SUMMARY
Inflammatory bowel diseases (IBD) and colorectal cancer (CRC) are heterogeneous intestinal diseases that threaten the health of an increasing number of individuals as their lifestyles become Westernized. New insights have been discovered with the development of various omics techniques, revealing that gut microbiota-derived metabolites play important roles in maintaining intestinal homeostasis and modulating the progression of intestinal diseases from both metabolic and immunological perspectives. Clinical metagenomic and metabolomic studies revealed a link between microbial bile acid (BA) metabolism, IBD and CRC progression. Several BA-derived metabolites have recently been demonstrated to play a role in intestinal immunity, providing fresh insights into how BAs affect the course of IBD and CRC. In this review, recent studies on the involvement of gut microbiota-derived BAs in intestinal immunity, inflammation and tumorigenesis are discussed, together with human omics data to provide prospective insights into future prevention and therapy of IBD and CRC.
Keywords: gut microbiota, bile acids, inflammatory bowel diseases, colorectal cancer, omics
In Brief
Cai et al. reviews recent studies that reveal novel mechanistic links between gut microbiota-derived bile acids and host immunity, intestinal inflammation, and tumorigenesis. Bile acid-related human omics data are summarized and bile acid-targeted therapeutics for the prevention and treatment of inflammatory bowel disease and colorectal cancer are discussed.
INTRODUCTION
The entire human gut microbiota system is now considered as a “microbial organ” that contributes to a variety of physiological processes including the metabolic function of fermenting non-digestible dietary fiber, the anaerobic metabolism of peptides and proteins, protective activity against pathogens, and even immune-system modulation (Guarner and Malagelada, 2003, Rooks and Garrett, 2016). The host and gut microbiota interact and produce a wide range of metabolites or components either from anaerobic fermentation of exogenous undigested dietary components, such as short-chain fatty acids (SCFAs), or endogenous substances generated by microorganisms and the host, such as bile acid (BA) metabolites (Jia et al., 2018). Studies in humans and mice have demonstrated that BAs, especially secondary BAs (SBAs) derived from bacterial metabolism of BAs produced in the liver, modulate intestinal inflammation (Schirmer et al., 2019, Lavelle and Sokol, 2020) and tumorigenesis (O’keefe, 2016, Wang et al., 2019). Of note, several studies recently defined the immunomodulatory properties of BAs in the intestine (Li et al., 2021, Song et al., 2020, Campbell et al., 2020). Crosstalk between gut microbiota, BAs, and the host, influence immunological functions, metabolic phenotypes, and risk factors for numerous diseases including obesity, diabetes, non-alcoholic fatty liver disease (NAFLD), inflammatory bowel disease (IBD) and various cancers (Schroeder and Backhed, 2016, Li et al., 2013, Sun et al., 2021).
Although many studies have shed light on the intricate and indispensable role that the gut microbiota plays in the physiological and pathological states of their hosts, much remains unexplored. With the emerging omics technologies that are integrated into gut microbiota-related studies, new insights have been generated that reveal critical associations between certain BAs derived from microbial metabolism, and human health. In this review, the interaction between gut microbiota and BA metabolism will be summarized, as well as the underlying mechanisms as revealed by contemporary human and mouse omics research. The role of gut microbiota and BA metabolites in intestinal immune response, inflammation and tumorigenesis will be discussed, with implications for the development of preventive strategies, new therapeutics and potential diagnostic tools for clinical applications.
Characterization of gut microbiota-BA axis
It is well established that the gut microbiota performs several BA biotransformative reactions and the composition and abundance of gut microbiota are in turn, also influenced by BAs. Here, we will highlight a number of studies that characterized this gut microbiota-BA axis and provide context for the later discussion on the role of gut bacteria and BAs in IBD and colorectal cancer (CRC).
Gut microbiota mediates BA metabolism in the host
BAs are produced in the liver by the oxidation of cholesterol catalyzed by a series of cytochromes P450 (CYPs) via two biosynthetic pathways: the classic and alternative pathways (Axelson et al., 2000). Gut microbiota influences the expression of cholesterol 7α-hydroxylase (Cyp7a1), oxysterol 7α-hydroxylase (Cyp7b1), and sterol 27-hydroxylase (Cyp27a1) in mice (Sayin et al., 2013). Bile acid-CoA synthetase (BACS) and bile acid-CoA: amino acid N-acyltransferase (BAAT) carry out the conjugation of primary BAs (PBAs) with either taurine or glycine in different proportions in the liver before they are transported into the gallbladder (Johnson et al., 1991, Killenberg, 1978). Conjugated BAs are secreted into the gallbladder from hepatocytes after meals and enter the intestine by bile flow where they function to facilitate dietary lipid and vitamin absorption (Gonzalez, 2012). BAs are reabsorbed via a combination of active transport in the distal ileum and passive absorption along the whole intestine, and then recirculated via the portal vein back to the liver (Chiang, 2013, Gonzalez, 2012). This process is known as enterohepatic circulation which happens about 4 to 12 times per day in humans (Chiang, 2013) (Figure 1).
Figure 1. Gut Microbiota Regulates BA Metabolism in the Enterohepatic Circulation.
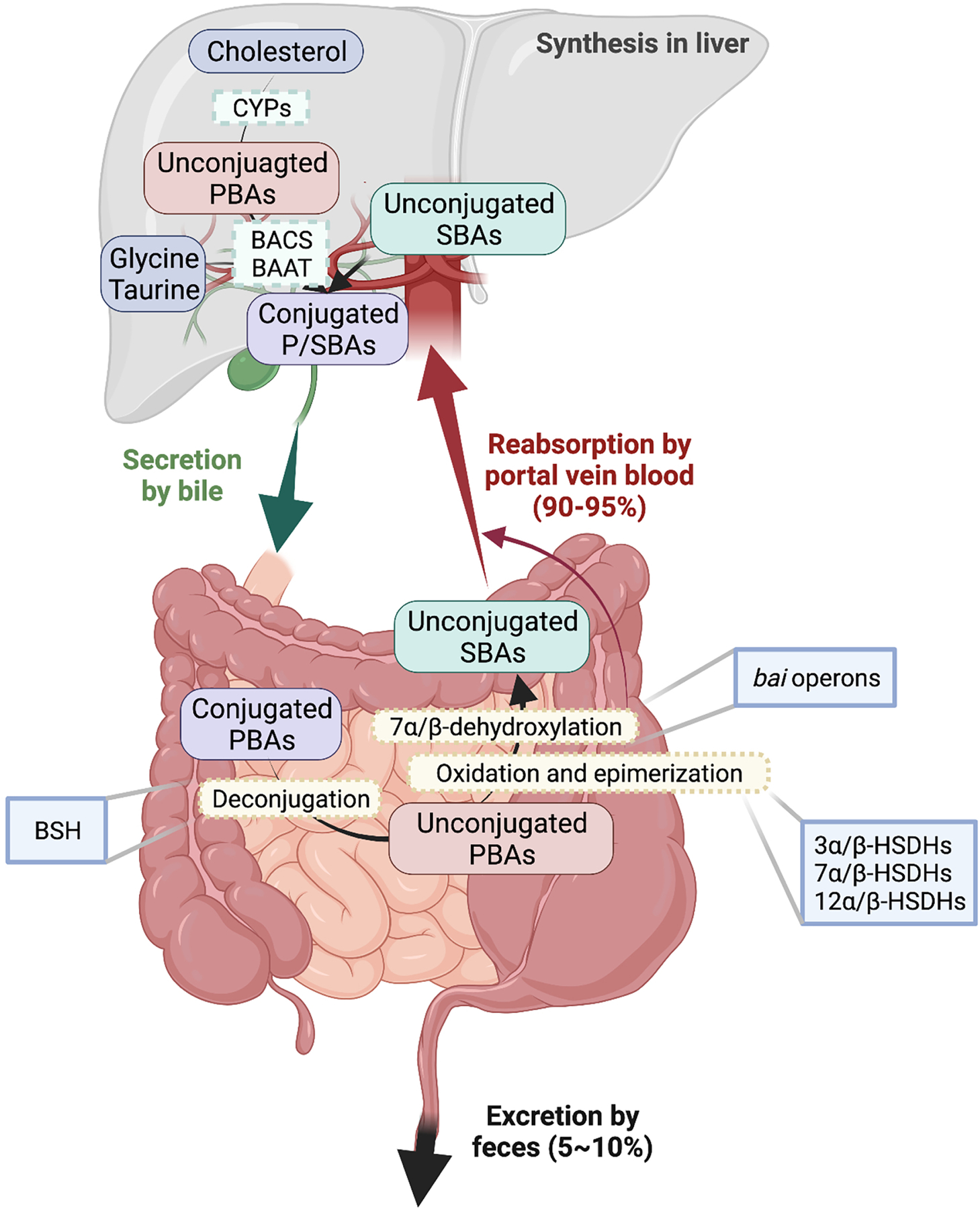
BAs are synthesized from cholesterol in the liver via oxidation catalyzed by cytochromes P450 (CYPs) and conjugation catalyzed by bile acid-CoA synthetase (BACS) and bile acid-CoA: amino acid N-acyltransferase (BAAT). Subsequently, BAs are secreted into intestine post prandial. Most BAs (90–95%) are reabsorbed into liver by portal vein blood and only a small portion (5–10%) are excreted into feces. Gut microbiota inhabiting the intestines carry out biotransformations to convert primary bile acids (PBAs) into secondary bile acids (SBAs) and convert conjugated BAs into unconjugated BAs. Major microbial biotransformation reactions include deconjugation mediated by bile salt hydrolases (BSH), 7α/β-dehydroxylation mediated by bai genes, and oxidation and epimerization mediated by hydroxysteroid dehydrogenases (HSDH).
