Abstract
Background
Immune responses to seasonal endemic coronaviruses might have a pivotal role in protection against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Those SARS-CoV-2-crossreactive T cells were recently described in immunocompetent individuals. Still, data on cross-reactive humoral and cellular immunity in kidney transplant recipients is currently lacking.
Methods
The pre-existing, cross-reactive antibody B and T cell immune responses against SARS-CoV-2 in unexposed adults with kidney transplantation (Tx, n = 14) and without (non-Tx, n = 12) sampled before the pandemic were compared with 22 convalescent patients with COVID-19 (Cp) applying enzyme-linked immunosorbent assay and flow cytometry.
Results
In both unexposed groups, SARS-CoV-2 IgG antibodies were not detectable. Memory B cells binding spike (S) protein SARS-CoV-2 were detected in unexposed individuals (64% among Tx; 50% among non-Tx) and higher frequencies after infection (80% Cp). The numbers of SARS-CoV-2-reactive T cells were comparable between patients who had undergone Tx and those who had not. SARS-CoV-2-reactive follicular T helper cells were present in 61% of the unexposed cohort in both patients who had undergone Tx and those who had not.
Conclusions
Cross-reactive memory B and T cells against SARS-CoV-2 exist also in transplanted adults, suggesting a primed adaptive immunity. The effect on the disease course may depend on the concomitant immunosuppressive drugs.
Pre-existing immunity to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), most likely resulting from cross-reactivity against seasonal coronaviruses has been addressed early in the pandemic as a potential immunomodulatory factor affecting the immune response after SARS-CoV-2 infection or vaccination [1], [2], [3], [4], [5]. Grifoni et al and Nelde et al found wild-type S-reactive CD4+ T cells in 40% to 60% and 80% of unexposed individuals, respectively, suggesting a SARS-T cell immunity that is cross-reactive with HCoV [6,7]. Despite the weak evidence of pre-existing SARS-CoV-2 cross-reactive serum antibodies in prepandemic donors [8,9], independent study groups have detected cross-reactive pre-existing SARS-CoV-2 B cell memory cells in healthy individuals unexposed to SARS-CoV-2 [8], [9], [10]. Pre-existing cross-reactive T memory cells are recalled and expanded on SARS-CoV-2 infection, reinforcing but not preventing a robust and persistent primary response to new epitopes of SARS-CoV-2 [8,11,12].
Nevertheless, data on pre-existing SARS-CoV-2 immunity among patients who had were immunosuppressed, such as transplant recipients, are currently scarce. In this study, we aimed to characterize SARS-CoV-2-reactive pre-existing SARS-CoV-2-reactive T and B cells in a cohort of patients who had undergone renal transplant under immunosuppressive therapy who were unexposed to SARS-CoV-2 in comparison to unexposed immunocompetent blood donors.
Methods
Study Participants
Peripheral blood mononuclear cells (PBMCs) from 26 individuals unexposed to SARS-CoV-2 were collected and cryopreserved between 2017 and January 2020 and from a control group of 22 patients convalescing from COVID-19. The unexposed cohort consisted of 2 populations, the immunocompetent study control group of healthy donors (non-Tx; n = 12) and immunocompromised kidney transplant recipients (Tx; n = 14). Samples from the unexposed individuals were collected between 2017 and January 2020 before the spread of SARS-COV-2 in Europe, excluding any possibility of SARS-CoV-2 infection. The first SARS-CoV-2 infection in Germany was documented on January 1, 2020 [13], followed by documentation of the first case in the state of North Rhine-Westphalia on February 26, 2020. The study was approved by the ethics committees of the Ruhr University Bochum (20-6886) and University Hospital Essen (20-9214-BO). Written informed consent was obtained from all participants. Demographic characteristics are provided in Tables 1 and 2 .
Table 1.
Demographic Characteristics of the Exposed and Unexposed Study Participants
| SARS-Cov-2 Unexposed Patients | ||||||||
|---|---|---|---|---|---|---|---|---|
| Number | Age, y (Transplant Cohort) | Women (%), Transplant Cohort (%) | Sample Collection Dates | Transplant (%) | Characterization of T Cells (Number of Transplant Participants) | Characterization of B Cells (Number of Participants) | ||
| 26 | N/A | 11 (42%)-5 (36%) | 2017-2020 | 14 (54%) | 18 (7) | 26 | ||
| Median | N/A | 69 (55) | N/A | N/A | N/A | N/A | N/A | |
| Minimum | N/A | 37 (37) | N/A | N/A | N/A | N/A | N/A | |
| Maximum | N/A | 91 (75) | N/A | N/A | N/A | N/A | N/A | |
| Convalescent COVID-19 Patients | ||||||||
|---|---|---|---|---|---|---|---|---|
| Age, y | Sex | SARS-CoV-2 PCR | Sample Collection Day (Day 1 = 1st Positive PCR) | Disease Severity | Disease Outcome | Characterization of T Cells | Characterization of B cells | |
| CoV1 | 64 | W | positive | 110 | severe | Recovery | x | x |
| CoV2 | 65 | W | positive | 121 | mild | Recovery | x | |
| CoV3 | 60 | M | positive | 121 | mild | Recovery | x | |
| CoV4 | 60 | M | positive | 121 | mild | Recovery | x | |
| CoV5 | 73 | M | positive | 79 | severe | Recovery | x | |
| CoV6 | 45 | W | positive | 45 | moderate | Recovery | x | |
| CoV7 | 67 | M | positive | 62 | severe | Recovery | x | |
| CoV8 | 89 | W | positive | 36 | severe | Recovery | x | |
| CoV9 | 52 | W | positive | 178 | mild | Recovery | x | |
| CoV10 | 75 | M | positive | 178 | mild | Recovery | x | |
| CoV11 | 83 | W | positive | 22 | severe | Recovery | x | |
| CoV12 | 28 | W | positive | 198 | mild | Recovery | x | |
| CoV13 | 61 | W | positive | 147 | mild | Recovery | x | |
| CoV14 | 55 | M | positive | 100 | unknown | Recovery | x | |
| CoV15 | 63 | M | positive | 113 | asymptomatic | Recovery | x | |
| CoV16 | 75 | M | positive | 133 | severe | Recovery | x | |
| CoV17 | 45 | M | positive | 32 | critical | Recovery | x | |
| CoV18 | 88 | W | positive | 34 | severe | Recovery | x | |
| CoV19 | 53 | M | positive | 30 | critical | Recovery | x | |
| CoV20 | 52 | W | positive | 23 | asymptomatic | Recovery | x | |
| CoV21 | 56 | W | positive | 120 | asymptomatic | Recovery | x | |
| CoV22 | 47 | W | positive | 103 | asymptomatic | Recovery | x | |
| No. (%) | NA | 12 (55%) | N/A | N/A | N/A | N/A | 9 | 14 |
| Median | 54,5 | N/A | N/A | 110 | N/A | N/A | N/A | N/A |
| Minimum | 28 | N/A | N/A | 22 | N/A | N/A | N/A | N/A |
| Maximum | 89 | N/A | N/A | 198 | N/A | N/A | N/A | N/A |
M, man; PCR, polymerase chain reaction; W, woman.
