Abstract
The proteases of the mitochondrial inner membrane are challenging yet highly desirable drug targets for complex, multifactorial diseases prevalent mainly in the elderly. Among them, OMA1 with its substrates OPA1 and DELE1 safeguards mitochondrial homeostasis at the intersection of energy metabolism and apoptosis, which may have relevance for neurodegeneration, malignancy and heart failure, among other diseases. Little is known about OMA1. Its structure has not been solved and we are just beginning to understand the enzyme’s context-dependent regulation. OMA1 appears dormant under physiological conditions as judged by OPA1’s processing pattern. The protease is rapidly activated, however, when cells experience stress or undergo apoptosis. Intriguingly, genetic OMA1 ablation can delay or even prevent apoptosis in animal models for diseases that can be broadly categorized as ischemia-reperfusion related disorders. Three groups have reported their efforts implementing OMA1 drug screens. This article reviews some of the technical challenges encountered in these assays and highlights what can be learned for future screening campaigns, and about the OMA1 protease more broadly. OMA1 does not exists in a vacuum and potent OMA1 inhibitors are needed to tease apart OMA1’s intricate interactions with the other mitochondrial proteases and enzymes. Furthermore, OMA1 inhibitors hold the promise of becoming a new class of cytoprotective medicines for disorders influenced by dysfunctional mitochondria, such as heart failure or Alzheimer’s Disease.
Keywords: mitochondria, membrane proteases, protease inhibitors, drug discovery, neurodegeneration, cancer
INTRODUCTION/BACKGROUND
Entering the realms of drug discovery, a former colleague of mine cautioned me, “Marcel, you will always just find what you screened for.” In light of his enlightening words, I would like to reflect on the advances in, and challenges of, implementing OMA1 drug screening campaigns. This is a highly relevant topic as three research groups (including my own “group”) have recently reported their efforts.
OMA1 is a conserved, Zn2+-dependent metalloendopeptidase in the inner membrane of mitochondria and a magnificent drug target, because so little is known about the enzyme. OMA1’s structure has not been solved and no inhibitors or chemical probes have been disclosed. OMA1’s function evolved over time with distinct substrates in invertebrates and mammals [1]. The enzyme was first described in yeast as part of a membrane protein disposal system [2]. Later, mammalian OMA1 was recognized to process the inner-membrane shaping protein OPA1 in the context of mitophagy, outer membrane permeabilization and apoptotic signaling [3–5]. And fairly recently, it was suggested OMA1 can cleave the signaling peptide DELE1, which evokes the integrated stress response and launches counter measures combating proteotoxicity [6, 7]. OMA1 functionally interacts with the eponymous m-AAA protease (the OMA1 acronym stands for overlapping activity with m-AAA protease 1) and other proteases and scaffolding proteins in the inner membrane.
OMA1 acts at the intersection of energy metabolism and stress signaling, which has relevance for a number of multifactorial, complex diseases, such as heart failure, neurodegeneration and cancer. For example, whole exome sequencing of 1,000 individuals with heart failure revealed an association with the coding polymorphism rs17117699 (p.Phe211Cys) [8]. Four non-coding markers in an intron of the DAB1 gene less than 1 MB downstream of OMA1 showed a weak association with coronary artery disease in an independent genome-wide association study (4 of a total of 1,035 markers across the genome with p<0.001: rs1524715, rs11207058, rs1524716, and rs1880443; HGVST75) [9]. More importantly, OMA1 ablation in three unrelated mouse models of heart failure could protect cardiomyocytes thus providing genetic target validation [10]. Intriguingly, also alterations of OMA1’s substrate OPA1 appear to be connected to heart failure [11–15]
OMA1 has also relevance for neurodegeneration, mainly through its substrates DELE1 and OPA1. Also certain PINK1 mutants pertaining to Parkinson’s Disease were found to be digested by OMA1 [16]. DELE1 can signal to the integrated stress response, which is active in post-mortem brains from individuals and animal models of cognitive and neurodegenerative disorders, including Alzheimer’s Disease, Parkinson’s Disease, Huntington Disease, amyotrophic lateral sclerosis, traumatic brain injury, Down syndrome, and Charcot-Marie-Tooth disease [17]. Mutations in OPA1 on the other hand can lead to dominant optic atrophy (ADOA), a rare form of childhood blindness [18], accompanied at times by more severe neurological ailments, including parkinsonism [19, 20]. Pathological processing of OPA1 was also documented in autopsy brain specimens from individuals with Alzheimer’s Disease [21, 22], who can experience optic nerve degeneration as well [23]. However, OMA1’s role during neurodegeneration and in particular OMA1’s connection to Alzheimer’s Disease are not very well understood. Five non-coding markers, again in the intronic sequence of DAB1, were associated with Alzheimer’s Disease (5 of 761 markers with p<0.001: rs1341316, rs1539053, rs12029004, rs10889061, and rs852807; HGVST318) [24]. A working hypothesis currently being investigated is whether proteotoxic Alzheimer’s proteins, such as amyloid plaques and neurofibrillary tangles, can activate OMA1. I refuted an earlier hypothesis linking OMA1 activity with tau phosphorylation by demonstrating that both can be modulated independently from one another [25].
Energy metabolism and stress signaling is also highly relevant for cancer. The model here is that proliferating cells in a stressful tumor environment characterized by hypoxia, inflammation and mutational burden, survive thanks in part by keeping a check on OMA1 [26]. Also different chemotherapies can evoke OMA1-dependent OPA1 proteolysis, which would align with such a model [27–29]. Accordingly, survival of patients with breast cancer, colorectal cancer, and lung cancer could be stratified retrospectively by OMA1 gene expression levels [26, 30, 31].
However, there is more to it, because OMA1 can reprogram the energy-metabolism and thus influence cell differentiation. For example, OMA1 knockout suppressed azoxymethane/dextran sodium sulfate-induced colorectal cancer development in mice and tumor growth in xenograft mouse models of colorectal cancer [30]. On the other hand, silencing OMA1 in patient-derived metastatic breast cancer cells significantly increased migratory properties of these cells, which promoted malignant progression with an unfavorable clinical outcome [31]. This means OMA1 inhibitors could benefit patients with colorectal cancer, while patients with breast cancer would presumably benefit from OMA1 activators. OMA1 is not a (proto-)oncogene nor a tumor suppressor gene but contributes to malignancy on multiple levels. For this reason, it is necessary to understand the workings of the OMA1 mechanism in a certain disease, and maybe even in an individual patient, before deciding on the specific type of intervention.
OMA1 mutations are not linked to a specific disease, and OMA1 knockout mice are viable and fertile with no apparent phenotype—discounting diet-induced obesity [32]. This suggests only limited on-target side-effects if OMA1 inhibitors were just specific enough. Drugging OMA1 therefore is merely an engineering task of developing highly potent/specific OMA1 inhibitors—still a quite formidable task though. Once specific OMA1 inhibitors become available, one can evaluate their efficacy in the different preclinical disease models thereby corroborating the rational for human proof-of-concept studies.
