Abstract
Introduction
Neoadjuvant chemotherapy (NAC) for muscle-invasive bladder cancer (MIBC) improves overall survival, but pathological response rates are low. Predictive biomarkers could select those patients most likely to benefit from NAC. Radiomics technology offers a novel, non-invasive method to identify predictive biomarkers. Our study aimed to develop a predictive radiomics signature for response to NAC in MIBC.
Methods
An institutional bladder cancer database was used to identify MIBC patients who were treated with NAC followed by radical cystectomy. Patients were classified into responders and non-responders based on pathological response. Bladder lesions on computed tomography images taken prior to NAC were contoured. Extracted radiomics features were used to train a radial basis function support vector machine classifier to learn a prediction rule to distinguish responders from non-responders. The discriminative accuracy of the classifier was then tested using a nested 10-fold cross-validation protocol.
Results
Nineteen patients who underwent NAC followed by radical cystectomy were found to be eligible for analysis. Of these, nine (47%) patients were classified as responders and 10 (53%) as non-responders. Nineteen bladder lesions were contoured. The sensitivity, specificity, and discriminative accuracy were 52.9±9.4%, 69.4±8.6%, and 62.1±6.1%, respectively. This corresponded to an area under the curve of 0.63±0.08 (p=0.20).
Conclusions
Our developed radiomics signature demonstrated modest discriminative accuracy; however, these results may have been influenced by small sample size and heterogeneity in image acquisition. Future research using novel methods for computer-based image analysis on a larger cohort of patients is warranted.
Introduction
Neoadjuvant chemotherapy (NAC) prior to radical cystectomy (RC) in muscle-invasive bladder cancer (MIBC) has led to an improvement in five-year overall survival (OS) rates by 5% as compared to RC alone, with preferential benefit for patients who achieve a pathological response (i.e., ≤ypT1).1–4 However, the majority of patients fail to respond to NAC, with evidence of muscle-invasive disease at the time of cystectomy.3 This low response rate, coupled with the significant risk of toxicities with recommended cisplatin-based chemotherapy regimens, raises concerns about the risk-benefit ratio of using NAC in patients with MIBC, particularly in patients who are older and have comorbidities.5,6 Accordingly, predictive biomarkers are critical to identify patients who will respond to NAC in order to limit potential toxicities from cytotoxic chemotherapy in patients who are unlikely to derive benefit.
Radiomics technology offers a promising method to identify predictive and prognostic biomarkers by extracting quantitative information from radiological imaging to describe tumor biology.7,8 As the analysis is not limited to the biopsied sample, radiomics analysis can assess for intra-tumoral heterogeneity, as well as the tumor microenvironment.9 Furthermore, as a non-invasive method, it provides negligible risk to the patient and can be easily used to assess for tumor response to therapy over time. The potential opportunity of radiomics technology as a predictive biomarker has been demonstrated across multiple solid tumor malignancies (i.e., head and neck, lung, esophageal, rectal, and testicular cancers), highlighting its potential as a predictive biomarker in MIBC.9–14
The Radiomic signature in predicting rEsponse to neoadjuVAnt chemotherapy in muscle-invasive bLadder cancer (REVeAL) study was thus conducted to develop and evaluate a radiomics signature that can predict the likelihood of response to NAC based on pre-treatment computed tomography (CT) for patients treated for MIBC at a single institution.
Methods
Cohort creation
All patients diagnosed with MIBC between January 1, 2001, and December 31, 2016, were identified from a retrospective review of the University Health Network Bladder Cancer database following institutional ethics approval. Eligible patients for inclusion included adult patients (age ≥18 years) with biopsy-proven urothelial carcinoma (squamous cell histology was excluded) who received NAC with cisplatin-based treatment. Patients were excluded if there were no CT images prior to initiation of NAC and if there was no surgical pathology available from cystectomy. This cohort was divided into two subgroups based on pathological response at the time of cystectomy: responders (i.e., pathological tumor [T] stage of ≤ypT1) and non-responders (i.e., >ypT1).
For all eligible patients, the following clinical data were extracted: patient demographics (i.e., age, sex); disease characteristics (i.e., TNM stage); details of NAC (including chemotherapy regimen, number of cycles completed); pathological response at time of cystectomy; and clinical outcomes (i.e., recurrence and survival).
Radiomics analysis
Radiological contouring of the primary bladder tumor was performed manually on the pre-treatment CT images by two reviewers in tandem (AP, AQ) using 3D Slicer software (3D Slicer version 4.10.2, available at: https://www.slicer.org/). A third investigator (AH) was available to adjudicate any disagreements. The region of interest (ROI) for image contouring was restricted to the primary bladder tumor, with exclusion of regional lymph nodes.