About 5–10% of BAs are secreted into the colon where they are mainly biotransformed by the gut microbiota or excreted into feces (Gonzalez, 2012). The BA biotransformations carried out by the gut microbiota include deconjugation, oxidation and epimerization of the 3-, 7- and 12-hydroxy groups, 7-dehydroxylation, esterification and desulfatation (Gerard, 2013, Ridlon et al., 2016, Ridlon et al., 2006) (Table 1). Bile salt hydrolases (BSH) mediate the gateway reaction of SBA production by hydrolyzing the C-24 N-acyl bond that binds BAs to taurine or glycine; these reactions occur mostly in the lower small intestine and proximal colon, coinciding with the major colonization site of BSH-producing bacterial populations (Ridlon et al., 2006). Gram-positive gut bacteria have the most diverse distribution of BSH, including Clostridium, Enterococcus, Bifidobacterium, and Lactobacillus, while the distribution of BSH in the Gram-negatives is only detected in members of the phylum Bacteroidetes (Jones et al., 2008, Patel et al., 2010, Yao et al., 2018). A more recent next-generation sequencing-based study on the human gut microbiome from 11 populations from six continents revealed the prevalence of BSH in 12 different phyla, including the two dominant phyla Bacteroidetes and Firmicutes in the human gut (Song et al., 2019).
Table 1.
Summary of BA Metabolism by Microbial Producers
| Enzymes catalyzing biotransformation reactions | BA transformations | Microbial producers | References |
|---|---|---|---|
| Bile salt hydrolases (BSH) | conjugated BAs→unconjugated BAs |
Clostridium Bacteroides Bifidobacterium Enterococcus Lactobacillaceae (family) |
(Coleman and Hudson, 1995, Kishinaka et al., 1994, Stellwag and Hylemon, 1976, Yao et al., 2018, Elkins et al., 2001, Tanaka et al., 2000, Wijaya et al., 2004) |
| BA-inducible (Bai) operon | CDCA→LCA CA→DCA |
Clostridium scindens Clostridium hylemonae Clostridium hiranonis (reclassified as Peptacetobacter hiranonis) Clostridium leptum |
(Doerner et al., 1997, Ridlon et al., 2016, Kitahara et al., 2000, Kitahara et al., 2001) |
| 3α-hydroxysteroid dehydrogenases (HSDH) | DCA→3-oxoDCA |
Eggerthella lenta
Ruminococcus gnavus Clostridium perfringens |
(Harris et al., 2018, Doden et al., 2021, Macdonald et al., 1976) |
| 3β-HSDH | 3-oxoDCA→isoDCA |
Eggerthella lenta Ruminococcus gnavus Peptostreptococcus productus (reclassified as Blautia producta) |
(Harris et al., 2018, Doden et al., 2021, Edenharder et al., 1989) |
| 7α-HSDH | CDCA→7-oxoLCA |
Eggerthella lenta
Clostridium scindens Escherichia coli |
(Harris et al., 2018, Doden et al., 2021) |
| 7β-HSDH | 7-oxoLCA→urso-DCA | Ruminococcus gnavus | (Doden et al., 2021) |
| 12α-HSDH | DCA→12-oxoLCA |
Clostridium scindens Clostridium hylemonae Clostridium hiranonis (reclassified as Peptacetobacter hiranonis) Clostridium leptum Eggerthella lenta |
(Harris et al., 2018, Harris and Hylemon, 1978, Doden et al., 2018) |
| 12β-HSDH | 12-oxoLCA→epiDCA | Clostridium paraputrificum | (Doden et al., 2021) |
CA, cholic acid; CDCA, chenodeoxycholic acid; DCA, deoxycholic acid; LCA, lithocholic acid.
Production of the SBAs, deoxycholic acid (DCA) and lithocholic acid (LCA), by removing the 7α/β-hyroxy group from PBAs, has been discovered in several strains, including C. scindens, C. hiranonis and C. hylemonae (Ridlon et al., 2016). Different from oxidation and epimerization, 7α/β-dehydroxylation is limited to free BAs and is a multistep pathway carried out by several BA-inducible (bai) genes (Funabashi et al., 2020). Recently, Faecalicatena contorta S122 (S122), which is more prevalent and abundant than C. hiranonis or C. scinden in humans, was shown to harbor putative bai operons and could convert cholic acid (CA)/chenodeoxycholic acid (CDCA) to DCA/LCA (Jin et al., 2022). Moreover, Eggerthella lenta is capable of oxidizing and epimerizing BA hydroxyl groups as revealed by several functional studies (Macdonald et al., 1979, Devlin and Fischbach, 2015, Harris et al., 2018) while “bai-like genes” have been identified in E. lenta (Devlin and Fischbach, 2015). Recently, bai genes were identified in E. lenta, however, without any further functional characterization (Lee et al., 2021). Collectively, it remains inconclusive whether E. lenta could produce DCA or LCA.
Pyridine nucleotide-dependent hydroxysteroid dehydrogenases (HSDH) encoded by the gut microbiota, carry out the oxidation/reduction of hydroxy groups at the 3-, 7- and 12- carbons of BAs (Doden and Ridlon, 2021). Epimerization (α↔β) requires two position-specific BA α- and β-HSDHs of intraspecies or interspecies origin that function together with the generation of a stable oxo-BA intermediate (Ridlon et al., 2006). Amide conjugation of the cholate backbone with the amino acid phenylalanine, tyrosine and leucine was recently identified as a novel mechanism of BA transformation that was strongly positively correlated with the presence of Clostridium bolteae in mice fed a high-fat diet (Quinn et al., 2020). The mechanism by which these amino acid-conjugated BAs are synthesized and what their physiobiological functions are in humans remain unknown.
BAs reshape the gut microbiota composition
Hydrophobic BAs at high concentrations can exert direct antimicrobial activities mainly through membrane damage. BAs can alter membrane lipid composition (Taranto et al., 2006), and increased BA concentrations can solubilize membranes and dissociate integral membrane proteins (Coleman et al., 1980). Gram-positive bacteria are often more sensitive to BAs such as oxbile(oxgall)than Gram-negative bacteria (Begley et al., 2005), whereas Gram-negatives, particularly Bacteroides, have been found to be more sensitive to free BAs (Devlin and Fischbach, 2015, Watanabe et al., 2017). Unconjugated BAs are generally more hydrophobic than the corresponding conjugated forms, and SBAs are more hydrophobic than the corresponding PBAs and thus unconjugated BAs and SBAs are more likely to cause noxious effects (Martinez-Augustin and Sanchez De Medina, 2008). BAs could also shape the composition of gut microbiota by indirect effects through BA receptors, especially farnesoid X receptor (FXR), encoded by NR1H4. FXR induces genes involved in enteroprotection to inhibit bacterial overgrowth and mucosal injury in the ileum (Inagaki et al., 2006). Mice lacking FXR have increased ileal levels of bacteria and a compromised epithelial barrier (Inagaki et al., 2006). In healthy volunteers, the FXR agonist obeticholic acid (OCA) inhibited the synthesis of endogenous BAs and significantly increased the abundances of several Gram-positive strains including Streptococcus thermophilus, Lactobacillus casei and paracasei, Bifidobacterium breve, and Lactococcus lactis, suggesting that FXR activation alters the small intestinal microbiota in response to changes in endogenous BA concentrations (Friedman et al., 2018).
Several studies found that administration of BAs could shift the composition of the BA pool and gut microbiota. For example, in Apcmin/+ mice, CA increased opportunistic pathogens such as Prevotella and Desulfovibrio, and decreased beneficial bacteria such as Ruminococcus, Lactobacillus and Roseburia (Wang et al., 2019). Similarly, diets supplemented with CA simplified the composition of the microbiota, resulting in the expansion of several strains in the classes Clostridia and Erysipelotrichi (Islam et al., 2011). CA was efficiently transformed to DCA by 7α-dehydroxylation as intestinal DCA concentrations are increased in rats (Islam et al., 2011), indicating that the shift of gut microbiota composition after CA administration could be due to the potent antimicrobial activity of DCA. In mice fed a DCA-supplemented diet, the abundances of Parabacteroides and Bacteroides were significantly increased while the BSH producers Lactobacillus, Clostridium XI, and Clostridium XIV were decreased compared to the control group (Xu et al., 2021a). Taken together, these data demonstrate that BA supplementation reshapes the composition of gut microbiota.
In addition, decreased BA levels caused by clinically- and experimentally-induced liver injuries could lead to intestinal bacterial overgrowth (Clements et al., 1996, Ilan, 2012, Wigg et al., 2001). For example, bile duct ligation resulted in an increase of bacteria in the small intestine, which enabled bacterial translocation across the epithelial barrier causing systematic infection that can be reversed by BA administration (Lorenzo-Zuniga et al., 2003, Fouts et al., 2012).
The role of microbial BAs in intestinal immunity and IBD
IBD, including Crohn’s disease (CD) and ulcerative colitis (UC), are multifactorial chronic diseases that are heterogeneous at the clinical, immunological, molecular, genetic, and microbial levels (Mulder et al., 2014). Globally, a rapid rise in the incidence and prevalence of IBD poses important challenges to the Western world and in newly industrialized countries (Molodecky et al., 2012, Kaplan, 2015). In this section, we will summarize recent use of BA-related human omics data in the study of experimental IBD and discuss the underlying mechanisms by which BAs and its microbial producers influence disease occurrence and progression.
Correlations between microbial BA metabolism and human IBD
Various omics platforms including metagenomics, metabolomics, proteomics, and transcriptomics have been applied to dissect the complex interactions between genetic makeup, microbiota composition, environmental factors, and mucosal immune responses in IBD. Of note, next generation sequencing techniques such as 16S rRNA sequencing and shotgun metagenomics were introduced to analyze microbial genomes, and targeted/untargeted metabolomics have been applied to the study the composition and abundance of gut metabolites (Table 2).
Table 2.
Bile Acids in Human IBD and CRC Metabolomics Studies.