Table 2.
Clinical Characteristics of Transplant Unexposed Participants
| Patient | Age, y | Sex | Transplanted organ(s) | Year of transplantation | Previous transplants | Donor type (Living/Deceased) | IS (Target Concentration, ng/mL) | Viral Infections/Coinfections (>1000 IU/mL) |
|---|---|---|---|---|---|---|---|---|
| Tx1 | 57 | M | kidney | 03.08.2019 | 0 | deceased | Tacrolimus (3-5) Everolimus (3-6) Prednisolon 10mg |
BK-Virus(BKV)+CMV |
| Tx2 | 55 | M | kidney | 19.12.2019 | 0 | deceased | Tacrolimus (4-7) MMF 360mg Prednisolon 7,5mg |
BKV |
| Tx3 | 51 | M | kidney | 06.12.2017 | 1 | deceased | Cyclosporin (80-120) MMF 1080mg Prednisolon 5mg |
EBV |
| Tx4 | 72 | M | kidney | 04.02.2017 | 0 | deceased | Tacrolimus (3-5) Everolimus(3-5) Prednisolon 5mg |
EBV+CMV+BKV |
| Tx5 | 55 | W | kidney | 07.05.2019 | 0 | deceased | Tacrolimus (4-6) Everolimus (4-6) Prednisolon 7,5mg |
CMV |
| Tx6 | 73 | M | kidney | 29.12.2016 | 0 | deceased | Tacrolimus (4-6) MMF 500mg Prednisolon 10mg |
CMV |
| Tx7 | 51 | M | kidney | 08.01.2020 | 1 | deceased | Cyclosporin (80-120) MMF 720mg Prednisolon 7,5mg |
CMV |
| Tx8 | 69 | W | kidney | 10.12.2016 | 0 | deceased | Tacrolimus (4-6) MMF 1440mg Prednisolon 5mg |
CMV |
| Tx9 | 68 | W | kidney | 11.04.2019 | 0 | deceased | Tacrolimus (6-8) Prednisolon 7,5mg MMF 720mg |
EBV |
| Tx10 | 52 | M | kidney | 04.01.2020 | 0 | living | Tacrolimus (5-8) MMF1440mg Prednisolon 7,5mg |
BKV |
| Tx11 | 53 | M | kidney | 06.12.2015 | 0 | deceased | Tacrolimus (2-4) Everolimus (4-6) Prednisolon 4mg |
EBV |
| Tx12 | 37 | W | Kidney Liver |
19.02.2009 11.12.2001 |
0 0 |
Deceased unknown |
Tacrolimus (4-6) Belatacept Prednisolon 7,5mg |
EBV |
| Tx13 | 75 | W | kidney | 20.02.2006 | 0 | deceased | Tacrolimus (5-7) MMF 1440mg Prednisolon 5mg |
EBV |
| Tx14 | 44 | M | liver | 25.07.1998 | 0 | unknown | Tacrolimus (5-7ng/mL) Prednisolon 5mg |
EBV |
BKV, BK-Virus; CMV, cytomelovirus; EBV, epstein barr virus; M, man; MMF, mycophenolate mofetil; W, woman.
Preparation of PBMCs
Peripheral blood was collected in S-Monovette K3 EDTA blood collection tubes (Sarstedt, Germany). Collected blood was prediluted in phosphate-buffered saline (PBS/BSA) (Gibco, USA) at a 1:1 ratio and underlaid with 15 mL of Ficoll-Paque Plus (GE Healthcare, Germany). Tubes were centrifuged at 800 g for 20 minutes at room temperature. Isolated PBMCs were washed twice with PBS/BSA and stored at -80°C. The cryopreserved PBMCs were thawed by incubating cryovials for 2 to 3 minutes at 37°C in a bead bath, washed twice in 37°C RPMI 1640 medium (Life Technologies, USA) supplemented with 1% penicillin-streptomycin-glutamine (Sigma–Aldrich, USA) and 10% fetal calf serum (PAN-Biotech, Germany), and incubated overnight at 37°C.