Quantitative radiomic features from the primary bladder lesion were extracted using computational algorithms. Each image voxel was quantized from a grey level continuum to a finite range of 32 grey levels. Each algorithm was initially described for two-dimensional (2D) pictures and then adapted for three-dimensional (3D) volumes by extrapolating from the four principal directions in 2D to the 13 principal directions in 3D to generate a volume of interest (VOI).9 The radiomic features were then computed in a step-wise fashion. First-order metrics specific to lesion intensity (i.e., the brightness or darkness of the imaging voxels) were computed from each VOI and classified into 11 percentiles, ranging from 0–100%. The intensities from each VOI were then divided into 32 equal-sized bins that spanned the range of image intensities between the 1st and 99th percentiles (percentiles chosen to minimize effect of outliers) prior to computing the 142 second-order texture metrics. The second-order texture analysis was based upon four classes of texture matrices that are well-described in the literature: 1) 13 Haralick features from the gray level co-occurrence matrix; 2) five features from the neighborhood gray tone difference matrix; 3) 10 features from the gray level run length matrix; and 4) 10 features from the gray level size zone matrix.15–18 Integration of these 142 radiomics features was used to characterize the radiomics signature for responders and non-responders.
Machine learning
Each radiomics signature was defined as a responder or non-responder using the final pathological response. This information was used to train a radial basis function (RBF) support vector machine (SVM) classifier to learn a prediction rule to distinguish responders from non-responders.19,20 In this regard, the SVM functioned as a learning model to optimally separate points within a feature space into responders vs. non-responders. In the setting of ambiguity, the RBF kernel was used to map the initial feature space into a higher dimensional feature space where they ultimately become linearly separable.
In stage 1, cross-validation was used to test and optimize the SVM classifier using a 10-fold nested cross-validation. This approach used the same data for training and testing but in separate iterations. Nested validation allows for two loops to be performed in parallel: an inner loop (hyperparameter tuning), which finds the optimal combination of classifier settings, and the outer loop, which tests the accuracy of the classifier.21 The hyper-parameters were manually set and included: 1) feature selection (fraction of the most highly associated features to retain for the prediction rule); 2) cost hyperparameter (determines the tradeoff for outliers); and 3) scale hyperparameter (determines the smoothness of the decision surface).22 For each of the 100 iterations of the outer loop, 10-fold cross-validation was used, with 90% of the data used in the inner loop and 10% of the data for testing. Within the inner loop, 10-fold cross-validation protocol was employed for each point in a 3D grid covering a range of fractions of the best features to retain (feature selection), values of the cost parameter, and values of the scale parameter. For each grid point searched, the inner loop cross-validation result was recorded and then the best-performing triple of hyperparameters was used to train a classifier using all the inner loop data. This classifier was then applied to the 10% data held for testing, and the results recorded as the accuracy.
In stage 2, the texture features, as per the binary classification by response, were inputted into the supervised machine learning (SML) RBF SVM classifier to train for the discrimination between the responders and non-responders. The accuracy of the classifier was then tested using a nested 10-fold cross-validation protocol.
Outcome measure
Description of the accuracy of the final predictive radiomics signature from the SVM classifier was reported by its sensitivity, specificity, area under the receiver operator curve (AUC), and statistical significance.
Statistical analysis
Descriptive statistics (mean, median, counts, and proportions) were used to summarize the clinical characteristics of the included cohort. All data was analyzed descriptively with proportions for categorical data or medians and interquartile ranges (IQR) for continuous data. The two patient groups were compared with the use of Pearson’s Chi-squared test for all categorical data. Continuous data was compared with a one-way analysis of variance. Data for time dependent variables were reported as medians with 95% confidence intervals (CI). The Kaplan-Meier method was used to estimate relapse-free survival (RFS) and OS, with corresponding 95% CI. RFS was defined as the time from pathological diagnosis to date of first recurrence, death, or censored at the date of last followup. OS was defined as the time from pathological diagnosis to date of death or censored at the date of last followup. Log-rank statistics were used to compare RFS between responders and non-responders. Statistical significance for the predictive accuracy of the radiomics signature from the SVM classifier was determined by permutation testing, at a significance level of 5%.