THE CHALLENGES
“It ain’t easy” is the common credo sung by all drug hunters (including Ziggy Stardust) and applies of course to OMA1 as well. OMA1’s physical interactions with lipids and proteins of the inner membrane together with the enzyme’s self-processing provide the biggest challenges for the isolation of functional OMA1 protein for in vitro assays and structural studies. OMA1 is presumably an integral membrane protease comparable in its function with PARL [33]. Interestingly, in yeast it is the mitochondrial rhomboid protein PCP1/RBD1 (i.e. PARL) instead of OMA1 that cleaves yeast’s OPA1 homolog MGM1 [1, 34]. OMA1 and PARL functionally and physically interact with the membrane anchored m-AAA and i-AAA proteases in a not very well understood mitochondrial membrane protein recycling machinery [35]. OMA1 and the i-AAA protease, which shares the OPA1 protein as substrate with OMA1, were suggested to regulate one another by reciprocal proteolysis [36, 37]. OMA1 is not active under physiological cell-culture conditions but becomes active when cells are challenged with stressors, such as the protonophore CCCP [3, 4]. The molecular basis for OMA1’s context-dependent activation, however, remains elusive. Once active, OMA1 starts to degrade through self-digestion and/or mediated by the i-AAA protease, though at a much slower pace than OPA1’s proteolysis [36–38] (see, for example, Figures 1A & B). The protease thus can limit its own function by removing itself quite similar to a timer switch. Interestingly, short deletions near OMA1’s carboxy terminus were shown to stabilize the protease, while still preserving its catalytic function [39]. OMA1 can bind to cardiolipin, the predominant lipid of the mitochondrial inner membrane, and so it was suggested that prohibitins can regulate OMA1’s turnover through the interaction with cardiolipin [40].
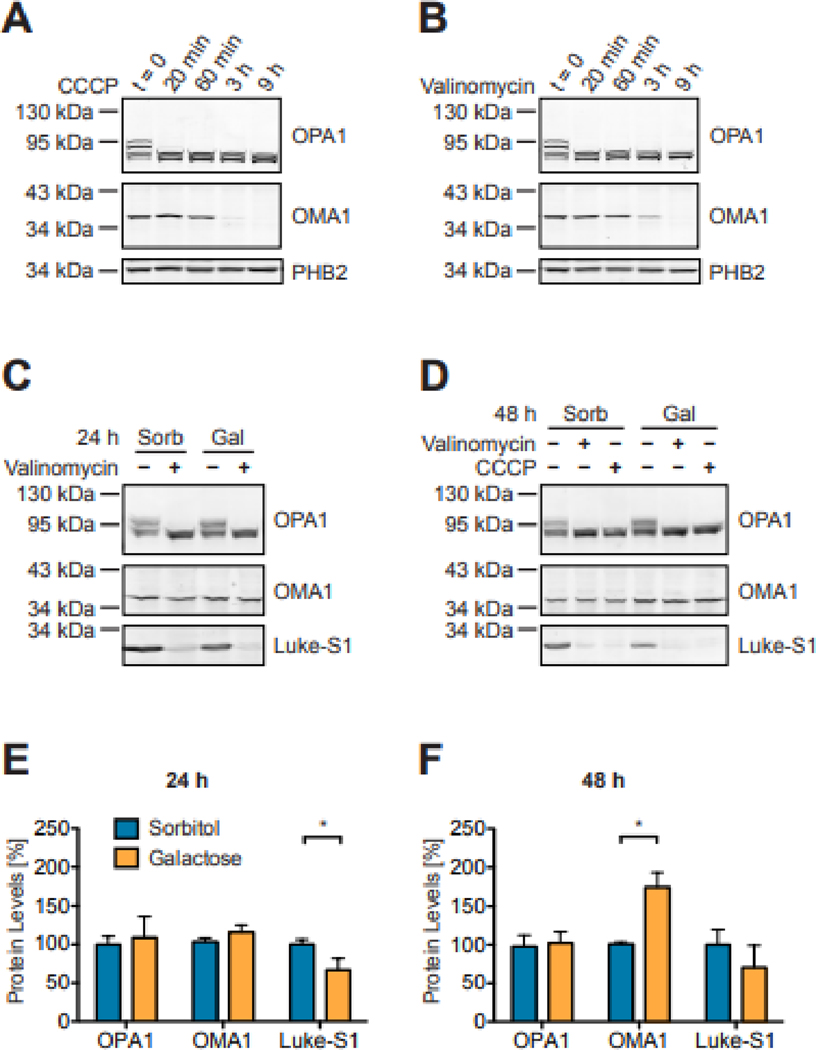
Figure 1: OMA1, OPA1 and Luke-S1 reporter protein turnover in Western blots.
OMA1 is activated within minutes upon exposure of HEK293T cells to 2 μM CCCP (A) or 100 nM Valinomycin (B) as evident by cleavage of the large OPA1 isoforms. OMA1 hydrolysis by contrast follows a slower kinetics. PHB2 is not an OMA1 substrate and remained stable even after 9 h of CCCP or valinomycin treatment. Luke-S1 reporter protein levels were significantly higher in HEK293T reporter cells cultured for 24 h in serum-free medium with 100 μM sorbitol than in cells cultured with 100 μM galactose (C). This effect was less pronounced after 48 h (D). OMA1 and OPA1 protein levels were about the same after 24 h, but OMA1 protein levels were significantly higher in cells maintained for 48 h in galactose medium. Panels E & F show the densitometric quantification of Western blots; n = 2 independent replicates; 2-way ANOVA: p<0.05 (*).
At the same time, OMA1’s active site appears not very specific for any one cleavage-site motif. For example, OMA1 cleaves the sequence T-A-F-R—A-T-D-R of OPA1 (P4-P3-P2-P1—P’1-P’2-P’3-P’4; with ‘—’ denoting the scissile bond; NP_056375.2) [41] but the sequence L-R-Q-H—I-L-P-S of DELE1 (NP_055588.3) [7]. Moreover, measuring hydrolysis of FITC-casein allowed for the study of OMA1-expressing yeast strains in in vitro assays, further demonstrating promiscuity of OMA1’s active site [42, 43]. An OPA1-based reporter was still processed when OPA1’s cleavage site was replaced with a TEV-cleavage site, though at a different position [27]. A chimeric protein tethering the inner and outer mitochondrial membranes by combining SCO1 with TOM20 domains was also cleaved by OMA1 [44]. And mitochondrial contact site and cristae organizing system (MICOS) subunit MIC19 appears to be cleaved by OMA1 upon SAMM50 knockdown in HCT116 cells [45]. These findings together suggest that the (miss-)placement of a protein in the inner membrane could suffice for its recognition by the OMA1 protease. This conclusion is further substantiated by the aforementioned report of misrouted PINK1 mutants that became an OMA1 substrate [16].