Flow Cytometry
Measurement of SARS-CoV-2-reactive T cells
In brief, as previously described [14], PBMCs were plated in 96-U-Well plates in RPMI 1640 medium (Life Technologies) and stimulated with SARS-CoV-2 S-peptide (Miltenyi Biotec) or left untreated as a control for 16 hours. For a positive control, cells were stimulated with staphylococcal enterotoxin B (1 μg/mL, Sigma–Aldrich). After 2 hours, brefeldin A was added. A detailed list of the antibody panel for general phenotyping and T cell activation ex vivo is shown in Table 3 . After stimulation overnight, the PBMCs were stained with optimal concentrations of antibodies for 10 minutes at room temperature in the dark. Stained cells were washed twice with PBS/BSA before preparation for intracellular staining with the Intracellular Fixation & Permeabilization Buffer Set (Thermo Fisher Scientific, USA) according to the manufacturer's instructions. Fixed and permeabilized cells were stained for 30 minutes at room temperature in the dark with an optimal dilution of antibodies against the intracellular antigen. All samples were immediately acquired on a CytoFLEX flow cytometer (Beckman Coulter, Germany). Quality control was performed daily using the recommended CytoFLEX daily QC fluorospheres (Beckman Coulter). No modification to the compensation matrices was required throughout the study. Antigen-reactive responses were considered positive after the nonreactive background was subtracted and greater than 0.01% was detectable. Negative values were set to zero. In a single exception to the abovementioned minimum limit of 0.01%, we evaluated all positive frequencies of CD4+CD154+CD137+CXCR5+ cells after the background was subtracted, because no large populations of T follicular helper (Tfh) cells were expected to be found in circulation.
Table 3.
Fluorochrome Coupled Antibodies and Fluorescent Dye for Analysis of SARS-Cov-2 Reactive T Cells
| Antibodies or Fluorescent Dye | Fluorochrome | Source | Cat No. |
|---|---|---|---|
| Fixable Viability-Dye | eFluor780 | eBioscience, USA | 65-0865-14 |
| Anti-CCR7 (clone G043H7) | PerCP-Cy5.5 | BioLegend, USA | 353220 |
| Anti-CD4 (clone OKT4) | A700 | BioLegend | 317426 |
| Anti-CD8 (clone RPA-T8) | V500 | BD Biosciences | 560775 |
| Anti-CD45RA (clone HI100) | BV605 | BioLegend | 304134 |
| Anti-granzyme B (clone GB11) | FITC | BioLegend | 515403 |
| Anti-IL2 (clone MQ1-17H12) | PE | BioLegend | 500307 |
| Anti-CD185(CXCR5) (clone MP4-25D2) | PE-Dazzle594 | BioLegend | 356927 |
| Anti-CD137 (4-1BB) (clone 4B4-1) | PE-Cy7 | BioLegend | 309818 |
| Anti-CD154 (CD40L) (clone 24-31) | A647 | BioLegend | 310818 |
| Anti-TNFα (clone MAb11) | eFluor450 | eBioscience | 48-7349-42 |
| Anti-IFNγ (clone 4S.B3) | BV650 | BioLegend | 502538 |
| Anti-CD3 (clone OKT3) | BV785 | BioLegend | 317330 |
Cat, catalog. SARS-Cov-2, severe acute respiratory syndrome coronavirus 2.
Measurement of SARS-CoV-2-reactive B cells
As previously described [15], SARS-CoV-2 S1/S2-protein (henceforth referred to as S-protein) (Sino Biological Inc.) was aliquoted into 3 samples. Sample 1 was left unlabeled for blocking, and samples 2 and 3 were coupled to fluorescein isothiocyanate (FITC) and Cy5 fluorochromes, respectively. PBMCs were divided into 3 samples: 1. blocked, 2. unblocked, and 3. negative control samples. Blocking was performed by using a 10 times excess of unlabeled protein. After blocking, PBMCs were surface-stained with fluorochrome-labeled antibodies, as described in Table 4 . Finally, mixed FITC- and Cy5-labeled protein was added. Cells were stained for 10 minutes at 4°C. After washing with PBS, the samples were stored at 4°C until measurement on a Cytoflex flow cytometer. Directly before analysis, the samples were stained with DAPI to differentiate live from dead cells. Antigen-reactive responses were considered positive after the blocked background was subtracted and greater than 0.001% was detectable. Negative values were set to zero.
Table 4.
Fluorochrome Coupled Antibodies and Fluorescent Dye for Analysis of SARS-Cov-2 Reactive B Cells
| Antibodies or Fluorescent Dye | Fluorochrome | Source | Cat No. |
|---|---|---|---|
| Anti-CD20 (clone 2H7) | BV510 | BioLegend | 302340 |
| IgD (clone IgD26) | VioBlue | Miltenyi Biotec | 130-123-319 |
| Anti-CD19 (clone HIB19) | BV605 | BioLegend | 302244 |
| Anti-CD3 (clone OKT3) | BV785 | BioLegend | 317330 |
| Anti-CD14 (clone M5E2) | APC-Cyanine 7 | BioLegend | 301820 |
| Anti-CD27 (cloneO232) | PE | BioLegend | 302808 |
| DAPI | N/A | ThermoScientific | 62248 |
Cat, catalog.
SARS-CoV-2 IgG antibody titers
Peripheral blood was collected in S-Monovette Z-Gel (Sarstedt). SARS-CoV-2 IgG titers were analyzed in purified serum using a SARS-CoV-2 IgG kit (Euroimmun, Lübeck, Germany). The test was performed according to the manufacturer's instructions. Briefly, serum samples were diluted 1:100 and added to plates coated with recombinant SARS-CoV-2 antigen. Bound SARS-CoV-2 S1 protein-reactive IgG was detected by horseradish peroxidase-conjugated anti-human IgG. The absorbance was assessed on a microplate reader at 450 nm with a reference at 620 nm and evaluated as the ratio of the absorbance of the sample to the absorbance of the internal standard.
Statistical Analysis
Flow cytometry data were analyzed using FlowJo version 10.6.2 (BD Biosciences, USA); gating strategies are presented in Fig 1, Fig 2 . For the analysis of anti-SARS-CoV-2 T and B cells, a threshold of 0.01% and 0.001% was employed respectively, to define a detectable response. Single stains and fluorescence-minus-1 controls were used for gating. Gates for each individual were adjusted according to the negative control. CD4+ T cells expressing CD154 and CD137 and CD8+ T cells expressing CD137 were defined as reactive T cells. Statistical analysis was performed using GraphPad Prism v7. Categorical variables are summarized as numbers and frequencies; quantitative variables are reported as medians and interquartile ranges. Normality tests were performed with D'Agostino and Pearson, Shapiro-Wilk, and Anderson-Darling tests. All applied statistical tests were two-sided. The frequencies of SARS-CoV-2-reactive B and T cells in patients who had recovered from COVID-19 and immunocompetent donors were compared using an exact 2-tailed Mann-Whitney test, and, for grouped data, the Mann-Whitney test was used. The age of patients who were unexposed and exposed was compared using an unpaired 2-tailed t test, and sex was compared using a 2-tailed Fisher exact test. P values below .050 were considered significant; only significant P values are reported in the figures. P values were not corrected for multiple testing, because this study was of an exploratory nature.