Results
Study cohort
Thirty-two patients with MIBC who underwent NAC were identified. Of these, eight patients were excluded due to lack of available pre-treatment CT images, four due to an inability to localize the bladder tumor, and one due to imaging artifact, leaving 19 patients eligible for further analysis. Of these, nine (47%) patients were classified as responders and 10 (53%) as non-responders based on their pathology at the time of cystectomy. Table 1 summarizes the clinical characteristics and outcomes of the included cohort. The majority of patients had their pre-treatment CT imaging following pathological sampling (n=13; 68%), with a median time from pathological diagnosis to CT imaging of 43 days (95% CI 20–106). Four patients (21%) had CT imaging prior to pathological sampling, and for two patients (11%), the date of pathological sampling is unknown. The median time from pre-treatment CT imaging to cystectomy was 157 days (95% CI 128–235).
Table 1.
Characteristics and outcomes of included patients
| Characteristic | Responder (n=9) | Non-responder (n=10) | p |
|---|---|---|---|
| Age, years; median (IQR) | 68 (62–73) | 66 (59–69) | 0.88 |
| Male sex (%) | 8 (89) | 6 (60) | 0.36 |
| Clinical nodal status1 (%) | 1 | ||
| 0 | 8 (89) | 8 (80) | |
| 1 | 1 (11) | 2 (20) | |
| Neoadjuvant chemotherapy2 | 0.962 | ||
| GC (%) | 8 (89) | 10 (100) | |
| Cycles, median (range) | 4 (2–6) | 4 (2–4) | |
| MVAC (%) | 1 (11) | 0 | |
| Cycles, median (range) | 4 | – | |
| Pathological stage (%) | |||
| Tumor stage | <0.05 | ||
| T0 | 7 (78) | 0 | |
| Ta | 1 (11) | 0 | |
| T1 | 1 (11) | 0 | |
| T2 | 0 | 5 (50) | |
| T3 | 0 | 3 (30) | |
| T4 | 0 | 2 (20) | |
| Nodal stage | 0.20 | ||
| N0 | 9 (100) | 7 (70) | |
| N1 | 0 | 2 (20) | |
| N2 | 0 | 1 (10) | |
| Outcomes (%) | |||
| Distant recurrence | 3 (33) | 6 (60) | 0.48 |
Aside from the expected difference in pathological stage, no statistically significant differences were noted between the two groups.
Nodal status was determined on pretreatment computed tomography imaging completed prior to neoadjuvant chemotherapy.
Statistical testing completed on proportion of patients who received each chemotherapy regimen.
GC: gemcitabine-cisplatin; IQR: interquartile range; MVAC: methotrexatevinblastine-doxorubicin-cisplatin.
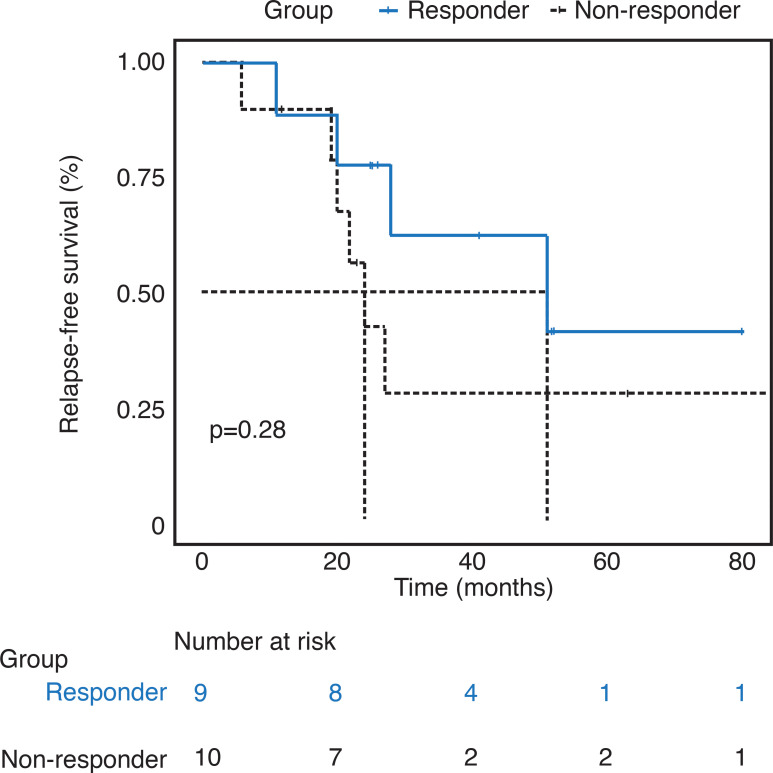
The median RFS for responders and non-responders was 51 months (95% CI 28–not reached) and 24 months (95% CI 20–not reached, p=0.28), respectively (Fig. 1). Median OS for the study cohort was not reached.
