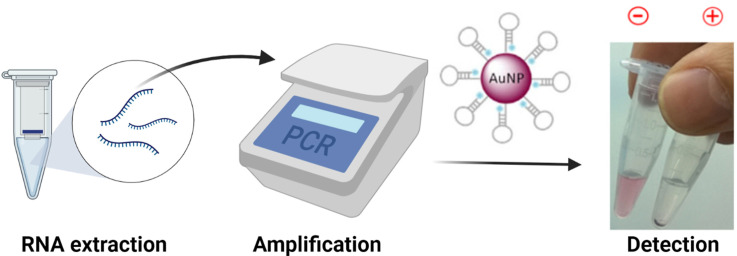
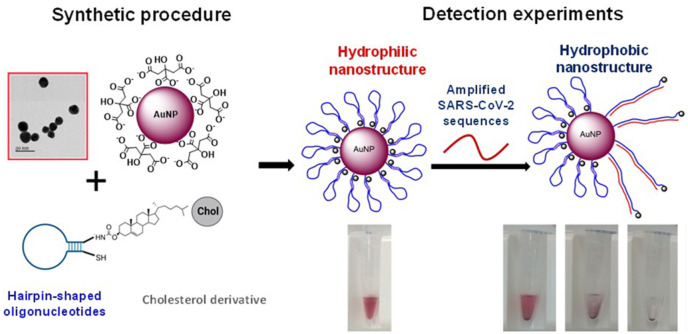
Abstract
We present a fast, reliable and easy to scale-up colorimetric sensor based on gold nanoparticles (AuNPs) to detect the sequences coding for the RdRp, E, and S proteins of SARS-CoV-2. The optimization of the system (so-called “the sensor”) includes the evaluation of different sizes of nanoparticles, sequences of oligonucleotides and buffers. It is stable for months without any noticeable decrease in its activity, allowing the detection of SARS-CoV-2 sequences by the naked eye in 15 min. The efficiency and selectivity of detection, in terms of significative colorimetric changes in the solution upon target recognition, are qualitatively (visually) and quantitatively (absorbance measurements) assessed using synthetic samples and samples derived from infected cells and patients. Furthermore, an easy and affordable amplification approach is implemented to increase the system's sensitivity for detecting high and medium viral loads (≥103 - 104 viral RNA copies/μl) in patient samples. The whole process (amplification and detection) takes 2.5 h. Due to the ease of use, stability and minimum equipment requirements, the proposed approach can be a valuable tool for the detection of SARS-CoV-2 at facilities with limited resources.
Keywords: SARS-CoV-2, Gold nanoparticles, Nucleic acids, Detection, PCR
Graphical abstract
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was identified in December 2019 in Wuhan (China) as the cause of the illness designated COVID-19 [1], and it has rapidly spread all around the globe [2]. As a matter of fact, the ongoing SARS-CoV-2 pandemic is far from being solved, and its continuous surveillance should be maintained to monitor its evolution. In this scenario, the relatively low seroprevalence, especially in low and medium-income countries, is insufficient to provide herd immunity [3]. Universal vaccination still will take a long time and faces the challenge of virus mutations, which can reduce or even prevent an adequate immunization. Therefore, fast, reliable, affordable, and efficient diagnostic methods for the rapid identification and isolation of infected patients, as well as contact tracing, are the foremost priorities to prevent the virus from spreading [4]. The detection of SARS-CoV-2 is currently done by three major procedures, where each one aims to detect an alternative target: i) viral RNA, ii) viral proteins and iii) host antibodies against the virus. The two first methods are suitable for detecting ongoing infections. The third approach is related to acquired immunity once exposed to the virus (Table S1). Based on all those strategies, companies and research centers are developing diagnostic systems worldwide. Since December 2019, more than 1.000 molecular tests, either commercially available or in development, have been registered in https://www.finddx.org/covid-19/. The latest advances have been reviewed in several articles, where the need for accurate and fast detection to prevent and control the COVID-19 spreading has been highlighted [[5], [6], [7]].
SARS-CoV-2 is an enveloped virus from the Coronaviridae family with a diameter of ∼100 nm [8]. Its genome is composed of a single-stranded positive-sense RNA that is around 30,000 nucleotides in length and encodes for 27 proteins, including the four structural proteins E (envelope), N (nucleocapsid), M (matrix) and S (spike), and RdRp or R (RNA-dependent RNA polymerase) [9,10].
The gold-standard method for SARS-CoV-2 detection is the reverse transcription quantitative polymerase chain reaction (RT-qPCR) test performed using nasopharyngeal swabs to target a variety of specific RNA viral sequences [11,12]. There are different proposed workflows [13,14], being the one described by Drosten lab [15] the most prevalently used in Europe [16]. The first step in their protocol consists of detecting all SARS-related viruses by targeting regions of the E gene, common for all Sarbecovirus subgenus members. If the test is positive, they propose detecting the R gene sequence, specific for SARS-CoV-2 [15]. The continuous emergence of single-nucleotide polymorphisms and deletions that might adversely affect RT-qPCRs justifies that at least two genomic regions should be targeted to avoid false negatives in the molecular diagnosis of SARS-CoV-2. Those genetic modifications can also be used as indicators for the screening of particular variants of concern (VOC), such as Omicron, with a negative or significantly weaker positive S-gene result (“S-gene dropout”) in multiplex RT-PCR assays, with positive results for the other targets [17].
RT-qPCR is widely used for its high specificity and sensitivity. However, this technique presents some disadvantages, such as the need for qualified personnel, sophisticated equipment (a thermocycler with fluorescent readout system), and expensive qPCR reagent kits (fluorescent probes), potentially limiting its application due to the current huge demand [18,19]. For these reasons, new, fast, low-cost and reliable point-of-care devices are needed [20].
To address this challenge, we have developed a colorimetric test based on gold nanoparticles (AuNPs) and an amplification procedure based on PCR. Particularly, we have optimized a sensor based on AuNPs to detect SARS-CoV-2 RNA by adapting an approach previously developed by us [21]. The sensor is the solution containing the AuNPs functionalized with the capture oligonucleotide (molecular beacon-like) in an optimized buffer formulation for the specific detection of the target. For the amplification method, we have implemented a PCR procedure, which just requires a modified primer and a standard thermocycler, and is completed in 1 h, after RNA extraction. Subsequent to the amplification step, T7 exonuclease digestion of one of the amplicon's strands leads to the ssDNA target for the direct and fast detection with our specific sensor.
Regarding the sensor, AuNPs were synthesized following the Turkevich method [22] and functionalized with cholesterol-modified oligonucleotides via thiol-Au bonds. These oligonucleotides, similar to molecular beacons (MB), have a hairpin-like structure with two main regions: the central loop, which is complementary to the target sequence, and the stem that is formed by a few nucleotides at each terminus, which are complementary to each other and maintain the molecule closed in the native state in the absence of its specific target. In this configuration, the cholesterol moieties are buried inside the nanostructure, and the solution has a reddish color. But when the target is present, the oligonucleotides unfold due to the hybridization between the target and the loop, and the cholesterol units become exposed to the aqueous media. The hydrophobic nature of cholesterol molecules leads to a color change in the solution because of the destabilization, aggregation and precipitation of AuNPs (Fig. 1 ).
Fig. 1.
Schematic representation of the sensor. Gold nanoparticles are modified with oligonucleotides containing a cholesterol moiety. The nanostructures are exposed to SARS-CoV-2 sequences, which induce the rearrangement of the hairpin structure of the oligonucleotides to an open structure. Thus, the cholesterol is exposed to the aqueous solution, causing the aggregation of the nanoparticles, detectable by the naked eye. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The selectivity and sensitivity of the sensors described were tested both qualitatively (visually) and quantitatively (absorbance measurements). It is worth mentioning that, compared to other approaches based on AuNPs’ aggregation [[23], [24], [25]], in this case, only one set of nanoparticles is needed. Those strategies require the precise equimolar combination of nanoparticles containing different oligonucleotides to target adjacent regions of the genomic nucleic acid. Thus, minimum changes in the ratio of the nanoparticles or the different accessibility to the target domains can reduce the sensitivity of the sensor. Therefore, using one set of nanoparticles provides a more reliable system. In addition, the multiplexed detection of two or three different but specific targets at a time, increases not only the sensibility for each individual gene but the trustability of the results. Also, although sensitive and fast colorimetric systems have been implemented recently for COVID-19 diagnosis, some of them require specialized knowledge or the use of additional ribonucleoproteins (such as CRISPR/Cas) [26,27].
