Abstract
DNA sequencing data showed that five clinical isolates of Escherichia coli with reduced susceptibility to ceftazidime, ceftriaxone, and cefotaxime contain an ampC gene that is preceded by a strong promoter. Transcription from the strong promoter was 8- to 18-fold higher than that from the promoter from a susceptible isolate. RNA studies showed that mRNA stability does not play a role in the control of AmpC synthesis.
The chromosomal cephalosporinase gene, ampC, of Escherichia coli is regulated by a weak promoter (18) and a transcriptional attenuator (13). Strains carrying the wild-type gene produce low basal amounts of AmpC and are inherently susceptible to ampicillin (15). Occasionally, however, this enzyme is overproduced in E. coli and these strains are resistant to cefoxitin and have reduced susceptibilities to the newer β-lactams such as oxyiminocephalosporins (2, 3, 10, 11, 15). Studies on the molecular basis of AmpC overproduction in E. coli have shown that some hyperproducers contain more than one copy of ampC (4, 5), while others contain mutations in the regulatory region and/or attenuator of ampC, resulting in more efficient transcription of the structural gene (13). Acquisition of a stronger promoter from Shigella spp. (13) or insertion of an insertion element containing promoter sequences (14) has also been proposed as the molecular basis of hyperproduction of AmpC in some E. coli strains.
During 1997, five nonrepetitive clinical isolates of E. coli, designated E1, E2, E3, E4, and E5, with antibiotic resistance profiles suggestive of the hyperproduction of AmpC were isolated from patients in hospitals in Cape Town, South Africa. Strain E1 was isolated from a patient in Victoria Hospital, Cape Town, and the remaining strains were from Groote Schuur Hospital, Cape Town. Two of the strains were isolated from urine, two were from wounds, and one was from an abscess. Isolation of the strains was sporadic, suggesting that they were not related to an outbreak. However, the isolates were not typed. Antibiotic disk susceptibility testing using National Committee for Clinical Laboratory Standards criteria (16, 17) showed that E1, E2, E3, E4, and E5 were resistant to cefoxitin and cefuroxime. Only one of the isolates (E2) showed reduced susceptibility to cefotaxime; the remainder were susceptible to this antibiotic. During the same period, a β-lactam-susceptible E. coli strain (E6) was isolated, and it was used as a susceptible isolate for comparisons. The MICs of ceftazidime, cefuroxime, ceftriaxone, and cefotaxime were determined by using E-strips (AB Biodisk, Solna, Sweden) and are presented in Table 1. Each of the antibiotics was considerably less active against E1, E2, E3, E4, and E5 than against the susceptible isolate, E6. The β-lactamase content of the E. coli strains was not investigated.
TABLE 1.
MICs for E. coli E1 to E6
| Strain | MIC (μg/ml) of:
|
|||
|---|---|---|---|---|
| Ceftazidime | Cefuroxime | Ceftriaxone | Cefotaxime | |
| E1 | 32 | >256 | 3 | 3 |
| E2 | 6 | >256 | 2 | 3 |
| E3 | 24 | >256 | 4 | 4 |
| E4 | 32 | >256 | 4 | 4 |
| E5 | 16 | >128 | 1.5 | 2 |
| E6 | 0.25 | 4 | 0.0047 | 0.0047 |
DNA sequence analysis of the regulatory regions of ampC genes.