Fig. 1.
Relapse-free survival for included cohort by subgroup.
Among the included study cohort, a total of 19 bladder lesions in 19 patients were analyzed. The majority of bladder tumors were visualized on contrast-enhanced CT scans (n=18; 95%) in the delayed (n=13; 72%) or venous (n=5; 28%) phase. CT images were acquired with 3 mm slice thickness for all but one patient (n=18; 95%). All but one included patient had a solitary bladder lesion that was included for image analysis. One patient in the non-responding subgroup was found to have multifocal lesions, all of which were included as one lesion in the ROI. Image analysis was restricted to the bladder lesion only. Thus, for the three patients identified to have node-positive disease, nodal disease was not included in the ROI. The anatomical location of the lesions across the cohort were found to be posterior-lateral (right: seven [37%], left: three [16%]), anterior-lateral (right: four [21%], left: one [5%]), two left-lateral (11%), one posterior (5%), one multifocal (5%). Median time from CT imaging to initiation of NAC for the cohort was 24 days (95% CI 10–43).
Radiomics analysis
Neuroimaging Informatics Technology Initiative (NIfTI) software libraries were used to extract lesion VOI contours. Series of lesion VOI that were specific to a subject were examined using the radiomics analysis software to confirm successful extraction of VOI contours from the NIfTI files and to confirm their positions matched those of a bladder lesion. The data set was found to be suitable for radiomics analysis. Batch processing of the total REVeAL data generated the set of texture features that were then used by the second stage radiomics SML application.
Machine learning
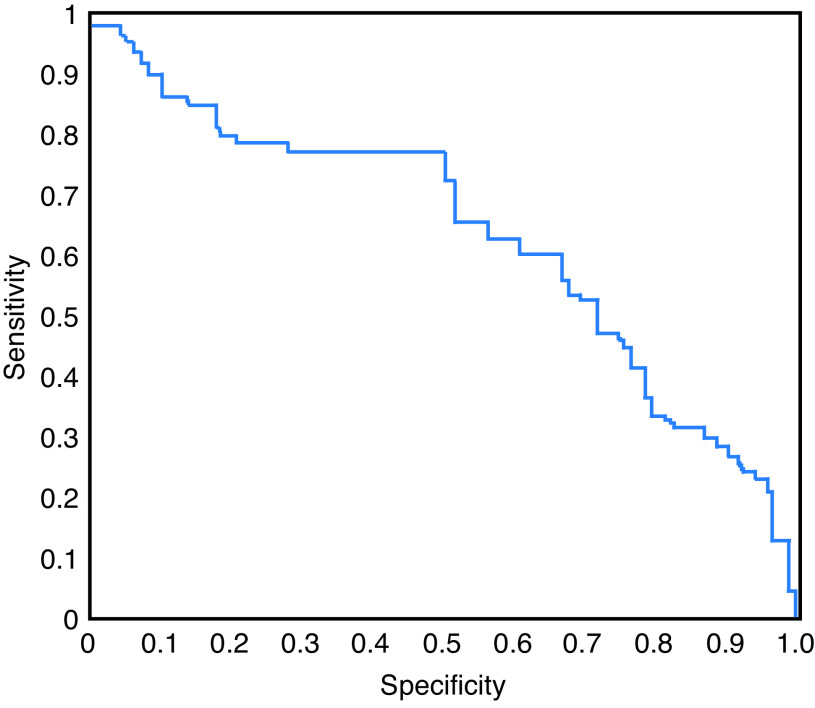
The receiver-operator characteristic (ROC) curve, based upon the iterative grid over the region of highest cross-validation accuracy, is represented in Fig. 2. The mean discriminative accuracy of the radiomics signature from the SVM classifier was found to be 62.1±6.1%. The sensitivity and specificity of the radiomics signature were found to be 52.9±9.4% and 69.4±8.6%, respectively. This corresponded to an AUC of 0.63±0.08 (p=0.20).
Fig. 2.
Receiver operating characteristic curve for radiomic classifier for discriminating responders from non-responders on pre-treatment computed tomography images.
Discussion
The REVeAL study aimed to develop a predictive radiomics signature to identify patients with MIBC who were likely to respond to NAC. To our knowledge, this is the first evaluation of a predictive radiomics signature in MIBC developed on pre-treatment CT images that benchmarked response as per pathological assessment. Our study demonstrated the feasibility of extracting radiomic features from pre-treatment CT imaging in MIBC for the development of a predictive radiomics signature. The discriminative accuracy of our predictive radiomics signature was modest at 62%.