To assess this approach and optimize the system, we tested different AuNP sizes (15–38 nm) and lengths of the oligonucleotide stems (4–6 nucleotides). Particularly, the oligonucleotides were designed to detect the R and E genes at the central region usually amplified in currently used protocols based on RT-qPCR [15]. Once the system was optimized, we explored the detection of insertions and single-point mutations in the RNA coding for the spike protein. This protein is located on the surface of the particle and is of great interest for the detection, but also from a clinical and immunological point of view [28,29]. The stability of this type of sensors was evaluated for more than 3 months in two alternative storage conditions: room temperature and 4 °C. We could observe that the system's functionality was not affected by the storage time at both temperatures for 15 days, being advisable its conservation at 4 °C for more extended periods.
To improve the sensitivity of the approach, we implemented an amplification process based on PCR. In this case, one of the primers must be modified with phosphorothioate groups at its 5 ‘end. Thus, after the amplification, the product can be digested with an exonuclease, degrading only one of the two chains (the one with the non-modified 5 ‘end), leaving the complementary one intact and accessible for detection by the gold nanoparticle-based sensor [30].
In view of this, we have developed sensors to detect specific loci of the genome of SARS-CoV-2 that can be easily produced, handled and modulated to detect different regions according to the interest.
2. Materials and methods
2.1. Oligonucleotides design, synthesis and evaluation
Molecular beacons were designed to directly recognized specific probe sequences of E and R genes [15], the insertion region of S protein [31] or the mutation D614G [32]. The mfold Web Server from the RNA Institute, College of Arts and Sciences, State University of New York at Albany, [33] was used to predict their folding structures and the thermodynamic parameters in the absence or presence of the targets in order to choose the best candidates. Oligonucleotides were either purchased from Integrated DNA Technologies (IDT, Belgium) or synthetized in a H8 DNA/RNA synthesizer (K&A Laborgeraete). Their purity and concentration were determined by UV–vis spectra (λ = 260 nm) in a Cary 5000 Spectrophotometer (Agilent Technologies). Scramble sequences used in the diagnosis of other human pathogenic Coronaviruses [34] (HKU1, NL63, 229E and OC43) or microorganisms associated with respiratory infections and co-infections in COVID-19 disease [35] (influenza virus A, influenza virus B [36], Mycoplasma pneumoniae [37] and Legionella pneumophila [38]) were used to challenge the selectivity of the system. The sequences are described in Table S2.
2.2. Synthesis of gold nanoparticles
AuNPs of different sizes (15–38 nm) were synthesized following the Turkevich method [22]. Briefly, a solution of hydrogen tetrachloroaurate (III) hydrate (FisherScientific) in RNAse free water was stirred and heated to reflux. Then, a solution of sodium citrate tribasic dehydrate, dissolved in the same water, was added and the mixture was stirred for 15 min. After that time, the solution was allowed to cool down at room temperature and the AuNPs obtained were filtered through a 20-40 µm glass filter plate with the help of a vacuum pump to get rid of large aggregates, followed by a second filtration with a PES syringe filter of 0.22 μM (Millipore). Depending on the size of the desired AuNPs, different concentrations of reagents were used with the same final volume (110 mL). In the case of the 15 nm AuNPs, 4.15 mM of citrate and 0.864 mM of gold (ratio of 4.8) were used. For 20–21 nm AnNPs, 3.62 mM of citrate and 0.23 mM of gold (ratio of 17.7). Finally, for the 38 nm AuNPs, 0.4 mM of citrate and 0.23 mM of gold (ratio of 1.75).
2.3. Characterization of gold nanoparticles
AuNPs’ size and shape were examined by transmission electron microscopy (TEM) and Atomic Force Microscopy (AFM). Gold nanoparticles were visualized using a 120 KeV JEM1400 Flash (Jeol, Japan) at the CBMSO-CSIC. Samples were prepared by placing a glow-discharged carbon-shadowed formvar-coated HEX 400-mesh copper grid (Agar Scientific) over one drop of AuNPs for 2 min and drying the excess of the sample. The shape and size distributions were determined through an automated analysis of randomly selected TEM images’ areas with Image J software. For AFM visualization, freshly cleaved mica was pre-treated with poly-l-Lysine (PLL) (15 μL) solution 0.1% (w/v) in H2O (Sigma-Aldrich) via 10 min incubation at room temperature. The surfaces were then rinsed twice with MilliQ water and dried. Once prepared, vortexed and diluted nanoparticles (15 μL) were absorbed into the substrate following a similar procedure as done with PLL. Topography images were obtained in tapping mode in air using a JPK Nanowizard 2 microscope and HQXSC11-D (Mikromash) cantilevers (nominal spring constant of 42 N/m and resonance frequency of 350 kHz). Their analysis was performed using WSxM program and its “flooding” tool with an N between the range of 100–1000 individual particles. The average and standard deviation of the particle size were determined using a Lorentz fitting implemented in the OriginPro software (OriginLab).
AuNPs’ size and concentration were determined from UV–vis spectra in a Cary 5000 Spectrophotometer (Agilent Technologies) according to a described procedure [39]. The size of the nanoparticles is determined by the ratio of the absorbance plasmon peak to the absorbance at 450 nm and TEM images. Based on the diameter obtained, the concentration is determined by using the proper extinction coefficient at the wavelength of 450 nm. This is only true for spherical nanoparticles (below 38 nm).
2.4. AuNP-based sensor preparation
Citrate-stabilized AuNPs were functionalized with different molecular beacon oligonucleotides (MB) described in Table S2 through the formation of stable thiol-Au bonds [40,41].
The amount of the molecular beacon that can be conjugated to the nanoparticles was determined using Equation (1):
| (1) |
where AAuNP is the surface area of the nanoparticle (4·π·r2), CAuNP is the molar concentration of AuNP in solution, D is the maximum oligonucleotide density on each particle (3.5 × 10−7 mol/m2) [42] and V is the volume of AuNPs in L.
According to this, AuNP of 15 nm at 15 nM can be functionalized with 2.23 pmol MB/μL of AuNP. The concentration of nanoparticles of different sizes was established in agreement with the concentration of MB mentioned before. However, the amount of MB added was twice the calculated (4.46 pmol MB/μL) to ensure that AuNP are covered with the maximum number of oligonucleotides.
Before proceeding with the functionalization, the MB were treated for 1 h at RT with a 100 times excess of tris(2-carboxyethyl) phosphine hydrochloride (TCEP) to remove the protecting groups of the thiol moieties [43]. Then, they were added to AuNP solution, vortexed, and shaken for 20 min for 15 nm AuNP or during 1 h for bigger particles (24 and 38 nm AuNPs). Then, NaCl 5 M was added every 30 min in 5 steps to get a final concentration in the solution of 0.3 M. The NaCl was mixed by quick vortexing. The AuNP were incubated overnight in darkness under shaking at RT. After that, they were washed by two cycles of centrifugation and redispersed in water. To determine the loading of oligonucleotides onto the nanoparticles, the absorbance of the supernatants was quantified (λ = 260 nm) with the Synergy H4 microplate reader, and the value was extrapolated from the corresponding calibration curve (Table S3).
