Abstract
Intrinsically disordered proteins are frequently involved in important regulatory processes in the cell thanks to their ability to bind several different targets performing sometimes even opposite functions. The PentUnFOLD algorithm is a physicochemical method that is based on new propensity scales for disordered, nonstable and stable elements of secondary structure and on the counting of stabilizing and destabilizing intraprotein contacts. Unlike other methods, it works with a PDB file, and it can determine not only those fragments of alpha helices, beta strands, and random coils that can turn into disordered state (the “dark” side of the disorder), but also nonstable regions of alpha helices and beta strands which are able to turn into random coils (the “light” side), and vice versa (H ↔ C, E ↔ C). The scales have been obtained from structural data on disordered regions from the middle parts of amino acid sequences only, and not on their expectedly disordered N- and C-termini. Among other tendencies we have found that regions of both alpha helices and beta strands that can turn into the disordered state are relatively enriched in residues of Ala, Met, Asp, and Lys, while regions of both alpha helices and beta strands that can turn into random coil are relatively enriched in hydrophilic residues, and Cys, Pro, and Gly. Moreover, PentUnFOLD has the option to determine the effect of secondary structure transitions on the stability of a given region of a protein. The PentUnFOLD algorithm is freely available at http://3.17.12.213/pent-un-fold and http://chemres.bsmu.by/PentUnFOLD.htm.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00726-022-03153-5.
Keywords: Intrinsically disordered proteins; Structural shifts, Computer algorithm, Amino acid substitution, Human prion protein
Introduction
A large fraction of the human proteome comprises proteins that, under physiological conditions, lack ordered 3D structures as a whole or have segments that are not likely to form a defined 3D structure (Dunker et al. 2000; Uversky 2010, 2011). These proteins and regions are referred to as intrinsically disordered proteins (IDPs) and intrinsically disordered protein regions (IDPRs), respectively. IDPs/IDPRs are present in the proteome of all organisms, but were found to be most common in eukaryotic sequences (Romero et al. 1998; Ward et al. 2004a; Peng et al. 2015; Xue et al. 2012; Oates et al. 2013). Today it is already known that approximately 30–40% of human proteins contain one or more structurally disordered region with a length of at least 30 amino acid residues, and about 25% are completely disordered (Dunker et al. 2008). These proteins carry out essential functions breaking the classical structure–function paradigm (Hazy and Tompa 2009; Wright and Dyson 1999). In fact, the percentage of proteins with IDPRs may be even higher, since many structures from PDB (Protein Data Bank) are protein complexes with different ligands, which may contribute to the shift from a disordered to an ordered state and vice versa (Dyson and Wright 2005). However, folding upon binding is limited, and many IDPs preserve a partial disordered state in the bound state (Hazy and Tompa 2009; Tompa and Fuxreiter 2008). About 50% of proteins from PDB have intrinsically disordered regions (Gall et al. 2007) that are situated mostly at N- and C-termini of polypeptide chains. In addition, the crystallization process itself can also help intrinsically disordered regions to acquire a definite 3D structure.
Disordered regions should not be confused with random coil regions. In this study under the term “random coil” we mean regions that do not form neither alpha helix, nor beta strand, but have a definite structure. Disordered regions are those that lack definite 3D structure. We classify regions of alpha helices and beta strands able to turn into random coil as nonstable (N), as well as random coils that can turn into beta strands and alpha helices. In contrast, regions of random coil, alpha helices, and beta strands able to turn into disordered state are classified as disordered (D). So, the aim of the PentUnFOLD algorithm is to distinguish between these two types of structural instability: between “N” and “D”.
IDPs/IDPRs are widely associated with numerous human diseases (Uversky et al. 2008). Every year, the number of diseases that involve IDPs/IDPRs or proteins that can make structural transitions increases. Without structural transitions, it would not be possible to assemble and spread for the Influenza virus (Luo et al. 2012) and for the coronaviruses (Barik 2020). Changes in the structure of proteins can be followed by their subsequent aggregation, which is the cause of the development of conformational diseases (Carrell and Lomas 1997; Kopito and Ron 2000). The most famous conformational diseases are human and animal prion diseases representing a fatal neurodegenerative disease in which conformational changes in the normal prion protein are considered a central pathophysiological event (Ironside et al. 2017). Alzheimer's disease is a conformational disease that is caused by aggregation of a peptide that is cleaved from the transmembrane protein that includes a hydrophobic part that is normally situated in a lipid bilayer, while Huntington's, and Parkinson's diseases are accompanied by extra- or intracellular accumulation of protein aggregates formed by normally water soluble cellular proteins (Steckmann et al. 2017). As in the case of prion diseases, the key event in the pathogenesis and progression of Alzheimer's and Parkinson's diseases is protein misfolding (Aguilar-Calvo et al. 2015). Disordered proteins are also involved in the mechanisms of development of various types of cancer and other malignancies (Santofimia-Castaño et al. 2020). For example, it was shown experimentally that 88 human proteins are involved in pathogenesis of prostate cancer, many of which are intrinsically disordered (Uversky et al. 2017). Diabetes and cardiovascular diseases also belong to conformational diseases (Uversky et al. 2008). This broad involvement of misbehaving IDPs/IDPRs in human diseases is known as «disorder in disorders» (or D2) concept (Uversky et al. 2008, 2014).
With the introduction of the concept of IDPs/IDPRs, methods for their prediction began to be developed. The earliest methods, such as SEG (Wootton 1994) and CAST (Promponas et al. 2000), just search for the sites with low complexity. There are methods based on the search for the least hydrophobic sections of proteins, and they include one of the first algorithms for the prediction of instability of proteins (Xie et al. 1998). There have also been attempts to use the B-factor of Cα atoms to predict unstructured protein regions (Vihinen et al. 1994; Dunker et al. 1998; Zoete et al. 2002; Radivojac et al. 2004).
To date, more than 70 predictors of unstructured protein regions have been developed, which are divided into three categories. The first category includes ab-initio predictors, which are based on some identified features of unstructured proteins, i.e., on the differences between disordered and ordered proteins. This category includes algorithms like FoldIndex© (Prilusky et al. 2005), Globplot (Linding et al. 2003a), IUPred (Dosztanyi et al. 2005). The classical approach is the charge–hydropathy plot, in which the net charge of the protein is plotted as a function of its net hydrophobicity. IDPRs have a low overall hydrophobicity and large net charge (Uversky et al. 2000; Uversky and Dunker 2010). The second group includes self-learning algorithms that use information from special databases. This group includes DisEMBL (Linding et al. 2003b), DISOPRED2 (Ward et al. 2004b), PrDOS (Ishida and Kinoshita 2007) and others algorithms. The third group, meta-predictors, combines the results obtained by several algorithms. Representatives of this group of algorithms are PONDR-FIT (Xue et al. 2010), DISOPRED3 (Jones and Cozzetto 2015), MobiDB-lite (Necci et al. 2017). Despite the existence of such a variety of methods for predicting unstructured fragments in proteins, from 10 to 30% of unstructured proteins are not recognized by those algorithms (Katuwawala et al. 2020).
In 2021, the last experiment on Critical Assessment of protein Intrinsic Disorder prediction (CAID) was finished. In this experiment, 43 methods for the prediction of IDPR were evaluated. The test set included 646 proteins from the DisProt database (Necci et al. 2021). The best methods use deep learning techniques and notably outperform physicochemical methods (Necci et al. 2021). The top disorder predictor fIDPnn has Fmax = 0.483 on the full dataset of proteins (Necci et al. 2021). Across the different performance measures, the methods SPOT-Disorder2, fIDPnn, RawMSA and AUCpreD are consistently found among the top five (Necci et al. 2021). The flDPnn predictor was designed and trained before the CAID experiment on the 176 proteins extracted from the DisProt database (Hu et al. 2021). Therefore, it is not surprising that the second-best predictor is a derivative of the flDPnn model, flDPlr (Hu et al. 2021). Authors of the SPOT-Disorder2 method also used 72 fully-disordered proteins from DisProt database in training, validation and test sets (Hanson et al. 2019).
IDPs/IDPRs are involved in various diseases and may also have untapped therapeutic potential (Babu 2016; Corbi-Verge and Kim 2016; Hu et al. 2016). Although the human genome contains a large number of potential drug targets, however, only about 2% of human proteins are known to interact with approved drugs (Overington et al. 2006).
In this study, we describe a probabilistic algorithm that works on a principle that is different from those of other methods. Also, we tested that method together with abovementioned ones on a new set of proteins that contain IDPRs according to the comparison of their different 3D structures. The novelty of our method lies in the following features: (i) propensity scales used by the algorithm are based on proteins of different structural classes, because it is known that proteins of different structural classes differ in the level of structural stability (Poboinev et al. 2018); (ii) our algorithm can work not only with amino acid sequence of investigated protein, but with 3D structure of the protein, because the formation of the secondary and tertiary structure is influenced not only by the amino acid composition, but also by other factors, such as distant interactions between different regions of the protein (this factor is not directly taken into account when prediction of the structural instability of proteins is based only on their amino acid sequences); (iii) the PentUnFOLD algorithm can find alpha helices and beta strands that are able to turn into random coils as well as regions of random coil that are able to form alpha helix or beta strand; (iv) our algorithm checks the ability of alpha helices, beta strands, and random coils to turn into the disordered state in the central part of a sequence, and not just in N- and C-termini; (v) PentUnFOLD algorithm can determine the effect of amino acid substitution on protein secondary structure stability not only in the elements of secondary structure that exist before a structural shift, but in elements of secondary structure that are formed after that shift.
