Abstract
Droplet digital PCR is a particularly valuable tool for ratiometric assays because it provides simultaneous absolute quantification of two target sequences in a single assay. This manuscript addresses a challenge in establishing a new ratiometric droplet digital PCR assay for use in sputum, the rRNA synthesis ratio. In principle, the methods established to evaluate precision and determine the limit of quantification for a single measurand cannot be applied to a ratiometric assay. The precision of a ratio depends on precision in both the numerator and denominator. Here, we evaluated the MOVER approximated coefficient of variation as indicator of assay precision that does not require technical replicates. We estimated the MOVER approximated coefficient of variation in dilution series and routine assays and evaluated its agreement with the traditional coefficient of variation. We found that the MOVER approximated coefficient of variation was able to recapitulate the traditional coefficient of variation without the requirement for replicate assays. We also demonstrated that the MOVER approximated coefficient of variation threshold can be used to define the limit of quantification of the rRNA synthesis Ratio. In conclusion, the MOVER approximated coefficient of variation may be useful not only for the rRNA synthesis ratio but for other assays that measure ratios via droplet digital PCR.
Keywords: Precision Measurement, Quantification Limit, Droplet Digital PCR Assay, Ratio Measurement
1. Introduction
Quantification of the ratio between two nucleic acid sequences is the basis of several clinical or research assays [1-5]. An advantage of ratiometric assays is that ratios are inherently “self-normalizing,” thereby controlling for between-sample variability in inconsistent samples like sputum.
Droplet digital polymerase chain reaction (dPCR) is a particularly valuable tool for ratiometric assays because it provides simultaneous absolute quantification of two target sequences in a single reaction. Unlike qPCR, dPCR enables direct calculation of the ratio between two targets without dependence on external standard curves. Additionally, dPCR is more sensitive than qPCR and less susceptible to PCR inhibition in complex matrices such as human or environmental samples [5-9]. The dPCR technique is currently used in an FDA-approved clinical assay that quantifies the ratio of BCR-ABL to ABL transcripts in human blood to monitor treatment response and relapse risk in patients with chronic myeloid leukemia (CML) [10].
This manuscript addresses the challenge of quantifying precision and establishing thresholds for data quality control in a ratiometric dPCR assay applied to a heterogenous sputum matrix. We have developed an assay called the RS ratio for use as a pharmacodynamic (PD) marker in tuberculosis (TB). By quantifying the abundance of M. tuberculosis (Mtb) precursor rRNA (pre-rRNA) relative to the burden of 23S rRNA, the RS ratio indicates the degree of ongoing bacterial ribosomal RNA (rRNA) synthesis in the sputum of TB patients [11]. The RS ratio represents a fundamentally new measure of treatment efficacy. Unlike conventional PD markers which enumerate the burden of Mtb capable of growth in culture [12], the RS ratio evaluates the effect of drugs and regimens on a basic cellular process of the pathogen. The RS ratio appears to indicate the treatment-shortening potency of drugs and regimens [11] and therefore promises to accelerate the development of new, shorter and more effective TB treatments [12].
In principle, the methods established to determine the limit of quantification (LOQ) for a single measurand (such as a single target nucleic acid sequence) are not applicable to ratiometric assays [13-16]. For a single measurand, absolute values of the measurand typically correlate with precision of measurements. The lower the value of the measurand, the greater the variability in measurement. A threshold value for the measurand can be established, below which results are insufficiently precise and classified as below the LOQ. By contrast, the value of a ratio alone does not indicate precision. The precision of a ratio depends on precision in both the numerator and denominator. Conceptually, it is possible for a low ratio to be highly precise and a high ratio to be imprecise.
For a ratiometric assay in a more homogenous sample type like blood, this concept may not be a practical concern. For example, in the blood CML assay, the abundance of ABL transcript per microgram of RNA is relatively consistent across samples, irrespective of disease state [17,18]. Because the denominator is functionally static, the value of the blood BCR-ABL/ABL ratio is directly correlated with precision, the same as it would for a single measurand. By contrast, in TB the denominator is highly inconsistent. Even prior to treatment, the sputum burden of culturable Mtb in patients with active disease may range from undetectable to 109 organisms per milliliter [12,19]. Since the denominator of the RS ratio (23S rRNA) is a measure of Mtb burden, variability in bacterial burden influences precision of the RS ratio. As a result, the value of the RS ratio alone is not directly correlated with precision as it is with the BCR-ABL/ABL assay.
Precision in molecular assays is usually expressed as the coefficient of variation (CV) of a series of measurements [16]. Although the level of precision needed can vary depending on the type of assay and intended use, a CV less than 30% is generally considered an acceptable level of precision [20]. One method for quantifying the precision of the RS ratio would be to routinely perform multiple technical replicates for every assay and calculate the conventional CV which this manuscript will refer to as the “True” CV (CVTrue). However, dPCR has been shown to be highly repeatable [21-24] and technical replicates are not routinely recommended [25]. Technical replicates add expense and labor and may not be feasible in low-abundance samples. The RS ratio therefore demanded an alternative indicator of assay precision that does not require technical replicates.