Different modified nanoparticles were produced containing one, two or three MB in the same nanostructure. Particularly, the sensors (AuNPs + MB) containing one MB were: E, R, S Ins, D614, G614. In the case of two MB the sensors were: E + R, E + S Ins, R + S Ins. For the preparation of this multifunctional nanoparticle, 2.23 pmol of each MB per μL of AuNP were added. The sensor combining three MB was E + R + S Ins, and in this case, the 1.49 pmol of each MB were added per μL of AuNP.
2.5. Colorimetric detection experiments
For the regular detection experiments (short targets), a solution containing (in order of addition) phosphate buffer solution (PBS) 1X (100 μL), NaCl 5 M (5 μL), the testing sample (5 μL) at the required concentration and functionalized AuNP (50 μL) was prepared for each replicate (final volume of 160 μl) in 1.5 mL tubes (LabCon, USA). For the detection of longer targets (RT-PCR experiments), the NaCl concentration in the mix was raised either to 0.6 M or 1.2 M.
To evaluate the functionality of the sensor, qualitative (photographs) and quantitative (absorbance measurements) studies were done at different times (from 15 min to 24 h). Two precipitation procedures (gravity or mild centrifugation at 5000 rpm/1–3 min) were studied. For the visual readout of the sensor, photographs of the tubes placed against a white background were taken. For the quantitative determination, part of the mix was removed slowly (70 μL) from the upper part of the tube and the UV–Vis spectra was recorded in a Synergy H4 microplate reader (Biotek) from λ = 400 nm to λ = 650 nm in 3 nm steps. Then, the maximum absorbance peaks were used for the calculation of the % absorbance, being the control sample (without target) considered 100% of the absorbance signal. The relative absorbance (%) was calculated according to Equation (2):
| (2) |
Also, additional characterization by TEM, UV and DLS of bare and functionalized AuNPs with or w/o 250 nM of the target has been performed. TEM and UV determinations were performed as previously described. The size of the particles (sensors) using 10 μl of the sample after 1 h of reaction was evaluated using DLS in a Zetasizer Nano-ZS device (Malvern Instruments).
2.6. Sensor stability evaluation
21 nM AuNP functionalized with 4.46 pmol of MB R (stem 5)/μL AuNPs as previously described were stored under two different conditions: RT and 4 °C. To evaluate the stability of the system, detection experiments after 1 h in the presence of 250 nM RNA short targets R (specific), E (nonspecific) or non-target (H2O) in the course of several days/months. The samples’ absorbance was measured by UV–Vis spectra in a Synergy H4 microplate reader (Biotek) from λ = 400 nm to λ = 650 nm in 3 nm steps.
2.7. Modified RT-PCR detection experiments
Total RNA extracted from infected MAD6 cell cultures (kindly provided by Drs. Isabel Sola and Sonia Zúñiga) or patient samples were amplified by RT-PCR using the SuperScript™ III One-Step RT-PCR System with Platinum™ Taq DNA Polymerase (Invitrogen). Briefly, several individual reactions (50 μL each) were prepared according to the manufacturer's instructions with the following modifications: total RNA (20 ng) as template, 1 μM forward primer (the first 6 nucleotides at the 5′ end contain phosphorothioate bond modifications), 1.2 μM reverse primer (Table S4), and 2 mM MgSO4. The cycling parameters were adjusted to the specific amplified region and are summarized in Table S4. Amplified RT-PCR products for the same target region were pooled together and loaded in a 3% agarose gel to check the success of the reaction. dsDNA products were treated with 0.4 U of T7 exonuclease (New England Biolabs; Cat. Nº M0263L) per μL of RT-PCR reaction during 45 min at 37 °C followed by 10 min at 80 °C to digest the unprotected (non-phosphorothioated) strand. Finally, when needed, ssDNA was purified employing Monarch PCR & Cleanup Kit (New England Biolabs; Cat. Nº #T1030). The purified ssDNA product was resuspended in PCR-grade water and used as target in detection experiments as described previously.
3. Results and discussion
3.1. Sensor assembly
The biosensor based on functionalized gold nanoparticles for SARS-CoV-2 RNA detection was designed, synthesized, characterized and systematically evaluated in terms of stability, specificity, sensitivity and functionality in detection experiments using synthetic DNA and RNA molecules, total RNA obtained from infected cell cultures, and patients samples.
The oligonucleotides employed contain two different domains. The central one is complementary to the selected target regions of interest in the SARS-CoV-2 genome [15,31,44]. This domain is flanked by a few nucleotides (4–6) that stabilize the formation of a hairpin structure (Table S2). According to the theoretical results obtained by mfold Web Server [33], our oligonucleotides are stable in the working conditions of the detection experiments and will selectively recognize the target sequences. These oligonucleotides contain a sulfur and a cholesterol derivative at each end. The first modification is required to conjugate the oligonucleotides to the nanoparticles, whereas the hydrophobic molecule is needed to modulate the aggregation of the nanostructures. On the other hand, we obtained AuNPs of different diameters (from 15 to 38 nm) synthesized following the Turkevich method with modifications [19]. Their characterization by UV-spectra, TEM and AFM confirmed that we obtained colloidal dispersions of spherical gold nanoparticles of the desired sizes by adjusting the HAuCl4: citrate ratio (Figure S1). The functionalization of AuNPs with the designed molecular beacons led to stable colloidal dispersions of nanoparticles. The final loading of the MB was determined for each AuNP size, and is summarized in Table S3.
3.2. MB stem length
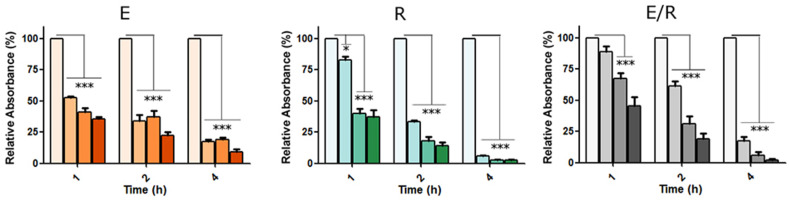
The performance of the MB in terms of time response and specificity depends not only on the sequence design, but on its thermodynamic behavior. One parameter that determines the stability and kinetics of the molecule is the beacons’ stem length. To test that, we prepared different versions of the MB using the same loop but varying the nucleotide length at the stem (4, 5 or 6 nt). The functionalization of 21 nm AuNPs with the different stem versions of the MBs E and R, alone or in combination, revealed that all sensors could recognize the DNA target sequence selectively (Fig. 2 and Figure S2). In terms of sensitivity, all the MB successfully detect the DNA target at 50 nM during the first hour. The stem of 5 nucleotides led to the fastest sensors detecting the targets [50 nM target induces a decrease in the absorbance in 2 h of 65.9% (E), 66.8% (R) and 40.1% (ER)] (Fig. 2). The 6 nt stem time-response is similar but less efficient when a single target is detected, and worst for the ER combination [33.5% (E), 44.5% (R) and 2.5% (ER) in the same conditions]. Surprisingly, the shortest stem (4 nucleotides) led to the slowest detection sensor [5.1% (E), 40.4% (R) and 0.1% (ER) in the same conditions], although the contrary was expected based on previous data on theoretical and experimental hybridization assays (Figure S2). Thus, based on these results, the stem with 5 nucleotides was selected for the following experiments.
Fig. 2.
Optimization of the MBs for the detection of target DNA sequences mimicking the E and R SARS-CoV-2 specific gene regions. AuNPs of 21 nm functionalized with the MBs E and R (alone or in combination) containing the 5 nt stem were tested using different concentrations of target (0 for the negative control, 50, 100 and 250 nM from left to right in the graphs). Data are presented as mean ± SD (n = 3) in terms of relative absorbance (%) vs negative control. Statistical analysis was performed using one-way ANOVA with Tukey's test (pairwise comparison). Signif. Codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘‘1.
