Abstract
The emergence of methicillin-resistant Staphylococcus aureus (MRSA) poses an important threat in human and animal health. In this study, we ask whether resistance and virulence genes in S. aureus are homogeneously distributed or constrained by different animal hosts. We carried out whole genome sequencing of 114 S. aureus isolates from ten species of animals sampled from four New England states (USA) in 2017–2019. The majority of the isolates came from cats, cows and dogs. The maximum likelihood phylogenetic tree based on the alignment of 89,143 single nucleotide polymorphisms of 1173 core genes reveal 31 sequence types (STs). The most common STs were ST5, ST8, ST30, ST133 and ST2187. Every genome carried at least eight acquired resistance genes. Genes related to resistance found in all genomes included norA (fluoroquinolone), arlRS (fluoroquinolone), lmrS (multidrug), tet(38) (tetracycline) and mepAR (multidrug and tigecycline resistance). The most common superantigen genes were tsst-1, sea and sec. Acquired antibiotic resistance (n = 10) and superantigen (n = 9) genes of S. aureus were widely shared between S. aureus lineages and between strains from different animal hosts. These analyses provide insights for considering bacterial gene sharing when developing strategies to combat the emergence of high-risk clones in animals.
Subject terms: Antimicrobial resistance, Genetic variation, Microbiology, Population genetics
Introduction
Staphylococcus aureus is notable for its ability to colonize a wide range of vertebrate hosts, with each host representing a distinct ecological niche. In humans, it is responsible for severe nosocomial and community-associated infections1–3. S. aureus infections in animals are most commonly reported as a cause of mastitis in dairy-producing animals (including cattle, sheep and goats), osteomyelitis in poultry, and skin abscesses, mastitis and septicemia in farmed rabbits4. S. aureus has been reported in companion animals (e.g., dogs, cats)5,6 and wildlife7,8. Animal-associated S. aureus is also known to carry diverse antibiotic resistance genes and can therefore act as a reservoir for transmission of resistant strains to humans9. The emergence and spread of multidrug-resistant and methicillin-resistant S. aureus (MRSA) are therefore not limited to hospital and community settings, but also pose an important threat in veterinary medicine, agricultural systems and food production. Therefore, understanding the population and genome dynamics of MRSA across multiple eukaryotic hosts is key to advancing a One Health approach to attain optimal health for people, domestic animals, wildlife, plants, and the environment10
The ability to switch and adapt to new hosts are a dynamic and a constant feature of the evolution of S. aureus. Some host transitions are particularly notable. Two lineages in the clonal complex (CC) 97 that cause epidemic community-associated MRSA in humans was inferred to have evolved from independent bovine-to-human host jumps more than 40 years ago11. Methicillin resistance in this clone was acquired after the host-jump, likely as a result of strong selective pressures imposed by the widespread use of antibiotics for treating human infections11. Host-jumping events can also happen from humans to animals, as in the case of sequence type 5 (ST5) in broiler chickens that jumped from humans in the early 1970s12. Another lineage, CC398, has jumped from humans to livestock and back to humans, with resistance to methicillin and tetracycline acquired after the introduction to livestock from humans13. The majority of healthcare-associated infections of livestock-associated MRSA CC398 in Denmark from 2014 to 2016 were shown to be the result of repeated spillover introductions from nearby pig farms into healthcare facilities14. Human-to-monkey jumps of S. aureus ST6 and ST15 that occurred 2700 years ago was facilitated by the loss of phage-carrying genes that played a role in human colonization15. Over the past 5000–6000 years, numerous instances of cross-species transmission events have occurred in S. aureus16. These host-species transitions have been associated with horizontal gene transfer (HGT) of genetic elements conferring traits required for survival in the new host16. Some of these cross-species jumps have resulted in the emergence of successful outbreak-causing and widely circulating S. aureus lineages in humans and animals16.
The generalist nature of a pathogen can provide many opportunities for mobilizable DNA to be disseminated more widely among strains that colonize different hosts. However, it remains unclear whether for multi-host bacteria such as S. aureus, horizontally transferred genes follow the distribution of the bacterial cell that harbor them or create their own distribution. In this study, we sought to elucidate the population genomic structure of clinical animal-associated S. aureus. We ask whether the gene pool of resistance and virulence in S. aureus is homogeneously distributed or constrained by different animal hosts. Using whole genome sequencing of 114 S. aureus isolates from ten animal species, we demonstrate that a remarkable array of acquired resistance genes and superantigen genes of S. aureus are widely shared between multiple S. aureus lineages and between strains from different animal hosts. This study provides important insights for considering bacterial gene sharing between different animal hosts in developing strategies to combat the emergence of high-risk clones in animals.
Results
Phylogenetic diversity of animal-associated S. aureus in New England
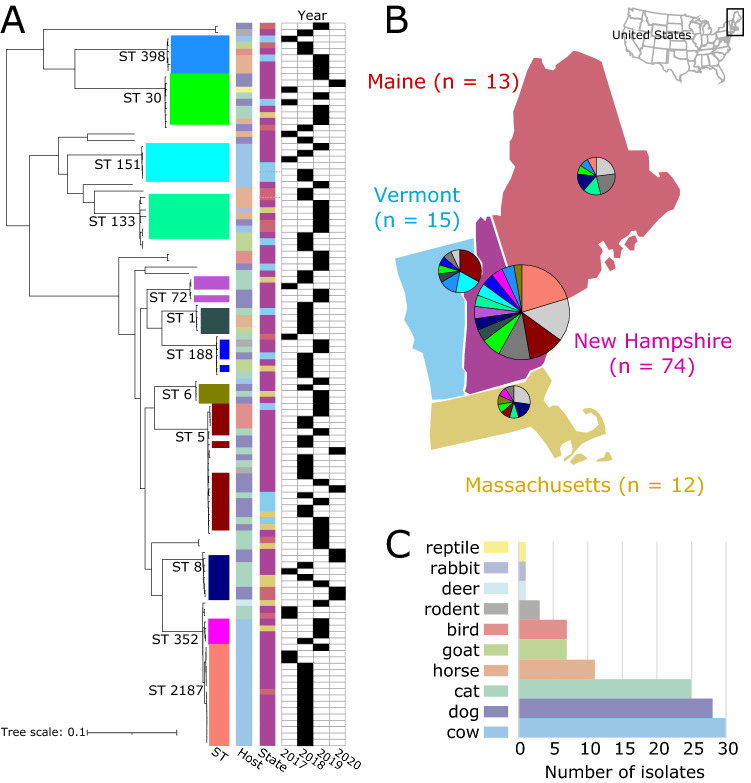
We obtained a total of 114 high quality draft genomes of S. aureus isolates obtained through routine diagnostic tests of clinical specimens from diseased animals. These were submitted to the New Hampshire Veterinary Diagnostic Laboratory (NHVDL) from October 2017 to October 2019 (Fig. 1A and Supplementary Table S1). We obtained the isolates from four states in the United States: New Hampshire (n = 74), Maine (n = 13), Massachusetts (n = 12) and Vermont (n = 15) (Fig. 1B). The majority of isolates came from cows (n = 30), dogs (n = 28) and cats (n = 25) (Fig. 1C). Other domestic animals from which we obtained isolates included horses, goats and rabbits, while wild animals included birds, rodents, deer, rabbits and a tortoise.