In the absence of contraindications, clinical practice guidelines recommend the use of NAC with cisplatin-based chemotherapy for patients with MIBC given data supporting a survival benefit as compared to cystectomy alone.2,23 Despite this, utilization is low, with population-level studies reporting less than 20% of patients with MIBC receive NAC.1,24 Due to the advanced age and high rate of comorbid conditions among patients with MIBC, it is postulated the low use of the NAC may be due to concerns for the risk of toxicities, coupled with an expected low benefit.5,6 As there is a preferential survival benefit from NAC seen among patients who achieve a pathological complete response, foregoing NAC may be appropriate in the 40–50% of patients who ultimately fail to receive benefit.3,25 However, with current evidence demonstrating the disproportionately low rates of NAC use, it is evident that there is likely a substantial proportion of patients in which curative treatment is underused. As such, there is a critical need for predictive biomarkers to allow for better precision in treatment decision-making.
Molecular profiling has been investigated for its use as a predictive biomarker for response to NAC. In particular, alterations in the DNA repair pathways have demonstrated potential in this regard, with loss of function alterations in ATM, RB1, FANCC, ERCC2, and BRCA1 being shown to predict pathological response and improve survival.26–29 Indeed, these genomic observations are consistent with the genotoxic mechanism of action of cisplatin, which is the back-bone of NAC regimens. Molecular subtyping based upon gene expression profiling has also generated enthusiasm for predicting response to NAC.30–33 Although early reports have demonstrated preferential response to NAC across the different molecular subtypes, they have not yet been validated as a predictive biomarker and would be challenging to integrate into routine clinical care for multiple reasons. Molecular subtyping may require additional tissue acquisition by cystoscopy with risk-associated biopsies, which are subject to sampling errors. Furthermore, the technology and expertise required to perform such analyses are usually restricted to research-focused academic laboratories.
There has been growing interest in the application of radiomics technology in oncology over the past decade. The early success of computer-based image analysis for clinical prediction has been established across a range of solid tumor malignancies. For instance, image analysis to predict histology and identify occult metastases, such as in regional lymph nodes, has shown early signs of promise.34,35 In addition, there is data supporting the use of this technology as a predictive biomarker in breast cancer, esophageal carcinoma, germ cell tumors, nasopharyngeal carcinoma, and rectal cancer, with discriminative accuracies greater than 70%.9,10,14,36–38 To date, computer-based image analysis has not been shown to outperform validated clinical prediction tools in settings whereby such tools exist; however, their combination with clinical predictors has been shown to improve discriminative accuracy.36 The utility of this technology in these early applications underscore the need for further study, particularly in settings where clinical predictors alone are ineffective.
Radiomics technology has the potential to serve as a much-needed predictive biomarker for NAC in MIBC. As there is no pathological assessment required, the need for additional invasive procedures is avoided, thereby minimizing risks to patients and also reducing healthcare resource use. Similarly, as patients routinely undergo radiological imaging prior to their treatment, no additional diagnostic imaging is required. As conducted through computer-based image analysis, the need for additional healthcare personnel to facilitate this predictive biomarker is greatly reduced. Consequently, the use of radiomics technology in this context promotes the most efficient use of our healthcare resources.
The use of radiomics technology as a predictive biomarker has been previously evaluated in MIBC.39,40 Cha et al evaluated the prediction accuracy of a radiomics signature on NAC treatment response as assessed by pre-and post-treatment CT images, with response classified as T0 tumor on post-treatment imaging. With over 80 patients in each of their training and validation cohorts, they demonstrated their radiomic signature to have an AUC of 0.69.40 In a later study, Cha et al evaluated a computerized-based decision-support system for treatment response that used a combination of both deep-learning neural networks and radiomic technology among 123 patients with response to NAC assessed by post-treatment CT imaging. In this analysis, the authors reported an AUC of 0.80, which outperformed clinician visual assessment alone (AUC 0.74).39 Although these two studies demonstrate the promise of radiomics to predict response to NAC, the use of post-treatment CT images as a surrogate for pathological response was an important limitation, as radiographic response does not always correlate with pathological outcomes.41 Unlike these prior studies in MIBC, the REVeAL study benchmarked response to treatment as per pathological assessment.