Materials and methods
The material for this study includes five initial sets of 3D structures of proteins that belong to: (1) alpha helical eukaryotic proteins; (2) beta structural eukaryotic proteins; (3) alpha + beta eukaryotic proteins; (4) alpha/beta eukaryotic proteins; (5) bacterial proteins; as well as a control set of proteins of different origins and structural classes (6). Each set contains no homologs, since the similarity between sequences was lower than 25% according to the Decrease Redundancy algorithm (https://web.expasy.org/decrease_redundancy/). Each protein has two to five different 3D structures in PDB. Those structures belong to proteins with 100% identity of amino acid sequences, but their secondary structures may be different. Thus, samples consisting of 100 alpha helical eukaryotic proteins and 378 structures, 100/355 beta structural eukaryotic proteins/structures, 100/387 alpha + beta eukaryotic proteins/structures, 100/386 alpha/beta eukaryotic proteins/structures, and 189/610 bacterial proteins/structures were formed. Average resolution of all X-ray structures of alpha helical proteins is 2.21 Å, of beta structural proteins—2.11 Å, of alpha + beta proteins—2.08 Å, of alpha/beta proteins—2.00 Å, of bacterial proteins—2.12 Å. The control set consists of 74/249 eukaryotic, bacterial and viral proteins/structures. Average resolution of all X-ray structures from control set of proteins is 2.29 Å. The IDs of all 3D structures in PDB, as well as a resolution are provided in Table 1S and Table 2S from the Supplementary Material. As we used a new control set of proteins we also provide information about amino acid sequences of all used 3D structures of proteins, information about their secondary structure and the results of all the algorithms described in the manuscript in Table 3S from Supplementary Material.
Secondary structure has been estimated with a help of the DSSP algorithm (Kabsch and Sander 1983). Finally, for each protein we found: those random coils (C), alpha helices (H), and beta strands (E) that stay the same in all identical structures; those residues of alpha helices that exist in random coil in some of the structures (HC); those residues of beta strands that exist in random coil in some structures (EC); absolutely disordered fragments that cannot be seen in any of the examined structures (0); random coils that can turn into the disordered state (0C); alpha helices that can turn into the disordered state (0H); beta strands that can turn into the disordered state (0E). Also, we found a significant number of cases in which an alpha helix turns into random coil in some structures, but in other structures turns into the disordered state (0HC), and those cases, in which a beta strand turns into random coil in some structures and in other structures it turns to the disordered state (0EC). Interestingly, the number of residues that can exist in both alpha helical and beta structural state is quite low. Thus, a total of 46,249 cases of H, 27,798—E, 59,023—C, 1260—0, 3960—HC, 2835—EC, 1596—0C, 274—0H, 69—0E, 106—0HC, 33—0EC, 4—HE, 2—HEC were analyzed. Disordered N-terminal and C-terminal parts of proteins have been ignored in all calculations, except the testing of the PentUnFOLD 1D algorithm on the control set.
The amino acid content of each of the abovementioned structural states has been calculated. Then, usages of amino acid residues in different structural states have been compared with each other by two-tailed t test for relative values, standard errors were provided in figures. In the same manner, we compared pentapeptide contents of those structural states. Pentapeptides were used with the aim to consider the influence of short-range interactions between amino acid residues and alternations of hydrophilic (P) and hydrophobic (H) residues on the probability of structural shifts. In those pentapeptides, amino acid residues are roughly divided into hydrophilic (Ser, Thr, Asp, Glu, Asn, Gln, His, Arg, Lys) and hydrophobic ones (Gly, Ala, Met, Leu, Ile, Val, Phe, Tyr, Trp, Cys, Pro) (Tina et al. 2007). The methodology of such comparisons has been described in details previously (Khrustalev et al. 2019).
Additionally, we calculated the amino acid content and pentapeptide content of first and last amino acid residues of alpha helices and beta strands with stable and nonstable N- and C-termini, as well as for flanking random coil residues. For alpha helices, we also considered second positions of both N- and C-termini.
Propensity scales (both amino acid and pentapeptide ones) have been created for each set of structural states. All those scales can be seen in Table 4S from Supplementary material. The workflow of the PentUnFOLD algorithm is described in the subsection of the Results and Discussion section, the manual is available as Supplementary material “PentUnFOLD-manual.pdf” file.
Secondary structure of each protein from all the six sets has been determined with the DSSP program (Kabsch and Sander 1983), tertiary structure of each protein from the control set has been studied with the help of the PIC server (Tina et al. 2007). Among intraprotein interactions we consider hydrogen bonds, hydrophobic contacts, ionic contacts, cation-pi interactions, aromatic-aromatic, and aromatic-sulfur interactions, as well as disulfide bonds. We use the same criteria for their consideration as the PIC server (Tina et al. 2007). The number of amino acid residues that make contacts with a given residue is calculated. Then the algorithm counts for every amino acid residue the number of contacts with stable residues, with nonstable residues, with disordered ones, and with completely disordered ones.
The information on 103 structures of human serum albumin can be found in Table 5S from the Supplementary Material file.
Three PentUnFOLD algorithms are available on the web server (http://3.17.12.213/pent-un-fold) and on the page of our university (http://chemres.bsmu.by/PentUnFOLD.htm). PentUnFOLD 1D, PentUnFOLD 2D, and PentUnFOLD 3D require PDB file as an input, while PentUnFOLD 1D can also work with an amino acid sequence. The output of those algorithms is provided as a downloadable MS Excel file. Predictions can be easily copied from those files. Moreover, new calculations or formatting can be easily performed directly in those MS Excel worksheets. The server uses an output of DSSP algorithm with the aim to determine secondary structure for PentUnFOLD 2D and PentUnFOLD 3D. In case of DSSP server failure, secondary structure is determined by our own JAVA script based on DSSP criterions (Kabsch and Sander 1983). For the 3D version of the algorithm our server finds all the possible intraprotein interactions according to the criterions of the PIC server using a new JAVA script.
Results
Comparison between completely disordered state, disordered random coil, and stable random coil
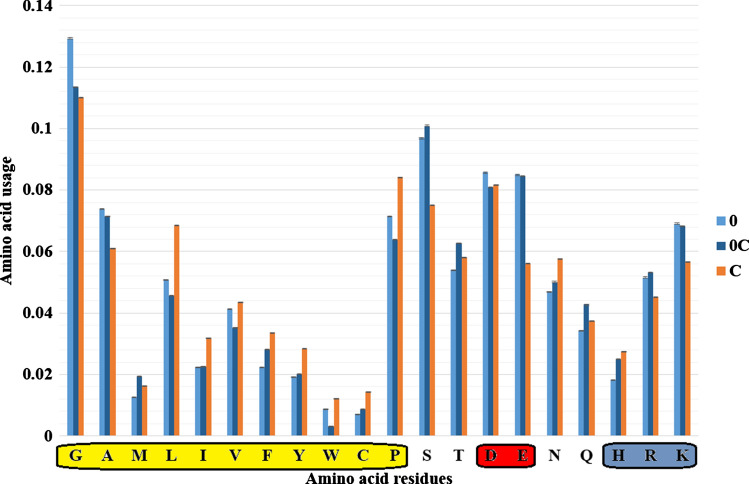
One of the fundamental questions of this study is to find out are there any differences between the regions of proteins that can never be seen in PDB files and those that can be seen in some PDB files, but “disappear” in others. In Fig. 1, we show amino acid content of completely disordered regions (excluding those that exist in N-termini and C-termini of proteins), amino acid content of regions that exist as disordered ones in some 3D-strutures and form random coil in other structures with 100% similarity of amino acid sequence, as well as amino acid content of those regions of random coil that stay in random coil state in all examined 3D structures of identical proteins. The differences between stable random coils and unstable ones are as follows: the usage of several hydrophobic residues is significantly higher in stable random coils (Leu, Ile, Val, Phe, Tyr, Trp, Cys, Pro), as well as the usage of some hydrophilic ones (Asp, Asn, His). The differences between stable random coils and completely disordered regions are quite similar to the previously described ones: the usage of several hydrophobic residues is significantly higher in stable random coils (Met, Leu, Ile, Val, Phe, Tyr, Trp, Cys, Pro), as well as the usage of some hydrophilic ones (Thr, Asn, Gln, His). Completely disordered regions are enriched in several amino acid residues relative to the unstable random coils: by Gly, Ala, Leu, Val, Trp, Pro, Asp, and Lys. However, the magnitude of differences in amino acid usage is higher than 25% for three amino acid residues only: Met, Trp, and His. At the same time, the differences between completely disordered state and stable random coils are higher than 25% for Ser, Glu, Leu, Ile, Tyr, Trp, Cys, Phe, and His. Among the differences between unstable and stable random coils with a magnitude higher than 25%, we find the same Ser, Glu, Leu, Ile, Tyr, Trp, Cys, but without Phe and His.
Fig. 1.
