Abstract
The sequences of the blaTEM genes encoding TEM-20, TEM-21, TEM-22, and TEM-29 extended-spectrum β-lactamases were determined. Analysis of the deduced amino acid sequences indicated that TEM-20 and TEM-29 were derived from TEM-1 and that TEM-21 and TEM-22 were derived from TEM-2. The substitutions involved were Ser-238 and Thr-182 for TEM-20; His-164 for TEM-29; Lys-104, Arg-153, and Ser-238 for TEM-21; and Lys-104, Gly-237, and Ser-238 for TEM-22. The promoter region of the blaTEM-22 gene was identical to that of blaTEM-3. High-level production of TEM-20 could result from a 135-bp deletion which combined the −35 region of the Pa promoter with the −10 region of the P3 promoter and a G→T transition in the latter motif.
Extended-spectrum β-lactamases TEM-20 and TEM-21 were detected in two nosocomial Klebsiella pneumoniae strains isolated in Tunis, Tunisia, in 1986 and 1988, respectively (5). A high level of resistance to cefotaxime, ceftriaxone, ceftazidime, and aztreonam was observed for both strains. TEM-20 and TEM-21 showed hydrolysis rates for cefotaxime and ceftriaxone similar to those of TEM-3 (22) and TEM-4 (18). In contrast to TEM-20, TEM-21 showed a significant rate of ceftazidime hydrolysis that was similar to those of TEM-3 (22) and TEM-4 (18). Isoelectric point determination (5.4 for TEM-20 and 6.4 for TEM-21) and hybridization studies led to the designation of the enzymes (5). Point mutations in the corresponding structural genes detected by PCR-restriction fragment length polymorphism (RFLP) (3) indicated the following substitutions in the deduced amino acid sequences. TEM-20 had a Glu at position 104 as in TEM-1 and the same amino acid substitution, Ser-238 for Gly, as in TEM-19 (6, 12, 15), but the corresponding gene differed from the blaTEM-19 gene by a G→A transversion at position 925. TEM-21 was derived from TEM-2 and had the same substitutions enhancing the substrate spectrum as TEM-3 and TEM-4 but differed from TEM-3 by an additional substitution (His-153 for Arg), resulting in a pI difference (6.4 versus 6.3). TEM-22 (pI 6.3) from a K. pneumoniae clinical isolate was previously characterized by biochemical analysis only (4). Donor, transconjugant, and transformant strains producing this enzyme have a higher level of resistance to aztreonam than to ceftazidime and cefotaxime (4). This very unusual resistance phenotype led to the designation TEM-22. TEM-29 (pI 5.4) from three Escherichia coli clinical strains isolated in 1987 was reported in 1995 (3).
We report the sequence of the structural genes for TEM-20, TEM-21, TEM-22, and TEM-29 and of the promoters of the genes encoding TEM-20, and TEM-22. The strains used were E. coli J53-2 (pUD30, TEM-20) (5), E. coli J53-2 (pUD31, TEM-21) (5), E. coli HB101 (TEM-22) (4) and the clinical isolate E. coli DEL (TEM-29) (3). PCR amplification of the blaTEM genes was performed using primers OT3 and OT4 (3) corresponding to positions 209 to 228 and 1047 to 1066, respectively, of the blaTEM-1 gene (26), which allow the amplification of the entire coding region. Both strands of the PCR products were sequenced (21) using the PCR primers, fluorescent dye-labeled dideoxynucleotides, thermal cycling with Taq polymerase, and an ABI 373A DNA sequencer (Applied Biosystems, Foster City, Calif.). In case of differences in the sequences obtained for the two strands, a PCR product obtained independently was subsequently sequenced. PCR amplification and DNA sequencing of the promoter and coding regions of the blaTEM-20 and blaTEM-22 genes were performed as described (16).