3.3. AuNP's size optimization
Next, we analyzed the influence of AuNP's size in the detection performance of the system using DNA target sequences of R and E (Figure S3). We observed that the sensors based on the biggest nanoparticle (38 nm) were less sensitive and more unstable than the sensors prepared with AuNPs of 21 nm. On the contrary, 15 nm AuNP were much more stable, but also required longer times for the detection. To confirm these results, several batches of AuNPs between 20 and 25 nm were prepared and tested, confirming that their stability and sensitivity were optimum for our purposes (Figure S4). Therefore, AuNP of this range of size were chosen for the detection of these sequences instead of smaller AuNP (slow detection) and the bigger AuNP (unstable and less sensitive at low target concentrations).
Then, we confirmed that our system could also be used for the detection of RNA sequences (Table S2). Particularly, when ssRNA sequences were used, similar results were obtained in terms of sensitivity, selectivity and detection speed (Figure S5).
3.4. Sensor selectivity
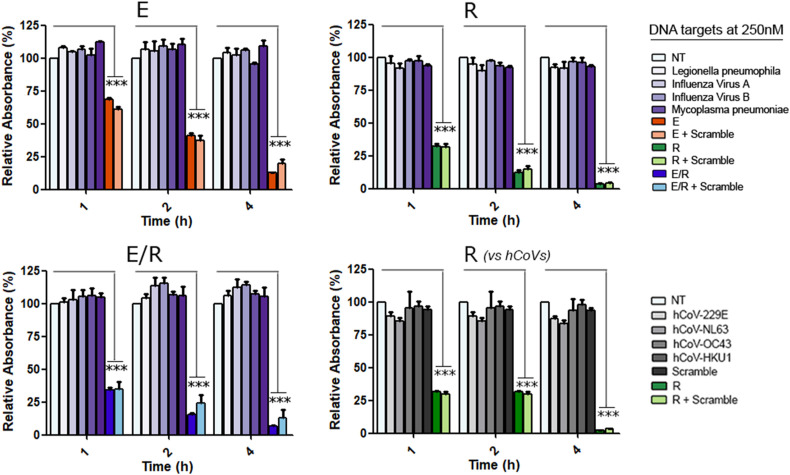
We also assessed the selectivity of the system since the presence of other respiratory pathogens (e.g., viruses, bacteria or fungi) in COVID-19 suspected patients might interfere with the proper diagnosis, treatment and prognosis [35,45,46]. From a detection point of view, the presence of those microorganisms, concomitant or not to SARS-CoV-2 in the samples, could affect or even impair the specific detection of the target virus. Thus, we evaluated if our systems present cross-reactivity with other genomic sequences among the most common pathogens that might be present in COVID-19 and non- COVID-19 patients. Particularly, representative sequences of the pathogenic HCoVs HKU1, NL63, 229E and OC43 [34], Influenza virus A, Influenza virus B [36], Mycoplasma pneumoniae [37] and Legionella pneumophila [38], were added to our sensors in the presence of the SARS-CoV-2 R and/or E target sequences (Fig. 3 ). Remarkably, only when the specific SARS target was present in the mixture, we observed the expected AuNP aggregation, confirming the high selectivity of the biosensors.
Fig. 3.
Gold Nanoparticles based sensors' selectivity for the detection of R and/or E RNA mimicking SARS-CoV-2 targets in the presence of other pathogens DNA sequences (Scramble). AuNPs of 23 nm were functionalized with MB E and/or R and tested using 250 nM target (final concentration independently if they are alone or in combination). Data are presented as mean ± SD (n = 3) in terms of relative absorbance (%) vs negative (NT) control. Statistical analysis was performed using one-way ANOVA with Tukey's test (pairwise comparison). Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.5. Sensor sensitivity
After these studies, we evaluated the detection limit using RNA sequences at different concentrations (1 nM–100 nM) (Fig. 4 and Figure S6). The system can detect the target sequences at low nanomolar, particularly the E sequence at 10 nM after 2 h, and the R at 20 nM after 4 h. Remarkably, when the system combined two sequences related to SARS-CoV-2 (E + R) the sensitivity was better (500 pM of each target, after 6 h), suggesting a cooperative effect in the functionality of the sensor. This can be exploited for the future design of sensors to improve their sensitivity and for the detection of multiple sequences.
Fig. 4.
Gold Nanoparticles based sensors' detection limit. Titration experiments using targets R and/or E RNA mimicking SARS-CoV-2. AuNPs of 21 nm functionalized with the MBs E and R (alone or in combination) containing the 5 nt stem were tested using different concentrations of target (0 for the negative control, 1, 10 and 20 nM from left to right in the graphs). Data are presented as mean ± SD (n = 3) in terms of relative absorbance (%) vs negative control. Statistical analysis was performed using one-way ANOVA with Tukey's test (pairwise comparison). Signif. Codes: 0 ‚***’ 0.001 ‚**’ 0.01 ‚*’ 0.05 ‚.’ 0.1 ‚ ‚ 1. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.6. Sensor kinetics
To optimize the detection time, we test our sensor (R) in the presence of different concentrations of the synthetic target R (Figure S7). After 15 min of incubation, samples were centrifuged at 5.000 rpm during 2–3 min. After that, the absorbance of the supernatant was measured. Results show that low-speed centrifugation (either in a centrifuge or spinner) accelerates the sedimentation process, yielding reliable and statistically significant results in minutes vs hours (when only gravity is involved).
3.7. Sensor stability
Then, we studied the sensor's activity overtime to ensure that the sensors could be produced and stored for months without losing their properties. Thus, the effect of the temperature during the storage for the R sensor (5 nt stem) to achieve a specific and sensitive detection of the R (RNA) target at 250 nM after 1 h was evaluated. In this case, absorbance measurements using the same sensor preparations but stored at different temperatures (RT or 4 °C) were carried out. The results showed that the system's functionality was not affected by the storage time at both temperatures during the first 15 days, being advisable its conservation at 4 °C for longer periods, such as 3 months (Figure S8).
3.8. Sensor biophysical characterization
Additionally to the above-mentioned results, a more biophysical characterization of the sensor's molecular behavior has been carried out. Specifically, UV visible spectra, TEM imaging and DLS data before and after 1 h in the presence of 250 nM RNA synthetic target R is reported (Figure S9). Nanoparticles aggregation is clearly shown as a decrease in the absorbance at ∼520 nm of the solution in the presence of 250 nM synthetic target, relative to the control without target (Figure S9A). Electron microscopy images clearly show how the nanoparticles are well dispersed when no target is present, and the cholesterol is hidden, while aggregates and precipitates in its presence, when cholesterol moiety is exposed to the aqueous media (Figure S9B). The structural changes at the nanoparticle surface are also evident in the DLS data, where the average size of the nanoparticles (i.e., their hydrodynamic radius) increases in the positive samples (∼70 nm) compared to the negative control (∼50 nm, Figure S9C). This size change could be due to the opening of the MBs upon target binding. The detection of bigger aggregates (as can be seen in TEM images) is not observed in any case, probably due to de resolution limitations of the DLS in the micrometric range. Altogether, the determinations suggest that the interaction between the loop and the target, followed by the cholesterol moiety exposure in the sensor's surface, is the molecular force driving the aggregation process.
3.9. Single point mutations detection
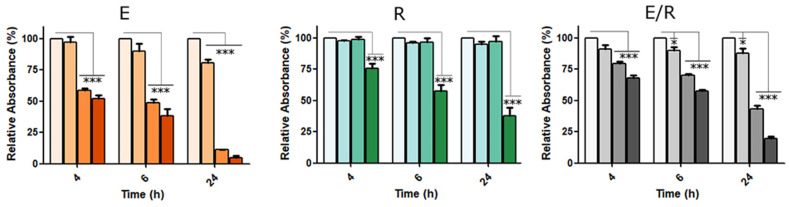
Once we confirm that our system is robust enough to ensure its future industrialization, we decided to assess its use to detect relevant mutations. Thus, we evaluated the sequence encoding for the spike protein, which is located on the surface of the virus particle and is of great interest from a clinical, epidemiological and immunological point of view. The spike protein is the ligand interacting with the cellular receptor ACE2, promoting entry in the cells and the virus spread through the body [47,48]. Also, the spike protein constitutes an essential antigen for the immune system recognition and activation [49,50], and specific mutations seem to play a crucial role in the patient outcome [44], reinfection [51] or vaccination efficacy [[52], [53], [54], [55]]. We selected as targets the insertion region (680SPRRAR↓SV687) and the mutation D614G. This last mutation is related to enhanced infectivity and stability of the virions, and has been one of the dominant forms [28,44]. Therefore, complementary MB loop sequences for this insertion (S Ins) or the D614G mutation were prepared and tested in detection experiments against DNA targets mimicking those genomic locations in the virus sequence. The results confirm the functionality of the sensor S Ins, similarly to E and R in previous experiments, responding at 50 nM DNA target concentration in 1 h and even at 1 nM after 24 h (Fig. 5 and Figure S10). What is more, our sensors were able to discriminate a single mismatch in the target sequence (D614G > gat to ggt), where the G614 sensor presented greater sensitivity and velocity than that of D614 (Figure S10).
