Abstract
In this review, we aim to provide a comprehensive summary of the various microRNAs (miRNAs) shown to be involved in glaucoma and intraocular pressure regulation. miRNAs are short, single-stranded, and noncoding RNAs that regulate gene expression in a number of physiological conditions and human diseases, including glaucoma. Numerous miRNAs display differential expression in glaucoma-affected tissues, such as aqueous humor, tears, trabecular meshwork, and retina analyzed from patients and animal models, suggesting their potential involvement in glaucoma pathogenesis. Several studies summarized here have also investigated the challenge of delivering intact miRNAs to target tissues in order to develop miRNA-based glaucoma therapies. We extend these reports by conducting an additional layer of analysis that integrates the interaction between glaucoma-related miRNAs and glaucoma-associated genes. We conclude with a comprehensive discussion of the therapeutic potential of miRNAs, the cellular pathways that link these miRNAs together, and the most promising miRNAs for future glaucoma research.
Keywords: Glaucoma, miRNA, aqueous humor, tears, trabecular meshwork, retina
1. Introduction
MicroRNAs (miRNAs) are widely studied for their role in numerous diseases, including cancer, polycystic kidney disease, post-myocardial infarction, vascular disease, cardiac fibrosis, amyotrophic lateral sclerosis (ALS), and glaucoma (Chakraborty et al., 2021; Gonzalez et al., 2014). Numerous miRNAs are differentially expressed in glaucoma patients compared to non-glaucoma controls and in in vitro and in vivo models of glaucoma. The purpose of this review is to provide a comprehensive summary of the different miRNAs involved in various subtypes of glaucoma. We also describe various methods used to study miRNA expression and conclude with a discussion on the potential of miRNA-based therapeutics and future research in the field.
1.1. MicroRNA
miRNAs are single-stranded, noncoding RNAs approximately 19-22 nucleotides in length. miRNAs were first discovered in 1993 in Caenorhabditis elegans, a nematode worm commonly used for biological studies. (Lee et al., 1993; Wightman et al., 1993). Since their discovery, miRNAs have been found in numerous other organisms, including humans, and have been associated with several human diseases (Chakraborty et al., 2021; Gonzalez et al., 2014). miRNAs are transcribed from noncoding genes or from untranslated regions of protein-coding genes as pre-miRNAs, then exported out of the nucleus by exportin-5 and processed by Dicer to remove the hairpin loop (Hammond, 2015). Mature miRNAs silence or suppress mRNAs by base-pairing with incomplete complementary sequences within mRNA molecules to cause cleavage, destabilization, or less efficient translation of the mRNA (Filipowicz et al., 2008). miRNAs regulate gene expression at the post-transcriptional mRNA level based on the changing requirements of the cell (Hammond, 2015). For example, let-7 expression increases a hundred fold during differentiation of embryonic cells while let-7 expression is blocked in embryonic cells (Nam et al., 2011; Thomson et al., 2006). Approximately 2600 miRNAs are encoded in the human genome, and each miRNA is predicted to regulate the expression of hundreds of target genes (Hammond, 2015; Lim et al., 2005). To complicate things even more, a single gene could be targeted by multiple miRNAs to regulate its expression. For more details about miRNA biogenesis, refer to reviews by Ha and Kim (2014) and Hammond (2015). For more details about miRNA translation inhibition, please refer to the review by Filipowicz et al. (2008).
1.2. MicroRNA Profiling Tools
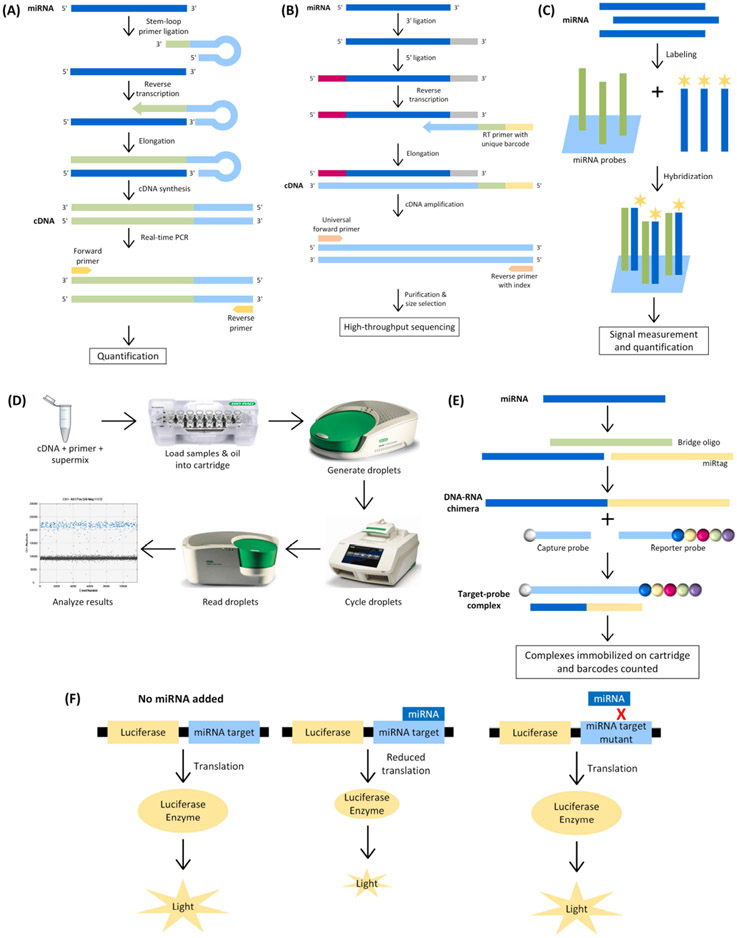
The ability of miRNAs to regulate gene expression and their involvement in many diseases make them an attractive candidate for their potential role in ocular disorders, including glaucoma. However, their short length of 19-22 nucleotides presents significant challenges for miRNA expression profiling. Traditional techniques for mRNA-based expression profiling, such as RT-PCR or arrays, need to be modified for miRNAs. For instance, miRNAs often need to be extended by ligating specially designed oligonucleotides to their 5’ and 3’ ends. Current experimental techniques to study miRNA expression include real-time RT-PCR, small RNA sequencing, miRNA arrays, droplet digital PCR (ddPCR), and NanoString nCounter-based miRNA assays (Figure 1). The advantages and disadvantages of each technique are discussed here and summarized in Table 1.
Fig. 1. Schematics of miRNA profiling tools.
(A) Real-time RT-PCR for miRNA. A small miRNA-specific stem-looped primer is ligated to the miRNA of interest then cDNA is reverse transcribed, amplified, and used in real-time PCR. Fluorescence is quantified to determine expression levels of the miRNA of interest. (B) Small RNA sequencing. Isolated miRNA is ligated to 3’ and 5’ adaptors then reverse transcribed with an RT primer linked to a unique barcode. The resulting cDNA is amplified using a universal forward primer and reverse primer with index. The cDNA then undergoes purification and size selection and is sequenced using a high-throughput method. (C) miRNA array. Isolated miRNAs are labeled with a fluorescent dye then hybridized to the microarray. Labeled miRNAs bind to complementary probes that are immobilized on the microarray. Fluorescence from each labeled miRNA at different positions on the array is detected and quantified. (D) ddPCR for miRNA. cDNA, miRNA specific primers, and supermix are combined and loaded into the sample wells on the ddPCR cartridge. Oil is loaded into the oil wells on the cartridge and then placed into the droplet generator. The oil and sample mixture is transferred to a 96-well plate and placed in the thermal cycler for amplification. The plate is then placed in the droplet reader for quantification and analysis. (E) NanoString nCounter miRNA assay. The miRNA of interest is ligated to a miRtag using a bridge oligo resulting in a DNA-RNA chimera. The chimera is paired with a capture probe on the 5’ end and a reporter probe on the 3’ end, resulting in a target-probe complex. The complexes are then immobilized on a cartridge and the unique barcodes are counted. (F) miRNA luciferase reporter assay. The luciferase gene is fused with the 3’ UTR of the predicted miRNA target gene. In the absence of miRNA, translation occurs normally, and the luciferase enzyme produces light. When miRNA binds to the target gene, translation is significantly reduced, and less light is produced. When the target gene is mutated, the miRNA cannot bind resulting in normal translation and light production.
Table 1.
Summary of the advantages and disadvantages of each miRNA profiling tool described in section 1.2.
| Technique | Advantages | Disadvantages |
|---|---|---|
| Real-time RT-PCR | Simple, easy to set up, inexpensive | Relative quantification, low throughput, requires reference genes and 3-4 duplicates in each run |
| Small RNA sequencing | Covers all small RNA species with new sequences, requires less RNA input, wide dynamic range, high sensitivity | Large data that requires bioinformatics expertise, relatively expensive |
| miRNA arrays | Profile thousands of miRNAs at once, inexpensive, large amount of RNA input | Less sensitive, relative quantification, limited dynamic range, restricted to known miRNAs |
| ddPCR | Precise, high sensitivity, absolute quantification, no duplicates needed, no need to include the reference genes in each run | Relatively expensive, requires specific ddPCR system, low throughput |
| NanoString nCounter-based miRNA assays | Fast and simple sample preparation and data analysis | Limited to known miRNAs, requires access to specific platform, relatively expensive for each run |
| Luciferase reporter assay | Inexpensive, simple, used in live cells | Requires miRNA mimic transfection which may cause cytotoxicity, not performed in disease relevant tissue/cells, requires overexpression of miRNA |
| miRNA-containing viral vectors | Long-term stable miRNA expression, does not cause cytotoxicity | Subject to host epigenetic silencing, relatively expensive to generate, biosafety hazard |
To perform real-time RT-PCR, miRNA-containing total RNA is extracted from the tissue or cells of interest, then a stem-looped primer or specific oligonucleotides are ligated to the 5’ and 3’ ends of the miRNAs. After adaptor ligation, the extended miRNAs are reverse-transcribed based on the ligated oligonucleotide sequences (Figure 1a). miRNA-specific primers or unique probes are designed to amplify and quantify the miRNA of interest. The inclusion of unique probes in real-time PCR leads to increased specificity of the miRNA assays compared to assays without the probe (Liu et al., 2020). The advantages of real-time RT-PCR are that it is simple, easy to set up, and relatively inexpensive. The disadvantages are its semi-quantification, low throughput, and its requirement to include reference genes and 3-4 duplicates per sample in each run (Table 1). However, the inclusion of a standard curve provides better quantification of the target miRNAs (Liu et al., 2020).
Small RNA sequencing (RNA-Seq) is used to isolate and sequence small RNA species, including miRNAs. To perform small RNA-Seq, 5’ and 3’ adaptors are ligated to isolated miRNAs, followed by reverse transcription using a primer containing a unique sample-index sequence. The cDNA is then amplified, size-selected, and used for high-throughput sequencing (Figure 1b). Small RNA-Seq can query the expression of thousands of miRNAs with high sensitivity, single-base resolution, and a wide dynamic range. The benefit of using small RNA-Seq instead of arrays is that sequencing covers all the small RNA and miRNA species without prior information of their sequences and requires less RNA input than arrays (Liu et al., 2020). Disadvantages of small RNA-Seq are that it generates a large amount of sequencing data that requires specific computational and bioinformatics expertise to analyze, and it is more expensive compared to other techniques (Liu et al., 2020) (Table 1).
miRNA arrays are a hybridization-based tool capable of monitoring the expression of thousands of select small noncoding RNAs at once. To perform a miRNA array, miRNA-containing total RNA is extended and reverse-transcribed, fluorescently labeled, then hybridized to complementary miRNA probes that are immobilized on a glass slide. After hybridization, the fluorescent signal is scanned and quantified to derive the expression of specific miRNAs based on their positions on the slide (Li & Ruan, 2009) (Figure 1c). The advantages of miRNA arrays are that they can profile the expression of thousands of miRNAs from many different samples at once and are relatively inexpensive. The disadvantages are that they are less sensitive, are semi-quantitative, have a limited dynamic range, and are restricted to known miRNAs (Li & Ruan, 2009) (Table 1).
For ddPCR, reverse transcription occurs in the same manner as described for real-time RT-PCR. Due to the design of ddPCR with Poisson distribution and digital droplets, the absolute copy numbers of miRNAs in a reaction are derived without a standard curve using tens of thousands of single droplets with nanoliter reaction volumes (Figure 1d). ddPCR performs digital PCR within each droplet based on water-oil emulsion droplet technology. This unique feature of ddPCR allows it to be more precise and sensitive so that low-level expressions in miRNA expression can be detected (Liu et al., 2020). The advantages of ddPCR are its absolute quantification, no requirement of technical duplicates or reference genes in each run, and high sensitivity with low RNA input. The disadvantages are its relatively high cost for each reaction, requirement of a specific system, and relatively low throughput (Table 1).
NanoString nCounter miRNA assays are hybridization-based digital assays that use molecular barcodes to detect individual miRNA molecules without reverse transcription or amplification. During NanoString assays, each miRNA is ligated to a miRtag using a bridge oligo to create a DNA-RNA chimera. The chimera is then paired with a unique complementary capture and reporter probe complex allowing for identification of the miRNAs (Figure 1e). The major advantage of this technique is that it allows for more efficient and simpler sample preparation and data analysis (Kulkarni, 2011). It also allows the digital detection and quantification of expression of up to 800 miRNAs and/or mRNAs simultaneously across all biological samples without amplification (Kulkarni, 2011). However, since this technique is hybridization-based, it is limited to the known miRNAs and mRNAs with unique nCounter Reporter Probes. It requires access to the expensive NanoString nCounter Analysis System (Table 1). In addition, miRNAs with highly similar sequences might not be quantified accurately.
During the study of miRNAs and their target genes, it is often necessary to confirm or validate these target genes. Among the different approaches, luciferase reporter assays are commonly used to validate the specific interaction between a miRNA and its target genes (Jin et al., 2013). Cells are first transfected with a construct containing the mutated or non-mutated 3’-UTR sequence of the predicted miRNA target gene fused with the 3’-UTR region of the luciferase gene. The cells are then transfected with the miRNA mimic of interest, and luciferase activity is measured (Figure 1f). Decreased luciferase activity in the non-mutated 3’-UTR sequence group compared to the mutated 3’-UTR sequence group suggests that the miRNA could be bound to the specific 3’-UTR sequence in the target gene and inhibit expression of the target gene. Advantages of this approach are that it is inexpensive, relatively simple to perform, and can be used in live cells. Disadvantages are that it requires transfection with miRNA mimics for overexpression, which may cause cytotoxicity, and it is often performed in immortalized cell lines instead of disease-relevant tissues or primary cells. Immortalized cells allow for easier and more reliable transient transfection with high efficiency while primary cells are difficult to be transfected and have limited passages for culture Table 1) (Keller et al., 2018).
Besides these expression profiling and target validation techniques, stable and consistent gain or loss of miRNA expression is often needed to study the function of miRNAs. siRNA or miRNA-containing viral vectors are often designed to knockdown or overexpress a miRNA of interest to study the functional effects of altered miRNA expression (Fan et al., 2020). Viral vectors allow for stable, long-term, regulated miRNA expression that could be used to observe the effects of a particular miRNA on cell or tissue function. On the other hand, miRNA mimic or inhibitory oligos can be transfected into immortalized or primary cells in vitro through chemical transduction or electroporation for transient overexpression or knockdown of miRNA. The significant advantage of the viral vector system over the use of miRNA mimics alone is that mimics may cause cytotoxicity and are suited only for short-term studies. However, viral vectors are relatively expensive to generate, could pose a biosafety hazard, and can be subject to host epigenetic silencing (Honda et al., 2016) Table 1). While many tools, including the CRISPR-Cas9 system, are available to study the function of miRNAs, significant work is still needed to make analysis more efficient and reliable (Ran et al., 2013).
1.3. Glaucoma
Glaucoma is a group of diseases that result in progressive loss of retinal ganglion cells (RGCs) and optic nerve degeneration, leading to visual field loss and blindness. Glaucoma is a leading cause of irreversible vision loss worldwide, affecting more than 80 million individuals (Tham et al., 2014). Several known risk factors for developing glaucoma include elevated intraocular pressure (IOP), advanced age, ethnicity, myopia, and a family history of glaucoma (Liu & Allingham, 2017). The most common type of glaucoma, accounting for approximately 75-90% of glaucoma cases, is primary open-angle glaucoma (POAG). It is characterized by an open iridocorneal angle and no identifiable cause. Based on the level of IOP, POAG could be categorized into two different sub-types: normal-tension glaucoma (NTG), with untreated maximal IOP ≤ 21 mmHg, and high-tension glaucoma (HTG), with untreated maximal IOP > 21 mmHg (Bailey et al., 2016; Bailey et al., 2018; Wiggs et al., 2012; Youngblood et al., 2019). In contrast to POAG, the other main type of glaucoma is primary angle-closure glaucoma (PACG) in which the iridocorneal angle is narrow, leading to blockage of drainage tissues by iris that acutely and dramatically elevates in IOP. Exfoliation glaucoma (XFG) is the most common secondary glaucoma and is caused by exfoliation syndrome (XFS) (Ritch, 2008; Ritch & Schlotzer-Schrehardt, 2001; Schlotzer-Schrehardt, 2009). XFS is an age-related disease characterized by deposition of extracellular fibrillar material in various ocular and non-ocular tissues, including trabecular meshwork, ciliary body, cornea, lens, skin, heart, and lung (Aboobakar & Allingham, 2014; Ritch, 2008; Schlotzer-Schrehardt, 2009). Compared to POAG, XFG is associated with higher IOPs, increased vision loss at diagnosis, and a more sluggish response to treatment (Aboobakar & Allingham, 2014). All currently available treatments for glaucoma work by reducing IOP to delay or prevent the progression of vision loss (Jonas et al., 2017; Weinreb et al., 2014; Youngblood et al., 2019). Even with all of the different IOP lowering treatments available, glaucoma progression and related visual field loss continue in many patients. Thus, more effective IOP management by targeting the tissues that regulate IOP, the conventional outflow tissues, is still needed to prevent vision loss in these patients.