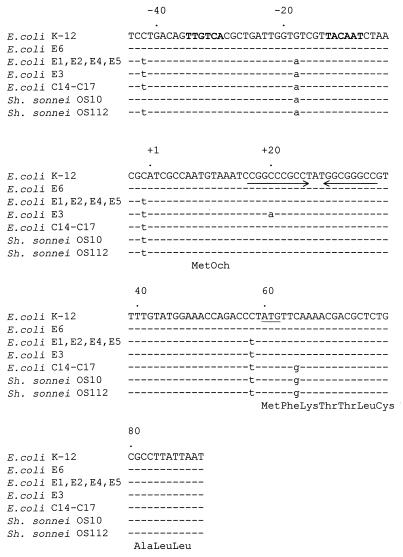
By using primers, 5′-CTACGGTCTGGCTGCTA-3′ and 5′-TGGAGCAAGAGGCGGTA-3′, which anneal to nucleotides −61 to −45 and +92 to +108 (12), respectively, of the region 5′ of the structural ampC gene, the regulatory region of ampC was amplified by PCR and sequenced directly. The sequences corresponding to nucleotides −44 to +91 (12) from E1, E2, E3, E4, E5, and E6 are shown in Fig. 1. The sequences from the resistant strains (E1, E2, E3, E4, and E5) contain a T at −42, a G at −18, a T at −1, and a T at +58, whereas C, A, T, and C are present in the corresponding positions in the sequence from susceptible strain E6. In addition, E3 contains a mutation in the attenuator at nucleotide +20. The mutation at −42 changes a C to a T, creating a hexamer with perfect homology to the consensus −35 sequence (TTGACA) recognized by the major RNA polymerase (Eς70) in E. coli (9). The mutation at −18 (G→A) generates a consensus −10 hexamer (TATCGT) that is separated from the novel −35 hexamer by 17 bp, creating a strong promoter in the ampC regulatory region. In this respect, the promoters of the resistant strains are the same as those of clinical isolates of E. coli (C11 and C14 to C17) and Shigella sonnei that over express AmpC (18). It was previously suggested that the E. coli strains had acquired the regulatory sequences from S. sonnei, generating a small E. coli subpopulation that contains the ampC regulatory region of Shigella spp., which have the potential to mutate to high levels of ampicillin resistance (18). Although E. coli does not readily mutate to high-level ampicillin resistance (18), the possibility that the mutations identified in the strains in this study resulted from antibiotic selective pressure, and not from the acquisition of sequences from Shigella spp., cannot be excluded. In this regard, the susceptible strain (E6), like E. coli K-12 (18), contains a C in the wobble position of the second amino acid of the signal sequence (+65). The E. coli hyperproducers in this study also contain a C at this position (Fig. 1). However, a G is present at the corresponding position in the sequences from the E. coli hyperproducers (C11 and C14 to 17), the S. sonnei hyperproducers, and wild-type S. sonnei (18).
FIG. 1.
Comparison of DNA sequences (135 bp) of the ampC control region and signal peptide from susceptible and AmpC-hyperproducing E. coli and Shigella strains. The sequences from E. coli K-12, hyperproducing strains C14 to C17, susceptible S. sonnei OS10, and hyperproducing S. sonnei OS112 are from reference 13. There is a dot above the sequence every 20 nucleotides. Identical nucleotides are indicated by dashes. Nucleotides that differ from the sequences from the susceptible E. coli strains (K-12 and E6) are in lowercase. The prototype −35 and −10 regions are in boldface lettering, and the attenuator is indicated by the arrows. The initiation codon is underlined, and the three-letter amino acid code is directly below the partial sequence of the signal peptide.
It is interesting to speculate on the role of the other mutations (C→T at −1 and +58, respectively) in the increased transcription of ampC. It has been shown that DNA sequences downstream of pause sites influence the pause half-life of RNA polymerase (8) and that a bound ribosome prevents the formation of stem-loop structures (19). It may be that the C→T mutations at −1 and +58, together with the attenuator, cause the RNA polymerase to pause, allowing the ribosome to stay close behind, thereby negating the formation of the attenuator stem and loop and facilitating transcriptional readthrough. Identical mutations were observed in the corresponding sequences from the E. coli hyperproducers C11 and C14 to C17 (18). Interestingly, the sequence from E. coli C13 does not contain the mutations at −1 and +58; rather, a mutation in the attenuator, which resulted in reduction of the thermodynamic strength of the formation of the stem-loop structure, was identified in this strain (18).
Expression of ampC.
To study further the expression from the strong promoters, the regulatory regions (nucleotides −61 to +59) from E1, E2, E3, E4, E5, and E6 were amplified by using primers 5′-TCCGAATTCCTACGGTCTGGCTGCTA-3′ and 5′-TTTGGATCCAGGGTCTGGTTTCCAT-3′, where the underscores represent EcoRI and BamHI restriction sites, respectively. These primers correspond to nucleotides −61 to −45 and +44 to +59 in the region upstream of ampC (12). PCR products were purified by silica gel membrane spin columns (Qiagen), digested with EcoRI and BamHI, and cloned into similarly restricted pUC19 (20) in the orientation opposite to that of the lac promoter. Subsequently, a BamHI-SalI fragment encoding luciferase was restricted from pGEM-luc (Promega) and cloned downstream of the promoters previously ligated into pUC19. The constructs (pLucE1, pLucE2, pLucE3, pLucE4, pLucE5, and pLucE6) were expressed in E. coli DH5α, and luciferase activity was measured. Cultures of E. coli were grown to mid-log phase (optical density at 600 nm, 0.7 to 0.8). One milliliter of cells was harvested, resuspended in 100 μl of 25 mM Tris-HCl (pH 7.8)–2 mM EDTA–10% glycerol–1% Triton X-100, sonicated in a Branson water bath sonicator for 20 s, and placed on ice for 20 s. Sonication and cooling were repeated, after which, the lysate was centrifuged for 5 min at 14,000 × g. The supernatant was collected, and the protein concentration was determined by using a Bio-Rad DC protein assay. Luciferase activity was measured in a BioOrbit 1253 luminometer after 100 μl of luciferin (Promega) had been mixed with 20 μl of cell extract.