Fig. 5.
Gold Nanoparticles based sensors' functionality targeting the insertion region in S gene (S Ins) or the S mutation D614G. For the detection, DNA mimics of the different S regions were prepared and confronted to the complementary loops included in MB S Ins, G614 or D614 used to functionalize 23 nm AuNPs. Data are presented as mean ± SD (n = 3) in terms of relative absorbance (%) vs negative control. The concentration of targets used (from left to right) were 0, 50 nM, 100 and 250 nM (for S Ins) or 0, 25, 50 and 100 nM (for G614 in blue and D614 in brown). Statistical analysis was performed using one-way ANOVA Tukey's test (pairwise comparison). Signif. Codes: 0 ‚***’ 0.001 ‚**’ 0.01 ‚*’ 0.05 ‚.’ 0.1 ‚ ‚ 1. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
We also assessed the sensors using RNA molecules, revealing a behavior similar to the one observed with DNA derivatives. Precisely, the sensors designed to detect the insertion or the G614 mutation were able to detect their corresponding target sequences at 50 nM after 1–2 h. On the other hand, the sensor D614 was less efficient, and the detection was achieved after 24 h at 100 nM. (Figure S11 and data not shown).
3.10. Multiplexed detection
We also evaluated the sensing capabilities of the system in a multiplexing approach as performed above, but including the spike. Particularly, the insertion region of the spike (S) and the E and R genes (Figure S12). Remarkably, the combined detection of E or R with S led to a higher reduction of the absorbance after 2 h, 73.5% and 83.8%, respectively. The detection of the three sequences also provided an excellent reduction of absorbance over this period (77.9%). Interestingly, this multiplexed approach increases the specificity of the sensor, which is required for proper diagnosis, and also the sensitivity. However, when this approach was evaluated using RNA sequences, the sensitivity was less pronounced. In this case, the reduction in absorbance after 2 h at 50 nM of RNA for the combined detection of E + S, R + S and E + R + S, was 33.3, 26.6 and 31.3%, respectively (Figure S13). Based on these results, we believe that the secondary structure formed by the RNA molecules might be preventing the efficient interaction with the oligonucleotides on the AuNPs, reducing their sensing capabilities. Thus, our system might be more suitable for the detection of DNA molecules instead of RNA. For these reasons, we explored procedures to transform the viral RNA into DNA to ease its detection, such as a modification of the standard PCR procedure described below.
3.11. Detection of the virus from infected samples
According to the existing data, the SARS-CoV-2 viral loads in the throat swab peaked at around 5–6 days after symptom onset, ranging from around 104 to 107 copies per mL during this time [56]. Our sensor lowest detection limit in the best formulations (E + R) using short RNA targets (24 nt R or 26 nt E) has been 1 nM (0.5 nM each), which represents around 7.8–9.6 × 107 virus copies per mL. Thus, our sensor could be used to detect the virus directly in samples with high loads, but not with samples from patients with low virus titers. Also, the wingspan of the viral RNA and their structural complexity might decrease the sensitivity by mediating a stabilization effect, probably due to the molecular crowding effect exerted by the huge genome molecule non-interacting with the MB loop. For those reasons, we decided to couple a modified PCR-based amplification after the required transcription of the RNA into DNA. The method provides short single-stranded amplicons after exonuclease digestion of the products, which can be used in the detection experiments.
3.12. Clinical samples testing optimization
For these experiments, we used RNA samples obtained from SARS-CoV-2 infected cells or COVID-19 nasopharyngeal patient samples with different viral loads. The clinical samples were provided by the Hospital Ramón y Cajal (Madrid, Spain) and Ct values from clinical samples were determined by RT-qPCR following the Thermo Fisher TaqPath COVID-19 protocol. The inclusion of the swabs in transportation buffer containing guanidine isothiocyanate makes RNA cleaning unavoidable in this particular case, due to the interference of denaturing agents with the subsequent enzymatic amplification process. The PCR conditions were finely tuned to avoid false positives (even when clinical samples contain other human pathogenic Coronaviruses) while keeping the direct correlation between the viral load in the samples and the amplicon performance (Figure S14). Particularly we adjusted: (1) RT-PCR primers design to produce short amplicons with an unprotected strand substrate for exonuclease digestion, (2) primer concentration, (3) cycling parameters, (4) Exonuclease digestion directly in the amplification buffer (5) NaCl concentration in the detection mix, (6) target sample volume (7) MB loop length. This optimization provides us with a method that can be used to detect the virus from patient samples accounting for different viral loads and without the need for amplicon purification. Based on these experiments, the optimal conditions were (1) amplicons of around 70bp or less, using (2) 1 μM and 1.2 μM of Forward (phosphorothioated) and Reverse primers, respectively, in a (3) 32 cycles PCR reaction (∼1 h) (4) directly treated with T7 exonuclease and (5) 1.2 M (purified amplicon) (Figures S15-S17) or 0.6 M NaCl (non-purified amplicon) in the sensor mix, using (6) 15 μL directly from the crude digested reaction and (7) an extended MB loop of 25 nt (Figures S18-S19).
3.13. Validation of the detection method using SARS-CoV-2 infected samples and patient samples
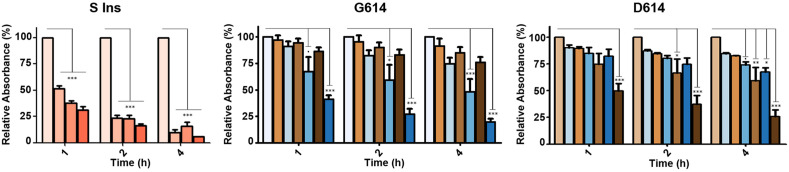
During the optimization phase, we found that amplicons of around 70 bp were recognizable by our sensor, promoting a successfully specific hybridization followed by the desired aggregation of the nanoparticles. To improve the simplicity, sensitivity and kinetics of the detection reactions, the increase of the NaCl concentration in the solution, the extension of the loop size in the MB and the volume of crude reactions were important aspects. To optimize the NaCl concentration, we first prepared and purified short single-stranded amplicons of the key gene regions R (74 bp), E (74 bp) and S (72 bp). The source for its production was the total RNA extracted from SARS-CoV-2 infected cells (containing the G614 mutation). Remarkably, in all cases, our system was able to detect the purified amplicons in a specific way with a sensitivity of 50 nM (E74), 100 nM (R74) or 250 nM (S72), once NaCl was adjusted to a final concentration of 1.2 M (Fig. 6 and Figures S15-S17). In all experiments, a 72 bp GFP gene-derived fragment was used as control, which did not produce any significant aggregation. The sensor with the best performance was the E, followed by the S and the R. The differences observed might be due to the different amplicon sequences obtained and their secondary structure. Based on these results, we decided to focus our efforts on the detection of the S gene, which is crucial for the identification of multiple variants of the virus, and our system could detect it in a few hours.
Fig. 6.