2. MicroRNAs in Glaucoma
MicroRNAs are involved in numerous human diseases (Chakraborty et al., 2021; Gonzalez et al., 2014). Recently, a role for miRNAs in glaucoma has become apparent (Drewry et al., 2018; Hubens et al., 2021; Jayaram et al., 2015; Jayaram et al., 2017; Liu & Allingham, 2017; Liu et al., 2018; Raga-Cervera et al., 2021). Many studies, which will be discussed here, have revealed numerous miRNAs that are differentially expressed in samples from glaucoma patients or in animal models of glaucoma. These studies have primarily focused on four major anatomic structural regions in the eye, including the aqueous humor (AH), the tears, trabecular meshwork (TM), and the retina. All miRNAs discussed here are divided into sections based on these four regions (Table 2).
Table 2.
Compiled list of all miRNAs discussed in this review listed in chronological order by the anatomic structural regions of the eye in which they were studied. The methods used to analyze the miRNAs in each study are listed in column 2. The experimental design, including sample size and grouping for each study, are listed in column 3. The conclusions of each study in context to miRNA expression or affected genes/pathways are listed in column 4. The average number of reads for each miRNA after miRNA-Seq using RNA from seven normal human TM samples are listed in column 5.
| Reference | Methods | Experimental Design |
Conclusion | Expression in HTM1 |
|---|---|---|---|---|
| AQUEOUS HUMOR | ||||
| Tanaka et al., 2014 | miRNA array (Toray Industries) | 10 control (cataract & epiretinal membrane) 10 glaucoma (1 PACG, 7 POAG, 2 PEX) |
↑ miR-1587, miR-486-3p, miR-3185, miR-940, miR-3652, miR-3135b, miR-5572, miR-4640-5p, miR-4259, miR-92a-2-5p, miR-4449 ↓ miR-4507, miR-3620-5p, miR-4484, miR-5001-5p, miR-6132, miR-6515-3p, miR-4467, miR-3663-3p, miR-187-3p, miR-4433-5p, miR-6717-5p, miR-6722-3p, miR-4725-3p, miR-1202, miR-3197, miR-4749-5p, miR-1260b, miR-4634 |
hsa-miR-486-3p (0.29); hsa-miR-940 (0.43); hsa-miR-3135b (0.57) |
| Liu et al., 2016 | Small RNA-Seq (Illumina MiSeq), ddPCR | NEIGHBORHO OD analysis: 33,480 control, 3853 POAG miRNA-Seq & ddPCR: 10 healthy donors |
↑ MIR182 point mutation = miR-182 | hsa-miR-182-5p (525.43) |
| Jayaram et al., 2017 | miRNA array (Applied Biosystems, Thermo Fisher Scientific) | 8 cataract control, 6 POAG | ↑ miR-518d, miR-143 ↓ miR-660 |
hsa-miR-143-3p (31562.00); hsa-miR-143-5p (4.86); hsa-miR-660-5p (33.14) |
| Drewry et al., 2018 | NanoString, ddPCR | NanoString: 11 cataract control, 12 POAG, 12 XFG ddPCR: 10 cataract control, 17 POAG, 14 XFG |
↑ miR-302d-3p, miR-451a, miR-122-5p, miR-3144-3p, miR-320e, miR-630 ↓ miR-125b-5p, miR-320a |
hsa-miR-125b-5p (1349.29); hsa-miR-302d-3p (0); hsa-miR-451a (13.86) |
| Liu et al., 2018 | Small RNA-Seq (Illumina) | 9 cataract control, 9 POAG | ↑ 15 miRNAs ↓ 73 miRNAs |
N/A |
| Yang et al., 2018 | Real-time RT-PCR, Luciferase reporter assay | 8 cataract control, 16 glaucoma | ↑ miR-211 = ↓ Frs2 | hsa-miR-211-3p (0); hsa-miR-211-5p (608.43) |
| Hindle et al., 2019 | Qiagen miRNA PCR Array | 3 control, 6 POAG | ↑ miR-4667-5p, miR-99b-3p, miR-637, miR-4490, miR-1253, miR-3190-3p, miR-3173-3p, miR-608, miR-4725-3p, miR-4448, miR-323b-5p, miR-4538, miR-3913-3p, miR-3159, miR-4663, miR-4767, miR-4724-5p, miR-1306-5p, miR-181b-3p, miR-433-3p | hsa-miR-99b-3p (1.29); hsa-miR-4448 (0); hsa-miR-323-5p (0); hsa-miR-3159 (0); hsa-miR-1306-5p (0.29); hsa-miR-181b-3p (0); hsa-miR-433-3p (0.57) |
| Hubens et al., 2021 | Small RNA-Seq (Illumina) | 10 cataract control, 9 POAG | ↑ miR-30a-3p, miR-143-3p, miR-211-5p, miR-221-3p ↓ miR-92a-3p, miR-451a, miR-486-5p |
hsa-miR-30a-3p (115.57); hsa-miR-143-3p (31562.00); hsa-miR-211-5p (608.43); hsa-miR-221-3p (48.43); hsa-miR-92a-3p (1258.00); hsa-miR-451a (13.86); hsa-miR-486-5p (438.00) |
| TEARS | ||||
| Tamkovich et al., 2019 | Small RNA-Seq (Illumina MiSeq) | 29 healthy control, 33 POAG | ↑ miR-146b, miR-16, miR-126 | hsa-miR-146b-3p (0.86); hsa-miR-146b-5p (27.57); hsa-miR-16-5p (505.43); hsa-miR-126-3p (153.86); hsa-miR-126-5p (2237.57) |
| Raga-Cervera et al., 2021 | Small RNA-Seq (Illumina) | 22 OHT control, 20 POAG | ↑ miR-26b-5p, miR-152-3p, miR-30e-5p, miR-125b-2-5p, miR-224-5p, miR-27a-3p ↓ miR-151a-3p, miR-1307-3p |
hsa-miR-26b-5p (287.86); hsa-miR-152-3p (6.86); hsa-miR-30e-5p (25.86); hsa-miR-224-5p (0.43); hsa-miR-151a-3p (375.00); hsa-miR-1307-3p (1.43); hsa-miR-27a-3p (142.57) |
| TRABECULAR MESHWORK | ||||
| Li et al., 2009 | miRNA array (Asuragen Inc.), real-time RT-PCR, Luciferase assay | Senescent HTM cells from 3 donors and matched non-senescent controls | ↑ miR-200c, miR-182, miR-139, miR-183, miR-192 ↓ miR-15a, miR-15b, miR-16, miR-17-5p, miR-18a, miR-20a, miR-92, miR-106a, miR-106b, miR-146b, miR-195, miR-199b, miR-204, miR-342 ↑ miR-106a = ↓ p21 and ↑ cell proliferation ↑ miR-182 = ↓ RARG and ↑ SA-beta-galactosidase activity |
hsa-miR-200c-3p (202.14); hsa-miR-200c-5p (0.14); hsa-miR-182-5p (525.43); hsa-miR-139-3p (1); hsa-miR-139-5p (16.71); hsa-miR-183-5p (22.43); hsa-miR-192-5p (430.43); hsa-miR-15a-5p (3.00); hsa-miR-15b-3p (0.14); hsa-miR-15b-5p (13.43); hsa-miR-16-5p (505.43); hsa-miR-17-5p (11.14); hsa-miR-18a-3p (0); hsa-miR-18a-5p (0.29); hsa-miR-20a-3p (0.29); hsa-miR-20a-5p (13.57); hsa-miR-92a-1-5p (0.57); hsa-miR-92a-3p (1258.00); hsa-miR-92b-3p (245.43); hsa-miR-106a-5p (0.29); hsa-miR-106b-3p (2.14); hsa-miR-106b-5p (3.57); hsa-miR-146b-3p (0.86); hsa-miR-146b-5p (27.57); hsa-miR-195-3p (3.71); hsa-miR-195-5p (62.29); hsa-miR-199b-3p (94.86); hsa-miR-199b-5p (175.14); hsa-miR-204-3p (0.43); hsa-miR-204-5p (2449.00); hsa-miR-342-3p (55.71); hsa-miR-342-5p (0.29) |
| Luna et al., 2009 | Luciferase assay, real-time RT-PCR | 3 sets of miR-29b mimic or mimic control of HTM cells from one donor without eye disease | ↑ miR-29b = ↓ ECM components, BMP1, ADAM12, and NKIRAS2 | hsa-miR-29b-1-5p (0.29); hsa-miR-29b-2-5p (0.43); hsa-miR-29b-3p (26.00) |
| Li et al., 2010b | Luciferase assay, real-time RT-PCR | 3-4 replicates of HTM cells from one donor | ↑ miR-183 = ↓ ITGB1, KIF2A ↓ cell adhesion ↑ phagocytosis |
hsa-miR-183-5p (22.43) |
| Li et al., 2010a | Real-time RT-PCR | HTM cells from two donors at p15 or p4 Or HTM cells from three donors transfected with miR-146a mimic or mimic control |
↑ miR-146a, miR-192, miR-329, miR-369-5p, miR-409-5p, miR-432, miR-493, miR-495 ↓ miR-204, miR-183, miR-182, miR-155, miR-92, miR-20a, miR-20b, miR-18a, miR-17–5p, miR-106a ↑ miR-146a = ↓ CCL2, CXCL3, IL11, SLC10A3, PTGS1, IL6, GALNT10, CCL20, CXCL6, PPP2R1B, SERPINE1, HAS1, IL8, IRAK1 During senescence: ↑ IL8, PAI-1, CXCL6, CCL20, IL6, CXCL3, CCL2 ↓ HAS1, PPP2R1B |
hsa-miR-146a-5p (126.29); hsa-miR-192-5p (430.43); hsa-miR-329-3p (0.14); hsa-miR-369-5p (0.14); hsa-miR-409-5p (0.14); hsa-miR-432-5p (5.29); hsa-miR-493-3p (3.00); hsa-miR-493-5p (10.86); hsa-miR-495-3p (0.43); hsa-miR-204-3p (0.43); hsa-miR-204-5p (2449.00); hsa-miR-183-5p (22.43); miR-182-5p (525.43); hsa-miR-155-5p (15.71); hsa-miR-92a-1-5p (0.57); hsa-miR-92a-3p (1258.00); hsa-miR-92b-3p (245.43); hsa-miR-20a-3p (0.29); hsa-miR-20a-5p (13.57); hsa-miR-20b-5p (0.57); hsa-miR-18a-3p (0); hsa-miR-18a-5p (0.29); hsa-miR-17-5p (11.14); hsa-miR-106a-5p (0.29) |
| Li et al., 2011 | Real-time RT-PCR, Luciferase assay | HTM cells from 2 donors transfected with miR-204 mimic or mimic control | ↑ miR-204 = ↓ AP1S2, Bcl2l2, BIRC2, EDEM1,EZR, FZD1, M6PR, RAB22A, RAB40B, SERP1, TCF12, TCF4 ↑ miR-204 = ↑ apoptosis, cytotoxicity, accumulation of carbonylated proteins, expression of ER stress markers and ↓ inflammatory factors |
hsa-miR-204-3p (0.43); hsa-miR-204-5p (2449.00) |
| Luna et al., 2011a | Real-time RT-PCR, luciferase reporter assay | Triplicates of miR-29 mimic or mimic control of HTM cells from 3 donors without eye disease | TGF-β2 = ↓ miR-29 and miR-29 = ↓ TGF-β2 |
hsa-miR-29a-3p (152.71); hsa-miR-29a-5p (0.14); hsa-miR-29b-1-5p (0.29); hsa-miR-29b-2-5p (0.43); hsa-miR-29b-3p (26.00); hsa-miR-29c-3p (88.00); hsa-miR-29c-5p (1.29) |
| Luna et al., 2011b | Real-time RT-PCR, Luciferase reporter assay | Triplicates of miR-24 mimic or mimic control of HTM cells from 2 donors without eye disease | CMS = ↑ miR-16, miR-27a, miR-27b, miR-7, let-7f, miR-26a, miR-24 and ↑ miR-24 = ↓ FURIN |
hsa-miR-16-5p (505.43); hsa-miR-27a-3p (142.57); hsa-miR-27b-3p (4044.29); hsa-miR-7-1-3p (0.43); hsa-miR-7-5p (0); hsa-let-7f-1-3p (2.71); hsa-let-7f-2-3p (5.29); hsa-let-7f-5p (4104.29); hsa-miR-26a-1-3p (0.57); hsa-miR-26a-2-3p (1.86); hsa-miR-26a-5p (16979.29); hsa-miR-24-2-5p (0.71); hsa-miR-24-3p (28.29); hsa-miR-24-2-5p (0.71) |
| Luna et al., 2012 | Real-time RT-PCR, Luciferase reporter assay | Triplicates of miR-200c mimic or mimic control of HTM cells from 2 donors without eye disease; 7 scramble mimic rats and 7 miR-200c mimic rats | ↑ miR-200c = ↓ ETAR, LPAR1, FHOD1, ZEB1, and ZEB2 and ↑ miR-200c = ↓ IOP |
hsa-miR-200c-3p (202.14); hsa-miR-200c-5p (0.14) |
| Paylakhi et al., 2013 | Real-time RT-PCR | Triplicates of miR-204 or miR-211 mimic or mimic control of HTM cells from 2 donors | ↑ miR-204 & miR-211 = ↓ FOXC1 | hsa-miR-204-3p (0.43); hsa-miR-204-5p (2449.00); hsa-miR-211-3p (0); hsa-miR-211-5p (608.43) |
| Shen et al., 2015 | Real-time RT-PCR, Luciferase reporter assay | Triplicates of HTM cells from one donor | Oxidative stress = ↓ miR-483-3p ↑ miR-483-3p = ↓ ECM genes and Smad4 |
hsa-miR-483-3p (0.14) |
| Wang et al., 2016 | Real-time RT-PCR | Triplicates of HTM cells from one donor transfected with pre-miR-93 or miR-93 sponge | ↓ miR-93 = ↑ NFE2L2 and ↓ apoptosis | hsa-miR-93-3p (0.14); hsa-miR-93-5p (31.49) |
| Li et al., 2017 | Real-time RT-PCR, Luciferase reporter assay | Triplicates of HTM cells from 3 donor without eye disease transfected with miR-143, miR-145, or control antagomiRs 23 wild-type mice, 24 miR-143/145 DKO mice |
↑ miR-143 and miR-145 in HTM miR-143/145 deletion = ↓ IOP in mice |
hsa-miR-143-3p (31562.00); hsa-miR-143-5p (4.86); hsa-miR-145-3p (0.14); hsa-miR-145-5p (615.57) |
| Ruibin et al., 2018 | Real-time RT-PCR, Luciferase reporter assay | Triplicates of HTM cells from 2 donors without eye disease transfected with miR-1298 mimic or miR-1298 inhibitor | COS = ↓ miR-1298 ↑ miR-1298 = ↓ EIF4E3 and cytotoxicity |
hsa-miR-1298-3p (0); hsa-miR-1298-5p (0.43) |
| Wang et al., 2018 | Real-time RT-PCR | Triplicates of HTM cells from one donor transfected with miR-181a mimic or si-miR-181a | ↑ miR-181a = ↓ NF-κB and JNK signaling and apoptosis | hsa-miR-181a-2-3p (58.14); hsa-miR-181a-3p (22.43); hsa-miR-181a-5p (2803.00) |
| Wang et al., 2019 | Real-time RT-PCR, Luciferase reporter assay | Triplicates of HTM cells from one donor transfected with miR- 17- 5p inhibitor or miR- 17- 5p mimic | H2O2 exposure = ↓ miR-17-5p ↑ miR-17-5p = ↓ PTEN, Bax, Bcl-xL, and Bcl-2 expression and apoptosis |
hsa-miR-17-5p (11.14) |
| Zhao et al., 2019 | Real-time RT-PCR | Triplicates of HTM cells from one donor transfected with miR-27a inhibitor or negative control and treated with 0–5 μM of Sal | Salidroside treatment = ↑ miR-27a = ↑ PI3K/AKT and Wnt/β-catenin pathways and protective effects | hsa-miR-27a-3p (142.57); hsa-miR-27a-5p (2.71) |
| Shen et al., 2020 | Real-time RT-PCR, Luciferase reporter assay | Nine replicates of HTM cells from one donor treated with 300 μM H2O2 then transfected with miR- 200c- 3p mimic or negative control mimic | ↑ miR-200c-3p = ↓ PTEN expression and apoptosis | hsa-miR-200c-3p (202.14) |
| Youngblood et al., 2020 | NanoString, ddPCR | HTM cells from 5 donors without eye disease split into 2 groups, each with 3 stretched and 3 not stretched | CMS = ↑ hsa-miR-4286, hsa-miR-29b-3p, hsa-miR-29a-3p, hsa-miR-140-5p, hsa-miR-100-5p, hsa-miR-136-5p, hsa-miR-32-5p, hsa-miR-24-3p, hsa-miR-151a-3p, hsa-miR-27a-3p, hsa-miR-4284, hsa-miR-222-3p, hsa-miR-93-5p, hsa-miR-3615, hsa-miR-21-5p, hsa-miR-376c-3p, hsa-miR-25-3p, hsa-miR-642a-3p, hsa-miR-27b-3p, hsa-miR-34a-5p, hsa-miR-127-3p, hsa-miR-31-5p, hsa-miR-377-3p, hsa-miR-22-3p, hsa-miR-15a-5p, hsa-miR-125b-5p, hsa-miR-181a-5p, hsa-miR-99b-5p, hsa-miR-30a-5p, hsa-miR-185-5p, hsa-miR-376a-3p, hsa-miR-378i, hsa-miR-574-3p, hsa-miR-337-5p, hsa-miR-125a-5p, hsa-miR-3690, hsa-miR-191-5p, hsa-miR-190a-5p, hsa-miR-181c-5p and ↓ hsa-miR-1275, hsa-miR-187-3p, hsa-miR-1323 |
hsa-miR-29b-3p (26.00); hsa-miR-29a-3p (152.71); hsa-miR-140-5p (6.86); hsa-miR-100-5p (563.29); hsa-miR-136-5p (0.71); hsa-miR-32-5p (0.29); hsa-miR-24-3p (28.29); hsa-miR-151a-3p (375.00); hsa-miR-27a-3p (142.57); hsa-miR-222-3p (16.43); hsa-miR-93-5p (31.49); hsa-miR-3615 (0.29); hsa-miR-21-5p (1692.86); hsa-miR-376c-3p (7.43); hsa-miR-25-3p (347.29); hsa-miR-27b-3p (4044.29); hsa-miR-34a-5p (19.86); hsa-miR-127-3p (851.00); hsa-miR-31-5p (0.43); hsa-miR-377-3p (1.00); hsa-miR-22-3p (8077.29); hsa-miR-15a-5p (3.00); hsa-miR-125b-5p (1349.29); hsa-miR-181a-5p (2803.00); hsa-miR-99b-5p (869.14); hsa-miR-30a-5p (268.71); hsa-miR-185-5p (0.86); hsa-miR-376a-3p (2.29); hsa-miR-574-3p (185.00); hsa-miR-337-5p (0.14); hsa-miR-125a-5p (279.14); hsa-miR-191-5p (3311.29); hsa-miR-190a-5p (0.86); hsa-miR-187-3p (0); hsa-miR-181c-5p (155.86) |
| RETINA | ||||
| Jayaram et al., 2015 | Real-time RT-PCR | 8 glaucoma model rats, 8 control rats | In glaucomatous retina from rats: ↓ miR-181c, miR-497, miR-204, let-7a, miR-29b, miR-16, miR-106b, miR-25 ↑ miR-27a |
hsa-miR-181c-3p (0); hsa-miR-181c-5p (155.86); hsa-miR-497-3p (1.00); hsa-miR-497-5p (34.57); hsa-miR-204-3p (0.43); hsa-miR-204-5p (2449.00); hsa-let-7a-2-3p (0.57); hsa-let-7a-3p (3.14); hsa-let-7a-5p (7622.29); hsa-miR-29b-1-5p (0.29); hsa-miR-29b-2-5p (0.43); hsa-miR-29b-3p (26.00); hsa-miR-16-5p (505.43); hsa-miR-106b-3p (2.14); hsa-miR-106b-5p (3.57); hsa-miR-25-3p (347.29); hsa-miR-25-5p (0); hsa-miR-27a-3p (142.57); hsa-miR-27a-5p (2.71) |