Figure 1.
Phylogenetic diversity and sources of animal-associated S. aureus in New England. (A) Midpoint-rooted maximum likelihood phylogenetic tree. Tree scale represent the number of substitutions per site. For visual clarity, only those STs with at least three isolates are labeled. (B) Geographical distribution of STs. Pie charts show the proportion of major STs shown in panel A. Light gray includes STs consisting of a single isolate and novel STs, while dark gray represents STs consisting of two isolates. The number in parentheses indicates the number of isolates from each state. Maps were created using QGIS v3.22 (https://www.qgis.org/en/site/) (C) Number of isolates from different animal hosts.
De novo assembly of the 114 genomes generated sequences of sizes ranging from 2.68 to 2.90 Mb (mean = 2.77 Mb). The number of predicted genes ranged from 2429 to 2739 per genome (mean = 2570) (Supplementary Table S1). The pan-genome of the New England S. aureus population consisted of 7500 orthologous gene families. The genes in the pan-genome were categorized into core genes (present in 99% of genomes), soft-core genes (present in 95% to < 99% of genomes), shell genes (present in 15% to < 95% of genomes), and cloud genes (present in < 15% of genomes). We identified 1773 core genes (present in 113–114 genomes), 116 soft core genes (present in 109–112 genomes), 1158 shell genes (present in 18–108 genomes) and 4453 cloud genes (present in 1–17 genomes) (Supplementary Table S2). The combined core and soft-core genes comprised 25.19% of the pan-genome, while the combined shell and cloud genes (which together make up the accessory genome) comprised 74.81% of the pan-genome. We identified 1716 genes, representing 22.88% of the species pan-genome, that are unique to a single strain.
The maximum likelihood phylogenetic tree based on the alignment of 89,143 SNPs of the core genes revealed many deep branching lineages consisting of 31 previously identified STs (Fig. 1A). The most common STs were ST5 (n = 15 genomes), ST8 (n = 7 genomes), ST30 (n = 8 genomes), ST133 (n = 7 genomes) and ST2187 (n = 16 genomes). Despite the unequal number of isolates from the four states (Fig. 1B), the phylogeny showed a lack of structure relative to the state from which the isolate originated.
Some STs were obtained from multiple animal host species (Fig. 1C), indicating a broad host range: ST5 in cat (n = 3), dog (n = 8), bird (n = 4); ST8 in cat (n = 4), dog (n = 3); ST30 in cat (n = 3), dog (n = 3), horse (n = 1), reptile (n = 1); ST133 in horse (n = 3), dog (n = 1), goat (n = 1), rabbit (n = 1), cow (n = 1); ST398 in horse (n = 3), bird (n = 1), cow (n = 1), goat (n = 1). STs of which all isolates were associated with only a single animal species included STs 151, 352 and 2187, which all originated from cows. However, this ST distribution among animal hosts may reflect the disproportionate number of isolates from cattle in our dataset and is not necessarily reflective of host-restricted niches.
Distribution of acquired genes related to antibiotic resistance
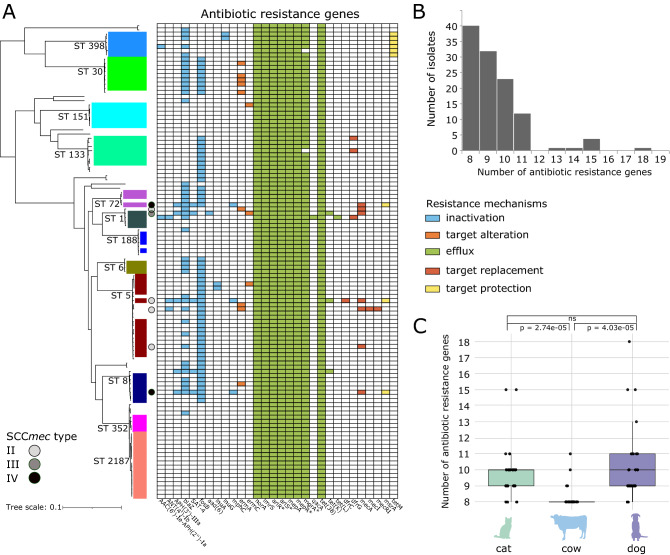
We determined the presence of acquired genes associated with resistance to different antibiotics. These genes represent a variety of resistance mechanisms (drug inactivation, target alteration, efflux, target replacement, target protection) based on definitions in the CARD database17. We identified a total of 30 resistance genes across the entire dataset (Fig. 2A and Supplementary Table S3).
Figure 2.
Antibiotic resistance profiles of animal-associated S. aureus in New England. (A) Distribution of acquired antibiotic resistance genes. The genes are color-coded according to resistance mechanisms. The type of SCCmec is also shown. Asterisks indicate genes associated with antibiotic resistance regulation. (B) Number of antibiotic resistance genes per genome. (C) Comparison of number of resistance genes per S. aureus genome in cats, cows and dogs. Significance was estimated using Welch’s t-test.