Gold Nanoparticles based sensors’ functionality targeting the gene regions E, R or S mutation D614G. For the detection, ssDNA purified amplicons were prepared by RT-PCR followed by T7 exonuclease digestion and confronted to the complementary loops included in MB E, R or S G614 (loop 25 nt) used functionalize 23 nm AuNPs. Data are presented as mean ± SD (n = 3) in terms of relative absorbance (%) vs negative control (without target). The concentration of targets used (from left to right) were 0, 50 nM, 100 and 250 nM. Statistical analysis was performed using one-way ANOVA Tukey's test (pairwise comparison). Signif. Codes: 0 ‚***’ 0.001 ‚**’ 0.01 ‚*’ 0.05 ‚.’ 0.1 ‚ ‚ 1. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Afterward, we applied the procedure to purified ssDNA amplicons obtained directly from COVID-19 infected and non-infected patients (Figure S19), confirming the feasibility of the method for direct determination of SARS-CoV-2 infections. Preliminary results indicate that purified amplicons, reacting during 20 min with our sensor, could be then centrifuged (5.000 rpm/2 min/RT), speeding up the detection process from hours to minutes (Figure S20).
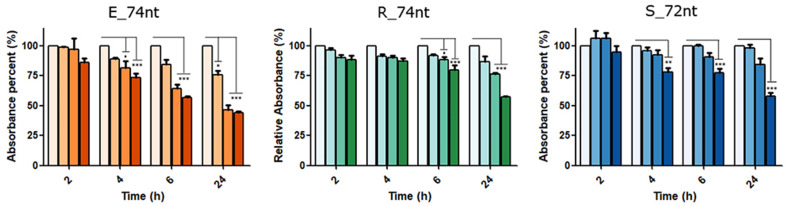
Then, once we confirmed that the approach could be used to detect the virus in patient samples, we optimized several parameters to increase usability. Specifically, we established the optimum volume of digested amplicon crude reaction (∼10–15 μl) and, due to the contribution of salts coming from the sample, 0.6 M NaCl was used in the sensor mix to avoid nonspecific sensor aggregation. In those conditions, we were capable of detecting the SARS-CoV-2 at different viral loads (Fig. 7 and Figure S21), demonstrating the potential use of our system as a reliable and easy-to-implement detection method for SARS-CoV-2 infections in patients with possibilities to infect others (Cts <30). This system can be easily extrapolated to target any genomic region in SARS-CoV-2 genome, as we demonstrate, for example, with the E gene (Figure S21), highlighting the potential and robustness of the approach for the specific detection of nucleic acids with diagnostic purposes.
Fig. 7.
Gold Nanoparticles based sensors’ functionality targeting the S gene mutation D614G region from non-infected and COVID-19 patient's samples with different viral loads. For the detection, crude reactions prepared by RT-PCR followed by T7 exonuclease digestion were confronted with the S G614 sensor. Data are presented as mean ± SD (n = 3) in terms of relative absorbance (%) vs negative control (without target). Statistical analysis was performed using one-way ANOVA with Tukey's test (pairwise comparison). Signif. Codes: 0 ‚***’ 0.001 ‚**’ 0.01 ‚*’ 0.05 ‚.’ 0.1 ‚ ‚ 1. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Conclusion
We have prepared colorimetric biosensors for the selective detection of sequences of SARS-CoV-2 based on gold nanoparticles. The evaluation of the influence of the stem's length of the molecular beacon as well as the size of AuNP, lead us to establish the best formulation for the detection of individual targets for E and R, but also for their combination. To expand the scope of action, we also designed a sensor to target the insertion region of Spike (S Ins), which works even better in terms of sensitivity than the previous formulations E and R. Surprisingly, E + R, E + S, R + S and E + R + S worked better than their individual options, so the possibility of targeting a wide variety of regions simultaneously could be further explored. Lastly, promising results were obtained for the detection of the single point mutation in D614G, suggesting that the system could be adjusted for the identification of different strains of this virus.
To use these sensors in clinical samples, a modified PCR procedure was implemented. Our approach was able to detect the presence of the virus in patients with different loads. The procedure is easy to implement, requiring a standard thermocycler and the sensor optimized herein to assess the presence of the viral genome in samples from patients.
Author contributions
Ciro RodríguezDíaz: Validation, Investigation, Writing – review & editing, Nuria Lafuente-Gómez: Validation, Investigation, Writing – original draft, Catarina Coutinho: Validation, Investigation, Demián Pardo: Validation, Investigation, Hernán Alarcón-Iniesta: Validation, Investigation, María López-Valls Validation, Investigation, Rocío Coloma Validation, Investigation, Paula Milán-Rois: Validation, Investigation, Mirian Domenech: Validation, Investigation, Melanie Abreu: Resources, Investigation, Rafael Cantón: Resources, Investigation, Juan Carlos Galán: Resources, Investigation, Rebeca Bocanegra: Validation, Investigation, Luis A. Campos: Validation, Investigation, Rodolfo Miranda: Project administration, Milagros Castellanos: Conceptualization, Methodology, Formal analysis, Validation, Investigation, Writing – review & editing, Supervision, Álvaro Somoza: Conceptualization, Methodology, Writing – review & editing, Supervision, Project administration.
Ethics approval
The samples were obtained with the consent all participants, and approved by “Comité de Ética de la Investigación con Medicamentos del Hospital Universitario Ramón y Cajal”. Reference: 127-21.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Drs. Isabel Sola and Sonia Zúñiga (CNB-CSIC) for the preparation of the SARS-CoV-2 infected cell cultures RNA extracts. This work was supported by the Instituto de Salud Carlos III (FONDO COVID19, proyectos COV20/00122 y 00144), and co-financed by European Structural and Investment Fund, the Spanish Ministry of Economy and Competitiveness (SAF2017-87305-R, PID2020-119352RB-I00), Comunidad de Madrid (IND2017/IND-7809; S2017/BMD-3867 RENIM-CM, NANOCOV-CM) and IMDEA Nanociencia. Also, this work was supported by a CRUE/Santander project (Fondo Supera COVID-19) and by the Spanish Ministerio de Ciencia, Innovación y Universidades project PGC2018-098613-B-C21 (SpOrQuMat), the European Union (H2020-NMP-2015/GA number 685795) and the Comunidad de Madrid, project NanoMagCOST-CM, S2018/NMAT-4321. IMDEA Nanociencia acknowledges support from the ‘Severo Ochoa’ Programme for Centers of Excellence (MINECO, SEV-2016-0686, CEX2020-001039-S). N. Lafuente-Gómez thanks the Spanish Education Ministry (FPU18/02323) and P. Milán-Rois thanks the Spanish Science and Innovation Ministry (BES-2017-082521) for their corresponding funding.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.talanta.2022.123393.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lai C.C., Shih T.P., Ko W.C., Tang H.J., Hsueh P.R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int. J. Antimicrob. Agents. 2020;55(3):105924. doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pollán M., Pérez-Gómez B., Pastor-Barriuso R., Otero J., Hernán M.A., Pérez-Olmeda M., Sanmartín J.L., Fernández-García A., Cruz I., Fernández de Larrea N., Molina M., Rodríguez-Cabrera F., Martín M., Merino-Amador P., León Paniagua J., Muñoz-Montalvo J.F., Blanco F., Yotti R., ENE-COVID Study Group Prevalence of SARS-CoV-2 in Spain (ENE-COVID): a nationwide, population-based seroepidemiological study. Lancet (Lond., Engl.) 2020;6736(20):1–11. doi: 10.1016/S0140-6736(20)31483-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Udugama B., Kadhiresan P., Kozlowski H.N., Malekjahani A., Osborne M., Li V.Y.C., Chen H., Mubareka S., Gubbay J.B., Chan W.C.W. Diagnosing COVID-19 : the disease and tools for detection. ACS Nano. 2020;14:3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- 5.Taleghani N., Taghipour F. Diagnosis of COVID-19 for controlling the pandemic: a review of the state-of-the-art. Biosens. Bioelectron. 