Summary
The exposome concept encourages holistic consideration of the non-genetic factors (environmental exposures including lifestyle) that influence an individual’s health over their life course. However, disconnect between the concept and practical application has promoted divergent interpretations of the exposome across disciplines and reinforced separation of the environmental (emphasizing exposures) and biological (emphasizing responses) research communities. In particular, while knowledge of biological responses can help to distinguish actual (i.e. experienced) from potential exposures, the inclusion of endogenous processes has generated confusion about the position of the exposome in a multi-omics systems biology context. We propose a reattribution of “exposome” to exclusively represent the totality of contact with external factors that a biological entity experiences, and introduce the term “functional exposomics” to denote the systematic study of exposure-phenotype interaction. This reoriented definition of the exposome allows a more readily integrable dataset for multi-omics and systems biology research.
Subject areas: Environmental health, Exposure assessment, Omics
Graphical Abstract
Highlights
-
•
Reattribution of exposome concept to exclusively represent environmental exposures
-
•
Generalized the exposome concept for all levels of biological organization
-
•
Functional exposome presented as the totality of exposure-phenotype interaction
Introduction
The exposome concept initially represented an individual human’s “life-course environmental exposures (including lifestyle factors), from the prenatal period onwards” (Wild, 2005). Presented as a complement to the genome, a shift toward more comprehensive characterization of exposure was advocated, aiming to raise the prioritization of exposure risk factors to a comparable level as for genetic risk factors. Notably, the terms “envirome” and “environome” refer to the same concept (Anthony et al., 1995; Sher et al., 2010; Toscano and Oehlke, 2005), but have not received the same recognition and use as the exposome.
Recognizing the critical role of underlying endogenous processes in the continuum from exposure to disease, practical application of the exposome concept often includes in-depth biological characterization across molecular omics layers to better understand mechanisms underlying diseases. As a result, the exposome was further elaborated to encompass the associated biological responses to exposures which are vital to understand environmental influence on human health (Wild, 2012). Building upon this, Miller and Jones redefined the exposome as “the cumulative measure of environmental influences and associated biological responses throughout the lifespan, including exposures from the environment, diet, behavior, and endogenous processes” (Miller and Jones, 2014). The re-definition emphasized the need for integrative study of genetic and environmental determinants of health and reflected progression of the exposome concept into a practicable research discipline.
The potential for comprehensive characterization of exposure to further understanding about the environmental influence on health has led to the exposome concept being embraced beyond epidemiology and public health domains, adopted within, e.g. personalized health and precision medicine, pharmacology, (eco)toxicology, and risk assessment (Escher et al., 2017; Niedzwiecki et al., 2019; Sillé, 2020; Vermeulen et al., 2020). Yet, adoption of the exposome concept by many scientific fields has led to developments framed by the different terminologies and theoretical perspectives of various disciplines, presenting a barrier for interdisciplinary research.
Integration of an exposome component into the multi-omics framework was recently encouraged, but came with the proviso that inclusion of an exposomics layer should harmonize with the framework of established molecular layers to capitalize upon the extensive wealth of biological information already captured in multi-omics applications (Miller, 2021).
Although domain-specific contextualization has advanced the exposome from concept to practical applications, it is recognized that many fundamental features of the exposome have remained ambiguous as the concept evolved in the past 15 years (Haddad et al., 2019), such as what constitutes exposure (Brunekreef, 2013) and what distinguishes response and effect?
Through contemplating these conceptual ambiguities, our perspective rationalizes depicting exposures as contact events and reattributing the exposome to exclusively represent these events, i.e. environmental exposures. By uncoupling exposure and response, a clear position for the exposome within an integrated molecular omics framework is outlined. The framework generalizes to all organisms and levels of biological organization. Moving from concept to application, we discuss emerging functional exposomics studies beginning to advance understanding about the environmental contribution to phenotype and health.
What constitutes exposure?
Strikingly, “exposure” is often ambiguously defined within scientific literature (Brunekreef, 2013) with 28 definitions compiled via the US EPA Terminology Service (United States Environmental Protection Agency, 2009). Of these, there are two prevailing interpretations:
-
i)
Contact between an agent and a target. Contact takes place at an exposure surface over an exposure period.
-
ii)
Concentration or amount of a particular agent that reaches a target organism, system, or (sub)population in a specific frequency for a defined duration.
The former definition, endorsed by the International Society of Exposure Science (Zartarian et al., 2005), represents a qualitative observation (presence of contact event) exclusive for a single occurrence (exposure period, i.e. continuous contact). The latter, widely used in chemical hazard/risk assessment (International Programme on Chemical Safety & Organisation for Economic Co-operation and Development, 2004), provides a quantitative characterization (agent concentration) over a time frame which can comprise multiple contact events (exposure duration). Adding further ambiguity is that the discourse of chemical exposure has commonly used the term “exposure” to refer to agents that one may or may not come into contact with, i.e. potential exposure (Everett et al., 2019; Rappaport and Smith, 2010). Though seemingly minor, these discrepancies have wider implications; for instance, the prioritization of the sum of exposure agent concentration above the spatiotemporal dynamics of exposure, or enabling an interpretation that the exposome refers to a universal measure of all potential exposure agents rather than related with actualized contact. We propose a refined definition of environmental exposure that builds upon the event of contact:
Environmental exposure: a contact between external factor(s) (agent) and a biological entity occurring at an (exposure) interface. A single exposure event (exposure period) is a continuous contact with an agent.