Our study is not without its limitations. Our radiomics signature was developed on a small pilot cohort of patients from a single institution, which is among the smaller sample sizes of radiomics analysis in the published literature.39–41 This small sample size limits the ability to make any conclusions on the utility of radiomics analysis as a predictive biomarker for response to NAC in MIBC. The retrospective design of this study led to variations in the images used in the analysis. For instance, not all images were obtained with intravenous contrast, with additional variations in the CT scanners and imaging protocols that introduced differences with respect to image resolution. Further, heterogeneity in the distensions of the bladder lesions could have introduced uncontrollable variation in the image analysis. In addition, the retrospective nature of this study also introduced the potential for time variation, as time of imaging prior to NAC was variable. Furthermore, the small sample size necessitated the use of a cross-validation approach, as opposed to independent validation on a separate cohort of patients. However, this approach is established in the machine-learning literature.42 Finally, the current study lacks a comparative analysis to address how the radiomics signature performs as compared with, or to complement, clinician prediction and/or molecular profiles.
Although the utility of our radiomics signature was limited, this represents a starting point for further evaluation of radiomics in MIBC. As the field of radiomics rapidly evolves, there is opportunity to introduce novel methods to improve the performance of the radiomics signature. This may include the use of more advanced supervised machine learning, such as with convolutional neural networks that learn predictive features in a more automated fashion and have been shown to improve diagnostic accuracy.43 In addition, methods such as unsupervised deep learning, automated volume delineation, or variational auto-encoders that allow for automated image analysis will allow for image analysis across a larger cohort of patients, leading to data that more confidently could inform treatment decision-making in MIBC.
Conclusions
Through the REVeAL study, we demonstrated the feasibility of developing a predictive radiomics signature to discriminate response to NAC for patients with MIBC that demonstrated modest accuracy. Future research using novel methods for computer-based image analysis on a larger cohort of patients is planned to improve the utility of radiomics as a predictive biomarker in MIBC.
Footnotes
Competing interests: Dr. Sim has received research funding from Abbvie and Bristol-Myers Squibb. Dr. Metser has served in a consulting or advisory role for Point Biopharm Inc. Dr. Hansen has received honoraria from AstraZeneca/MedImmune, Bristol-Myers Squibb, GlaxoSmithKline, Merck, Novartis, and Pfizer; has served in a consulting or advisory role for Boehringer Ingelheim, Boston Biomedical, Bristol-Myers Squibb, GlaxoSmithKline, Merck, Novartis, and Roche/Genetech; and has received research funding from Bristol-Myers Squibb, Boehringer Ingelheim, GlaxoSmithKline, Janssen, Karyopharm Therapeutics, Merck, Novartis, and Roche/Genetech. The remaining authors report no personal or financial conflicts related to this work.
This paper has been peer-reviewed.
References
- 1.Booth CM, Siemens DR, Li G, et al. Perioperative chemotherapy for muscle-invasive bladder cancer: A population-based outcomes study. Cancer. 2014;120:1630–8. doi: 10.1002/cncr.28510. [DOI] [PubMed] [Google Scholar]
- 2.Grossman HB, Natale RB, Tangen CM, et al. Neoadjuvant chemotherapy plus cystectomy compared with cystectomy alone for locally advanced bladder cancer. N Engl J Med. 2003;349:859–66. doi: 10.1056/NEJMoa022148. [DOI] [PubMed] [Google Scholar]
- 3.Lee FC, Harris W, Cheng HH, et al. Pathologic response rates of gemcitabine/cisplatin versus methotrexate/vinblastine/adriamycin/cisplatin neoadjuvant chemotherapy for muscle-invasive urothelial bladder cancer. Adv Urol. 2013;2013:317190. doi: 10.1155/2013/317190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Winquist E, Kirchner TS, Segal R, et al. Neoadjuvant chemotherapy for transitional cell carcinoma of the bladder: A systematic review and meta-analysis. J Urol. 2004;171:561–9. doi: 10.1097/01.ju.0000090967.08622.33. [DOI] [PubMed] [Google Scholar]