THE DIFFERENT APPROACHES
The first attempts to design an OMA1 assay made use of a peptide resembling OPA1’s cleavage site coupled to a fluorescence resonance energy transfer (FRET) pair [46]. The basic idea behind this assay is that the FRET acceptor would quench the FRET donor until the peptide is cleaved and the FRET pair separated, whereby fluorescence is emitted (see also Table 1). Such FRET-based protease assays are popular for their ease of use and because the fluorescence signal correlates directly with protease activity. They allow for measuring accurate kinetic parameters in a straightforward, one-step reaction. The FRET reporter peptide and the protease are combined and fluorescence recorded. Still there are some inherent limitations to such protease assays stemming for example from the specific FRET pair, as well as additional constrains introduced by the protease. In case of OMA1, an eight-amino acid peptide of the rat OPA1 cleavage site (A-F-R—A-T-D-G-H) was coupled to the fluorophore MCA (7-Methoxycoumarin-4-ylacetyl) on the amino end and the quencher DNP (2,4-Dinitrophenyl) on the carboxy terminus [46]. The chemical nature of the MCA-DNP pair limits its use to in vitro assays. The inventors of this assay hence used whole cell lysates from rat kidney cells and mouse embryonic fibroblasts for their assays. Another group used whole cell lysate from HEK293T cells that overexpress OMA1 [8]. Interestingly, both groups appear to measure only basal OMA1 activity in their assays, because OMA1 was not activated by stress challenges before harvesting the cells for analyses. Unfortunately, it is not understood what impact detergents, such as SDS in the solubilization buffer have on OMA1 activity. Solubilization of cells usually does not per se result in OMA1 activation—at least as judged by OPA1 cleavage [3, 4]. Another open question is how efficiently the FRET peptide would reach the active site of the membrane protease. Overall, this assay showed little specificity, because OMA1 knockdown and OMA1 knockout cells considerably amplified the fluorescence signal of the reporter, but still to a lesser extent than control cells [46]. In silico predictions indicate that the eight-amino acid peptide can be hydrolyzed by other proteases as well, which is why purified and functional OMA1 protease would be required to improve OMA1 specificity of this assay. But then again, it is not easy to purify functional OMA1, even more so at scale for high-throughput drug screening. These limitations of the FRET reporter peptide render it impractical for drug screening.
Table 1.
Main features of the 3 OMA1 assays side by side.
| FRET peptide | PINK1C125G-EYFP reporter | Luke-S1 reporter | |
|---|---|---|---|
| Assay type: | In vitro assay | Cellular reporter assay | Target-based cellular reporter assay |
| Principle: | FRET pair serves as | Fluorescent protein | Luciferase targeted to the inner |
| OMA1 substrate | targeted to the inner membrane serves as OMA1 and PARL substrate | membrane serves as OMA1 substrate | |
| Correlation with OMA1 activity: | Direct* | No | Indirect |
| Advantages: | + Straightforward assay set up + Plate reader compatibility + Direct correlation of emitted signal and OMA1 activity |
+ Additional readout of cell permeability and cell toxicity + Direct correlation of OMA1 inhibition and emitted signal |
+ High specificity though spatial confinement of the reporter to the inner membrane + High sensitivity through enzymatic signal amplification + Additional readout of cell permeability and cell toxicity + Plate reader compatibility + Indirect correlation of emitted signal and OMA1 activity |
| Disadvantages: | − Requires purified and functional OMA1 protease for specificity − Potentially confounded by autofluorescence of test molecules |
− Requires high content imaging system − OMA1-independent events can also generate signal − Limited to cell permeable and non-toxic compounds − Potentially confounded by autofluorescence of test molecules |
− Limited to cell permeable and non-toxic compounds − Potentially confounded by luciferase modulators |
| Reference: | Tobacyk et al. 2019 [46] | Houston et al. 2021 [49] | Alavi 2021 [27] |
The FRET peptide is presumably recognized by other proteases as well.
Another attempt to measure OMA1 protease activity utilized said PINK1 mutant processed by OMA1 [16]. The native PINK1 protein is usually imported into healthy mitochondria, where it is processed by PARL in the inner membrane and subsequently degraded via the proteasome. But PINK1 is stabilized in dysfunctional mitochondria destined for mitophagy and recruits PARKIN to the mitochondrial surface to initiate the formation of the autophagosome [47, 48]. A set of experiments led thereby to the identification of the PINK1 p.C125G mutant. CCCP can stabilize PINK1 by impeding its import. The proteasome inhibitor MG132 on the other hand can block PINK1 degradation after processing by PARL, whereby a cleaved form of PINK1 is retained. Simultaneous treatment with CCCP and MG132 stabilized full-length PINK1 in agreement with a model in which CCCP frustrated PINK1’s import and thus prevented PINK1 from reaching the PARL protease in the inner membrane [16]. The PINK1C125G mutant behaved differently in these experiments, because a PINK1C125G-EYFP fusion protein was not stabilized by CCCP, but still degraded [49]. Simultaneous treatment with CCCP and MG132 also stabilized a cleaved form of PINK1C125G-EYFP as opposed to full-length PINK-EYFP. Follow-up experiments with OMA1 knockout cells yielded full-length PINK1C125G-EYFP upon CCCP treatment, which established that PINK1C125G-EYFP is processed by OMA1. Motivated by this result, the authors of the study reasoned that PINK1C125G-EYFP could be used for drug screening, whereby the PINK1C125G-EYFP fluorescence would inversely correlate with OMA1 activity in CCCP-treated cells [49]. However, the complex dependencies of the reporter in this assay limit its utility. Stabilization of the PINK1C125G-EYFP reporter can be confounded by interference with its import, its processing, and its degradation. Not surprisingly, the chemicals that were identified in a pilot screen of 1,100 FDA-approved drugs were either proteasome inhibitors or chlorhexidine and alexidine, which shattered the inner membrane. The complexity of the various interactions of this reporter could even be higher. In OMA1 knockdown cells, for example, MG132 still stabilized cleaved PINK1C125G-EYFP [49]. This cleavage event was attributed to PARL. So both OMA1 and PARL can process PINK1C125G-EYFP apparently compensating for one another. It is also not understood how PINK1C125G-EYFP could reach the inner membrane in CCCP-treated cells. Protein transport across membranes requires energy, which is usually provided by the mitochondrial membrane potential Δψ. When CCCP disrupts Δψ, then other means, such as chaperones, would need to provide that energy, which is another potential confounding factor for PINK1C125G-EYFP-based screens. All these uncertainties increase the likelihood of finding false positives in PINK1C125G-EYFP reporter assays and reduce the chances of identifying true OMA1 inhibitors.