CA, cholic acid; CDCA, chenodeoxycholic acid; DCA, deoxycholic acid; GCA, glycocholic acid; GCDCA, glycochenodeoxycholic acid; GDCA, glycodeoxycholic acid; LCA, lithocholic acid; TCA, taurocholic acid; TCDCA, taurochenodeoxycholic acid; TDCA, taurodeoxycholic acid; UDCA, ursodeoxycholic acid
As part of the Integrative Human Microbiome Project, an IBD multi-omics database (IBDMDB) was introduced to leverage a systematic view of the etiology of the IBD-associated gut microbiome (Lloyd-Price et al., 2019). BAs were featured predominantly in the network as overall chemical classes. PBAs, such as unconjugated CA, its taurine and glycine conjugates (taurocholic acid and glycocholic acid; TCA and GCA, respectively) and glycochenodeoxycholate (GCDCA), were enriched in dysbiotic samples from CD patients, whereas SBAs, such as LCA and DCA, were obviously reduced, implying that SBA-producing bacteria are depleted in IBD-related dysbiosis (Lloyd-Price et al., 2019). Members of the Roseburia genus were metatranscriptionally and metagenomically associated with BAs, implying its essential participation in the BA dysregulation observed in IBD (Lloyd-Price et al., 2019).
In another study, untargeted LC-MS metabolomic profiling and shotgun metagenomic sequencing were performed with cross-sectional stool samples from discovery and validation cohorts of CD, UC, and non-IBD control subjects in an integrated multi-omics framework (Franzosa et al., 2019). One hundred twenty-two robust microbiota-metabolite associations were identified and validated with the largest metabolite cluster enriched in IBD containing 12 putative BAs, including matches to CA, CDCA and their structural variants (Franzosa et al., 2019). The PBAs CA and CDCA were enriched in IBD patients along with potential microbial mechanistic associations, while the SBAs DCA and LCA were depleted in CD, but without meeting the threshold for false discovery rate significance (Franzosa et al., 2019). Similarly, levels of Ruminococcaceae and the bai genes were found to be significantly lower in UC pouch patients compared with noninflamed familial adenomatous polyposis pouch patients (Sinha et al., 2020). As a result, CDCA levels were significantly increased, whereas levels of DCA and LCA were decreased in UC pouch patients (Sinha et al., 2020). Taken together, PBAs are overabundant while SBAs are reduced/deleted in IBD, consistent with disrupted BA transformation activities.
BAs are not only highly associated with disease occurrence but also able to predict early remission in IBD. In a recently published study, fecal metagenomics, serum metabolomics and proteomics were performed to reveal microbial determinants that predict differential responses to anti-cytokine and anti-integrin therapies in IBD. Serum SBAs such as glycolithocholate, glycodeoxycholate and ursodeoxycholate (UDCA) enrichment predicted early remission in IBD patients undergoing anti-cytokine therapies (Lee et al., 2021). Furthermore, the diversity of microbial species and the abundance of bai genes were positively correlated with the likelihood of remission associated with anti-cytokine response, suggesting that microbial signatures could preferentially favor responses to biologic therapies in IBD (Lee et al., 2021).
Other than BAs, a metagenomic analysis of the human microbiome revealed that BSH abundance was associated with human IBD. The abundance of cluster BSH genes predominantly from phylum Proteobacteria were increased and the abundance of those predominantly from phylum Firmicutes were decreased in human IBD (Jia et al., 2020). These data are consistent with the findings of increased levels of phylum Proteobacteria and deceased levels of phylum Firmicutes in IBD patients (Huttenhower et al., 2014), indicating an association between BSH abundance and IBD occurrence.
BAs control the differentiation of T cells and skew macrophage polarization
T lymphocytes, blood cells that originate in the bone marrow, mature in the thymus and function in peripheral tissues, are divided into three subsets, naïve T cells, memory T cells and regulatory T (Treg) cells (Dong, 2021, Kumar et al., 2018). Naïve T cells respond to new antigens and differentiate into effector T cells such as helper CD4+ T cells and cytotoxic CD8+ T cells to defend against infection, whereas Treg cells suppress the immune response and prevent autoimmunity (Kumar et al., 2018, Dong, 2021). Unique lymphocyte populations function cooperatively to maintain the intestinal barrier system, among which forkhead box protein P3 (FOXP3)-expressing Treg cells are involved in maintaining homeostasis of the intestinal immune system (Tanoue et al., 2016). About 65% of the Treg cell population in the colon and 35% in the small intestine express retinoic acid receptor-related orphan receptor γt (RORγt, an isoform of RORγ) (Tanoue et al., 2016). RORγt is one of the key regulators of pro-inflammatory T helper 17 (TH17) cells, while interleukin 10 (IL-10) produced by Treg cells could suppress pro-inflammatory TH17 cell differentiation in the intestine (Sefik et al., 2015, Tanoue et al., 2016). Hence, a balanced ratio of TH17 and Treg cell population is critical for maintenance of the intestinal immune system.
LCA and DCA derivatives act as important signaling molecules that regulate the differentiation of TH17 and Treg cells, which further remodel intestinal inflammation (Figure 2A, B) (Hang et al., 2019, Li et al., 2021, Campbell et al., 2020). Distinct derivatives of LCA and DCA including iso-, 3-oxo-LCA/DCA, allo-, 3-oxoallo- and isoalloLCA are produced by the cooperation of 5α/β-reductase and 3α/β-HSDH (Devlin and Fischbach, 2015, Sato et al., 2021). 3-OxoLCA directly binds to RORγt and inhibits the differentiation of TH17 cells, while isoalloLCA enhances the differentiation of anti-inflammatory Treg cells through the production of mitochondrial reactive oxygen species that increase FOXP3 expression (Hang et al., 2019). Different from the previously identified bacterial metabolite butyrate that increases Treg differentiation dependent on the intronic Foxp3 conserved noncoding sequence (CNS) 1 enhancer (Arpaia et al., 2013), isoalloLCA-enhanced Treg differentiation requires CNS3 (Hang et al., 2019). The nuclear receptors vitamin D receptor (VDR) and FXR did not contribute to the promoting activities of isoalloLCA on Treg cells or the suppressive activities on TH17 cells (Hang et al., 2019). A later study further revealed that isoalloLCA increased the binding of NR4A1 at the Foxp3 locus, resulting in enhanced Foxp3 gene transcription and thus Treg cell differentiation (Li et al., 2021). A biosynthetic gene cluster in phylum Bacteroidetes consisting of a 5β-reductase, 5α-reductase, and 3β-HSDH was identified to convert 3-oxoLCA to isoalloLCA (Li et al., 2021). Both levels of isoalloLCA and related biosynthetic genes were significantly reduced in IBD patients, suggesting the importance of isoalloLCA and this biosynthetic pathway in maintaining intestinal homeostasis (Li et al., 2021).
Figure 2. Gut Microbiota-derived BAs Regulate Immune Response in Colon.
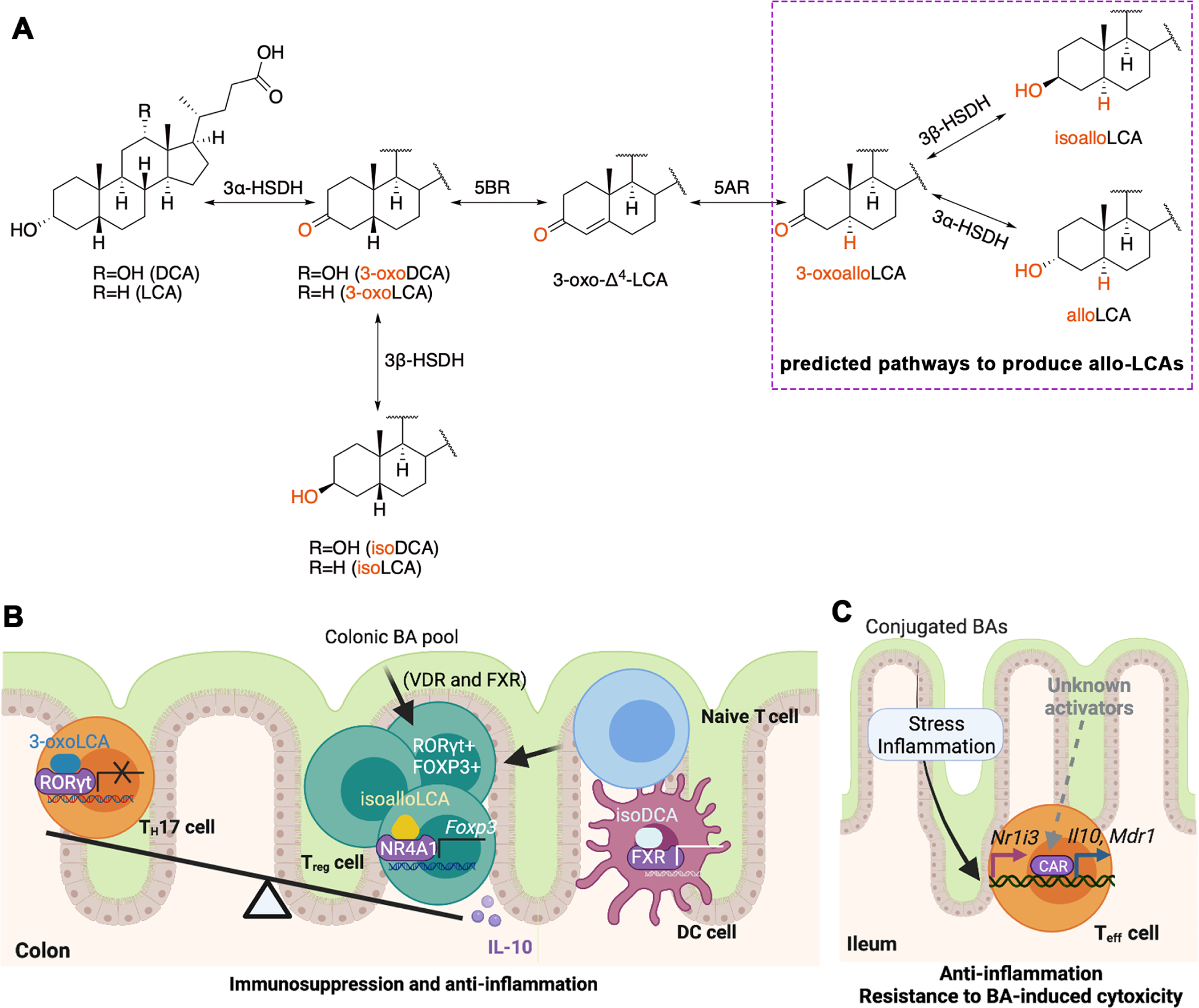
(A) Bacterial epimerization and isomerization generate distinct BAs derivatives. 3-Oxodeoxycholic acid (DCA)/lithocholic acid (LCA) is generated from DCA/LCA, and isoDCA/LCA is generated by 3-oxoDCA/LCA by the actions of 3α-HSDH and 3β-HSDH respectively. Furthermore, allo-, 3-oxoallo-, isoalloLCA are produced by the cooperation of 5α/β-reductase (5AR/BR) and 3α/β-HSDH. (B) BA metabolites control colon T cell differentiation and dendritic cell (DC) activity. 3-OxoLCA binds to retinoic acid receptor-related orphan receptor γt (RORγt) and directly inhibits the differentiation of T helper 17 (TH17) cells, while isoalloLCA enhances the differentiation of anti-inflammatory regulatory T (Treg) cells by inducing production of mitochondrial reactive oxygen species and increasing binding of nuclear hormone receptor NR4A1 at the Foxp3 locus, leading to enhanced Foxp3 gene transcription. Moreover, isoDCA inhibits immunostimulatory properties of DC dependent on farnesoid X receptor (FXR), resulting in the expansion of peripheral Treg cells. (C) In small intestine lamina propria CD4+ T effector (Teff) cells, the nuclear receptor constitutive androstane receptor (CAR) regulates multidrug resistance protein 1(Mdr1) expression and induces anti-inflammatory cytokine interleukin 10 (IL-10) to detoxify BAs exposure in the lumen. VDR, vitamin D receptor.