The BLASTN (1) program at the National Center for Biotechnology Information was used for database searches, and the Clustal V program (11) was used to align multiple protein sequences. The EMBL accession numbers for the nucleotide sequences are Y17581 for blaTEM-20, Y17582 for blaTEM-21, Y17583 for blaTEM-22, and Y17584 for blaTEM-29. The changes in the genes are shown in Table 1, and those in the deduced amino acid sequences are shown in Table 2.
TABLE 1.
Nucleotide substitutions among the blaTEM genes from Tn1 (blaTEM-2), Tn2 (blaTEM-1B), and Tn3 (blaTEM-1A) and those encoding TEM-3, TEM-20, TEM-21, TEM-22, and TEM-29
| Positiona | Gene
|
|||||||
|---|---|---|---|---|---|---|---|---|
| blaTEM-1A | blaTEM-1B | blaTEM-2 | blaTEM-3 | blaTEM-20 | blaTEM-21 | blaTEM-22 | blaTEM-29 | |
| Promoter region | ||||||||
| 32 | C | C | T | T | Deleted | NDb | T | ND |
| 147 | T | T | T | A | Deleted | ND | A | ND |
| 162 | G | G | G | G | T | ND | G | ND |
| 175 | A | G | A | A | A | ND | A | ND |
| Coding region | ||||||||
| 226c | C | T | C | C | C | C | C | C |
| 317 | C | C | A | A | C | A | A | C |
| 346c | A | A | G | G | G | G | G | A |
| 436c | C | T | T | T | T | T | T | T |
| 512 | G | G | G | A | G | A | A | G |
| 604c | G | T | G | G | G | G | G | T |
| 660 | A | A | A | A | A | G | A | A |
| 682c | T | T | C | C | C | C | C | T |
| 693 | G | G | G | G | G | G | G | A |
| 747 | T | T | T | T | C | T | T | T |
| 912 | C | C | C | C | C | C | G | C |
| 914 | G | G | G | A | A | A | A | G |
| 925c | G | G | A | A | A | A | A | G |
Numbering according to Sutcliffe (26).
ND, not determined.
Position at which only silent mutations occur.
TABLE 2.
Amino acid substitutions in TEM-type β-lactamases
| β-lactamase | Amino acid located at positiona:
|
||||||
|---|---|---|---|---|---|---|---|
| 39 | 104 | 153 | 164 | 182 | 237 | 238 | |
| TEM-1 | Gln | Glu | His | Arg | Met | Ala | Gly |
| TEM-2 | Lys | ||||||
| TEM-3 | Lys | Lys | Ser | ||||
| TEM-11 | Lys | His | |||||
| TEM-19 | Ser | ||||||
| TEM-20 | Thr | Ser | |||||
| TEM-21 | Lys | Lys | Arg | Ser | |||
| TEM-22 | Lys | Lys | Gly | Ser | |||
| TEM-29 | His | ||||||
Numbering according to Ambler et al. (2). Amino acids indicated in bold extend the spectrum of β-lactamase activity.
The sequence of blaTEM-20 revealed that this gene was derived from the blaTEM-2-like group (silent mutations of blaTEM-2 and C at position 317) (7) whereas we proposed, following PCR-RFLP, that TEM-20 was derived from TEM-1 (3). Two mutations, T747→C and G914→A, resulting in amino acid substitutions Met-182→Thr and Gly-238→Ser, were found. Substitution Ser for Gly at position 238 has been observed in TEM-19 (12) and TEM-25 (8) and extends the substrate profile of the enzyme to cefotaxime (14), as for TEM-25 (19). Similar levels of resistance to expanded-spectrum cephalosporins were observed in the clinical strains producing these enzymes. The second substitution (Thr for Met at position 182) in TEM-20, which was not detected by PCR-RFLP (3), distinguished the enzyme from TEM-19 and TEM-25. The same substitution occurs in TEM-32 (IRT-3), TEM-43 (6), and TEM-52 (20). Alone, this substitution does not seem to extend the substrate profile; however, when combined with Lys-104 and Ser-238, Thr-182 confers moxalactam resistance, as observed for TEM-52 (20).
