M, Matschiner M, Tørresen OK, Star B, Snipen LG, Hansen TF, Baalsrud HT, Nederbragt AJ, Hanel R, Salzburger W, Stenseth NC, Jakobsen KS, Jentoft S, Evolution of the immune system influences speciation rates in teleost fishes. Nature Genetics. 48, 1204–1210 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Schiffels S, Durbin R, Inferring human population size and separation history from multiple genome sequences. Nat Genet. 46, 919–925 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cui R, Medeiros T, Willemsen D, Iasi LNM, Collier GE, Graef M, Reichard M, Valenzano DR, Relaxed Selection Limits Lifespan by Increasing Mutation Load. Cell. 178, 385–399.e20 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Nikolaev SI, Montoya-Burgos JI, Popadin K, Parand L, Margulies EH, of H NI. I. S. C. C. S. Program, Antonarakis SE, Life-history traits drive the evolutionary rates of mammalian coding and noncoding genomic elements. PNAS. 104, 20443–20448 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Popadin K, Polishchuk LV, Mamirova L, Knorre D, Gunbin K, Accumulation of slightly deleterious mutations in mitochondrial protein-coding genes of large versus small mammals. PNAS. 104, 13390–13395 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rolland J, Schluter D, Romiguier J, Vulnerability to Fishing and Life History Traits Correlate with the Load of Deleterious Mutations in Teleosts. Mol Biol Evol. 37, 2192–2196 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pianka ER, On r- and K-Selection. The American Naturalist. 104, 592–597 (1970). [Google Scholar]
- 32.Deevey ES, Life Tables for Natural Populations of Animals. The Quarterly Review of Biology. 22, 283–314 (1947). [DOI] [PubMed] [Google Scholar]
- 33.Barneche DR, Robertson DR, White CR, Marshall DJ, Fish reproductive-energy output increases disproportionately with body size. Science. 360, 642–645 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Sayres MAW, Venditti C, Pagel M, Makova KD, Do Variations in Substitution Rates and Male Mutation Bias Correlate with Life-History Traits? A Study of 32 Mammalian Genomes. Evolution. 65, 2800–2815 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Martin AP, Palumbi SR, Body size, metabolic rate, generation time, and the molecular clock. PNAS. 90, 4087–4091 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jónsson H, Sulem P, Kehr B, Kristmundsdottir S, Zink F, Hjartarson E, Hardarson MT, Hjorleifsson KE, Eggertsson HP, Gudjonsson SA, Ward LD, Arnadottir GA, Helgason EA, Helgason H, Gylfason A, Jonasdottir A, Jonasdottir A, Rafnar T, Frigge M, Stacey SN, Magnusson OT, Thorsteinsdottir U, Masson G, Kong A, Halldorsson BV, Helgason A, Gudbjartsson DF, Stefansson K, Parental influence on human germline de novo mutations in 1,548 trios from Iceland. Nature. 549, 519–522 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Ségurel L, Wyman MJ, Przeworski M, Determinants of Mutation Rate Variation in the Human Germline. Annu. Rev. Genom. Hum. Genet. 15, 47–70 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, Boutselakis H, Cole CG, Creatore C, Dawson E, Fish P, Harsha B, Hathaway C, Jupe SC, Kok CY, Noble K, Ponting L, Ramshaw CC, Rye CE, Speedy HE, Stefancsik R, Thompson SL, Wang S, Ward S, Campbell PJ, Forbes SA, COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 47, D941–D947 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harris K, Pritchard JK, Rapid evolution of the human mutation spectrum. eLife. 6, e24284 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DOI: 10.5281/zenodo.5535007. [DOI]
- 41.Love MS, Certainly More Than You Want to Know about the Fishes of the Pacific Coast: A Postmodern Experience (Really Big Press, 2011). [Google Scholar]
- 42.Ishimaru T, Tateda Y, Tsumune D, Aoyama M, Hamajima Y, Kasamatsu N, Yamada M, Yoshimura T, Mizuno T, Kanda J, Mechanisms of radiocesium depuration in Sebastes cheni derived by simulation analysis of measured 137Cs concentrations off southern Fukushima 2014–2016. Journal of Environmental Radioactivity. 203, 200–209 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Love MS, McCrea M, Kui L, Aspects of the Life Histories of Pinkrose Rockfish (Sebastes simulator) and Swordspine Rockfish (Sebastes ensifer) with Notes on the Subgenus Sebastomus. soca. 117, 64–76 (2018). [Google Scholar]
- 44.Harris JP, Hutchinson C, Wildes S, Using otolith morphometric analysis to improve species discrimination of blackspotted rockfish (Sebastes melanostictus) and rougheye rockfish (S. aleutianus). FB. 117, 234–244 (2019). [Google Scholar]
- 45.Fields R, Spatial and Temporal Variation in Rosy Rockfish (Sebastes Rosaceus) Life History Traits. Capstone Projects and Master’s Theses. 562 (2016) (available at https://digitalcommons.csumb.edu/caps_thes/562). [Google Scholar]