| Gao et al., 2016 | GeneChip miRNA array (Affymetrix), Real-time RT-PCR | 30 wild-type mice, 30 OPTN (E50K) transgenic mice |
OPTN (E50K) mutation = ↑ miR-141, miR-200a, miR-200b, miR-200c, miR-429 |
hsa-miR-141-3p (37.43); hsa-miR-141-5p (11.29); hsa-miR-200a-3p (25.14); hsa-miR-200a-5p (7.29); hsa-miR-200b-3p (15.29); hsa-miR-200b-5p (2.00); hsa-miR-200c-3p (202.14); hsa-miR-200c-5p (0.14); hsa-miR-429 (18.86) |
| He et al., 2018 | RT-PCR, Luciferase reporter assay | 5 replicates of Müller cells from rat retinas transfected with miR-124, anti-miR-124, or respective controls | ↑ miR-124 = ↑ RGC axon growth | hsa-miR-124-3p (0.86); hsa-miR-124-5p (0.14) |
| Li et al., 2018 | Real-time RT-PCR, Luciferase reporter assay | 5 sets of RGCs from rat transfected with miR-93-5p mimic or negative control and treated with NMDA | ↑ miR-93-5p = ↓ RGC autophagy via ↓ PTEN | hsa-miR-93-5p (31.49) |
| Nie et al., 2018 | Real-time RT-PCR, Luciferase reporter assay | 5 sets of RGCs exposed to elevated IOP transfected with miR-149 mimic, miR-149 inhibitor, or negative controls | ↓ miR-149 = ↓ RGC apoptosis | hsa-miR-149-5p (11.43) |
| Mak et al., 2020 | miRNA array (Agilent Technologies), Real-time RT-PCR, Luciferase reporter assay | 60 AAV-miR-19a-EGFP-transduced human adult RGCs, 61 AAV-EGFP-transduced human adult RGCs and triplicates of rat RGCs transfected with miR-19a mimic or scramble |
↑ miR-19a = ↑ axon regeneration via ↓ PTEN | hsa-miR-19a-3p (3.71) |
| Ou-Yang et al., 2020 | Luciferase reporter assay | Triplicates of rabbit RGCs transfected with miR-223 mimic, inhibitor, or control | ↑ miR-223 = ↑ apoptosis via ↓ HSP-70 | hsa-miR-223-3p (3.71); hsa-miR-223-5p (0) |
| Wang et al., 2021 | Real-time RT-PCR, Luciferase reporter assay | 3 control rats, 3 AOH rats, 3 agomir negative control + AOH rats, 3 miR-93 agomir + AOH rats | ↑ miR-93 = ↓ microglia activation and RGC death via ↓ STAT3 in rats | hsa-miR-93-3p (0.14); hsa-miR-93-5p (31.49) |
2.1. Aqueous Humor
There have been numerous studies to identify differentially expressed miRNAs in human AH samples. One of the first studies by Tanaka et al. (2014) compared 10 glaucoma patients with 3 different subtypes of glaucoma to 10 cataract controls using a miRNA array. This study identified 11 significantly upregulated and 18 significantly down-regulated miRNAs in AH from glaucoma patients that target genes involved in cell signaling and cell cycle checkpoint control (Tanaka et al., 2014). In a subsequent study, Liu et al. (2016) used the NEIGHBORHOOD dataset containing 3,853 POAG patients and 33,480 controls to assess the genetic association of 85 common variants within 76 MIR genes with POAG. The authors identified an association between POAG and a common variant, rs76481776, in the MIR182 gene, which was validated using ddPCR and small RNA-Seq (Liu et al., 2016). Another study compared miRNA expression between 6 POAG patients and 8 cataract controls using a miRNA array (Jayaram et al., 2017). This study showed that miR-518d and miR-143 were significantly upregulated, and miR-660 was significantly downregulated in AH of POAG patients (Jayaram et al., 2017). These miRNAs were predicted to negatively modulate pathways involved in cell proliferation, cell survival, extracellular matrix (ECM) remodeling, and adherents junction function (Jayaram et al., 2017).
Three additional studies were performed in 2018, comparing miRNA expression between glaucoma and control groups. One study examined expression changes of 800 pre-selected miRNAs between 12 POAG, 12 XFG patients, and 11 cataract controls using NanoString nCounter miRNA digital assays (Drewry et al., 2018). NanoString analysis revealed that miR-125b-5p, miR-302d-3p, and miR-451a were differentially expressed in POAG compared to controls, and miR-122-5p, miR-3144-3p, miR-320a, miR-320e, and miR-630 were differentially expressed in XFG compared to controls (Drewry et al., 2018). These results were validated using ddPCR in additional AH samples. Pathway analysis showed that these miRNAs are involved in potential glaucoma-related pathways, including focal adhesion, tight junctions, and TGF-β signaling (Drewry et al., 2018). In a second study, Liu et al. (2018) used small RNA-Seq to examine differences in miRNA expression between 9 POAG patients and 9 cataract controls. This study revealed 15 upregulated and 73 downregulated miRNAs in POAG patients, predicted to regulate thiamine and purine metabolism and transcriptional dysregulation, which may play a role in the development of glaucoma (Liu et al., 2018). Lastly, Yang et al. (2018) used real-time RT-PCR and a luciferase reporter assay to evaluate changes in miR-211 expression between 16 glaucoma patients and 8 cataract controls. The data showed that miR-211 was significantly upregulated in AH samples from glaucoma patients and targeted the Frs2 gene (Yang et al., 2018).
Two recent studies have also assessed differences in overall miRNA expression in AH samples taken from glaucoma patients and controls. Hindle et al. (2019) compared 6 POAG patients with 3 controls using a miRNA array and found 20 miRNAs that were significantly upregulated in POAG (Table 2). The other study compared 9 POAG patients and 10 cataract controls using small RNA-Seq (Hubens et al., 2021). This study found that miR-30a-3p, miR-143-3p, miR-211-5p, and miR-221-3p were upregulated, while miR-92a-3p, miR-451a, and miR-486-5p were downregulated in AH from POAG patients (Hubens et al., 2021). Some of these miRNAs were shown to be involved in glaucoma-related pathways such as TGF-β and neurotrophin signaling (Hubens et al., 2021).
Despite many studies on miRNA profiling with aqueous humor samples, no shared miRNAs with similar differential expression have been identified across these different studies. This discrepancy is likely due to experimental design issues, including but not limited to 1) small sample size of each study, 2) varying experimental techniques and kits to isolate and profile miRNAs, 3) clinical differences in glaucoma definition and subtypes, 4) different medicinal and surgical treatments to lower and maintain IOP levels in glaucoma patients involved in each study, 5) difference in sex, age, and ethnicity of the individuals in each study. To eliminate these issues, we recommend performing a large miRNA expression profiling study with hundreds of AH samples from glaucoma patients and controls with consistent clinical inclusion/exclusion criteria, detailed demographic information for stratified analysis, and clinical treatment history. It would also be necessary to use the same profiling technique, optimally small RNA-Seq, within the same facility with quality control measurements in place.
2.2. Tears
The study of miRNAs from tears is a relatively new development in the field of glaucoma. It provides a significant advantage compared to aqueous humor or tissue samples since tears are less invasive and easier to collect. However, the content of tear samples may not relate to glaucoma-affected ocular tissues and may not reflect the clinical status or progression of glaucoma in patients. Tear samples are subject to confounders such as ocular surface disease such as dry eye or corneal infection, but most relevant to glaucoma is effect of topical glaucoma drops on ocular surface health. Due to these limitations, only two studies have assessed miRNA expression changes in tears from glaucoma patients.
The first study compared expression of three selected miRNAs (miR-146b, miR-16, and miR-126), previously shown to be involved in several ophthalmic diseases, between 33 POAG patients and 29 controls using TaqMan-probe based real-time RT-PCR (Tamkovich et al., 2019). miR-146b, miR-16, and miR-126 were found to be significantly upregulated in POAG patients and may be involved in glaucoma progression (Tamkovich et al., 2019). The second study compared expression of 95 miRNAs between 20 POAG patients and 22 non-glaucoma individuals with ocular hypertension (OHT) but without neurodegeneration using small RNA-Seq (Raga-Cervera et al., 2021). The results showed that miR-26b-5p, miR-152-3p, miR-30e-5p, miR-125b-2-5p, miR-224-5p, and miR-27a-3p were upregulated and miR-151a-3p and miR-1307-3p were downregulated in tears from POAG patients compared to OHT controls (Raga-Cervera et al., 2021). Gene ontology (GO) analysis suggests that these miRNAs were linked to target genes controlling IOP, AH outflow, ECM function, apoptosis, and several other pathways (Raga-Cervera et al., 2021) (Table 2). Many of these target genes are associated with glaucoma or IOP, such as MYOC, OPTN, FOXC1, TGFβ, FAS/TNFR, TP53, CASPs, BCL2, MMPs, and DIABLO (Gharahkhani et al., 2021; Liu & Allingham, 2017; Wiggs & Pasquale, 2017; Xu et al., 2021). It should be noted that miRNAs were isolated from processed tear pellets by Tamkovich et al. (2019) or from total tears by Raga-Cervera et al. (2021), making it difficult to compare these two studies directly.
2.3. Trabecular Meshwork Cells
In addition to biological fluid samples from human patients, numerous studies have been performed using primary TM cells in culture from human donors or TM tissue from mice or rats (Table 2). Many of these studies use various stressors including senescence, cyclic mechanical stress (CMS), and oxidative stress, to mimic physiological or pathophysiological conditions. For example, oxidative stress has been shown to contribute to dysfunction of AH outflow by inducing apoptosis or senescence of TM cells (Ruibin et al., 2018; Wang et al., 2019). Senescence of TM cells may contribute to a pathological decrease in AH outflow leading to elevated IOP and glaucoma (Li et al., 2009). In terms of normal physiology, TM cells are exposed to CMS due to ocular pulse created by the cardiac cycle, CMS is hypothesized to be involved in homeostatic feedback loop that to regulate AH outflow in (Luna et al., 2011a; Madekurozwa et al., 2021). Introduction of these stressors can make studies more physiologically or pathophysiological relevant and provide insights into specific disease mechanisms.
The leading studies investigating miRNA expression in human TM cells were done by Dr. Pedro Gonzalez’s group at Duke University over the span of four years. The first study in 2009 examined changes in miRNA expression associated with stress-induced premature senescence (SIPS), a type of proliferative arrest due to various stresses, such as oxidative stress or DNA damage (Li et al., 2009). The miRNA array using human TM (HTM) cells from three donors showed that 38 miRNAs were significantly differentially expressed in senescent HTM cells compared to non-senescent controls (Li et al., 2009). Real-time RT-PCR confirmed these results for 19 miRNAs (Li et al., 2009) (Table 2). Luciferase assays showed that miR-106a and miR-106b target p21 and miR-182 targets RARG (Li et al., 2009). Transfection of HTM cells with miR-106a mimic increased cell proliferation and transfection of HTM cells with miR-182 mimic increased SA-beta-galactosidase activity (Li et al., 2009). Another study examined the effects of miR-29b in HTM cells from one donor using real-time RT-PCR and validated its target genes using a luciferase assay (Luna et al., 2009). The luciferase assay showed that miR-29b directly targets BMP1, ADAM12, and NKIRAS2 and real-time RT-PCR after transfection of HTM cells with miR-29b mimic confirmed these results (Luna et al., 2009). Transfection of HTM cells with miR-29b mimic also resulted in downregulation of several other ECM components including six collagens, LAMC1, and FBN which may lead to decreased AH outflow and elevated IOP in glaucoma (Luna et al., 2009).
The subsequent study by this group focused on the effects of miR-183 differential expression in HTM cells that were identified in their first study (Li et al., 2010a). Luciferase assays showed that miR-183 targets ITGB1 and KIF2A (Li et al., 2010a). Transfection of HTM cells with the miR-183 mimic resulted in decreased cell adhesion and increased phagocytic activity (Li et al., 2010a). The authors concluded that miR-183 could play a role in the function of neurosensory organs, such as the retina, the inner ear, and the olfactory system, and contribute to functional alterations associated with cellular senescence in HTM cells (Li et al., 2010a). Another study from 2010 examined changes in miRNA expression during replicative senescence in HTM cells from two donors (Li et al., 2010a). Real-time RT-PCR indicated that expression of 18 miRNAs was altered, including upregulation of miR-146a (Li et al., 2010a). Transfection of HTM cells from three additional donors with miR-146a mimic resulted in downregulation of CCL2, CXCL3, IL11, SLC10A3, PTGS1, IL6, GALNT10, CCL20, CXCL6, PPP2R1B, SERPINE1, HAS1, IL8, and IRAK1 (Li et al., 2010a). Of these 14 genes, seven were upregulated and two were downregulated in senescent HTM cells from two donors (Li et al., 2010a) (Table 2). The next study examined the effect of miR-204 on gene expression and cell function in normal HTM cells (Li et al., 2011). Real-time RT-PCR of HTM cells from two donors showed downregulation of 28 genes after miR-204 mimic transfection (Li et al., 2011) (Table 2). Luciferase assays confirmed that miR-204 targets 12 of these genes including AP1S2, BCL2L2, BIRC2, EDEM1, EZR, FZD1, M6PR, RAB22A, RAB40B, SERP1, TCF12, and TCF4 (Li et al., 2011). The study also showed that miR-204 increased apoptosis, cytotoxicity, accumulation of carbonylated proteins, and expression of ER stress markers and decreased inflammatory factors in HTM cells (Li et al., 2011).
