Abstract
Current RNA vaccines against severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) are limited by instability of both the RNA and the lipid nanoparticle delivery system, requiring storage at −20°C or −70°C and compromising universally accessible vaccine distribution. This study demonstrates the thermostability and adaptability of a nanostructured lipid carrier (NLC) delivery system for RNA vaccines that has the potential to address these concerns. Liquid NLC alone is stable at refrigerated temperatures for ≥1 year, enabling stockpiling and rapid deployment by point-of-care mixing with any vaccine RNA. Alternatively, NLC complexed with RNA may be readily lyophilized and stored at room temperature for ≥8 months or refrigerated temperature for ≥21 months while still retaining the ability to express protein in vivo. The thermostability of this NLC/RNA vaccine delivery platform could significantly improve distribution of current and future pandemic response vaccines, particularly in low-resource settings.
Keywords: RNA vaccine, nanostructured lipid carrier (NLC), thermostability, lyophilization, RNA vaccine platform, saRNA, stability
Graphical abstract
RNA vaccines face challenges during storage and distribution due to their need for frozen storage. In this work, the authors utilize a nanostructured lipid carrier (NLC) delivery system for RNA that can be lyophilized in order to achieve long-term storage stability at refrigerated or room temperatures.
Introduction
RNA-based vaccines show great promise to effectively address existing and emerging infectious diseases,1, 2, 3 including the ongoing pandemic caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). RNA vaccines can be rapidly adapted to new targets and manufactured using sequence-independent operations, thus reducing the cost and time to develop new vaccines, particularly in pandemic settings.4 The Emergency Use Authorization granted to two safe and highly effective mRNA vaccines targeting SARS-CoV-2, less than 1 year after sequencing the novel coronavirus, highlights the power of this new technology.5,6 However, one of the biggest challenges facing these extraordinary new vaccines is the ability to successfully distribute them widely in the face of a pandemic. Cold chain storage is required for both authorized vaccines (−70°C and −20°C for the SARS-CoV-2 RNA vaccines produced by Pfizer/BioNTech and Moderna, respectively). Frozen shipping and storage even at standard freezer conditions poses difficulties in settings with well-established medical infrastructure, challenges greatly compounded in areas with limited resources.7, 8, 9
Lack of stability in RNA vaccines is a critical issue, but the physicochemical reasons behind this are under-studied and poorly understood.9, 10, 11 However, several facts are clear. First, vaccine RNA molecules are prone to cleavage by ubiquitous ribonucleases (i.e., RNases). Engineering of the RNA has previously been done in order to stabilize it, as reviewed by Sahin and colleagues,12 but stability problems remain. Second, due to its size, negative charge, and hydrophilicity, RNA alone cannot easily cross a cell membrane to enter target cells upon injection.13 Thus, RNA delivery formulations are needed to stabilize and protect RNA molecules from degradation (reviewed by Kowalski et al.14 and Guan et al.15). The current system of choice for delivering RNA vaccines, including all SARS-CoV-2 vaccines in clinical trials to date, is a lipid nanoparticle (LNP) delivery system5,16, 17, 18, 19 in which the negatively charged RNA molecule is encapsulated within a multicomponent lipid system. This results in 70–100 nm diameter RNA/LNP complexes that protect the RNA from RNase degradation and allow successful endocytosis by the cell.18,20 However, stability of both the RNA and LNP remain an issue,9, 10, 11 with sensitivity to frozen temperatures resulting in detrimental impacts to their colloidal stability after freeze/thaw.21,22 A number of recent studies have reported on improvements to the long-term thermostability of RNA vaccines at non-frozen temperatures;23, 24, 25 however, all currently authorized RNA vaccines available in the United States still require frozen storage.26,27
A number of alternative lipid-based delivery systems have been proposed and developed to deliver RNA vaccines.28, 29, 30 Here, we demonstrate the ability of a lyophilizable, thermostable nanostructured lipid carrier (NLC) system to effectively deliver replicating RNA-based vaccines by intramuscular (i.m.) injection. This NLC delivery system can also be complexed and lyophilized with mRNA. The liquid NLC alone maintains stability for at least 1 year of storage at refrigerated temperatures, while lyophilized NLC/RNA complexes have been shown to retain their biophysical properties and ability to induce protein expression in vivo after at least 8 months of room temperature storage and at least 21 months at refrigerated temperatures.
Results
Refrigerated stability of NLC delivery system as a liquid
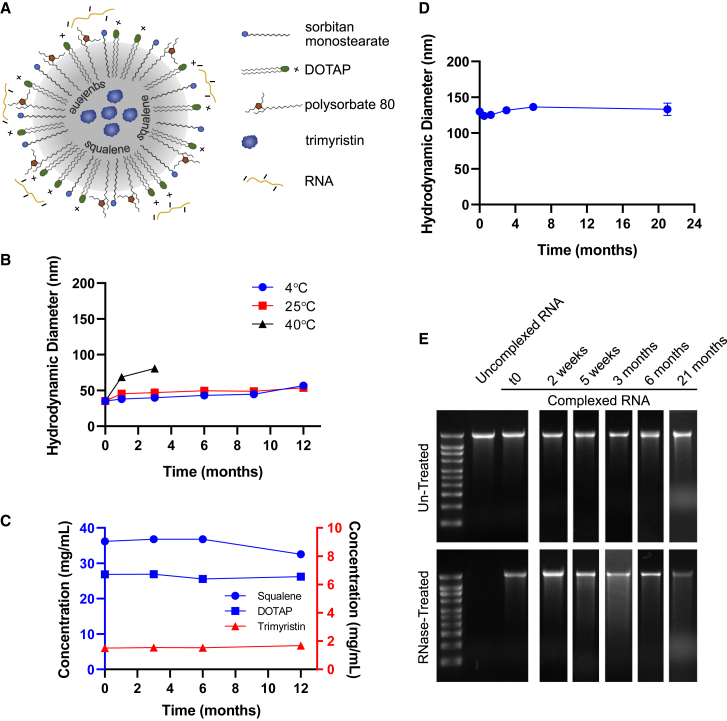
The NLC delivery system (described by Erasmus et al.29) consists of an oil core composed of solid (trimyristin) and liquid (squalene) lipids surrounded by surfactants (sorbitan monostearate and polysorbate 80) and a cationic lipid (1,2-dioleoyl-3-trimethylammonium-propane [DOTAP]) (Figure 1A). This oil-in-water system is prepared in the absence of RNA. The NLC system itself (i.e., in the absence of RNA) displays long-term stability as a liquid at 4°C, maintaining its particle size and component concentrations for at least 1 year (Figures 1B, 1C, and S1). To prepare a vaccine, vaccine RNA is simply mixed with the NLC, and the NLC/RNA complexes form spontaneously through electrostatic interactions between the negatively charged phosphate groups in the RNA backbone and the positively charged amine group in the DOTAP component of the NLC (Figure 1A). With this complexing method, the RNA remains at the oil-water interface of the NLC particle (Figure S2). Using a self-amplifying RNA (saRNA) expressing the reporter protein secreted alkaline phosphatase (SEAP), the NLC is shown to retain its ability to complex with the RNA after storage of the liquid NLC component for at least 21 months at refrigerated temperatures (Figure 1D). The complexed RNA is protected from degradation by RNases despite being outside the NLC, whereas uncomplexed RNA is not (Figure 1E). This is consistent with protection of RNA complexed on the outside of LNPs as demonstrated by others.30 Also, this protection from RNase degradation is maintained even after long-term storage (at least 21 months) of the NLC component as a liquid prior to complexing (Figure 1E). Due to this long-term stability, NLC is suitable for stockpiling for pandemic preparedness applications; RNA targeting a specific pathogen can be rapidly produced in response to a pandemic and complexed with pre-manufactured and stockpiled NLC.