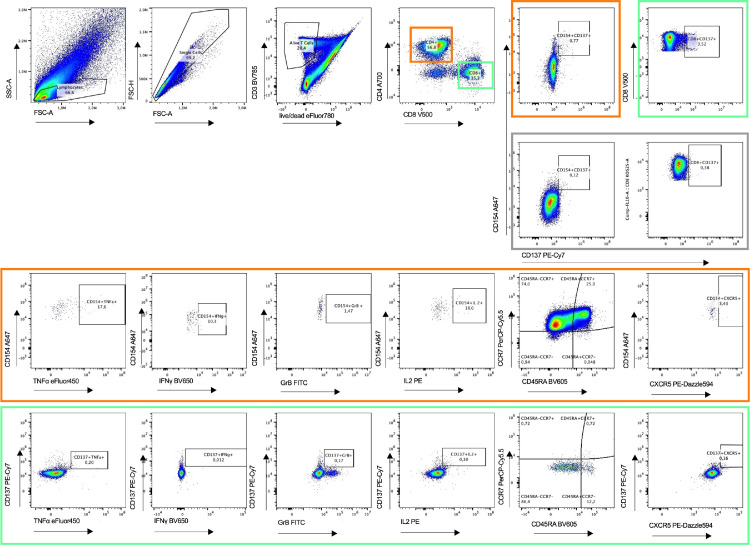
Fig 1.
Flow cytometry gating strategy for identification and quantification of severe acute respiratory syndrome coronavirus 2 S-peptide. After 2 hours, Brefeldin A was added to the culture to block secretion of cytokines and effector molecules. Living single lymphocytes were analyzed for expression of CD3, CD4, and CD8. CD4+ T cells (oranges boxes) were analyzed for the expression of CD154 and CD137. CD8+ T cells (green boxes) were analyzed for expression of CD137. Both CD4+ and CD8+ T cells were further analyzed for production of cytokines IFNγ, TNFα, IL-2 and GrB. The gray box includes untreated samples. Furthermore, CD4+CD154+CD137+ and CD8+CD137+ cells were analyzed for the expression of CXCR5. Representative example of 26 unexposed and 14 convalescent patients. Plots of an unexposed study subject are being presented.
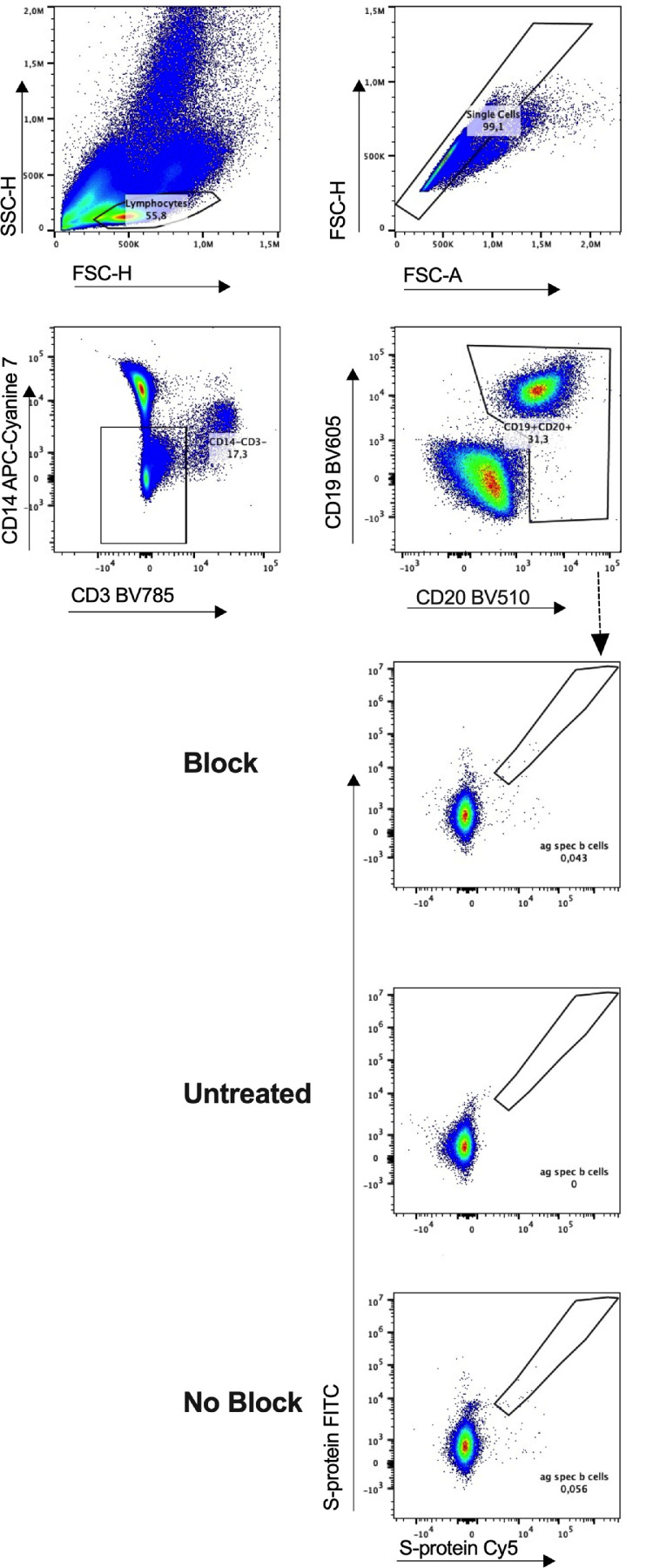
Fig 2.
