Abstract
Introduction
Genomic surveillance of the SARS-CoV-2 virus is important to assess transmissibility, disease severity, and vaccine effectiveness. The SARS-CoV-2 genome consists of approximately 30 kb single-stranded RNA that is too large to analyze the whole genome by Sanger sequencing. Thus, in this study, we performed Sanger sequencing following long-range RT-PCR of the entire SARS-CoV-2 S-gene and analyzed the mutational dynamics.
Methods
The 4 kb region, including the S-gene, was amplified by two-step long-range RT-PCR. Then, the entire S-gene sequence was determined by Sanger sequencing. The amino acid mutations were identified as compared with the reference SARS-CoV-2 genome.
Results
The S:D614G mutation was found in all samples. The R.1 variants were detected after January 2021. Alpha variants started to emerge in April 2021. Delta variants replaced Alpha in July 2021. Then, Omicron variants were detected after December 2021. These mutational dynamics in samples collected in the Chiba University Hospital were similar to those in Japan.
Conclusion
The emergence of variants of concern (VOC) has been reported by the entire S-gene analysis. As the VOCs have unique mutational patterns of the S-gene region, analysis of the entire S-gene will be useful for molecular surveillance of the SARS-CoV-2 in clinical laboratories.
Keywords: Surveillance, SARS-CoV-2, S-gene, Long-range RT-PCR, Sanger sequencing
1. Introduction
The global coronavirus disease 2019 (COVID-19) pandemic is a serious health problem caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [1]. The emergence of the variants of concern (VOC) and the variants of interest (VOI), which increase transmissibility, were reported [2], [3]. Genomic surveillance is important to investigate virus transmission dynamics, detect the novel genetic variants and, assess the impact of mutations on the performance of molecular diagnostic methods, antiviral drugs, and the effectiveness of the vaccine [2], [4], [5].
The SARS-CoV-2 genome consists of 29,903 bases long single-stranded RNA [6]. The S-gene is composed of 3,822 bases and encodes spike proteins (1,273 amino acids) covering the surface of SARS-CoV-2 [6]. This spike protein binds to the host cell receptor and cell membranes before the release of the virus genome. The spike protein is essential for transmissibility and targeting vaccine development [4], [5]. Additionally, VOC and VOI have unique mutational patterns of the spike protein [7]. Thus, the determination of the S-gene sequence will be useful for the estimation of SARS-CoV-2 variants.
We have mainly two sequencing options: Sanger sequencing or massively parallel sequencing (MPS). The MPS has been commonly used for the genomic surveillance of SARS-CoV-2 [8]. The MPS allows multiple samples to be sequenced together; however, it is a less cost-efficient method when it comes to detecting mutations in smaller samples. As compared with MPS, Sanger sequencing is a gold standard method, easy-to-use, cost-effective if few targets are required, and available at many clinical laboratories [8].
The polymerase chain reaction (PCR)-based target enrichment strategy such as multiplex tiling PCR has been widely used for genomic sequencing (the ARTIC Network, https://artic.network/ncov-2019). The major limitation of PCR-based target enrichment is that the amplification failure may occur as a result of mutations in primer binding sites [8]. In contrast to conventional PCR, long-range PCR requires fewer primers to amplify the target regions. Moreover, long-amplicon is suitable for Sanger sequencing, which can obtain relatively longer sequence reads.
In this study, we analyzed the whole length of the S-gene for tracking SARS-CoV-2 variants by using long-range reverse transcription-polymerase chain reaction (RT-PCR) followed by Sanger sequencing. The lineage of VOCs was estimated by the mutational pattern of the S-gene. Then, the prevalence of the VOCs in Chiba University Hospital over time was observed and compared with the respective duration in Japan to examine the usefulness of this method.
2. Materials and methods
2.1. Specimens
In this study, the SARS-CoV-2 RNA positive samples (>100 copies/uL, Cq > 30) from November 2020, to January 2022, at Chiba University Hospital were included.
2.2. Detection and quantification of SARS-CoV-2 RNA
The SARS-CoV-2 RNA detection test was performed by using the real-time RT-PCR kit (Ampdirect 2019-nCoV detection kit, Shimadzu, Kyoto, Japan). Then, positive samples were quantified by using the previously described multiplex RT-qPCR methodology [9].