We favor describing exposure as a contact event because it emphasizes the many dynamic properties required for exposure characterization, e.g. spatial (exposure interface) and temporal (exposure period) dimensions, quality (type), and quantity of external factor(s). Plus, it permits assessment of mixed (i.e. numerous types of external factors) and/or multiple (i.e. multiple contact events) exposures, needed to investigate real-world scenarios (Tipton, 2012). Other common misconceptions are that exposure only pertains to negative effects (Miller and Jones, 2014). Therefore, we have elaborated “agent” in the hopes to emphasize the breadth of exposure science beyond assessment of the toxic chemical domain to encompass physical (e.g. ambient light), biological (e.g. probiotics), and psychosocial (e.g. social integration) factors. A more balanced recognition of beneficial exposure effects, alongside awareness of harmful effects, will be paramount to understand wellbeing and good health beyond pathological states.
The intersect of exposure-response
We fully support the notion that the influence of exposures cannot be interpreted without considering biological response (Dennis et al., 2016). Overall, the exposure-response relationship is bidirectional, i.e. each mutually act on one another, and so it is unnecessary to establish temporal order to directly characterize exposure-response interaction (Arora et al., 2020). However, the measurement of contact events is often indirect (Committee on Human and Environmental Exposure Science in the 21st Century; Board on Environmental Studies and Toxicology; Division on Earth and Life Studies; National Research Council, 2012; Rappaport, 2011) and incidence inferred. Moreover, within the continuum of exposure-response relationships, each individual event operates is a unidirectional forward relationship and is unique, i.e. an exposure occurs prior to a response and individual events cannot be undone nor repeated (Comunidad Los Horcones, 2005). For indirect measures, exposure and response are distinct, even if linked and their boundaries impossible to discern. Building upon our refined definition of exposure, we propose reattributing the exposome to exclusively represent environmental exposure:
Exposome: the totality of environmental exposures, i.e. the totality of contact between external factors (agents) and a biological entity.
Our definition encapsulates the totality of contact events, and a distinction between exposure and response enables measurements related to contact with external factors to be isolated from the cascades of endogenous processes, e.g. DNA mutations arising from the imperfect repair of DNA polymerase errors. While the term “endogenous exposome” has been used to describe these cascades (Nakamura et al., 2014), we consider them as indirect outcomes from endogenous processes and not representing an environmental exposure as a contact event with an external factor. The separation is beneficial for molecular multi-omics communication because these non-canonical outcomes of biological processes are inherently captured within measures of other factors, e.g. metabolomics, proteomics etc.
We acknowledge that distinction between exposure and response is difficult to apply for psychosocial factors. Various social factors can be considered external factors (e.g. access to green space) and psychological characteristics predominantly response (e.g. emotional reactions), but in many cases the intersection and traversal of external-internal boundaries is often hard to categorize (e.g. subjection to stressors–stressor perception).
Notably, molecular omics characterization has been adopted in clinical settings to measure allostatic load, whereby multiple biomarkers indicative of physiological dysregulation are used in combination to represent the cumulative burden of stress (Fava et al., 2019). Similarly to exposure, there are multiple interpretations of stress in different disciplines (Fink, 2016; Mason, 1975; Selye, 1975). It was recently posited that the most generic definition (“stress is the non-specific response of the body to any demand”) (Selye, 1976) could be unassailable by disregarding “non-specific” (Fink, 2016). Therefore, the measurement of allostatic load (stress) can be viewed as a characterization of the (physiological) responses following contact between stressors (external factors) and a biological entity over a defined time-period. It is effectively the characterization of response to exposures, and thus complementary to exposomics studies that focus upon characterizing exposures, i.e. contact between external factors and a biological entity over a defined time-period.
While some specific responses can act as an indirect measure of specific exposure, the composite measurement of allostatic load/stress provides a crude proxy of multiple exposures. Establishing a relationship between complex biomarkers of physiological dysregulation and individual exposures remains challenging (Logan et al., 2018). Therefore, increased characterization (i.e. compositional, spatiotemporal, and quantitative) of exposure is needed to identify those that are modifiable and specific for disease prevention and improving wellbeing.
Many exposures do not elicit readily observable biochemical responses (Peters et al., 2012) and molecular multi-omics characterizations can be limited in scope to investigate exposure compared to response, which is more easily represented via cellular processes or biosignals indicative of, e.g. behavioral (Beauchaine, 2012) and emotional responses (Behnke et al., 2022; Zaehringer et al., 2020). Molecular characterizations need to complement, and be complemented with, structural and psychosocial characterization at individual and macro-level (Peters et al., 2021). The greater integration of psychosocial factors and sociological expertise has been advocated to shape the development of exposome studies. This will be particularly critical to reduce risk of individualized multi-omics approaches proceeding toward incidental associations of limited significance to public health (Kohane et al., 2006; Senier et al., 2017) and to increase recognition of overarching factors underlying health disparities, which are often unrecognized or not deemed modifiable in public health strategies, e.g. economic and racial marginalization (Braveman and Gottlieb, 2014; Juarez et al., 2014; Stringhini et al., 2017).