The third attempt to design an OMA1 protease screening assay combined a split luciferase enzyme with two OPA1 domains in a reporter, which can be expressed in mammalian cells [25, 27]. This reporter, dubbed Luke-S1, establishes OMA1 specificity through its spatial confinement to the inner membrane, where it acts as artificial OMA1 substrate. The bioluminescence emitted by this reporter inversely correlates with OMA1 activity. HEK293T reporter cells produced readily detectable bioluminescence when cultured under standard conditions. OMA1 activation by CCCP or valinomycin on the other hand led to reporter cleavage and elimination of its bioluminescence [27]. Cellular protease assays preserve the native environment of the enzyme, which seems particularly advantageous when investigating mitochondrial membrane proteases. Cell permeability and cytotoxicity, however, are additional constrains of cellular protease assays, which should be considered when designing drug screening campaigns. The increased stringency might reduce the hit rate of such drug screens on one hand, but could also lead to hits with more desirable physiochemical properties precisely because of the additional readout for cell permeability and cytotoxicity. Assay interference, that is molecules engaging the luciferase, is another potentially confounding factor. For example, one study reported that 2.7% of about 42,000 compounds of a chemical diversity set were potential luciferase inhibitors [50]. Though this is less of an issue when screening for OMA1 inhibitors, because of the inverse correlation of the bioluminescence with OMA1 activity. OMA1 cleaves the luciferase and kills its signal. OMA1 inhibitors hence would sustain bioluminescence. And luciferase inhibitors would just add to the number of negative scoring molecules, but not lead to false positive hits in this assay. A screening campaign of about 20,000 molecules for OMA1 inhibitors had a hit rate of 0.03% [27].
ADDITIONAL CONSIDERATIONS
What has been learned from the screens? Testing 166 FDA-approved cancer drugs at a single dose with the cellular luciferase-based Luke-S1 reporter assay identified mainly kinase inhibitors as potential OMA1 triggers [27]. 30 of the 166 cancer drugs reduced the bioluminescence by more than 37.5% after incubating OMA1 reporter cells for 60 min with 10 μM of the drug. 19 of these drugs reduced also the bioluminescence of control cells, which expressed the unaltered, native luciferase. Remarkably, the dose-response curves in OMA1 reporter cells and in control cells were almost identical, thus confirming that these 19 drugs primarily engaged the luciferase. 10 of the 11 remaining drugs were kinase inhibitors (and #11 was tamoxifen). However, there was a disconnect between the results of the luciferase assays and Western blots, because only 2 of the 10 tested kinase inhibitors (i.e. ceritinib and sorafenib) demonstrated OPA1 cleavage in Western blots—and only after 3 h of incubation with 5-times the concentration of the drug (i.e. 50 μM). Western blots against the luciferase reporter matched the OPA1 blots in that only ceritinib and sorafenib led to reporter cleavage [27].
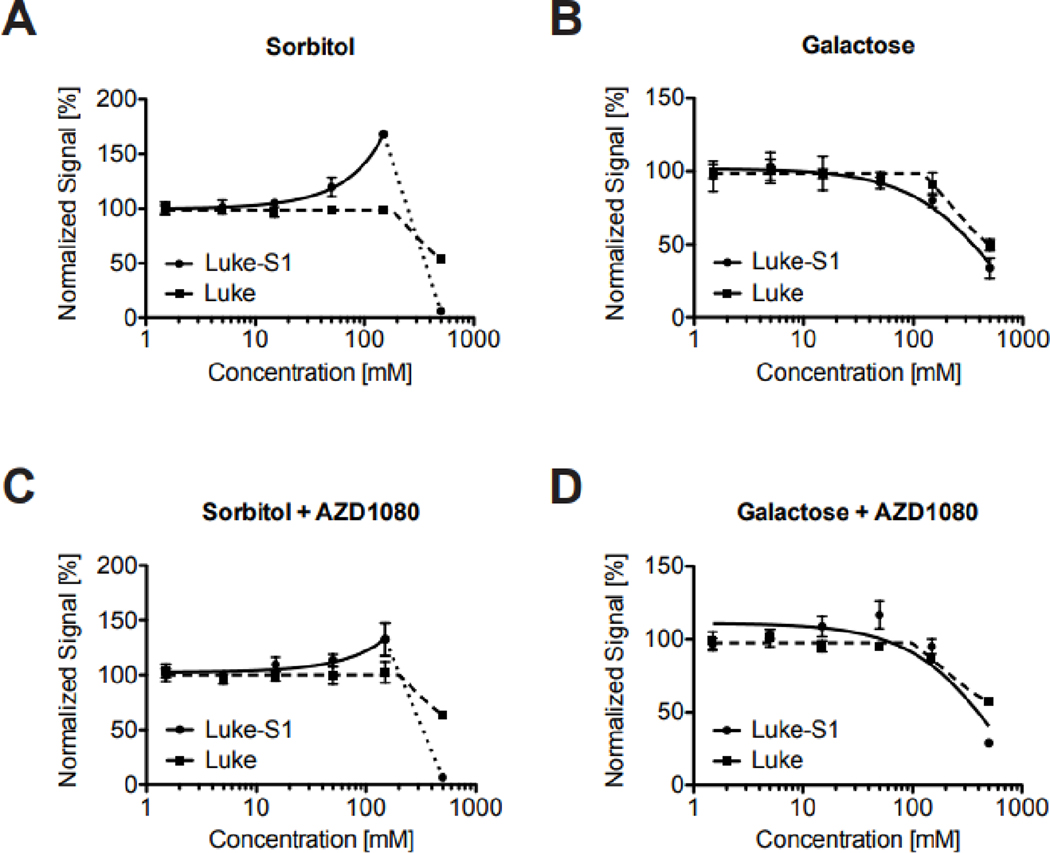
The discovery that 8 kinase inhibitors lowered bioluminescence without apparent reporter cleavage could point to posttranslational modifications of OMA1, OPA1 and/or the reporter. Protein phosphorylation could regulate either protein levels or function of either the luciferase or the protease. Investigations into the influence of the energy-metabolic state on the OMA1 reporter supported the former. Alternative carbon sources in the growth media can force cells to alter their energy metabolism [51, 52]. OMA1 reporter cells cultured for 24 h with sorbitol as the sole carbon source had significantly higher Luke-S1 reporter protein levels compared to cells cultured in galactose (Figure 1C—F). This effect leveled off after 48 h, but OMA1 protein levels significantly increased in galactose at this time. Luciferase assays also showed increasing bioluminescence after 24 h with increasing sorbitol concentrations, but not with galactose (Figures 2A & B). This effect was specific to the OMA1 reporter and not rooted in higher cell growth, because luciferase control cells maintained a steady signal. The GSK3 inhibitor AZD1080 alleviated some of sorbitol’s effect on bioluminescence (Figures 2C & D), suggesting that reporter turnover could be regulated by phosphorylation. An independent study reported that the GSK3 inhibitor SB216763 stabilized OMA1 (and restored OPA1 processing) in leptin-treated human mesenchymal stem cells [53], whereas higher glucose concentrations reduced OPA1 protein levels through posttranslational modifications in neonatal cardiomyocytes [54]. Mitochondrial NAD-dependent protein deacetylase sirtuin-3 knockout mice and hypertrophic mouse hearts displayed OPA1 hyperacetylation [55]. And another study on oxidative stress in cardiomyocytes implied protein kinase A signaling upstream of sirtuin-3 in the regulation of OPA1 [56]. Moreover, skin fibroblasts infected with human cytomegalovirus also showed temporal changes in OPA1’s acetylation pattern, which were ascribed to OPA1’s interaction with sirtuin-3 [57]. Together, these findings suggest that posttranslational modifications can influence OMA1 and OPA1—how exactly, in what cell types, and whether directly or indirectly still needs to be determined. It is also not clear if the same or different mechanisms regulate the turnover of the Luke-S1 reporter.