In addition to T lymphocytes, macrophages also function as an important compartment of the immune system and exhibit either a classically activated pro-inflammatory M1 phenotype or an alternatively activated anti-inflammatory M2 phenotype (Murray and Wynn, 2011). DCA was enriched in a high-fat diet-induced colon inflammation mouse model as a result of the increased percentage of Gram-positive bacteria, especially genus Clostridium. Mechanistically, DCA promoted macrophage polarization toward the M1 phenotype at least partially through Toll-like receptor 2 transactivated by M2 muscarinic acetylcholine receptor, resulting in increased production of proinflammatory cytokines (Wang et al., 2020).
BA receptors mediate the regulation of intestinal inflammation
BA receptors have been reported to be involved in the regulation of intestinal inflammation by BAs. Another DCA derivative, isoDCA, increased Foxp3 induction by inhibiting the immunostimulatory properties of dendritic cells (DC), resulting in the expansion of peripheral Treg cells in the colon (Campbell et al., 2020). Transplantation of an engineered isoDCA-producing consortia to germ-free mice promoted colon RORγt+ Treg cell generation in a CNS1 dependent manner (Campbell et al., 2020). Ablating FXR in DCs led to a similar transcriptional profile with isoDCA and potentiated peripheral Treg cell generation, indicating an interaction between FXR and isoDCA in maintaining the anti-inflammatory phenotype in DCs (Campbell et al., 2020). In germ-free mice, transplantation of Bacteroides species induced colon RORγ+ Treg cells via VDR and FXR, while genetic disruption of the BA deconjugation enzyme, BSH, in Bacteroides species, significantly abolished this induction (Song et al., 2020). Restoration of the intestinal BA pool increased the RORγ+ Treg population and ameliorated host susceptibility to inflammatory colitis under minimal-diet treatment (reduced nutrients) with a decreased effect of dietary components on this Treg population (Song et al., 2020). These studies establish that BA receptors mediate the effect of BAs on the regulation of Treg cell populations.
The role of FXR in IBD has been investigated in both mice and humans. A novel engineered FGF19 analogue FGF19-M52 protected mice from intestinal inflammation coupled with modulation of the microbiota composition and this anti-inflammatory activity was completely abolished in Fxr-null mice (Gadaleta et al., 2020). In CD patients, circulating FGF19 levels were reduced which indicates that the FGF19 analogue could be a therapeutic agent in the treatment of intestinal inflammation with concomitant disrupted BA homeostasis (Gadaleta et al., 2020). These results suggest that FXR and its target gene encoding FGF19 play a protective role in IBD. However, amide conjugation of the cholate with the amino acids, phenylalanine and tyrosine (Phe-chol and Tyr-chol), were identified as potent FXR agonists and found to be enriched in the dysbiotic state associated with CD patients (Quinn et al., 2020). These compounds boosted the expression of intestinal FXR target genes Fgf15 and Shp while decreasing the expression of hepatic Cyp7a1 and Cyp8b1 in mice (Quinn et al., 2020). Further studies are needed to explore whether Phe-chol and Tyr-chol contribute to intestinal inflammation caused by microbial dysbiosis and whether FXR is involved in the mechanisms of action.
In response to BA-induced cytotoxicity, CD4+ T effector (Teff) cells also showed anti-inflammatory properties dependent on the constitutive androstane receptor (CAR, encoded by NR1I3)-multidrug resistance protein 1 (MDR1)-IL10 axis (Cao et al., 2017, Chen et al., 2021) (Figure 2C). In mouse ileum, Teff cells upregulated MDR1 which mitigated oxidative stress and maintained homeostasis in Teff cells exposed to conjugated BA toxicity (Cao et al., 2017). Furthermore, blocking ileal conjugated BA reabsorption in transferred Rag1−/− mice restored MDR1-deficient Teff cell homeostasis and suppressed CD-like small bowel inflammation (Cao et al., 2017). Mechanistically, the nuclear xenobiotic receptor CAR was recently identified as a regulator of MDR1 expression which induces the expression of detoxifying enzymes and drug transporters in small intestine lamina propria Teff cells, and the key anti-inflammatory cytokine IL-10 (Chen et al., 2021). CAR deficiency in Teff cells exacerbated BA-driven ileitis in T cell-reconstituted Rag1−/− or Rag2−/− mice, whereas it was suppressed by pharmacological activation of CAR, suggesting that CAR activation in T cells as a novel strategy to treat CD (Chen et al., 2021).
G protein-coupled bile acid receptor 1 (GPBAR1, also known as TGR5), another important BA receptor, was found to be involved in the pathogenesis of IBD by mediating the effects of SBAs (Figure 3A). LCA and DCA promoted regeneration of the intestinal organoids via activation of TGR5 in intestinal stem cells (ISCs) through activation of the SRC/yes-associated protein 1 (YAP) regenerative machinery (Sorrentino et al., 2020). Mice with TGR5 disruption in ISCs developed more severe colitis than the wild-type group, and TGR5 activation by the TGR5 agonist INT-777 promoted stem cell reconstitution, suggesting that TGR5 is required for intestinal epithelium regeneration (Sorrentino et al., 2020). LCA treatment lost its beneficial effects in mice with TGR5-deficient immune cells, thus highlighting the important role of TGR5 in immune cell signaling (Sinha et al., 2020). In another study, LCA inhibited NLRP3 inflammasome activation via the TGR5-cAMP-Protein kinase A (PKA) axis, leading to improved inflammation management (Guo et al., 2016). TGR5 activation by LCA induced PKA kinase activation, which phosphorylated and ubiquitinated NLRP3 and blocked NLRP3 inflammasome-dependent inflammation in bone marrow-derived macrophages and mice, suggesting TGR5 as a potential target for NLRP3 inflammasome-related inflammation like IBD (Guo et al., 2016).
Figure 3. BA Receptors in IBD and CRC.
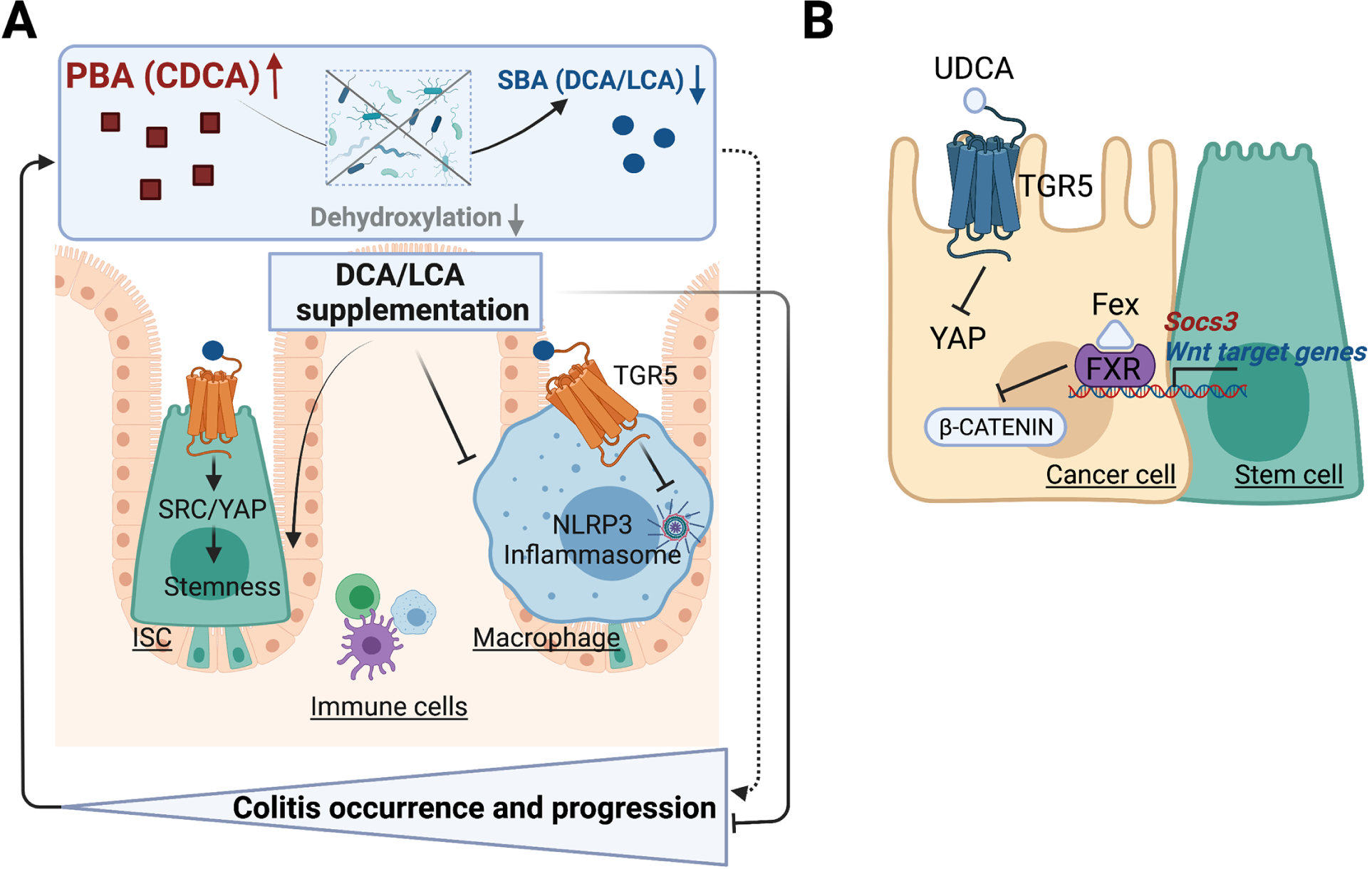
(A) G protein-coupled receptor 1 (TGR5) activation by secondary bile acids (SBAs) drive intestinal epithelium regeneration and prevents intestinal inflammation. Primary BAs (PBAs) are biotransformed to SBA via a series of gut microbiota-mediated reactions. Microbial dysbiosis disrupts biotransformation and alters the composition of intestinal BA pool, leading to colitis occurrence and progression. In intestinal stem cells (ISCs), TGR5 activation activates SRC/yes-associated protein 1 (YAP) signaling, triggering ISC renewal and proliferation. In macrophage, TGR5 activation blocks NLRP3 inflammasome phosphorylation and ubiquitination, thus inhibiting intestinal inflammation. Therefore, deoxycholic acid (DCA)/lithocholic acid (LCA) supplementation promotes ISCs stemness and inhibits NLRP3 inflammasome activity, alleviating colitis occurrence. (B) BA receptors FXR and TGR5 are activated by BA derivatives or other agonists, stimulating targeted pathways to regulate intestinal stemness, inflammation and tumorigenesis. FXR activation reshapes the BA profiles, antagonizes Wnt/β-catenin signaling and suppresses transactivation of suppressor of cytokine signaling 3 (SOCS3) gene, thus curtailing CRC progression. TGR5 activation by UDCA suppresses YAP signaling, which inhibits cell proliferation and malignant tumor progression. CDCA, chenodeoxycholic acid; Fex, fexaramine; UDCA, ursodeoxycholic acid.