2021;174:112830. doi: 10.1016/j.bios.2020.112830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rai P., Kumar B.K., Deekshit V.K., Karunasagar I., Karunasagar I. Detection technologies and recent developments in the diagnosis of COVID-19 infection. Appl. Microbiol. Biotechnol. 2021;105(2):441–455. doi: 10.1007/s00253-020-11061-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jayamohan H., Lambert C.J., Sant H.J., Jafek A., Patel D., Feng H., Beeman M., Mahmood T., Nze U., Gale B.K. SARS-CoV-2 pandemic: a review of molecular diagnostic tools including sample collection and commercial response with associated advantages and limitations. Anal. Bioanal. Chem. 2021;413(1):49–71. doi: 10.1007/s00216-020-02958-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bar-On Y.M., Flamholz A., Phillips R., Milo R. Sars-cov-2 (Covid-19) by the numbers. Elife. 2020;9 doi: 10.7554/eLife.57309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J., Sheng J., Quan L., Xia Z., Tan W. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27(3):325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nalla A.K., Casto A.M., Huang M.L.W., Perchetti G.A., Sampoleo R., Shrestha L., Wei Y., Zhu H., Jerome K.R., Greninger A.L. Comparative performance of SARS-CoV-2 detection assays using seven different primer-probe sets and one assay kit. J. Clin. Microbiol. 2020;58(6):1–6. doi: 10.1128/JCM.00557-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sethuraman N., Jeremiah S.S., Ryo A. Interpreting diagnostic tests for SARS-CoV-2. JAMA. 2020;323(22):2249–2251. doi: 10.1001/jama.2020.8259. [DOI] [PubMed] [Google Scholar]
- 13.Chu D.K.W., Pan Y., Cheng S.M.S., Hui K.P.Y., Krishnan P., Liu Y., Ng D.Y.M., Wan C.K.C., Yang P., Wang Q., Peiris M., Poon L.L.M. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin. Chem. 2020;7:1–7. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan J.F.W., Yip C.C.Y., To K.K.W., Tang T.H.C., Wong S.C.Y., Leung K.H., Fung A.Y.F., Ng A.C.K., Zou Z., Tsoi H.W., Choi G.K.Y., Tam A.R., Cheng V.C.C., Chan K.H., Tsang O.T.Y., Yuen K.Y. Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID19-RdRP/Hel real-time reverse transcription-PCR assay validated in vitro and with clinical specimens. J. Clin. Microbiol. 2020;58(5):1–10. doi: 10.1128/JCM.00310-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G.J.C., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romett J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P.G., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3):1–8. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reusken C.B.E.M., Broberg E.K., Haagmans B., Meijer A., Corman V.M., Papa A., Charrel R., Drosten C., Koopmans M., K. Leitmeyer and on behalf of EVD-LabNet and ERLI-Net Laboratory readiness and response for novel coronavirus (2019-nCoV) in expert laboratories in 30 EU/EEA countries, January 2020. Euro Surveill. 2020;25(6):1–6. doi: 10.2807/1560-7917.ES.2020.25.6.2000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Korukluoglu G., Kolukirik M., Bayrakdar F., Ozgumus G.G., Altas A.B., Cosgun Y., Kolukirik C.Z.K. 40 minutes RT-qPCR assay for screening Spike N501Y and HV69-70del mutations. bioRxiv. 2021 doi: 10.1101/2021.01.26.428302. [DOI] [Google Scholar]
- 18.Paper O., Lippi G., Simundic A., Plebani M. Potential preanalytical and analytical vulnerabilities in the laboratory diagnosis of coronavirus disease 2019 (COVID-19) Clin. Chem. Lab. Med. 2020;58(7):1070–1076. doi: 10.1515/cclm-2020-0285. [DOI] [PubMed] [Google Scholar]
- 19.Russo A., Minichini C., Starace M., Astorri R., Caló F., Coppola N. Current status of laboratory diagnosis for COVID-19: a narrative review. Infect. Drug Resist. 2020;13:2657–2665. doi: 10.2147/IDR.S264020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cui F., Zhou H.S. Biosensors and Bioelectronics Diagnostic methods and potential portable biosensors for coronavirus disease 2019. Biosens. Bioelectron. 2020;165:112349. doi: 10.1016/j.bios.2020.112349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Latorre A., Posch C., Garcimartín Y., Ortiz-Urda S., Somoza Á. Single-point mutation detection in RNA extracts using gold nanoparticles modified with hydrophobic molecular beacon-like structures. Chem. Commun. 2014;50(23):3018. doi: 10.1039/c3cc47862a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghosh D., Sarkar D., Girigoswami A., Chattopadhyay N. A fully standardized method of synthesis of gold nanoparticles of desired dimension in the range 15 nm-60 nm. J. Nanosci. Nanotechnol. 2011;11(2):1141–1146. doi: 10.1166/jnn.2011.3090. [DOI] [PubMed] [Google Scholar]
- 23.Ma L., Yin L., Li X., Chen S., Peng L., Liu G., Ye S., Zhang W., Man S. A smartphone-based visual biosensor for CRISPR-Cas powered SARS-CoV-2 diagnostics. Biosens. Bioelectron. 2022;195:113646. doi: 10.1016/j.bios.2021.113646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alafeef M., Moitra P., Dighe K., Pan D. RNA-extraction-free nano-amplified colorimetric test for point-of-care clinical diagnosis of COVID-19. Nat. Protoc. 2021;16(6):3141–3162. doi: 10.1038/s41596-021-00546-w. [DOI] [PubMed] [Google Scholar]
- 25.Cao Y., Wu J., Pang B., Zhang H., Le X.C. CRISPR/Cas12a-mediated gold nanoparticle aggregation for colorimetric detection of SARS-CoV-2. Chem. Commun. 2021;57(56):6871–6874. doi: 10.1039/d1cc02546e. [DOI] [PubMed] [Google Scholar]
- 26.Gao Y., Han Y., Wang C., Qiang L., Gao J., Wang Y., Liu H., Han L., Zhang Y. Rapid and sensitive triple-mode detection of causative SARS-CoV-2 virus specific genes through interaction between genes and nanoparticles. Anal. Chim. Acta. 2021;1154:338330. doi: 10.1016/j.aca.2021.338330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang W.S., Pan J., Li F., Zhu M., Xu M., Zhu H., Yu Y., Su G. Reverse transcription recombinase polymerase amplification coupled with CRISPR-Cas12a for facile and highly sensitive colorimetric SARS-CoV-2 detection. Anal. Chem. 2021;93(8):4126–4133. doi: 10.1021/acs.analchem.1c00013. [DOI] [PubMed] [Google Scholar]
- 28.Plante J.A., Liu Y., Liu J., Xia H., Johnson B.A., Lokugamage K.G., Zhang X., Muruato A.E., Zou J., Fontes-Garfias C.R., Mirchandani D., Scharton D., Bilello J.P., Ku Z., An Z., Kalveram B., Freiberg A.N., Menachery V.D., Xie X., Plante K.S., Weaver S.C., Shi P.Y. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2021;592:116–121. doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo Y., Huang L., Zhang G., Yao Y., Zhou H., Shen S., Shen B., Li B., Li X., Zhang Q., Chen M., Chen D., Wu J., Fu D., Zeng X., Feng M., Pi C., Wang Y., Zhou X., Lu M., Li Y., Fang Y., Lu Y.Y., Hu X., Wang S., Zhang W., Gao G., Adrian F., Wang Q., Yu F., Peng Y., Gabibov A.G., Min J., Wang Y., Huang H., Stepanov A., Zhang W., Cai Y., Liu J., Yuan Z., Zhang C., Lou Z., Deng F., Zhang H., Shan C., Schweizer L., Sun K., Rao Z. A SARS-CoV-2 neutralizing antibody with extensive Spike binding coverage and modified for optimal therapeutic outcomes. Nat. Commun. 2021;12(1) doi: 10.1038/s41467-021-22926-2. article number 2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nikiforov T.T., Rendle R.B., Kotewicz M.L., Rogers Y.H. The use of phosphorothioate primers and exonuclease hydrolysis for the preparation of single-stranded PCR products and their detection by solid-phase hybridization. PCR Methods Appl. 1994;3(5):285–291. doi: 10.1101/gr.3.5.285. [DOI] [PubMed] [Google Scholar]