Flow cytometry gating strategy for identification and quantification of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) reactive B cells. Representative example for the detection of dual-labeled SARS-CoV-2 S-protein binding B-cells and quantification of antigen-reactive B cell subsets. Comparison of samples without fluorochrome-coupled SARS-CoV-2 protein (untreated) and SARS-CoV-2 S-protein in and without excess unlabeled protein to block B cell. Representative example of 26 unexposed and 14 convalescent patients. Plots of a convalescent COVID-19 patient are presented.
Results
Baseline Characteristics of the Study Cohort
We analyzed 26 individuals unexposed to SARS-CoV-2, of whom 14 had undergone Tx, 12 had not undergone Tx, and 22 were Cp (see Table 1). The median age of the unexposed individuals at the time of study inclusion was 69 years, with participant ages ranging from 37 to 91 years, with 58% men and 42% women. Patients who had undergone Tx (12 kidney transplant, 1 liver transplant, 1 combined kidney/liver transplant recipient) were significantly younger, with a median age of 55 years (range, 37-75; P = .0069) compared to the patients who had not undergone Tx with a median age of 73 years (range, 49-91). We compared the patients who had undergone Tx to immunocompetent Cp at a median time of 110 days after diagnosis or onset of symptoms (range, 22-198 days). All included Cp were confirmed to be SARS-CoV-2-positive by PCR. The median age of the Cp group was 54.5 years (range, 28-89) and not significantly different from that of the unexposed group (median age, 69 years; range, 37-91) (P = .5182; 2-tailed unpaired t test). There were no significant differences regarding sex between the unexposed and patients with COVID-19 (P = .5626; 2-tailed Fisher exact test). Demographic characteristics are provided in Tables 1 and 2.
Presence of Pre-existing SARS-CoV-2–Reactive T Cells in Unexposed Study Participants
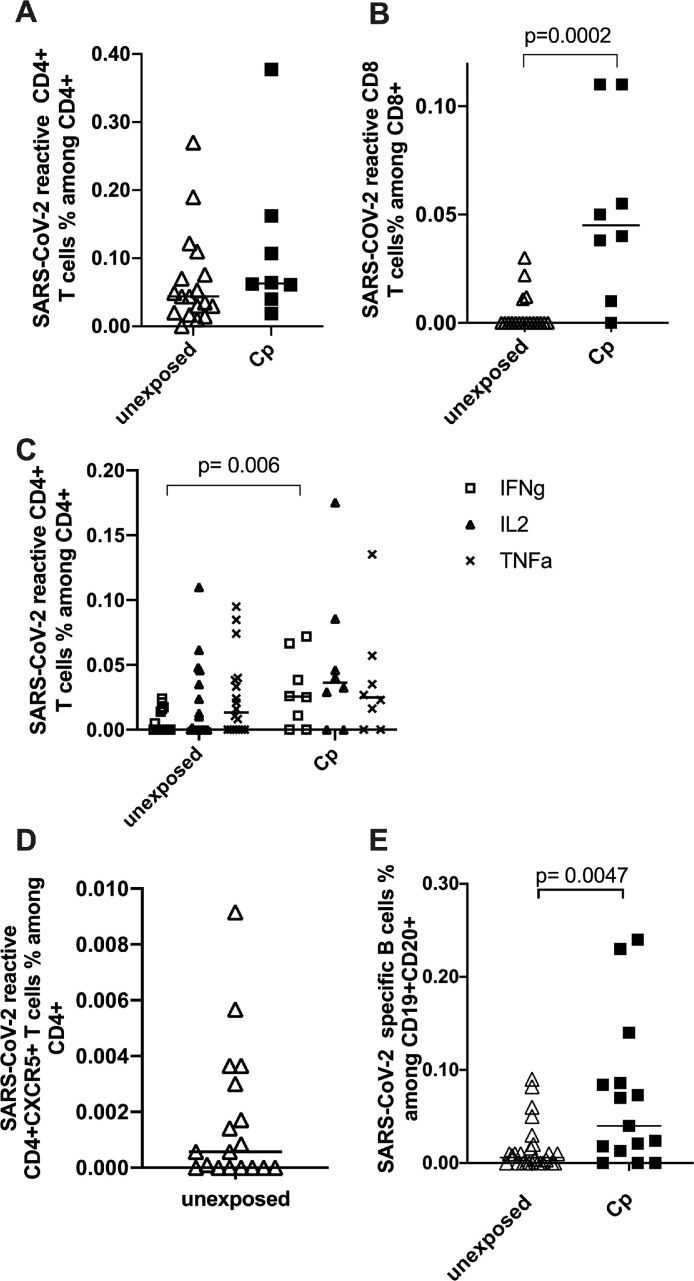
As applied in previous studies [14,16], antigen-reactive T cell responses were considered positive after the background was subtracted and greater than 0.01% was detectable. The exception to this rule was the detection of circulating follicular CD4+ T helper cells, for which no minimum numerical limit was set due to the extremely low number of this cell population normally in circulation. We found detectable SARS-CoV-2 S-protein-reactive CD4+ and CD8+ T cells in 94% and 22% of unexposed individuals, respectively. The frequencies of SARS-CoV-2-reactive CD4+ T cells in the exposed cohort were higher, without statistical significance regarding SARS-CoV-2-reactive CD4+ T cells (P = .19; Mann-Whitney test) (Fig 3 A). However, the frequencies of SARS-CoV-2-reactive CD8+ cells were significantly higher in the Cp cohort (P = .0002; Mann–Whitney test) (Fig 3 B). Inferferon γ–producing S-protein-reactive CD4+ T cells showed significantly higher frequencies in Cp than in unexposed individuals (P = .006). For all other cytokines determined in our study, no significant differences were observed regarding the frequencies of cytokine-producing CD4+ T cells between the Cp and unexposed study participants (Fig 3 C).
Fig 3.