Several earlier IBD investigations in mouse models revealed that a detrimental role for SBAs in intestinal inflammation through disrupting the intestinal mucosal barrier, increasing proinflammatory cytokines production and contributing to dysbiosis (Liu et al., 2018, Xu et al., 2021a, Xu et al., 2021b) (Figure 3A). In murine colitis models, fecal SBAs were increased and DCA supplementation induced intestinal inflammation, which was associated with BA metabolic disturbances and intestinal dysbiosis (Xu et al., 2021a, Xu et al., 2021b). In contrast, DCA/LCA supplementation via rectal administration showed anti-inflammatory effects partly dependent on TGR5 signaling in three murine colitis models (Sinha et al., 2020), which is consistent with the depletion of SBAs in IBD patients (Lloyd-Price et al., 2019, Sinha et al., 2020) . Curiously, DCA exerted both pro-inflammatory and anti-inflammatory effects in these studies. This might result from different routes of administration and different types of colitis models. The inconsistent results from mouse studies suggests that findings from experimental mouse IBD studies should be evaluated and the findings translated to human IBD with caution.
The role of microbial BAs in CRC
In the United States, CRC is the third leading cause of cancer deaths in men and women according to 2020 American Cancer Society statistics (Siegel et al., 2020). IBD and CRC are closely associated and IBD is considered a major risk factor for CRC (Schmitt and Greten, 2021). In this section, we will first summarize recent BAs-related human omics findings in CRC and then delve into the underlying mechanisms based on our earlier discussion of how BAs operate as intestinal immune system regulators.
Correlations between microbial BA metabolism and human CRC
Similar to IBD, CRC is also a heterogeneous disease closely correlated with the gut microbiota. Given the link between gut microbiota and colon tumorigenesis along with the heterogeneity of CRC, multi-omics techniques have been applied to understand the etiology of the disease, discover prognostic or predictive biomarkers, and develop therapeutic or preventive agents (Table 2).
The abundance of SBA synthesis-related genes is significantly associated with human CRC. In an unbiased meta-analysis of eight studies of CRC from seven different countries, the bai genes were highly enriched in stool from CRC patients at both the genomic and transcriptomic levels, which could potentially be used as a surrogate microbiome marker for CRC diagnosis (Wirbel et al., 2019). In another worldwide metagenomic study of the human microbiome, the abundance of BSH genes predominantly from phylum Firmicutes is negatively associated with CRC (Jia et al., 2020) in agreement with previous studies showing the depletion of Firmicutes in CRC patients (Ahn et al., 2013), indicating a relationship between BSH abundance and human CRC.
In a whole-genome shotgun metagenomics and metabolomics study on fecal samples obtained from patients with different stages of colorectal neoplasia, microbiome and metabolome shifts were investigated during multistep tumorigenesis. DCA was significantly increased in patients with multiple polypoid adenomas and the only species found to be significantly correlated with DCA in this study was Bilophila wadsworthia (Yachida et al., 2019), which utilizes taurine-conjugated BAs for sulphite reduction and promotes colitis in genetically-susceptible Il10−/− mice (Devkota et al., 2012).
In a nested case-control study within the European Prospective Investigation into Cancer and Nutrition (EPIC) cohort, pre-diagnostic plasma levels of seven conjugated BA metabolites including the PBAs GCA, TCA, GCDCA, taurochenodeoxycholic acid (TCDCA) and glycohyocholic acid (GHCA) and the SBAs glycodeoxycholic acid (GDCA) and taurodeoxycholic acid (TDCA) were found to be positively correlated with risk of colon cancer while unconjugated BAs and tertiary BAs were not associated with cancer risk (Kuhn et al., 2020). Additional studies found that SBAs were elevated in the adenoma group and positively correlated with Bacteroides in a fecal metabolomics study (Weir et al., 2013) while UDCA was decreased in stool samples in CRC patients compared to healthy controls (Weir et al., 2013). Thus, several studies demonstrate that DCA may contribute to CRC pathogenesis while additional studies are needed to establish the role of other BAs, especially primary BAs, in human CRC.
Mechanism actions of BAs in intestinal tumorigenesis
SBA derivatives 3-oxoLCA, isoalloLCA and isoDCA were identified as T cell regulators by inhibiting the differentiation of TH17 cells or increasing the differentiation of Treg cells with increased expression of Foxp3 in intestinal lamina propria (Hang et al., 2019, Campbell et al., 2020). Although the increasing population of FOXP3+ Treg cells is beneficial for alleviating intestinal inflammation, it impedes the effectiveness of anti-tumor immunity and reduces the efficacy of immunotherapies as increased number of tumor-infiltrating Treg cells is associated with poor prognosis in various cancer types (Tanaka and Sakaguchi, 2017). However, the correlation between Treg cell infiltration and CRC remains inconclusive as several clinical studies showed controversial results (Tanaka and Sakaguchi, 2017, Saito et al., 2016). Two FOXP3+CD4+ Treg cell subpopulations impact CRC diagnosis in distinct ways as CRC patients with abundant infiltration of FOXP3lo Treg cells (unstable FOXP3 and non-expression of native T cell marker CD45RA) was correlated with better prognosis than those with predominantly FOXP3hi (stable FOXP3 and expression of native T cell marker CD45RA) Treg cells (Saito et al., 2016). The role of gut microbiota-derived BA metabolites in immune response during CRC progression needs to be further investigated.
Secondary hydrophobic BAs generated from microbial reactions, including DCA, LCA and UDCA, are the most widely studied BA metabolites in CRC. Increased DCA and LCA concentrations in the colon stimulate cellular responses including activation of the Wnt/β-catenin and NF-κB signaling pathways, leading to DNA oxidative damage and enhanced mitotic activities (Nusse and Clevers, 2017, Jia et al., 2018), and activation of intrinsic apoptotic pathways including mitochondrial oxidative stress, cytochrome C release and cytosolic caspases (Gadaleta et al., 2017, Jia et al., 2018). SBAs also regulate intestine tumorigenesis by activating the BA receptors FXR and TGR5 (Figure 3B). Colon FXR was found to play a protective role in carcinogenesis in several studies (Sun et al., 2021). Compared to FXR, the role of TGR5 in CRC is less studied. TGR5 is overexpressed in colon tumor tissues in azoymethane/dextran sulfate sodium (AOM/DSS)-induced CRC mouse model, and activation of TGR5 by UDCA inhibited malignant tumor progression by suppressing YAP signaling and inhibiting cell proliferation (Zhang et al., 2021). Interestingly, TGR5 activation by INT-777 in ISCs promoted YAP activation and protracted YAP-driven intestinal regeneration (Sorrentino et al., 2020), whereas TGR5 activation by UDCA and INT-777 in CRC cell lines suppressed YAP expression and inhibited cell growth (Zhang et al., 2021). Mice with depletion of TGR5 in ISCs were more susceptible to colitis compared to controls (Sorrentino et al., 2020). However, in vivo studies are lacking to validate the role of TGR5 in intestinal cancer cells.
Gut microbiota-BA axis as a novel target for the treatment of IBD and CRC
Manipulation of SBA production-related microbial elements including BSH and bai genes are under explored as targets for the gut microbiota-BA axis. BSH abundance or activity is correlated with multiple human diseases including IBD and CRC (Jia et al., 2020). A targeted chemoproteomic strategy was developed to profile the altered BSH activity both in vitro and within the gut microbiota in a mouse model of colitis, which may prove useful for the future diagnosis of diseases associated with altered BSH activity (Parasar et al., 2019). Efforts have also been made to discover potent and specific BSH inhibitors. A high-throughput screening based on a high-purity BSH from a chicken Lactobacillus salivarius strain was performed and several potent BSH inhibitors such as riboflavin and phenethyl caffeate (CAPE) were identified (Smith et al., 2014). In a subsequent study, a potent, selective, covalent pan-BSH inhibitor, Compound 7, was reported using a rational design approach and its inhibitory activity was confirmed both in vitro and in vivo (Adhikari et al., 2020). More recently, a second-generation gut-restricted BSH inhibitor compound AAA-10 was developed with enhanced potency, reduced off-target effects and durable in vivo efficacy (Adhikari et al., 2021). These inhibitors are far from ready to be put into clinical application as new drug candidates, but they can still be used as chemical probes to better understand the effects of BAs on host metabolism.