Figure 2: Influence of the energy-metabolic state on the Luke-S1 OMA1 reporter.
Luke-S1 reporter cells exhibit a sorbitol-dose dependent increase in bioluminescence compared to Luke cells, which express just the native luciferase (A). No signal changes were observed in cells cultured for 24 h with galactose (B). The GSK3 inhibitor AZD1080 mitigated sorbitol’s effect (C) and had no other effects on cells cultured with galactose (D). All cells were cultured for 24 h in 96-well plates in serum-free RPMI 1640 medium without glucose, supplemented with sorbitol or galactose before luciferase substrate was added and bioluminescence measured.
The higher sensitivity of the OMA1 luciferase assays for ceritinib and sorafenib than the readout of OPA1 by Western blots could be due to the enzymatic signal amplification by the luciferase. It is also possible that higher OMA1 substrate concentrations in the reporter cells lowered the detection limits by better exploiting OMA1’s full capacity. Another explanation is that OMA1 does not always engage in OPA1 cleavage. For example, OMA1 could show a preference for the reporter or other substrates because of a higher binding affinity, less steric hindrance or conformational changes.
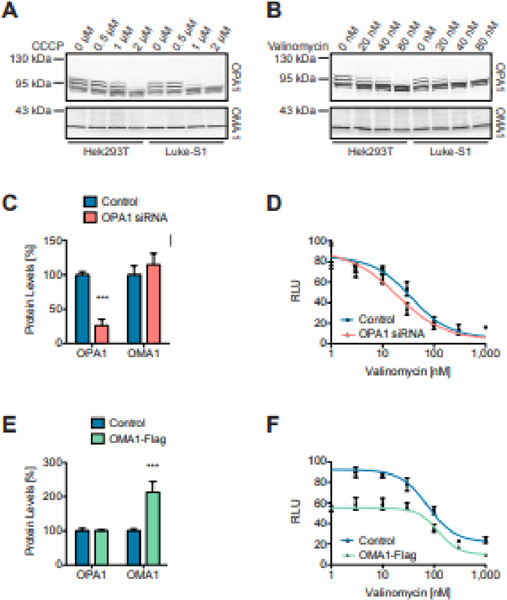
Comparing OPA1 cleavage in native HEK293T cells and Luke-S1 reporter cells by Western blotting revealed an about 20%-reduced half maximal effective concentrations (EC50) of CCCP and valinomycin in the latter (Figures 3A&B). So, a higher OMA1 substrate abundance could indeed lower the detection limit or the threshold for OMA1 activation. Such an effect could be due to the formation of an inactive, intermediary enzyme-substrate complex that required lower activation energy than the protease alone. Non-denaturing electrophoresis of yeast and mammalian cell extracts actually uncovered higher molecular weight OMA1 complexes [2, 38, 42, 43]. More importantly, sucrose-density fractions and 2D blue-native PAGE revealed that OMA1 and OPA1 comigrate together in higher molecular weight complexes [38, 40]. It is still unclear, however, whether OMA1 and OPA1 interact directly or indirectly. For example, both proteins can bind to the MICOS complex [44, 58, 59]. And yet, siRNA-mediated OPA1 knockdown also reduced valinomycin’s EC50 in the luciferase assays (Figures 3C&D). This result argues against altered substrate levels as primary determinants of the detection limit. An alternative explanation could be a limiting stoichiometric OPA1-to-OMA1 ratio regulating the function of the alleged OMA1-OPA1 complexes. OMA1 knockdown did not alter the valinomycin-dose response relationship (data not shown). But OMA1 overexpression diminished the baseline bioluminescence, leading to a less pronounced response to valinomycin (Figures 3E&F). This finding, while in line with a critical OPA1-to-OMA1 ratio, could also indicate that OMA1 has some basal activity (tying back to the findings with the FRET peptide). Evidence for constitutive OMA1 activity comes from OMA1 knockout mouse embryonic fibroblasts, which were lacking certain OPA1 isoforms [38]. On the other hand, exogenously expressed OMA1 was misrouted to the endoplasmic reticulum in earlier studies [60], which could result in higher proteotoxicity and overall pronounced OMA1 activity.
Figure 3: OMA1 substrate levels and OMA1 protein levels can modulate the protease’s response to CCCP and valinomycin.
Luke-S1 reporter cells showed a higher CCCP (A) and valinomycin (B) sensitivity in OPA1 Western blots than native HEK293T cells. OPA1 knockdown lowered valinomycin’s EC50 in luciferase assays. OPA1 protein levels were significantly reduced in Luke-S1 cells treated for 36 h with OPA1 siRNA (C); these cells showed a higher valinomycin sensitivity in luciferase assays (D). Ectopic OMA1-Flag expression reduced the baseline fluorescence, which diminished the valinomycin response but did not change valinomycin’s EC50. OMA1 protein levels were significantly increased and more than doubled in Luke-S1 cells 24 h post transfection with an OMA1-Flag plasmid (E). The OMA1-Flag expressing reporter cells showed about 40% reduced bioluminescence at low valinomycin concentrations but still responded to higher valinomycin concentrations with the same EC50 as non-transfected cells (F). All cells were treated for 30 minutes with CCCP or valinomycin in serum-free DMEM/F12 media before the analysis. Panels C & E show the densitometric quantification of Western blots; n = 3 independent replicates; 2-way ANOVA: p<0.001 (***).
Another screening campaign with the Luke-S1 assay tested 20,000 chemicals in search of OMA1 inhibitors. To this end, OMA1 was activated by valinomycin. An independent cellular luciferase-based screen searched for molecules that would prevent valinomycin-induced MFN1 degradation mediated by PARKIN recruitment to the mitochondrial outer membrane [61]. The investigators of that study expressed a MFN1-luciferase fusion protein in HeLa cells, which was degraded by the proteasome when cells were exposed to valinomycin. MFN1 is among the proteins that are ubiquitinated once PINK1 has recruited PARKIN to the outer membrane [62, 63]. This particular assay relied on the correct function of the proteasome for readout and presumably led to the identification of proteasome inhibitors. The OMA1 assay is superior in that signal changes are a direct result of target engagement without any mediators. That is, OMA1 inactivates the luciferase by cleaving it. This reduces the likelihood of identifying false positives. But mammalian cells still accommodate enough pathways with the potential to skew outcomes when using cellular assays for drug screening. One such example are efflux pumps. HEK293T cells express the ATP-dependent translocase ABCB1, aka multidrug resistance 1 (MDR1) or P-glycoprotein (P-gp), which can carry valinomycin across membranes [64]. Molecules that modulate ABCB1 could in theory show up in a screen. Also the different, in part cell-type dependent posttranslational modifications of OMA1, OPA1 and the reporter can create biases. On the positive side, the OMA1 protease is activated within minutes (Figure 1) [27]. Careful timing thus can help mitigate some of the more profound processes potentially skewing screening results, such as changes in gene regulation, protein turnover, OMA1 (self-)degradation, and apoptotic signaling. It ain’t easy and you will always just find what you screened for.