Characterization of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) S-reactive T and B specific cells in unexposed and exposed subjects. Blood samples of 18 unexposed patients (7 out of 18 patients who had undergone transplant) and 9 control patients convalescing from COVID-19 were stimulated with SARS-CoV-2 S-protein and analyzed by flow cytometry. (A) Frequencies of CD4+CD154+CD137+ and (B) CD8+CD137+. (C) Frequencies of mono- and bifunctional SARS-CoV-2 reactive CD4+ T cells expressing granzyme B (GrB), IFNγ, interleukin 2 or tumor necrosis factor α (TNFα). (D) SARS-CoV-2 reactive CD4+CD154+CD137+ cells of unexposed donors were analyzed for CXCR5 positivity. (E) Correlation of fluorochrome labelled SARS-CoV-2 S-protein binding B cells in 26 unexposed and 14 patients convalescing from COVID-19. Analysis was performed exact two-tailed Mann-Whitney Test. SARS-CoV-2 T cells are defined the CD4+CD154+CD137+ and CD8+CD137+ cells are defined as reactive SARS-CoV-2 T cells. Negative controls were subtracted from reactive stimulated samples to exclude unreactive activation. Statistical comparison was done with Mann-Whitney-test. P < .05 was considered significant, and only significant P values are documented in the figures.
Positive Frequencies of SARS-CoV-2–Reactive Follicular CD4+ T Cells in the Unexposed Cohort
Tfh cells directly interact with B cells, indicate maturation of the humoral immune response, and are crucial for the establishment of antigen-reactive B memory cells, which provide long-term immunity [17]. We characterized circulating SARS-CoV-2 S-protein-reactive Tfh cells in the unexposed cohort by the expression of CXCR5 (Fig 3 D). In a single exception to the abovementioned minimum limit of 0.01%, we evaluated all positive frequencies of CD4+CD154+CD137+CXCR5+ cells after the background was subtracted, because no large populations of Tfh cells were expected to be found in circulation. Among the total population of unexposed individuals, 11 of them (61%) showed positive frequencies for CD4+CXCR5+ T cells.
Detection of Pre-existing SARS-CoV-2 S-Protein-Reactive B Cells in 58% of Unexposed Individuals
To explore whether B cells reactive against SARS-CoV-2 S-protein were detectable in unexposed individuals, we analyzed the frequencies of SARS-CoV-2 S-protein-reactive B cells by flow cytometry using FITC- and Cy5-labeled S-protein, as previously described [15]. Specificity was controlled by blocking with excess unlabeled SARS-CoV-2 S-protein (Fig 2) [18,19]. Double-positive S-protein-FITC- and S-protein-Cy5-reactive B cells were considered to specifically bind to the S-protein when the frequency was above 0.001% after the frequency of the blocked sample was subtracted.
We observed detectable S-reactive B cells in 80% of the Cp control group and in 58% of the unexposed individuals. The control group of Cp showed significantly higher frequencies of SARS-CoV-2-reactive B cells compared to unexposed individuals (P = .0047; exact 2-tailed Mann–Whitney Test) (Fig 3 E). Out of the 18 unexposed individuals with characterized T and B cell responses, 11 individuals presented pre-existing SARS-CoV-2-specific Tfh cells, 7 of whom had a detectable SARS-CoV-2-specific B cell response.
Similar Frequencies of Pre-existing T and B Cell Subsets Among Tx and Non-Tx Individuals
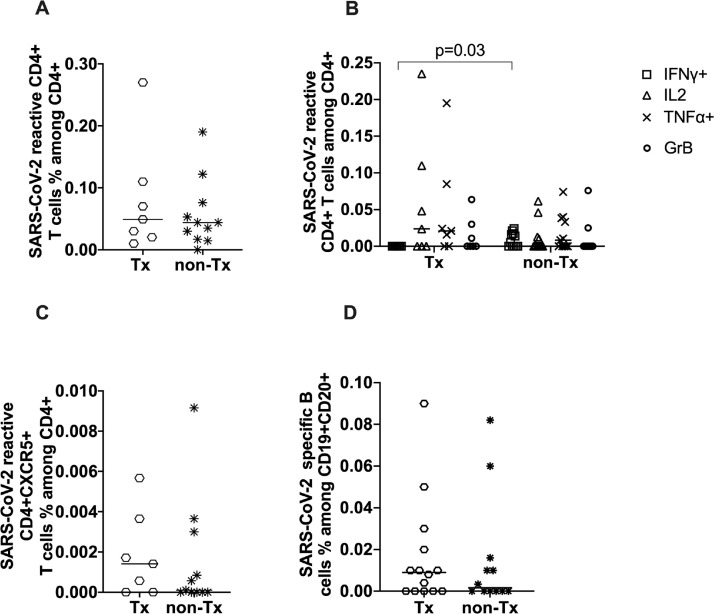
The frequencies of SARS-CoV-2-reactive and cytokine-producing CD4+ T cells were similar among unexposed patients who had undergone Tx and patients who had not (Fig 4 A). Similarly, there was no significant difference in CD4+CXCR5+ cells among patients who had undergone Tx and those who had not (Fig 4 C). The frequencies of SARS-CoV-2-reactive B cells in the Tx group were not significantly different compared to the immunocompetent participants (Fig 4 D) (P = .1588). SARS-CoV-2-reactive B cells were found more frequently among patients who had undergone Tx, with 9 patients who had undergone Tx (64%) showing SARS-CoV-2-reactive B cells compared with 6 participants who had not undergone Tx (50%) showing SARS-CoV-2-reactive B cell immunity.
Fig 4.
Characterization of severe acute respiratory syndrome coronavirus 2 reactive T and B cells in transplant and non-transplant donors. (A) CD4+CD154+CD137+ showed no significant statistical difference among transplant and non-transplant patients. (B) Analysis of the monofunctional CD4+CD154+CD137+ cells. (C) Correlation of CD4+CD154+CD137+CXCR5+ among patients who had undergone transplant those who had not. (D) Analysis of fluorochrome severe acute respiratory syndrome coronavirus 2 S-protein binding B cells among the unexposed cohort of patients who had undergone transplant vs patients who were immunocompetent (P = .1588). Statistical comparison was done with the Mann-Whitney test. P < .05 was considered significant, and only significant P values are documented in the figures.