Compared with BSH, our knowledge of the bai genes is more limited and efforts to better understanding the role of bai genes on host physiology are underway. A model for the 7α-dehydroxylation pathway in C. scindens based on the characterization of native or recombinant enzymes encoded by the bai operon was proposed in which CA was characterized as a substrate (Ridlon et al., 2016). A recent study reconstituted 7α-dehydroxylation in vitro, and then transferred this pathway into a nonproducing commensal bacterial species C. sporogenes to establish a complete eight-step conversion of CA to DCA catalyzed by a set of six necessary and sufficient enzymes (Funabashi et al., 2020). In comparison to SPF mice colonized with WT Faecalicatena contorta S122, SPF mice colonized with baiH mutant S122 demonstrated reduced intestinal inflammation, indicating the potential of baiH as a drug target in colitis treatment (Jin et al., 2022). These studies expand our understanding of 7α-dehydroxylation and yielded novel insights into target bai genes to manipulate the gut microbiota. To our knowledge, no inhibitors of the Bai proteins have been described.
Manipulation of BAs receptors FXR and TGR5 signaling is under investigation as a therapeutic strategy to treat intestinal inflammation and tumorigenesis. OCA, a BA analogue and potent FXR agonist that was approved by the FDA for the treatment of primary biliary cholangitis (PBC), exerted a protective effect in DSS- and 2,4,6-trinitrobenzene sulfonic acid (TNBS)-induced colitis mouse models with decreased pro-inflammatory cytokine production and preserved epithelial barrier function (Gadaleta et al., 2011). UDCA, a therapeutic BA and TGR5 agonist approved by the FDA for the treatment of PBC (Floreani and Mangini, 2018), reduced the risk for advanced adenoma in men but not in women by shifting the microbial community composition (Pearson et al., 2019). An ongoing clinical trial has included UDCA as part of a combination chemotherapy for metastatic colorectal cancer (clinicaltrials.gov identifier: NCT00873275).
CONCLUSION AND OUTLOOK
Gut microbiota and its SBA metabolites interact with the host and help maintain intestinal homeostasis through different BA receptors and cell signaling pathways. Recent human omics studies have validated what was found in murine models and deepened our understanding of the associations between BAs and their microbial producers, and intestinal inflammation and tumorigenesis. However, most human omics studies either focus on metagenomics or BA-related metabolomics instead of integrating these data and clarifying the connections between the change of abundance or/and function of gut microbiota and BA concentrations. The field is just beginning to explore how we can manipulate the gut microbiota for therapeutic purposes. Notably, BSH inhibitors have been developed without further in vivo validation in animal models whereas recombinant Bai proteins have been purified without small molecular ligands having been developed.
Of great interest, the newly discovered immunomodulatory properties of SBA derivatives expanded our knowledge of BAs as cell signaling factors. However, most studies focused on the effects of SBA derivatives on the differentiation of Treg and TH17 cells without including a thorough examination of other T cell subtypes. More studies need to be done to define the role of BAs in the adaptive immune system and thus better facilitate the development of therapeutic agents or diagnostic tools on IBD and CRC.
In summary, targeting gut microbiota-BA axis has tremendous potential in clinical prognosis and therapeutics for CRC and IBD but substantial efforts must be devoted to deepening our knowledge of the mechanistic links and translation into clinical practice.
ACKNOWLEDGEMENTS
Supported by the National Cancer Institute Intramural Research Program.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adhikari AA, Ramachandran D, Chaudhari SN, Powell CE, Li W, Mccurry MD, Banks AS & Devlin AS (2021). A gut-restricted lithocholic acid analog as an inhibitor of gut bacterial bile salt hydrolases. ACS Chem Biol, 16, 1401–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikari AA, Seegar TCM, Ficarro SB, Mccurry MD, Ramachandran D, Yao L, Chaudhari SN, Ndousse-Fetter S, Banks AS, Marto JA, et al. (2020). Development of a covalent inhibitor of gut bacterial bile salt hydrolases. Nat Chem Biol, 16, 318–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn J, Sinha R, Pei Z, Dominianni C, Wu J, Shi J, Goedert JJ, Hayes RB & Yang L (2013). Human gut microbiome and risk for colorectal cancer. J Natl Cancer Inst, 105, 1907–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpaia N, Campbell C, Fan X, Dikiy S, Van Der Veeken J, Deroos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, et al. (2013). Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature, 504, 451–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelson M, Ellis E, Mork B, Garmark K, Abrahamsson A, Bjorkhem I, Ericzon BG & Einarsson C (2000). Bile acid synthesis in cultured human hepatocytes: support for an alternative biosynthetic pathway to cholic acid. Hepatology, 31, 1305–1312. [DOI] [PubMed] [Google Scholar]
- Begley M, Gahan CGM & Hill C (2005). The interaction between bacteria and bile. Fems Microbiology Reviews, 29, 625–651. [DOI] [PubMed] [Google Scholar]
- Brown DG, Borresen EC, Brown RJ & Ryan EP (2017). Heat-stabilised rice bran consumption by colorectal cancer survivors modulates stool metabolite profiles and metabolic networks: a randomised controlled trial. Br J Nutr, 117, 1244–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell C, Mckenney PT, Konstantinovsky D, Isaeva OI, Schizas M, Verter J, Mai C, Jin WB, Guo CJ, Violante S, et al. (2020). Bacterial metabolism of bile acids promotes generation of peripheral regulatory T cells. Nature, 581, 475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W, Kayama H, Chen ML, Delmas A, Sun A, Kim SY, Rangarajan ES, Mckevitt K, Beck AP, Jackson CB, et al. (2017). The xenobiotic transporter Mdr1 enforces T cell homeostasis in the presence of intestinal bile acids. Immunity, 47, 1182–1196 e1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ML, Huang X, Wang H, Hegner C, Liu Y, Shang J, Eliason A, Diao H, Park H, Frey B, et al. (2021). CAR directs T cell adaptation to bile acids in the small intestine. Nature, 593, 147–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang JY (2013). Bile acid metabolism and signaling. Compr Physiol, 3, 1191–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements WD, Parks R, Erwin P, Halliday MI, Barr J & Rowlands BJ (1996). Role of the gut in the pathophysiology of extrahepatic biliary obstruction. Gut, 39, 587–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman JP & Hudson LL (1995). Cloning and characterization of a conjugated bile acid hydrolase gene from Clostridium perfringens. Appl Environ Microbiol, 61, 2514–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman R, Lowe PJ & Billington D (1980). Membrane lipid composition and susceptibility to bile salt damage. Biochim Biophys Acta, 599, 294–300. [DOI] [PubMed] [Google Scholar]
- Cross AJ, Moore SC, Boca S, Huang WY, Xiong X, Stolzenberg-Solomon R, Sinha R & Sampson JN (2014). A prospective study of serum metabolites and colorectal cancer risk. Cancer, 120, 3049–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, Antonopoulos DA, Jabri B & Chang EB (2012). Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature, 487, 104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin AS & Fischbach MA (2015). A biosynthetic pathway for a prominent class of microbiota-derived bile acids. Nat Chem Biol, 11, 685–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doden H, Sallam LA, Devendran S, Ly L, Doden G, Daniel SL, Alves JMP & Ridlon JM (2018). Metabolism of oxo-bile acids and characterization of recombinant 12α-hydroxysteroid dehydrogenases from bile acid 7α-dehydroxylating human gut bacteria. Appl Environ Microbiol, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doden HL & Ridlon JM (2021). Microbial hydroxysteroid dehydrogenases: From α to omega. Microorganisms, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doden HL, Wolf PG, Gaskins HR, Anantharaman K, Alves JMP & Ridlon JM (2021). Completion of the gut microbial epi-bile acid pathway. Gut Microbes, 13, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerner KC, Takamine F, Lavoie CP, Mallonee DH & Hylemon PB (1997). Assessment of fecal bacteria with bile acid 7α-dehydroxylating activity for the presence of bai-like genes. Appl Environ Microbiol, 63, 1185–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C (2021). Cytokine regulation and function in T cells. Annu Rev Immunol, 39, 51–76. [DOI] [PubMed] [Google Scholar]
- Edenharder R, Pfützner A & Hammann R (1989). Characterization of NAD-dependent 3α-and 3β-hydroxysteroid dehydrogenase and of NADP-dependent 7β-hydroxysteroid dehydrogenase from Peptostreptococcus productus. Biochimica et Biophysica Acta (BBA)-Lipids and Lipid Metabolism, 1004, 230–238. [DOI] [PubMed] [Google Scholar]
- Elkins CA, Moser SA & Savage DC (2001). Genes encoding bile salt hydrolases and conjugated bile salt transporters in Lactobacillus johnsonii 100–100 and other Lactobacillus species. Microbiology (Reading), 147, 3403–3412. [DOI] [PubMed] [Google Scholar]
- Floreani A & Mangini C (2018). Primary biliary cholangitis: Old and novel therapy. Eur J Intern Med, 47, 1–5. [DOI] [PubMed] [Google Scholar]