In silico detection of the mecA gene from the genome sequences revealed seven MRSA isolates and 107 methicillin-susceptible S. aureus (MSSA) isolates. The mecA gene encodes an extra penicillin-binding protein (PBP2a) that has low affinity to virtually all beta-lactam antibiotics18,19. The mecA gene is carried by a mobile chromosomal cassette SCCmec and is classified into types based on the combination of the ccr and mec complexes they carry19. To date, there are 14 structurally distinct SCCmec types (I–XIV) that have been described19–21. SCCmec typing of the New England S. aureus population revealed the presence of types II (n = 4 genomes), III (n = 1 genomes) and IV (n = 2 genomes) (Fig. 2A and Supplementary Table S1). SCCmec type II was detected in ST 5, type III in ST1 and type IV in STs 8 and 72. We did not detect the gene mecC, which is a divergent form of mecA and also mediates beta-lactam resistance22. We found a slight discrepancy between the in silico detection of the mecA gene and in vitro phenotypic testing for methicillin resistance. There were two isolates (3659B and 4052C) whose genomes did not contain the mecA gene but were phenotypically tested as MRSA. These alternative mechanisms warrant further study.
Resistance-related genes associated with antibiotic efflux systems were ubiquitous in our dataset. We identified 11 such acquired genes, of which seven were found in all 114 genomes. These seven genes included norA (fluoroquinolone resistance), tet(38) (tetracycline resistance), arlRS (fluoroquinolone resistance), lmrS (multidrug resistance) and mepAR (multidrug and tigecycline resistance). The expression of norA is affected by the two-component regulatory system ArlRS23. lmrS confers resistance to aminoglycosides, macrolides, phenicols, diaminopyrimidine and oxazolidinone24. mepA is an efflux protein regulated by mepR and part of the mepRAB cluster, which makes up the multidrug and toxin extrusion (MATE) mechanism in S. aureus25. Also widely detected was mgrA (also known as norR), which is a regulator for norA and tet(38)26. The mgrA gene was detected in all genomes except two.
Other resistance genes were less frequently detected. The gene fosB (fosfomycin resistance) was detected in 60 genomes and was present in some genomes from STs 5, 6, 8, 30, 72 and 133. We detected the gene blaZ (beta-lactam resistance) in 43 genomes and was present in some genomes from STs 5, 6, 8, 30, 72 and 398. In all, every genome carried at least eight resistance-related genes, of which seven genomes have at least 13 resistance related genes (Fig. 2B). Among these less common genes, their distribution across disparate parts of the phylogeny indicates the potential for multiple independent HGT events across different genetic backgrounds, rather than the spread of resistance through clonal expansion.
We next compared the frequency of resistance-related genes among isolates from different animal species. To maintain consistency in our comparison, we focused only on those animals with the highest number of isolates (cats, cows, dogs). We detected a median of ten resistance-related genes (range = 8–15) in S. aureus from cats, ten resistance-related genes (range = 8–18) in S. aureus from dogs and eight resistance-related genes (range = 8–11) in S. aureus from cows (Fig. 2C). The aad(6) gene (aminoglycoside resistance) was found only in one isolate from a cat, whereas the resistance genes dfrC (diaminopyrimidine resistance), lnuG (lincosamide resistance), mecI and mecR1 (beta-lactam resistance), and tetM (tetracycline resistance) appeared only in isolates from dogs. Several other resistance genes including mecA, mphC (macrolide resistance), msrA (erythromycin and streptogramin resistance), tetK (tetracycline resistance), APH(3’)-IIIa (aminoglycoside resistance), ermA (resistance to streptogramin, macrolide and lincosamide) and SAT-4 (nucleoside resistance) were present in S. aureus from dogs and cats, but not from cows. Three STs (STs 151, 352, 2187) which were isolated exclusively from cows exhibited a lack of accessory antimicrobial resistance genes, except for ermC found in a single isolate in those three STs. All other resistance genes found in these three STs were core genes found across all other genomes in the study. The total number of resistance genes varied between isolates from cats and cows, as well as between isolates from dogs and cows (p = 2.74e-05 and 4.03e-05, respectively; Welch’s t-test), but not between isolates from dogs and cats (p = 0.527; Welch’s t-test) (Fig. 2C).
Distribution of staphylococcal virulence genes
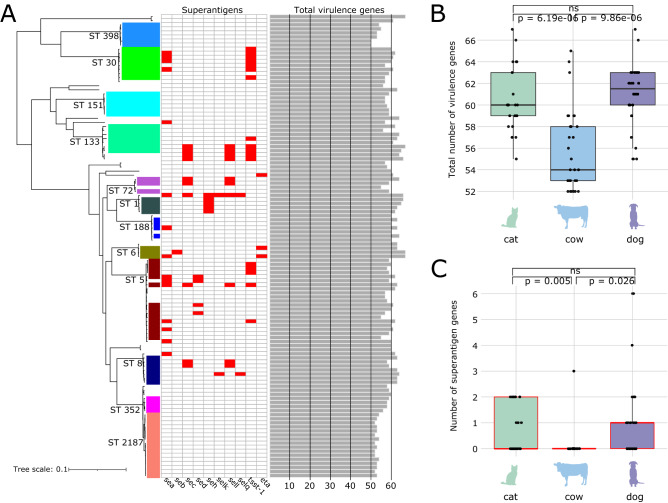
Animal-associated S. aureus carry numerous virulence-related genes. We detected 80 virulence genes in all 114 genomes and of which ten are known as superantigens (Fig. 3A and Supplementary Table S4). Superantigens constitute a family of secreted toxins that trigger excessive non-specific T-cell activation and proliferation, resulting in the overproduction of cytokines27. These potent toxins cause a variety of human diseases from transient food poisoning to lethal toxic shock27. To date, there are at least 24 known superantigens in S. aureus28. Of the 114 genomes in our dataset, 39 genomes harbor 1–6 distinct superantigens that were distributed in divergent parts of the phylogeny. Similar to the phylogenetic distribution of the antibiotic resistance genes described above, the distribution of the superantigens across diverse STs or lineages suggest multiple independent HGT events. The most common superantigen genes were tsst-1 (n = 17 genomes in STs 5, 30 and 133), sea (n = 15 genomes in STs 5, 6, 30 and 188) and sec (n = 10 genomes in STs 5, 8, 72 and 133). STs 151, 352, and 2187 which were isolated exclusively from cows contained no superantigen genes, while ST 2187 exhibited fewer virulence genes compared to all other genomes. ST 398, collected from diverse animal hosts, also showed fewer virulence genes and was additionally void of superantigen genes. A notable virulence factor in S. aureus is the Panton-Valentine Leukocidin (PVL) pore-forming cytotoxin assembled by the genes lukF-PV and lukS-PV29. PVL toxin-producing strains causes leukocytolysis and tissue necrosis and are often associated with community acquired MRSA infections in humans30. In our dataset, we detected 96 and 3 genomes that carry lukF-PV and lukS-PV, respectively.