Discussion
Here, we report cross-reactive and pre-existing B and T cell immunity to SARS-CoV-2 in a cohort of unexposed individuals, including immunocompetent individuals and renal transplant recipients. Our study suggests that renal transplant patients can generate a pre-existing SARS-CoV-2 response that is comparable to immunocompetent adults.
Low frequencies of circulating pre-existing Tfh cells could be detected in Tx and non-Tx participants to a comparable extent. Accumulating data show that SARS-CoV-2-reactive circulating Tfh cells play a key role in effective immunity and the generation of B cell memory and are consistent with the observation that the frequency of total circulating Tfh cells increases at the time of SARS-CoV-2 clearance and the detection of robust Tfh cell responses [20], [21], [22], [23]. Lipsitch et al imply in a theoretical model the potential contribution of SARS-CoV-2 pre-existing Tfh cells in accelerating antibody production in the case of SARS-CoV-2 infection [24]. The presence of cross-reactive Tfh cells in the unexposed cohort, including immunosuppressed adults, may be of particular clinical importance and in theory could boost the protective role of pre-existing SARS-CoV-2 immunity. In line, a detailed recent study shows robust vaccination-induced immune responses in patients who had not undergone Tx, depending on the concomitant medication, especially the absence of Rituximab [25].
We detected SARS-CoV-2-specific B cells among the unexposed individuals, and surprisingly, we observed a slight predominance in the patients who had undergone Tx in the formation of pre-existing B cells compared with individuals who had not undergone Tx. The predominance of patients who had undergone Tx in the formation of pre-existing B cells might be explained by the higher incidence of different viral, bacterial, or fungal coinfections, because patients who had undergone Tx demonstrate a broader antigenic experience with a higher chance of generating cross-reactive cellular immunity than the immunocompetent population. Bacher et al and others have suggested that pre-existing SARS-CoV-2 memory is the result of a diverse memory pool that may not be of viral origin at all, which accumulates in humans throughout life and might contain T cell receptors specific for neoantigens similar to the naive T cell pool, with a broad range of affinities [12,26,27].
Study limitations
A limitation of our study was the small number of patients, which makes robust assumptions challenging. Subsequent studies should enroll larger patient cohorts with a greater demographic variability to include individuals of all ages from different social levels and environments. Also, multi-center design should be performed to exclude a local bias (ethnicity, environmental/seasonal corona viruses, treatment). The significant age gap between the Tx and non-Tx cohorts should also be taken into consideration.
Conclusions
Overall, our study demonstrates pre-existing SARS-CoV-2 immunity among the transplant cohort, which is comparable to the immunocompetent study group. Independent working groups demonstrate the poor immune response and waning of antibodies after SARS-CoV-2 infection or vaccination among transplant recipients [28,29]. Taking into consideration the emerging SARS-CoV-2 variants of concern understanding the influence of pre-existing cross-reactive immunity to SARS-CoV-2 on the adaptive immune response is also of critical importance.
Compliance with ethical Standarts
The study was approved by the Ethics Committee of the Ruhr University Bochum (20-6886) and University Hospital Essen (20-9214-BO). Written informed consent was obtained from all participants. The authors have no relevant financial or non-financial interests to disclose.
Author contributions
Conceptualization: Krystallenia Paniskaki, Nina Babel; Data curation and sample acquisition: Krystallenia Paniskaki, Margarethe J. Konik; Methodology: Krystallenia Paniskaki, Moritz Anft, Sarah Skrzypczyk, Mikalai Nienen, Anna Stittrich; Writing – original draft preparation: Krystallenia Paniskaki, Moritz Anft; Writing- review and editing: Krystallenia Paniskaki, Nina Babel, Toralf Roch, Guido Heine, Constantin J. Thieme, Sebastian Dolff; Funding acquisition and Resources: Nina Babel, Oliver Witzke, Ulrik Stervbo, Timm H. Westhoff.
Declaration of Competing Interest
None.
Footnotes
This work was supported by grants of Mercator Foundation, the BMBF e:KID (01ZX1612A), and BMBF NoChro (FKZ 13GW0338B).
Data Availability
Data will be made available on request.