- Fouts DE, Torralba M, Nelson KE, Brenner DA & Schnabl B (2012). Bacterial translocation and changes in the intestinal microbiome in mouse models of liver disease. Journal of Hepatology, 56, 1283–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzosa EA, Sirota-Madi A, Avila-Pacheco J, Fornelos N, Haiser HJ, Reinker S, Vatanen T, Hall AB, Mallick H, Mciver LJ, et al. (2019). Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol, 4, 293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman ES, Li Y, Shen TD, Jiang J, Chau L, Adorini L, Babakhani F, Edwards J, Shapiro D, Zhao C, et al. (2018). FXR-dependent modulation of the human small Intestinal microbiome by the bBile acid derivative obeticholic acid. Gastroenterology, 155, 1741–1752 e1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabashi M, Grove TL, Wang M, Varma Y, Mcfadden ME, Brown LC, Guo C, Higginbottom S, Almo SC & Fischbach MA (2020). A metabolic pathway for bile acid dehydroxylation by the gut microbiome. Nature, 582, 566–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadaleta RM, Garcia-Irigoyen O, Cariello M, Scialpi N, Peres C, Vetrano S, Fiorino G, Danese S, Ko B, Luo J, et al. (2020). Fibroblast growth factor 19 modulates intestinal microbiota and inflammation in presence of farnesoid X receptor. EBioMedicine, 54, 102719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadaleta RM, Garcia-Irigoyen O & Moschetta A (2017). Bile acids and colon cancer: Is FXR the solution of the conundrum? Mol Aspects Med, 56, 66–74. [DOI] [PubMed] [Google Scholar]
- Gadaleta RM, Van Erpecum KJ, Oldenburg B, Willemsen EC, Renooij W, Murzilli S, Klomp LW, Siersema PD, Schipper ME, Danese S, et al. (2011). Farnesoid X receptor activation inhibits inflammation and preserves the intestinal barrier in inflammatory bowel disease. Gut, 60, 463–472. [DOI] [PubMed] [Google Scholar]
- Gerard P (2013). Metabolism of cholesterol and bile acids by the gut microbiota. Pathogens, 3, 14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez FJ (2012). Nuclear receptor control of enterohepatic circulation. Compr Physiol, 2, 2811–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarner F & Malagelada JR (2003). Gut flora in health and disease. Lancet, 361, 512–519. [DOI] [PubMed] [Google Scholar]
- Guo C, Xie S, Chi Z, Zhang J, Liu Y, Zhang L, Zheng M, Zhang X, Xia D, Ke Y, et al. (2016). Bile acids control Inflammation and metabolic disorder through inhibition of NLRP3 inflammasome. Immunity, 45, 802–816. [DOI] [PubMed] [Google Scholar]
- Hang S, Paik D, Yao L, Kim E, Trinath J, Lu J, Ha S, Nelson BN, Kelly SP, Wu L, et al. (2019). Bile acid metabolites control TH17 and Treg cell differentiation. Nature, 576, 143–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris JN & Hylemon PB (1978). Partial purification and characterization of NADP-dependent 12α-hydroxysteroid dehydrogenase from Clostridium leptum. Biochim Biophys Acta, 528, 148–157. [DOI] [PubMed] [Google Scholar]
- Harris SC, Devendran S, Mendez-Garcia C, Mythen SM, Wright CL, Fields CJ, Hernandez AG, Cann I, Hylemon PB & Ridlon JM (2018). Bile acid oxidation by Eggerthella lenta strains C592 and DSM 2243(T). Gut Microbes, 9, 523–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenhower C, Kostic AD & Xavier RJ (2014). Inflammatory bowel disease as a model for translating the microbiome. Immunity, 40, 843–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilan Y (2012). Leaky gut and the liver: a role for bacterial translocation in nonalcoholic steatohepatitis. World J Gastroenterol, 18, 2609–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki T, Moschetta A, Lee YK, Peng L, Zhao GX, Downes M, Yu RT, Shelton JM, Richardson JA, Repa JJ, et al. (2006). Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proceedings of the National Academy of Sciences of the United States of America, 103, 3920–3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam KBMS, Fukiya S, Hagio M, Fujii N, Ishizuka S, Ooka T, Ogura Y, Hayashi T & Yokota A (2011). Bile acid Is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology, 141, 1773–1781. [DOI] [PubMed] [Google Scholar]
- Jacobs JP, Goudarzi M, Singh N, Tong M, Mchardy IH, Ruegger P, Asadourian M, Moon BH, Ayson A, Borneman J, et al. (2016). A disease-associated microbial and metabolomics state in relatives of pediatric inflammatory bowel disease patients. Cell Mol Gastroenterol Hepatol, 2, 750–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson J, Willing B, Lucio M, Fekete A, Dicksved J, Halfvarson J, Tysk C & Schmitt-Kopplin P (2009). Metabolomics reveals metabolic biomarkers of Crohn’s disease. PLoS One, 4, e6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia B, Park D, Hahn Y & Jeon CO (2020). Metagenomic analysis of the human microbiome reveals the association between the abundance of gut bile salt hydrolases and host health. Gut Microbes, 11, 1300–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia W, Xie G & Jia W (2018). Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat Rev Gastroenterol Hepatol, 15, 111–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin WB, Li TT, Huo D, Qu S, Li XV, Arifuzzaman M, Lima SF, Shi HQ, Wang A, Putzel GG, et al. (2022). Genetic manipulation of gut microbes enables single-gene interrogation in a complex microbiome. Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MR, Barnes S, Kwakye JB & Diasio RB (1991). Purification and characterization of bile acid-CoA:amino acid N-acyltransferase from human liver. J Biol Chem, 266, 10227–10233. [PubMed] [Google Scholar]
- Jones BV, Begley M, Hill C, Gahan CG & Marchesi JR (2008). Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Natl Acad Sci U S A, 105, 13580–13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan GG (2015). The global burden of IBD: from 2015 to 2025. Nat Rev Gastroenterol Hepatol, 12, 720–727. [DOI] [PubMed] [Google Scholar]
- Killenberg PG (1978). Measurement and subcellular distribution of choloyl-CoA synthetase and bile acid-CoA:amino acid N-acyltransferase activities in rat liver. J Lipid Res, 19, 24–31. [PubMed] [Google Scholar]
- Kishinaka M, Umeda A & Kuroki S (1994). High concentrations of conjugated bile acids inhibit bacterial growth of Clostridium perfringens and induce its extracellular cholylglycine hydrolase. Steroids, 59, 485–489. [DOI] [PubMed] [Google Scholar]
- Kitahara M, Takamine F, Imamura T & Benno Y (2000). Assignment of Eubacterium sp. VPI 12708 and related strains with high bile acid 7α -dehydroxylating activity to Clostridium scindens and proposal of Clostridium hylemonae sp. nov., isolated from human faeces. Int J Syst Evol Microbiol, 50 Pt 3, 971–978. [DOI] [PubMed] [Google Scholar]
- Kitahara M, Takamine F, Imamura T & Benno Y (2001). Clostridium hiranonis sp. nov., a human intestinal bacterium with bile acid 7α-dehydroxylating activity. Int J Syst Evol Microbiol, 51, 39–44. [DOI] [PubMed] [Google Scholar]
- Kuhn T, Stepien M, Lopez-Nogueroles M, Damms-Machado A, Sookthai D, Johnson T, Roca M, Husing A, Maldonado SG, Cross AJ, et al. (2020). Prediagnostic plasma bile acid levels and colon cancer risk: A prospective study. J Natl Cancer Inst, 112, 516–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar BV, Connors TJ & Farber DL (2018). Human T cell development, localization, and function throughout life. Immunity, 48, 202–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavelle A & Sokol H (2020). Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nature Reviews Gastroenterology & Hepatology, 17, 223–237. [DOI] [PubMed] [Google Scholar]
- Lee JWJ, Plichta D, Hogstrom L, Borren NZ, Lau H, Gregory SM, Tan W, Khalili H, Clish C, Vlamakis H, et al. (2021). Multi-omics reveal microbial determinants impacting responses to biologic therapies in inflammatory bowel disease. Cell Host Microbe, 29, 1294–1304 e1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Jiang C, Krausz KW, Li Y, Albert I, Hao H, Fabre KM, Mitchell JB, Patterson AD & Gonzalez FJ (2013). Microbiome remodelling leads to inhibition of intestinal farnesoid X receptor signalling and decreased obesity. Nat Commun, 4, 2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Hang S, Fang Y, Bae S, Zhang Y, Zhang M, Wang G, Mccurry MD, Bae M, Paik D, et al. (2021). A bacterial bile acid metabolite modulates Treg activity through the nuclear hormone receptor NR4A1. Cell Host Microbe, 29, 1366–1377 e1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Dong W, Wang S, Zhang Y, Liu T, Xie R, Wang B & Cao H (2018). Deoxycholic acid disrupts the intestinal mucosal barrier and promotes intestinal tumorigenesis. Food Funct, 9, 5588–5597. [DOI] [PubMed] [Google Scholar]
- Lloyd-Price J, Arze C, Ananthakrishnan AN, Schirmer M, Avila-Pacheco J, Poon TW, Andrews E, Ajami NJ, Bonham KS, Brislawn CJ, et al. (2019). Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature, 569, 655–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo-Zuniga V, Bartoli R, Planas R, Hofmann AF, Vinado B, Hagey LR, Hernandez JM, Mane J, Alvarez MA, Ausina V, et al. (2003). Oral bile acids reduce bacterial overgrowth, bacterial translocation, and endotoxemia in cirrhotic rats. Hepatology, 37, 551–557. [DOI] [PubMed] [Google Scholar]
- Macdonald IA, Jellett JF, Mahony DE & Holdeman LV (1979). Bile salt 3α- and 12α-hydroxysteroid dehydrogenases from Eubacterium lentum and related organisms. Appl Environ Microbiol, 37, 992–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald IA, Meier EC, Mahony DE & Costain GA (1976). 3α-, 7α-And 12α-hydroxysteroid dehydrogenase activities from Clostridium perfringens. Biochimica et Biophysica Acta (BBA)-Lipids and Lipid Metabolism, 450, 142–153. [DOI] [PubMed] [Google Scholar]