Figure 3.
Virulence profiles of animal-associated S. aureus in New England. (A) Phylogenetic distribution of superantigen genes (shown in presence [red] or absence [white] matrix) and total number of virulence genes (shown in bar plots). (B) Number of virulence genes per S. aureus genome in cats, cows and dogs. (C) Number of superantigen genes per S. aureus genome in cats, cows and dogs. Significance was estimated using Welch’s t-test.
We also compared the number of virulence genes among isolates from the three most common animal hosts. Staphylococcal virulence genes were unevenly distributed among the three animal hosts. Isolates from cats harbored a median of 60 virulence genes (range = 55–67), 62 (range = 55–67) in dogs and 54 (range = 52–65) in cows (Fig. 3B). The total number of virulence genes varied between strains from cats and cows, as well as between isolates dogs and cows (p = 6.19e-06 and 9.86e-06 respectively; Welch’s t-test), but not between isolates from dogs and cats (p = 0.331; Welch’s t-test) (Fig. 3B). When superantigens were considered, the number of superantigen genes carried by a genome ranged from 0–2, 0–3 and 0–6 in isolates from cats, cows and dogs, respectively (Fig. 3C). There was also no significant difference in the number of superantigen genes between strains from dogs and cats (p = 0.366; Welch’s t-test). Three superantigen genes (sec, sell, tsst-1) were found in isolates from dogs, cats and cows. Six superantigen genes (eta, sea, sed, seh, selk, selq) were found only in dogs and cats, but not cows. Lastly, seb was found only in a single dog.
Widespread gene sharing between animal-associated S. aureus
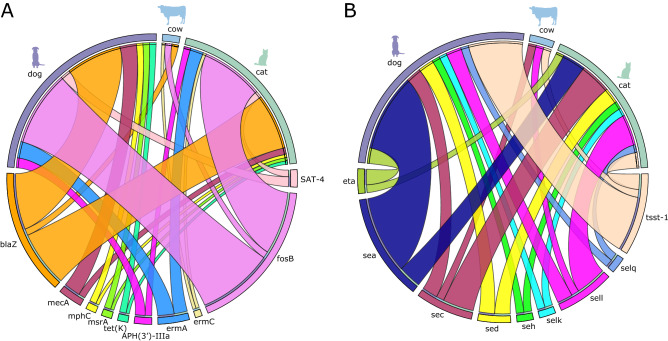
Evidence for HGT in S. aureus and its contributions to adaptation in animal hosts has been demonstrated previously16,31,32. However, little is known about the spread of potentially transferrable resistance and superantigen genes among the animal hosts of S. aureus. We mapped the presence or absence of these genes shared between isolates from the three most common animal hosts (cats, cows, dogs). For the 15 resistance genes found in more than one isolate, the genes blaZ and fosB were present in isolates from all three animal hosts (Fig. 4A). We detected blaZ in 2, 17 and 17 isolates from cows, cats and dogs, respectively. We detected fosB in 2, 19 and 23 isolates from cows, cats and dogs, respectively. The genes which were detected in isolates from both cats and dogs, but not cows, were APH(3')-IIIa, ermA, SAT-4, mecA, mphC, msrA, and tet(K). The gene ermC was present in isolates from cats and cows, but not dogs. Lastly, there were no genes that were shared between dogs and cows, but not cats.
Figure 4.
Distribution of antibiotic resistance (A) and superantigen (B) genes in S. aureus from cats, cows, and dogs. Only genes present in more than one isolate are shown and only the three most common animals were included. The outer ring of the upper half of circos plots represent the number of isolates sampled from dogs (purple), cows (light blue), and cats (light green). The outer ring of the bottom half of the circos plots represent the number of isolates that carry the shared antibiotic resistance genes (panel A) and superantigen genes (panel B). Connecting lines between the specific gene and the animal host are shown if the gene was detected in isolates from any of the three animal hosts.
Staphylococcal superantigens genes are often located in mobile genetic elements, such as prophages, transposons, plasmids and pathogenicity islands33,34, thus facilitating their mobilization. We found that superantigen genes were widely distributed among isolates from different animal hosts (Fig. 4B). We detected sec in 1, 4, and 2 isolates from cows, cats and dogs, respectively. We detected tsst-1 in 1, 2, and 7 isolates from cows, cats and dogs, respectively. The genes sea, sed, seh, selk, selg, and eta were present in isolates from both dogs and cats, but not cows. No superantigens were shared between isolates from dogs and cows, but not cats, Lastly, no superantigen genes were shared between isolates from cats and cows, but not dogs. Overall, these results showed widespread gene sharing between S. aureus gene pools from different host species.
Discussion
While humans are considered its primary reservoir, S. aureus can readily cross species barriers and infect new hosts16. Host-jumping events of bacterial pathogens are likely amplified by agricultural intensification, habitat encroachment and animal domestication35,36. In the case of S. aureus, the range of eukaryotic species that it can colonize as a commensal or opportunistic pathogen remains unclear. Regardless, the remarkable capacity of S. aureus to adapt to new or multiple hosts thus makes it a formidable bacterium that can threaten animal health, agriculture and the economy. Host-jumping events are often associated with acquisition of genetic elements from host-specific gene pools that confer traits required for survival and adaptation in the new host niche16. These traits include a variety of virulence factors such as superantigens that can be used to manipulate innate and adaptive immune responses28. For example, HGT in S. aureus mediated by mobile genetic elements occurs rapidly after a host-jumping event, potentially affecting the innate immune response of the new host16. During animal colonization, frequent HGT is facilitated by few genetic barriers to HGT in vivo (e.g., restriction–modification systems) and the successful replication and integration of different mobile elements37,38.