In this paper, we establish and evaluate a new precision metric for ratiometric dPCR assays called the method of variance estimates recovery (MOVER) approximated CV (CVMA). The CVMA has been chosen to describe precision in a way similar to the traditional CVTrue without the requirement for replicate assays. The CVMA threshold provides a useful new measure of uncertainty around a ratio estimate and can be used to define the LOQ of the RS ratio. The CVMA may be useful not only for the RS ratio but for other assays that measure ratios via dPCR.
2. Materials and methods
2.1. Conceptual approach
Two distinct approaches were used to evaluate CVMA as surrogate for CVTrue. First, the relationship between CVMA and CVTrue was interrogated in a 10,000-fold dilution series using two sputum RNA samples. Second, the same relationship was evaluated in sputum RNA samples that underwent RS ratio assays in our routine workflow. The RS ratio assay was previously described [11] and has been summarized in Supplemental Material.
2.2. Description of sample set
This analysis used RNA-preserved human sputa collected longitudinally in Study 31, an international, multicenter, randomized, open-label, phase 3, noninferiority trial conducted at sites of the Centers for Disease Control and Prevention Tuberculosis Trials Consortium and the National Institutes of Health AIDS Clinical Trials Group [26]. Participants provided written informed consent for the use of their sputa for research. Details of supervising institutional review boards is provided in Supplemental Material.
2.3. Description of dilution series
Serial dilutions were performed using two Mtb-infected sputum RNA samples, one known to have a high RS ratio, and another known to have a low RS ratio. The low and high RS ratio samples were serially diluted 2 to 200-fold and 4 to 10,000-fold, respectively, generating up nine dilution levels. The RS ratio was assayed in the dilutions in sextuplicate.
2.4. Evaluation in routine workflow
In our routine workflow, we first conducted a screening qPCR assay to estimate the burden of Mtb 23S rRNA then quantified the RS ratio in a singlicate dPCR assay. RNA extraction and quantification of 23S rRNA and RS ratio are described in Supplemental Material. dPCR was conducted with replicates if the screening qPCR showed the burden of rRNA was very low (i.e., < 200 copies) or if the CVMA in the singlicate assay was >30%. These replicate assays are the basis for our comparison of CVMA and CVTrue in routine use. The usefulness of the CVMA to define the LOQ of the RS ratio was evaluated in routine samples.
2.5. Description of dPCR statistics for duplex assays
dPCR partitions each sample into ~20,000 droplets that undergo endpoint PCR and are individually classified as having presence or absence of the target sequence. Using these ~20,000 binary events, the QuantaSoft software package (Bio-Rad, AP v1.0) applies Poisson statistics to estimate the absolute copy number of each target in the sample with a 95% confidence interval (CI). QuantaSoft calculates the ratio between the two targets and uses the method of variance estimates recovery (MOVER) Fieller's theorem to calculate a 95% CI around the ratio (Personal Communication, Bio-Rad) [27-29]. The 95% CI alone is of limited usefulness for evaluating precision because it is not “scaled” relative to the ratio. To avoid leading zeros, the ratio provided by QuantaSoft is multiplied by 10,000, resulting in the RS ratio.
2.6. Calculation of CVTrue and CVMA
CVTrue was calculated in replicate assays using the mean and standard deviation of the ratios. Calculation of CVMA uses the ratio and 95% CI as presented in situ in QuantaSoft. To calculate CVMA the half-width of the 95% CI around the ratio is first divided by the appropriate standard normal value (1.96), resulting in a term that is the conceptual equivalent of the standard deviation. This term is then divided by the value of the ratio to give the CVMA. Calculation of CVMA does not require technical replicates. Therefore, the median CVMA across technical replicates was selected to evaluate the relationship between CVMA and CVTrue.
2.7. Relationship between CVMA and CVTrue
Pearson correlation and Bland-Altman analyses were used to evaluate the relationship between CVMA and CVTrue. The practical impact of using CVMA to define the LOQ of the RS ratio was tested based on the CVMA rather than the CVTrue as follows. Applying a conventional approach, samples with RS ratio CVTrue ≤ 30% or > 30% were classified as quantifiable and non-quantifiable, respectively. This classification based on CVTrue served as our reference standard. We then tested whether a CVMA of 30% would provide the same classification as CVTrue. Receiver operating characteristic (ROC) analysis was further used to test the practical effect of using CVMA thresholds other than 30% to classify samples as quantifiable versus non-quantifiable. In the context of ROC analysis, sensitivity was defined as the proportion of samples that were quantifiable based on CVTrue and were correctly classified as quantifiable based on CVMA. Specificity was defined as the proportion of samples that were non-quantifiable based on CVTrue and were correctly classified as non-quantifiable based on CVMA. Both CVTrue and CVMA were reported as percentages. A p-value < 0.05 was considered as sufficient evidence for a real association. Statistical analysis was conducted in R (v 3.5.3; R Development Core Team, Vienna, Austria).