The potential of greater characterization of exposure to psychosocial factors was recently demonstrated, where the twenty-year trajectories of >3000 individuals' neighborhood deprivation and social capital were classified into groups via latent class growth analysis (Prior, 2021). The groups showed graded-associations with later measures of allostatic load, e.g. histories of greater and more severe exposure to structural disadvantage were related to higher allostatic load. The study evidenced that the future allostatic load of different demographics may be predicted from dynamic measures of exposure history (Prior, 2021).
What distinguishes biological response and effect?
In the context of molecular multi-omics characterization, we find it useful to distinguish between molecular and higher-order phenotypic traits. Similar distinction is present in the concept of allostatic load, whereby physiological dysregulation (molecular trait) is separated from clinical disease (higher-order phenotypic trait). We suggest that actualized exposure events can be divided into those with a molecular-level response but no discernible effect on higher-order phenotype (i.e. promote the shift of homeodynamic equilibrium but not beyond the window of tolerance/contribute to allostatic load) and those resulting in an observable phenotypic effect (i.e. disturb homeodynamic equilibrium beyond its window of tolerance/allostatic overload (McEwen and Wingfield, 2003)). This framing provides clarity for the use of molecular signatures as biomarkers of exposure and biomarkers of effect (Mayeux, 2004; U.S. Food and Drug Administration - National Institutes of Health, Biomarker Working Group, 2016; World Health Organization & International Programme on Chemical Safety, 1993). For example, biomarkers of exposure would include presence of external factors in direct contact (e.g. surface microbes, internalized chemicals etc.), alongside responses indicative of a prior contact event (e.g. biotransformation products of chemical agents, presence of specific antibodies, and DNA methylations). Along the same lines, biomarkers of phenotypic effect may be implicit of the trait (e.g. single-gene mutations) or specifically indicative (e.g. CD4 T cell count and Hemoglobin A1c concentration). The phrasing is potentially useful for molecular toxicology, able to portray a no-observed-effect level (World Health Organization & International Programme on Chemical Safety, 1994) (i.e. molecular response but no effect on higher-order phenotype), and may be beneficial for increasing the use of omics technologies in next-generation toxicology (Barouki et al., 2021) and risk assessment (Canzler et al., 2020; Sillé, 2020).
Chemical biomarkers: internal chemical exposome & metabolome
As previously mentioned, when chemical exposures are considered, it is common to use external factor characterization as proxy for exposure (Everett et al., 2019; Rappaport and Smith, 2010), which has often led to ambiguity surrounding the difference between metabolomics and internal chemical exposomics. Herein, we present a perspective divide: chemical exposomics focuses on characterizing contact with chemical agents, while metabolomics focuses on characterizing biochemical reactions of a biological entity. The interpretation of internal chemicals measurements therefore greatly differs between fields.
Internal chemical exposomics research focuses on mapping markers of chemical exposure to external sources and origins, reverse dosimetry (internal dose to predict external amount) to estimate exposure risk, detoxification kinetics, and toxicity effects , whereas metabolomics involves characterization of pathway perturbations, metabolic flux, and regulation of biological responses. From another angle, internal chemical exposomics characterizes contact with chemicals that promote deviation from homeodynamic equilibrium while metabolomics characterizes the biochemical processes regulating homeodynamic equilibrium. Practically, the fields converge at the interface of measuring chemical exposure-biochemical response. Each –ome, including the metabolome, operates in the continuum of endogenous-xenobiotic processes (Nicholson and Wilson, 2003) and externally derived chemicals are constituents of the metabolome (Athersuch and Keun, 2015). Yet, a separate term is beneficial to convey the differing perspectives regarding the subsequent interpretation and raises awareness of the specific challenges faced to profile endogenous or exogenous components (David et al., 2021). As such, we propose:
Internal chemical exposome: the totality of internal contact between environmentally derived chemicals (chemical agents including biotransformation products) and a biological entity.
The definition explicitly couples internalized chemical agents with their metabolic products to encompass cellular chemical fate. In doing so, characterization of the internal chemical exposome naturally integrates the toxicokinetics of exposure agents with the toxicodynamics of exposure contact (Ghosh, 2019), and underlines the role exposomics can play in next-generation exposure risk assessment.
Generalization of the exposome concept
The exposome concept can easily be extrapolated to other organisms and complex biological systems. All living organisms have a phenotype that is the result of interactions between genetic and environmental factors, encompassing ecological and psychosocial conditions. The exposome concept has been applied to study other organisms and cells (Broadrup et al., 2019; David et al., 2017; Teixeira et al., 2011; Xu, 2016) and implementation of systems biology approaches to understand individual phenotype (Toscano and Oehlke, 2005) and ecosystem health (Purdy et al., 2010) hailed for many years.