Another study by Dr. Gonzalez’s group continued their 2009 work on miR-29 by examining the effect of TGF-β1 or TGF-β2 treatment on miR-29 expression (Luna et al., 2011a). Real-time RT-PCR with primary HTM cells showed that TGF-β2 significantly decreased expression of miR-29 while TGF-β1 had no uniform effect, sometimes increasing and other times not affecting expression of miR-29 (Luna et al., 2011a). Luciferase assays showed that miR-29b significantly decreased TGF-β2 mRNA expression in HTM cells from all three donors and decreased TGF-β2 protein expression in HTM cells from two out of three donors (Luna et al., 2011a). Another study by Luna et al. (2011a) examined the effect of miR-24 transfection on HTM cells and changes in miRNA expression in response to CMS. miRNA PCR arrays showed that miR-16, miR-27a, miR-27b, miR-7, let-7f, miR-26a, and miR-24 were upregulated in HTM cells from three donors after CMS (Luna et al., 2011a). Luciferase assays confirmed that miR-24 directly targets FURIN, a gene known to play a significant role in processing TGF-β1 (Luna et al., 2011a). Likewise, real-time RT-PCR from primary HTM cells showed that TGF-β1 significantly increased miR-24 expression (Luna et al., 2011a). In 2012, Luna et al. examined the effects of miR-200c overexpression on HTM cells and found that miR-200c downregulates the expression of ETAR, LPAR1, FHOD1, ZEB1, and ZEB2, genes involved in TM cell contraction (Luna et al., 2012). The luciferase assay confirmed that miR-200c targets ETAR, LPAR1 and RHOA. The study also showed that transfection with miR-200c significantly reduces cell contractility and traction stress in HTM cells and lowers IOP in rats (Luna et al., 2012). In summary, Dr. Gonzalez’s group first screened for miRNAs that could be involved in glaucoma using a miRNA array, then validated several of these miRNAs in subsequent studies in various physiological conditions. Some of the miRNAs identified by this group could serve as the focus of continued research.
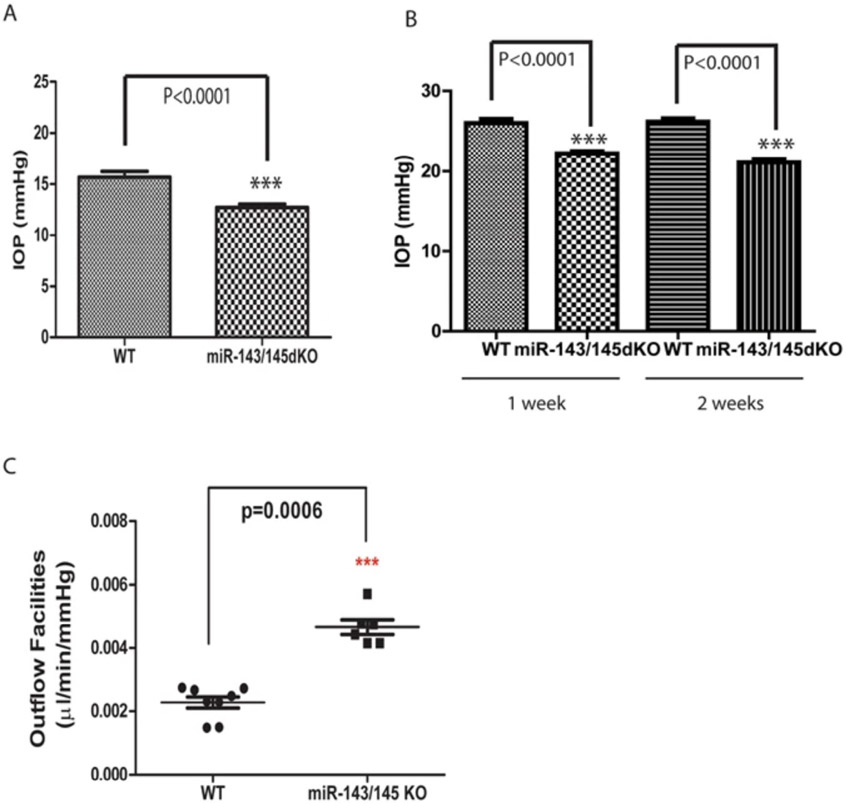
In addition to these studies, numerous other studies have investigated the role of candidate miRNAs in TM cellular functions. One study investigated how miR-204 and miR-211 regulate FOXC1 expression in primary HTM cells from two donors (Paylakhi et al., 2013). Real-time RT-PCR showed that transfection of HTM cells with the miR-204 and miR-211 mimetics significantly reduced FOXC1 expression, a transcription factor involved in glaucoma (Paylakhi et al., 2013). Another study in 2015 examined the effect of miR-483-3p on ECM production in primary HTM cells from one donor (Shen et al., 2015). Real-time RT-PCR showed that miR-483-3p expression was decreased after H2O2-induced oxidative stress (300 μM) and that overexpression of miR-483-3p inhibited the expression of several ECM genes including fibronectin, laminin, and collagen I (Shen et al., 2015). Luciferase assays confirmed that miR-483-3p directly targets SMAD4, leading to decreased ECM production (Shen et al., 2015). A 2016 study by Wang et al. aimed to determine the effect of miR-93 on apoptosis in glaucomatous HTM cells. Real-time RT-PCR showed that miR-93 was significantly upregulated 4-fold in glaucomatous HTM cells compared to control HTM cells (Wang et al., 2016). Inhibition of miR-93 expression led to increased cell viability and suppression of apoptosis in glaucomatous TM cells via increased NFE2L2 protein expression (Wang et al., 2016). Another study showed that miR-143 and miR-145 are highly expressed in primary HTM cells from three donors using real-time RT-PCR (Li et al., 2017). The luciferase assays showed that miR-143/145 directly targets genes involved in actin dynamics and contractility of TM cells including ARPC-2, −3 and −5, PDGFRA, ABLIM-2 and MLCK (Li et al., 2017). Figure 2 adapted from Li et al. (2017) shows that targeted deletion of miR-143/145 in mice resulted in significantly reduced IOP and increased AH outflow (Li et al., 2017). Altogether, these studies identify a wide range of effects due to differential expression of several miRNAs, including gene expression, ECM production, and apoptosis.
Fig. 2. Reduced IOP and increased outflow facilities in miR-143/145 dKO mice.
(A) Reduced intraocular pressure (IOP) in miR-143/145 dKO mice. ***p < 0.0001; (B) Reduced intraocular pressure in miR-143/145 dKO mice in an experimental glaucoma model. The IOP of the mice was measured at 1 and 2 weeks after microbeads injection into the eye. ***p < 0.0001; (C) Significant increase in outflow facilities in 16-month-old miR-143/145 dKO mice compared to that in WT control mice. ***p < 0.0006. Figure adapted from Li et al. (2017) with permission.
The following few studies focused on the effects of several miRNAs on TM cells under oxidative stress. The first study by Ruibin et al. (2018) examined the role of miR-1298 in TM cells under chronic oxidative stress (COS). COS was induced by incubating the cells in a 40% oxygen mixture with 5% CO2 for 4–5 days (Ruibin et al., 2018). Real-time RT-PCR showed that miR-1298 was significantly downregulated in HTM cells after COS (Ruibin et al., 2018). The luciferase assay in primary HTM cells from one donor showed that miR-1298 directly targets EIF4E3 to reduce its expression, which was confirmed by real-time RT-PCR (Ruibin et al., 2018). Transfection of primary HTM cells from two donors with miR-1298 mimics decreased cell cytotoxicity induced by COS, potentially by inhibiting the TGF-β2/Smad4 pathway and activating the canonical Wnt pathway (Ruibin et al., 2018). Another group investigated the effect of miR-181a on H2O2-induced apoptosis in primary HTM cells using increasing concentrations of H2O2 (0, 50, 100, 200, 300 μM) (Wang et al., 2018). Real-time RT-PCR with HTM from one donor showed that miR-181a expression decreased after H2O2 treatment. Overexpression of miR-181a improved TM cell survival and suppressed H2O2-induced apoptosis by inhibiting the NF-κB and JNK signaling pathways (Wang et al., 2018).
A study in 2019 investigated the effects of miR-17-5p on proliferation and apoptosis in primary HTM cells from one donor in response to oxidative stress. Oxidative stress was induced by exposing HTM cells to 300 μM of H2O2 for 3-7 hours (Wang et al., 2019). Real-time RT-PCR showed that H2O2 exposure decreased the expression of miR-17-5p and increased cell apoptosis in HTM cells (Wang et al., 2019). Luciferase assays showed that miR-17-5p directly targets PTEN (Wang et al., 2019). Subsequently, overexpression of miR-17-5p in HTM cells induced a significant increase in cell proliferation and a decrease in apoptosis by reducing the expression of PTEN and the apoptosis-related proteins BAX, BCL- XL, and BCL- 2 (Wang et al., 2019). Another study in 2019 investigated how Salidroside (Sal) treatment affected HTM cells exposed to oxidative stress induced by exposing cells to increasing concentrations of H2O2 (0, 50, 100, 200, 300, 400 μM) (Zhao et al., 2019). Real-time RT-PCR showed that Sal treatment upregulated miR-27a in cells both with and without H2O2 exposure (Zhao et al., 2019). The study also found that transfection of HTM cells with miR-27a inhibitor decreased cell viability and increased apoptosis. Likewise, cells treated with Sal after H2O2 exposure exerted protective effects in HTM cells by upregulating miR-27a, which activated the PI3K/AKT and Wnt/β-catenin pathways (Zhao et al., 2019). Likewise, suppression of miR-27a significantly reversed the protective effect of Sal (Zhao et al., 2019). Shen et al. (2020) investigated the effects of miR-200c-3p in primary HTM cells from one donor after oxidative stress, induced by treatment with 300 μM H2O2 for 2 hours. Real-time RT-PCR showed that miR-200c-3p was downregulated in HTM cells under oxidative stress (Shen et al., 2020). Like Wang et al. (2019), luciferase assays showed that miR-200c-3p targeted PTEN and overexpression of miR-200c-3p led to decreased PTEN expression, inhibition of apoptosis, and promotion of cell proliferation (Shen et al., 2020). Collectively these studies on oxidative stress show that various miRNAs can protect HTM cells from cytotoxicity and apoptosis due to oxidative stress. However, one study by Liu et al. (2021) described contrasting effects in which miR-29b-3p is not protective and may induce apoptosis in HTM cells. It would still be interesting to determine if some shared regulators mediate these protective effects. It is also necessary to validate these findings using consistent oxidative stress models.
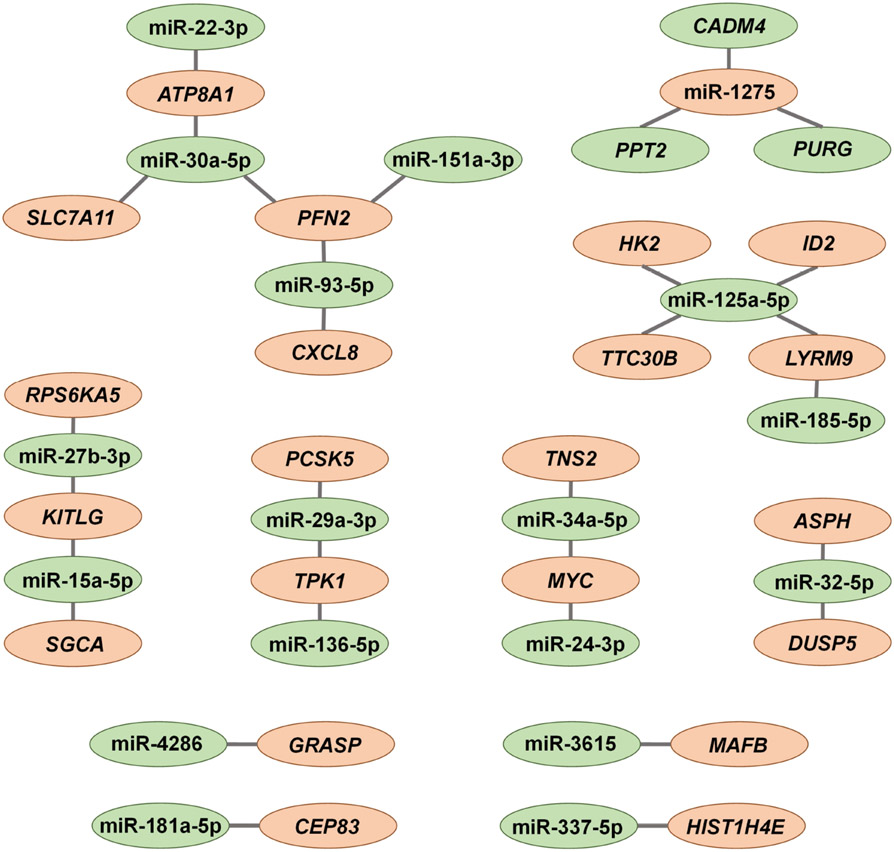
The most recent study in 2020 investigated changes in miRNA expression after CMS in primary HTM cells from five donors. NanoString-based miRNA assays identified 39 upregulated and 3 downregulated miRNAs in HTM cells after CMS (Youngblood et al., 2020) (Table 2). Three miRNAs, miR-27a-3p, miR-29a-3p, and miR-181c-5p, were confirmed to be upregulated by ddPCR (Youngblood et al., 2020). Further analysis using Ingenuity Pathway Analysis (IPA) revealed that 18 of the stretch responsive miRNAs target 24 mRNA genes with differential expression in the opposite direction compared to miRNA expression changes, summarized in Figure 3 adapted from Youngblood et al. (2020). This integrative analysis of miRNA-mRNAs from the same samples provides direct evidence on their regulatory interaction.
Fig. 3.
Gene network of stretch-responsive miRNAs (an absolute FC > 1.3, P < 0.05) and their validated target genes (an absolute FC > 2, P < 0.05). Upregulated RNAs appear in green and downregulated RNAs appear in orange. Figure adapted from Youngblood et al. (2020) with permission.
In comparison to miRNA screening studies in the area of aqueous humor or tear samples, these TM cell-based miRNA studies focused on cellular responses or phenotypes relevant to glaucoma such as cellular phagocytosis, oxidative stress, and contractility as well as in vivo studies using transgenic mice (Gonzalez et al., 2014; Li et al., 2017). These studies provide critical insights into relevant molecular pathways such as apoptosis, cell survival, or ECM maintenance (Gonzalez et al., 2014). The in vivo studies also provide important direction on how to determine their clinical relevance (Li et al., 2017).
2.4. Retina
Since glaucoma is defined by RGC loss and optic nerve cupping, the retina is a critical ocular region where the role of miRNA in disease progression has been studied. However, it is essential to note that the proportion of RGC is minimal compared to other cell types in the retina (Menon et al., 2019; Shekhar & Sanes, 2021; Yamagata et al., 2021; Yan et al., 2020). The first study in 2015 aimed to identify differentially expressed miRNAs in glaucomatous retina compared to non-glaucomatous retina from rats (Jayaram et al., 2015). Real-time RT-PCR with 17 preselected miRNA candidates using retina samples from 8 glaucoma and 8 control rats identified that eight miRNAs (miR-181c, miR-497, miR-204, let-7a, miR-29b, miR-16, miR-106b, and miR-25) were downregulated and miR-27a was upregulated significantly in glaucomatous retina compared to control retina (Jayaram et al., 2015). The next study in 2016 investigated the role of miRNAs in POAG induced by a mutation in OPTN (E50K) using 30 transgenic and 30 control mice with miRNA arrays (Gao et al., 2016). This mouse study identified five miRNAs that were upregulated (miR-141, miR-200a, miR-200b, miR-200c, and miR-429) in the retina of transgenic mice with the OPTN (E50K) mutation, followed by real-time RT-PCR confirmation (Gao et al., 2016) (Table 2).
In contrast to these two screening studies, several other studies investigated the functional role of a single specific miRNA in glaucoma. He et al. (2018) studied the role of miR-124 in axon growth of RGCs re-differentiated from rat Müller cells. RT-PCR showed that two genes involved in axon growth, Sema3A and Nrp-1, were significantly upregulated in miR-124 transfected cells suggesting that miR-124 indirectly promotes axon growth (He et al., 2018). Luciferase assays confirmed that miR-124 targets COREST, a protein that suppresses Nrp-1 transcription (He et al., 2018). Therefore, miR-124 was hypothesized to promote RGC axon growth by downregulating COREST and increasing transcription of Nrp-1 (He et al., 2018). Another study in 2018 focused on the role of miR-93-5p in a less commonly used NMDA-induced rat glaucoma model (Li et al., 2018). The study found that miR-93-5p expression decreased with the treatment of increased concentrations of NMDA by targeting PTEN through the AKT/mTOR pathway (Li et al., 2018). Overexpression of miR-93-5p in rat RGCs resulted in suppressed autophagy of the RGCs after NMDA treatment (Li et al., 2018). In another study by Nie et al. (2018) tested the effects of miR-149 on RGCs in a mouse model of glaucoma with elevated IOP induced by episcleral venous occlusion with cauterization. Luciferase assays confirmed that miR-149 targets BTC (betacellulin) (Nie et al., 2018). Real-time RT-PCR showed that downregulation of miR-149 increased expression of Btc, Pi3k, and Akt, leading to increased viability and decreased apoptosis of RGCs (Nie et al., 2018).