3. Results
3.1. RS ratio in dilution series
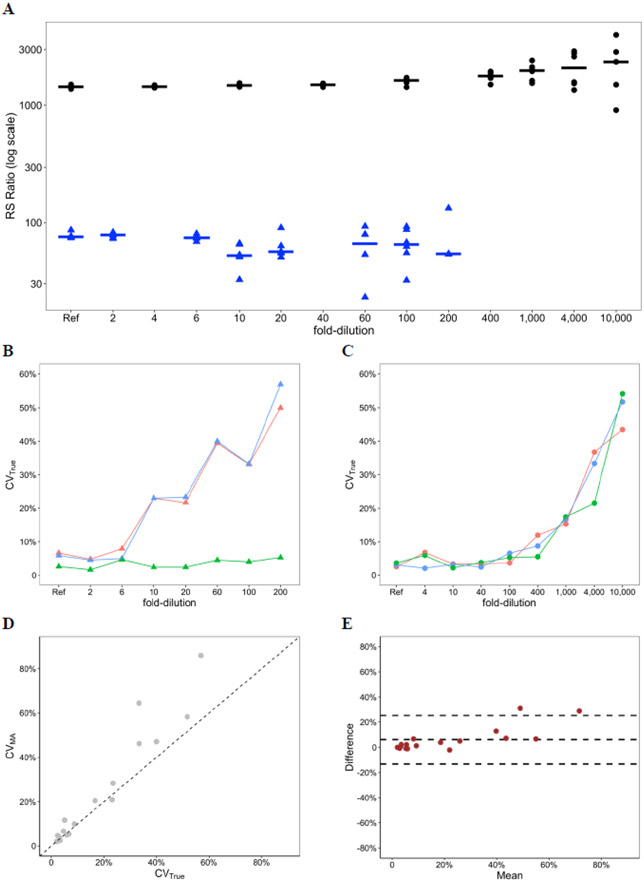
RS ratio results in up to 10,000-fold dilution of two sputum specimens are shown in Table 1 and Fig. 1a. For both the low and high RS ratio samples, quantification of RS ratio was highly precise (CVTrue <10%) in minimally diluted samples (<10-fold dilution for the low RS ratio sample and <1000-fold for the high RS ratio sample). At progressively higher dilutions, precision decreased (reflected by increasing CVTrue). Precision decreased more rapidly with higher dilutions for the low RS ratio sample than for the high RS ratio sample. Consistent with the established performance characteristics of dPCR [25], the CVTrue for individual pre-rRNA and 23S rRNA targets increased substantially once the absolute copy number of either target was less than 50. The low RS ratio sample showed that if the CVTrue of the denominator was low (i.e., the denominator was estimated with precision), then precision in the RS ratio followed precision of the numerator (i.e., pre-rRNA) (Fig. 1b). By contrast, in the high RS ratio sample, precision depended on precision of both the numerator and the denominator (i.e., pre-rRNA and 23S rRNA, respectively) (Fig. 1c).
Table 1.
Droplet digital PCR results from a dilution series of RNA from two human sputum samples selected for their low and high RS ratio estimates. Mean copy numbers and CVTrue. for the pre-rRNA numerator and 23S rRNA denominator illustrate the effect of decreasing target abundance on precision and effect on CVMA and CVTrue for the RS ratio. Six replicates were used for the calculation of each mean and CVTrue except for the 200-fold low RS ratio dilution (N=3) and the 10,000-fold high RS ratio dilution (N=5).
| Sample | Dilution | Pre-rRNA |
23S rRNA |
RS ratio |
||||
|---|---|---|---|---|---|---|---|---|
| Mean | CVTrue | Mean | CVTrue | Mean | CVTrue | CVMA | ||
| Low RS ratio sample | Ref. | 614 | 7% | 79,472 | 3% | 77 | 6% | 5% |
| 2-fold | 301 | 5% | 38,612 | 2% | 78 | 5% | 7% | |
| 6-fold | 106 | 8% | 14,234 | 5% | 74 | 5% | 12% | |
| 10-fold | 30 | 23% | 5,725 | 2% | 53 | 23% | 21% | |
| 20-fold | 18 | 22% | 2,904 | 2% | 62 | 23% | 28% | |
| 60-fold | 7 | 39% | 1,081 | 5% | 63 | 40% | 47% | |
| 100-fold | 3 | 33% | 432 | 4% | 66 | 33% | 65% | |
| 200-fold | 2* | 50% | 222 | 5% | 80† | 57%† | 86%† | |
| High RS ratio sample | Ref. | 23,921 | 3% | 166,174 | 4% | 1,440 | 3% | 4% |
| 4-fold | 7,153 | 7% | 49,491 | 6% | 1,445 | 2% | 2% | |
| 10-fold | 2,780 | 3% | 18,702 | 2% | 1,487 | 3% | 2% | |
| 40-fold | 744 | 3% | 4,978 | 4% | 1,495 | 2% | 5% | |
| 100-fold | 518 | 4% | 3,220 | 5% | 1,612 | 7% | 5% | |
| 400-fold | 161 | 12% | 911 | 6% | 1,768 | 9% | 10% | |
| 1,000-fold | 34 | 15% | 177 | 17% | 1,946 | 17% | 20% | |
| 4,000-fold | 9 | 37% | 42 | 22% | 2,127 | 33% | 46% | |
| 10,000-fold | 3‡ | 43% | 14 | 54% | 2,323§ | 52%§ | 58%§ | |
3 of 6 samples had zero pre-rRNA copies so RS ratio could not be calculated.