CONCLUDING REMARKS
Tremendous progress has been made in recent years and we now have arrived at a point where drug screening for OMA1 modulators became a reality. A target-based cellular reporter assay finally did the trick. This assay allowed for testing of OMA1-triggering chemicals simply by exposing cells to the chemicals and then measuring changes in luciferase activity. In living cells that is, because the luciferase substrate is cell permeable and non-toxic. One can even envision transgenic mice that express the reporter. This would be extremely useful for in vivo monitoring of OMA1 in the context of diseases, such as heart failure, neurodegeneration or cancer. Moreover, it would allow to investigate the behavior of the OMA1 protease across different cell types. Over the past years it became clear, OMA1 does not exists in a vacuum but presumably relies on interactions with other enzymes and proteases for proper function and/or regulation. Too little is known about the impact of posttranslational modifications on OMA1, OPA1 and other substrates, such as DELE1, and how these are influenced by the energy-metabolism and other factors. The molecular basis for OMA1’s regulation is also still just a plain white space, which is unacceptable considering recent technological advances in cryo-electron microscopy and artificial intelligence approaches for protein structure predictions. The first specific OMA1 modulators will be critical facilitating complementary studies. In addition, OMA1 inhibitors hold the promise of becoming a new class of cytoprotective medicines for a range of diseases, which are influenced by dysfunctional mitochondria.
HIGHLIGHTS.
Potent OMA1 inhibitors might advance research and could help address unmet medical needs.
OMA1 is a magnificent drug target validated by knockdown in mouse disease models.
Target-based cellular reporter assays are a viable option for OMA1 drug screens.
OMA1 does not exists in a vacuum but is modulated by protein-protein interactions.
Paying close attention to timing and energy metabolism can minimize biases.
ACKNOWLEDGMENTS
Thank you for the generous support of my research by the National Institute on Aging (NIA) of the National Institutes of Health (NIH) under Award Number R43AG063642. I am grateful for the very insightful comments by the anonymous reviewers of this article.
Footnotes
Chemical Compounds: AZD1080 (PubChem CID: 135564570);
CCCP (PubChem CID: 2603);
Ceritinib (PubChem CID: 57379345);
MG132 (PubChem CID: 462382);
SB216763 (PubChem CID: 176158);
Sorafenib (PubChem CID: 216239);
Tamoxifen (PubChem CID: 2733526);
Valinomycin (PubChem CID: 3000706);
CRediT author statement
Marcel V. Alavi: Conceptualization, Formal analysis, Investigation, Data curation, Writingˇ original draft preparation, Writing - review & editing, Project administration, Funding acquisition
CONFLICT OF INTEREST
Dr. Marcel V. Alavi is shareholder of 712 North Inc., a California-based pharmaceutical company.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Duvezin-Caubet S, Jagasia R, Wagener J, Hofmann S, Trifunovic A, Hansson A, Chomyn A, Bauer MF, Attardi G, Larsson NG, Neupert W, Reichert AS, Proteolytic processing of OPA1 links mitochondrial dysfunction to alterations in mitochondrial morphology, J Biol Chem 281 (2006) 37972–37979. [DOI] [PubMed] [Google Scholar]
- [2].Kaser M, Kambacheld M, Kisters-Woike B, Langer T, Oma1, a novel membrane-bound metallopeptidase in mitochondria with activities overlapping with the m-AAA protease, J Biol Chem 278 (2003) 46414–46423. [DOI] [PubMed] [Google Scholar]
- [3].Ehses S, Raschke I, Mancuso G, Bernacchia A, Geimer S, Tondera D, Martinou JC, Westermann B, Rugarli EI, Langer T, Regulation of OPA1 processing and mitochondrial fusion by m-AAA protease isoenzymes and OMA1, J Cell Biol 187 (2009) 1023–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Head B, Griparic L, Amiri M, Gandre-Babbe S, van der Bliek AM, Inducible proteolytic inactivation of OPA1 mediated by the OMA1 protease in mammalian cells, J Cell Biol 187 (2009) 959–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, Alroy J, Wu M, Py BF, Yuan J, Deeney JT, Corkey BE, Shirihai OS, Fission and selective fusion govern mitochondrial segregation and elimination by autophagy, EMBO J 27 (2008) 433–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Fessler E, Eckl EM, Schmitt S, Mancilla IA, Meyer-Bender MF, Hanf M, Philippou-Massier J, Krebs S, Zischka H, Jae LT, A pathway coordinated by DELE1 relays mitochondrial stress to the cytosol, Nature 579 (2020) 433–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Guo X, Aviles G, Liu Y, Tian R, Unger BA, Lin YT, Wiita AP, Xu K, Correia MA, Kampmann M, Mitochondrial stress is relayed to the cytosol by an OMA1-DELE1-HRI pathway, Nature 579 (2020) 427–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hu D, Li S, Hu S, Sun Y, Xiao L, Li C, Wang J, Wang Y, Ni L, Zhao C, Wang DW, A Common Missense Variant in OMA1 Associated with the Prognosis of Heart Failure, Cardiovasc Drugs Ther 34 (2020) 345–356. [DOI] [PubMed] [Google Scholar]
- [9].Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, Dixon RJ, Meitinger T, Braund P, Wichmann HE, Barrett JH, Konig IR, Stevens SE, Szymczak S, Tregouet DA, Iles MM, Pahlke F, Pollard H, Lieb W, Cambien F, Fischer M, Ouwehand W, Blankenberg S, Balmforth AJ, Baessler A, Ball SG, Strom TM, Braenne I, Gieger C, Deloukas P, Tobin MD, Ziegler A, Thompson JR, Schunkert H, Genomewide association analysis of coronary artery disease, N Engl J Med 357 (2007) 443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Acin-Perez R, Lechuga-Vieco AV, Del Mar Munoz M, Nieto-Arellano R, Torroja C, Sanchez-Cabo F, Jimenez C, Gonzalez-Guerra A, Carrascoso I, Beninca C, Quiros PM, Lopez-Otin C, Castellano JM, Ruiz-Cabello J, Jimenez-Borreguero LJ, Enriquez JA, Ablation of the stress protease OMA1 protects against heart failure in mice, Sci Transl Med 10 (2018). [DOI] [PubMed] [Google Scholar]
- [11].Adaniya SM, Cypress OUJ,MW, Kusakari Y, Jhun BS, Posttranslational modifications of mitochondrial fission and fusion proteins in cardiac physiology and pathophysiology, Am J Physiol Cell Physiol 316 (2019) C583–C604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ong SB, Kalkhoran SB, Hernandez-Resendiz S, Samangouei P, Ong SG, Hausenloy DJ, Mitochondrial-Shaping Proteins in Cardiac Health and Disease - the Long and the Short of It!, Cardiovasc Drugs Ther 31 (2017) 87–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Knowlton AA, Liu TT, Mitochondrial Dynamics and Heart Failure, Compr Physiol 6 (2015) 507–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Burke N, Hall AR, Hausenloy DJ, OPA1 in Cardiovascular Health and Disease, Curr Drug Targets 16 (2015) 912–920. [DOI] [PubMed] [Google Scholar]