While ecosystem applicability of the exposome concept has been described, phrasing of the “eco-exposome” vision was complex and did not place the concept in a wider biological framework (Committee on Human and Environmental Exposure Science in the 21st Century; Board on Environmental Studies and Toxicology; Division on Earth and Life Studies; National Research Council, 2012). The vision was subsequently explained and positioned within systems toxicology (Escher et al., 2017) at the interface of aggregate exposure pathway (AEP) and adverse outcome pathway (AOP) frameworks, yet the definition remained abstruse. A further re-definition narrowed the eco-exposome to represent “the totality of internal exposure over a lifetime to individuals of a given species” (Scholz et al., 2022). The definition was presented as exclusive for chemicals and has limited applicability to ecological communities and holobionts, both in phrasing and the disregard of psychosocial (and physical) factors.
Recently, the integration of exposome and One Health concepts was noted along the need for closer alignment of environmental chemistry and environmental toxicology disciplines (Gao, 2021). Expanding the exposome from an anthropocentric view to all ecological levels of organization (Table 1) demonstrates their synergy: in the same way that the influence of individual exposures cannot be interpreted without considering response, the environment of an individual or population can only be contextualized within the broader community/ecosystem network (Arah, 2009; Myers et al., 2013).
Table 1.
Scheme of major systems biology disciplines, expanded to include the exposome
| Prefix | individual scale | population/species scale | community scale |
ecosystem scale |
non-naturala |
Endogenous |
exogenous |
|---|---|---|---|---|---|---|---|
| meta- | eco- | xeno- | endo- | exo- | |||
| gen(o)me | totality of [all types of] heritable genetic material (genes) of a biological entity | genomes of an entity population/species | genomes of a community of entities | all genes of an ecosystem | non-natural genes | inherited genes | non-inherited genes |
| transcript(o)me | totality of transcribed genetic material (transcripts) of a biological entity | transcriptomes of an entity population/species | transcriptomes of a community of entities | all transcripts of an ecosystem | non-natural transcripts | native transcripts | non-native transcripts |
| prote(o)me | totality of proteins/peptides of a biological entity | proteomes of an entity population/species | proteomes of a community of entities | all proteins/peptides of an ecosystem | non-natural proteins/peptides | natively expressed proteins/peptides | non-native proteins/peptides |
| metabol(o)me | totality of substances involved in metabolic processes (metabolites) of a biological entity | metabolomes of an entity population/species | metabolomes of a community of entities | all metabolites of an ecosystem | non-natural metabolites i.e derived from artificial substances | native metabolites | non-native metabolites i.e. acquired |
| microbi(o)me | totality of microorganisms in direct interaction with a biological entity | microbiomes of an entity population/species | microbiomes of a community of entities | all microorganisms of an ecosystem | N/A | indigenous microorganisms | non-indigenous microorganisms |
| phen(o)me | totality of traits/characteristics displayed by a biological entity | phenomes of an entity population/species | phenomes of a community of entities | all characteristics an ecosystem displaysb | N/A | N/A | N/A |
| expos(o)me | totality of contact between an external factor and a biological entity | exposomes of an entity population/species | exposomes of a community of entities | all contact between an external factor and an ecosystemc | contact with non-natural external factors i.e. artificial substances | N/A | N/A |
Prior to point in time, e.g. human activity in ecosystem.
i.e. Description of biophysical environment.
i.e. Ecosystem-ecosystem interaction. If generalized to ecosphere, the external factors are fundamental forces.
Elucidating the temporal dynamics of genotype and environment interplay is critical to understand phenotype trajectories (Boyce et al., 2020), and establishing chronology across multiple time scales and generations can advance the exposome from an individual to an evolutionary framework. Notably, the temporal dynamics of an exposure event, and thus the biologically meaningful time frame for characterization of the exposome depends upon the biological entity under study (Assmus et al., 2006; Jackson et al., 2021). For example, the time frame to study an individual infected with an infectious agent is shorter than time frame needed to study a population. Similarly, studying the exposome of a single cell would require a shorter measurement period than studying a multicellular organism with longer life cycle. Greater consideration of the coarse graining of time and biological levels of organization in studies and confronting the problems this simplification imposes on our understanding of biological function within evolving systems has been deemed imperative (Bergelson et al., 2021).
While Gao described the exposome and One Health concepts as multiple stressor and multiple receptor approaches, respectively (Gao, 2021), we view each as describing the same multiple interactions from the mirrored perspectives of multi-level exposure and response characterization.