Amino acid content of completely disordered regions (“0”), amino acid content of regions that exist as disordered ones in some 3D-strcutures and form random coil in other structures with 100% similarity of amino acid sequence (“0C”), and amino acid content of those regions of random coil that stay in random coil state (“C”) in all examined 3D structures of identical proteins. Amino acids from N-termini and C-termini of proteins were excluded. Standard errors are provided in the barcharts. Names of hydrophobic residues are in the yellow bar, names of negatively charged residues are in the red bar, while names of positively charged residues are in the blue bar
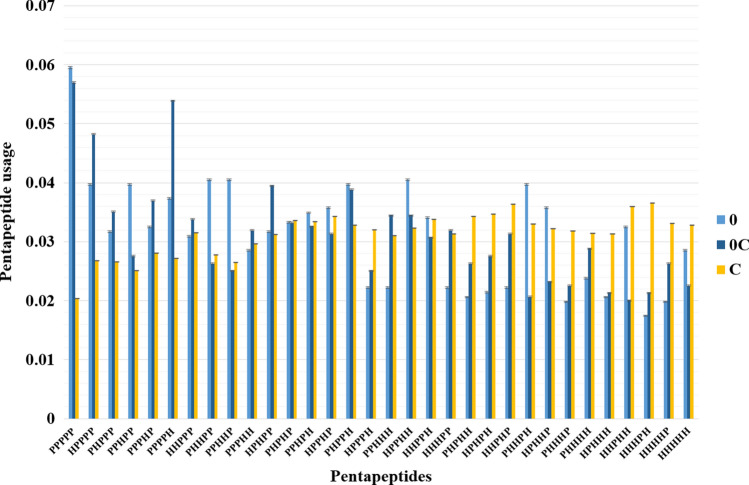
In Fig. 2, we show the same comparison, but for 32 pentapeptides. The tendency of the enrichment of both completely disordered regions and unstable random coils by hydrophilic amino acid residues and pentapeptides composed of them is clear. However, there are some pentapeptides that are landmarks of the unstable random coil and not completely disordered regions: HPPPP; PPPPH; PHPPP; PPPHP; HHPPP; PPPHH; PPHHH; HHHPP; HPHPP (P is a hydrophilic amino acid residue, H is a hydrophobic amino acid residue). In the same way, there are a few quite frequent pentapeptides exactly in completely disordered regions: PHHPP; PPHHP; PPHPP; PHHPH; HPHHP. Interestingly, five last pentapeptides are known as alpha helical ones (Khrustalev and Barkovsky 2012).
Fig. 2.
Pentapeptide content of completely disordered regions (“0”), pentapeptide content of regions that exist as disordered ones in some 3D-strcutures and form random coil in other structures with 100% similarity of amino acid sequence (“0C”), and pentapeptide content of those regions of random coil that stay in random coil state (“C”) in all examined 3D structures of identical proteins. Pentapeptides from N-termini and C-termini of proteins were excluded. “P” is a hydrophilic amino acid residue, “H” is a hydrophobic amino acid residue. Standard errors are provided in the barcharts. The pentapeptides are arranged in order of increasing hydrophobicity from left to right
On one hand, amino acid and pentapeptide content of completely disordered regions and unstable random coils are closer to each other, than to those of stable random coils. But on the other hand, there are some sharp differences between them, especially, if we consider combinations of hydrophobic and hydrophilic residues in pentapeptides. Because of these reasons, we included two methods to find disordered regions in random coils of proteins. In the first, we use a combined scale based on average characteristics of both completely disordered regions and unstable random coils. In the other method, we distinguish completely disordered regions from unstable random coils.
Comparison between alpha helices that can turn into the disordered state, those that can turn into random coil, and stable alpha helices
In this study we have found out that alpha helices that can turn into the disordered state are different from those that can turn into random coil. As one can see in Fig. 3, alpha helices able to form random coil are enriched in all nine hydrophilic amino acid residues, as well as in Cys, Pro, and Gly relative to stable alpha helices. In contrast to the last ones, alpha helices able to turn into the disordered state, and not to the random coil, are enriched in Ala and Met, and depleted in Gly, Pro, Thr, Asn, His, and Arg residues, relative to stable alpha helices. Even more surprisingly, alpha helices that are able to form disordered state are significantly enriched in Ala, Met, Ile, Val, Tyr, Asp, Glu, Gln, and Lys relative to those that can turn into the random coil. Especially prominent differences are evident (Fig. 3) for Ala, Asp, Glu, Gln, and Lys residues. These residues (except Asp) are well-known helix formers (Chou and Fasman 1978), but their usages are higher in those alpha helices that can turn into disordered state than in stable alpha helices and in those that can form random coil.
Fig. 3.
Amino acid content of alpha helices that can turn into the disordered state (“0H”), amino acid content of alpha helices that can turn into random coil (“HC”), and amino acid content of those regions of alpha helices that stay in alpha helical state (“H”) in all examined 3D structures of identical proteins. Amino acids from N-termini and C-termini of proteins were excluded. Standard errors are provided in the barcharts. Names of hydrophobic residues are in the yellow bar, names of negatively charged residues are in the red bar, while names of positively charged residues are in the blue bar
In Fig. 4, one can see that alpha helices prone to form disordered fragments of proteins are enriched in hydrophilic pentapeptides, as well as by a few less hydrophilic ones: PPHPH; HPPHP; PHPHH; HHHHP. Stable alpha helices are enriched in the most of hydrophobic pentapeptides. Those alpha helices that are prone to form random coil demonstrate higher usage of hydrophilic pentapeptides than stable alpha helices, but not as high as those alpha helices that can turn into disordered state. An opposite situation is there with some hydrophobic pentapeptides. Several pentapeptides with an average usage of hydrophilic residues are more frequently used in alpha helices prone to form random coil than in two other types, for example: HHPPP; HPHPP; HPPPH.
Fig. 4.
Pentapeptide content of alpha helices that can turn into the disordered state (“0H”), pentapeptide content of alpha helices that can turn into random coil (“HC”), and pentapeptide content of those regions of alpha helices that stay in alpha helical state (“H”) in all examined 3D-structures of identical proteins. Pentapeptides from N-termini and C-termini of proteins were excluded. “P” is a hydrophilic amino acid residue, “H” is a hydrophobic amino acid residue. Standard errors are provided in the barcharts. The pentapeptides are arranged in order of increasing hydrophobicity from left to right
In the PentUnFOLD algorithm, we check whether a fragment of an alpha helix is prone to form random coil, and then we check can it turn into the disordered region using separate propensity scales (Table 4S).
Comparison between beta strands that can turn into the disordered state, those that can turn into random coil, and stable beta strands
Absolutely in the same way, as with alpha helices, beta strands able to form random coil are enriched in all nine hydrophilic amino acid residues, as well as in Cys, Pro, and Gly relative to stable beta strands (Fig. 5). At the same time, amino acid contents of alpha helices and beta strands (both able and unable to turn into random coil) are quite different. Beta strands prone to form disordered regions are enriched in several amino acids relatively to two other types of beta strands: Ala, Met, Tyr, Asp, His, Lys. Interestingly, alpha helices prone to form disordered regions are also enriched in Ala, Met, Asp, and Lys relative to two other types of alpha helices. It seems like being enriched in the same types of amino acid residues, both beta strands and alpha helices are becoming prone to form a disordered region, while being enriched in other types of amino acid residues they are both becoming able to turn into random coil. However, both transitions (to the disordered state and to the random coil) are becoming possible for beta strands and for alpha helices when they still have quite different amino acid contents.
Fig. 5.
Amino acid content of beta strands that can turn into the disordered state (“0E”), amino acid content of beta strands that can turn into random coil (“EC”), and amino acid content of stable beta strands (“E”). Amino acids from N-termini and C-termini of proteins were excluded. Standard errors are provided in the barcharts. Names of hydrophobic residues are in the yellow bar, names of negatively charged residues are in the red bar, while names of positively charged residues are in the blue bar
It is not surprising that beta strands able to turn into random coil are enriched in more hydrophilic pentapeptides, while stable beta strands are more hydrophobic (Fig. 6). Surprisingly, beta strands that are prone to turn into disordered state are especially enriched with several concrete pentapeptides: PPPPH; PPPHH; PHPHP; PPHHH; PHHHP; HHHPH. This information is used by the PentUnFOLD algorithm to check if a beta strand fragment can turn into random coil, and if it is prone to turn into the disordered state.
Fig. 6.
Pentapeptide content of beta strands that can turn into the disordered state (“0E”), pentapeptide content of beta strands that can turn into random coil (“EC”), and pentapeptide content of stable beta strands (“E”). Pentapeptides from N-termini and C-termini of proteins were excluded. “P” is a hydrophilic amino acid residue, “H” is a hydrophobic amino acid residue. Standard errors are provided in the barcharts. The pentapeptides are arranged in order of increasing hydrophobicity from left to right
The information on instability of N- and C-termini of alpha helices and beta strands
Most of helix to coil and beta sheet to coil transitions have been found by us in N- and C-termini of alpha helices and beta strands, respectively. These transitions have been studied separately from cases of complete helix to coil and sheet to coil transformations. Actually, in previous sections we described amino acid content of alpha helices and beta strands that are able to turn into random coil completely. Here, we compare amino acid content of N-termini of stable alpha helices and amino acid content of instable N-termini of alpha helices. Since in N-caps of alpha helices a residue situated before an alpha helix usually makes hydrogen bonds with residues from an alpha helix, those residues for stable and instable N-termini of helices have been compared as well. We also considered helix to coil transitions made by a single N-terminal residue separately from such transition made by two N-terminal residues. The same comparisons have been made for C-termini of alpha helices.
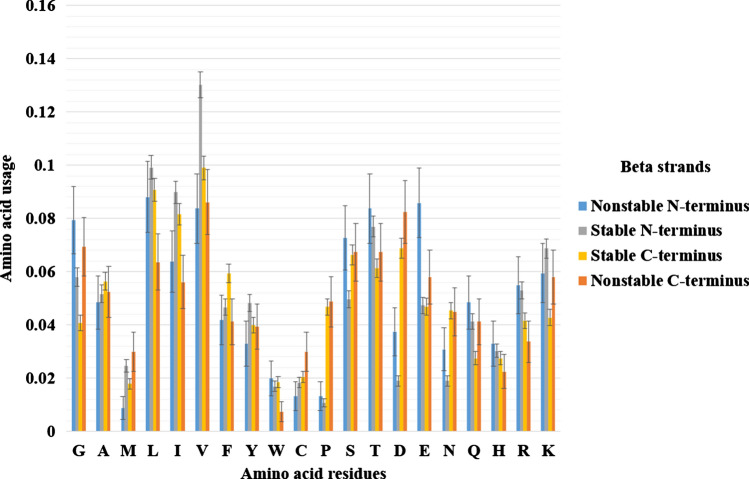
In Fig. 7, one can observe that proline in the first position can significantly stabilize N-terminus of an alpha helix, while aspartic acid is usually making the first position of an alpha helix prone to turn into random coil. C-termini of alpha helices are stable if Val, Tyr or Asp is situated there. Nonstable C-terminal residues are Ser and Arg. The pentapeptide that stabilizes N-termini of alpha helices better than others is HPHPP (alpha helix starts from the third position).