- 31.Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir. Res. 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Becerra-Flores M., Cardozo T. SARS-CoV-2 viral spike G614 mutation exhibits higher case fatality rate. Int. J. Clin. Pract. 2020;74(8) doi: 10.1111/ijcp.13525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Canducci F., Debiaggi M., Sampaolo M., Marinozzi M.C., Berrè S., Terulla C., Gargantini G., Cambieri P., Romero E., Clementi M. Two-year prospective study of single infections and co-infections by respiratory syncytial virus and viruses identified recently in infants with acute respiratory disease. J. Med. Virol. 2008;80(4):716–723. doi: 10.1002/jmv.21108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xing Q.S., Li G.J., Xing Y.H., Chen T., Li W.J., Ni W., Deng K., Gao R.Q., Chen C.Z., Gao Y., Li Q., Yu G.L., Tong J.N., Li W., Hao G.L., Sun Y., Zhang A., Wu Q., Li Z.P., Pan S.L. Precautions are needed for COVID-19 patients with coinfection of common respiratory pathogens. medRxiv. 2020 doi: 10.1101/2020.02.29.20027698. 2020.02.29.20027698. [DOI] [Google Scholar]
- 36.Ho Y.I.I., Wong A.H., Lai R.W.M. Comparison of the cepheid xpert xpress Flu/RSV assay to in-house Flu/RSV triplex real-time RT-PCR for rapid molecular detection of influenza A, influenza B and respiratory syncytial virus in respiratory specimens. J. Med. Microbiol. 2018;67(11):1576–1580. doi: 10.1099/JMM.0.000841. [DOI] [PubMed] [Google Scholar]
- 37.Dumke R., Schurwanz N., Lenz M., Schuppler M., Lück C., Jacobs E. Sensitive detection of Mycoplasma pneumoniae in human respiratory tract samples by optimized real-time PCR approach. J. Clin. Microbiol. 2007;45(8):2726–2730. doi: 10.1128/JCM.00321-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chaudhry R., Valavane A., Sreenath K., Choudhary M., Sagar T., Shende T., Varma-Basil M., Mohanty S., Kabra S.K., Dey A.B., Thakur B. Detection of Mycoplasma pneumoniae and Legionella pneumophila in patients having community-acquired pneumonia: a multicentric study from New Delhi, India. Am. J. Trop. Med. Hyg. 2017;97(6):1710–1716. doi: 10.4269/ajtmh.17-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haiss W., Thanh N.T.K., Aveyard J., Fernig D.G. Determination of size and concentration of gold nanoparticles from UV-vis spectra. Anal. Chem. 2007;79(11):4215–4221. doi: 10.1021/ac0702084. [DOI] [PubMed] [Google Scholar]
- 40.Jin R., Wu G., Li Z., Mirkin C.A., Schatz G.C. What controls the melting properties of DNA-linked gold nanoparticle assemblies? J. Am. Chem. Soc. 2003;125(6):1643–1654. doi: 10.1021/JA021096V. [DOI] [PubMed] [Google Scholar]
- 41.Mirkin C.A., Letsinger R.L., Mucic R.C., Storhoff J.J. A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature. 1996;382(6592):607–609. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 42.Demers L.M., Mirkin C.A., Mucic R.C., Reynolds R.A., Letsinger R.L., Elghanian R., Viswanadham G. A fluorescence-based method for determining the surface coverage and hybridization efficiency of thiol-capped oligonucleotides bound to gold thin films and nanoparticles. Anal. Chem. 2000;72(22):5535–5541. doi: 10.1021/AC0006627. [DOI] [PubMed] [Google Scholar]
- 43.Hinman S.S., McKeating K.S., Cheng Q. DNA linkers and diluents for ultrastable gold nanoparticle bioconjugates in multiplexed assay development. Anal. Chem. 2017;89(7):4272–4279. doi: 10.1021/acs.analchem.7b00341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Becerra-Flores M., Cardozo T. SARS-CoV-2 viral spike G614 mutation exhibits higher case fatality rate. Int. J. Clin. Pract. 2020;74(8) doi: 10.1111/ijcp.13525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lansbury L., Lim B., Baskaran V., Lim W.S. Co-infections in people with COVID-19: a systematic review and meta-analysis. J. Infect. 2020;81(2):266–275. doi: 10.1016/j.jinf.2020.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nickbakhsh S., Ho A., Marques D.F.P., McMenamin J., Gunson R.N., Murcia P.R. Epidemiology of seasonal Coronaviruses: establishing the context for the emergence of coronavirus disease 2019. J. Infect. Dis. 2020;222(1):17–25. doi: 10.1093/infdis/jiaa185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 48.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., Marrama D., de Silva A.M., Frazier A., Carlin A.F., Greenbaum J.A., Peters B., Krammer F., Smith D.M., Crotty S., Sette A. Targets of T Cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181(7):1489–1501. doi: 10.1016/j.cell.2020.05.015. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Premkumar L., Segovia-Chumbez B., Jadi R., Martinez D.R., Raut R., Markmann A., Cornaby C., Bartelt L., Weiss S., Park Y., Edwards C.E., Weimer E., Scherer E.M., Rouphael N., Edupuganti S., Weiskopf D., Tse L.V., Hou Y.J., Margolis D., Sette A., Collins M.H., Schmitz J., Baric R.S., de Silva A.M. The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci. Immunol. 2020;5(48):eabc841. doi: 10.1126/sciimmunol.abc8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.To K.K.W., Hung I.F.N., Daniel Ip J., Chu A.W.H., Chan W.M., Tam A.R., Fong C.H.Y., Yuan S., Tsoi H.W., Ng A.C.K., Lee L.L.Y., Wan P., Tso E.Y.K., To W.K., Tsang D.N.C., Chan K.H., Huang J.D., Kok K.H., Cheng V.C.C., Yuen K.Y. Coronavirus disease 2019 (COVID-19) Re-infection by a phylogenetically distinct severe acute respiratory syndrome coronavirus 2 strain confirmed by whole genome sequencing. Clin. Infect. Dis. 2020;73(9):e2946–e2951. doi: 10.1093/cid/ciaa1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Andreano E., Piccini G., Licastro D., Casalino L., Johnson N.V., Paciello I., Dal Monego S., Pantano E., Manganaro N., Manenti A., Manna R., Casa E., Hyseni I., Benincasa L., Montomoli E., Amaro R.E., McLellan J.S., Rappuoli R. SARS-CoV-2 escape in vitro from a highly neutralizing COVID-19 convalescent plasma. bioRxiv Prepr. Serv. Biol. 2020 doi: 10.1101/2020.12.28.424451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Greaney A.J., Loes A.N., Crawford K.H.D., Starr T.N., Malone K.D., Chu H.Y., Bloom J.D. PREPRINT: comprehensive mapping of mutations to the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human serum antibodies. Cell Host Microbe. 2021;29(3):463–476. doi: 10.1016/j.chom.2021.02.003. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Williams T.C., Burgers W.A. SARS-CoV-2 evolution and vaccines: cause for concern? Lancet Respir. Med. 2021;9(4):333–335. doi: 10.1016/S2213-2600(21)00075-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alenquer M., Ferreira F., Lousa D., Valério M., Medina-Lopes M., Bergman M.L., Gonçalves J., Demengeot J., Leite R.B., Lilue J., Ning Z., Penha-Gonçalves C., Soares H., Soares C.M., Amorim M.J. Signatures in SARS-CoV-2 spike protein conferring escape to neutralizing antibodies. PLoS Pathog. 2021;17(8) doi: 10.1371/journal.ppat.1009772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pan Y., Zhang D., Yang P., Poon L.L.M., Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect. Dis. 2020;20(4):411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.