Based on the 3 replicates for which RS ratio could be calculated.
1 of 6 samples had zero pre-rRNA copies so an RS ratio could not be calculated.
based on the 5 replicates for which RS ratio could be calculated.
Copy number refers to copies per 20uL well.
Fig. 1. Evaluation of CVMA as alternative for CVTrue in dilution series.
A, RS ratio results in a dilution series of a human sputum sample with high RS ratio (black circles) and a sputum sample with low RS ratio (blue triangle). For the low RS ratio sample, variability increased at a lower level of dilution than for the high RS ratio sample. Horizontal lines show median. B, Precision of the low RS ratio sample in dilution series. For the low RS ratio sample, precision of the RS ratio (blue) was primarily driven by precision of the numerator, (i.e., pre-rRNA, red). 23S rRNA (green) was relatively less variable. C, Precision of the high RS ratio sample in dilution series. For the high RS ratio sample, precision of the RS ratio (blue) depended on precision of both the numerator (i.e., pre-rRNA, red) and the denominator (i.e., 23S rRNA, green). D, Correlation between CVMA and CVTrue in both dilution series. A scatterplot showed a strong correlation between the two variables (Pearson correlation = 0.96). The 45-degree diagonal dashed line is a reference line that shows perfect equality between the two variables. E, Agreement between CVMA and CVTrue in both dilution series. A Bland-Altman plot showed a high agreement between CVMA and CVTrue. The middle dashed line represents the bias (mean difference) between CVMA and CVTrue. The upper and lower dashed lines represent the 95% limits of agreement.
3.2. Relationship between CVMA and CVTrue in dilution series
CVMA and CVTrue were strongly correlated (Pearson correlation = 0.96) in dilution series (Fig. 1d), suggesting a strong agreement between the two metrics. At higher dilutions, CVMA slightly over-estimated variability relative to CVTrue. A Bland-Altman plot further confirmed the agreement between CVMA and CVTrue (Fig. 1e). The Bland-Altman bias (i.e., mean difference between CVMA and CVTrue) was 6% (95% CI: −13% to 25%), again illustrating that on average, CVMA tends to slightly overestimate CVTrue.
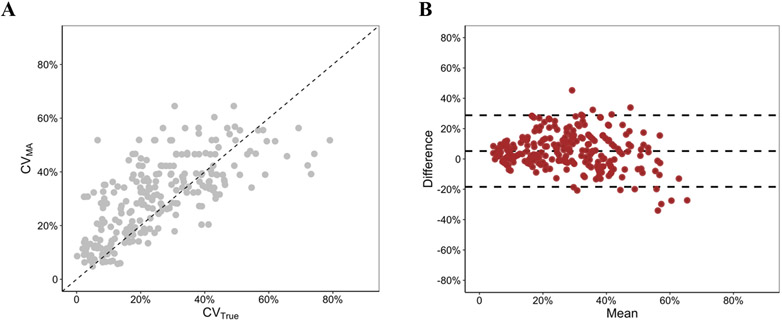
3.3. Relationship between CVMA and CVTrue in routine workflow
In our routine testing of 2,239 sputa, 244 (10.9%) samples met quality criteria to be tested in replicate assays and were therefore available for use in this analysis. In this practical evaluation, CVMA and CVTrue were also strongly correlated (Pearson correlation = 0.70) (Fig. 2a). Consistent with the dilution series results, a Bland-Altman analysis also showed a high agreement between CVMA and CVTrue (Fig. 2b). The Bland-Altman bias was 5% (95% CI: −18% to 29%).
Fig. 2. Evaluation of CVMA as surrogate of CVTrue in routine assays.
A, Correlation between CVMA and CVTrue in routine assays. A scatterplot confirmed a strong correlation between the 2 variables (Pearson correlation = 0.70, P < 0.01). The 45-degree diagonal dashed line is a reference line that shows perfect equality between the two variables. B, Agreement between CVMA and CVTrue in routine assays. A Bland-Altman plot confirmed a high agreement between CVMA and CVTrue. The middle dashed line represents the bias (mean difference) between CVMA and CVTrue. The upper and lower dashed lines represent the 95% limits of agreement.