Fig. 7.
Amino acid content of stable and instable N- and C-termini of alpha helices. Standard errors are provided in the barcharts
N-terminal residues of beta strands that prevent those N-termini from turning into random coil are Met, Ile, and Val (Fig. 8). If a beta strand starts from Asp or Glu, it is prone to become shorter from its N-terminus. C-termini of beta strands are significantly stabilized by Leu, Ile, and Trp. If Gly is situated in the C-terminal position of a beta strand, that strand has a high chance to become shorter from its C-terminus (Fig. 8). The pentapeptide that stabilizes N-termini of beta strands better than others has the following sequence: HPHHH. The best stabilizer of the C-terminus of a beta strand is the HHHPP pentapeptide.
Fig. 8.
Amino acid content of stable and instable N- and C-termini of beta strands. Standard errors are provided in the barcharts
The information on amino acid residues that make N- and C-termini of alpha helices and beta strands more or less stable is used by the PentUnFOLD to consider the stability of those caps. Users are able to change the positions of N-termini and C-termini of alpha helices and beta strands to search for their most stable (and so most expected) positions in the PentUnFOLD 2D version.
The principles of the PentUnFOLD algorithms
There are three versions of the PentUnFOLD algorithm: 1D version predicts based on just an amino acid sequence, 2D version uses both amino acid sequence and the data on secondary structure; while 3D version uses amino acid sequence, secondary structure, and the map of intraprotein contacts between amino acid residues.
The PentUnFOLD algorithm predicts fragments of alpha helices and beta strands that can turn into random coil (referred to as “N” residues) separately from those fragments of alpha helices, beta strands, and random coils that can turn into completely disordered state (referred to as “D” residues). A fragment of a protein may be both “N” and “D”.
The PentUnFOLD algorithm requires description of a polypeptide chain from a PDB file, the results of the evaluation of its secondary structure from the DSSP algorithm, and the information on intraprotein contacts between amino acid residues as an input. So, amino acid sequence is extracted from the lines “ATOM” of the PDB file. However, users of the PentUnFOLD 2D version can introduce unlimited number of amino acid substitutions in a sequence. The information on the secondary structure is extracted from the output of the DSSP algorithm, but users can change secondary structure manually in the 2D version of the algorithm.
The algorithm solves the following problems: (i) it checks stability of alpha helices and beta strands in terms of their ability to turn into random coil; (ii) it checks ability of alpha helices, beta strands, and random coils to turn into the disordered state; (iii) it finds regions of random coil that are able to form alpha helix or beta strand. PentUnFOLD 1D version also predicts secondary structure for a protein considering that it can completely turn into a disordered state and fold back.
The first problem requires two separate calculations: the estimation of the stability of N- and C-termini of alpha helices and beta strands, and the estimation of the stability of central regia. For termini of beta strands, the algorithm uses four scales: amino acid and pentapeptide propensity scales for the first (last) residue in a beta strand and for the flanking residue from the random coil. If the average value from these four scales is higher than 0.5, the terminal residue is considered to be stable. Terminal residue is also considered to be stable if a beta strand is predicted there by both amino acid and pentapeptide scales. Amino acid residues from the body of a beta strand are judged by the amino acid and pentapeptide scales first, and by the average results for the scales (amino acid and pentapeptide ones) for stable and nonstable bodies of beta strands next. Finally, the algorithm shows residues of beta strands that have a low probability to turn into random coil (ES), and those residues that have a high probability to turn into random coil (EN).
The algorithm works with alpha helices in the same way as with beta strands, while the algorithm for their N- and C-termini is more complicated, since it includes also a calculation featuring the second residue from the N-terminus and the second residue from the C-terminus. The first (last) residue in an alpha helix is considered to be stable if it is predicted to be stable by both methods (the one featuring just the first residue, and the second considering two N-terminal or C-terminal residues). The second residue in an alpha helix is considered to be stable with a help of the method that includes the average result for six scales: amino acid scales and pentapeptide scales for the first and the second residues in an alpha helix, and for the residue in random coil before (after) the alpha helix.
The ability of a random coil to turn into the disordered state is considered by the comparison between four scales: combined scale for completely disordered state and for random coil that can turn into the disordered state, stable random coil, combined scale for alpha helices that can turn into the disordered state and those that can turn into both disordered state and random coil, combined scale for beta strands that can turn into the disordered state and those that can turn into both disordered state and random coil. In case if random coil is considered to be disordered by this method, it is also judged by the method that features just two scales: for the completely disordered state and for random coils that can turn into the disordered state. With the first method the algorithm can also consider fragments of random coil that can turn into alpha helix or beta strand.
Alpha helices are considered to be prone to turn into disordered state using a method that features amino acid and pentapeptide scales that include two options each: the scale for stable alpha helices, and the combined scale for alpha helices that can turn into the disordered state and those that can turn into both disordered state and random coil. If a residue is considered to be disordered by this method, it is also judged by the next one that compares completely disordered state with the combined scale for alpha helices that can turn into the disordered state and those that can turn into both disordered state and random coil. In the same manner the algorithm finds disordered residues in beta strands, and chooses absolutely disordered residues (V) among them.
Finally, the algorithm predicts unstable alpha helices and beta strands that can appear in the place of random coil or disordered state. This prediction may be useful for consideration of the structure of those proteins that can completely turn into the disordered state and fold back.
At the end of the 2D prediction step amino acid residues are classified into five categories: “V” means completely disordered residues; “D” means disordered residues; “N” means nonstable residues of alpha helices and beta strands that can turn into random coil or vice versa; “Z” means neither stable, nor nonstable residues of random coil; “S” means stable residues.
The purpose of the 3D prediction step is to consider the influence of stabilizing and destabilizing contacts between amino acid residues. Only if a residue is predicted to be completely disordered (V), we ignore any contacts it can make to other parts of the protein. If a residue is predicted to be just disordered (D) by the 2D algorithm, it stays disordered only if the number of its contacts with stable (S) residues is less than 3. If a residue is predicted to be nonstable (N), it is classified as disordered one only if it makes less than 4 contacts with stable (S) residues. If a residue of a random coil is neither stable, nor nonstable (Z), it is classified as disordered one if it makes no contacts with other residues at all, or if the sum of its contacts with V, D, and N residues (actually, 0.5 · N) is higher than the number of its contacts with stable residues. Even stable (S) residue can become disordered if the sum of the numbers of its contacts with “V”, 0.5 · “D”, and 0.25 · “N” residues is higher than the number of its contacts with other “S” residues. Taking together, 3D prediction finds those residues that are situated in the disordered 2D environment, but surrounded by stable residues in the 3D space, as well as residues in the stable 2D environment, that are destabilized by contacts with other residues. Also, residues that are not making any contacts are considered to be disordered, and that is the case mostly for unfolded N- and C-termini of proteins.
At the final step, residues surrounded by disordered ones in the primary sequence are considered to be disordered (DXD = DDD), and then residues surrounded by ordered ones (O) are considered to be ordered (OXO = OOO).
Performance of PentUnFOLD algorithms
In the test set, we classified residues as disordered ones in case if they were missing at least in one of the structures with 100% identity of sequences, compared to the randomly selected initial one. There were 1782 disordered residues in the whole test set. As one can see in Table 1, the PentUnFOLD 3D showed the highest sensitivity (71.44%) compared to nine other methods (from 3.59% for the Depicter to 43.60% for the Foldindex). This increased sensitivity has been reached thanks to the 3D step of prediction. Indeed, for the PentUnFOLD 1D sensitivity is equal to 12.21%, and for the PentUnFOLD 2D algorithm it is equal to 39.25%. Interestingly, if we consider just those residues that are classified as prone to turn into the disordered state (“D” and “V”) by the PentUnFOLD 2D algorithm, its sensitivity is equal to 11.34%, while if consider just structurally nonstable residues (N), the sensitivity is even higher (22.18%). These results prove that transitions of helix to coil and beta to coil (and vice versa) make a significant contribution into the “disappearance” of protein fragments from 3D structures. Provided data also shows that interactions between amino acid residues is the third key to open up the door to the understanding of the nature of intrinsically disordered regions of proteins, while the second key is the actual secondary structure, and the first key is their amino acid content and composition.
Table 1.