- 46.Bracewell R, Chatla K, Nalley MJ, Bachtrog D, Dynamic turnover of centromeres drives karyotype evolution in Drosophila. eLife. 8, e49002 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramani V, Cusanovich DA, Hause RJ, Ma W, Qiu R, Deng X, Blau CA, Disteche CM, Noble WS, Shendure J, Duan Z, Mapping 3D genome architecture through in situ DNase Hi-C. Nature Protocols. 11, 2104–2121 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meng G, Li Y, Yang C, Liu S, MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Research. 47, e63–e63 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chin C-S, Peluso P, Sedlazeck FJ, Nattestad M, Concepcion GT, Clum A, Dunn C, O’Malley R, Figueroa-Balderas R, Morales-Cruz A, Cramer GR, Delledonne M, Luo C, Ecker JR, Cantu D, Rank DR, Schatz MC, Phased diploid genome assembly with single-molecule real-time sequencing. Nat Methods. 13, 1050–1054 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM, Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruan J, Li H, Fast and accurate long-read assembly with wtdbg2. Nature Methods. 17, 155–158 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vaser R, Sović I, Nagarajan N, Šikić M, Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 27, 737–746 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guan D, McCarthy SA, Wood J, Howe K, Wang Y, Durbin R, Identifying and removing haplotypic duplication in primary genome assemblies. Bioinformatics. 36, 2896–2898 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ghurye J, Rhie A, Walenz BP, Schmitt A, Selvaraj S, Pop M, Phillippy AM, Koren S, Integrating Hi-C links with assembly graphs for chromosome-scale assembly. PLOS Computational Biology. 15, e1007273 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dudchenko O, Batra SS, Omer AD, Nyquist SK, Hoeger M, Durand NC, Shamim MS, Machol I, Lander ES, Aiden AP, Aiden EL, De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science. 356, 92–95 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Durand NC, Robinson JT, Shamim MS, Machol I, Mesirov JP, Lander ES, Aiden EL, Juicebox Provides a Visualization System for Hi-C Contact Maps with Unlimited Zoom. Cell Systems. 3, 99–101 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rhie A, Walenz BP, Koren S, Phillippy AM, Merqury: reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biology. 21, 245 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Waterhouse RM, Seppey M, Simão FA, Manni M, Ioannidis P, Klioutchnikov G, Kriventseva EV, Zdobnov EM, BUSCO Applications from Quality Assessments to Gene Prediction and Phylogenomics. Mol Biol Evol. 35, 543–548 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zimin AV, Marçais G, Puiu D, Roberts M, Salzberg SL, Yorke JA, The MaSuRCA genome assembler. Bioinformatics. 29, 2669–2677 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR, STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Iwata H, Gotoh O, Benchmarking spliced alignment programs including Spaln2, an extended version of Spaln that incorporates additional species-specific features. Nucleic Acids Res. 40, e161 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hoff K, Lomsadze A, Borodovsky M, Stanke M, Whole-Genome Annotation with BRAKER. Methods Mol Biol. 1962, 65–95 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lechner M, Findeiß S, Steiner L, Marz M, Stadler PF, Prohaska SJ, Proteinortho: Detection of (Co-)orthologs in large-scale analysis. BMC Bioinformatics. 12, 124 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Buchfink B, Xie C, Huson DH, Fast and sensitive protein alignment using DIAMOND. Nat Methods. 12, 59–60 (2015). [DOI] [PubMed] [Google Scholar]
- 65.Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R, IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol Biol Evol. 37, 1530–1534 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mai U, Mirarab S, TreeShrink: fast and accurate detection of outlier long branches in collections of phylogenetic trees. BMC Genomics. 19, 272 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Flouri T, Jiao X, Rannala B, Yang Z, Species Tree Inference with BPP Using Genomic Sequences and the Multispecies Coalescent. Molecular Biology and Evolution. 35, 2585–2593 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Revell LJ, phytools: an R package for phylogenetic comparative biology (and other things). Methods in Ecology and Evolution. 3, 217–223 (2012). [Google Scholar]
- 69.Smith MD, Wertheim JO, Weaver S, Murrell B, Scheffler K, Kosakovsky Pond S. L., Less Is More: An Adaptive Branch-Site Random Effects Model for Efficient Detection of Episodic Diversifying Selection. Molecular Biology and Evolution. 32, 1342–1353 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yu G, Wang L-G, Han Y, He Q-Y, clusterProfiler: an R Package for Comparing Biological Themes Among Gene Clusters. OMICS: A Journal of Integrative Biology. 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wertheim JO, Murrell B, Smith MD, Kosakovsky Pond SL, Scheffler K, RELAX: Detecting Relaxed Selection in a Phylogenetic Framework. Mol Biol Evol. 32, 820–832 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hua X, Cowman P, Warren D, Bromham L, Longevity Is Linked to Mitochondrial Mutation Rates in Rockfish: A Test Using Poisson Regression. Mol Biol Evol. 32, 2633–2645 (2015). [DOI] [PubMed] [Google Scholar]