Figure 1.
Nanostructured lipid carrier formulation alone is stable as a liquid at 4°C, allowing stockpiling
(A) Schematic of RNA electrostatically binding to the outside of the NLC. (B) Particle size of NLC alone after storage at indicated temperatures. n = 3 replicate measurements. (C) Concentration of NLC components after long-term 4°C storage. Concentration data after storage at 25°C and 40°C are in Figure S1. n = 3 replicate measurments. (D) Ability of 4°C-stored NLC to complex with SEAP reporter saRNA and produce complexes of consistent particle size after the indicated storage time of the NLC. n = 3 replicate measurements. Error bars represent the standard deviation. (E) Ability of long-term 4°C-stored NLC to protect complexed SEAP reporter saRNA from RNase degradation. Time points represent the length of time that the liquid NLC was stored at 4°C prior to complexing with the SEAP reporter saRNA. Full RNA gel electrophoresis images are in Figure S4.
Lyophilization of Zika NLC/saRNA vaccine
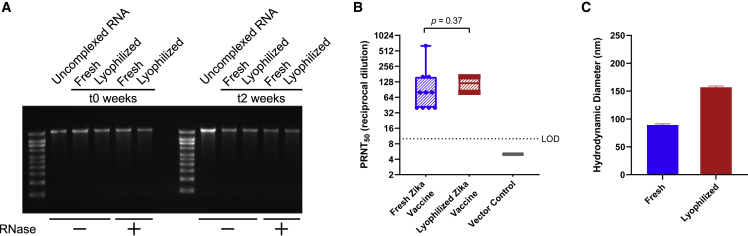
Previously, we demonstrated the utility of an NLC/saRNA vaccine against Zika virus (ZIKV) that induced high levels of neutralizing antibodies and protected mice against viral challenge.29 Here, we demonstrate that the next generation of this Zika NLC/saRNA vaccine (Figure S3A) can be successfully lyophilized for potential long-term storage (Figure 2) with the addition of 10% w/v sucrose as a lyoprotectant. The presence of sucrose promotes the formation of a dense, white, lyophilized cake and also serves to protect the components of the system against the stresses encountered during freezing, drying, and reconstitution.
Figure 2.
Comparison of lyophilized Zika NLC/saRNA vaccine with freshly complexed vaccine
(A) Integrity of Zika saRNA under fresh or lyophilized/reconstituted conditions after it has been extracted from the NLC (RNase –) and protection of Zika saRNA from RNase degradation after it has been treated with RNase and then extracted from the NLC (RNase +). The fresh and lyophilized/reconstituted vaccines were also evaluated for retention of RNA integrity and NLC-mediated protection from RNases after 2 weeks of storage at 4°C. Densitometry analysis of the gel bands is in Figure S5A. (B) In vivo immunogenicity equivalence of fresh and lyophilized/reconstituted Zika vaccine by PRNT at t0. SEAP NLC/saRNA was used as an in vivo negative vector control. n=10 mice in all groups. (C) Hydrodynamic diameter of fresh and lyophilized/reconstituted vaccine by dynamic light scattering (DLS). n = 3 replicate measurements. Error bars represent the standard deviation.
RNA integrity and NLC-mediated protection from RNase degradation is maintained after lyophilization/reconstitution as shown by agarose gel electrophoresis of RNA extracted from NLC/RNA complexes (Figures 2A and S5A). Furthermore, both the freshly complexed liquid and the lyophilized/reconstituted vaccines are stable for at least 2 weeks at refrigerated temperatures (Figures 2A and S5A), retaining their ability to protect the RNA from RNase degradation compared with both freshly mixed and freshly reconstituted lyophilized vaccine. Upon reconstitution and i.m. injection into C57BL/6 mice, the lyophilized Zika saRNA vaccine is able to induce neutralizing (Figure 2B) antibody titers identical to freshly complexed, un-lyophilized vaccine at the same 1 μg dose, indicating that the lyophilization and reconstitution processes do not affect immunogenicity of this vaccine. The size of the complex does increase post lyophilization and reconstitution (Figure 2C); however, this does not appear to affect in vivo efficacy. Thus, beyond the utility of NLC for stockpiling, NLC/RNA vaccines are readily lyophilizable, which has the potential to significantly ease the challenges of distributing RNA vaccines in both pandemic and non-pandemic situations.
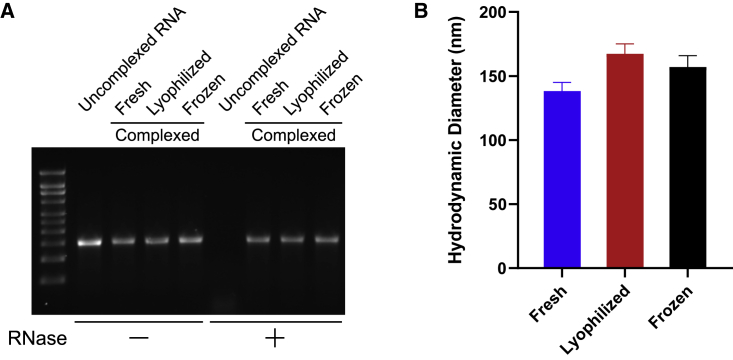
Lyophilization of OVA NLC/mRNA model vaccine
As mRNA-based vaccines have been the frontrunners in coronavirus disease 2019 (COVID-19) response, the ability of a thermostable delivery system to effectively deliver vaccine mRNA is a critical need now and in the future. The flexibility and utility of this NLC-based system is shown by complexing it with commercially available mRNA encoding ovalbumin (OVA). Biophysical characterization of the NLC/mRNA complexes shows protection of the mRNA against degradation by RNase (Figures 3A and S5B) as with the Zika NLC/saRNA complexes. Increasing the concentration of the lyoprotectant to 20% w/v sucrose minimizes the lyophilization-induced increase in particle size that was seen with the Zika NLC/saRNA vaccine containing only 10% w/v sucrose (compare Figures 3B with Figure 2C), resulting in a particle size post lyophilization and reconstitution that is similar to that observed in complexes that are frozen and subsequently thawed. This demonstrates proof of concept for lyophilization of NLC/mRNA complexes, resulting in similar biophysical characteristics to those of frozen NLC/mRNA complexes.
Figure 3.