The results of the testing of the PentUnFOLD and other algorithms on the test set of 74 proteins
| Algorithms | Sensitivity, % | Specificity, % | Accuracy | MCC | F1 |
|---|---|---|---|---|---|
| Depicter | 3.59 | 9.22 | 0.890 | 0.006 | 0.052 |
| Foldindex | 43.60 | 9.69 | 0.612 | 0.037 | 0.159 |
| GlobPlot | 7.24 | 4.25 | 0.785 | -0.061 | 0.054 |
| PONDR VL-XT | 22.05 | 9.38 | 0.756 | 0.018 | 0.132 |
| PONDR VSL2 | 23.91 | 10.01 | 0.756 | 0.029 | 0.141 |
| flDPnn | 7.30 | 14.79 | 0.887 | 0.048 | 0.098 |
| AUCpreD | 8.59 | 27.77 | 0.905 | 0.114 | 0.131 |
| DISOPRED3 | 6.90 | 29.29 | 0.908 | 0.107 | 0.112 |
| DISOPRED3 (disordered) | 3.09 | 26.57 | 0.912 | 0.065 | 0.055 |
| PentUnFOLD 1D | 12.22 | 14.14 | 0.853 | 0.052 | 0.131 |
| PentUnFOLD 2D | 39.25 | 7.99 | 0.570 | − 0.012 | 0.133 |
| PentUnFOLD 2D (D) | 11.34 | 8.04 | 0.817 | − 0.005 | 0.094 |
| PentUnFOLD 2D (N) | 22.18 | 7.66 | 0.711 | − 0.015 | 0.114 |
| PentUnFOLD 3D | 71.44 | 9.05 | 0.374 | 0.034 | 0.161 |
Interestingly, nine tested algorithms, show low (Table 1) specificity (from 4.25 to 29.29%). It means that all of them largely overpredict disordered regions. PentUnFOLD is not an exception in terms of specificity: it is equal to 9.04% for its 3D version and 7.99% for its 2D version, but it is higher (14.14%) for its 1D version. Does that mean that we cannot trust in such predictions, or does it mean that the definition of disordered regions in the current test was too strict? To answer this question, we studied a set of 103 structures of human serum albumin (HSA). Indeed, the higher the number of 3D structures are available for a given protein, the higher the percent of residues that are missing from at least one of them. Actually, for human serum protein 59% of amino acid residues are disordered, according to our definition.
As one can see in Table 2, the levels of specificity for all tested algorithms are much higher for human serum protein, than for proteins with a few known 3D structures. Actually, for Depicter, the level of specificity is even equal to 100%, since it has predicted just a single disordered residue, and that N-terminal residue may indeed disappear from 3D structures. For the other algorithms, specificity varies from 35.71% (DISOPRED3, disordered) to 75.00% (fIDPnn). The specificity for the PentUnFOLD 3D is equal to 60.87%, while its sensitivity again showed the highest level among other tested algorithms (65.50%). As in the test set of proteins, PentUnFOLD 1D showed worse sensitivity than PentUnFOLD 2D (7.02% vs. 48.83%), while their specificities were comparable with each other (53.33% vs. 58.19%).
Table 2.
The results of the testing of the PentUnFOLD and other algorithms on the human serum albumin (HSA) with 103 available 3D structures
| Algorithms | Sensitivity, % | Specificity, % | Accuracy | MCC | F1 |
|---|---|---|---|---|---|
| Depicter | 0.29 | 100.00 | 0.411 | 0.022 | 0.006 |
| Foldindex | 47.95 | 65.60 | 0.544 | 0.011 | 0.554 |
| GlobPlot | 2.05 | 46.67 | 0.408 | − 0.010 | 0.039 |
| PONDR VL-XT | 11.40 | 55.71 | 0.423 | − 0.003 | 0.189 |
| PONDR VSL2 | 38.30 | 70.43 | 0.541 | 0.014 | 0.496 |
| flDPnn | 0.88 | 75.00 | 0.413 | 0.010 | 0.017 |
| DISOPRED3 | 7.89 | 65.85 | 0.432 | 0.005 | 0.141 |
| DISOPRED3 (disordered) | 1.46 | 35.71 | 0.402 | − 0.022 | 0.028 |
| PentUnFOLD 1D | 7.02 | 53.33 | 0.415 | − 0.005 | 0.124 |
| PentUnFOLD 2D | 48.83 | 58.19 | 0.491 | − 0.002 | 0.531 |
| PentUnFOLD 2D (D) | 18.42 | 62.38 | 0.453 | 0.003 | 0.284 |
| PentUnFOLD 2D (N) | 42.98 | 57.65 | 0.476 | − 0.002 | 0.492 |
| PentUnFOLD 3D | 65.50 | 60.87 | 0.547 | 0.006 | 0.631 |
The results provided above show that proteins usually have a lot of regions that can change their structure or turn into the disordered state. Even subtle changes in conditions or the binding of specific ligands can change the network of intraprotein amino acid contacts, and release disordered regions from stabilizing interactions.
Evaluation of the consequences of amino acid substitutions in the most disordered region of human major prion protein by the PentUnFOLD algorithm
According to the results of the PentUnFOLD algorithm, there are several disordered areas in the human major prion protein. We used NMR structure with PDB ID: 1HJM as an input. According to the 2D predictions, the first alpha helix (144–152) contains disordered N-terminal part (3 residues) and disordered C-terminal residue. There are just two stable residues in that alpha helix: 149–150. The long second alpha helix (174–194) is completely instable and just two residues are stable (Phe175 and Val180). C-terminal part of the second alpha helix is disordered (190–194), and one of those residues (Thr192) is absolutely disordered. The third alpha helix (200–225) contains disordered N-terminal part (5 residues), disordered residue 215 and one more disordered region (218–223). At the same time, just 10 out of 26 residues from the third alpha helix are predicted as nonstable. Residue 196 situated in the random coil between the second and the third alpha helices is predicted to be disordered.
Results of the PentUnFOLD algorithm are in agreement with previous works that showed that the region containing the C-terminal part of the second alpha helix, the N-terminal part of the third alpha helix, and random coil between them is prone to form beta structure (Khrustalev et al. 2016).
With the help of our new algorithm we estimated consequences of amino acid substitutions in this region that are known to be associated with prion diseases.
H187R. This amino acid substitution is associated with Gerstmann-Straussler disease (GSD). As a result, two amino acid residues before R187 are becoming disordered. If we consider that there is a beta strand in place of the second alpha helix, there are no changes after the amino acid substitution (Table 3).
Table 3.
Consequences of amino acid substitutions associated with the development of human prion diseases according to the original PentUnFOLD and other algorithms (2D predictions)
| Amino acid substitution | PentUnFOLD | GlobPlot 2.3 | FoldIndex© | PONDR® VL-XT | PONDR® VSL2 | PrDOS | DEPICTER |
|---|---|---|---|---|---|---|---|
| H187R | H: O → D (Lys185-Gln186); E: no changes | No changes |
O → D (Ile184-Thr188); |
O → D (Lys194-Asn197) |
O → D (Val189; Val210) |
No changes | No changes |
| F198S | O → D (Glu196-Glu200) | No changes |
No changes U: -0.085 → -0.096 |
O → D (Glu196-Asn197) |
O → D (Thr188-Val189; Val210) |
No changes |
O → D (191–203) |
| D202N | H: No changes; E: nonstable Lys204 → stable Lys204 |
No changes |
O → D Asn202 |
D → O (Phe198) |
O → D (Val189; Val210) |
No changes | No changes |
| Q212P | H: nonstable state → stable state (Gln212-Met213); E: stable state → nonstable state (Gln212-Met213) | No changes |
No changes U: -0.085 → -0.080 |
O → D (Gln217-Tyr218) |
D → O (Arg208-Val209; Glu211-Gln212; Cys214) |
No changes | No changes |
| E196K | H: No changes; E (2nd and 3rd): D → O (Lys194) | No changes |
No changes U: -0.085 → -0.067 |
D → O (Phe198-Glu200) |
D → O (Val209); O → D (Val189) |
No changes | No changes |
| E200K | H: No changes; E: nonstable Glu200 → stable Glu200 | No changes |
No changes U: -0.085 → -0.067 |
D → O (Phe198-Glu200) |
D → O (Val209) |
No changes | No changes |
| V203I | H: No changes; E: nonstable Lys204 → stable Lys204 | No changes |
No changes U: -0.085 → -0.084 |
No changes |
D → O (Met205-Val209) |
No changes | No changes |
| R208H | H, E: No changes | No changes |
No changes U: -0.085 → -0.091 |
D → O (Phe198-Thr199; Gln212-Thr216; Glu219) |
D → O (Glu207-Val209; Glu211-Cys214) |
No changes | No changes |
| V210I | H: No changes; E: nonstable Arg208 → stable Arg208; stable Glu211 → nonstable Glu211 | No changes |
No changes U: -0.085 → -0.084 |
D → O (Glu219) |
D → O (Met205-Val209; Glu211-Gln212) |
No changes | No changes |
| E211Q | H, E: No changes | No changes |
No changes U: -0.085 → -0.076 |
D → O (Thr216; Glu219) |
D → O (Val209) |
No changes | No changes |
H alpha helix, E beta strand, D disordered state, O ordered state, U unfoldability
F198S. This substitution is associated with atypical GSD. As a result, the long fragment of random coil (196–199) becomes disordered, while the first residue in the third alpha helix (200) becomes absolutely disordered (Table 3).
D202N. This is a GSD substitution. Such amino acid replacement does not lead to any consequences if we consider a native secondary structure of prion protein. However, if we consider that there is a beta strand in place of the third alpha helix, D202N results in the appearance of a stable residue 204. Interestingly, beta strand in place of the first half of the third alpha helix (200–214) is nonstable, except stabilized residues 211 and 214. Moreover, it has two long disordered regions (201–203 and 205–211). So, D202N replacement makes this disordered beta strand a little bit more stable.
Q212P. This is a GSD substitution. Obviously, this replacement destabilizes the third alpha helix: nonstable residues appear in positions 212 and 213. More interestingly, if we consider a beta strand in place of the third alpha helix, Q212P substitution makes it more stable (positions 212 and 213 are stabilized).
E196K. This substitution is described in patients with Creutzfeldt-Jakob disease (CJD). There are no significant changes revealed by the algorithm. However, random coil between the second and the third alpha helices becomes a little closer to the disordered state, according to the calculations, while the potentials to form disordered state are still less than 0.5. If we consider that there are beta strands in place of second and third alpha helices, E196K replacement makes C-terminus of the 1st beta strand a little bit more ordered.