- 5.Bellmunt J, Mottet N, De Santis M. Urothelial carcinoma management in elderly or unfit patients. EJC Suppl. 2016;14:1–20. doi: 10.1016/j.ejcsup.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galsky MD, Hahn NM, Rosenberg J, et al. Treatment of patients with metastatic urothelial cancer “unfit” for cisplatin-based chemotherapy. J Clin Oncol. 2011;29:2432–8. doi: 10.1200/JCO.2011.34.8433. [DOI] [PubMed] [Google Scholar]
- 7.Lambin P, Rios-Velazquez E, Leijenaar R, et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48:441–6. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumar V, Gu Y, Basu S, et al. Radiomics: The process and the challenges. Magn Reson Imaging. 2012;30:1234–48. doi: 10.1016/j.mri.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tixier F, Le Rest CC, Hatt M, et al. Intratumor heterogeneity characterized by textural features on baseline 18F-FDG PET images predicts response to concomitant radiochemotherapy in esophageal cancer. J Nucl Med. 2011;52:369–78. doi: 10.2967/jnumed.110.082404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang H, Tan S, Chen W, et al. Modelling pathological response of esophageal cancer to chemoradiation therapy using spatial-temporal 18F-FDG PET features, clinical parameters, and demographics. Int J Radiat Oncol Biol Phys. 2014;88:195–203. doi: 10.1016/j.ijrobp.2013.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yip SS, Coroller TP, Sanford NN, et al. Relationship between the temporal changes in positron emission tomography imaging-based textural features and pathological response and survival in esophageal cancer patients. Front Oncol. 2016;6:72. doi: 10.3389/fonc.2016.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Rossum PS, Fried DV, Zhang L, et al. The incremental value of subjective and quantitative assessment of 18F-FDG PET for the prediction of pathologic complete response to preoperative chemoradiotherapy in esophageal cancer. J Nucl Med. 2016;57:691–700. doi: 10.2967/jnumed.115.163766. [DOI] [PubMed] [Google Scholar]
- 13.Cook GJ, Yip C, Siddique M, et al. Are pre-treatment 18F-FDG PET tumor textural features in non-small cell lung cancer associated with response and survival after chemoradiotherapy? J Nucl Med. 2013;54:19–26. doi: 10.2967/jnumed.112.107375. [DOI] [PubMed] [Google Scholar]
- 14.Metser U. Multiparameteric PET-MR assessment of response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer: PET, MR, PET-MR, and tumor texture analysis: A pilot study. Adv Mol Imag. 2015;5:49–60. doi: 10.4236/ami.2015.53005. [DOI] [Google Scholar]
- 15.Haralick R, Shanmugam K, Dinstein I. Textural features for image classification. IEEE Transact Syst Man Cybernetics. 1973;3:610–21. doi: 10.1109/TSMC.1973.4309314. [DOI] [Google Scholar]
- 16.Lou HH, Leu JG, Luo RC. The analysis of natural textures using run length features. IEEE Transactions Indust Electronics. 1988;35:323–8. doi: 10.1109/41.192665. [DOI] [Google Scholar]
- 17.Thibault G, Fertil B, Navarro C, et al. Texture indexes and gray level size zone matrix: Application to cell nuclei classification. Patt Recogn Info Process. 2009:140–5. https://www.thibault.biz/Doc/Publications/TextureIndexesAndGrayLevelSizeZoneMatrixApplicationToCellNucleiClassification_PRIP2009.pdf . [Google Scholar]
- 18.Amadasun M, King R. Textural features corresponding to textural properties. IEEE Transact Syst Man Cybernetics. 1989;19:1264–74. doi: 10.1109/21.44046. [DOI] [Google Scholar]
- 19.Chang CC, Lin CJ. A library for support vector machines. ACM Trans Intell Syst Technol. 2011;2:1–27. doi: 10.1145/1961189.1961199. [DOI] [Google Scholar]
- 20.Boser BE, Guyon IM, Vapnik VN. A training algorithm for optimal margin classifiers. Proceedings of the fifth annual workshop on computational learning theory. COLT ’92. 1992:144–52. doi: 10.1145/130385.130401. [DOI] [Google Scholar]
- 21.Cawley GC, Talbot NLC. On over-fitting in model selection and subsequent selection bias in performance evaluation. J Mach Learn Res. 2010;11:2079–2107. https://www.jmlr.org/papers/volume11/cawley10a/cawley10a.pdf . [Google Scholar]
- 22.Parmar C, Grossmann P, Bussink J, et al. Machine learning methods for quantitative radiomic biomarkers. Sci Rep. 2015;5:13087. doi: 10.1038/srep13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kulkarni GS, Black PC, Sridhar SS, et al. Canadian Urological Association guideline: Muscle-invasive bladder cancer. Can Urol Assoc J. 2019;13:230–8. doi: 10.5489/cuaj.5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zaid HB, Patel SG, Stimson CJ, et al. Trends in the utilization of neoadjuvant chemotherapy in muscle-invasive bladder cancer: Results from the National Cancer Database. Urology. 2014;83:75–80. doi: 10.1016/j.urology.2013.07.072. [DOI] [PubMed] [Google Scholar]