2.3. Long-range RT-PCR
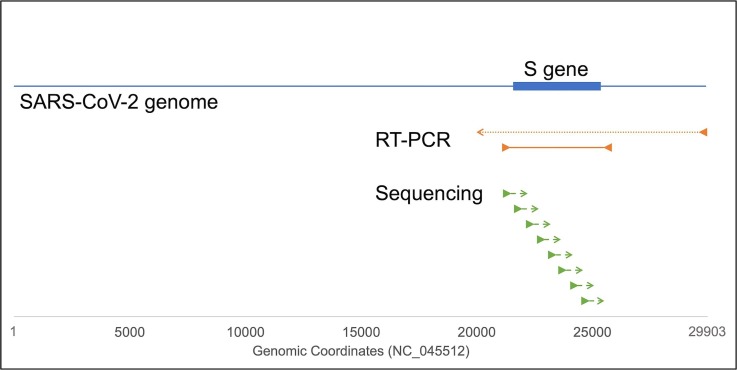
The reverse transcription (RT) was performed using a PrimeScript IV 1st strand cDNA Synthesis Mix (Takara, Shiga, Japan). Each 5 μL reaction mixture contained 1 ul of PrimeScript IV cDNA synthesis mix (oligo dT primer included) and 4 μL of extracted viral RNA. The RT reaction was done at 42 °C for 20 min followed by enzyme inactivation at 70 °C for 15 min. Then, the 4 kb region including the entire S-gene of SARS-CoV-2 was amplified by long-range PCR (Fig. 1 ). The 20 μL of PCR reaction included 10 μL of 2 × Gflex PCR buffer, 0.4 μL of Tks Gflex DNA polymerase (Final 0.5 U), 0.2 µM of each forward and reverse primers (Table 1 ), 1 μL of 20 × EvaGreen Dye (Biotium, CA, USA), 2 μL of the cDNA, and 4.6 μL of nuclease-free water. The real-time PCR was performed using a LightCycler Nano instrument (Roche Diagnostics, Mannheim, Germany). The PCR cycling program was as follows: pre-incubation at 94 °C for 1 min; followed by 40 cycles at 98 °C for 10 s and 65 °C for 180 s (signal acquisition); melting at 98 °C for 30 s, 65 °C for 30 s, and a continuous increase in temperature from 65 °C to 98 °C at the rate of 0.5 °C/s with signal acquisitions. After amplification check (Supplementary Fig. 1), the amplicons were purified using an equal volume of AMPure XP (Beckman coulter, Brea, CA, USA), according to the manufacturer’s instruction. Purified amplicons were eluted in 50 uL nuclease-free water.
Fig. 1.
Method overview of the long-range RT-PCR followed by Sanger sequencing. The SARS-CoV-2 genome consists of 29,903 bases (blue line). The S-gene is encoded 21,563–25,384 position of the genome (blue box). “RT-PCR,” dotted line and solid lines indicate first-strand cDNA and following PCR amplification region, respectively. Sequencing, dashed lines indicate sequencing regions. Triangles indicate primer binding sites.
Table 1.
Primers used to amplify the cDNA.
| Name | Sequence (5′ > 3′) | Genomic coordinates(NC_045512) |
|---|---|---|
| For PCR | ||
| SC2-S-4kbF | aggggtactgctgttatgtcttt | 21,421–21,443 |
| SC2-S-4kbR | aggcttgtatcggtatcgttgc | 25,489–25,510 |
| For Sanger sequencing | ||
| SC2-S-SeqF1 | tgatatgattttatctcttcttagtaaagg | 21,462–21,491 |
| SC2-S-SeqF2 | tgtgaatttcaattttgtaatgatcc | 21,953–21,978 |
| SC2-S-SeqF3 | agtgatcgttgaaatccttcactg | 22,461–22,484 |
| SC2-S-SeqF4 | ttgtttaggaagtctaatctcaaacc | 22,925–22,950 |
| SC2-S-SeqF5 | aagtccctgttgctattcatgc | 23,418–23,439 |
| SC2-S-SeqF6 | aactggaatagctgttgaacaagac | 23,863–23,887 |
| SC2-S-SeqF7 | attcaagactcactttcttccacag | 24,362–24,386 |
| SC2-S-SeqF8 | ggcacacactggtttgtaacac | 24,857–24,878 |
2.4. Sanger sequencing
The BigDye Terminator v3.1 cycle sequencing kit (Thermo Fisher Scientific, MA, USA) was used for the cycle sequencing reaction. Each 10 µL reaction mixture contained 1 µL of BigDye Terminator ready reaction mix, 2 µL of 5 × sequencing buffer, 0.4 µM of each sequencing primer (Table 1), and 40 ng of purified amplicons; the cycling conditions were as follows: 96 °C for 1 min, followed by 25 cycles at 96 °C for 10 s, 50 °C for 5 s, and 60 °C for 1 min. The cycle sequencing products were purified using the BigDye XTerminator purification kit (Thermo Fisher Scientific), according to the manufacturer’s protocol. Then, capillary electrophoresis was performed using the DS3000 compact sequencer (Hitachi High-Tech Corporation, Tokyo, Japan). The consensus sequence of S-gene was generated by using Unipro UGENE software (version 37) [10].