Here, we sought to elucidate the population genomic structure of 114 S. aureus isolates sampled from diseased animals in New England, USA from 2017–2019. We found that many multidrug resistant STs were detected in multiple wild and domestic animals. Notable were STs 5, 8 and 30, which are major S. aureus clones that are implicated in nosocomial and community-associated infections in humans3,39. ST133 isolates has been identified in healthy donkeys destined for food consumption in Tunisia40, caprine and ovine animals in Australia41, and the gut of healthy humans in Spain42. ST2187 has a long history of association with cows, and more specifically their milk43,44. Although some S. aureus lineages are specifically adapted to a narrow host range on a short evolutionary time scale45, as in the case of STs 151, 352 and 2187 in our study, this may be due to uneven and scarce sampling done in animals, particularly of wildlife species. Also notable is that 43 out of 114 genomes from six STs presented blaZ. For comparison, high penicillin susceptibility rate has been reported in MSSA from bloodstream infections in humans, primarily from CCs 5 and 39846. Penicillin may be considered a therapeutic option in the treatment of animal infections, but ST identity should be carefully considered as blaZ appears to be more phylogenetically widespread in animal-associated S. aureus. As for the two isolates whose genomes did not contain the mecA gene but were phenotypically tested as MRSA, a similar finding has been reported in four isolates from the Scottish MRSA Reference Laboratory and which has been posited to indicate the existence of alternative mechanisms of beta-lactam resistance47. Whole genome sequencing also revealed that those isolates phenotypically tested as MSSA often harbor numerous antibiotic resistance determinants. This result highlights the need to further investigate resistance characteristics beyond methicillin resistance in animal-associated S. aureus, which are often overlooked in many surveillance studies. In addition, such efforts will be instrumental in advancing the One Health concept, focused on the interconnectedness of animal, human and environmental well-being10.
Genes that encode for antibiotic resistance and superantigens were shared not only between divergent genetic backgrounds (or STs) of S. aureus but also between the animal hosts in which they reside. Hence, the ability of S. aureus to colonize multiple animal hosts means that mobilizable DNA may be disseminated more widely, creating a shared pool of resistance and virulence that is not limited by the animal hosts that harbor them. Our study also showed that cats, cows and dogs were frequent carriers of numerous S. aureus clones, each with distinct repertoire of resistance and superantigen genes. Hence, these animals are a major reservoir of clinically relevant genes and high-risk clones that can be transmitted to humans through frequent close contact. Overlapping ecological niches and/or physical proximity between animal hosts (e.g., pets in the same household, livestock animals in the same or nearby farms, interactions at the interface between wild and domestic animals) can certainly promote multi-host colonization and frequent gene sharing between S. aureus isolates. Domestic animals can therefore act as melting pots whereby genetic elements from various S. aureus lineages are combined in new genetic backgrounds. A variety of genetic assortments can promote the rapid emergence of high-risk clones with novel phenotypic characteristics. Large-scale genomic changes derived through HGT can generate “hopeful monsters” that may potentially cause public and animal health threats in ways that are hard to predict48,49. Because S. aureus can also acquire DNA from other Staphylococcus species with which it shares its niche with31,50, its gene pool may be further augmented with DNA that can confer additional adaptive or pre-adaptive features.
There are limitations in our study that need to be acknowledged. We recognize the sampling bias in our dataset that heavily favored isolates from cats, cows and dogs. Moreover, most isolates in our dataset were collected in New Hampshire where NHVDL is located, resulting in comparatively low sample sizes from the surrounding regions. Such bias did not allow us to carry out a more systematic analyses of the host distribution of STs and patterns of gene sharing between animal hosts that included other eukaryotes. The limited number of eukaryotic hosts means that those STs identified as host-restricted may in fact be found in multiple animal species. This also means that certain STs were overrepresented and rare ones were overlooked. Wildlife-associated S. aureus may likely harbor novel genetic variants or mobile genetic elements that pose an unknown level of risk to humans, companion animals and livestock. The structure and gene content of mobile genetic elements, such as pathogenicity islands and phages, and how they shape patterns of gene sharing in animal-associated S. aureus should also be investigated. Future work should therefore include a broader surveillance of S. aureus in other less commonly studied domestic animals and wildlife species, especially those species that often interface with livestock and/or exist at the junction of urban and natural landscapes. It is likely that the known range of species that S. aureus can colonize will expand as studies continue to examine S. aureus in wild animals. Our dataset also included only isolates from disease cases; hence, we do not have data to describe the extent of S. aureus carriage in animals and its contributions to the overall population genetic structure. Comparison of S. aureus in carriage and infections is critical to understanding the genetic basis of pathogenicity and hence, reduce the threats to animal health. Lastly, our study encompassed only three years of bacterial sampling, which was not sufficient to elucidate the long-term evolution of highly virulent and/or multidrug resistant lineages. Future investigations therefore necessitate close monitoring of high-risk clones and the underlying reasons that contribute to their persistence or expansion in the population.
In summary, our findings highlight the role of animals in disseminating resistance and virulence determinants of S. aureus. The remarkable ability of S. aureus as a versatile, multi-host pathogen lies partly on its ability to acquire and disseminate genetic material between lineages and between animal hosts within a short period of time. This study reveals widespread gene sharing between bacterial strains colonizing different animal hosts and highlights the need for routine surveillance to capture the dynamic genetic context of S. aureus.
Methods
Sample collection in New England
The New England S. aureus collection consisted of 133 isolates that were retrospectively sampled from September 2017 through March 2020. Isolates were obtained as culture swabs from routine clinical specimen submissions to the New Hampshire Veterinary Diagnostic Laboratory (NHVDL), New Hampshire, USA. The clinical specimens were received from multiple veterinary practices from the states of Connecticut, New Hampshire, Maine, Massachusetts and Vermont. These states are located in the northeastern part of the country. All isolates were from animals with confirmed clinical infections. No live vertebrates were used in this study; hence, the NHVDL was exempt from the IACUC approval process. Pure isolates were cultured in commercially prepared tryptic soy agar with 10% sheep red blood cells and brain heart infusion broth. Initial species identification was carried out using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) using the Bruker Biotyper instrument and following manufacturer’s protocols. Species assignments were made by comparing mass spectra of our samples to two libraries of reference spectra RUO library 6903(V6) and 7311(V7) available in the Bruker MBT Compass (Bruker Daltonics, Bremen, Germany). The most common sites of isolation included skin, ears, nasal passages, milk and wounds. Associated metadata information for each isolate are included in Supplementary Table S1. All isolates were stored in DMSO solution in − 80 °C.
Methicillin susceptibility screening
In vitro screening for cefoxitin and oxacillin resistance was carried out using the Kirby Bauer disc diffusion technique. We followed the breakpoint guidelines of the most current Clinical and Laboratory Standards Institute using cefoxitin, which is used as the official predictor of methicillin resistance for S. aureus51. For isolates identified as methicillin resistant, we determined the presence of the penicillin-binding protein PBP2 using a commercial latex agglutination test (MASTALEX MRSA Latex Kit, MAST, UK) following manufacturer's guidelines. We confirmed the presence of the mecA gene by screening the genome sequence of each isolate (described below).