Analysis of the sequence of blaTEM-21 showed that this gene derived from blaTEM-2 (A at position 317). The deduced TEM-21 protein had the same substitutions responsible for resistance to broad-spectrum cephalosporins and aztreonam as TEM-3 (24) and TEM-4 (derived from TEM-1) (25). The enzyme had a His-153→Arg substitution recently found in a TEM-21 produced by a clinical isolate of Morganella morganii (27). The consequence of this substitution on the catalytic activity of the β-lactamase is unknown and possibly insignificant since residue 153 is far from the β-lactam binding site (13).
blaTEM-22 differs from blaTEM-3 by a C→G mutation at position 912 leading to a Gly for Ala substitution at position 237, probably involved in the high-level aztreonam resistance of the host. Replacement of Ala-237 by threonine, as in TEM-5 and TEM-24 (6), increases the affinity of the enzyme for extended-spectrum β-lactams (14). Replacement by saturated mutagenesis of Ala-237 by Asp or Thr but not by Gly increases the catalytic activity of TEM-1 against cephems (10). The effect of these substitutions on the catalytic efficiency against aztreonam has not been studied.
The sequence of blaTEM-29 showed that this gene was derived from blaTEM-1B and differed by a silent mutation, T226→C, as in blaTEM-1A. An additional mutation, G693→A (also detected by PCR-RFLP) (3), leads to the amino acid substitution His for Arg at position 164, which increases ceftazidime resistance (14). However, Ser for Arg is the most common substitution at this position in TEM variants from clinical isolates (6). The Gly-for-Arg substitution confers ceftazidime resistance in TEM-1 mutants obtained by random mutagenesis (17). In clinical isolates, this substitution is rarely observed alone, except for TEM-11 (CAZ-lo) derived from TEM-2 (28). The low-level resistance to ceftazidime resulting from this substitution may account for the difficulty in detecting such variants.
Our results confirm that sequencing is the only truly definitive method for DNA analysis, even if PCR-RFLP is a useful approach for epidemiological studies.
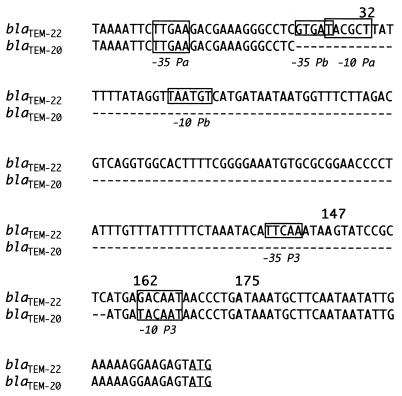
Analysis of the promoter regions of blaTEM-20 and blaTEM-22 indicated that blaTEM-22 had the same T-for-C change at position 32 as that observed in Tn1, blaTEM-3, blaTEM-4, and blaTEM-5 (Fig. 1) (24, 25). This change converts the weak P3 promoter of blaTEM-1 (Tn3) into the two overlapping Pa + Pb promoters and results in a large increase in β-lactamase production (9). There was a 135-bp deletion in the promoter region of blaTEM-20. This deletion was associated with a point mutation, T for G in the −10 region of P3, leading to greater similarity to the consensus promoter sequence, as previously observed for blaTEM-1 in Shigella flexneri (23). Combination of the deletion and the mutation, leading to the association of the −35 region of Pa with the more efficient −10 region of P3, may account for high-level resistance to oxyiminocephalosporins of the strain producing TEM-20 due to high-level enzyme production (5).
FIG. 1.