E200K. This is a CJD substitution. The resulting consequence is the formation of a stable N-terminus of a beta strand that appears in place of the third alpha helix.
V203I. This is a CJD substitution. Once again, the consequence of this substitution is significant only for the beta strand, but not for the third alpha helix. As a result, a stable residue 204 appears in the beta strand.
R208H. This is a CJD substitution. There are no significant changes revealed by the algorithm in both alpha helical and beta structural states of that region. However, the alpha helix is becoming less disordered, but the beta strand is becoming more disordered according to the calculations.
V210I. This is a CJD substitution. As a result, residue 211 becomes nonstable, but residue 208 becomes more stable if we consider a beta strand in the place of the third alpha helix.
E211Q. This is a CJD substitution. There are no significant changes revealed by the algorithm in both alpha helical and beta structural states of that region. For both states the degree of disorder is becoming lower in case of this substitution.
Taken together, substitutions associated with hereditary prion diseases in the second half of the second alpha helix and in the loop between the second and the third alpha helices are leading to the increase of the disorder (H187R, F198S). Several substitutions (E200K, V203I, D202N, V210I, Q212P, E196K) stabilize a beta strand that appears in the place of that helix in PrPSc. Substitutions like R208H and E211Q are not directly associated with stabilization of a beta strand and destabilization of an alpha helix. Using the PentUnFOLD algorithm, 8 out of 10 known substitutions leading to the prion disease development in the region known to form beta structure in a corresponding peptide can be linked with the shift of equilibrium from alpha helical and random coil states to the disordered state and from the random coil state to the beta structural state.
After determining the impact of amino acid substitutions associated with the development of human prion diseases using the original PentUnFOLD algorithm, we evaluated their impact using the algorithms GlobPlot 2.3 (Linding et al. 2003a), FoldIndex© (Prilusky et al. 2005), PONDR VL-XT (Romero et al. 2001), PONDR VSL2 (Peng et al. 2006), PrDOS (Ishida and Kinoshita 2007) and one of the newest methods for the prediction of intrinsically disordered regions, DEPICTER (DisorderEd PredictIon CenTER). GlobPlot 2.3 and FoldIndex© are ab-initio algorithms (1st group); PONDR VL-XT, PONDR VSL2 and PrDOS are self-learning algorithms (2nd group); DEPICTER is a meta-predictor, based on prediction of IUPredL, IUPredS, SPOT-Disorder (Barik et al. 2020). Results of all mentioned algorithms are provided in Table 3. The PentUnFOLD algorithm has not determined the effect on secondary structure stability of human prion protein only for two amino acid substitutions. Algorithms from the 1st (GlobPlot 2.3) and from the 2nd (PrDOS) group have not determined the impact of any amino acid substitutions on the stability of protein structure. Meta-predictor Depicter has determined that only one amino acid substitution (F198S) can affect stability of human prion protein. The result of Depicter algorithm is consistent with PONDR VL-XT, PONDR VSL2 and with our algorithm: amino acid substitution F198S increases the instability of prion protein. FoldIndex© has shown that there are no changes in secondary structure stability after this amino acid substitution, but unfoldability has decreased. As the results of amino acid substitution H187R three out of six algorithms as well as PentUnFOLD determined the transition from ordered state to disordered. As the results of amino acid substitution D202N PentUnFOLD algorithm determined that there is no change in stability of protein with native conformation and slight increase of stability of the 2nd beta strand in place of the third alpha helix. It can explain the confusion between other algorithms: FoldIndex© and PONDR VSL2 determined the O → D shift, while PONDR VL-XT determined the D → O shift. The same situation is observed as the result of Q212P substitution. This replacement destabilizes the third alpha helix and stabilizes the 2nd beta strand in place of the third alpha helix. PONDR VL-XT determined the O → D shift, but PONDR VSL2 determined the D → O shift. Two amino acids substitutions (E196K and E200K) according to PONDR VL-XT and PONDR VSL2 lead to the increase of stability of prion protein. Applying our algorithm, we can say that the stabilization occurs only after the structural transition from the alpha helical to the beta structural state (E196K) means from the 3rd alpha helix to the beta strand (E200K). After amino acid substitution V203I five out of six algorithms did not recognize any changes. Only PONDR VSL2 and PentUnFOLD algorithms determined the D → O shift. Valine can also be replaced with isoleucine at position 210. In this case the D → O shift is also observed, which is determined by PONDR VL-XT and PONDR VSL2 algorithms. PentUnFOLD algorithm shows more specific results: after amino acids substitution V210I stabilization of one part of beta strand (Arg208) and destabilization of another part of it (Glu211) is observed.
If we consider 3D step of prediction, then the most of the human prion protein from the 1HJM PDB structure would be classified as disordered. Among ordered regions there are: two fragments of random coil (156—157; 165—166), and a long fragment of the 3rd alpha helix (205—213). The latter fragment is considered to be stable on its own, and it makes more contacts with stable residues, than with disordered or nonstable ones. Among stable residues that should keep the structure of the 3rd alpha helix there are residues Tyr149 and Tyr150 from the 1st alpha helix that are involved in hydrophobic and aromatic-sulfur interactions. In the absence of the 1st helix an isolated sequence of amino acid residues from the 3rd alpha helix may form beta sheet. Indeed, the CC36 peptide with the original sequence (residues 179–214) failed to be synthesized because of the formation of beta sheet by its C-terminus (Khrustalev et al. 2016).
Prediction of intrinsically disordered protein regions from amino acid sequence by the PentUnFOLD 1D algorithm
A lot of proteins have intrinsically disordered regions at N-terminus or at C-terminus of chain and most of algorithms easily find these regions. More difficult task is to find a very short unstructured region in the center of a protein. In sulfotransferase 1A3 there are disordered regions at positions Gly64–Val77, Ser91–Leu93, Pro216–Ala261, as well as disordered N-terminus (amino acid residues 1–7) and C-terminus (amino acid residues 294–295) (Bidwell et al. 1999). We have determined the structural instability of sulfotransferase 1A3 using the amino acid sequence, and not 3D structure, of this protein by PentUnFOLD 1D and other algorithms. Results of structural stability/instability of sulfotransferase 1A3 are provided in Table 6S in the Supplementary Material.
In Protein Data Bank there are two structures of this protein: 1CJM and 2A3R. In 1CJM structure only sulfate ions are present. In 2A3R structure there are two ligands: A3P (adenosine 3'-phosphate-5'-phosphate) and LDP (dopamine) and the only one disordered region (at the N-terminus) is present. Using PLIP program (Salentin et al. 2015) we have determined amino acids forming interactions with these ligands. A3P forms hydrogen bonds with Lys48, Ser49, Gly50, Thr51, Thr52, Ser138, Thr227, Phe255, Arg257, Lys258, Gly259; water bridges with Phe229, Arg257, Met260; π-π interactions with Trp53, Phe229; salt bridges with Lys48, Arg130, Arg257. As one can see, A3P forms interactions with the ordered part of that protein and with N- and C-termini of disordered region Pro216–Ala261, but not with central part of it. The second ligand (LDP) does not form interactions with amino acids from parts of sulfotransferase 1A3 known to be disordered.
We can say that intrinsically disordered fragment Pro216–Ala261 has only a few key amino acids «responsible for» its disordered state. Most of algorithms predicted disordered region in boarders Pro216–Ala261. PONDR VSL2 found IDPR in boarders Leu215–Asp249. PONDR VL-XT determined disordered state between Ile204 and Met220. FoldIndex© found IDPRs in boarders Arg213–Thr219, Lys230–Asn239, Gln245–His250, Pro254–Gly259, Ala261–Thr266. GlobPlot 2.3 did not predict disordered state in known boarders. Predictions of PentUnFOLD algorithm are very specific: it showed that only a few amino acids at N- and C-termini of IDPR 216—261 are “producers” of the disordered state of the whole region (Gln225, Phe229, Lys230, Glu231, Met232, Met256, Arg257).
Intrinsically disordered region in boarders Gly64–Val77 was found only by FoldIndex©, but its prediction is not specific. PentUnFOLD algorithm predicted concrete amino acids responsible for the disordered state in this region of the protein: Ile61, Tyr62, Lys69, Cys70, Phe74.
Intrinsically disordered region Ser91–Leu93 was found by all the algorithms, but the results were very nonspecific: all algorithms predicted this disordered fragment in much wider borders. The PentUnFOLD 1D algorithm did not predict the instability of this region of the protein. Meta-predictors like Depicter and MetaDisorder did not find the IDR in the known borders of investigated protein, even though MetaDisorder assembles 13 disordered predictors: DisEMBL, DISOPRED2, DISpro, Globplot, iPDA, IUPred, Pdisorder, Poodle-s, Poodle-L, PrDOS, Spritz, DisPSSMP, RONN (Li et al. 2015).
Discussion
Comparison of performance of different predicting methods and computer algorithms is not something completely straightforward and objective. There are many different criterions to evaluate their ability to predict that usually show different results. From this point of view, it is important to discuss advantages and disadvantages of those algorithms to understand when and why they become suitable, and to identify conditions in which they are becoming misleading.