DNA extraction and whole genome sequencing
DNA extraction was carried out using the Zymo Research Quick-DNA Fungal/Bacterial Miniprep Kit (Irvine, California) following the manufacturer's protocol. We quantified DNA concentration using a Qubit fluorometer (Invitrogen, Grand Island, NY). We prepared DNA libraries using the Nextera XT protocol with 1 ng of genomic DNA per isolate. Samples were sequenced using the Illumina HiSeq platform (San Diego, California) to produce 250 bp paired end reads. Sequencing was carried out at the Hubbard Center for Genome Studies at the University of New Hampshire.
Genome assembly and annotation
We assembled all genomes using Shovill v1.1.0 (https://github.com/tseemann/shovill). Shovill is a series of methods that includes subsampling read depth down to 150X, trimming adapters, correcting sequencing errors and assembling using SPAdes v3.13.052. We used QUAST v5.0.253 and CheckM v1.1.354 to assess the quality of our sequences and exclude genomes with < 90% completeness and > 5% contamination. We also excluded assemblies with > 200 contigs and an N50 < 40,000 bp. Only 19 out of 133 genomes did not pass these quality thresholds and were therefore excluded from all downstream analyses. The remaining 114 genomes were annotated using Prokka v1.14.655. Genomes were compared to the S. aureus reference genome (NCBI Reference Sequence: Accession No. NC_007795.1) with the program FastANI v1.32 to confirm species identification using the > 95% average nucleotide identity (ANI) threshold56.
Pan-genome analysis and phylogenetic reconstruction
We used Roary v3.13.057 to characterize core genes and accessory genes that make up the pan-genome57. To balance the tradeoff between inferring robust phylogenetic relationships versus accounting for assembly errors, we included core genes if they were present in ≥ 99% of the genomes. Nucleotide sequences of each orthologous gene family were aligned using MAFFT v7.47558. Aligned core genes were concatenated to generate a core genome alignment. Phylogenetically informative single nucleotide polymorphisms (SNPs) in the core genome alignment were extracted using SNP-sites59. We used the core SNP alignment to construct a maximum likelihood phylogenetic tree using RAxML v8.2.1260 employing a general time-reversible nucleotide substitution model61 and four gamma categories for rate heterogeneity. Phylogenetic trees were visualized using the online platform Interactive Tree of Life (IToL)62.
In silico sequence typing, detection of resistance genes, virulence genes and SCCmec
Using the contig files, we determined the multilocus sequence types (ST) for all genomes used in this study using the program mlst v2.19.0 (https://github.com/tseemann/mlst). STs pertain to allelic profiles that characterize nucleotide differences in partial sequences of seven housekeeping genes63. In S. aureus, these seven genes consist of arcC, aroE, glpF, gmk, pta, tpi and yqiL64. Allelic profiles of the genomes used in this study were compared to those in the S. aureus MLST database (https://pubmlst.org/)65. We screened for the presence of horizontally acquired antibiotic resistance genes and virulence factors using ABRicate v1.0.1 (https://github.com/tseemann/abricate) utilizing the Comprehensive Antibiotic Resistance Database (CARD)17 and the Virulence Factor Database (VFDB)66. Finally, we used SCCmecFinder67 implemented in SCCion v0.1 (https://github.com/esteinig/sccion) to determine the presence and type of the mobile genetic element SCCmec. We used the minimum thresholds of > 60% for sequence coverage and > 90% sequence identity to identify the SCCmec. Visualization of the distribution of resistance and virulence genes was carried out using Circos68.
We used the default parameters for each program unless indicated otherwise.
Statistical tests
We used Welch’s t-test to compare the number of resistance and virulence genes of isolates from different animal hosts. Results were considered significant when p < 0.05.
Supplementary Information
Acknowledgements
The authors thank the UAlbany Research Technology Services where all bioinformatics analyses were carried out. We are grateful to the Hubbard Center for Genome Studies at the University of New Hampshire for sequencing support. We are also grateful to the staff of NHVDL for laboratory support.
Author contributions
C.P.A. and S.A.B. designed the work. S.A.B. performed all bioinformatics analyses. J.T.S., J.L.M., J.B., D.N., and R.G. performed all sampling, culturing, and DNA extractions. C.P.A. and S.A.B. wrote the manuscript with contributions from all authors. C.P.A. guided the work. All authors read and approved the final manuscript.
Funding
The study was supported by the National Institutes of Health (NIH) Award No. 1R35GM142924 and startup research funds from the College of Arts and Sciences, University at Albany, State University of New York to CPA. JTS was supported by the University of New Hampshire 2020 Graduate School Dissertation Year Fellowship. The funders had no role in study design, data collection and analysis, decision to publish and preparation of the manuscript and the findings do not necessarily reflect views and policies of the authors’ institutions and funders.
Data availability
Raw reads of the 114 S. aureus genomes have been deposited in the Sequence Read Archive (SRA) in the National Center for Biotechnology Information (NCBI) under BioProject accession No. PRJNA741582. Sample accession numbers and associated metadata are listed in Supplementary Table S1.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Spencer A. Bruce, Email: sbruce@albany.edu
Cheryl P. Andam, Email: candam@albany.edu
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-08230-z.