- 25.Lane G, Risk M, Fan Y, et al. Persistent muscle-invasive bladder cancer after neoadjuvant chemotherapy: An analysis of Surveillance, Epidemiology and End Results-Medicare data. BJU Int. 2019;123:818–25. doi: 10.1111/bju.14529. [DOI] [PubMed] [Google Scholar]
- 26.Plimack ER, Dunbrack RL, Brennan TA, et al. Defects in DNA repair genes predict response to neoadjuvant cisplatin-based chemotherapy in muscle-invasive bladder cancer. Eur Urol. 2015;68:959–67. doi: 10.1016/j.eururo.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Allen EM, Mouw KW, Kim P, et al. Somatic ERCC2 mutations correlate with cisplatin sensitivity in muscle-invasive urothelial carcinoma. Cancer Discov. 2014;4:1140–53. doi: 10.1158/2159-8290.CD-14-0623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu D, Plimack ER, Hoffman-Censits J, et al. Clinical validation of chemotherapy response biomarker ERCC2 in muscle-invasive urothelial bladder carcinoma. JAMA Oncol. 2016;2:1094–6. doi: 10.1001/jamaoncol.2016.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Font A, Taron M, Gago JL, et al. BRCA1 mRNA expression and outcome to neoadjuvant cisplatin-based chemotherapy in bladder cancer. Ann Oncol. 2011;22:139–44. doi: 10.1093/annonc/mdq333. [DOI] [PubMed] [Google Scholar]
- 30.Seiler R, Ashab HA, Erho N, et al. Impact of molecular subtypes in muscle-invasive bladder cancer on predicting response and survival after neoadjuvant chemotherapy. Eur Urol. 2017;72:544–54. doi: 10.1016/j.eururo.2017.03.030. [DOI] [PubMed] [Google Scholar]
- 31.Kardos J, Chai S, Mose LE, et al. Claudin-low bladder tumors are immune infiltrated and actively immune-suppressed. JCI Insight. 2016;1:e85902. doi: 10.1172/jci.insight.85902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi W, Porten S, Kim S, et al. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell. 2014;25:152–65. doi: 10.1016/j.ccr.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cancer Genome Atlas Research N. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507:315–22. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coroller TP, Agrawal V, Huynh E, et al. Radiomic-based pathological response prediction from primary tumors and lymph nodes in NSCLC. J Thorac Oncol. 2017;12:467–76. doi: 10.1016/j.jtho.2016.11.2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu W, Parmar C, Grossmann P, et al. Exploratory study to identify radiomics classifiers for lung cancer histology. Front Oncol. 2016;6:71. doi: 10.3389/fonc.2016.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lewin J, Dufort P, Halankar J, et al. Applying radiomics to predict pathology of post-chemotherapy retroperitoneal nodal masses in germ cell tumors. JCO Clin Cancer Inform. 2018;2:1–12. doi: 10.1200/CCI.18.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu J, Mao Y, Li Z, et al. Use of texture analysis based on contrast-enhanced MRI to predict treatment response to chemoradiotherapy in nasopharyngeal carcinoma. J Magn Reson Imaging. 2016;44:445–55. doi: 10.1002/jmri.25156. [DOI] [PubMed] [Google Scholar]
- 38.Ahmed A, Gibbs P, Pickles M, et al. Texture analysis in assessment and prediction of chemotherapy response in breast cancer. J Magn Reson Imaging. 2013;38:89–101. doi: 10.1002/jmri.23971. [DOI] [PubMed] [Google Scholar]
- 39.Cha KH, Hadjiiski LM, Cohan RH, et al. Diagnostic accuracy of CT for prediction of bladder cancer treatment response with and without computerized decision support. Acad Radiol. 2019;26:1137–45. doi: 10.1016/j.acra.2018.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cha KH, Hadjiiski L, Chan HP, et al. Bladder cancer treatment response assessment in CT using radiomics with deep-learning. Sci Rep. 2017;7:8738. doi: 10.1038/s41598-017-09315-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fukui T, Matsui Y, Umeoka S, et al. Predictive value of radiological response rate for pathological response to neoadjuvant chemotherapy and post-cystectomy survival of bladder urothelial cancer. Jpn J Clin Oncol. 2016;46:560–567. doi: 10.1093/jjco/hyw025. [DOI] [PubMed] [Google Scholar]
- 42.Petrick N, Sahiner B, Armato SG, 3rd, et al. Evaluation of computer-aided detection and diagnosis systems. Med Phys. 2013;40:087001. doi: 10.1118/1.4816310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Truhn D, Schrading S, Haarburger C, et al. Radiomic versus convolutional neural networks analysis for classification of contrast-enhancing lesions at multiparametric breast MRI. Radiology. 2019;290:290–7. doi: 10.1148/radiol.2018181352. [DOI] [PubMed] [Google Scholar]