Comparison of lyophilized or frozen OVA NLC/mRNA with freshly complexed material
(A) Integrity of OVA mRNA under fresh, frozen, or lyophilized conditions after it has been extracted from the NLC complex (RNase –) and protection of OVA mRNA integrity after it has been treated with RNase and then extracted from the NLC complex (RNase +). Densitometry analysis of the gel bands is in Figure S5B. (B) Hydrodynamic diameter of fresh, frozen, and lyophilized complexes by DLS. n = 3 replicate measurements. Error bars represent the standard deviation.
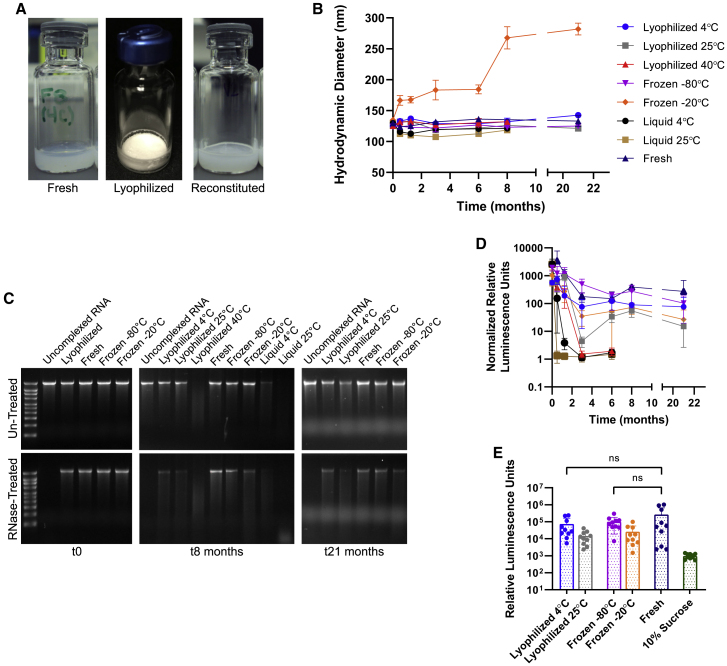
Long-term stability of lyophilized SEAP NLC/saRNA complexes
Finally, we demonstrate the long-term thermostability of the NLC-based RNA vaccine platform using a self-amplifying RNA antigen expression reporter system expressing secreted alkaline phosphatase (SEAP-saRNA) (Figure S3B), which allows sensitive mouse serum detection of i.m.-injected saRNA. Lyophilized SEAP NLC/saRNA complexes with 20% w/v sucrose as a lyoprotectant stored at 4°C, 25°C, and 40°C are compared with frozen complexes stored at −80°C and −20°C, liquid complexes stored at 4°C and 25°C, and freshly made complexes prepared each analysis day. All lyophilized samples maintain an elegant, white cake throughout the study with no discoloration or cracking and minimal cake shrinkage. All lyophilized samples readily reconstitute with nuclease-free water into the milky white solution typically observed for the NLC/RNA complexes (Figure 4A).
Figure 4.
SEAP NLC/saRNA under lyophilized, frozen, or liquid storage conditions in comparison with freshly complexed material
(A) Vial images of freshly complexed, lyophilized, and reconstituted material at t0. (B) Hydrodynamic diameter of the complexes over time compared with a freshly complexed control. n = 3 replicate measurements. Error bars represent the standard deviation. (C) RNA integrity of the stored samples by agarose gel electrophoresis at t0, t8 months, and t21 months, and protection after treatment with RNase at each time point. Gel images at all time points have been shown in Figure S4, and densitometry analysis is shown in Figure S5C. (D) Normalized in vivo SEAP expression for lyophilized, frozen, or liquid stored samples in comparison with freshly complexed material after long-term storage. SEAP expression of each sample was normalized to the SEAP expression of the 10% sucrose solution negative control at each time point. Error bars represent the standard deviation. (E) Comparable in vivo SEAP expression at 21 months for lyophilized vaccine stored at 4°C, frozen vaccine stored at −80°C, and freshly prepared vaccine; the 10% sucrose solution group serves as a negative control and contains no SEAP NLC/saRNA. ns, non-significant difference (p>0.05).
Initially, all NLC/saRNA complexes (Figure 4B) measure 125 ± 10 nm in diameter, including liquid, frozen, and lyophilized versions. Differences of less than 15% are observed between the initial and final time points for all conditions except for frozen material stored at −20°C. This demonstrates the excellent colloidal stability of NLC/RNA complexes, allowing them to withstand the stresses of the lyophilization process and long-term storage, even at elevated temperatures (40°C for lyophilized and 25°C for liquid storage). It is interesting to note that, while size stability is not maintained for complexes stored at −20°C, this did not affect the ability of the NLC/saRNA complex to drive protein expression in vivo upon i.m. injection (Figures 4D and 4E).
RNA integrity in the NLC/saRNA complexes is again retained after lyophilization and after freeze/thaw as demonstrated by agarose gel electrophoresis after extraction of the RNA from the stored NLC complexes, and this integrity is maintained after long-term storage (Figures 4C, S4, and S5C). Samples not treated with RNase show band intensity comparable with the freshly complexed sample for complexes stored lyophilized at 25°C for 8 months, and for complexes stored lyophilized at 4°C and frozen at −80°C and −20°C out to 21 months (Figure S5C). Under these same storage conditions after treatment with RNase, the main RNA band is still present, although its intensity is diminished, whereas no RNA band is present after RNase treatment for samples stored lyophilized at 40°C or liquid at 4°C and 25°C after 8 months of storage. Under accelerated conditions, loss of the main RNA band after RNase treatment is first observed at 2 weeks for the liquid 25°C condition, at 5 weeks for the liquid 4°C condition, and at 3 months for the lyophilized 40°C condition (Figures S4 and S5C).
The ability of stored NLC/saRNA to express protein in vivo is demonstrated by injection of 100 ng of NLC/SEAP-saRNA complex i.m. into C57BL/6 mice, followed by collection of mouse sera 5 days post injection and analysis of SEAP content by enzymatic assay (Figure 4D). At each time point, a group receiving freshly prepared (i.e., not stored) complex was included as a positive control, and a group that received an injection of a 10% sucrose solution was a negative control. SEAP expression at each time point was normalized to this negative control in Figure 4D. Overall, saRNA-mediated protein expression in vivo correlates well with the RNA integrity assessed by gel electrophoresis. Comparable band intensity to the freshly complexed sample in the absence of RNase treatment and presence of a main RNA band after RNase treatment predict SEAP expression in vivo (Figures S4 and S5C). Loss of SEAP expression under accelerated conditions is observed at 2 weeks for the liquid 25°C condition, at 5 weeks for the liquid 4°C condition, and at 3 months for the lyophilized 40°C condition, consistent with the characterization by agarose gel. After 8 months of storage, complex stored lyophilized at 4°C and 25°C and frozen at −80°C and −20°C demonstrated clear serum SEAP expression in vivo (Figure 4D). At the 21-month time point, the lyophilized 25°C-stored and frozen −20°C-stored complexes show a decrease in SEAP expression relative to the freshly complexed material; however, SEAP expression still remains significantly above baseline levels for both conditions (Figure 4E). A key observation is that, after 21 months of storage, no significant difference was detected in the level of in vivo expressed SEAP for the lyophilized 4°C and frozen −80°C-stored complexes when compared with the freshly complexed saRNA/NLC material (Figure 4E). Given the comparability of these stored samples at 21 months with the freshly complexed sample, the decrease in overall luminescence across all samples at later time points seen in Figure 4D is attributed to a change in luminometer instrumentation—with a substantially different photomultiplier detection system—rather than a reflection of the activity of the stored samples. This is shown by the same decrease in luminescence seen in the freshly complexed control sample, which was prepared each analysis day, as with the stored samples. Therefore, the ability of these NLC/saRNA complexes to drive in vivo SEAP expression after storage demonstrates the potential for long-term thermostability of this delivery system.