Constructs containing the regulatory sequences from the hyperproducers (pLucE1, pLucE2, pLucE3, pLucE4, and pLucE5) resulted in 8- to 18-fold increases in luciferase activity, compared to that obtained with a pLuc construct (pLucE6) containing the regulatory sequences from the susceptible isolate.
DNA-RNA hybridizations.
To study the stability of the ampC transcript, the degradation of the mRNA species after the addition of rifampin was examined. Total RNA was extracted (7), and 15 μg was electrophoresed in 1.2% agarose–0.66 M formaldehyde with 40 mM morpholinepropanesulfonic acid–10 mM sodium acetate–1 mM EDTA (pH 7.7) and transferred to Hybond N+ (Amersham International) in 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Primers 5′-TACTGGCGTGCTTGGTG-3′ and 5′GACTCTCGCTGGATTGG-3′, corresponding to nucleotides +314 to +330 and +1141 to +1157, respectively, were used to amplify an 844-bp internal portion of ampC (12). The amplicon was purified by using a Qiagen spin column and labelled with [α-32P]dCTP using Ready-To-Go DNA Labelling Beads (Pharmacia Biotech). Similarly, an internal fragment of the 16S rRNA gene of E. coli was amplified by using the universal primers (6) and labelled. The prehybridization and hybridization procedures used and the conditions used for posthybridization washes were those recommended by the manufacturer of Hybond.
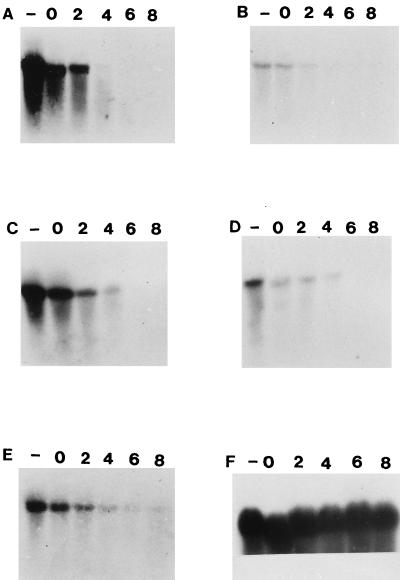
When the membranes were probed with an internal portion of ampC, no transcripts were detected in the susceptible isolate, E6; however, a strong signal with the 16S rRNA probe was obtained with RNA from this strain (Fig. 2), indicating that RNA had been transferred to the membrane. The ampC probe hybridized to a transcript of the expected size (1.2 kb) in each of the resistant strains (Fig. 2), signifying an increase in the rate of transcription of ampC in the hyperproducers. The signal obtained with RNAs from E1, E3, and E4 was more intense than the signal obtained with the RNAs from E2 and E5, suggesting a greater abundance of ampC transcripts in the total RNAs isolated from E1, E3, and E5. However, this did not affect the level of β-lactam resistance. The amounts of transcripts began to decrease immediately after the addition of rifampin. Four minutes after the addition of this antibiotic, the signal was barely visible (Fig. 2), indicating that the half-life of the ampC transcript was typical of E. coli messages (1). These studies show that mRNA stability is not responsible for the increase in AmpC, implying that the gene is transcriptionally regulated.
FIG. 2.
Hybridization of the Northern blots prepared from RNAs extracted from E. coli E1 to E6 and probed with either the ampC or the 16S rRNA probe. The strains were cultured until exponential phase. At this point (lane −), a 10-ml aliquot was removed and RNA was extracted. Rifampin (0.2 mg/ml) was added, a 10-ml aliquot was withdrawn immediately (lane 0), and RNA was extracted. Additional aliquots were removed after 2, 4, 6, and 8 min, and RNA was extracted. Panels: A, RNA from E1 probed with the ampC probe; B, RNA from E2 probed with the ampC probe; C, RNA from E3 probed with the ampC probe; D, RNA from E4 probed with the ampC probe; E, RNA from E5 probed with the ampC probe; F, RNA from E6 probed with 16S rRNA probe.
In conclusion, we have shown that the mechanism of cephalosporin resistance in five clinical isolates of E. coli from hospitals in Cape Town, South Africa, is due to increased transcription of ampC.
Nucleotide sequence accession numbers.
The sequences from E1 to E5 have been deposited in GenBank under accession no. BankIt247416 AF119769, BankIt247443 AF119770, BankIt247445 AF119771, BankIt247446 AF119772, and BankIt247447 AF119773, respectively.