2.5. Identification of amino acid mutations
The amino acid mutations and types of VOC were determined by Nextclade (v1.12.0, https://clades.nextstrain.org) [11]. The Wuhan-Hu-1/2019 (genbank: MN908947) was used as a reference genome.
2.6. Statistical analysis
Statistical analysis were performed using R version 4.1.2 statistical software [12]. Spearman rank-order correlation test were performed using the “stats” package.
3. Results
3.1. Long-range RT-PCR
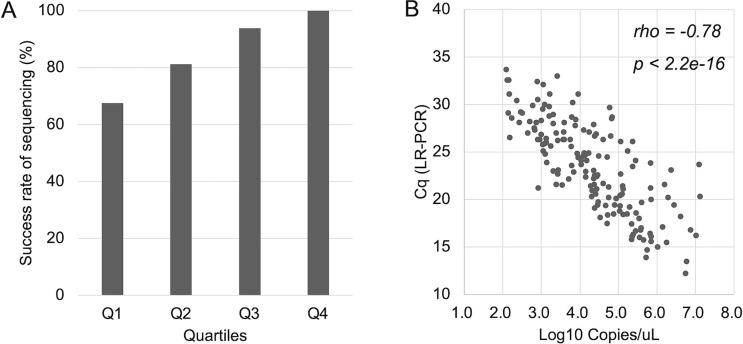
Total 183 samples were included for long-rage RT-PCR analysis. The samples were stratified into quartile groups according to viral RNA concentration: Q1 (equal or lower than 3.0 log10 copies/uL, Cq > 26), Q2 (>3.0–≤4.0 log10 copies/uL, Cq 23–26), Q3 (>4.0–≤5.0 log10 copies/uL, Cq 20–23), and Q4 (>5.0 log10 copies/uL, Cq < 20). Among them, 158 samples (86.3 %) were successfully amplified and sequenced the entire S-gene. The success rate of sequencing was correlated with viral RNA concentration: Q1, 68% (27/40); Q2, 81% (39/48); Q3, 94% (46/49); and Q4, 100% (46/46) (Fig. 2 A). Moreover, the Cq values of long-range RT-PCR were also negatively correlated with viral RNA concentration (Fig. 2B, r = –0.77).
Fig. 2.
Correlations of viral RNA concentration, the success rate of sequencing, and the Cq values of long-range RT-PCR. (A) viral RNA concentration vs. success rate of sequencing. Quartile groups: Q1 (≤3.0 log10 copies/uL), Q2 (>3.0–≤4.0 log10 copies/uL), Q3 (>4.0–≤5.0 log10 copies/uL), and Q4 (>5.0 log10 copies/uL). The success rate of sequencing: Q1, 68% (27/40); Q2, 81% (39/48); Q3, 94% (46/49); and Q4, 100% (46/46). (B) viral RNA concentration vs. Cq values of long-range RT-PCR. The Cq values of long-range RT-PCR were negatively correlated with viral RNA concentration. The p-value was determined by Spearman rank-order correlation test.