Discussion
RNA vaccines are important tools to combat existing and emerging infectious diseases, including SARS-CoV-2, due to their rapid adaptability to new target pathogens.1, 2, 3, 4, 5, 6 However, strict cold chain requirements for current RNA vaccine formulations greatly complicate global distribution and increase cost, leading to calls for rapid advances in the stability of RNA vaccine formulations.9,10 We demonstrate that a safe and effective NLC-based RNA vaccine delivery system29 has potential to enable greatly increased thermostability relative to current LNP formulations. The liquid NLC alone is stable at refrigerated temperatures for greater than 1 year. NLC complexed with mRNA or saRNA is able to be lyophilized with both lyophilized and frozen forms of SEAP NLC/saRNA showing stability after storage for extended periods of time. Moreover, upon reconstitution, NLC-formulated RNA vaccine retains its integrity by agarose gel electrophoresis for at least 2 weeks of storage at refrigerated temperatures. This NLC-based delivery technology may have significant applications for RNA vaccine manufacture, storage, distribution, and overall cost due to its thermostable properties.
We hypothesize multiple mechanisms behind the improved thermostability of NLC-based delivery formulations relative to LNP-based formulations. First, the robust physical stability of the NLC allows minimal growth in particle size, retention of constituent components, and maintenance of complexing compatibility for at least 1 year under refrigerated storage. While refrigerated storage stability data for the SARS-CoV-2 mRNA/LNP vaccines is limited in the literature, Moderna recommends storage of unopened vials for up to 1 month at 2-8°C,26 and Pfizer/BioNTech recommends up to 10 weeks of 2-8°C storage.27 Other reports suggest that liquid LNP formulations may be stable for multiple months at refrigerated temperatures with non-mRNA-based systems.21,31 Thus, the limited stability of liquid mRNA/LNP vaccines at refrigerated temperatures is more attributable to limited mRNA stability, even with the RNA fully encapsulated in the delivery vehicle, than instability of the delivery vehicle itself.10
Second, the NLC system provides excellent protection to RNA against RNases. Currently authorized RNA vaccines encapsulate the RNA in the core of the LNP. However, this encapsulation is not strictly necessary to protect and deliver RNA to cells.30 With the NLC delivery system, the electrostatic interaction between RNA’s negatively charged phosphate backbone and the positively charged amine group of the NLC’s DOTAP component drives NLC/RNA complex formation and allows improved protection of the NLC-complexed RNA from cleavage by RNases during long-term storage and after administration compared with uncomplexed RNA.
Third and most importantly, the physical characteristics of this NLC-based RNA vaccine formulation allow lyophilization, a technique commonly used to stabilize vaccines and biologics and eliminate a cold chain requirement.7,8,32, 33, 34, 35 In lyophilized drug products, non-reducing sugars (such as sucrose) act as lyoprotectants through multiple proposed mechanisms, such as replacing water in hydrogen bonding with the components of the system or enclosing the system within the rigid sugar matrix of the dried state where enzymatic or other degradation is limited.36 As noted above, mRNA stability appears to be the limiting factor in the shelf-life of current mRNA/LNP vaccines. To address that concern, RNA molecules alone have been shown to be amenable to lyophilization, with Jones et al.33 reporting that lyophilized RNA retained its ability to drive protein expression after storage at ≤4°C for up to 10 months. However, lyophilization of liposome-like formulations has been pursued for decades (reviewed by Franze et al.36 and Wang et al.37) but can be difficult due to the liposome’s physical structure (i.e., a lipid bilayer surrounding a core aqueous phase). The freezing, drying, and reconstitution steps of lyophilization may result in bilayer rupture, drug leakage, and/or colloidal instability.36,37 While the exact structure of mRNA-loaded LNPs is still being investigated38, 39, 40 and may vary based on composition and production process, their hypothesized core-shell structure10 may still be susceptible to the same rupture, leakage, and instability as liposomes. Recent published attempts at RNA/LNP vaccine lyophilization have been semi-successful, but either were not evaluated after long-term storage21 or showed significant loss of RNA activity after long-term storage even with the addition of lyoprotectants.22 While optimization of LNP lyophilization may yet be attempted (reviewed by Chen and colleagues41), the technical challenge of redesigning and clinically testing lyophilizable liposome-based RNA vaccine delivery formulations is significant and without guaranteed success. In contrast, the structure of the NLC delivery system is more similar to an oil-in-water emulsion than to an LNP. Bilayer rupture and/or drug leakage are not a concern with this system because the RNA is complexed to the surface of the NLC, and maintenance of RNA integrity and colloidal stability have been demonstrated in our study. Furthermore, lyophilized vaccines containing squalene-based adjuvant systems have previously demonstrated potential for long-term vaccine thermostability.42
Another potential advantage of the NLC delivery system for pandemic response is its straightforward and scalable manufacturing process. This employs processes and equipment similar to the oil-in-water emulsion technology already used in licensed vaccines, key manufacturing properties to support large-scale pandemic response. The NLC system also does not require the use of specially designed, proprietary ionizable lipids to generate an appropriate immune response as is the case with LNP-based formulations.43 Rather, the system relies on the presence of squalene—in combination with innate immune-stimulating double-stranded RNA (dsRNA) intermediates produced by saRNAs—to stimulate robust immune responses. Therefore, the NLC can use the commercially available cationic lipid DOTAP as the source of positive charge. Furthermore, in contrast to currently authorized RNA/LNP vaccines, the NLC delivery system is manufactured separately from the RNA. For pandemic preparedness, the long-term refrigerator-stable NLC alone could be stockpiled to enable rapid response. Because it is manufactured separately, RNA of different lengths or with multiple genetic variations may be rapidly synthesized and complexed on the outside of the NLC, allowing rapid vaccine adaptation to evolving viral variants or emerging pathogens.
We do note that the presented long-term stability data of the lyophilized NLC system is with RNA expressing a reporter protein (i.e., SEAP) rather than a vaccine antigen. While use of a reporter protein is a common approach,21,30,44 a demonstration of long-term stability with an actual vaccine construct, including maintenance of vaccine immunogenicity, is a clear next step for development of this technology. However, it is likely that vaccine antigen expression would be similar to that of the reporter protein expression demonstrated here. Additionally, long-term stability of the NLC formulation alone is relevant for application to any vaccine target. Finally, while this specific NLC-based formulation has not yet been clinically tested, safety concerns are low. Squalene, polysorbate 80, and sorbitan monostearate are already in US Food and Drug Administration (FDA)-licensed drug products. Trimyristin is commonly used in the cosmetics industry and is closely related to tristearin, found in licensed drug products, and the cationic lipid DOTAP has been successfully evaluated in multiple clinical trials.