3.4. Determination of LOQ based on CVMA in routine samples
A CVMA threshold of 30% roughly recapitulated the reference classification based on CVTrue. This means, in a single dPCR assay, samples would be classified as quantifiable and non-quantifiable when CVMA <=30% and CVMA >30%, respectively. With this threshold, 91% of non-quantifiable samples based on CVTrue were correctly classified as non-quantifiable based on CVMA (i.e., specificity = 91%, Table 2). The 30% CVMA threshold also achieved a high classification accuracy (74%) while keeping the false positive rate reasonably low (9%). None of the other CVMA thresholds tested resulted in a meaningfully better recapitulation of the reference classification. Relative to a CVMA threshold of 30%, a CVMA threshold of 35% achieved a slightly greater classification accuracy (76%), but the specificity was 19% lower (72%), and the false positive rate was three times higher (28%).
Table 2.
CVMA thresholds with their ability to correctly identify quantifiable versus non-quantifiable samples in routine assays.
| CVMA thresholds |
Sensitivity | FN rate | Specificity | FP rate | Accuracy |
|---|---|---|---|---|---|
| 20% | 42% | 58% | 99% | 1% | 62% |
| 25% | 51% | 49% | 95% | 5% | 67% |
| 30% | 65% | 35% | 91% | 9% | 74% |
| 35% | 78% | 22% | 72% | 28% | 76% |
| 40% | 89% | 11% | 45% | 55% | 73% |
FN = false negative; FP = false positive.
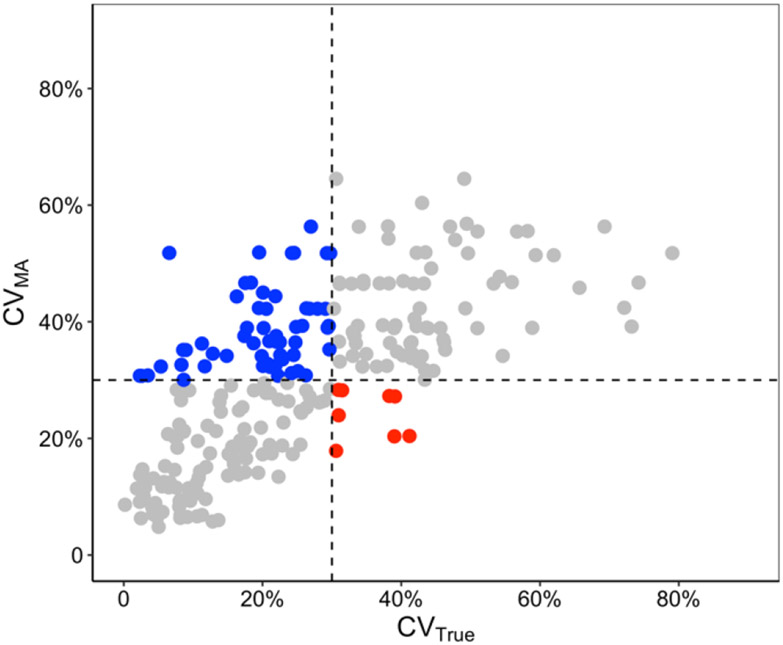
Fig. 3 illustrates the implications of applying a CVMA threshold of 30% in our routine workflow. A small number of samples were false positive (N = 8; 3% of total samples), meaning they were erroneously classified as quantifiable based on CVMA but were non-quantifiable based on CVTrue. Among these eight samples that CVMA classified as quantifiable, CVTrue was only modestly >30% (median = 35%, min = 31%, max = 41%). A larger number of samples were false negative (N = 56; 23% of total samples), meaning that they were erroneously classified as non-quantifiable based on CVMA but were quantifiable based on CVTrue. The majority of samples had consistent classification with both CVMA and CVTrue (N = 180; 74% of total samples).
Fig. 3. Classification of routine assays based on CVMA and CVTrue.
Red and blue dots represent false positives and false negatives, respectively. Gray dots represent true positives (bottom left quadrant) and true negatives (top right quadrant). Dashed lines indicate 30% thresholds for both CVMA and CVTrue. The proportions of false positive, false negative and correctly classified samples are 9%, 35% and 74%, respectively.
4. Discussion
This work addressed the need for a practical measure of precision for a ratiometric dPCR assay in sputum that does not require multiple technical replicates. Our results showed that CVMA approximates CVTrue. CVMA slightly over-estimated actual variation relative to CVTrue, making it a conservative alternative measure. CVMA offers a practical solution to a challenge in use of a powerful new technology.