3.2. Tracking SARS-CoV-2 variants
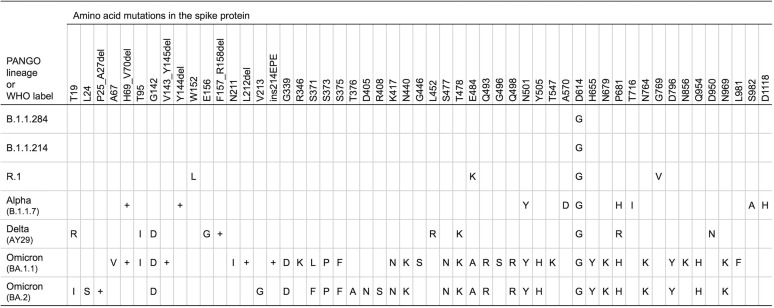
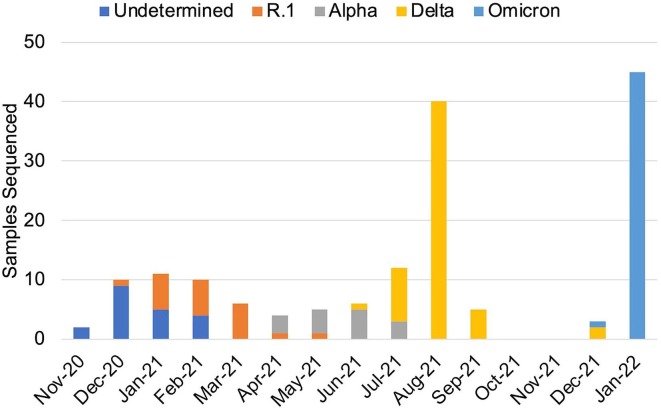
According to the outbreak information webpage (https://outbreak.info/), the frequently observed lineages of the SARS-CoV-2 in Japan were B.1.1.284, B.1.1.214, R.1, Alpha, Delta, and Omicron (Supplementary Fig. 2) [13]. World Health Organization (WHO) classified R.1 as formerly monitored variants, and Alpha, Delta, and Omicron variants were listed as VOCs. The mutational patterns of spike protein in these SARS-CoV-2 lineages were shown in Fig. 3 . Samples that could not determine the lineage of SARS-CoV-2 because of the lack of specific mutation patterns except for S:D614G were classified as “Undetermined.” To assess the dynamic of circulating SARS-CoV-2, we compared our data with the reported lineages in Chiba University Hospital (Fig. 4 ). All of the sequenced samples had S:D614G mutation. Some samples collected from November 2020 to February 2021 showed a few mutations, but could not be determined lineages. Time to time analysis revealed that R.1 lineage was the dominant variant among the samples collected from January to March 2021. The Alpha variants replaced R.1 and were dominant from April to June 2021. Delta variants were increased and replaced Alpha variants from July to September 2021 (Fig. 4). Many sub-lineages in Delta variants have been reported [11]. Almost all Delta variants analyzed in this study were estimated AY.29 sub-lineage because of detection of the S:T95I and S:G142D mutations. Then, Omicron variants were detected and increased after December 2021. Almost all Omicron variants analyzed in this study were estimated BA.1.1 sub-lineage because of detection of the S:R346K mutations. Notably, a BA.2 sub-lineage of Omicron variant was also detected in January 2022. These mutational dynamics in Chiba University Hospital were similar to those in Japan [13].
Fig. 3.
The mutational pattern of spike protein among the frequently observed SARS-CoV-2 variants in Japan.
Fig. 4.
Tracking SARS-CoV-2 variants by the spike protein mutation. “Undetermined” includes various lineages of SARS-CoV-2 which could not be determined the lineages because of the lack of specific mutation patterns except for S:D614G.
4. Discussion
In this study, we developed long-range RT-PCR followed by Sanger sequencing for surveillance of SARS-CoV-2 variants. More than 80% of the samples were able to analyze the entire S-gene sequence. In addition, current long-range RT-PCR products can be also available for high-throughput analysis using nanopore sequencing (Supplementary Fig. 3). Therefore, our method will be useful for tracking SARS-CoV-2 variants in many clinical laboratories equipped with conventional Sanger sequencing instruments.
Recently, real-time PCR-based screening methods were used for mutational analysis [14]. These methods were simple to rapidly determine S-gene mutation. However, this method could not identify the novel mutations. Thus, a sequencing-based approach will be also required for the determination of SARS-CoV-2 variants, such as VOCs and VOIs.
Similarly, the MPS is widely used for the whole genome sequencing of SARS-CoV-2. Since bioinformatics analysis is an essential step for MPS, expertise is required for data analysis [8]. Additionally, the running cost of MPS is relatively high in the analysis of a small number of samples. In these situations, Sanger sequencing is useful for molecular surveillance.
The major limitation of our Sanger sequencing approach is the lineage estimation by the sequences of the S-gene only, which is unable to conclude lineage exactly. Therefore, whole genome sequencing to identify the novel mutations will be required for correct lineage identification. Moreover, the mutations at primer binding sites will affect the result of sequencing.
In conclusion, the emergence of VOCs, which have increased transmissibility, has been reported worldwide. These VOCs have unique mutational patterns of the S-gene. Therefore, the sequencing and analysis of the entire S-gene is a potential tool for molecular surveillance of the SARS-CoV-2 at clinical laboratories.
CRediT authorship contribution statement
Mirai Matsubara: Investigation, Writing – original draft. Yuri Imaizumi: Investigation. Tatsuki Fujikawa: Investigation. Takayuki Ishige: Conceptualization, Investigation, Writing – review & editing. Motoi Nishimura: Conceptualization, Writing – review & editing. Akiko Miyabe: Investigation. Shota Murata: Investigation. Kenji Kawasaki: Supervision. Toshibumi Taniguchi: Resources, Supervision. Hidetoshi Igari: Resources, Supervision, Project administration. Kazuyuki Matsushita: Writing – review & editing, Supervision, Project administration.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cca.2022.03.014.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.World Health Organization (WHO), WHO Coronavirus (COVID-19) Dashboard. https://covid19.who.int, 2021 (accessed 16 December 2021).