Functional exposomics: concept to application
Disparity between the holistic exposome concept and current measurement capabilities is often highlighted. The feasibility of lifetime internal exposure measurement is called into question (Sabbioni et al., 2020), the plausibility for biomarker signatures to reflect all relevant exposures is doubted (Peters et al., 2012), and concerns raised about increasingly personalized exposure measures potentially hindering recognition of population-level or social elements (Canali, 2020; Senier et al., 2017). External factors are highly diverse and characterizing the composition of exposure (i.e. the type(s) of factor) is therefore more challenging than compositional characterization of other molecular omics. That said, the (lack of) feasibility for absolute -ome measurement can be debated for all omics disciplines (Prohaska and Stadler, 2011), particularly regarding quantitative measures and spatiotemporal characterization (Mahner and Kary, 1997). For example, decades after qualitative whole genome sequencing became viable, the assessment of the accumulation of DNA mutations during aging is a burgeoning field (Schumacher et al., 2021; Vijg and Montagna, 2017). Techniques for spatial and/or temporal characterization of molecular omics layers are emerging, though approaches for in situ molecular omics analysis (i.e. real-time direct on living systems) remain limited.
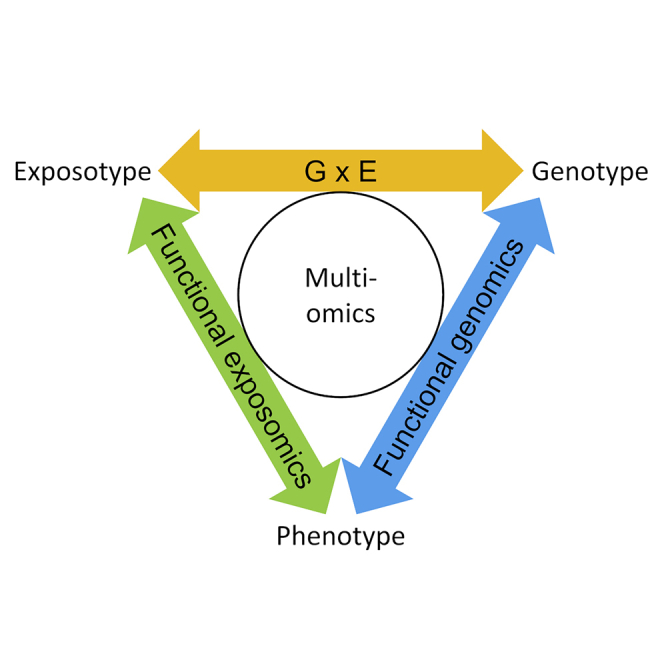
Previously, Chung et al. defined the functional exposome as “the life-course exposures to both endogenous and exogenous biologically active molecules” and “the totality of the biologically active exposures relevant to disease development” (Chung et al., 2021). However, environmental exposure was limited to external agents that physically interact with human host (Chung et al., 2021). We favor a universal definition of the term “functional exposomics” to embody a practical approach to advance understanding of all external factors contributing to phenome (i.e. the totality of traits/characteristics displayed by a biological entity):
Functional exposomics: the systematic and comprehensive study of environmental exposure-phenotype interaction over a defined time-period.
The phrase is intuitive and makes clear that it is essential to understand the complex interrelationship of exposure and endogenous processes. Each of the -omes inherently comprise influence from environmental factors and contextualizing molecular multi-omics with environmental measures places functional exposomics as a cornerstone of integrative biology (Figure 1). Functional exposomics focuses on the need to further elucidate exposure-phenotype interaction and both exposure-wide (Ioannidis, 2016) and outcome-wide approaches (Braun et al., 2019; VanderWeele, 2017) will be required. There is an analogy to earlier functional genomics efforts that focused on expressed genes, i.e. those that encoded proteins and were directly linked to function. Similarly, functional exposomics focuses effort on those exposures that have a demonstrable impact on phenotype.
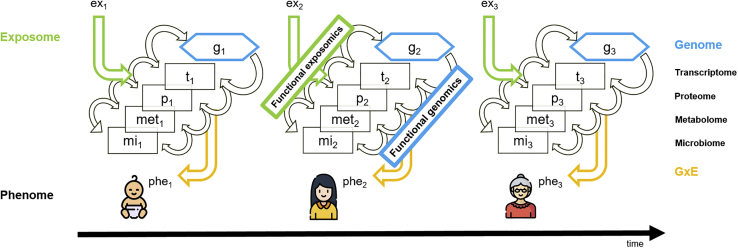
Figure 1.
Example of integrating the exposome in a human multi-omics approach and complementing genotype-to-phenotype (functional genomics) with exposotype-to-phenotype (functional exposomics) mapping
The chronological analysis of expressed genes, exposures experienced, and their interactions (GxE) can elucidate the genetic and environmental influence on phenotype development and health. Created using icons from https://icons8.com.
Appropriate time-periods for functional exposomics investigations should be defined that enable the study of causal relations between exposure and outcomes (Fang et al., 2021). Yet identification of relevant periods is challenging, especially due to latency between exposure and phenotypic effect (Balbus et al., 2013; Gillman, 2005).
Acknowledging the regulated trajectory of dynamic phenotype development (Lenart et al., 2019), Lenart et al. applied functional linear models to predict future phenotypic traits from previous observations and to identify critical windows of exposure vulnerability (i.e. periods where external factors show greater influence) (Lenart et al., 2021). In another example, Wagner et al. estimated trajectories of association between BMI history and cognitive decline using a landmark approach, identifying two critical windows (Wagner et al., 2021). Application of these approaches to prospective cohorts could be valuable for investigating latent health outcomes.