- [15].Nan J, Zhu W, Rahman MS, Liu M, Li D, Su S, Zhang N, Hu X, Yu H, Gupta MP, Wang J, Molecular regulation of mitochondrial dynamics in cardiac disease, Biochim Biophys Acta Mol Cell Res 1864 (2017) 1260–1273. [DOI] [PubMed] [Google Scholar]
- [16].Sekine S, Wang C, Sideris DP, Bunker E, Zhang Z, Youle RJ, Reciprocal Roles of Tom7 and OMA1 during Mitochondrial Import and Activation of PINK1, Mol Cell 73 (2019) 1028–1043 e1025. [DOI] [PubMed] [Google Scholar]
- [17].Costa-Mattioli M, Walter P, The integrated stress response: From mechanism to disease, Science 368 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Alavi MV, Fuhrmann N, Dominant optic atrophy, OPA1, and mitochondrial quality control: understanding mitochondrial network dynamics, Mol Neurodegener 8 (2013) 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lynch DS, Loh SHY, Harley J, Noyce AJ, Martins LM, Wood NW, Houlden H, Plun-Favreau H, Nonsyndromic Parkinson disease in a family with autosomal dominant optic atrophy due to OPA1 mutations, Neurol Genet 3 (2017) e188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Carelli V, Musumeci O, Caporali L, Zanna C, La Morgia C, Del Dotto V, Porcelli AM, Rugolo M, Valentino ML, Iommarini L, Maresca A, Barboni P, Carbonelli M, Trombetta C, Valente EM, Patergnani S, Giorgi C, Pinton P, Rizzo G, Tonon C, Lodi R, Avoni P, Liguori R, Baruzzi A, Toscano A, Zeviani M, Syndromic parkinsonism and dementia associated with OPA1 missense mutations, Ann Neurol 78 (2015) 21–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Manczak M, Calkins MJ, Reddy PH, Impaired mitochondrial dynamics and abnormal interaction of amyloid beta with mitochondrial protein Drp1 in neurons from patients with Alzheimer’s disease: implications for neuronal damage, Human molecular genetics 20 (2011) 2495–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wang X, Su B, Lee HG, Li X, Perry G, Smith MA, Zhu X, Impaired balance of mitochondrial fission and fusion in Alzheimer’s disease, The Journal of neuroscience : the official journal of the Society for Neuroscience 29 (2009) 9090–9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hinton DR, Sadun AA, Blanks JC, Miller CA, Optic-nerve degeneration in Alzheimer’s disease, N Engl J Med 315 (1986) 485–487. [DOI] [PubMed] [Google Scholar]
- [24].Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, Jones N, Thomas C, Stretton A, Morgan AR, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Morgan K, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Love S, Kehoe PG, Hardy J, Mead S, Fox N, Rossor M, Collinge J, Maier W, Jessen F, Schurmann B, Heun R, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Hull M, Rujescu D, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Muhleisen TW, Nothen MM, Moebus S, Jockel KH, Klopp N, Wichmann HE, Carrasquillo MM, Pankratz VS, Younkin SG, Holmans PA, O’Donovan M, Owen MJ, Williams J, Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease, Nat Genet 41 (2009) 1088–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Alavi MV, Tau phosphorylation and OPA1 proteolysis are unrelated events: Implications for Alzheimer’s Disease, Biochim Biophys Acta Mol Cell Res (2021) 119116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Alavi MV, Targeted OMA1 therapies for cancer, Int J Cancer 145 (2019) 2330–2341. [DOI] [PubMed] [Google Scholar]
- [27].Alavi MV, OMA1 High-Throughput Screen Reveals Protease Activation by Kinase Inhibitors, ACS Chem Biol 16 (2021) 2202–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kong B, Wang Q, Fung E, Xue K, Tsang BK, p53 is required for cisplatin-induced processing of the mitochondrial fusion protein L-Opa1 that is mediated by the mitochondrial metallopeptidase Oma1 in gynecologic cancers, J Biol Chem 289 (2014) 27134–27145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Zhao X, Tian C, Puszyk WM, Ogunwobi OO, Cao M, Wang T, Cabrera R, Nelson DR, Liu C, OPA1 downregulation is involved in sorafenib-induced apoptosis in hepatocellular carcinoma, Lab Invest 93 (2013) 8–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wu Z, Zuo M, Zeng L, Cui K, Liu B, Yan C, Chen L, Dong J, Shangguan F, Hu W, He H, Lu B, Song Z, OMA1 reprograms metabolism under hypoxia to promote colorectal cancer development, EMBO Rep 22 (2021) e50827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Daverey A, Levytskyy RM, Stanke KM, Viana MP, Swenson S, Hayward SL, Narasimhan M, Khalimonchuk O, Kidambi S, Depletion of mitochondrial protease OMA1 alters proliferative properties and promotes metastatic growth of breast cancer cells, Sci Rep 9 (2019) 14746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Quiros PM, Ramsay AJ, Sala D, Fernandez-Vizarra E, Rodriguez F, Peinado JR, Fernandez-Garcia MS, Vega JA, Enriquez JA, Zorzano A, Lopez-Otin C, Loss of mitochondrial protease OMA1 alters processing of the GTPase OPA1 and causes obesity and defective thermogenesis in mice, EMBO J 31 (2012) 2117–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Alavi MV, OMA1-An integral membrane protease?, Biochim Biophys Acta Proteins Proteom 1869 (2020) 140558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Leroy I, Khosrobakhsh F, Diot A, Daloyau M, Arnaune-Pelloquin L, Cavelier C, Emorine LJ, Belenguer P, Processing of the dynamin Msp1p in S. pombe reveals an evolutionary switch between its orthologs Mgm1p in S. cerevisiae and OPA1 in mammals, FEBS Lett 584 (2010) 3153–3157. [DOI] [PubMed] [Google Scholar]
- [35].Leonhard K, Herrmann JM, Stuart RA, Mannhaupt G, Neupert W, Langer T, AAA proteases with catalytic sites on opposite membrane surfaces comprise a proteolytic system for the ATP-dependent degradation of inner membrane proteins in mitochondria, EMBO J 15 (1996) 4218–4229. [PMC free article] [PubMed] [Google Scholar]