Promoter sequence of the blaTEM-20 and blaTEM-22 genes. The differences in the promoter region of Tn2 are shown in bold and numbered according to Sutcliffe (26). The −35 and −10 regions of Pa, Pb, and P3 are boxed. The start codon is underlined. Deleted nucleotides are indicated by dashes.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Ambler R P, Coulson F W, Frère J M, Ghuysen J M, Joris B, Forsman M, Levesque R C, Tiraby G, Waley S G. A standard numbering scheme for the class A β-lactamases. Biochem J. 1991;276:269–272. doi: 10.1042/bj2760269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arlet G, Brami G, Decré D, Flippo A, Gaillot O, Lagrange P H, Philippon A. Molecular characterization by PCR-restriction length polymorphism of TEM β-lactamases. FEMS Microbiol Lett. 1995;134:203–208. doi: 10.1111/j.1574-6968.1995.tb07938.x. [DOI] [PubMed] [Google Scholar]
- 4.Arlet G, Rouveau M, Fournier G, Lagrange P H, Philippon A. Novel, plasmid-encoded, TEM-derived extended-spectrum β-lactamase in Klebsiella pneumoniae conferring higher resistance to aztreonam than to extended-spectrum cephalosporins. Antimicrob Agents Chemother. 1993;37:2020–2023. doi: 10.1128/aac.37.9.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ben Redjeb S, Fournier G, Mabilat C, Ben Hassen A, Philippon A. Two novel transferable extended-spectrum β-lactamases from Klebsiella pneumoniae in Tunisia. FEMS Microbiol Lett. 1990;67:33–38. doi: 10.1016/0378-1097(90)90163-k. [DOI] [PubMed] [Google Scholar]
- 6.Bush K, Jacoby G. Nomenclature of TEM β-lactamases. J Antimicrob Chemother. 1997;39:1–3. doi: 10.1093/jac/39.1.1. [DOI] [PubMed] [Google Scholar]
- 7.Canica M M M, Lu C Y, Krishnamoorthy R, Paul G C. Molecular diversity and evolution of blaTEM genes encoding β-lactamases resistant to clavulanic acid in clinical E. coli. J Mol Evol. 1997;44:57–65. doi: 10.1007/pl00006121. [DOI] [PubMed] [Google Scholar]
- 8.Chanal C, Sirot D, Malaure H, Poupart M C, Sirot J. Sequences of CAZ-3 and CTX-2 extended-spectrum β-lactamases genes. Antimicrob Agents Chemother. 1994;38:2452–2453. doi: 10.1128/aac.38.10.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen S T, Clowes R C. Two improved promoter sequences for the β-lactamase expression arising from a single base-pair substitution. Nucleic Acids Res. 1984;7:3219–3234. doi: 10.1093/nar/12.7.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Healey W J, Labgold M R, Richards J H. Substrate specificities in class A β-lactamases: preference for penams vs. cephems. The role of residue 237. Proteins Struct Funct Genet. 1989;6:275–283. doi: 10.1002/prot.340060310. [DOI] [PubMed] [Google Scholar]
- 11.Higgins D G, Bleasby A J, Fuchs R. Clustal V: improved software for multiple alignments. Comput Appl Biosci. 1992;8:189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- 12.Jacoby G A, Han P, Alvarez M, Tenover F. Program and abstracts of the 35th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1995. Survey of extended-spectrum β-lactamase (ESBL) production in U.S. clinical isolates, abstr. C40; p. 46. [Google Scholar]
- 13.Jelsch C, Mourey L, Masson J M, Samama J P. Crystal structure of Escherichia coli TEM-1 β-lactamase at 1.8 Å resolution. Proteins Struct Funct Genet. 1993;16:364–383. doi: 10.1002/prot.340160406. [DOI] [PubMed] [Google Scholar]