The sequence of exposure is critical (Ashauer et al., 2017; Jackson et al., 2021) and the chronological profile of exposure events can be deemed a regulated trajectory, meaning periodic characterization of exposure profiles within relevant time windows should enable future exposures to be predicted, and potentially altered. Expanding from single traits to composite profiles will require application of multivariate functional models which are able to handle the greater sparsity and variability of exposure measures, compared to biological effect (Li et al., 2019b; Peters et al., 2021).
The genetic constraints of phenotype development can also be used as an anchor to elucidate environmental influence. For example, Teixeira et al. applied a constrained partial least-squares regression-based approach to explain 90% of the observed variance in metabolic profiles of mammalian cell culture over time through temporal measures of extracellular variables (Teixeira et al., 2011).
Mapping environment-to-phenotype has already been shown as an effective theoretical approach to study population adaptation strategies (Xue et al., 2019), new frameworks to model complex gene-environment interaction show promise for more accurate disease risk estimation (Li et al., 2019a), and advanced approaches to integrate multimodal (Pan et al., 2022) and longitudinal multi-omics are emerging (Bodein et al., 2021; Kaur et al., 2020; Li et al., 2020). However, extending functional exposomics models to complex organisms and societies entails challenges, including i) the difficulty to incorporate measures of psychosocial and perceived factors alongside biological and physical factors, ii) the complexity to integrate individualized molecular measures with community-level data, iii) the fusion of exposure, response, and effect measures in interpretable dynamic networks and most critically, iv) the translation of knowledge to operable practices that improve wellbeing.
Functional exposomics investigations leveraging comprehensive characterization of environmental factors are emerging and a template for longitudinal outcome-wide association studies has been developed (VanderWeele et al., 2020). Recently, the longitudinal molecular multi-omics and exposure profiles of an individual (human) were integrated (Gao et al., 2021) and thousands of internal biomarkers correlated with external measures. These associations were cross-validated with clinical data, showcasing the potential for precision environmental health monitoring (Gao et al., 2021). In another first, the molecular multi-omics profiles of 1301 mother-child pairs were integrated with personal and community-level environmental metrics for two time-periods (Maitre et al., 2021). Thousands of associations between biological response and exposure were identified, including unique signatures for e.g. indoor air quality and weather conditions. Notably, recent childhood exposures were shown to be associated with features from all omics layers, while changes to the DNA methylome best captured in utero exposure (Maitre et al., 2021).
Conclusion
We have positioned the exposome within a multi-omics framework, generalizing the concept to be applicable to all levels of the complex biological organization. Deeming exposure as a contact event emphasizes the dynamic relationship between external factors and biological response. Characterization of both sides of this interface will require extensive collaboration across the environmental (exposure focused) and biological (response focused) research communities. Functional exposomics to associate environmental exposure to phenotype complements functional genomics linking genotype to phenotype. More extensive characterization of the exposome and integration with molecular multi-omics profiling is set to advance the factoring of phenotypic variance into genetic and environmental components. Studying their interplay places genotype-environment interaction at the center of integrative biology toward deepening our understanding of phenotype development and adaptation to further personal, population, and ecosystem health.
Summary of definitions
Environmental exposure: a contact between external factor(s) (agent) and a biological entity occurring at an (exposure) interface. A single exposure event (exposure period) is a continuous contact with a unique agent.
Exposome: the totality of environmental exposures, i.e. the totality of contact between external factors (agents) and a biological entity.
Internal chemical exposome: the totality of internal contact between environmentally derived chemicals (chemical agents including biotransformation products) and a biological entity.
Functional exposomics: the systematic and comprehensive study of environmental exposure-phenotype interaction over a defined time-period.
Acknowledgments
E.J.P. acknowledges support from the Czech Operational Programme Research, Development and Education – Project MSCAfellow4@MUNI (No. CZ.02.2.69/0.0/0.0/20_079/0017045) and Project Postdoc@MUNI (No. CZ.02.2.69/ 0.0/0.0/16_027/0008360). E.J.P., C.M.V., and J.K. acknowledge the RECETOX research infrastructure supported by the Ministry of Education, Youth and Sports of the Czech Republic (LM2018121) and funding from the CETOCOEN EXCELLENCE Teaming two project supported by Horizon 2020 (857560) and the Ministry of Education, Youth and Sports of the Czech Republic (02.1.01/0.0/0.0/18_046/0015975). G.W.M., D.I.W., and J.K. acknowledge support from the European Commission (EU-H2020 874627). R.B., K.A., and X.C. acknowledge support from INSERM and Université de Paris to Unit 1124. D.I.W. acknowledges support from the National Institutes of Health (U2C-ES030859).
Author contributions
All authors contributed to the work presented in this manuscript, provide final approval and are accountable.
Declaration of interest
Dr. Miller receives royalties for his books The Exposome: A Primer and The Exposome: A New Paradigm for the Environment and Health. He has no other conflicts of interest. The other authors declare they have no actual or potential competing financial interests.