- 73.Zhou X, Dou Q, Fan G, Zhang Q, Sanderford M, Kaya A, Johnson J, Karlsson EK, Tian X, Mikhalchenko A, Kumar S, Seluanov A, Zhang ZD, Gorbunova V, Liu X, Gladyshev VN, Beaver and Naked Mole Rat Genomes Reveal Common Paths to Longevity. Cell Reports. 32, 107949 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim EB, Fang X, Fushan AA, Huang Z, Lobanov AV, Han L, Marino SM, Sun X, Turanov AA, Yang P, Yim SH, Zhao X, Kasaikina MV, Stoletzki N, Peng C, Polak P, Xiong Z, Kiezun A, Zhu Y, Chen Y, Kryukov GV, Zhang Q, Peshkin L, Yang L, Bronson RT, Buffenstein R, Wang B, Han C, Li Q, Chen L, Zhao W, Sunyaev SR, Park TJ, Zhang G, Wang J, Gladyshev VN, Genome sequencing reveals insights into physiology and longevity of the naked mole rat. Nature. 479, 223–227 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Keane M, Semeiks J, Webb AE, Li YI, Quesada V, Craig T, Madsen LB, van Dam S, Brawand D, Marques PI, Michalak P, Kang L, Bhak J, Yim H-S, Grishin NV, Nielsen NH, Heide-Jørgensen MP, Oziolor EM, Matson CW, Church GM, Stuart GW, Patton JC, George JC, Suydam R, Larsen K, López-Otín C, O’Connell MJ, Bickham JW, Thomsen B, de Magalhães JP, Insights into the Evolution of Longevity from the Bowhead Whale Genome. Cell Rep. 10, 112–122 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Quesada V, Freitas-Rodríguez S, Miller J, Pérez-Silva JG, Jiang Z-F, Tapia W, Santiago-Fernández O, Campos-Iglesias D, Kuderna LFK, Quinzin M, Álvarez MG, Carrero D, Beheregaray LB, Gibbs JP, Chiari Y, Glaberman S, Ciofi C, Araujo-Voces M, Mayoral P, Arango JR, Tamargo-Gómez I, Roiz-Valle D, Pascual-Torner M, Evans BR, Edwards DL, Garrick RC, Russello MA, Poulakakis N, Gaughran SJ, Rueda DO, Bretones G, Marquès-Bonet T, White KP, Caccone A, López-Otín C, Giant tortoise genomes provide insights into longevity and age-related disease. Nature Ecology & Evolution. 3, 87–95 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sedlazeck FJ, Rescheneder P, von Haeseler A, NextGenMap: fast and accurate read mapping in highly polymorphic genomes. Bioinformatics. 29, 2790–2791 (2013). [DOI] [PubMed] [Google Scholar]
- 78.Jones P, Binns D, Chang H-Y, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, Pesseat S, Quinn AF, Sangrador-Vegas A, Scheremetjew M, Yong S-Y, Lopez R, Hunter S, InterProScan 5: genome-scale protein function classification. Bioinformatics. 30, 1236–1240 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Numanagić I, Gökkaya AS, Zhang L, Berger B, Alkan C, Hach F, Fast characterization of segmental duplications in genome assemblies. Bioinformatics. 34, i706–i714 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mendes FK, Vanderpool D, Fulton B, Hahn MW, CAFE 5 models variation in evolutionary rates among gene families. Bioinformatics. 36, 5516–5518 (2020). [DOI] [PubMed] [Google Scholar]
- 81.Pockrandt C, Alzamel M, Iliopoulos CS, Reinert K, GenMap: ultra-fast computation of genome mappability. Bioinformatics. 36, 3687–3692 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schiffels S, Durbin R, Inferring human population size and separation history from multiple genome sequences. Nat Genet. 46, 919–925 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Finch C, Pike M, Witten M, Slow mortality rate accelerations during aging in some animals approximate that of humans. Science. 249, 902–905 (1990). [DOI] [PubMed] [Google Scholar]
- 84.Then AY, Hoenig JM, Hall NG, Hewitt DA, Evaluating the predictive performance of empirical estimators of natural mortality rate using information on over 200 fish species. ICES J Mar Sci. 72, 82–92 (2015). [Google Scholar]
- 85.Dick EJ, Beyer S, Mangel M, Ralston S, A meta-analysis of fecundity in rockfishes (genus Sebastes). Fisheries Research. 187, 73–85 (2017). [Google Scholar]
- 86.Felsenstein J, Inbreeding and Variance Effective Numbers in Populations with Overlapping Generations. Genetics. 68, 581–597 (1971). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bonsall MB, Longevity and ageing: appraising the evolutionary consequences of growing old. Philosophical Transactions of the Royal Society B: Biological Sciences. 361, 119–135 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data have been depositing in the European Nucleotide Archive under accession PRJEB42258. Complete methods are described in the supplementary materials (9) and all code required to recapitulate analyses, including BUSCO gene alignments and gene trees, has been posted at https://zenodo.org/record/5535007#.YVOk9y1h3wo (40).