We anticipate that CVMA will be particularly useful for ratiometric assays in sample types such as sputum in which the abundance of targets is highly variable. Our results demonstrated that it is possible to generate imprecise ratios under two circumstances: (1) when the absolute copy number for the numerator approaches single digits, irrespective of the value of the denominator (e.g., in a sample with a low ratio) or (2) when the absolute copy number of both the numerator and denominator values are low (e.g., in a sample with a high ratio but low abundance of both targets). We confirmed that the value of the ratio itself is not a proxy for precision. A low ratio may be measured with a high degree of precision and a high ratio may be measured imprecisely. The CVMA provides a practical solution to assure that ratiometric assay results meet an acceptable precision threshold. Although calculation of the CVMA may not be essential for ratiometric assays such as the BCR-ABL test that have highly consistent denominators, it may nonetheless be a useful adjunctive confirmation for all ratiometric dPCR assays.
These findings highlight two types of misclassified results: (1) false positives that the CVMA erroneously classified as quantifiable and (2) false negatives that the CVMA erroneously classified as non-quantifiable. False positives are problematic because they over-estimate the precision with which a ratio is estimated. However, analysis of our routine workflow showed that when samples are erroneously classified as quantified, their CVTrue is typically only marginally higher than the acceptable 30% variability threshold, suggesting that this misclassification may be of limited consequence.
With a CVMA threshold of 30%, we found a false negative rate of 35%. Importantly, this does not indicate that 35% of all results are erroneously rejected. The analysis presented here included only the small subset of samples that had features suggesting imprecision (10.9% of all samples tested). We consider it appropriate to be conservative in assessment of this subset of potentially problematic samples. Additionally, erroneous rejection in a singlicate assay does not mean the sample necessarily is unreportable because a “rescue strategy” exists. In routine practice, when a single assay has CVMA >30%, suggesting insufficient precision, it is repeated with technical replicates which are merged in QuantaSoft to enhance precision [25], frequently resulting in CVTrue <30% and acceptance of the result. For the RS ratio, we therefore selected an LOQ (CVMA =30%) that favored specificity over sensitivity, resulting in very few false positives. This strategy and workflow assure that assays meet a consistent standard for precision while minimizing the number of replicate assays that must be performed.
This work has several limitations. First, while our goal is a practical measure of precision to be used routinely for all samples, the set of routine assays analyzed here had a disproportionate over-representation of samples with very low Mtb abundance. Our routine workflow results in replicate assays primarily for the most variable samples. Despite an unusually variable sample set that makes this a “worst case” analysis, a high agreement was observed between CVMA and CVTrue. Second, the selection of a CVMA threshold of 30% as the LOQ for the RS ratio requires a judgement about the relative importance of sensitivity and specificity. A conservative approach was used to maximize specificity and minimize false positives, but other thresholds could also be acceptable. Finally, this analysis addresses precision only at the dPCR step of the RS ratio, not variability that might in the process of RNA extraction or reverse transcription. An advantage of a ratiometric assay is that variation in RNA recovery does not systematically affect the RS ratio unless there is differential loss of the pre-rRNA or 23S rRNA which we have not observed.
In summary, we have used the output of a single dPCR assay (i.e., tens of thousands of binary results) to create a proxy for the traditional CV without the requirement for replicate assays. This is an important step for the development of the RS ratio as a marker of treatment response for TB regimen development since it shows that we now have a reliable measure of precision for this important marker. The CVMA may be a practical tool not only for the RS ratio but for other assays that measure ratios via dPCR.
Supplementary Material
Highlights.
The precision of a ratio depends on precision in both the numerator and denominator
The value of a ratio alone does not indicate precision
A low ratio value can be highly precise and a high ratio value can be imprecise
Precision methods for a single measurand cannot be applied to ratio measurements
Established methods for quantification limit are not applicable to ratio assays
Acknowledgment
The authors wish to acknowledge the TB Trials Consortium and AIDS Clinical Trials study teams and participants in the Study 31 trial that provided samples used for practical evaluation.
Funding
NDW acknowledges funding from Veterans Affairs (1IK2CX000914-01A1 and 1I01BX004527-01A1) and from the Doris Duke Charitable Foundation Clinical Scientist Development Award. NDW, PPJP, RSM, and PN acknowledge funding from the US National Institutes of Health (1R01AI127300-01A1) and the Bill & Melinda Gates Foundation (OPP1213947).