References
- Anthony J.C., Eaton W.W., Henderson A.S. Looking to the future in psychiatric epidemiology. Epidemiol. Rev. 1995;17:240–242. doi: 10.1093/oxfordjournals.epirev.a036182. [DOI] [PubMed] [Google Scholar]
- Arah O.A. On the relationship between individual and population health. Med. Heal. Care Philos. 2009;12:235–244. doi: 10.1007/s11019-008-9173-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora M., Giuliani A., Curtin P. Biodynamic interfaces are essential for human–environment interactions. Bioessays. 2020;42:2000017. doi: 10.1002/bies.202000017. [DOI] [PubMed] [Google Scholar]
- Ashauer R., O’Connor I., Escher B.I. Toxic mixtures in time - the sequence makes the poison. Environ. Sci. Technol. 2017;51:3084–3092. doi: 10.1021/acs.est.6b06163. [DOI] [PubMed] [Google Scholar]
- Assmus H.E., Herwig R., Cho K.H., Wolkenhauer O. Dynamics of biological systems: role of systems biology in medical research. Expert Rev. Mol. Diagn. 2006;6:891–902. doi: 10.1586/14737159.6.6.891. [DOI] [PubMed] [Google Scholar]
- Athersuch T.J., Keun H.C. Metabolic profiling in human exposome studies. Mutagenesis. 2015;30:755–762. doi: 10.1093/mutage/gev060. [DOI] [PubMed] [Google Scholar]
- Balbus J.M., Barouki R., Birnbaum L.S., Etzel R.A., Gluckman S.P.D., Grandjean P., Hancock C., Hanson M.A., Heindel J.J., Hoffman K., et al. Early-life prevention of non-communicable diseases. Lancet. 2013;381:3–4. doi: 10.1016/S0140-6736(12)61609-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barouki R., Audouze K., Becker C., Blaha L., Coumoul X., Karakitsios S., Klanova J., Miller G.W., Price E.J., Sarigiannis D. The exposome and toxicology: a win–win collaboration. Toxicol. Sci. 2021:kfab149. doi: 10.1093/toxsci/kfab149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchaine T.P. Physiological markers of emotion and behavior dysregulation in externalizing psychopathology. Monogr. Soc. Res. Child Dev. 2012;77:79–86. doi: 10.1111/j.1540-5834.2011.00665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnke M., Buchwald M., Bykowski A., Kupiński S., Kaczmarek L.D. Psychophysiology of positive and negative emotions, dataset of 1157 cases and 8 biosignals. Sci. Data. 2022;9:1–15. doi: 10.1038/s41597-021-01117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergelson J., Kreitman M., Petrov D.A., Sanchez A., Tikhonov M. Functional biology in its natural context: a search for emergent simplicity. Elife. 2021;10:1–12. doi: 10.7554/eLife.67646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodein A., Scott-Boyer M.-P., Perin O., Lê Cao K.-A., Droit A. Interpretation of network-based integration from multi-omics longitudinal data. Nucleic Acids Res. 2021:gkab1200. doi: 10.1093/nar/gkab1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce W.T., Sokolowski M.B., Robinson G.E. Genes and environments, development and time. Proc. Natl. Acad. Sci. U S A. 2020;117:23235–23241. doi: 10.1073/pnas.2016710117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun J.M., Kalloo G., Kingsley S.L., Li N. Using phenome-wide association studies to examine the effect of environmental exposures on human health. Environ. Int. 2019;130:104877. doi: 10.1016/j.envint.2019.05.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braveman P., Gottlieb L. The social determinants of health: it’s time to consider the causes of the causes. Public Health Rep. 2014;129:19–31. doi: 10.1177/00333549141291s206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadrup R.L., Mayack C., Schick S.J., Eppley E.J., White H.K., Macherone A. Honey bee (apis mellifera) exposomes and dysregulated metabolic pathways associated with nosema ceranae infection. PLoS One. 2019;14:1–18. doi: 10.1371/journal.pone.0213249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunekreef B. Exposure science, the exposome, and public health. Environ. Mol. Mutagen. 2013;54:596–598. doi: 10.1002/em.21767. [DOI] [PubMed] [Google Scholar]
- Canali S. What is new about the exposome? Exploring scientific change in contemporary epidemiology. Int. J. Environ. Res. Public Health. 2020;17:2879. doi: 10.3390/ijerph17082879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canzler S., Schor J., Busch W., Schubert K., Rolle-Kampczyk U.E., Seitz H., Kamp H., von Bergen M., Buesen R., Hackermüller J. Prospects and challenges of multi-omics data integration in toxicology. Arch. Toxicol. 2020;94:371–388. doi: 10.1007/s00204-020-02656-y. [DOI] [PubMed] [Google Scholar]