Coming back to Table 1 one can see that the highest accuracy belongs to the DISOPRED3 (disordered) algorithm (91.16%). Intriguingly, the closest value of accuracy among our algorithms (85.31%) belongs to the PentUnFOLD 1D. The second best of our algorithms in terms of accuracy (81.70%) is the PentUnFOLD 2D in case if we consider only “D” and “V” residues. However, PentUnFOLD 3D has the value of accuracy equal to just 37.42%. The reason of this difference in accuracy is in the common style of disorder prediction for PentUnFOLD 1D, DISOPRED3 (disordered), DISOPRED3, AUCpreD, and flDPnn. All abovementioned algorithms have low sensitivity to the disordered residues, but high sensitivity to ordered residues. The fraction of ordered residues is higher than the fraction of disordered ones. That is why, taken together, the ratio between the sum of true positive and true negative residues and the sum of all residues is so high. One may choose those algorithms to find sequences with a high tendency to turn into the disordered state, as well as regions that are usually ordered. Such ability is well reflected by the MCC (Mathew’s correlation coefficient). The highest values of MCC, that are, actually, still far from 1, are there for AUCpred, DISOPRED3, and DISOPRED3 (disordered) (Table 1). Among PentUnFOLD algorithms, only the PentUnFOLD 1D has MCC value that is close to the one of DISOPRED3 (disordered). However, both in Tables 1 and 2 MCC values are somewhere near 0 reflecting that there is still a need of new ideas and approaches from the side of software for disordered regions prediction developers.
In Table 2, accuracy values for all algorithms never rich as high values, as in Table 1. Indeed, a lot of residues predicted to be ordered are really disordered at least in some structures of HSA. So, three other algorithms show highest accuracy values in the set of HSA structures: PentUnFOLD 3D, Foldindex, and VSL2. Those algorithms largely overpredict disorder in the test set (Table 1), but perform much better in the set with increased percent of disordered residues (Table 2). So, abovementioned algorithms are recommended in case if one wants to find all the regions that have a chance (even a low one) to turn into the disordered state. Indeed, if we consider F1 index, that is largely focused on the ability of algorithms to find true positives, we will see that such ability has the highest values in PentUnFOLD 3D, Foldindex, and VSL2. Notice that the values of F1 for these algorithms are much higher in Table 2 than in Table 1.
Taken together, the test of performance of current algorithms in a new set of proteins showed that they can be classified into two groups: those that are good in identification of regions that have high probability to turn into the disordered state, and those that are good in identification of regions that have high, average or even low probability to become disordered.
Conclusions
Due to the enormous functional and medical importance of IDPs/IDPRs, prediction of intrinsic protein disorder from amino acid sequence has become an area of active research. Such proteins are frequently involved in some of the most important regulatory functions in the cell, and the intrinsic lack of structure can confer functional advantages on a protein, including the ability to bind to several different targets performing sometimes even opposite functions. A lot of diseases are associated with different structural transitions. That is why approaches to creating new predictive algorithms are being developed.
Our algorithm, PentUnFOLD, is based on the newly obtained propensity scales and it can determine not only fragments of alpha helices, beta strands, and random coils that can turn into the completely disordered state, but also regions of alpha helices and beta strands which are able to turn into random coils, and vice versa (H ↔ C, E ↔ C) not just at the N- and C-termini of proteins, but in the middle of their sequences. Moreover, PentUnFOLD has the option not only to determine the effect of amino acid substitutions, but also secondary structure transitions on the stability of a given region in unmodified or modified protein.
Prediction of disordered regions from the 3D structure brings some benefits compared to the prediction from amino acid sequence. At first, amino acid content of alpha helices, beta strands and random coils prone to turn into the disordered state have some differences. So, it is better to know the secondary structure of a given fragment of polypeptide chain to consider its ability to turn into random coil or disordered state. At second, interactions between amino acid residues may decrease or increase the possibility of a given fragment transition to the disordered state.
Our web server (http://3.17.12.213/pent-un-fold) processes one PDB file or amino acid sequence at a time. The algorithm itself is incorporated into the MS Excel spreadsheet. So, all the data are inserted into the spreadsheet automatically by the JAVA scripts from our server. Then a user has to download resulting file and open it with either MS Excel or LibreOffice Calc. Users are also welcome to perform those operations manually with original spreadsheets (http://chemres.bsmu.by/PentUnFOLD.htm).
Supplementary Information
Below is the link to the electronic supplementary material.
Author contributions
VVP: conceptualization, methodology, investigation, formal analysis, writing—original draft preparation. VVK: investigation, formal analysis, writing—original draft preparation. TAK: investigation, supervision, writing—review and editing. TEK: creation of the web server, the development of the script for secondary structure determination. VDP: the development of the script for intraprotein interactions investigation.
Funding
Not applicable.
Data availability
All the data used to write this article are available in the "Supplementary material" section.
Code availability
Not applicable.
Declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Ethics approval
Animals and human biomaterials were not used when writing this article.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Aguilar-Calvo P, Garcia C, Espinosa JC, Andreoletti O, Torres JM. Prion and prion-like diseases in animals. Virus Res. 2015;207:82–93. doi: 10.1016/j.virusres.2014.11.026. [DOI] [PubMed] [Google Scholar]
- Babu MM. The contribution of intrinsically disordered regions to protein function, cellular complexity, and human disease. Biochem Soc Trans. 2016;44:1185–1200. doi: 10.1042/BST20160172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barik S. Genus-specific pattern of intrinsically disordered central regions in the nucleocapsid protein of coronaviruses. Comput Struct Biotechnol J. 2020;18:1884–1890. doi: 10.1016/j.csbj.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barik A, Katuwawala A, Hanson J, Paliwal K, Zhou Y, Kurgan L. DEPICTER: Intrinsic Disorder and Disorder Function Prediction Server. J Mol Biol. 2020;432:3379–3387. doi: 10.1016/j.jmb.2019.12.030. [DOI] [PubMed] [Google Scholar]
- Bidwell LM, McManus ME, Gaedigk A, Kakuta Y, Negishi M, Pedersen L, Martin JL. Crystal structure of human catecholamine sulfotransferase. J Mol Biol. 1999;293:521–530. doi: 10.1006/jmbi.1999.3153. [DOI] [PubMed] [Google Scholar]
- Carrell RW, Lomas DA. Conformational disease. Lancet. 1997;350:134–138. doi: 10.1016/S0140-6736(97)02073-4. [DOI] [PubMed] [Google Scholar]
- Chou PY, Fasman GD. Empirical predictions of protein conformation. Annu Rev Biochem. 1978;47:251–276. doi: 10.1146/annurev.bi.47.070178.001343. [DOI] [PubMed] [Google Scholar]
- Corbi-Verge C, Kim PM. Motif mediated protein-protein interactions as drug targets. Cell Commun Signal. 2016;14:8. doi: 10.1186/s12964-016-0131-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosztanyi Z, Csizmok V, Tompa P, Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- Dunker AK, Obradovic Z, Romero P, Garner EC, Brown CJ. Intrinsic protein disorder in complete genomes. Genome Inform Ser Workshop Genome Inform. 2000;11:161–171. [PubMed] [Google Scholar]
- Dunker AK, Silman I, Uversky VN, Sussman JL. Function and structure of inherently disordered proteins. Curr Opin Struct Biol. 2008;18:756–764. doi: 10.1016/j.sbi.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Dunker AK, Garner E, Guilliot S, Romero P, Albrecht K, Hart J, Obradovic Z, Kissinger C, Villafranca JE (1998) Protein disorder and the evolution of molecular recognition: theory, predictions and observations. Pac Symp Biocomput. pp 473–484. [PubMed]
- Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- Hanson J, Paliwal KK, Litfin T, Zhou Y. SPOT-Disorder 2: Improved Protein Intrinsic Disorder Prediction by Ensembled Deep Learning. Genomics Proteomics Bioinformatics. 2019;17:645–656. doi: 10.1016/j.gpb.2019.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazy E, Tompa P. Limitations of induced folding in molecular recognition by intrinsically disordered proteins. ChemPhysChem. 2009;10:1415–1419. doi: 10.1002/cphc.200900205. [DOI] [PubMed] [Google Scholar]
- Hu G, Wu Z, Wang K, Uversky VN, Kurgan L. Untapped potential of disordered proteins in current druggable human proteome. Curr Drug Targets. 2016;17:1198–1205. doi: 10.2174/1389450116666150722141119. [DOI] [PubMed] [Google Scholar]
- Hu G, Katuwawala A, Wang K, Wu Z, Ghadermarzi S, Gao J, Kurgan L. flDPnn: Accurate intrinsic disorder prediction with putative propensities of disorder functions. Nat Commun. 2021;12:4438. doi: 10.1038/s41467-021-24773-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ironside JW, Ritchie DL, Head MW. Prion diseases. Handb Clin Neurol. 2017;145:393–403. doi: 10.1016/B978-0-12-802395-2.00028-6. [DOI] [PubMed] [Google Scholar]