- [36].Anand R, Wai T, Baker MJ, Kladt N, Schauss AC, Rugarli E, Langer T, The i-AAA protease YME1L and OMA1 cleave OPA1 to balance mitochondrial fusion and fission, J Cell Biol 204 (2014) 919–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Rainbolt TK, Lebeau J, Puchades C, Wiseman RL, Reciprocal Degradation of YME1L and OMA1 Adapts Mitochondrial Proteolytic Activity during Stress, Cell reports 14 (2016) 2041–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Baker MJ, Lampe PA, Stojanovski D, Korwitz A, Anand R, Tatsuta T, Langer T, Stress-induced OMA1 activation and autocatalytic turnover regulate OPA1-dependent mitochondrial dynamics, EMBO J 33 (2014) 578–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Zhang K, Li H, Song Z, Membrane depolarization activates the mitochondrial protease OMA1 by stimulating self-cleavage, EMBO Rep 15 (2014) 576–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Anderson CJ, Kahl A, Fruitman H, Qian L, Zhou P, Manfredi G, Iadecola C, Prohibitin levels regulate OMA1 activity and turnover in neurons, Cell Death Differ 27 (2020) 1896–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ishihara N, Fujita Y, Oka T, Mihara K, Regulation of mitochondrial morphology through proteolytic cleavage of OPA1, EMBO J 25 (2006) 2966–2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Bohovych I, Donaldson G, Christianson S, Zahayko N, Khalimonchuk O, Stress-triggered activation of the metalloprotease Oma1 involves its C-terminal region and is important for mitochondrial stress protection in yeast, J Biol Chem 289 (2014) 13259–13272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Khalimonchuk O, Jeong MY, Watts T, Ferris E, Winge DR, Selective Oma1 protease-mediated proteolysis of Cox1 subunit of cytochrome oxidase in assembly mutants, J Biol Chem 287 (2012) 7289–7300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Viana MP, Levytskyy RM, Anand R, Reichert AS, Khalimonchuk O, Protease OMA1 modulates mitochondrial bioenergetics and ultrastructure through dynamic association with MICOS complex, iScience 24 (2021) 102119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Tang J, Zhang K, Dong J, Yan C, Hu C, Ji H, Chen L, Chen S, Zhao H, Song Z, Sam50-Mic19-Mic60 axis determines mitochondrial cristae architecture by mediating mitochondrial outer and inner membrane contact, Cell Death Differ 27 (2020) 146–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Tobacyk J, Parajuli N, Shrum S, Crow JP, MacMillan-Crow LA, The first direct activity assay for the mitochondrial protease OMA1, Mitochondrion 46 (2019) 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Sekine S, Youle RJ, PINK1 import regulation; a fine system to convey mitochondrial stress to the cytosol, BMC Biol 16 (2018) 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Bader V, Winklhofer KF, PINK1 and Parkin: team players in stress-induced mitophagy, Biol Chem 401 (2020) 891–899. [DOI] [PubMed] [Google Scholar]
- [49].Houston R, Sekine Y, Larsen MB, Murakami K, Mullett SJ, Wendell SG, Narendra DP, Chen BB, Sekine S, Discovery of bactericides as an acute mitochondrial membrane damage inducer, Mol Biol Cell (2021) mbcE21040191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ho PI, Yue K, Pandey P, Breault L, Harbinski F, McBride AJ, Webb B, Narahari J, Karassina N, Wood KV, Hill A, Auld DS, Reporter enzyme inhibitor study to aid assembly of orthogonal reporter gene assays, ACS Chem Biol 8 (2013) 1009–1017. [DOI] [PubMed] [Google Scholar]
- [51].Marroquin LD, Hynes J, Dykens JA, Jamieson JD, Will Y, Circumventing the Crabtree effect: replacing media glucose with galactose increases susceptibility of HepG2 cells to mitochondrial toxicants, Toxicol Sci 97 (2007) 539–547. [DOI] [PubMed] [Google Scholar]
- [52].Ghelli A, Zanna C, Porcelli AM, Schapira AH, Martinuzzi A, Carelli V, Rugolo M, Leber’s hereditary optic neuropathy (LHON) pathogenic mutations induce mitochondrial-dependent apoptotic death in transmitochondrial cells incubated with galactose medium, J Biol Chem 278 (2003) 4145–4150. [DOI] [PubMed] [Google Scholar]
- [53].Yang F, Wu R, Jiang Z, Chen J, Nan J, Su S, Zhang N, Wang C, Zhao J, Ni C, Wang Y, Hu W, Zeng Z, Zhu K, Liu X, Hu X, Zhu W, Yu H, Huang J, Wang J, Leptin increases mitochondrial OPA1 via GSK3-mediated OMA1 ubiquitination to enhance therapeutic effects of mesenchymal stem cell transplantation, Cell death & disease 9 (2018) 556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Makino A, Suarez J, Gawlowski T, Han W, Wang H, Scott BT, Dillmann WH, Regulation of mitochondrial morphology and function by O-GlcNAcylation in neonatal cardiac myocytes, Am J Physiol Regul Integr Comp Physiol 300 (2011) R1296–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Samant SA, Zhang HJ, Hong Z, Pillai VB, Sundaresan NR, Wolfgeher D, Archer SL, Chan DC, Gupta MP, SIRT3 deacetylates and activates OPA1 to regulate mitochondrial dynamics during stress, Molecular and cellular biology 34 (2014) 807–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Signorile A, Santeramo A, Tamma G, Pellegrino T, D’Oria S, Lattanzio P, De Rasmo D, Mitochondrial cAMP prevents apoptosis modulating Sirt3 protein level and OPA1 processing in cardiac myoblast cells, Biochim Biophys Acta Mol Cell Res 1864 (2017) 355–366. [DOI] [PubMed] [Google Scholar]
- [57].Sheng X, Cristea IM, The antiviral sirtuin 3 bridges protein acetylation to mitochondrial integrity and metabolism during human cytomegalovirus infection, PLoS Pathog 17 (2021) e1009506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Barrera M, Koob S, Dikov D, Vogel F, Reichert AS, OPA1 functionally interacts with MIC60 but is dispensable for crista junction formation, FEBS Lett 590 (2016) 3309–3322. [DOI] [PubMed] [Google Scholar]
- [59].Glytsou C, Calvo E, Cogliati S, Mehrotra A, Anastasia I, Rigoni G, Raimondi A, Shintani N, Loureiro M, Vazquez J, Pellegrini L, Enriquez JA, Scorrano L, Soriano ME, Optic Atrophy 1 Is Epistatic to the Core MICOS Component MIC60 in Mitochondrial Cristae Shape Control, Cell reports 17 (2016) 3024–3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Bao YC, Tsuruga H, Hirai M, Yasuda K, Yokoi N, Kitamura T, Kumagai H, Identification of a human cDNA sequence which encodes a novel membrane-associated protein containing a zinc metalloprotease motif, DNA Res 10 (2003) 123–128. [DOI] [PubMed] [Google Scholar]
- [61].Shiba-Fukushima K, Inoshita T, Sano O, Iwata H, Ishikawa KI, Okano H, Akamatsu W, Imai Y, Hattori N, A Cell-Based High-Throughput Screening Identified Two Compounds that Enhance PINK1-Parkin Signaling, iScience 23 (2020) 101048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Gegg ME, Cooper JM, Chau KY, Rojo M, Schapira AH, Taanman JW, Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy, Human molecular genetics 19 (2010) 4861–4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Tanaka A, Cleland MM, Xu S, Narendra DP, Suen DF, Karbowski M, Youle RJ, Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin, J Cell Biol 191 (2010) 1367–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Kimura Y, Kioka N, Kato H, Matsuo M, Ueda K, Modulation of drug-stimulated ATPase activity of human MDR1/P-glycoprotein by cholesterol, Biochem J 401 (2007) 597–605. [DOI] [PMC free article] [PubMed] [Google Scholar]