- 14.Knox J R. Extended-spectrum and inhibitor-resistant TEM-type β-lactamases: mutations, specificity, and three-dimensional structure. Antimicrob Agents Chemother. 1995;39:2593–2601. doi: 10.1128/aac.39.12.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mabilat C, Courvalin P. Development of “oligotyping” for characterization and molecular epidemiology of TEM β-lactamases in members of the family Enterobacteriaceae. Antimicrob Agents Chemother. 1990;34:2210–2216. doi: 10.1128/aac.34.11.2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mabilat C, Goussard S. PCR detection and identification of genes for extended-spectrum β-lactamases. In: Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology: principles and applications. Washington, D.C: American Society for Microbiology; 1993. pp. 553–559. [Google Scholar]
- 17.Palzkill T, Le Q-Q, Venkatachalam K V, LaRocco M, Ocera H. Evolution of antibiotic resistance: several different amino acid substitutions in an active site loop alter the substrate profile of β-lactamase. Mol Microbiol. 1994;12:217–229. doi: 10.1111/j.1365-2958.1994.tb01011.x. [DOI] [PubMed] [Google Scholar]
- 18.Paul G C, Gerbaud G, Buré A, Philippon A, Pangon B, Courvalin P. TEM-4, a new plasmid-mediated β-lactamase that hydrolyzes broad-spectrum cephalosporins in a clinical isolate of Escherichia coli. Antimicrob Agents Chemother. 1989;33:1958–1963. doi: 10.1128/aac.33.11.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poupart M C, Chanal C, Sirot D, Labia R, Sirot J. Identification of CTX-2, a novel cefotaximase from a Salmonella mbandaka isolate. Antimicrob Agents Chemother. 1991;35:1498–1500. doi: 10.1128/aac.35.7.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poyart C, Mugnier P, Quesne G, Berche P, Trieu-Cuot P. A novel extended-spectrum TEM-type β-lactamase (TEM-52) associated with decreased susceptibility to moxalactam in Klebsiella pneumoniae. Antimicrob Agents Chemother. 1998;42:108–113. doi: 10.1128/aac.42.1.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sirot D, Sirot J, Labia R, Morand A, Courvalin P, Darfeuille-Michaud A, Perroux R, Cluzel R. Transferable resistance to third-generation cephalosporins in clinical isolates of Klebsiella pneumoniae. Identification of CTX-1, a novel beta-lactamase. J Antimicrob Chemother. 1987;20:323–334. doi: 10.1093/jac/20.3.323. [DOI] [PubMed] [Google Scholar]
- 23.Siu L K, Ho P L, Yuen K Y, Wong S S, Chau P Y. Transferable hyperproduction of TEM-1 β-lactamase in Shigella flexneri due to a point mutation in the Pribnow box. Antimicrob Agents Chemother. 1997;41:468–470. doi: 10.1128/aac.41.2.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sougakoff W, Goussard S, Courvalin P. The TEM-3 β-lactamase, which hydrolyzes broad-spectrum cephalosporins, is derived from the TEM-2 penicillinase by two amino acid substitutions. FEMS Microbiol Lett. 1988;56:343–348. [Google Scholar]
- 25.Sougakoff W, Petit A, Goussard S, Sirot D, Buré A, Courvalin P. Characterization of the plasmid genes blaT-4 and blaT-5 which encode the broad-spectrum β-lactamases TEM-4 and TEM-5 in Enterobacteriaceae. Gene. 1989;78:339–348. doi: 10.1016/0378-1119(89)90236-9. [DOI] [PubMed] [Google Scholar]
- 26.Sutcliffe J G. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc Natl Acad Sci USA. 1978;75:3737–3741. doi: 10.1073/pnas.75.8.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tessier F, Arpin C, Allery A, Quentin C. Molecular characterization of a TEM-21 β-lactamase in a clinical isolate of Morganella morganii. Antimicrob Agents Chemother. 1998;42:2125–2127. doi: 10.1128/aac.42.8.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vuye A, Verschraegen G, Claeys G. Plasmid-mediated β-lactamases in clinical isolates of Klebsiella pneumoniae and Escherichia coli resistant to ceftazidime. Antimicrob Agents Chemother. 1989;33:757–761. doi: 10.1128/aac.33.5.757. [DOI] [PMC free article] [PubMed] [Google Scholar]