- Ishida T, Kinoshita K. PrDOS: prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 2007;35:460–464. doi: 10.1093/nar/gkm363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Cozzetto D. DISOPRED3: precise disordered region predictions with annotated protein-binding activity. Bioinformatics. 2015;31:857–863. doi: 10.1093/bioinformatics/btu744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Katuwawala A, Oldfield CJ, Kurgan L. Accuracy of protein-level disorder predictions. Brief Bioinform. 2020;21:1509–1522. doi: 10.1093/bib/bbz100. [DOI] [PubMed] [Google Scholar]
- Khrustalev VV, Barkovsky EV. Stabilization of secondary structure elements by specific combinations of hydrophilic and hydrophobic amino acid residues is more important for proteins encoded by GC-poor genes. Biochimie. 2012;94:2706–2715. doi: 10.1016/j.biochi.2012.08.008. [DOI] [PubMed] [Google Scholar]
- Khrustalev VV, Khrustaleva TA, Szpotkowski K, Poboinev VV, Kakhanouskaya KY. The part of a long beta hairpin from the scrapie form of the human prion protein is reconstructed in the synthetic CC36 protein. Proteins: Structure. Function and Bioinformatics. 2016;84:1462–1479. doi: 10.1002/prot.25090. [DOI] [PubMed] [Google Scholar]
- Khrustalev VV, Poboinev VV, Stojarov AN, Khrustaleva TA. Microenvironment of tryptophan residues in proteins of four structural classes: applications for fluorescence and circular dichroism spectroscopy. Eur Biophys J. 2019;48:523–537. doi: 10.1007/s00249-019-01377-0. [DOI] [PubMed] [Google Scholar]
- Kopito RR, Ron D. Conformational disease. Nat Cell Biol. 2000;2:207–209. doi: 10.1038/35041139. [DOI] [PubMed] [Google Scholar]
- Le Gall T, Romero PR, Cortese MS, Uversky VN, Dunker AK. Intrinsic disorder in the Protein Data Bank. J Biomol Struct Dyn. 2007;24:325–342. doi: 10.1080/07391102.2007.10507123. [DOI] [PubMed] [Google Scholar]
- Li J, Feng Y, Wang X, Li J, Liu W, Rong L, Bao J. An Overview of Predictors for Intrinsically Disordered Proteins over 2010–2014. Int J Mol Sci. 2015;16:23446–23462. doi: 10.3390/ijms161023446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linding R, Russell RB, Neduva V, Gibson TJ. GlobPlot: exploring protein sequences for globularity and disorder. Nucleic Acids Res. 2003;31:3701–3708. doi: 10.1093/nar/gkg519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linding R, Jensen LJ, Diella F, Bork P, Gibson TJ, Russell RB. Protein disorder prediction: implications for structural proteomics. Structure. 2003;11:1453–1459. doi: 10.1016/j.str.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Luo M. Influenza Virus Entry. In: Rossmann MG, Rao VB, editors. the Viral Molecular Machines. Boston, MA: Springer; 2012. pp. 201–221. [Google Scholar]
- Necci M, Piovesan D, Dosztanyi Z, Tosatto SCE. MobiDB-lite: fast and highly specific consensus prediction of intrinsic disorder in proteins. Bioinformatics. 2017;33:1402–1404. doi: 10.1093/bioinformatics/btx015. [DOI] [PubMed] [Google Scholar]
- Necci M, Piovesan D, Tosatto SCE. Critical assessment of protein intrinsic disorder prediction. Nat Methods. 2021;18:472–481. doi: 10.1038/s41592-021-01117-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oates ME, Romero P, Ishida T, Ghalwash M, Mizianty MJ, Xue B, Dosztányi Z, Uversky VN, Obradovic Z, Kurgan L, Dunker AK, Gough J. D2P2: database of disordered protein predictions. Nucleic Acids Res. 2013;41:508–516. doi: 10.1093/nar/gks1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- Peng K, Radivojac P, Vucetic S, Dunker AK, Obradovic Z. Length-dependent prediction of protein intrinsic disorder. BMC Bioinformatics. 2006;7:208. doi: 10.1186/1471-2105-7-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Z, Yan J, Fan X, Mizianty MJ, Xue B, Wang K, Hu G, Uversky VN, Kurgan L. Exceptionally abundant exceptions: comprehensive characterization of intrinsic disorder in all domains of life. Cell Mol Life Sci. 2015;72:137–151. doi: 10.1007/s00018-014-1661-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poboinev VV, Khrustalev VV, Stojarov AN, Khrustaleva TA (2018) Thermodynamic characteristics of the structure stability of four classes of proteins. Molecular, membrane and cellular bases of functioning of biosystems: international scientific conference; The Thirteenth Congress of the Belarusian public association of photobiologists and biophysicists: book of abstracts. p 34. (in Russian).
- Prilusky J, Felder CE, Zeev-Ben-Mordehai T, Rydberg EH, Man O, Beckmann JS, Silman I, Sussman JL. FoldIndex©: a simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics. 2005;21:3435–3438. doi: 10.1093/bioinformatics/bti537. [DOI] [PubMed] [Google Scholar]
- Promponas VJ, Enright AJ, Tsoka S, Kreil DP, Leroy C, Hamodrakas S, Sander C, Ouzounis CA. CAST: An iterative algorithm for the complexity analysis of sequence tracts. Bioinformatics. 2000;16:915–922. doi: 10.1093/bioinformatics/16.10.915. [DOI] [PubMed] [Google Scholar]
- Radivojac P, Obradovic Z, Smith DK, Zhu G, Vucetic S, Brown CJ, Lawson JD, Dunker AK. Protein flexibility and intrinsic disorder. Protein Sci. 2004;13:71–80. doi: 10.1110/ps.03128904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero P, Obradovic Z, Li X, Garner EC, Brown CJ, Dunker AK. Sequence complexity of disordered protein. Proteins. 2001;42:38–48. doi: 10.1002/1097-0134(20010101)42:1<38::AID-PROT50>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Romero P, Obradovic Z, Kissinger CR, Villafranca JE, Garner E, Guilliot S, Dunker AK (1998) Thousands of proteins likely to have long disordered regions. Pac Symp Biocomput. p 437–448. [PubMed]
- Salentin S, Schreiber S, Haupt VJ, Adasme MF, Schroeder M. PLIP: fully automated protein–ligand interaction profiler. Nucleic Acids Res. 2015;43:443–447. doi: 10.1093/nar/gkv315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santofimia-Castaño P, Rizzuti B, Xia Y, Abian O, Peng L, Velázquez-Campoy A, Neira JL, Iovanna J. Targeting intrinsically disordered proteins involved in cancer. Cell Mol Life Sci. 2020;77:1695–1707. doi: 10.1007/s00018-019-03347-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steckmann T, Bhandari YR, Chapagain PP, Gerstman BS (2017) Cooperative structural transitions in amyloid-like aggregation. J Chem Phys. Article 135103. [DOI] [PubMed]
- Tina KG, Bhadra R, Srinivasan N. PIC: Protein Interactions Calculator. Nucleic Acids Res. 2007;35:W473–W476. doi: 10.1093/nar/gkm423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa P, Fuxreiter M. Fuzzy complexes: polymorphism and structural disorder in protein-protein interactions. Trends Biochem Sci. 2008;33:2–8. doi: 10.1016/j.tibs.2007.10.003. [DOI] [PubMed] [Google Scholar]
- Uversky VN. Intrinsically disordered proteins from A to Z. Int J Biochem Cell Biol. 2011;43:1090–1103. doi: 10.1016/j.biocel.2011.04.001. [DOI] [PubMed] [Google Scholar]
- Uversky VN, Dunker AK. Understanding Protein Non-Folding. Biochim Biophys Acta. 2010;1804:1231–1264. doi: 10.1016/j.bbapap.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN, Gillespie JR, Fink AL. Why are "natively unfolded" proteins unstructured under physiologic conditions? Proteins. 2000;41:415–427. doi: 10.1002/1097-0134(20001115)41:3<415::AID-PROT130>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Uversky VN, Oldfield CJ, Dunker AK. Intrinsically disordered proteins in human diseases: introducing the D2 concept. Annu Rev Biophys. 2008;37:215–246. doi: 10.1146/annurev.biophys.37.032807.125924. [DOI] [PubMed] [Google Scholar]
- Uversky VN, Dave V, Iakoucheva LM, Malaney P, Metallo SJ, Pathak RR, Joerger AC. Pathological unfoldomics of uncontrolled chaos: Intrinsically disordered proteins and human diseases. Chem Rev. 2014;114:6844–6879. doi: 10.1021/cr400713r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN, Na I, Landau KS, Schenck RO. Highly Disordered Proteins in Prostate Cancer. Curr Protein Pept Sci. 2017;18:453–481. doi: 10.2174/1389203717666161028145848. [DOI] [PubMed] [Google Scholar]
- Uversky VN (2010) The mysterious unfoldome: Structureless, underappreciated, yet vital part of any given proteome. J Biomed Biotechnol. Article 568068. [DOI] [PMC free article] [PubMed]
- Vihinen M, Torkkila E, Riikonen P. Accuracy of protein flexibility predictions. Proteins. 1994;19:141–149. doi: 10.1002/prot.340190207. [DOI] [PubMed] [Google Scholar]
- Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Ward JJ, McGuffin LJ, Bryson K, Buxton BF, Jones DT. The DISOPRED server for the prediction of protein disorder. Bioinformatics. 2004;20:2138–2139. doi: 10.1093/bioinformatics/bth195. [DOI] [PubMed] [Google Scholar]
- Wootton JC. Non-globular domains in protein sequences: Automated segmentation using complexity measures. Comput Chem. 1994;18:269–285. doi: 10.1016/0097-8485(94)85023-2. [DOI] [PubMed] [Google Scholar]
- Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- Xie Q, Arnold GE, Romero P, Obradovic Z, Garner E, Dunker AK. The sequence attribute method for determining relationships between sequence and protein disorder. Genome Inform Ser Workshop Genome Inform. 1998;9:193–200. [PubMed] [Google Scholar]
- Xue B, Dunbrack RL, Williams RW, Dunker AK, Uversky VN. PONDR-FIT: a metapredictor of intrinsically disordered amino acids. Biochim Biophys Acta. 2010;1804:996–1010. doi: 10.1016/j.bbapap.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue B, Dunker AK, Uversky VN. Orderly order in protein intrinsic disorder distribution: disorder in 3500 proteomes from viruses and the three domains of life. J Biomol Struct Dyn. 2012;30:137–149. doi: 10.1080/07391102.2012.675145. [DOI] [PubMed] [Google Scholar]
- Zoete V, Michielin O, Karplus M. Relation between sequence and structure of HIV-1 protease inhibitor complexes: a model system for the analysis of protein flexibility. J Mol Biol. 2002;315:21–52. doi: 10.1006/jmbi.2001.5173. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data used to write this article are available in the "Supplementary material" section.
Not applicable.