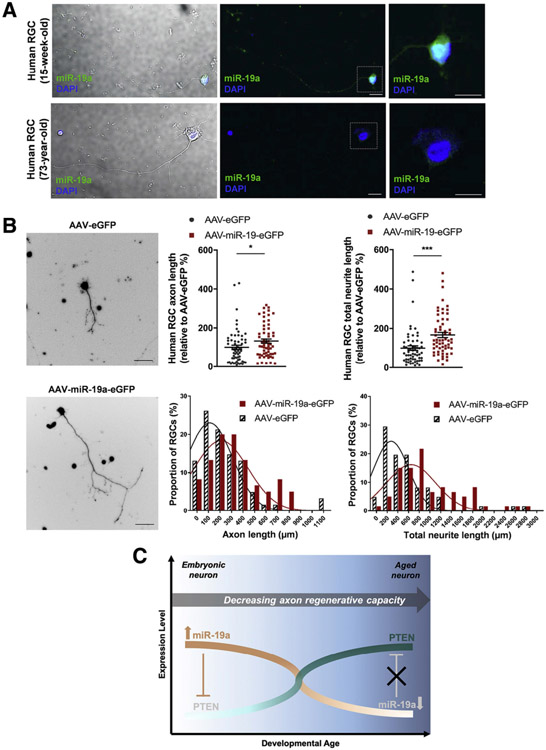
Additionally, a 2020 study aimed to determine the role of miR-19a in axon regeneration (Mak et al., 2020). Using a miRNA array, this study identified that miR-19a expression declined in parallel with decreased axon regenerative capacity in RGCs during development (Mak et al., 2020). Luciferase assays revealed that miR-19a targets Pten (Mak et al., 2020). Furthermore, injection of miR-19a mimic into the vitreous cavity in mice promotes axon regeneration after optic nerve crush in vivo. Figure 4A, adapted from Mak et al. (2020), shows that miR-19a is expressed at a higher level in human fetal RGCs compared to aged human RGCs suggesting that miR-19a is involved in RGC and axon development. Figure 4B shows that miR-19a overexpression in aged RGCs increases axon regeneration and total neurite length compared to controls (Mak et al., 2020). Figure 4C summarizes the results by showing that miR-19a expression increases axon regenerative capacity through suppression of Pten, and this effect decreases with age (Mak et al., 2020).
Fig. 4. miR-19a promotes axon regeneration in human adults RGCs.
(A) In situ hybridization images of endogenous miR-19a expression (green) in purified fetal (15-week-old) and adult (73-year-old) human RGCs at 7 days in vitro. Images on the right are magnifications of boxed areas. miR-19a was most prominently detected in the cytoplasm and decreased in expression from fetal to adult human RGCs. Scale bars, 10 μm. (B) Fluorescence images of purified human adult RGCs transduced with AAV-EGFP or AAV-miR-19a-EGFP (left). Scale bars, 25 μm. Single-cell analysis showed that AAV-miR-19a-EGFP-transduced human adult RGCs (n = 60 RGCs; 2 experimental replicates) had longer axon lengths and total neurite lengths compared with AAV-EGFP-transduced human adult RGCs (n = 61 RGCs; 2 experimental replicates) at 14 days in vitro (top) (axon length, p = 0.041; total neurite length, p = 0.0002; total n = 121 RGCs purified from two donors aged 69 years and 75 years). Unpaired two-tailed Student’s t test was used for all comparisons; *p < 0.05; ***p < 0.001. All values are shown as mean ± SEM. The proportions of RGCs by axon length and total neurite length between AAV-EGFP transduced and AAV-miR-19a-EGFP-transduced human RGCs are represented by histograms (bottom). The Gaussian distribution of AAV-EGFP and AAV-miR-19a- EGFP is indicated by an overlapping curve. (C) A schematic illustrating the reciprocal relationship of endogenous expression of miR-19a and PTEN in RGCs during development and their association with the decline in axon regenerative capacity. Figure adapted from Mak et al. (2020) with permission.
Another study examined the effect of miR-223 on rabbit RGC apoptosis (Ou-Yang et al., 2020). The carbomer-induced rabbit glaucoma model showed thinning of the retina, reduced numbers of RGCs, and increased RGC apoptosis (Ou-Yang et al., 2020). Glaucoma rabbits injected with antagomiR-223 showed the opposite, with normal retinal layers, densely arranged RGCs, and decreased RGC apoptosis (Ou-Yang et al., 2020). A luciferase assay showed that HSP-70 is a potential target of miR-223, suggesting that miR-223 promotes apoptosis of RGCs by inhibiting expression of HSP-70 (Ou-Yang et al., 2020). The most recent study investigated the role of miR-93 in modulating microglial reactivity and protecting RGCs in a rat model of acute ocular hypertension (AOH) (Wang et al., 2021). Real-time RT-PCR of rat retinas (n=3) showed that miR-93 was downregulated, and the inflammatory cytokines, TNF-α and IL-1β, were upregulated, indicating microglial activation in the AOH rats (Wang et al., 2021). Injection of a miR-93 agomir into the vitreous body of AOH rats significantly reduced cytokine expression, microglial activation, inflammation, and RGC death (Wang et al., 2021). Luciferase assays confirmed that miR-93 might target STAT3 to exert its effect on RGC death (Wang et al., 2021) (Table 2).
Due to the limited availability of glaucoma-affected postmortem human donor eyes, almost all the retina-related miRNA studies were conducted in various animal models. For example, Morrison et al. (2016) assessed gene expression changes after controlled acute elevation of IOP in rats, finding changes consistent with a chronic rat glaucoma model induced by episcleral vein injection of hypertonic saline. This type of validation of animal models is essential for future miRNA studies to ensure that the miRNA variation is not due to other factors such as sex, age, species, and genetic background. It will also be necessary to validate the role of the identified retinal miRNAs in human glaucoma patients using population-based glaucoma genetics data and functional genomics data in human retina tissue. A similar effort has been made in the research area of age-related macular degeneration (AMD), where expression quantitative trait loci (eQTL) and transcriptome-wide association analyses (TWAS) were integrated with single-nucleus RNA-Seq in human retina and retinal pigment epithelium to elucidate genetic pathways and potential therapeutic target for AMD (Menon et al., 2019; Morgan & DeAngelis, 2014; Orozco et al., 2020; Ratnapriya et al., 2019). It will be important to generate a relevant resource of glaucoma-affected human donor retina with gene expression data from bulk or single-cell RNA-Seq and small RNA-Seq (Liu & Allingham, 2017).
3. Therapeutic Applications of MicroRNAs
There are currently 16 miRNA-based therapies in the development phase for treatment of numerous diseases including cancer, hepatitis C virus infection, polycystic kidney disease, and ALS across the world (Chakraborty et al., 2021). Several preliminary studies indicate that miRNA-based therapy may be a promising treatment strategy for glaucoma. Current treatments for glaucoma, both pharmacological or surgical, often only delay or prevent the further progression of glaucoma (Youngblood et al., 2020). For a detailed review of general glaucoma treatment, please refer to a previous review article by Weinreb et al. (2014). miRNA-based therapies may provide an additional treatment option for glaucoma that could target the underlying molecular cause directly. This is because miRNA-based therapies are designed to target multiple genes in the same pathway and miRNA mimic oligos are relatively easily synthesized in large quantities with high purity (Chakraborty et al., 2021). Some disadvantages of miRNA-based therapies are potential off-target effects, activation of the immune system, and degradation of the miRNA prior to and during the delivery to the target tissue (Chakraborty et al., 2021).
There have been several studies to address the challenge of miRNA protection and delivery for the treatment of glaucoma. Most studies focus on the use of nanoparticles for miRNA delivery. In two consecutive studies, Tan et al. found that miR-21-5p was successfully delivered to the eye using polydopamine-polyethylenimine nanoparticles (PDA/PEI NPs), which significantly reduced IOP by modulating the expression of SMAD7 and FGF18, genes involved in AH outflow function (Tan et al., 2021; Tan et al., 2020). In a separate study, Li et al. (2020) found that miR-124 delivery using nanoparticles containing brinzolamide lowered IOP and prevented RGC damage. It remains unknown how this nanoparticle-based treatment compares to brinzolamide treatment alone without the use of nanoparticles. Another study injected miRNA containing exosomes collected from bone marrow-derived mesenchymal stem cells (BMSC) into the vitreous, which resulted in improved RGC survival and axonal regeneration in a rat optic nerve crush model (Mead et al., 2018; Mead & Tomarev, 2017). All of these studies demonstrate the plausibility of using nanoparticles or exosomes to deliver miRNAs to the target tissue as a potential glaucoma treatment.
There is significant evidence that miRNAs play an important role in glaucoma pathogenesis, as presented in this review. In the TM, miRNAs have been shown to affect ECM maintenance, apoptosis, and senescence, all of which could alter AH outflow homeostasis and lead to elevated IOP and glaucoma (Li et al., 2009, 2010a, 2011; Luna et al., 2009; Shen et al., 2020; Wang et al., 2019; Wang et al., 2016; Wang et al., 2018). In the retina, miRNAs may affect RGC death and axon regeneration, which could contribute to the visual field loss seen in glaucoma patients (He et al., 2018; Li et al., 2018; Mak et al., 2020; Nie et al., 2018; Ou-Yang et al., 2020; Wang et al., 2021). Many of these miRNAs could be a target for a miRNA-based therapy for glaucoma once the challenge of delivery is solved. However, one additional obstacle in developing a miRNA-based therapy for glaucoma is the lack of overlap in the miRNAs shown to be involved in glaucoma. In this review, there were only eight miRNAs that were identified by 2 or more studies that were not part of a targeted study. These miRNAs include miR-143, miR-221, miR-486, miR-4725, miR-125b, miR-451a, miR-92a, and miR-99b. . This could be due to the wide variety of animal models used, biological variations of the animals used (sex, age, and strains), small sample size, demographic differences in human samples, clinical differences in glaucoma types, or patient use of glaucoma treatments. POAG is a complex disorder that likely has many forms. More research is needed to identify the miRNAs that are most significantly involved in glaucoma pathogenesis and the effectiveness of targeting these miRNAs in human patients from various ethnic backgrounds.
4. Future Directions
Numerous miRNAs appear to be involved in the pathogenesis of glaucoma and need additional follow-up for further functional validation and replication. In an effort to further establish their potential connections with glaucoma- or IOP-associated genes identified through GWAS, we performed a miRNA-mRNA interaction analysis using IPA software. We derived glaucoma- or IOP-associated genes from three recent GWAS studies of POAG and IOP (Gharahkhani et al., 2021; Khawaja et al., 2018; MacGregor et al., 2018) and examined whether these POAG- or IOP-associated genes could be targeted by the identified miRNAs listed in Table 2. We identified a number of glaucoma-/IOP-associated genes that could be targeted by the reported miRNAs in the literature (Table 3). These genes include AFAP1, ATXN2, CAV2, ETS1, FNDC3B, FOXC1, GAS7, GLIS3, LMX1B, TMCO1, TNXB, and TXNRD2. Several of the identified miRNAs are highly expressed in non-glaucoma HTM tissues, with a normalized total read number above 500 in Table 2. These miRNAs include miR-182-5p, miR-143-3p, miR-125b-5p, miR-211-5p, miR-92a-3p, miR-16-5p, miR-126-5p, miR-204-5p, miR-21-5p, miR-145-5p, miR-181a-5p, miR-100-5p, and miR-7a-5p. It would be interesting to investigate whether the expression level of these miRNAs is changed in glaucomatous TM tissue and how these changes may affect TM function in vivo or ex vivo. If specific miRNAs are identified to alter TM function in a way that contributes to glaucoma pathogenesis, those miRNAs represent a potential therapeutic target for the treatment of glaucoma. Table 3 could also serve as a guide for determining which miRNAs may be of interest for further research in the TM, the retina, or other glaucoma-related tissues.
Table 3.
This table presents the results from IPA. We cross-referenced all the miRNAs from Table 2 to the genes shown to be associated with POAG or IOP from GWAS studies performed by Khawaja, et al., 2018 MacGregor, et al., 2018, and Gharahkhani et al., 2021. Column 4 lists the pathways affected by the corresponding miRNAs in column 3. Superscripts indicate whether the miRNAs have been experimentally observed2 to affect the listed pathways or whether IPA predicts with high confidence1 that the miRNAs affect the listed pathways.
| Gene Symbol |
Gene Name | miRNAs | Pathways Effected |
|---|---|---|---|
| ADAM12 | ADAM metallopeptidase domain 12 | hsa-miR-1491 | Axonal Guidance Signaling, Inhibition of Matrix Metalloproteases |
| AFAP1 | actin filament associated protein 1 | hsa-miR-6371 | Not in a pathway |
| ANAPC1 | anaphase promoting complex subunit 1 | hsa-miR-518d1 | Kinetochore Metaphase Signaling Pathway, Mitotic Roles of Polo-Like Kinase, Protein Kinase A Signaling, Protein Ubiquitination Pathway, Senescence Pathway |
| ANTXR1 | ANTXR cell adhesion molecule 1 | hsa-miR-3173-3p1, hsa-miR-12751 | Not in a pathway |
| ARHGAP20 | Rho GTPase activating protein 20 | hsa-miR-5001-5p1 | Not in a pathway |
| ARHGEF12 | Rho guanine nucleotide exchange factor 12 | hsa-miR-1821 | Actin Cytoskeleton Signaling, Axonal Guidance Signaling, Breast Cancer Regulation by Stathmin1, Molecular Mechanisms of Cancer, Phospholipase C Signaling, Reelin Signaling in Neurons, RHOA Signaling, RHOGDI Signaling, Semaphorin Neuronal Repulsive Signaling Pathway, Semaphorin Signaling in Neurons, Signaling by Rho Family GTPases, Thrombin Signaling |
| ATXN2 | Ataxin-2 | hsa-miR-92a-2-5p1, hsa-miR-6081, hsa-miR-46631, hsa-miR-161 | Not in a pathway |
| BCAS3 | BCAS3 microtubule associated cell migration factor | hsa-miR-4725-3p1, hsa-miR-127-3p1 | Not in a pathway |
| BTBD3 | BTB domain containing 3 | hsa-miR-44671, hsa-miR-152-3p1 | Not in a pathway |
| CAPZA1 | capping actin protein of muscle Z-line subunit alpha 1 | hsa-miR-61321 | Not in a pathway |
| CAV2 | caveolin 2 | hsa-miR-29b1,2 | G Beta Gamma Signaling |
| CDH11 | cadherin 11 | hsa-miR-2112 | Gα12/13 Signaling, RHOGDI Signaling, Signaling by Rho Family GTPases, Synaptogenesis Signaling Pathway |
| CLIC5 | chloride intracellular channel 5 | hsa-miR-4640-5p1, hsa-miR-1306-5p1, hsa-miR-34a-5p1,2 | Not in a pathway |
| COL4A1 | collagen type IV alpha 1 chain | hsa-miR-29b1,2, hsa-miR-30e-5p2, hsa-miR-6371, hsa-miR-1241 | GP6 Signaling Pathway, Hepatic Fibrosis / Hepatic Stellate Cell Activation |
| COL8A2 | collagen type VIII alpha 2 chain | hsa-miR-4749-5p1 | GP6 Signaling Pathway,Hepatic Fibrosis / Hepatic Stellate Cell Activation |
| CPXM1 | carboxypeptidase X, M14 family member 1 | hsa-miR-3663-3p1 | Not in a pathway |
| DLL1 | delta like canonical Notch ligand 1 | hsa-miR-34a-5p1,2, hsa-miR-161 | Epithelial Adherens Junction Signaling, Notch Signaling, Th1 and Th2 Activation Pathway, Th1 Pathway, Th2 Pathway |
| EMCN | endomucin | hsa-miR-486-3p1 | Not in a pathway |
| ETS1 | ETS proto-oncogene 1, transcription factor | hsa-miR-152-3p1, hsa-miR-1260b1, hsa-miR-377-3p1, hsa-miR-187-3p1 | B Cell Receptor Signaling, ERK/MAPK Signaling, GM-CSF Signaling, HER-2 Signaling in Breast Cancer, HGF Signaling, MSP-RON Signaling In Cancer Cells Pathway, Regulation Of The Epithelial Mesenchymal Transition By Growth Factors Pathway, Regulation of the Epithelial-Mesenchymal Transition Pathway, Renal Cell Carcinoma Signaling, Senescence Pathway, Sumoylation Pathway, Telomerase Signaling |
| EXOC2 | exocyst complex component 2 | hsa-miR-36901 | CDC42 Signaling, Remodeling of Epithelial Adherens Junctions |
| FANCA | FA complementation group A | hsa-miR-5001-5p1 | Hereditary Breast Cancer Signaling, Role of BRCA1 in DNA Damage Response |
| FERMT2 | FERM domain containing kindlin 2 | hsa-miR-433-3p1 | ILK Signaling |
| FMNL2 | formin like 2 | hsa-miR-22-3p1 | Not in a pathway |
| FNDC3B | fibronectin type III domain containing 3B | hsa-miR-1432, hsa-miR-162 | Not in a pathway |
| FOXC1 | forkhead box C1 | hsa-miR-2111 | Transcriptional Regulatory Network in Embryonic Stem Cells |