References
- 1.Klein EY, Sun L, Smith DL, Laxminarayan R. The changing epidemiology of methicillin-resistant Staphylococcus aureus in the United States: a national observational study. Am. J. Epidemiol. 2013;177:666–674. doi: 10.1093/aje/kws273. [DOI] [PubMed] [Google Scholar]
- 2.Tong SYC, Davis JS, Eichenberger E, Holland TL, Fowler VG. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015;28:603–661. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turner NA, et al. Methicillin-resistant Staphylococcus aureus: an overview of basic and clinical research. Nat. Rev. Microbiol. 2019;17:203–218. doi: 10.1038/s41579-018-0147-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fitzgerald JR. Livestock-associated Staphylococcus aureus: origin, evolution and public health threat. Trends Microbiol. 2012;20:192–198. doi: 10.1016/j.tim.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 5.Faires MC, Traverse M, Tater KC, Pearl DL, Weese JS. Methicillin-resistant and -susceptible Staphylococcus aureus infections in dogs. Emerg. Infect. Dis. 2010;16:69–75. doi: 10.3201/eid1601.081758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loncaric I, et al. Increased genetic diversity of methicillin-resistant Staphylococcus aureus (MRSA) isolated from companion animals. Vet. Microbiol. 2019;235:118–126. doi: 10.1016/j.vetmic.2019.06.013. [DOI] [PubMed] [Google Scholar]
- 7.Monecke S, et al. Diversity of Staphylococcus aureus isolates in European wildlife. PLoS ONE. 2016;11:e0168433. doi: 10.1371/journal.pone.0168433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heaton CJ, Gerbig GR, Sensius LD, Patel V, Smith TC. Staphylococcus aureus epidemiology in wildlife: a systematic review. Antibiotics (Basel) 2020;9:89. doi: 10.3390/antibiotics9020089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haag AF, Fitzgerald JR, Penadés JR. Staphylococcus aureus in animals. Microbiol. Spectr. 2019;7:11. doi: 10.1128/microbiolspec.gpp3-0060-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zinsstag J, Schelling E, Waltner-Toews D, Tanner M. From ‘one medicine’ to ‘one health’ and systemic approaches to health and well-being. Prev. Vet. Med. 2011;101:148–156. doi: 10.1016/j.prevetmed.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spoor LE, et al. Livestock origin for a human pandemic clone of community-associated methicillin-resistant Staphylococcus aureus. MBio. 2013;4:e00356-13. doi: 10.1128/mBio.00356-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lowder BV, et al. Recent human-to-poultry host jump, adaptation, and pandemic spread of Staphylococcus aureus. Proc Natl Acad Sci U S A. 2009;106:19545–19550. doi: 10.1073/pnas.0909285106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Price LB, et al. Staphylococcus aureus CC398: host adaptation and emergence of methicillin resistance in livestock. MBio. 2012;3:e0030511. doi: 10.1128/mBio.00305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sieber RN, et al. Genome investigations show host adaptation and transmission of LA-MRSA CC398 from pigs into Danish healthcare institutions. Sci Rep. 2019;9:18655. doi: 10.1038/s41598-019-55086-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Senghore M, et al. Transmission of Staphylococcus aureus from humans to green monkeys in the Gambia as revealed by whole-genome sequencing. Appl. Environ. Microbiol. 2016;82:5910–5917. doi: 10.1128/AEM.01496-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richardson EJ, et al. Gene exchange drives the ecological success of a multi-host bacterial pathogen. Nat. Ecol. Evol. 2018;2:1468–1478. doi: 10.1038/s41559-018-0617-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jia B, et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucl. Acids Res. 2017;45:D566–D573. doi: 10.1093/nar/gkw1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katayama Y, Ito T, Hiramatsu K. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 2000;44:1549–1555. doi: 10.1128/aac.44.6.1549-1555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (IWG-SCC). Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob Agents Chemother53, 4961–4967 (2009). [DOI] [PMC free article] [PubMed]
- 20.Lakhundi S, Zhang K. Methicillin-resistant Staphylococcus aureus: molecular characterization, evolution, and epidemiology. Clin Microbiol Rev. 2018;31(4):e00020-18. doi: 10.1128/CMR.00020-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Urushibara N, Aung MS, Kawaguchiya M, Kobayashi N. Novel staphylococcal cassette chromosome mec (SCCmec) type XIV (5A) and a truncated SCCmec element in SCC composite islands carrying speG in ST5 MRSA in Japan. J. Antimicrob. Chemother. 2020;75:46–50. doi: 10.1093/jac/dkz406. [DOI] [PubMed] [Google Scholar]
- 22.García-Álvarez L, et al. Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. Lancet Infect. Dis. 2011;11:595–603. doi: 10.1016/S1473-3099(11)70126-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fournier B, Aras R, Hooper DC. Expression of the multidrug resistance transporter NorA from Staphylococcus aureus is modified by a two-component regulatory system. J. Bacteriol. 2000;182:664–671. doi: 10.1128/jb.182.3.664-671.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Floyd JL, Smith KP, Kumar SH, Floyd JT, Varela MF. LmrS is a multidrug efflux pump of the major facilitator superfamily from Staphylococcus aureus. Antimicrob. Agents Chemother. 2010;54:5406–5412. doi: 10.1128/AAC.00580-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaatz GW, McAleese F, Seo SM. Multidrug resistance in Staphylococcus aureus due to overexpression of a novel multidrug and toxin extrusion (MATE) transport protein. Antimicrob. Agents Chemother. 2005;49:1857–1864. doi: 10.1128/AAC.49.5.1857-1864.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Truong-Bolduc QC, Dunman PM, Strahilevitz J, Projan SJ, Hooper DC. MgrA is a multiple regulator of two new efflux pumps in Staphylococcus aureus. J. Bacteriol. 2005;187:2395–2405. doi: 10.1128/JB.187.7.2395-2405.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu SX, McCormick JK. Staphylococcal superantigens in colonization and disease. Front Cell Infect. Microbiol. 2012;2:52. doi: 10.3389/fcimb.2012.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tuffs SW, Haeryfar SMM, McCormick JK. Manipulation of innate and adaptive immunity by staphylococcal superantigens. Pathogens. 2018;7:53. doi: 10.3390/pathogens7020053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Melles DC, et al. Panton-Valentine leukocidin genes in Staphylococcus aureus. Emerg. Infect. Dis. 2006;12:1174–1175. doi: 10.3201/eid1207.050865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boyle-Vavra S, Daum RS. Community-acquired methicillin-resistant Staphylococcus aureus: the role of Panton-Valentine leukocidin. Lab Invest. 2007;87:3–9. doi: 10.1038/labinvest.3700501. [DOI] [PubMed] [Google Scholar]