Acknowledgments
This work was supported by grants from the University of Cape Town, the Foundation for Research and Development (FRD), and the Medical Research Council to B.G.E., and E.C.N. is the recipient of an FRD bursary.
REFERENCES
- 1.Belasco J G, Nilsson G, von Gabain A, Cohen S N. The stability of E. coli gene transcripts is dependent on determinants localised to specific mRNA segments. Cell. 1986;46:245–251. doi: 10.1016/0092-8674(86)90741-5. [DOI] [PubMed] [Google Scholar]
- 2.Bergström S, Normark S. β-Lactam resistance in clinical isolates of Escherichia coli caused by elevated production of the ampC-mediated chromosomal β-lactamase. Antimicrob Agents Chemother. 1979;16:427–433. doi: 10.1128/aac.16.4.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cooksey R, Swenson J, Clark N, Gay E, Thornsberry C. Patterns and mechanisms of β-lactam resistance among isolates of Escherichia coli from hospitals in the United States. Antimicrob Agents Chemother. 1990;34:739–745. doi: 10.1128/aac.34.5.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edlund T, Grundström T, Normark S. Isolation and characterisation of DNA repetitions carrying the chromosomal β-lactamase gene of Escherichia coli K-12. Mol Gen Genet. 1979;173:115–125. doi: 10.1007/BF00330301. [DOI] [PubMed] [Google Scholar]
- 5.Edlund T, Normark S. Recombination between short DNA homologies causes tandem duplication. Nature. 1981;292:269–271. doi: 10.1038/292269a0. [DOI] [PubMed] [Google Scholar]
- 6.Edwards U, Rogall T, Blöcker H, Emde M, Böttger E. Isolation and direct complete nucleotide determination of entire genes. Characterization of gene coding for 16S ribosomal RNA. Nucleic Acids Res. 1989;17:7843–7853. doi: 10.1093/nar/17.19.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elisha B G, Steyn L M. Cloning of AAC(3) and AAD(2") genes from Acinetobacter: differential expression in the host strain. Curr Microbiol. 1991;22:259–263. [Google Scholar]
- 8.Feng G, Lee D L, Wang D, Chan C L, Landick R. GreA-induced transcript cleavage in transcription complexes containing Escherichia coli RNA polymerase is controlled by multiple factors, including nascent transcription and structure. J Biol Chem. 1994;269:22282–22294. [PubMed] [Google Scholar]
- 9.Hawley D K, McClure W R. Compilation and analysis of Escherichia coli promoter sequences. Nucleic Acids Res. 1983;11:2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacoby G A, Sutton L. β-Lactamases and β-lactam resistance in Escherichia coli. Antimicrob Agents Chemother. 1985;28:703–705. doi: 10.1128/aac.28.5.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jaffé A, Chabbert Y A, Derlot E. Selection and characterization of β-lactam-resistant Escherichia coli K-12 mutants. Antimicrob Agents Chemother. 1983;23:622–625. doi: 10.1128/aac.23.4.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jaurin B, Grundström T. ampC cephalosporinase of Escherichia coli K-12 has a different evolutionary origin from that of β-lactamase of the penicillinase type. Proc Natl Acad Sci USA. 1981;78:4897–4901. doi: 10.1073/pnas.78.8.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaurin B, Grundström T, Edlund T, Normark S. The E. coli β-lactamase attenuator mediates growth rate-dependent regulation. Nature. 1981;290:221–225. doi: 10.1038/290221a0. [DOI] [PubMed] [Google Scholar]
- 14.Jaurin B, Normark S. Insertion of IS2 creates a novel ampC promoter in Escherichia coli. Cell. 1983;32:809–816. doi: 10.1016/0092-8674(83)90067-3. [DOI] [PubMed] [Google Scholar]
- 15.Livermore D M. β-Lactamases in laboratory and clinical resistance. Clin Microbiol Rev. 1995;8:557–584. doi: 10.1128/cmr.8.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.National Committee for Clinical Laboratory Standards. Methods for dilution of antimicrobial susceptibility tests for bacteria that grow aerobically. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1998. [Google Scholar]
- 17.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial disk susceptibility tests. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1998. [Google Scholar]
- 18.Olsson O, Bergström S, Lindberg F P, Normark S. ampC β-lactamase hyperproduction in Escherichia coli: natural ampicillin resistance generated by horizontal chromosomal DNA transfer from Shigella. Proc Natl Acad Sci USA. 1983;80:7556–7560. doi: 10.1073/pnas.80.24.7556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Platt T. Transcription termination and the regulation of gene expression. Annu Rev Biochem. 1986;55:339–372. doi: 10.1146/annurev.bi.55.070186.002011. [DOI] [PubMed] [Google Scholar]
- 20.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]