While the current study demonstrates the excellent thermostability of an NLC/RNA complex, this was a proof-of-concept attempt for this system and did not include optimization of the formulation matrix (i.e., buffer, pH, or additional excipients) or lyophilization cycle. Further work will be conducted to optimize this system to push the limits of manufacturing, storage, and use conditions and then to demonstrate its utility with an actual vaccine product. Future development of a spray-dried RNA vaccine, for example, could potentially harness the advantages of a dried system in terms of stability, as seen in this study, while decreasing potential manufacturing bottlenecks that lyophilization can pose. Additionally, further optimization of the NLC/RNA drug product formulation may also allow greater liquid stability, leading to significantly easier manufacturing, storage, and distribution. Widespread vaccine administration in the face of a pandemic remains challenging. With its flexibility and enhanced stability, the NLC delivery system provides an additional tool for RNA vaccine delivery to improve global vaccine distribution both now and in the future.
Materials and methods
saRNA DNA templates
DNA templates for saRNA encoding the Zika pre-membrane (prM) and envelope (E) proteins were produced as previously described.29 Briefly, sequences for the ZIKV signal peptide at the N-terminal end of the capsid protein through the prM and E genes were taken from ZIKV strain H/PF/2013 (GenBank: KJ776791), codon optimized for mammalian expression, and subcloned into a T7-TC83 plasmid. The resulting plasmid pT7-VEE-Zika-prME contains the 5′ UTR, 3′ UTR, and non-structural proteins derived from the attenuated TC-83 strain of Venezuelan equine encephalitis virus (VEEV), with the aforementioned ZIKV genes replacing the VEEV structural proteins downstream of a subgenomic promoter (Figure S3A). Plasmid pT7-VEE-Zika-prME varies slightly from the previously published Zika vaccine plasmid29 with a change of the antibiotic resistance gene from ampicillin to kanamycin to allow manufacture according to current Good Manufacturing Practices (cGMP) and an optimization of the subgenomic promoter for antigen expression enhancement.
Similarly, DNA templates for self-amplifying RNA encoding the SEAP protein were constructed in two different versions (Figures S3B and S3C). The first, pT7-VEEV-SEAP-V1, is identical to that published by Erasmus et al.29 and was used as the template for all SEAP-saRNA used in the long-term stability studies shown in Figure 4. An updated version (pT7-VEEV-SEAP-V2) reflects the same antibiotic resistance gene and subgenomic promoter changes described above to allow optimal comparison with pT7-VEE-Zika-prME in the vaccine immunogenicity studies in Figure 2. All plasmid sequences were confirmed using Sanger sequencing. DNA templates were amplified in Escherichia coli and isolated using maxi or gigaprep kits (Qiagen) and linearized by NotI restriction digest (New England Biolabs). Linearized DNA was purified by phenol-chloroform extraction.
RNA production and purification
Generation of saRNA stocks was achieved by T7 promoter-mediated in vitro transcription using NotI-linearized DNA template. In vitro transcription was performed using an in-house-optimized protocol using T7 polymerase, RNase inhibitor, and pyrophosphatase enzymes procured from Aldevron. DNA plasmid was digested away (DNase I, Aldevron) and cap0 structures were added to the transcripts by vaccinia capping enzyme, GTP, and S-adenosyl-methionine (Aldevron). RNA was then purified from the transcription and capping reaction components by chromatography using a CaptoCore 700 resin (GE Healthcare) followed by diafiltration and concentration using tangential flow filtration. The saRNA material was terminally filtered with a 0.22 μm polyethersulfone filter and stored at −80°C until use. All saRNA was characterized by agarose gel electrophoresis and quantified both by UV absorbance (NanoDrop 1000) and Ribogreen assay (Thermo Fisher). OVA-expressing mRNA was obtained from a commercial vendor (TriLink CleanCap OVA mRNA, L-7610).
NLC formulation production
The NLC formulation was prepared as described previously.29 Briefly, squalene (Sigma), sorbitan monostearate (Sigma), DOTAP (Corden), and trimyristin (IOI Oleochemical) were mixed and heated at 70°C in a bath sonicator. Separately, polysorbate 80 (Fisher Scientific) was diluted in 10 mM sodium citrate trihydrate and also heated to 70°C in a bath sonicator. After all components were dissolved, the oil and aqueous phases were mixed at 7,000 rpm in a high-speed laboratory emulsifier (Silverson Machines). The mixture was then processed by high-shear homogenization to further decrease particle size. Using an M-110P microfluidizer (Microfluidics), the colloid mixture was processed at 30,000 psi for 11 discrete microfluidization passes. The NLC product was terminally filtered with a 0.22 μm polyethersulfone filter and stored at 2–8°C until use.
NLC formulation component assay
The concentrations of DOTAP, squalene, and trimyristin in the NLC were determined by high-performance liquid chromatography (HPLC). Samples were prepared in triplicate, diluted 1:20 in HPLC mobile phase B (50 μL of sample into 950 μL of mobile phase B), injected at 10 μL injection volume, then analyzed using an Agilent 1100 quaternary pump HPLC system in combination with a Corona Veo charged aerosol detector (CAD). The method utilized a Phenomenex Synergi Hydro RP C18 80 A column (4 μm 4.6 × 250 mm) with a two-solvent system gradient consisting of a mixture of 75:15:10 methanol:chloroform:water (mobile phase A) and a 1:1 mixture of methanol:chloroform (mobile phase B), with both mobile phases containing 20 mM ammonium acetate and 1% acetic acid. The system was held at 35°C and run at a flow rate of 1 mL/min. DOTAP, trimyristin, and squalene were dissolved in mobile phase B, and the injection volume was varied to create a five-point standard curve.
NLC/RNA complexing
NLC/RNA complexes were prepared at a nitrogen:phosphate (N:P) ratio of 15 for all cases. Fresh complexes were prepared by mixing RNA 1:1 by volume with NLC prepared in a buffer containing 10 mM sodium citrate and 20% w/v sucrose (RNase-free) to achieve a final complex containing 200 ng/μL RNA in an isotonic 5 mM sodium citrate and 10% w/v sucrose aqueous buffer. Complexes for lyophilization were prepared with 10% or 20% w/v sucrose as noted in the text without additional sodium citrate. Complexes were incubated on ice for 30 min after mixing to ensure complete complexing.
NLC/RNA complex lyophilization
Lyophilized complex was prepared using a VirTis AdVantage 2.0 EL-85 bench-top freeze dryer controlled by the microprocessor-based Wizard 2.0 software. The lyophilization cycle consisted of a freezing step at −50°C, a primary drying step at −30°C and 50 mTorr, and a secondary drying step at 25°C and 50 mTorr. At the completion of the cycle, samples were brought to atmospheric pressure, blanketed with high-purity nitrogen, and stoppered prior to being removed from the freeze-dryer chamber. Lyophilized material was reconstituted using nuclease-free water and gently swirled. Reconstituted material was diluted to 5 mM sodium citrate and 10% w/v sucrose (for isotonicity) prior to any in vivo experiments.