- Martinez-Augustin O & Sanchez De Medina F (2008). Intestinal bile acid physiology and pathophysiology. World J Gastroenterol, 14, 5630–5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molodecky NA, Soon IS, Rabi DM, Ghali WA, Ferris M, Chernoff G, Benchimol EI, Panaccione R, Ghosh S, Barkema HW, et al. (2012). Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology, 142, 46–54 e42; quiz e30. [DOI] [PubMed] [Google Scholar]
- Mulder DJ, Noble AJ, Justinich CJ & Duffin JM (2014). A tale of two diseases: the history of inflammatory bowel disease. J Crohns Colitis, 8, 341–348. [DOI] [PubMed] [Google Scholar]
- Murray PJ & Wynn TA (2011). Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol, 11, 723–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusse R & Clevers H (2017). Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell, 169, 985–999. [DOI] [PubMed] [Google Scholar]
- O’keefe SJD (2016). Diet, microorganisms and their metabolites, and colon cancer. Nature Reviews Gastroenterology & Hepatology, 13, 691–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parasar B, Zhou H, Xiao X, Shi Q, Brito IL & Chang PV (2019). Chemoproteomic profiling of gut microbiota-associated bile salt hydrolase activity. ACS Cent Sci, 5, 867–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AK, Singhania RR, Pandey A & Chincholkar SB (2010). Probiotic bile salt hydrolase: current developments and perspectives. Appl Biochem Biotechnol, 162, 166–180. [DOI] [PubMed] [Google Scholar]
- Pearson T, Caporaso JG, Yellowhair M, Bokulich NA, Padi M, Roe DJ, Wertheim BC, Linhart M, Martinez JA, Bilagody C, et al. (2019). Effects of ursodeoxycholic acid on the gut microbiome and colorectal adenoma development. Cancer Med, 8, 617–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn RA, Melnik AV, Vrbanac A, Fu T, Patras KA, Christy MP, Bodai Z, Belda-Ferre P, Tripathi A, Chung LK, et al. (2020). Global chemical effects of the microbiome include new bile-acid conjugations. Nature, 579, 123–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridlon JM, Harris SC, Bhowmik S, Kang DJ & Hylemon PB (2016). Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes, 7, 22–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridlon JM, Kang DJ & Hylemon PB (2006). Bile salt biotransformations by human intestinal bacteria. J Lipid Res, 47, 241–259. [DOI] [PubMed] [Google Scholar]
- Rooks MG & Garrett WS (2016). Gut microbiota, metabolites and host immunity. Nat Rev Immunol, 16, 341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Nishikawa H, Wada H, Nagano Y, Sugiyama D, Atarashi K, Maeda Y, Hamaguchi M, Ohkura N, Sato E, et al. (2016). Two FOXP3(+)CD4(+) T cell subpopulations distinctly control the prognosis of colorectal cancers. Nat Med, 22, 679–684. [DOI] [PubMed] [Google Scholar]
- Sato Y, Atarashi K, Plichta DR, Arai Y, Sasajima S, Kearney SM, Suda W, Takeshita K, Sasaki T, Okamoto S, et al. (2021). Novel bile acid biosynthetic pathways are enriched in the microbiome of centenarians. Nature, 599, 458–464. [DOI] [PubMed] [Google Scholar]
- Sayin SI, Wahlström A, Felin J, Jäntti S, Marschall HU, Bamberg K, Angelin B, Hyötyläinen T, Orešič M & Bäckhed F (2013). Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-β-muricholic acid, a naturally occurring FXR antagonist. Cell Metab, 17, 225–235. [DOI] [PubMed] [Google Scholar]
- Schirmer M, Garner A, Vlamakis H & Xavier RJ (2019). Microbial genes and pathways in inflammatory bowel disease. Nature Reviews Microbiology, 17, 497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt M & Greten FR (2021). The inflammatory pathogenesis of colorectal cancer. Nat Rev Immunol, 21, 653–667. [DOI] [PubMed] [Google Scholar]
- Schroeder BO & Backhed F (2016). Signals from the gut microbiota to distant organs in physiology and disease. Nat Med, 22, 1079–1089. [DOI] [PubMed] [Google Scholar]
- Sefik E, Geva-Zatorsky N, Oh S, Konnikova L, Zemmour D, Mcguire AM, Burzyn D, Ortiz-Lopez A, Lobera M, Yang J, et al. (2015). MUCOSAL IMMUNOLOGY. Individual intestinal symbionts induce a distinct population of RORγ(+) regulatory T cells. Science, 349, 993–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, Cercek A, Smith RA & Jemal A (2020). Colorectal cancer statistics, 2020. CA Cancer J Clin, 70, 145–164. [DOI] [PubMed] [Google Scholar]
- Sinha SR, Haileselassie Y, Nguyen LP, Tropini C, Wang M, Becker LS, Sim D, Jarr K, Spear ET, Singh G, et al. (2020). Dysbiosis-induced secondary bile acid deficiency promotes intestinal inflammation. Cell Host Microbe, 27, 659–670 e655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K, Zeng X & Lin J (2014). Discovery of bile salt hydrolase inhibitors using an efficient high-throughput screening system. PLoS One, 9, e85344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Sun X, Oh SF, Wu M, Zhang Y, Zheng W, Geva-Zatorsky N, Jupp R, Mathis D, Benoist C, et al. (2020). Microbial bile acid metabolites modulate gut RORγ (+) regulatory T cell homeostasis. Nature, 577, 410–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z, Cai Y, Lao X, Wang X, Lin X, Cui Y, Kalavagunta PK, Liao J, Jin L, Shang J, et al. (2019). Taxonomic profiling and populational patterns of bacterial bile salt hydrolase (BSH) genes based on worldwide human gut microbiome. Microbiome, 7, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrentino G, Perino A, Yildiz E, El Alam G, Bou Sleiman M, Gioiello A, Pellicciari R & Schoonjans K (2020). Bile acids signal via TGR5 to activate intestinal stem cells and epithelial regeneration. Gastroenterology, 159, 956–968 e958. [DOI] [PubMed] [Google Scholar]
- Stellwag EJ & Hylemon PB (1976). Purification and characterization of bile salt hydrolase from Bacteroides fragilis subsp. fragilis. Biochim Biophys Acta, 452, 165–176. [DOI] [PubMed] [Google Scholar]
- Sun L, Cai J & Gonzalez FJ (2021). The role of farnesoid X receptor in metabolic diseases, and gastrointestinal and liver cancer. Nat Rev Gastroenterol Hepatol, 18, 335–347. [DOI] [PubMed] [Google Scholar]
- Tanaka A & Sakaguchi S (2017). Regulatory T cells in cancer immunotherapy. Cell Res, 27, 109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Hashiba H, Kok J & Mierau I (2000). Bile salt hydrolase of Bifidobacterium longum - biochemical and genetic characterization. Applied and Environmental Microbiology, 66, 2502–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanoue T, Atarashi K & Honda K (2016). Development and maintenance of intestinal regulatory T cells. Nat Rev Immunol, 16, 295–309. [DOI] [PubMed] [Google Scholar]
- Taranto MP, Perez-Martinez G & Font De Valdez G (2006). Effect of bile acid on the cell membrane functionality of lactic acid bacteria for oral administration. Res Microbiol, 157, 720–725. [DOI] [PubMed] [Google Scholar]
- Wang L, Gong Z, Zhang X, Zhu F, Liu Y, Jin C, Du X, Xu C, Chen Y, Cai W, et al. (2020). Gut microbial bile acid metabolite skews macrophage polarization and contributes to high-fat diet-induced colonic inflammation. Gut Microbes, 12, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Dong W, Liu L, Xu M, Wang Y, Liu T, Zhang Y, Wang B & Cao H (2019). Interplay between bile acids and the gut microbiota promotes intestinal carcinogenesis. Mol Carcinog, 58, 1155–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Fukiya S & Yokota A (2017). Comprehensive evaluation of the bactericidal activities of free bile acids in the large intestine of humans and rodents. J Lipid Res, 58, 1143–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir TL, Manter DK, Sheflin AM, Barnett BA, Heuberger AL & Ryan EP (2013). Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS One, 8, e70803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigg AJ, Roberts-Thomson IC, Dymock RB, Mccarthy PJ, Grose RH & Cummins AG (2001). The role of small intestinal bacterial overgrowth, intestinal permeability, endotoxaemia, and tumour necrosis factor α in the pathogenesis of non-alcoholic steatohepatitis. Gut, 48, 206–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijaya A, Hermann A, Abriouel H, Specht I, Yousif NM, Holzapfel WH & Franz CM (2004). Cloning of the bile salt hydrolase (bsh) gene from Enterococcus faecium FAIR-E 345 and chromosomal location of bsh genes in food enterococci. J Food Prot, 67, 2772–2778. [DOI] [PubMed] [Google Scholar]
- Wirbel J, Pyl PT, Kartal E, Zych K, Kashani A, Milanese A, Fleck JS, Voigt AY, Palleja A, Ponnudurai R, et al. (2019). Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat Med, 25, 679–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Cen M, Shen Y, Zhu Y, Cheng F, Tang L, Hu W & Dai N (2021a). Deoxycholic acid-induced gut dysbiosis disrupts bile acid Enterohepatic circulation and promotes intestinal Inflammation. Dig Dis Sci, 66, 568–576. [DOI] [PubMed] [Google Scholar]
- Xu M, Shen Y, Cen M, Zhu Y, Cheng F, Tang L, Zheng X, Kim JJ, Dai N & Hu W (2021b). Modulation of the gut microbiota-farnesoid X receptor axis improves deoxycholic acid-induced intestinal inflammation in mice. J Crohns Colitis, 15, 1197–1210. [DOI] [PubMed] [Google Scholar]
- Yachida S, Mizutani S, Shiroma H, Shiba S, Nakajima T, Sakamoto T, Watanabe H, Masuda K, Nishimoto Y, Kubo M, et al. (2019). Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat Med, 25, 968–976. [DOI] [PubMed] [Google Scholar]
- Yao L, Seaton SC, Ndousse-Fetter S, Adhikari AA, Dibenedetto N, Mina AI, Banks AS, Bry L & Devlin AS (2018). A selective gut bacterial bile salt hydrolase alters host metabolism. Elife, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Xu H, Zhang C, Tang Q & Bi F (2021). Ursodeoxycholic acid suppresses the malignant progression of colorectal cancer through TGR5-YAP axis. Cell Death Discov, 7, 207. [DOI] [PMC free article] [PubMed] [Google Scholar]