- 31.Méric G, et al. Ecological overlap and horizontal gene transfer in Staphylococcus aureus and Staphylococcus epidermidis. Genome Biol. Evol. 2015;7:1313–1328. doi: 10.1093/gbe/evv066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murray S, et al. Recombination-mediated host adaptation by avian Staphylococcus aureus. Genome Biol. Evol. 2017;9:830–842. doi: 10.1093/gbe/evx037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Novick RP. Mobile genetic elements and bacterial toxinoses: the superantigen-encoding pathogenicity islands of Staphylococcus aureus. Plasmid. 2003;49:93–105. doi: 10.1016/s0147-619x(02)00157-9. [DOI] [PubMed] [Google Scholar]
- 34.Alibayov B, Zdenkova K, Sykorova H, Demnerova K. Molecular analysis of Staphylococcus aureus pathogenicity islands (SaPI) and their superantigens combination of food samples. J. Microbiol. Methods. 2014;107:197–204. doi: 10.1016/j.mimet.2014.10.014. [DOI] [PubMed] [Google Scholar]
- 35.Jones BA, et al. Zoonosis emergence linked to agricultural intensification and environmental change. Proc. Natl. Acad. Sci. U S A. 2013;110:8399–8404. doi: 10.1073/pnas.1208059110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mourkas E, et al. Agricultural intensification and the evolution of host specialism in the enteric pathogen Campylobacter jejuni. Proc. Natl. Acad. Sci. U S A. 2020;117:11018–11028. doi: 10.1073/pnas.1917168117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roberts GA, et al. Impact of target site distribution for Type I restriction enzymes on the evolution of methicillin-resistant Staphylococcus aureus (MRSA) populations. Nucl. Acids Res. 2013;41:7472–7484. doi: 10.1093/nar/gkt535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCarthy AJ, et al. Extensive horizontal gene transfer during Staphylococcus aureus co-colonization in vivo. Genome Biol. Evol. 2014;6:2697–2708. doi: 10.1093/gbe/evu214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Monecke S, et al. A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS ONE. 2011;6:e17936. doi: 10.1371/journal.pone.0017936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gharsa H, et al. High diversity of genetic lineages and virulence genes in nasal Staphylococcus aureus isolates from donkeys destined to food consumption in Tunisia with predominance of the ruminant associated CC133 lineage. BMC Vet. Res. 2012;8:203. doi: 10.1186/1746-6148-8-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McMillan K, Moore SC, McAuley CM, Fegan N, Fox EM. Characterization of Staphylococcus aureus isolates from raw milk sources in Victoria Australia. BMC Microbiol. 2016;16:169. doi: 10.1186/s12866-016-0789-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benito D, Lozano C, Gómez-Sanz E, Zarazaga M, Torres C. Detection of methicillin-susceptible Staphylococcus aureus ST398 and ST133 strains in gut microbiota of healthy humans in Spain. Microb. Ecol. 2013;66:105–111. doi: 10.1007/s00248-013-0240-1. [DOI] [PubMed] [Google Scholar]
- 43.Naushad, S., Nobrega, D. B., Naqvi, S. A., Barkema, H. W. & De Buck, J. Genomic analysis of bovine Staphylococcus aureus isolates from milk to elucidate diversity and determine the distributions of antimicrobial and virulence genes and their association with mastitis. mSystems5, (2020). [DOI] [PMC free article] [PubMed]
- 44.Thomas A, et al. Prevalence and distribution of multilocus sequence types of Staphylococcus aureus isolated from bulk tank milk and cows with mastitis in Pennsylvania. PLoS ONE. 2021;16:e0248528. doi: 10.1371/journal.pone.0248528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matuszewska M, Murray GGR, Harrison EM, Holmes MA, Weinert LA. The evolutionary genomics of host specificity in Staphylococcus aureus. Trends Microbiol. 2020;28:465–477. doi: 10.1016/j.tim.2019.12.007. [DOI] [PubMed] [Google Scholar]
- 46.Mama OM, et al. Penicillin susceptibility among invasive MSSA infections: a multicentre study in 16 Spanish hospitals. J. Antimicrob. Chemother. 2021;76:2519–2527. doi: 10.1093/jac/dkab208. [DOI] [PubMed] [Google Scholar]
- 47.Ba X, et al. Novel mutations in penicillin-binding protein genes in clinical Staphylococcus aureus isolates that are methicillin resistant on susceptibility testing, but lack the mec gene. J. Antimicrob. Chemother. 2014;69:594–597. doi: 10.1093/jac/dkt418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Croucher NJ, Klugman KP. The emergence of bacterial ‘hopeful monsters’. MBio. 2014;5:e01550–e1514. doi: 10.1128/mBio.01550-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Coyle NM, et al. A hopeful sea-monster: a very large homologous recombination event impacting the core genome of the marine pathogen Vibrio anguillarum. Front. Microbiol. 2020;11:1430. doi: 10.3389/fmicb.2020.01430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.John J, George S, Nori SRC, Nelson-Sathi S. Phylogenomic analysis reveals the evolutionary route of resistant genes in Staphylococcus aureus. Genome Biol. Evol. 2019;11:2917–2926. doi: 10.1093/gbe/evz213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.CLSI Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. VET01. (Clinical and Laboratory Standards Institute, 2018).
- 52.Bankevich A, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 56.Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018;9:5114. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Page AJ, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Katoh K, Asimenos G, Toh H. Multiple alignment of DNA sequences with MAFFT. Methods Mol. Biol. 2009;537:39–64. doi: 10.1007/978-1-59745-251-9_3. [DOI] [PubMed] [Google Scholar]
- 59.Page AJ, et al. SNP-sites: rapid efficient extraction of SNPs from multi-FASTA alignments. Microb. Genom. 2016;2:e000056. doi: 10.1099/mgen.0.000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tavaré S. Some probabilistic and statistical problems in the analysis of DNA sequences. Am. Math. Soc. Lect. Math. Life Sci. 1986;17:57–86. [Google Scholar]
- 62.Letunic I, Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucl. Acids Res. 2019;47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maiden MC, et al. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. U S A. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 2000;38:1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jolley KA, Bray JE, Maiden MCJ. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018;3:124. doi: 10.12688/wellcomeopenres.14826.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu B, Zheng D, Jin Q, Chen L, Yang J. VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucl. Acids Res. 2019;47:D687–D692. doi: 10.1093/nar/gky1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kaya H, et al. SCCmecFinder, a web-based tool for typing of staphylococcal cassette chromosome mec in Staphylococcus aureus using whole-genome sequence data. mSphere. 2018;3:e0061217. doi: 10.1128/mSphere.00612-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Krzywinski M, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw reads of the 114 S. aureus genomes have been deposited in the Sequence Read Archive (SRA) in the National Center for Biotechnology Information (NCBI) under BioProject accession No. PRJNA741582. Sample accession numbers and associated metadata are listed in Supplementary Table S1.