Particle size characterization
Hydrodynamic diameter (particle size) of both the NLC formulation alone and the NLC/RNA complex was determined using dynamic light scattering (Zetasizer Nano ZS, Malvern Instruments). Samples were diluted 1:100 in nuclease-free water in triplicate preparations and measured in a disposable polystyrene cuvette with the following parameters: material refractive index (RI) = 1.59, dispersant RI (water) = 1.33, T = 25°C, viscosity (water) = 0.887 centipoise (cP), measurement angle = 173° backscatter, measurement position = 4.65 mm, automatic attenuation.
NLC/RNA complex RNase protection assay
Integrity of RNA after complexing and protection against degradation by RNase was evaluated by agarose gel electrophoresis. Fresh, frozen/thawed, or lyophilized/reconstituted samples were diluted to a final RNA concentration of 40 ng/μL in nuclease-free water. For RNase-treated samples, the diluted RNA was incubated with RNase A (Thermo Scientific) for 30 min at room temperature at amounts sufficient to completely degrade uncomplexed RNA (ratios of 1:40 RNase:SEAP-RNA and 1:200 RNase:Zika-RNA). This was followed by treatment with recombinant Proteinase K (Thermo Scientific) at a ratio of 1:100 RNase A:Proteinase K for 10 min at 55°C. For both treated and un-treated samples, RNA was then extracted from the complexes by adding 25:24:1 phenol:chloroform:isoamyl alcohol (Invitrogen) to the complex 1:1 by volume, vortexing, and centrifuging at 17,000 × g for 15 min. The supernatant for each sample was mixed 1:1 by volume with Glyoxal load dye (Invitrogen) and incubated at 50°C for 20 min. For each complex, 200 ng of RNA was loaded and run on a denatured 1% agarose gel at 120 V for 45 min in Northern Max Gly running buffer (Invitrogen). Uncomplexed RNA under RNase-treated and un-treated conditions was included in each gel as a control. Millenium RNA marker (Thermo Fisher) was included on each gel with markers at 0.5, 1, 1.5, 2, 2.5, 3, 4, 5, 6, and 9 kilobases. Gels were imaged using an ethidium bromide protocol on a ChemiDoc MP imaging system (BioRad). Densitometry analysis of the gel images was performed using Image Lab software version 6.1.0 and comparing sample RNA band intensity with the band intensity of an RNA loading control or a freshly complexed (i.e., not stored) control as appropriate for each experiment.
Solubility of RNA in squalene
The solubility of RNA in squalene was evaluated in the presence and absence of DOTAP. Zika saRNA was diluted in nuclease-free water to a concentration of 200 ng/μL, and DOTAP was solubilized in squalene to a concentration of 0.6% w/v. The saRNA solution was added 1:1 by volume to tubes containing either squalene alone or DOTAP in squalene, then vortexed for 5 s to mix. Controls containing saRNA only and squalene only were used undiluted. All samples and controls were centrifuged at 17,000 × g for 15 min to separate the oil and aqueous layers. After separation, samples were taken from the oil layer, the aqueous layer, and the aqueous layer plus the interface for both the RNA + squalene sample and the RNA + DOTAP + squalene sample. Samples taken from the oil layer were mixed 1:1 by volume with water in preparation for extraction. All other samples were diluted to 40 ng/μL RNA, based on a theoretical concentration of 200 ng/μL. Phenol-chloroform extraction, as described above, was performed for all samples and controls in order to remove the squalene and DOTAP prior to agarose gel electrophoresis conducted using the same method and parameters already described.
Mouse studies
C57BL/6J mice between 4 and 8 weeks of age at study onset obtained from The Jackson Laboratory were used for all animal studies in this work. All animal work was done under the oversight of the Infectious Disease Research Institute (IDRI) Institutional Animal Care and Use Committee and/or the Bloodworks Northwest Research Institute’s Institutional Animal Care and Use Committee. All animal work was in compliance with all applicable sections of the Final Rules of the Animal Welfare Act regulations (9 CFR parts 1, 2, and 3) and the NIH Guide for the Care and Use of Laboratory Animals: Eighth Edition.
Zika NLC/saRNA in vivo immunogenicity
To compare immunogenicity of lyophilized/reconstituted versus freshly complexed Zika NLC/saRNA vaccines, mice (n = 10/group) were immunized with 1 μg of freshly complexed Zika NLC/saRNA vaccine, 1 μg of lyophilized/reconstituted Zika NLC/saRNA vaccine, or 10 μg of SEAP NLC/saRNA complex as a negative control. Vaccine was injected i.m. in 50 μL volumes in both rear quadriceps muscles of each mouse for a total of 100 μL of vaccine per mouse. Injections sites were monitored for signs of reactogenicity for the 3 days post injection, with no such signs noted. Blood samples were taken from all immunized mice 14 days post immunization by the retro-orbital route for serum antibody assays by PRNT.
ZIKV PRNT
Plaque reduction neutralization test (PRNT) assays were performed on mouse serum samples to quantify neutralizing antibody titers. Briefly, Vero (ATCC CCL-81) cells were cultured at standard conditions (37°C, 5% CO2) in antibiotic-free high-glucose DMEM supplemented with GlutaMax (Gibco) and 10% v/v heat-inactivated FBS (HyClone). Cells were plated at a density of 5 × 105 cells/well in six-well plates (Corning) and incubated overnight to form 90% confluent monolayers. Mouse serum samples were serially diluted 1:2 in DMEM containing 1% heat-inactivated FBS. All serum dilutions were then diluted 1:2 with 100 PFU of ZIKV strain FSS13025 and incubated at 37°C for 1 h. Cell supernates were removed and replaced with 200 μL of the virus/serum dilutions and allowed to incubate at culture conditions for 1 h with gentle rocking every 20 min. Two milliliters of overlay medium composed of DMEM containing 1% agarose (SeaKem), GlutaMax, and 1% v/v FBS was added to each well, allowed to solidify, and plates were incubated for 3 days at standard culture conditions. Cells were then fixed in 10% formalin (Fisher Scientific) for 20 min and stained with crystal violet for plaque visualization and counting.
In vivo functionality of stored SEAP NLC/saRNA
To verify the in vivo functionality of long-term stored SEAP NLC/saRNA complexes, mice (n = 5 for t0 to t8 months and n = 10 for t21 months) received a total dose of 100 ng of RNA in a single 50 μL i.m. injection in one hind leg. A control group of mice received a 50 μL i.m. injection of 10% sucrose solution in a hind leg. Blood samples were taken from all immunized mice on days 3, 5, and 7 post injection by the retro-orbital route. Serum samples were assayed for SEAP expression using the NovaBright Phospha-Light EXP Assay Kit for SEAP (Thermo Fisher) according to the manufacturer’s directions. Relative luminescence was measured using a Biotek Synergy2 plate reader. At each time point, SEAP expression for the sample at each storage condition was normalized to the SEAP expression of the 10% sucrose solution control.