- 2.Volz E., Mishra S., Chand M., Barrett J.C., Johnson R., Geidelberg L., Hinsley W.R., Laydon D.J., Dabrera G., O’Toole Á., Amato R., Ragonnet-Cronin M., Harrison I., Jackson B., Ariani C.V., Boyd O., Loman N.J., McCrone J.T., Gonçalves S., Jorgensen D., Myers R., Hill V., Jackson D.K., Gaythorpe K., Groves N., Sillitoe J., Kwiatkowski D.P., Flaxman S., Ratmann O., Bhatt S., Hopkins S., Gandy A., Rambaut A., Ferguson N.M. Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature. 2021;593(7858):266–269. doi: 10.1038/s41586-021-03470-x. [DOI] [PubMed] [Google Scholar]
- 3.Singh J., Rahman S.A., Ehtesham N.Z., Hira S., Hasnain S.E. SARS-CoV-2 variants of concern are emerging in India. Nat. Med. 2021;27:1131–1133. doi: 10.1038/s41591-021-01397-4. [DOI] [PubMed] [Google Scholar]
- 4.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., Bailey R., Swanson K.A., Roychoudhury S., Koury K., Li P., Kalina W.V., Cooper D., Frenck R.W., Jr, Hammitt L.L., Türeci Ö., Nell H., Schaefer A., Ünal S., Tresnan D.B., Mather S., Dormitzer P.R., Şahin U., Jansen K.U., Gruber W.C. C4591001 Clinical Trial Group. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., McGettigan J., Khetan S., Segall N., Solis J., Brosz A., Fierro C., Schwartz H., Neuzil K., Corey L., Gilbert P., Janes H., Follmann D., Marovich M., Mascola J., Polakowski L., Ledgerwood J., Graham B.S., Bennett H., Pajon R., Knightly C., Leav B., Deng W., Zhou H., Han S., Ivarsson M., Miller J., Zaks T., COVE Study Group Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oude Munnink B.B., Worp N., Nieuwenhuijse D.F., Sikkema R.S., Haagmans B., Fouchier R.A.M., Koopmans M. The next phase of SARS-CoV-2 surveillance: real-time molecular epidemiology. Nat. Med. 2021;27:1518–1524. doi: 10.1038/s41591-021-01472-w. [DOI] [PubMed] [Google Scholar]
- 8.World Health Organization . World Health Organization; 2021. Genomic sequencing of SARS-CoV-2: a guide to implementation for maximum impact on public health, 8 January 2021.https://apps.who.int/iris/handle/10665/338480 [Google Scholar]
- 9.Ishige T., Murata S., Taniguchi T., Miyabe A., Kitamura K., Kawasaki K., Nishimura M., Igari H., Matsushita K. Highly sensitive detection of SARS-CoV-2 RNA by multiplex rRT-PCR for molecular diagnosis of COVID-19 by clinical laboratories. Clin. Chim. Acta. 2020;507:139–142. doi: 10.1016/j.cca.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okonechnikov K., Golosova O., Fursov M. UGENE team. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics. 2012;28:1166–1167. doi: 10.1093/bioinformatics/bts091. [DOI] [PubMed] [Google Scholar]
- 11.Aksamentov I., Roemer C., Hodcroft E.B., Neher R.A. Nextclade: clade assignment, mutation calling and quality control for viral genomes. J. Open Source Softw. 2021;6:3773. [Google Scholar]
- 12.R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2021. R: A language and environment for statistical computing.https://www.R-project.org/ [Google Scholar]
- 13.Julia L. Mullen, Ginger Tsueng, Alaa Abdel Latif, Manar Alkuzweny, Marco Cano, Emily Haag, Jerry Zhou, Mark Zeller, Emory Hufbauer, Nate Matteson, Kristian G. Andersen, Chunlei Wu, Andrew I. Su, Karthik Gangavarapu, Laura D. Hughes, and the Center for Viral Systems Biology outbreak.info. Available online: https://outbreak.info/ (2020).
- 14.Wang H., Jean S., Eltringham R., Madison J., Snyder P., Tu H., Jones D.M., Leber A.L. Mutation-specific SARS-CoV-2 PCR screen: rapid and accurate detection of variants of concern and the identification of a newly emerging variant with spike L452R mutation. J. Clin. Microbiol. 2021;59 doi: 10.1128/JCM.00926-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.