| GAB2 | GRB2 associated binding protein 2 | hsa-miR-4725-3p1, hsa-miR-36521, hsa-miR-46631, hsa-miR-4667-5p1, hsa-miR-5001-5p1, hsa-miR-42861 | B Cell Receptor Signaling, Chronic Myeloid Leukemia Signaling, Fcγ Receptor-mediated Phagocytosis in Macrophages and Monocytes, FLT3 Signaling in Hematopoietic Progenitor Cells, ICOS-ICOSL Signaling in T Helper Cells, IL-3 Signaling, Molecular Mechanisms of Cancer, MSP-RON Signaling In Macrophages Pathway, Phagosome Formation, PI3K/AKT Signaling, Thrombopoietin Signaling |
| GAS7 | growth arrest specific 7 | hsa-miR-29b2, hsa-miR-61321, hsa-miR-6371 | Not in a pathway |
| GLIS3 | GLIS family zinc finger 3 | hsa-miR-302d-3p1, hsa-miR-931 | Not in a pathway |
| GMDS | GDP-mannose 4,6-dehydratase | hsa-miR-4749-5p1, hsa-miR-26b-5p1 | Colanic Acid Building Blocks Biosynthesis, GDP-L-fucose Biosynthesis I (from GDP-D-mannose) |
| HHEX | hematopoietically expressed homeobox | hsa-miR-2111, hsa-miR-1451 | Not in a pathway |
| HLA-DQA1 | major histocompatibility complex, class II, DQ alpha 1 | hsa-miR-4640-5p1, hsa-miR-6717-5p1 | Allograft Rejection Signaling, Altered T Cell and B Cell Signaling in Rheumatoid Arthritis, Antigen Presentation Pathway, Autoimmune Thyroid Disease Signaling, B Cell Development, Calcium-induced T Lymphocyte Apoptosis, CD28 Signaling in T Helper Cells, CDC42 Signaling, Dendritic Cell Maturation, Glucocorticoid Receptor Signaling, Graft-versus-Host Disease Signaling, ICOS-ICOSL Signaling in T Helper Cells, IL-4 Signaling, MSP-RON Signaling In Macrophages Pathway, Neuroinflammation Signaling Pathway, +13 |
| IGF1 | insulin like growth factor 1 | hsa-miR-483-3p1,2, hsa-miR-27a-3p2, hsa-miR-162, hsa-miR-12751 | Amyotrophic Lateral Sclerosis Signaling, Autophagy, Axonal Guidance Signaling, Breast Cancer Regulation by Stathmin1, Cardiac Hypertrophy Signaling, Cardiac Hypertrophy Signaling (Enhanced), Clathrin-mediated Endocytosis Signaling, CREB Signaling in Neurons, Estrogen Receptor Signaling, Estrogen-Dependent Breast Cancer Signaling, Glioblastoma Multiforme Signaling, STAT3 pathway, Glucocorticoid Receptor Signaling, Growth Hormone Signaling, Hepatic Fibrosis / Hepatic Stellate Cell Activation, + 13 |
| IL34 | interleukin 34 | hsa-miR-31-5p1 | Neuroinflammation Signaling Pathway |
| KALRN | kalirin RhoGEF kinase | hsa-miR-92a-3p1 | Axonal Guidance Signaling, Ephrin B Signaling, Ephrin Receptor Signaling, Synaptogenesis Signaling Pathway |
| KBTBD8 | kelch repeat and BTB domain containing 8 | hsa-miR-6601, hsa-miR-100-5p1, hsa-miR-19a1 | Not in a pathway |
| KLF5 | Kruppel like factor 5 | hsa-miR-1451,2, hsa-miR-152-3p1 | Adipogenesis pathway |
| LIN28B | lin-28 homolog B | hsa-miR-125b-5p1, hsa-miR-30e-5p1 | Not in a pathway |
| LMO4 | LIM domain only 4 | hsa-miR-1431 | Not in a pathway |
| LMO4 | LIM domain only 4 | hsa-miR-1306-5p1 | Not in a pathway |
| LMO7 | LIM domain 7 | hsa-miR-44901 | Not in a pathway |
| LMX1B | LIM homeobox transcription factor 1 beta | hsa-miR-4640-5p1, hsa-miR-486-3p1, hsa-miR-4667-5p1, hsa-miR-61321, hsa-miR-6081, hsa-miR-6371, hsa-miR-92a-2-5p1, hsa-miR-5001-5p1 | Not in a pathway |
| LPP | LIM domain containing preferred translocation partner in lipoma | hsa-miR-31851, hsa-miR-1821, hsa-miR-12531, hsa-miR-3190-3p1, hsa-miR-31591, hsa-miR-34a-5p1, hsa-miR-31-5p1, hsa-miR-190a-5p1, hsa-miR-13231, hsa-miR-961, hsa-miR-1241 | Not in a pathway |
| MAFB | MAF bZIP transcription factor B | hsa-miR-44491, hsa-miR-152-3p1, hsa-miR-36151 | Not in a pathway |
| MCPH1 | microcephalin 1 | hsa-miR-146b2 | Not in a pathway |
| MFRP | membrane frizzled-related protein | hsa-miR-12021, hsa-miR-4640-5p1, hsa-miR-185-5p1 | Not in a pathway |
| MKI67 | marker of proliferation Ki-67 | hsa-miR-483-3p2 | Not in a pathway |
| NPAS2 | neuronal PAS domain protein 2 | hsa-miR-931 | Not in a pathway |
| NSF | N-ethylmaleimide sensitive factor, vesicle fusing ATPase | hsa-miR-92a-3p1, hsa-miR-31-5p1 | GABA Receptor Signaling, Huntington's Disease Signaling, Insulin Secretion Signaling Pathway, Phagosome Maturation, Synaptogenesis Signaling Pathway, Tight Junction Signaling |
| OXR1 | oxidation resistance 1 | hsa-miR-302d-3p1 | Not in a pathway |
| PARD3B | par-3 family cell polarity regulator beta | hsa-miR-36151 | Not in a pathway |
| PCSK5 | proprotein convertase subtilisin/kexin type 5 | hsa-miR-29b1 | Not in a pathway |
| PDE7B | phosphodiesterase 7B | hsa-miR-27a-3p1 | cAMP-mediated signaling, Cardiac Hypertrophy Signaling (Enhanced), Cardiac β-adrenergic Signaling, G-Protein Coupled Receptor Signaling, Protein Kinase A Signaling, Relaxin Signaling, tRNA Splicing |
| PKIA | cAMP-dependent protein kinase inhibitor alpha | hsa-miR-46631, hsa-miR-27a-3p1, hsa-miR-931 | cAMP-mediated signaling, Cardiac β-adrenergic Signaling |
| PLEKHA7 | pleckstrin homology domain containing A7 | hsa-miR-3135b1 | Not in a pathway |
| PTPN1 | protein tyrosine phosphatase non-receptor type 1 | hsa-miR-486-3p1 | 3-phosphoinositide Biosynthesis, 3-phosphoinositide Degradation, Caveolar-mediated Endocytosis Signaling, D-myo-inositol (1,4,5,6)-Tetrakisphosphate Biosynthesis, D-myo-inositol (3,4,5,6)-tetrakisphosphate Biosynthesis, D-myo-inositol-5-phosphate Metabolism, Insulin Receptor Signaling, JAK/STAT Signaling, Protein Kinase A Signaling, Role of JAK2 in Hormone-like Cytokine Signaling, Superpathway of Inositol Phosphate Compounds |
| PTPRR | protein tyrosine phosphatase receptor type R | hsa-miR-161, hsa-miR-12751 | Protein Kinase A Signaling |
| RARB | retinoic acid receptor beta | hsa-miR-30e-5p1 | Aryl Hydrocarbon Receptor Signaling, Glucocorticoid Receptor Signaling, Non-Small Cell Lung Cancer Signaling, RAR Activation, Retinoic acid Mediated Apoptosis Signaling, Sirtuin Signaling Pathway, Small Cell Lung Cancer Signaling, White Adipose Tissue Browning Pathway, WNT/β-catenin Signaling |
| RUNX2 | RUNX family transcription factor 2 | hsa-miR-30e-5p1,2 | BMP signaling pathway, Estrogen Receptor Signaling, Osteoarthritis Pathway, Role of Osteoblasts, Osteoclasts and Chondrocytes in Rheumatoid Arthritis, TGF-β Signaling, VDR/RXR Activation |
| SCAMP1 | secretory carrier membrane protein 1 | hsa-miR-181b-3p1 | Not in a pathway |
| SEMA3C | semaphorin 3C | hsa-miR-181b-3p1 | Axonal Guidance Signaling |
| SEMA3F | semaphorin 3F | hsa-miR-6722-3p1 | Axonal Guidance Signaling, RHOA Signaling, Semaphorin Neuronal Repulsive Signaling Pathway |
| SIX3 | SIX homeobox 3 | hsa-miR-4725-3p1, hsa-miR-6081, hsa-miR-4667-5p1, hsa-miR-185-5p1, hsa-miR-12751 | Transcriptional Regulatory Network in Embryonic Stem Cells |
| SOS2 | SOS Ras/Rho guanine nucleotide exchange factor 2 | hsa-miR-152-3p1 | Actin Cytoskeleton Signaling, Actin Nucleation by ARP-WASP Complex, Acute Phase Response Signaling, Adrenomedullin signaling pathway, Aldosterone Signaling in Epithelial Cells, Axonal Guidance Signaling, B Cell Receptor Signaling, Breast Cancer Regulation by Stathmin1, Cholecystokinin/Gastrin-mediated Signaling, CREB Signaling in Neurons, EGF Signaling, EIF2 Signaling, IL-2, 4, 6, & 7 Signaling, JAK/STAT Signaling, PI3K/AKT Signaling, TGF-β Signaling, + 72 |
| SYN3 | Synapsin III | hsa-miR-9401, hsa-miR-127-3p1 | Synaptogenesis Signaling Pathway |
| SYT13 | Synaptotagmin-13 | hsa-miR-3913-3p1 | Synaptogenesis Signaling Pathway |
| TCF7L2 | transcription factor 7 like 2 | hsa-miR-5001-5p1, hsa-miR-92a-2-5p1, hsa-miR-1831, hsa-miR-961 | Acute Myeloid Leukemia Signaling, Basal Cell Carcinoma Signaling, BEX2 Signaling Pathway, Colorectal Cancer Metastasis Signaling, Endocannabinoid Cancer Inhibition Pathway, Epithelial Adherens Junction Signaling, FAT10 Cancer Signaling Pathway, Hepatic Fibrosis Signaling Pathway, HOTAIR Regulatory Pathway, Human Embryonic Stem Cell Pluripotency, Mouse Embryonic Stem Cell Pluripotency, MSP-RON Signaling In Cancer Cells Pathway, Opioid Signaling Pathway, Osteoarthritis Pathway, WNT/β-catenin Signaling, +14 |
| TFEC | transcription factor EC | hsa-miR-6722-3p1, hsa-miR-12531, hsa-miR-3913-3p1 | Not in a pathway |
| TLL1 | tolloid like 1 | hsa-miR-29b1, hsa-miR-961 | Not in a pathway |
| TMCO1 | transmembrane and coiled-coil domains 1 | hsa-miR-30e-5p2, hsa-miR-61321, hsa-miR-187-5p1, hsa-miR-13231 | Not in a pathway |
| TMEM119 | transmembrane protein 119 | hsa-miR-6722-3p1, hsa-miR-5001-5p1, hsa-miR-12751 | Not in a pathway |
| TMEM181 | transmembrane protein 181 | hsa-miR-30e-5p1 | Not in a pathway |
| TNS1 | tensin 1 | hsa-miR-4725-3p1, hsa-miR-36521 | Epithelial Adherens Junction Signaling, FAK Signaling |
| TNXB | tenascin XB | hsa-miR-1491, hsa-miR-36151 | Not in a pathway |
| TRIOBP | TRIO and F-actin binding protein | hsa-miR-4640-5p1, hsa-miR-92a-2-5p1, hsa-miR-6081 | Not in a pathway |
| TSPAN18 | tetraspanin 18 | hsa-miR-5001-5p1, hsa-miR-4725-3p1, hsa-miR-4640-5p1, hsa-miR-6371, hsa-miR-161, hsa-miR-4667-5p1, hsa-miR-34a-5p1, hsa-miR-185-5p1, hsa-miR-3620-5p1 | Not in a pathway |
| TXNRD2 | thioredoxin reductase 2 | hsa-miR-61321, hsa-miR-6722-3p1, hsa-miR-486-3p1, hsa-miR-47671, hsa-miR-44671 | Antioxidant Action of Vitamin C, Mitochondrial Dysfunction, Thioredoxin Pathway, Vitamin-C Transport |
| UBIAD1 | UbiA prenyltransferase domain containing 1 | hsa-miR-1260b1 | Not in a pathway |
| VEGFC | vascular endothelial growth factor C | hsa-miR-27a-3p1 | Amyotrophic Lateral Sclerosis Signaling, Axonal Guidance Signaling, BEX2 Signaling Pathway, Bladder Cancer Signaling, Breast Cancer Regulation by Stathmin1, Clathrin-mediated Endocytosis Signaling, Colorectal Cancer Metastasis Signaling, Endocannabinoid Cancer Inhibition Pathway, eNOS Signaling, Estrogen Receptor Signaling, Hepatic Fibrosis Signaling Pathway, HIF1α Signaling, IL-8 & 17 Signaling, ILK Signaling, VEGF Signaling, +13 |
| WNT3 | Wnt family member 3 | hsa-miR-61321, hsa-miR-376c-3p1 | Axonal Guidance Signaling, Basal Cell Carcinoma Signaling, Cardiac Hypertrophy Signaling (Enhanced), Colorectal Cancer Metastasis Signaling, Glioblastoma Multiforme Signaling, Hepatic Fibrosis Signaling Pathway, HOTAIR Regulatory Pathway, Human Embryonic Stem Cell Pluripotency, Molecular Mechanisms of Cancer, Ovarian Cancer Signaling, PCP (Planar Cell Polarity) Pathway, Regulation Of The Epithelial Mesenchymal Transition In Development Pathway, Regulation of the Epithelial-Mesenchymal Transition Pathway, Role of Macrophages, WNT/β-catenin Signaling, +6 |
| ZNF516 | zinc finger protein 516 | hsa-miR-187-3p1 | Not in a pathway |
| ZNF652 | zinc finger protein 652 | hsa-miR-190a-5p1 | Not in a pathway |
Highly predicted: cumulative weighted context score as defined by TargetScan is less than or equal to −0.4, indicating that a microRNA represses a particular mRNA target by at least 25% relative to the normal level
Experimentally Observed
To provide guidance for future research, we performed an additional IPA using only the miRNAs from Table 2 that were identified in more than one screening study and linked to relevant cellular pathways. We did not include any miRNAs that were part of a targeted study. The miRNAs we identified were miR-143, miR-221, miR-486, miR-4725, miR-125b, miR-451a, miR-92a, and miR-99b. We performed a miRNA target filter analysis using IPA to identify 401 target genes for all these miRNAs, except miR-4725, filtering for only experimentally observed targets or targets predicted with high confidence in humans. IPA’s criteria for high confidence is defined as a cumulative weighted context score, as defined by TargetScan, less than or equal to −0.4, indicating that a miRNA represses a particular mRNA target by at least 25% relative to the normal level. We then used these target genes to perform an expression analysis using IPA to link these target genes to cellular pathways. Numerous pathways were significantly linked to these target genes, but the most relevant pathways that we highlight here are autophagy, apoptosis, senescence, and neuroinflammation (Figure 5). This analysis could help address the issue of the lack of overlapping miRNAs from the studies discussed in this review by providing some insight into which cellular pathways may be important for glaucoma pathology.
Fig, 5. miRNAs involved in glaucoma are linked to autophagy, apoptosis, senescence, and neuroinflammation pathways.
Eight miRNAs were identified in more than one screening study. Target genes for seven of these miRNAs were identified and then linked to cellular pathways using IPA. The pathways are shown here with each linked miRNA shown underneath the pathway name.
5. Conclusion
We aimed to provide a comprehensive overview of work performed to date on miRNAs involved in glaucoma pathology. Numerous studies discussed in this review and our IPA analysis (Table 3) revealed that miRNAs associated with glaucoma often have connections to cell survival or apoptosis pathways. It is important to note that some IPA predicted pathways still need to be validated functionally in vitro, ex vivo, or in vivo. Several studies specifically found Pten to be a target of various anti-apoptotic miRNAs (Li et al., 2018; Mak et al., 2020; Shen et al., 2020; Wang et al., 2019). Cell survival is an important factor in glaucoma, specifically in the TM and the retina. In the TM, accelerated cell death is a cornerstone feature of glaucomatous eyes (Alvarado et al., 1984). Similarly, loss of RGCs is accelerated dramatically in glaucoma, which results in progressive vision loss. Cells in both the TM and the RGC typically do not regenerate under normal conditions, so inhibition of cell death in these tissues with miRNA-based therapeutics could be one potential method for glaucoma treatment and prevention.
In addition to cell survival pathways, several other studies indicate that glaucoma-associated miRNAs regulate genes involved in ECM maintenance, TM contractility, and consequently AH outflow function (Luna et al., 2009; Shen et al., 2015). These two pathways are interconnected and are important for homeostasis in the TM (Acott et al., 2021). For example, if ECM is not properly maintained or becomes fibrotic, homeostatic responses to elevated IOP are compromised. These pathways also relate to the cell survival pathways discussed above because TM cell death can lead to ECM dysregulation and decreased AH outflow.
Overall, there has been immense progress studying miRNAs in glaucoma, but further research is still needed to better understand their specific targets and their mechanism of action. The lack of shared miRNAs with similar differential expression in the current studies should also be addressed. This issue may be solved by increasing sample size, using more uniform experimental techniques or consistent animal models, accounting for clinical differences in glaucoma types, medicinal or surgical intervention, and demographic differences of humans from which the samples were derived. Consistent identification of miRNAs associated with glaucoma across studies would better elucidate the role of miRNAs in glaucoma. Furthermore, a comprehensive understanding of miRNA targets and an optimized delivery mechanism will lead to more effective prevention and therapeutic options for glaucoma patients.
Highlights.
Brief summary of current, commonly used methods to study miRNAs
Discussion and literature review on the involvement of miRNAs in glaucoma
Summary of the potential therapeutic applications of miRNAs in glaucoma
Summary of the pathways that the identified miRNAs may target
Acknowledgments
Funding:
This research was funded by the BrightFocus Foundation; The Glaucoma Foundation; the Glaucoma Research Foundation; and by the National Institutes of Health, P30EY031631, R01EY023242, and R21EY028671.