Statistical analyses
Comparability of PRNT titers between lyophilized and freshly complexed vaccine presentations for the saRNA Zika vaccine (Figure 2B) were conducted by a two-tailed homoscedastic t test on natural log-transformed PRNT titers. Log-transformed data were visually assessed for normality prior to analysis. Comparability of SEAP expression levels at t21 months for each stored sample with a freshly complexed control (Figure 4E) was conducted using Dunnett’s multiple comparisons test on the data prior to normalization.
Acknowledgments
The authors would like to thank Jacob Archer, Julie Bakken, Peter Battisti, Stacey Ertel, Jasmine Fuerte-Stone, Brian Granger, and Jasneet Singh for technical assistance and Valerie Soza for editing of the manuscript. This project has been funded in whole or in part with federal funds from both the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services under contract no. 75N93019C00059 (C.C.) and from the Defense Advanced Research Projects Agency under cooperative agreement HR0011-18-2-0001. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Defense Advanced Research Projects Agency.
Author contributions
A.G., E.V., M.A., N.V.H., R.K., and C.C. conceived the study. A.G., E.V., N.V.H., R.K., C.F., and C.C. supervised the research. N.V.H. and C.C. acquired funding. A.G., E.V., S.R., E.L., and M.A. contributed to the investigation. A.G., E.V., S.R., E.L., and M.A. contributed to methodology. A.G., E.V., S.R., E.L., and M.A. analyzed the data. A.G. and E.V. visualized the data. A.G. and E.V. wrote the first draft of the manuscript. A.G., E.V., S.R., E.L., M.A., R.K., C.F., and C.C. reviewed and edited the manuscript.
Declaration of interests
C.F. and N.V.H. are co-inventors on patent applications relating to PCT/US2018/37,783, “Nanostructured lipid carriers and stable emulsions and uses thereof.” M.A., A.G., E.V., and R.K. are co-inventors on US patent application nos. PCT/US21/40,388; 63/075,032; and 63/107,383, “Co-lyophilized RNA and nanostructured lipid carrier” and 63/144,169, “A thermostable, flexible RNA vaccine delivery platform for pandemic response.” All other authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtm.2022.03.009.
Contributor Information
Alana Gerhardt, Email: alana.gerhardt@idri.org.
Emily Voigt, Email: emily.voigt@idri.org.
Supplemental information
References
- 1.Deering R.P., Kommareddy S., Ulmer J.B., Brito L.A., Geall A.J. Nucleic acid vaccines: prospects for non-viral delivery of mRNA vaccines. Expert Opin. Drug Deliv. 2014;11:885–899. doi: 10.1517/17425247.2014.901308. [DOI] [PubMed] [Google Scholar]
- 2.Rauch S., Jasny E., Schmidt K.E., Petsch B. New vaccine technologies to combat outbreak situations. Front Immunol. 2018;9:1963. doi: 10.3389/fimmu.2018.01963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang C., Maruggi G., Shan H., Li J. Advances in mRNA vaccines for infectious diseases. Front Immunol. 2019;10:594. doi: 10.3389/fimmu.2019.00594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J., et al. An mRNA vaccine against SARS-CoV-2 - preliminary report. N. Engl. J. Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Perez Marc G., Moreira E.D., Zerbini C., et al. Safety and efficacy of the BNT162b2 mRNA covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumru O.S., Joshi S.B., Smith D.E., Middaugh C.R., Prusik T., Volkin D.B. Vaccine instability in the cold chain: mechanisms, analysis and formulation strategies. Biologicals. 2014;42:237–259. doi: 10.1016/j.biologicals.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 8.Chen D., Zehrung D. Desirable attributes of vaccines for deployment in low-resource settings. J. Pharm. Sci. 2013;102:29–33. doi: 10.1002/jps.23352. [DOI] [PubMed] [Google Scholar]
- 9.Crommelin D.J.A., Anchordoquy T.J., Volkin D.B., Jiskoot W., Mastrobattista E. Addressing the cold reality of mRNA vaccine stability. J. Pharm. Sci. 2021;110:997–1001. doi: 10.1016/j.xphs.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schoenmaker L., Witzigmann D., Kulkarni J.A., Verbeke R., Kersten G., Jiskoot W., Crommelin D.J.A. mRNA-lipid nanoparticle COVID-19 vaccines: structure and stability. Int. J. Pharm. 2021;601:120586. doi: 10.1016/j.ijpharm.2021.120586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uddin M.N., Roni M.A. Challenges of storage and stability of mrna-based covid-19 vaccines. Vaccines (Basel) 2021;9:1033. doi: 10.3390/vaccines9091033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sahin U., Kariko K., Tureci O. mRNA-based therapeutics--developing a new class of drugs. Nat. Rev. Drug Discov. 2014;13:759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 13.Whitehead K.A., Langer R., Anderson D.G. Knocking down barriers: advances in siRNA delivery. Nat. Rev. Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kowalski P.S., Rudra A., Miao L., Anderson D.G. Delivering the messenger: advances in technologies for therapeutic mRNA delivery. Mol. Ther. 2019;27:710–728. doi: 10.1016/j.ymthe.2019.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guan S., Rosenecker J. Nanotechnologies in delivery of mRNA therapeutics using nonviral vector-based delivery systems. Gene Ther. 2017;24:133–143. doi: 10.1038/gt.2017.5. [DOI] [PubMed] [Google Scholar]
- 16.Tam Y.Y., Chen S., Cullis P.R. Advances in lipid nanoparticles for siRNA delivery. Pharmaceutics. 2013;5:498–507. doi: 10.3390/pharmaceutics5030498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao Y., Huang L. Lipid nanoparticles for gene delivery. Adv. Genet. 2014;88:13–36. doi: 10.1016/B978-0-12-800148-6.00002-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reichmuth A.M., Oberli M.A., Jaklenec A., Langer R., Blankschtein D. mRNA vaccine delivery using lipid nanoparticles. Ther. Deliv. 2016;7:319–334. doi: 10.4155/tde-2016-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bahl K., Senn J.J., Yuzhakov O., Bulychev A., Brito L.A., Hassett K.J., Laska M.E., Smith M., Almarsson O., Thompson J., et al. Preclinical and clinical demonstration of immunogenicity by mRNA vaccines against H10N8 and H7N9 influenza viruses. Mol. Ther. 2017;25:1316–1327. doi: 10.1016/j.ymthe.2017.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hassett K.J., Benenato K.E., Jacquinet E., Lee A., Woods A., Yuzhakov O., Himansu S., Deterling J., Geilich B.M., Ketova T., et al. Optimization of lipid nanoparticles for intramuscular administration of mRNA vaccines. Mol. Ther. Nucleic Acids. 2019;15:1–11. doi: 10.1016/j.omtn.2019.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ball R.L., Bajaj P., Whitehead K.A. Achieving long-term stability of lipid nanoparticles: examining the effect of pH, temperature, and lyophilization. Int. J. Nanomedicine. 