Footnotes
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Tantiwetrueangdet A, Panvichian R, Wongwaisayawan S, Sueangoen N, Lertsithichai P, Droplet digital PCR using HER2/EIF2C1 ratio for detection of HER2 amplification in breast cancer tissues, Med. Oncol 35 (2018) 149. 10.1007/s12032-018-1210-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Selvaraj V, Maheshwari Y, Hajeri S, Chen J, McCollum TG, Yokomi R, Development of a duplex droplet digital PCR assay for absolute quantitative detection of “Candidatus Liberibacter asiaticus,” PLOS ONE. 13 (2018) e0197184. 10.1371/journal.pone.0197184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Cheng X, Sun L, Zhao Q, Mi Z, Yu G, Wang Z, Sun Y, Wang C, Man C, Fu F, Liu H, Zhang F, Development and evaluation of a droplet digital PCR assay for the diagnosis of paucibacillary leprosy in skin biopsy specimens, PLoS Negl. Trop. Dis 13 (2019) e0007284. 10.1371/journal.pntd.0007284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Jennings LJ, George D, Czech J, Yu M, Joseph L, Detection and Quantification of BCR-ABL1 Fusion Transcripts by Droplet Digital PCR, J. Mol. Diagn 16 (2014) 174–179. 10.1016/j.jmoldx.2013.10.007. [DOI] [PubMed] [Google Scholar]
- [5].Wang W-J, Zheng C-F, Liu Z, Tan Y-H, Chen X-H, Zhao B-L, Li G-X, Xu Z-F, Ren F-G, Zhang Y-F, Chang J-M, Wang H-W, Droplet digital PCR for BCR/ABL(P210) detection of chronic myeloid leukemia: A high sensitive method of the minimal residual disease and disease progression, Eur. J. Haematol 101 (2018) 291–296. 10.1111/ejh.13084. [DOI] [PubMed] [Google Scholar]
- [6].Bizouarn F, Clinical Applications Using Digital PCR, in: Biassoni R, Raso A (Eds.), Quant. Real-Time PCR, Springer New York, New York, NY, 2014: pp. 189–214. 10.1007/978-1-4939-0733-5_16. [DOI] [PubMed] [Google Scholar]
- [7].Li H, Bai R, Zhao Z, Tao L, Ma M, Ji Z, Jian M, Ding Z, Dai X, Bao F, Liu A, Application of droplet digital PCR to detect the pathogens of infectious diseases, Biosci. Rep 38 (2018) BSR20181170. 10.1042/BSR20181170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Devonshire AS, O’Sullivan DM, Honeyborne I, Jones G, Karczmarczyk M, Pavšič J, Gutteridge A, Milavec M, Mendoza P, Schimmel H, Van Heuverswyn F, Gorton R, Cirillo DM, Borroni E, Harris K, Barnard M, Heydenrych A, Ndusilo N, Wallis CL, Pillay K, Barry T, Reddington K, Richter E, Mozioğlu E, Akyürek S, Yalçınkaya B, Akgoz M, Žel J, Foy CA, McHugh TD, Huggett JF, The use of digital PCR to improve the application of quantitative molecular diagnostic methods for tuberculosis, BMC Infect. Dis 16 (2016) 366. 10.1186/s12879-016-1696-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Devonshire AS, Honeyborne I, Gutteridge A, Whale AS, Nixon G, Wilson P, Jones G, McHugh TD, Foy CA, Huggett JF, Highly Reproducible Absolute Quantification of Mycobacterium tuberculosis Complex by Digital PCR, Anal. Chem 87 (2015) 3706–3713. 10.1021/ac5041617. [DOI] [PubMed] [Google Scholar]
- [10].Chung H-J, Hur M, Yoon S, Hwang K, Lim H-S, Kim H, Moon H-W, Yun Y-M, Performance Evaluation of the QXDx BCR-ABL %IS Droplet Digital PCR Assay, Ann. Lab. Med 40 (2020) 72–75. 10.3343/alm.2020.40.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Walter ND, Born SEM, Robertson GT, Reichlen M, Dide-Agossou C, Ektnitphong VA, Rossmassler K, Ramey ME, Bauman AA, Ozols V, Bearrows SC, Schoolnik G, Dolganov G, Garcia B, Musisi E, Worodria W, Huang L, Davis JL, Nguyen NV, Nguyen HV, Nguyen ATV, Phan H, Wilusz C, Podell BK, Sanoussi ND, de Jong BC, Merle CS, Affolabi D, McIlleron H, Garcia-Cremades M, Maidji E, Eshun-Wilson F, Aguilar-Rodriguez B, Karthikeyan D, Mdluli K, Bansbach C, Lenaerts AJ, Savic RM, Nahid P, Vásquez JJ, Voskuil MI, Mycobacterium tuberculosis precursor rRNA as a measure of treatment-shortening activity of drugs and regimens, Nat. Commun 12 (2021) 2899. 10.1038/s41467-021-22833-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Dooley KE, Phillips PPJ, Nahid P, Hoelscher M, Challenges in the clinical assessment of novel tuberculosis drugs, Adv. Drug Deliv. Rev 102 (2016) 116–122. 10.1016/j.addr.2016.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Armbruster DA, Pry T, Limit of blank, limit of detection and limit of quantitation, Clin. Biochem. Rev 29 Suppl 1 (2008) S49–52. [PMC free article] [PubMed] [Google Scholar]