Abbreviations:
- miRNA
microRNA
- POAG
primary open angle glaucoma
- IOP
intraocular pressure
- NTG
normal tension glaucoma
- HTG
high tension glaucoma
- PACG
primary angle closure glaucoma
- AH
aqueous humor
- ECM
extracellular matrix
- CMS
cyclic mechanical stretch
- TM
trabecular meshwork
- HTM
human trabecular meshwork
- RGC
retinal ganglion cells
- AOH
acute ocular hypertension
Footnotes
Competing interest statement: The authors declare no competing financial interests.
Submission declaration and verification: The authors verify that the work in this manuscript has not been published previously.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aboobakar IF, & Allingham RR (2014, Fall). Genetics of exfoliation syndrome and glaucoma. Int Ophthalmol Clin, 54(4), 43–56. 10.1097/IIO.0000000000000042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acott TS, Vranka JA, Keller KE, Raghunathan V, & Kelley MJ (2021, May). Normal and glaucomatous outflow regulation. Prog Retin Eye Res, 82, 100897. 10.1016/j.preteyeres.2020.100897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarado J, Murphy C, & Juster R (1984, Jun). Trabecular meshwork cellularity in primary open-angle glaucoma and nonglaucomatous normals. Ophthalmology, 91(6), 564–579. 10.1016/s0161-6420(84)34248-8 [DOI] [PubMed] [Google Scholar]
- Bailey JN, Loomis SJ, Kang JH, Allingham RR, Gharahkhani P, Khor CC, Burdon KP, Aschard H, Chasman DI, Igo RP Jr., Hysi PG, Glastonbury CA, Ashley-Koch A, Brilliant M, Brown AA, Budenz DL, Buil A, Cheng CY, Choi H, Christen WG, Curhan G, De Vivo I, Fingert JH, Foster PJ, Fuchs C, Gaasterland D, Gaasterland T, Hewitt AW, Hu F, Hunter DJ, Khawaja AP, Lee RK, Li Z, Lichter PR, Mackey DA, McGuffin P, Mitchell P, Moroi SE, Perera SA, Pepper KW, Qi Q, Realini T, Richards JE, Ridker PM, Rimm E, Ritch R, Ritchie M, Schuman JS, Scott WK, Singh K, Sit AJ, Song YE, Tamimi RM, Topouzis F, Viswanathan AC, Verma SS, Vollrath D, Wang JJ, Weisschuh N, Wissinger B, Wollstein G, Wong TY, Yaspan BL, Zack DJ, Zhang K, Study, E. N., Consortium, A., Weinreb RN, Pericak-Vance MA, Small K, Hammond CJ, Aung T, Liu Y, Vithana EN, MacGregor S, Craig JE, Kraft P, Howell G, Hauser MA, Pasquale LR, Haines JL, & Wiggs JL (2016, Feb). Genome-wide association analysis identifies TXNRD2, ATXN2 and FOXC1 as susceptibility loci for primary open-angle glaucoma. Nat Genet, 48(2), 189–194. 10.1038/ng.3482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JNC, Gharahkhani P, Kang JH, Butkiewicz M, Sullivan DA, Weinreb RN, Aschard H, Allingham RR, Ashley-Koch A, Lee RK, Moroi SE, Brilliant MH, Wollstein G, Schuman JS, Fingert JH, Budenz DL, Realini T, Gaasterland T, Scott WK, Singh K, Sit AJ, Igo RP Jr., Song YE, Hark L, Ritch R, Rhee DJ, Vollrath D, Zack DJ, Medeiros F, Vajaranant TS, Chasman DI, Christen WG, Pericak-Vance MA, Liu Y, Kraft P, Richards JE, Rosner BA, Hauser MA, Craig JE, Burdon KP, Hewitt AW, Mackey DA, Haines JL, MacGregor S, Wiggs JL, Pasquale LR, Australian, & New Zealand Registry of Advanced Glaucoma, C. (2018, Feb 1). Testosterone Pathway Genetic Polymorphisms in Relation to Primary Open-Angle Glaucoma: An Analysis in Two Large Datasets. Invest Ophthalmol Vis Sci, 59(2), 629–636. 10.1167/iovs.17-22708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty C, Sharma AR, Sharma G, & Lee SS (2021, Feb). Therapeutic advances of miRNAs: A preclinical and clinical update. JAdv Res, 28, 127–138. 10.1016/j.jare.2020.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewry M, Helwa I, Allingham RR, Hauser MA, & Liu Y (2016, Jul 1). miRNA Profile in Three Different Normal Human Ocular Tissues by miRNA-Seq. Invest Ophthalmol Vis Sci, 57(8), 3731–3739. 10.1167/iovs.16-19155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewry MD, Challa P, Kuchtey JG, Navarro I, Helwa I, Hu Y, Mu H, Stamer WD, Kuchtey RW, & Liu Y (2018, Apr 1). Differentially expressed microRNAs in the aqueous humor of patients with exfoliation glaucoma or primary open-angle glaucoma. Hum Mol Genet, 27(7), 1263–1275. 10.1093/hmg/ddy040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Feng Y, Zhang R, Zhang W, Shu Y, Zeng Z, Huang S, Zhang L, Huang B, Wu D, Zhang B, Wang X, Lei Y, Ye Z, Zhao L, Cao D, Yang L, Chen X, Liu B, Wagstaff W, He F, Wu X, Zhang J, Moriatis Wolf J, Lee MJ, Haydon RC, Luu HH, Huang A, He TC, & Yan S (2020, Jun). A simplified system for the effective expression and delivery of functional mature microRNAs in mammalian cells. Cancer Gene Ther, 27(6), 424–437. 10.1038/s41417-019-0113-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, & Sonenberg N (2008, Feb). Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet, 9(2), 102–114. 10.1038/nrg2290 [DOI] [PubMed] [Google Scholar]
- Gao L, Jiang BO, Lei D, Zhou X, & Yuan H (2016, Feb). Expression profiling of microRNAs in optineurin (E50K) mutant transgenic mice. Biomed Rep, 4(2), 193–196. 10.3892/br.2015.565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharahkhani P, Jorgenson E, Hysi P, Khawaja AP, Pendergrass S, Han X, Ong JS, Hewitt AW, Segre AV, Rouhana JM, Hamel AR, Igo RP Jr., Choquet H, Qassim A, Josyula NS, Cooke Bailey JN, Bonnemaijer PWM, Iglesias A, Siggs OM, Young TL, Vitart V, Thiadens A, Karjalainen J, Uebe S, Melles RB, Nair KS, Luben R, Simcoe M, Amersinghe N, Cree AJ, Hohn R, Poplawski A, Chen LJ, Rong SS, Aung T, Vithana EN, consortium, N., consortium, A., Biobank Japan, p., FinnGen, s., Eye, U. K. B., Vision, C., group, G. s., Me Research, T., Tamiya G, Shiga Y, Yamamoto M, Nakazawa T, Currant H, Birney E, Wang X, Auton A, Lupton MK, Martin NG, Ashaye A, Olawoye O, Williams SE, Akafo S, Ramsay M, Hashimoto K, Kamatani Y, Akiyama M, Momozawa Y, Foster PJ, Khaw PT, Morgan JE, Strouthidis NG, Kraft P, Kang JH, Pang CP, Pasutto F, Mitchell P, Lotery AJ, Palotie A, van Duijn C, Haines JL, Hammond C, Pasquale LR, Klaver CCW, Hauser M, Khor CC, Mackey DA, Kubo M, Cheng CY, Craig JE, MacGregor S, & Wiggs JL (2021, Feb 24). Genome-wide meta-analysis identifies 127 open-angle glaucoma loci with consistent effect across ancestries. Nat Commun, 12(1), 1258. 10.1038/s41467-020-20851-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez P, Li G, Qiu J, Wu J, & Luna C (2014, Mar-Apr). Role of microRNAs in the trabecular meshwork. J Ocul Pharmacol Ther, 30(2-3), 128–137. 10.1089/jop.2013.0191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M, & Kim VN (2014, Aug). Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol, 15(8), 509–524. 10.1038/nrm3838 [DOI] [PubMed] [Google Scholar]
- Hammond SM (2015, Jun 29). An overview of microRNAs. Adv Drug Deliv Rev, 87, 3–14. 10.1016/j.addr.2015.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Li HB, Li X, Zhou Y, Xia XB, & Song WT (2018). MiR-124 Promotes the Growth of Retinal Ganglion Cells Derived from Muller Cells. Cell Physiol Biochem, 45(3), 973–983. 10.1159/000487292 [DOI] [PubMed] [Google Scholar]
- Hindle AG, Thoonen R, Jasien JV, Grange RMH, Amin K, Wise J, Ozaki M, Ritch R, Malhotra R, & Buys ES (2019, Jan 2). Identification of Candidate miRNA Biomarkers for Glaucoma. Invest Ophthalmol Vis Sci, 60(1), 134–146. 10.1167/iovs.18-24878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda T, Yamamoto Y, Daito T, Matsumoto Y, Makino A, & Tomonaga K (2016, May 18). Long-term expression of miRNA for RNA interference using a novel vector system based on a negative-strand RNA virus. Sci Rep, 6, 26154. 10.1038/srep26154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubens WHG, Krauskopf J, Beckers HJM, Kleinjans JCS, Webers CAB, & Gorgels T (2021, Jun 1). Small RNA Sequencing of Aqueous Humor and Plasma in Patients With Primary Open-Angle Glaucoma. Invest Ophthalmol Vis Sci, 62(7), 24. 10.1167/iovs.62.7.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaram H, Cepurna WO, Johnson EC, & Morrison JC (2015, Dec). MicroRNA Expression in the Glaucomatous Retina. Invest Ophthalmol Vis Sci, 56(13), 7971–7982. 10.1167/iovs.15-18088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaram H, Phillips JI, Lozano DC, Choe TE, Cepurna WO, Johnson EC, Morrison JC, Gattey DM, Saugstad JA, & Keller KE (2017, Jun 1). Comparison of MicroRNA Expression in Aqueous Humor of Normal and Primary Open-Angle Glaucoma Patients Using PCR Arrays: A Pilot Study. Invest Ophthalmol Vis Sci, 58(7), 2884–2890. 10.1167/iovs.17-21844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Chen Z, Liu X, & Zhou X (2013). Evaluating the microRNA targeting sites by luciferase reporter gene assay. Methods Mol Biol, 936, 117–127. 10.1007/978-1-62703-083-0_10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas JB, Aung T, Bourne RR, Bron AM, Ritch R, & Panda-Jonas S (2017, Nov 11). Glaucoma. Lancet, 390(10108), 2183–2193. 10.1016/S0140-6736(17)31469-1 [DOI] [PubMed] [Google Scholar]
- Keller KE, Bhattacharya SK, Borras T, Brunner TM, Chansangpetch S, Clark AF, Dismuke WM, Du Y, Elliott MH, Ethier CR, Faralli JA, Freddo TF, Fuchshofer R, Giovingo M, Gong H, Gonzalez P, Huang A, Johnstone MA, Kaufman PL, Kelley MJ, Knepper PA, Kopczynski CC, Kuchtey JG, Kuchtey RW, Kuehn MH, Lieberman RL, Lin SC, Liton P, Liu Y, Lutjen-Drecoll E, Mao W, Masis-Solano M, McDonnell F, McDowell CM, Overby DR, Pattabiraman PP, Raghunathan VK, Rao PV, Rhee DJ, Chowdhury UR, Russell P, Samples JR, Schwartz D, Stubbs EB, Tamm ER, Tan JC, Toris CB, Torrejon KY, Vranka JA, Wirtz MK, Yorio T, Zhang J, Zode GS, Fautsch MP, Peters DM, Acott TS, & Stamer WD (2018, Jun). Consensus recommendations for trabecular meshwork cell isolation, characterization and culture. Exp Eye Res, 171, 164–173. 10.1016/j.exer.2018.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khawaja AP, Cooke Bailey JN, Wareham NJ, Scott RA, Simcoe M, Igo RP Jr., Song YE, Wojciechowski R, Cheng CY, Khaw PT, Pasquale LR, Haines JL, Foster PJ, Wiggs JL, Hammond CJ, Hysi PG, Eye, U. K. B., Vision, C., & Consortium, N. (2018, Jun). Genome-wide analyses identify 68 new loci associated with intraocular pressure and improve risk prediction for primary open-angle glaucoma. Nat Genet, 50(6), 778–782. 10.1038/s41588-018-0126-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni MM (2011, Apr). Digital multiplexed gene expression analysis using the NanoString nCounter system. Curr Protoc Mol Biol, Chapter 25, Unit25B 10. 10.1002/0471142727.mb25b10s94 [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, & Ambros V (1993, Dec 3). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell, 75(5), 843–854. 10.1016/0092-8674(93)90529-y [DOI] [PubMed] [Google Scholar]
- Li G, Luna C, Qiu J, Epstein DL, & Gonzalez P (2009, Nov-Dec). Alterations in microRNA expression in stress-induced cellular senescence. Mech Ageing Dev, 130(11-12), 731–741. 10.1016/j.mad.2009.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Luna C, Qiu J, Epstein DL, & Gonzalez P (2010a, Jun). Modulation of inflammatory markers by miR-146a during replicative senescence in trabecular meshwork cells. Invest Ophthalmol Vis Sci, 51(6), 2976–2985. 10.1167/iovs.09-4874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Luna C, Qiu J, Epstein DL, & Gonzalez P (2010b, Feb 19). Targeting of integrin beta1 and kinesin 2alpha by microRNA 183. J Biol Chem, 285(8), 5461–5471. 10.1074/jbc.M109.037127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Luna C, Qiu J, Epstein DL, & Gonzalez P (2011, May 6). Role of miR-204 in the regulation of apoptosis, endoplasmic reticulum stress response, and inflammation in human trabecular meshwork cells. Invest Ophthalmol Vis Sci, 52(6), 2999–3007. 10.1167/iovs.10-6708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Jin Y, Li Q, Sun X, Zhu H, & Cui H (2018, Apr). MiR-93-5p targeting PTEN regulates the NMDA-induced autophagy of retinal ganglion cells via AKT/mTOR pathway in glaucoma. Biomed Pharmacother, 100, 1–7. 10.1016/j.biopha.2018.01.044 [DOI] [PubMed] [Google Scholar]
- Li T, Wang Y, Chen J, Gao X, Pan S, Su Y, & Zhou X (2020, Dec). Co-delivery of brinzolamide and miRNA-124 by biodegradable nanoparticles as a strategy for glaucoma therapy. Drug Deliv, 27(1), 410–421. 10.1080/10717544.2020.1731861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, & Ruan K (2009, Jun). MicroRNA detection by microarray. Anal Bioanal Chem, 394(4), 1117–1124. 10.1007/s00216-008-2570-2 [DOI] [PubMed] [Google Scholar]
- Li X, Zhao F, Xin M, Li G, Luna C, Li G, Zhou Q, He Y, Yu B, Olson E, Gonzalez P, & Wang S (2017, Apr 19). Regulation of intraocular pressure by microRNA cluster miR-143/145. Sci Rep, 7(1), 915. 10.1038/s41598-017-01003-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, & Johnson JM (2005, Feb 17). Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature, 433(7027), 769–773. 10.1038/nature03315 [DOI] [PubMed] [Google Scholar]
- Liu CG, Calin GA, Volinia S, & Croce CM (2008). MicroRNA expression profiling using microarrays. Nat Protoc, 3(4), 563–578. 10.1038/nprot.2008.14 [DOI] [PubMed] [Google Scholar]
- Liu H, Xiu Y, Zhang Q, Xu Y, Wan Q, & Tao L (2021, Jun). Silencing microRNA29b3p expression protects human trabecular meshwork cells against oxidative injury via upregulation of RNF138 to activate the ERK pathway. Int J Mol Med, 47(6). 10.3892/ijmm.2021.4934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Tong H, Li T, Wang X, & Chen Y (2020). Research progress in molecular biology related quantitative methods of MicroRNA. Am J Transl Res, 12(7), 3198–3211. https://www.ncbi.nlm.nih.gov/pubmed/32774694 [PMC free article] [PubMed] [Google Scholar]
- Liu Y, & Allingham RR (2017, Jul). Major review: Molecular genetics of primary open-angle glaucoma. Exp Eye Res, 160, 62–84. 10.1016/j.exer.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Bailey JC, Helwa I, Dismuke WM, Cai J, Drewry M, Brilliant MH, Budenz DL, Christen WG, Chasman DI, Fingert JH, Gaasterland D, Gaasterland T, Gordon MO, Igo RP Jr., Kang JH, Kass MA, Kraft P, Lee RK, Lichter P, Moroi SE, Realini A, Richards JE, Ritch R, Schuman JS, Scott WK, Singh K, Sit AJ, Song YE, Vollrath D, Weinreb R, Medeiros F, Wollstein G, Zack DJ, Zhang K, Pericak-Vance MA, Gonzalez P, Stamer WD, Kuchtey J, Kuchtey RW, Allingham RR, Hauser MA, Pasquale LR, Haines JL, & Wiggs JL (2016, Aug 1). A Common Variant in MIR182 Is Associated With Primary Open-Angle Glaucoma in the NEIGHBORHOOD Consortium. Invest Ophthalmol Vis Sci, 57(10), 4528–4535. 10.1167/iovs.16-19688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Chen Y, Wang Y, Zhang X, Gao K, Chen S, & Zhang X (2018, Jun 1). microRNA Profiling in Glaucoma Eyes With Varying Degrees of Optic Neuropathy by Using Next-Generation Sequencing. Invest Ophthalmol Vis Sci, 59(7), 2955–2966. 10.1167/iovs.17-23599 [DOI] [PubMed] [Google Scholar]