2017;12:305–315. doi: 10.2147/IJN.S123062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao P., Hou X., Yan J., Du S., Xue Y., Li W., Xiang G., Dong Y. Long-term storage of lipid-like nanoparticles for mRNA delivery. Bioact Mater. 2020;5:358–363. doi: 10.1016/j.bioactmat.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stitz L., Vogel A., Schnee M., Voss D., Rauch S., Mutzke T., Ketterer T., Kramps T., Petsch B. A thermostable messenger RNA based vaccine against rabies. Plos Negl. Trop. Dis. 2017;11:e0006108. doi: 10.1371/journal.pntd.0006108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang N.N., Li X.F., Deng Y.Q., Zhao H., Huang Y.J., Yang G., Huang W.J., Gao P., Zhou C., Zhang R.R., et al. A thermostable mRNA vaccine against COVID-19. Cell. 2020;182:1271–1283.e1216. doi: 10.1016/j.cell.2020.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao H., Wang T.C., Li X.F., Zhang N.N., Li L., Zhou C., Deng Y.Q., Cao T.S., Yang G., Li R.T., et al. Long-term stability and protection efficacy of the RBD-targeting COVID-19 mRNA vaccine in nonhuman primates. Signal Transduct Target Ther. 2021;6:438. doi: 10.1038/s41392-021-00861-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spikevax E.M.A. EPAR - Product Information; 2021. (previously COVID-19 Vaccine Moderna) [Google Scholar]
- 27.Comirnaty E.M.A. 2021. EPAR - Product Information. [Google Scholar]
- 28.Brito L.A., Chan M., Shaw C.A., Hekele A., Carsillo T., Schaefer M., Archer J., Seubert A., Otten G.R., Beard C.W., et al. A cationic nanoemulsion for the delivery of next-generation RNA vaccines. Mol. Ther. 2014;22:2118–2129. doi: 10.1038/mt.2014.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Erasmus J.H., Khandhar A.P., Guderian J., Granger B., Archer J., Archer M., Gage E., Fuerte-Stone J., Larson E., Lin S., et al. A nanostructured lipid carrier for delivery of a replicating viral RNA provides single, low-dose protection against Zika. Mol. Ther. 2018;26:2507–2522. doi: 10.1016/j.ymthe.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blakney A.K., McKay P.F., Yus B.I., Aldon Y., Shattock R.J. Inside out: optimization of lipid nanoparticle formulations for exterior complexation and in vivo delivery of saRNA. Gene Ther. 2019;26:363–372. doi: 10.1038/s41434-019-0095-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki Y., Hyodo K., Tanaka Y., Ishihara H. siRNA-lipid nanoparticles with long-term storage stability facilitate potent gene-silencing in vivo. J. Control Release. 2015;220:44–50. doi: 10.1016/j.jconrel.2015.10.024. [DOI] [PubMed] [Google Scholar]
- 32.Fonte P., Reis S., Sarmento B. Facts and evidences on the lyophilization of polymeric nanoparticles for drug delivery. J. Control Release. 2016;225:75–86. doi: 10.1016/j.jconrel.2016.01.034. [DOI] [PubMed] [Google Scholar]
- 33.Jones K.L., Drane D., Gowans E.J. Long-term storage of DNA-free RNA for use in vaccine studies. Biotechniques. 2007;43:675–681. doi: 10.2144/000112593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petsch B., Schnee M., Vogel A.B., Lange E., Hoffmann B., Voss D., Schlake T., Thess A., Kallen K.J., Stitz L., et al. Protective efficacy of in vitro synthesized, specific mRNA vaccines against influenza A virus infection. Nat. Biotechnol. 2012;30:1210–1216. doi: 10.1038/nbt.2436. [DOI] [PubMed] [Google Scholar]
- 35.Alberer M., Gnad-Vogt U., Hong H.S., Mehr K.T., Backert L., Finak G., Gottardo R., Bica M.A., Garofano A., Koch S.D., et al. Safety and immunogenicity of a mRNA rabies vaccine in healthy adults: an open-label, non-randomised, prospective, first-in-human phase 1 clinical trial. Lancet. 2017;390:1511–1520. doi: 10.1016/S0140-6736(17)31665-3. [DOI] [PubMed] [Google Scholar]
- 36.Franze S., Selmin F., Samaritani E., Minghetti P., Cilurzo F. Lyophilization of liposomal formulations: still necessary, still challenging. Pharmaceutics. 2018;10:139. doi: 10.3390/pharmaceutics10030139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y., Grainger D.W. Lyophilized liposome-based parenteral drug development: reviewing complex product design strategies and current regulatory environments. Adv. Drug Deliv. Rev. 2019;151-152:56–71. doi: 10.1016/j.addr.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 38.Viger-Gravel J., Schantz A., Pinon A.C., Rossini A.J., Schantz S., Emsley L. Structure of lipid nanoparticles containing siRNA or mRNA by dynamic nuclear polarization-enhanced NMR spectroscopy. J. Phys. Chem. B. 2018;122:2073–2081. doi: 10.1021/acs.jpcb.7b10795. [DOI] [PubMed] [Google Scholar]
- 39.Yanez Arteta M., Kjellman T., Bartesaghi S., Wallin S., Wu X., Kvist A.J., Dabkowska A., Szekely N., Radulescu A., Bergenholtz J., et al. Successful reprogramming of cellular protein production through mRNA delivered by functionalized lipid nanoparticles. Proc. Natl. Acad. Sci. U S A. 2018;115:E3351–E3360. doi: 10.1073/pnas.1720542115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leung A.K., Hafez I.M., Baoukina S., Belliveau N.M., Zhigaltsev I.V., Afshinmanesh E., Tieleman D.P., Hansen C.L., Hope M.J., Cullis P.R. Lipid nanoparticles containing siRNA synthesized by microfluidic mixing exhibit an electron-dense nanostructured core. J. Phys. Chem. C Nanomater. Inter. 2012;116:18440–18450. doi: 10.1021/jp303267y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen C., Han D., Cai C., Tang X. An overview of liposome lyophilization and its future potential. J. Control Release. 2010;142:299–311. doi: 10.1016/j.jconrel.2009.10.024. [DOI] [PubMed] [Google Scholar]
- 42.Kramer R.M., Archer M.C., Orr M.T., Dubois Cauwelaert N., Beebe E.A., Huang P.D., Dowling Q.M., Schwartz A.M., Fedor D.M., Vedvick T.S., et al. Development of a thermostable nanoemulsion adjuvanted vaccine against tuberculosis using a design-of-experiments approach. Int. J. Nanomedicine. 2018;13:3689–3711. doi: 10.2147/IJN.S159839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alameh M.G., Tombacz I., Bettini E., Lederer K., Sittplangkoon C., Wilmore J.R., Gaudette B.T., Soliman O.Y., Pine M., Hicks P., et al. Lipid nanoparticles enhance the efficacy of mRNA and protein subunit vaccines by inducing robust T follicular helper cell and humoral responses. Immunity. 2021;54:2877–2892.e2877. doi: 10.1016/j.immuni.2021.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Linares-Fernandez S., Moreno J., Lambert E., Mercier-Gouy P., Vachez L., Verrier B., Exposito J.Y. Combining an optimized mRNA template with a double purification process allows strong expression of in vitro transcribed mRNA. Mol. Ther. Nucleic Acids. 2021;26:945–956. doi: 10.1016/j.omtn.2021.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.