References
- 1.Zhao H, Lu X, Deng Y, Tang Y, Lu J. COVID-19: asymptomatic carrier transmission is an underestimated problem. Epidemiol Infect. 2020;148:e116. doi: 10.1017/S0950268820001235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakajo K, Nishiura H. Transmissibility of asymptomatic COVID-19: data from Japanese clusters. Int J Infect Dis. 2021;105:236–238. doi: 10.1016/j.ijid.2021.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nishiura H, Kobayashi T, Miyama T, Suzuki A, Jung S, Hayashi K, et al. Estimation of the asymptomatic ratio of novel coronavirus infections (COVID-19) Int J Infect Dis. 2020;94:154–155. doi: 10.1016/j.ijid.2020.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braun J, Loyal L, Frentsch M, Wendisch D, Georg P, Kurth F, et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587:270–274. doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- 5.Mateus J, Dan JM, Zhang Z, Rydyznski Moderbacher C, Lammers M, Goodwin B, et al. Low-dose mRNA-1273 COVID-19 vaccine generates durable memory enhanced by cross-reactive T cells. Science. 2020;374:eabj9853. doi: 10.1126/science.abj9853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181 doi: 10.1016/j.cell.2020.05.015. 1489-1501.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nelde A, Bilich T, Heitmann JS, Maringer Y, Salih HR, Roerden M, et al. SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nature. 2020;22:74–85. doi: 10.1038/s41590-020-00808-x. [DOI] [PubMed] [Google Scholar]
- 8.Sakharkar M, Rappazzo CG, Wieland-Alter WF, Hsieh C-L, Wrapp D, Esterman ES, et al. Prolonged evolution of the human B cell response to SARS-CoV-2 infection. Sci Immunol. 2021;6:eabg6916. doi: 10.1126/sciimmunol.abg6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song G, He W, Callaghan S, Anzanello F, Huang D, Ricketts J, et al. Cross-reactive serum and memory B-cell responses to spike protein in SARS-CoV-2 and endemic coronavirus infection. Nat Com. 2021;12:2938. doi: 10.1038/s41467-021-23074-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nguyen-Contant P, Embong AK, Kanagaiah P, Chaves FA, Yang H, Branche AR, et al. S protein-reactive IgG and memory B cell production after human SARS-CoV-2 infection includes broad reactivity to the S2 subunit. mBio. 2021;11:11. doi: 10.1128/mBio.01991-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loyal L, Braun J, Henze L, Kruse B, Dingeldey M, Reimer U, et al. Cross-reactive CD4+ T cells enhance SARS-CoV-2 immune responses upon infection and vaccination. Science. 2021:eabh1823. doi: 10.1126/science.abh1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Low JS, Vaqueirinho D, Mele F, Foglierini M, Jerak J, Perotti M, et al. Clonal analysis of immunodominance and cross-reactivity of the CD4 T cell response to SARS-CoV-2. Science. 2021;372:1336. doi: 10.1126/science.abg8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schilling J, Lehfeld AS, Schumacher D, Diercke M, Buda S, Haas W, et al. Disease severity during the first COVID-19 wave in Germany based on the national german registry. J Health Monit. 2020;5:1–17. doi: 10.25646/7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anft M, Paniskaki K, Blazquez-Navarro A, Doevelaar A, Seibert FS, Hölzer B, et al. COVID-19-induced ARDS is associated with decreased frequency of activated memory/effector T cells expressing CD11a++ Mol Therapy. 2020;28:2691–2702. doi: 10.1016/j.ymthe.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thieme C, Abou-el-Enein M, Fritsche E, Anft M, Skrzypczyk S, Doevelaar A-A-N, et al. Detection of SARS-CoV-2 specific memory B cells to delineate long-term COVID-19 immunity. Allergy. 2020;76:2595–2599. doi: 10.1111/all.14827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thieme CJ, Anft M, Paniskaki K, Blazquez-Navarro A, Doevelaar A, Seibert FS, et al. The magnitude and functionality of SARS-CoV-2 reactive cellular and humoral immunity in transplant population is similar to the general population despite immunosuppression. Transplantation. 2021;105:2156–2164. doi: 10.1097/TP.0000000000003755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cox RJ, Brokstad KA. Not just antibodies: B cells and T cells mediate immunity to COVID-19. Nat Rev Immunol. 2021;20:581–582. doi: 10.1038/s41577-020-00436-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boonyaratanakornkit J, Taylor JJ. Techniques to study antigen-specific B cell responses. Front Immunol. 2019;10:1694. doi: 10.3389/fimmu.2019.01694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cossarizza A, Chang H-D, Radbruch A, Akdis M, Andrä I, Annunziato F, et al. Guidelines for the use of flow cytometry and cell sorting in immunological studies. Eur J Immun. 2017;47:1584–1797. doi: 10.1002/eji.201646632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mathew D, Giles JR, Baxter AE, Oldridge DA, Greenplate AR, Wu JE, et al. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369:eabc8511. doi: 10.1126/science.abc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neidleman J, Luo X, Frouard J, Xie G, Gill G, Stein ES, et al. SARS-CoV-2-specific T cells exhibit phenotypic features of helper function, lack of terminal differentiation, and high proliferation potential. Cell Rep Med. 2020;1 doi: 10.1016/j.xcrm.2020.100081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wowk K. Rapid generation of durable B cell memory to SARS-CoV-2 spike and nucleocapsid proteins in COVID-19 and convalescence. Sci Immunol. 2020;5:eabf8891. doi: 10.1126/sciimmunol.abf8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Juno JA, Tan H-X, Lee WS, Reynaldi A, Kelly HG, Wragg K, et al. Humoral and circulating follicular helper T cell responses in recovered patients with COVID-19. Nat Med. 2020;2623dc:1428–1434. doi: 10.1038/s41591-020-0995-0. [DOI] [PubMed] [Google Scholar]
- 24.Lipsitch M, Grad YH, Sette A, Crotty S. Cross-reactive memory T cells and herd immunity to SARS-CoV-2. Nat Rev Immunol. 2020;20:709–713. doi: 10.1038/s41577-020-00460-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rincon-Arevalo H, Choi M, Stefanski A-L, Halleck F, Weber U, Szelinski F, et al. Impaired humoral immunity to SARS-CoV-2 BNT162b2 vaccine in kidney transplant recipients and dialysis patients. Sci Immunol. 2021;6:eabj1031. doi: 10.1126/sciimmunol.abj1031. [DOI] [PubMed] [Google Scholar]
- 26.Bacher P, Rosati E, Esser D, Martini GR, Saggau C, Schiminsky E, et al. Low-avidity CD4+ T cell responses to SARS-CoV-2 in unexposed individuals and humans with severe COVID-19. Immunity. 2020;53:1258–1271. doi: 10.1016/j.immuni.2020.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ladner JT, Henson SN, Boyle AS, Engelbrektson AL, Fink ZW, Rahee F, et al. Epitope-resolved profiling of the SARS-CoV-2 antibody response identifies cross-reactivity with endemic human coronaviruses. Cell Rep Med. 2021;2 doi: 10.1016/j.xcrm.2020.100189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boyarsky BJ, Werbel WA, Avery RK, Tobian AAR, Massie AB, Segev DL, et al. Immunogenicity of a single dose of SARS-CoV-2 messenger RNA vaccine in solid organ transplant recipients. JAMA. 2021;325:1784–1786. doi: 10.1001/jama.2021.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kemlin D, Lemy A, Pannus P, Desombere I, Gemander N, Goossens ME, et al. Hybrid immunity to SARS-CoV-2 in kidney transplant recipients and hemodialysis patients. Am J Transplant. 2022;22:994–995. doi: 10.1111/ajt.16853. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.