- Luna C, Li G, Huang J, Qiu J, Wu J, Yuan F, Epstein DL, & Gonzalez P (2012). Regulation of trabecular meshwork cell contraction and intraocular pressure by miR-200c. PLoS One, 7(12), e51688. 10.1371/journal.pone.0051688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna C, Li G, Qiu J, Epstein DL, & Gonzalez P (2009, Nov 28). Role of miR-29b on the regulation of the extracellular matrix in human trabecular meshwork cells under chronic oxidative stress. Mol Vis, 15, 2488–2497. https://www.ncbi.nlm.nih.gov/pubmed/19956414 [PMC free article] [PubMed] [Google Scholar]
- Luna C, Li G, Qiu J, Epstein DL, & Gonzalez P (2011a, Jun 1). Cross-talk between miR-29 and transforming growth factor-betas in trabecular meshwork cells. Invest Ophthalmol Vis Sci, 52(6), 3567–3572. 10.1167/iovs.10-6448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna C, Li G, Qiu J, Epstein DL, & Gonzalez P (2011b, May). MicroRNA-24 regulates the processing of latent TGFbeta1 during cyclic mechanical stress in human trabecular meshwork cells through direct targeting of FURIN. J Cell Physiol, 226(5), 1407–1414. 10.1002/jcp.22476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGregor S, Ong JS, An J, Han X, Zhou T, Siggs OM, Law MH, Souzeau E, Sharma S, Lynn DJ, Beesley J, Sheldrick B, Mills RA, Landers J, Ruddle JB, Graham SL, Healey PR, White AJR, Casson RJ, Best S, Grigg JR, Goldberg I, Powell JE, Whiteman DC, Radford-Smith GL, Martin NG, Montgomery GW, Burdon KP, Mackey DA, Gharahkhani P, Craig JE, & Hewitt AW (2018, Aug). Genome-wide association study of intraocular pressure uncovers new pathways to glaucoma. Nat Genet, 50(8), 1067–1071. 10.1038/s41588-018-0176-y [DOI] [PubMed] [Google Scholar]
- Madekurozwa M, Stamer WD, Reina-Torres E, Sherwood JM, & Overby DR (2021, Apr 1). The ocular pulse decreases aqueous humor outflow resistance by stimulating nitric oxide production. Am J Physiol Cell Physiol, 320(4), C652–C665. 10.1152/ajpcell.00473.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak HK, Yung JSY, Weinreb RN, Ng SH, Cao X, Ho TYC, Ng TK, Chu WK, Yung WH, Choy KW, Wang CC, Lee TL, & Leung CK (2020, Sep 4). MicroRNA-19a-PTEN Axis Is Involved in the Developmental Decline of Axon Regenerative Capacity in Retinal Ganglion Cells. Mol Ther Nucleic Acids, 21, 251–263. 10.1016/j.omtn.2020.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead B, Amaral J, & Tomarev S (2018, Feb 1). Mesenchymal Stem Cell-Derived Small Extracellular Vesicles Promote Neuroprotection in Rodent Models of Glaucoma. Invest Ophthalmol Vis Sci, 59(2), 702–714. 10.1167/iovs.17-22855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead B, & Tomarev S (2017, Apr). Bone Marrow-Derived Mesenchymal Stem Cells-Derived Exosomes Promote Survival of Retinal Ganglion Cells Through miRNA-Dependent Mechanisms. Stem Cells Transl Med, 6(4), 1273–1285. 10.1002/sctm.16-0428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon M, Mohammadi S, Davila-Velderrain J, Goods BA, Cadwell TD, Xing Y, Stemmer-Rachamimov A, Shalek AK, Love JC, Kellis M, & Hafler BP (2019, Oct 25). Single-cell transcriptomic atlas of the human retina identifies cell types associated with age-related macular degeneration. Nat Commun, 10(1), 4902. 10.1038/s41467-019-12780-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan DJ, & DeAngelis MM (2014, Oct 23). Differential Gene Expression in Age-Related Macular Degeneration. Cold Spring Harb Perspect Med, 5(8), a017210. 10.1101/cshperspect.a017210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison JC, Cepurna WO, Tehrani S, Choe TE, Jayaram H, Lozano DC, Fortune B, & Johnson EC (2016, Dec 1). A Period of Controlled Elevation of IOP (CEI) Produces the Specific Gene Expression Responses and Focal Injury Pattern of Experimental Rat Glaucoma. Invest Ophthalmol Vis Sci, 57(15), 6700–6711. 10.1167/iovs.16-20573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam Y, Chen C, Gregory RI, Chou JJ, & Sliz P (2011, Nov 23). Molecular basis for interaction of let-7 microRNAs with Lin28. Cell, 147(5), 1080–1091. 10.1016/j.cell.2011.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie XG, Fan DS, Huang YX, He YY, Dong BL, & Gao F (2018, Dec 1). Downregulation of microRNA-149 in retinal ganglion cells suppresses apoptosis through activation of the PI3K/Akt signaling pathway in mice with glaucoma. Am J Physiol Cell Physiol, 315(6), C839–C849. 10.1152/ajpcell.00324.2017 [DOI] [PubMed] [Google Scholar]
- Orozco LD, Chen HH, Cox C, Katschke KJ Jr., Arceo R, Espiritu C, Caplazi P, Nghiem SS, Chen YJ, Modrusan Z, Dressen A, Goldstein LD, Clarke C, Bhangale T, Yaspan B, Jeanne M, Townsend MJ, van Lookeren Campagne M, & Hackney JA (2020, Jan 28). Integration of eQTL and a Single-Cell Atlas in the Human Eye Identifies Causal Genes for Age-Related Macular Degeneration. Cell Rep, 30(4), 1246–1259 e1246. 10.1016/j.celrep.2019.12.082 [DOI] [PubMed] [Google Scholar]
- Ou-Yang Y, Liu ZL, Xu CL, Wu JL, Peng J, & Peng QH (2020, Jan 15). miR-223 induces retinal ganglion cells apoptosis and inflammation via decreasing HSP-70 in vitro and in vivo. J Chem Neuroanat, 104, 101747. 10.1016/j.jchemneu.2020.101747 [DOI] [PubMed] [Google Scholar]
- Paylakhi SH, Moazzeni H, Yazdani S, Rassouli P, Arefian E, Jaberi E, Arash EH, Gilani AS, Fan JB, April C, Amin S, Suri F, & Elahi E (2013, Jun). FOXC1 in human trabecular meshwork cells is involved in regulatory pathway that includes miR-204, MEIS2, and ITGbeta1. Exp Eye Res, 111, 112–121. 10.1016/j.exer.2013.03.009 [DOI] [PubMed] [Google Scholar]
- Peng H, Sun YB, Hao JL, Lu CW, Bi MC, & Song E (2019, Feb). Neuroprotective effects of overexpressed microRNA-200a on activation of glaucoma-related retinal glial cells and apoptosis of ganglion cells via downregulating FGF7-mediated MAPK signaling pathway. Cell Signal, 54, 179–190. 10.1016/j.cellsig.2018.11.006 [DOI] [PubMed] [Google Scholar]
- Raga-Cervera J, Bolarin JM, Millan JM, Garcia-Medina JJ, Pedrola L, Abellan-Abenza J, Valero-Vello M, Sanz-Gonzalez SM, O'Connor JE, Galarreta-Mira D, Bendala-Tufanisco E, Mayordomo-Febrer A, Pinazo-Duran MD, & Zanon-Moreno V (2021, May 21). miRNAs and Genes Involved in the Interplay between Ocular Hypertension and Primary Open-Angle Glaucoma. Oxidative Stress, Inflammation, and Apoptosis Networks. J Clin Med, 10(11). 10.3390/jcm10112227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, & Zhang F (2013, Nov). Genome engineering using the CRISPR-Cas9 system. Nat Protoc, 8(11), 2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnapriya R, Sosina OA, Starostik MR, Kwicklis M, Kapphahn RJ, Fritsche LG, Walton A, Arvanitis M, Gieser L, Pietraszkiewicz A, Montezuma SR, Chew EY, Battle A, Abecasis GR, Ferrington DA, Chatterjee N, & Swaroop A (2019, Apr). Retinal transcriptome and eQTL analyses identify genes associated with age-related macular degeneration. Nat Genet, 51(4), 606–610. 10.1038/s41588-019-0351-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritch R (2008). The management of exfoliative glaucoma. Prog Brain Res, 173, 211–224. 10.1016/S0079-6123(08)01115-1 [DOI] [PubMed] [Google Scholar]
- Ritch R, & Schlotzer-Schrehardt U (2001, Jan-Feb). Exfoliation syndrome. Surv Ophthalmol, 45(4), 265–315. 10.1016/s0039-6257(00)00196-x [DOI] [PubMed] [Google Scholar]
- Ruibin W, Zheng X, Chen J, Zhang X, Yang X, & Lin Y (2018, Apr). Micro RNA-1298 opposes the effects of chronic oxidative stress on human trabecular meshwork cells via targeting on EIF4E3. Biomed Pharmacother, 100, 349–357. 10.1016/j.biopha.2018.02.001 [DOI] [PubMed] [Google Scholar]
- Schlotzer-Schrehardt U (2009, Apr). Molecular pathology of pseudoexfoliation syndrome/glaucoma--new insights from LOXL1 gene associations. Exp Eye Res, 88(4), 776–785. 10.1016/j.exer.2008.08.012 [DOI] [PubMed] [Google Scholar]
- Shekhar K, & Sanes JR (2021, Sep 15). Generating and Using Transcriptomically Based Retinal Cell Atlases. Annu Rev Vis Sci, 7, 43–72. 10.1146/annurev-vision-032621-075200 [DOI] [PubMed] [Google Scholar]
- Shen W, Han Y, Huang B, Qi Y, Xu L, Guo R, Wang X, & Wang J (2015, Dec). MicroRNA-483-3p Inhibits Extracellular Matrix Production by Targeting Smad4 in Human Trabecular Meshwork Cells. Invest Ophthalmol Vis Sci, 56(13), 8419–8427. 10.1167/iovs.15-18036 [DOI] [PubMed] [Google Scholar]
- Shen Y, Zhu Y, & Rong F (2020, Aug). miR200c3p regulates the proliferation and apoptosis of human trabecular meshwork cells by targeting PTEN. Mol Med Rep, 22(2), 1605–1612. 10.3892/mmr.2020.11198 [DOI] [PubMed] [Google Scholar]
- Tamkovich S, Grigor'eva A, Eremina A, Tupikin A, Kabilov M, Chernykh V, Vlassov V, & Ryabchikova E (2019, Aug). What information can be obtained from the tears of a patient with primary open angle glaucoma? Clin Chim Acta, 495, 529–537. 10.1016/j.cca.2019.05.028 [DOI] [PubMed] [Google Scholar]
- Tan C, Jia F, Zhang P, Sun X, Qiao Y, Chen X, Wang Y, Chen J, & Lei Y (2021, Apr 21). A miRNA stabilizing polydopamine nano-platform for intraocular delivery of miR-21-5p in glaucoma therapy. J Mater Chem B, 9(15), 3335–3345. 10.1039/d0tb02881a [DOI] [PubMed] [Google Scholar]
- Tan C, Song M, Stamer WD, Qiao Y, Chen X, Sun X, Lei Y, & Chen J (2020, Nov). miR-21-5p: A viable therapeutic strategy for regulating intraocular pressure. Exp Eye Res, 200, 108197. 10.1016/j.exer.2020.108197 [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Tsuda S, Kunikata H, Sato J, Kokubun T, Yasuda M, Nishiguchi KM, Inada T, & Nakazawa T (2014, May 28). Profiles of extracellular miRNAs in the aqueous humor of glaucoma patients assessed with a microarray system. Sci Rep, 4, 5089. 10.1038/srep05089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tham YC, Li X, Wong TY, Quigley HA, Aung T, & Cheng CY (2014, Nov). Global prevalence of glaucoma and projections of glaucoma burden through 2040: a systematic review and meta-analysis. Ophthalmology, 121(11), 2081–2090. 10.1016/j.ophtha.2014.05.013 [DOI] [PubMed] [Google Scholar]
- Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, & Hammond SM (2006, Aug 15). Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev, 20(16), 2202–2207. 10.1101/gad.1444406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LJ, Wang XZ, Li ZM, Kou D, Zhang D, & Xu ZZ (2020, Sep). MiR-126 facilitates apoptosis of retinal ganglion cells in glaucoma rats via VEGF-Notch signaling pathway. Eur Rev Med Pharmacol Sci, 24(17), 8635–8641. 10.26355/eurrev_202009_22800 [DOI] [PubMed] [Google Scholar]
- Wang X, Li Z, Bai J, Song W, & Zhang F (2019, Apr). miR175p regulates the proliferation and apoptosis of human trabecular meshwork cells by targeting phosphatase and tensin homolog. Mol Med Rep, 19(4), 3132–3138. 10.3892/mmr.2019.9973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Chen S, Wang J, Liu Y, Chen Y, Wen T, Fang X, Vidal-Sanz M, Jonas JB, & Zhang X (2021, Jan 4). MicroRNA-93/STAT3 signalling pathway mediates retinal microglial activation and protects retinal ganglion cells in an acute ocular hypertension model. Cell Death Dis, 12(1), 41. 10.1038/s41419-020-03337-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li F, & Wang S (2016, Dec). MicroRNA93 is overexpressed and induces apoptosis in glaucoma trabecular meshwork cells. Mol Med Rep, 14(6), 5746–5750. 10.3892/mmr.2016.5938 [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhou H, Liu X, Han Y, Pan S, & Wang Y (2018, May). MiR-181a inhibits human trabecular meshwork cell apoptosis induced by H(2)O(2) through the suppression of NF-kappaB and JNK pathways. Adv Clin Exp Med, 27(5), 577–582. 10.17219/acem/69135 [DOI] [PubMed] [Google Scholar]
- Weinreb RN, Aung T, & Medeiros FA (2014, May 14). The pathophysiology and treatment of glaucoma: a review. JAMA, 311(18), 1901–1911. 10.1001/jama.2014.3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiggs JL, & Pasquale LR (2017, Aug 1). Genetics of glaucoma. Hum Mol Genet, 26(R1), R21–R27. 10.1093/hmg/ddx184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiggs JL, Yaspan BL, Hauser MA, Kang JH, Allingham RR, Olson LM, Abdrabou W, Fan BJ, Wang DY, Brodeur W, Budenz DL, Caprioli J, Crenshaw A, Crooks K, Delbono E, Doheny KF, Friedman DS, Gaasterland D, Gaasterland T, Laurie C, Lee RK, Lichter PR, Loomis S, Liu Y, Medeiros FA, McCarty C, Mirel D, Moroi SE, Musch DC, Realini A, Rozsa FW, Schuman JS, Scott K, Singh K, Stein JD, Trager EH, Vanveldhuisen P, Vollrath D, Wollstein G, Yoneyama S, Zhang K, Weinreb RN, Ernst J, Kellis M, Masuda T, Zack D, Richards JE, Pericak-Vance M, Pasquale LR, & Haines JL (2012). Common variants at 9p21 and 8q22 are associated with increased susceptibility to optic nerve degeneration in glaucoma. PLoS Genet, 8(4), e1002654. 10.1371/journal.pgen.1002654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman B, Ha I, & Ruvkun G (1993, Dec 3). Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell, 75(5), 855–862. 10.1016/0092-8674(93)90530-4 [DOI] [PubMed] [Google Scholar]
- Xu Z, Hysi P, & Khawaja AP (2021, Sep 15). Genetic Determinants of Intraocular Pressure. Annu Rev Vis Sci, 7, 727–746. 10.1146/annurev-vision-031021-095225 [DOI] [PubMed] [Google Scholar]
- Yamagata M, Yan W, & Sanes JR (2021, Jan 4). A cell atlas of the chick retina based on single-cell transcriptomics. Elife, 10. 10.7554/eLife.63907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W, Peng YR, van Zyl T, Regev A, Shekhar K, Juric D, & Sanes JR (2020, Jun 17). Cell Atlas of The Human Fovea and Peripheral Retina. Sci Rep, 10(1), 9802. 10.1038/s41598-020-66092-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Wang N, & Luo X (2018, Sep 18). Intraocular miR-211 exacerbates pressure-induced cell death in retinal ganglion cells via direct repression of FRS2 signaling. Biochem Biophys Res Commun, 503(4), 2984–2992. 10.1016/j.bbrc.2018.08.082 [DOI] [PubMed] [Google Scholar]
- Youngblood H, Cai J, Drewry MD, Helwa I, Hu E, Liu S, Yu H, Mu H, Hu Y, Perkumas K, Aboobakar IF, Johnson WM, Stamer WD, & Liu Y (2020, May 11). Expression of mRNAs, miRNAs, and lncRNAs in Human Trabecular Meshwork Cells Upon Mechanical Stretch. Invest Ophthalmol Vis Sci, 61(5), 2. 10.1167/iovs.61.5.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngblood H, Hauser MA, & Liu Y (2019, Nov). Update on the genetics of primary open-angle glaucoma. Exp Eye Res, 188, 107795. 10.1016/j.exer.2019.107795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Du X, Wang M, Yang P, & Zhang J (2019, Dec). Salidroside mitigates hydrogen peroxide-induced injury by enhancement of microRNA-27a in human trabecular meshwork cells. Artif Cells Nanomed Biotechnol, 47(1), 1758–1765. 10.1080/21691401.2019.1608222 [DOI] [PubMed] [Google Scholar]