- [14].Shrivastava A, Gupta V, Methods for the determination of limit of detection and limit of quantitation of the analytical methods, Chron. Young Sci 2 (2011) 21. 10.4103/2229-5186.79345. [DOI] [Google Scholar]
- [15].Shabir GA, Step-by-step analytical methods validation and protocol in the quality system compliance industry, in: 2004. [Google Scholar]
- [16].Burd EM, Validation of Laboratory-Developed Molecular Assays for Infectious Diseases, Clin. Microbiol. Rev 23 (2010) 550–576. 10.1128/CMR.00074-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Beillard E, Pallisgaard N, van der Velden VHJ, Bi W, Dee R, van der Schoot E, Delabesse E, Macintyre E, Gottardi E, Saglio G, Watzinger F, Lion T, van Dongen JJM, Hokland P, Gabert J, Evaluation of candidate control genes for diagnosis and residual disease detection in leukemic patients using ‘real-time’ quantitative reverse-transcriptase polymerase chain reaction (RQ-PCR) – a Europe against cancer program, Leukemia. 17 (2003) 2474–2486. 10.1038/sj.leu.2403136. [DOI] [PubMed] [Google Scholar]
- [18].Arora R, Press RD, Measurement of BCR-ABL1 transcripts on the International Scale in the United States: current status and best practices, Leuk. Lymphoma 58 (2017) 8–16. 10.1080/10428194.2016.1190974. [DOI] [PubMed] [Google Scholar]
- [19].Davies GR, Early clinical development of anti-tuberculosis drugs: Science, statistics and sterilizing activity, Tuberculosis. 90 (2010) 171–176. 10.1016/j.tube.2010.03.007. [DOI] [PubMed] [Google Scholar]
- [20].Cui ZC, Allowable limit of error in clinical chemistry quality control, Clin. Chem 35 (1989) 630–631. [PubMed] [Google Scholar]
- [21].Morisset D, Štebih D, Milavec M, Gruden K, Žel J, Quantitative Analysis of Food and Feed Samples with Droplet Digital PCR, PLoS ONE. 8 (2013) e62583. 10.1371/journal.pone.0062583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Shehata HR, Li J, Chen S, Redda H, Cheng S, Tabujara N, Li H, Warriner K, Hanner R, Droplet digital polymerase chain reaction (ddPCR) assays integrated with an internal control for quantification of bovine, porcine, chicken and turkey species in food and feed, PLOS ONE. 12 (2017) e0182872. 10.1371/journal.pone.0182872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lu C, Wu J, Wang H, Wang S, Diao N, Wang F, Gao Y, Chen J, Shao L, Weng X, Zhang Y, Zhang W, Novel Biomarkers Distinguishing Active Tuberculosis from Latent Infection Identified by Gene Expression Profile of Peripheral Blood Mononuclear Cells, PLoS ONE. 6 (2011) e24290. 10.1371/journal.pone.0024290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Deprez L, Corbisier P, Kortekaas A-M, Mazoua S, Beaz Hidalgo R, Trapmann S, Emons H, Validation of a digital PCR method for quantification of DNA copy number concentrations by using a certified reference material, Biomol. Detect. Quantif 9 (2016) 29–39. 10.1016/j.bdq.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Droplet Digital™ PCR (ddPCR™) Technology ∣ LSR ∣ Bio-Rad, (n.d.). https://www.bio-rad.com/en-us/life-science/learning-center/introduction-to-digital-pcr/what-is-droplet-digital-pcr?ID=MDV31M4VY (accessed September 24, 2019).
- [26].Dorman SE, Nahid P, Kurbatova EV, Phillips PPJ, Bryant K, Dooley KE, Engle M, Goldberg SV, Phan HTT, Hakim J, Johnson JL, Lourens M, Martinson NA, Muzanyi G, Narunsky K, Nerette S, Nguyen NV, Pham TH, Pierre S, Purfield AE, Samaneka W, Savic RM, Sanne I, Scott NA, Shenje J, Sizemore E, Vernon A, Waja Z, Weiner M, Swindells S, Chaisson RE, Four-Month Rifapentine Regimens with or without Moxifloxacin for Tuberculosis, N. Engl. J. Med 384 (2021) 1705–1718. 10.1056/NEJMoa2033400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Dube S, Qin J, Ramakrishnan R, Mathematical Analysis of Copy Number Variation in a DNA Sample Using Digital PCR on a Nanofluidic Device, PLoS ONE. 3 (2008) e2876. 10.1371/journal.pone.0002876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Li H-Q, Tang M-L, Wong W-K, Confidence intervals for ratio of two Poisson rates using the method of variance estimates recovery, Comput. Stat 29 (2014) 869–889. 10.1007/s00180-013-0467-9. [DOI] [Google Scholar]
- [29].Lievens A, Jacchia S, Kagkli D, Savini C, Querci M, Measuring Digital PCR Quality: Performance Parameters and Their Optimization, PloS One. 11 (2016) e0153317. 10.1371/journal.pone.0153317. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.