- Chung M.K., Rappaport S.M., Wheelock C.E., Nguyen V.K., van der Meer T.P., Miller G.W., Vermeulen R., Patel C.J. Utilizing a biology-driven approach to map the exposome in health and disease: an essential investment to drive the next generation of environmental discovery. Environ. Health Perspect. 2021;129:085001. doi: 10.1289/EHP8327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Committee on Human and Environmental Exposure Science in the 21st Century; Board on Environmental Studies and Toxicology; Division on Earth and Life Studies; National Research Council . National Academies Press; 2012. Exposure Science in the 21st Century: A Vision and a Strategy. [DOI] [PubMed] [Google Scholar]
- Comunidad Los Horcones Types of relationships between events: their implication in the stimulus-response relationship. Behav. Dev. Bull. 2005;12:55–61. doi: 10.1037/h0100561. [DOI] [Google Scholar]
- David A., Chaker J., Price E.J., Bessonneau V., Chetwynd A.J., Vitale C.M., Klánová J., Walker D.I., Antignac J., Barouki R., Miller G.W. Towards a comprehensive characterisation of the human internal chemical exposome: challenges and perspectives. Environ. Int. 2021;156:106630. doi: 10.1016/j.envint.2021.106630. [DOI] [PubMed] [Google Scholar]
- David A., Lange A., Abdul-Sada A., Tyler C.R., Hill E.M. Disruption of the prostaglandin metabolome and characterization of the pharmaceutical exposome in fish exposed to wastewater treatment works effluent as revealed by nanoflow-nanospray mass spectrometry-based metabolomics. Environ. Sci. Technol. 2017;51:616–624. doi: 10.1021/acs.est.6b04365. [DOI] [PubMed] [Google Scholar]
- Dennis K.K., Auerbach S.S., Balshaw D.M., Cui Y., Fallin M.D., Smith M.T., Spira A., Sumner S., Miller G.W. The importance of the biological impact of exposure to the concept of the exposome. Environ. Health Perspect. 2016;124:1504–1510. doi: 10.1289/EHP140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escher B.I., Hackermüller J., Polte T., Scholz S., Aigner A., Altenburger R., Böhme A., Bopp S.K., Brack W., Busch W., et al. From the exposome to mechanistic understanding of chemical-induced adverse effects. Environ. Int. 2017;99:97–106. doi: 10.1016/j.envint.2016.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett J.R., Holmes E., Veselkov K.A., Lindon J.C., Nicholson J.K. A unified conceptual framework for metabolic phenotyping in diagnosis and prognosis. Trends Pharmacol. Sci. 2019;40:763–773. doi: 10.1016/j.tips.2019.08.004. [DOI] [PubMed] [Google Scholar]
- Fang M., Hu L., Chen D., Guo Y., Liu J., Lan C., Gong J., Wang B. Exposome in human health: utopia or wonderland? Innovation (N Y) 2021;2:100172. doi: 10.1016/j.xinn.2021.100172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fava G.A., McEwen B.S., Guidi J., Gostoli S., Offidani E., Sonino N. Clinical characterization of allostatic overload. Psychoneuroendocrinology. 2019;108:94–101. doi: 10.1016/j.psyneuen.2019.05.028. [DOI] [PubMed] [Google Scholar]
- Fink G. Stress: concepts, definition and history. Neurosci. Biobehav. Psychol. 2016:549–555. doi: 10.1016/B978-0-12-809324-5.02208-2. [DOI] [Google Scholar]
- Gao P. The exposome in the era of one health. Environ. Sci. Technol. 2021;55:2790–2799. doi: 10.1021/acs.est.0c07033. [DOI] [PubMed] [Google Scholar]
- Gao P., Shen X., Zhang X., Jiang C., Zhang S., Zhou X., Miryam S. Precision environmental health monitoring by longitudinal exposome and multi-omics profiling. bioRxiv. 2021 doi: 10.1101/2021.05.05.442855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh C. Relative matrix effects: a step forward using standard line slopes and ANOVA analysis. Arab. J. Chem. 2019;12:1378–1386. doi: 10.1016/j.arabjc.2014.11.019. [DOI] [Google Scholar]
- Gillman M.W. Developmental origins of health and disease. N. Engl. J. Med. 2005;353:1848–1850. doi: 10.1056/NEJMe058187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad N., Andrianou X.D., Makris K.C. A scoping review on the characteristics of human exposome studies. Curr. Pollut. Rep. 2019;5:378–393. doi: 10.1007/s40726-019-00130-7. [DOI] [Google Scholar]
- International Programme on Chemical Safety & Organisation for Economic Co-operation and Development . WHO; 2004. IPCS Risk Assessment Terminology.https://apps.who.int/iris/handle/10665/42908 [Google Scholar]
- Ioannidis J.P.A. Exposure-wide epidemiology: revisiting Bradford Hill. Stat. Med. 2016;35:1749–1762. doi: 10.1002/sim.6825. [DOI] [PubMed] [Google Scholar]
- Jackson M.C., Pawar S., Woodward G. The temporal dynamics of multiple stressor effects: from individuals to ecosystems. Trends Ecol. Evol. 2021;36:402–410. doi: 10.1016/j.tree.2021.01.005. [DOI] [PubMed] [Google Scholar]
- Juarez P.D., Matthews-Juarez P., Hood D.B., Im W., Levine R.S., Kilbourne B.J., Langston M.A., Al-Hamdan M.Z., Crosson W.L., Estes M.G., et al. The public health exposome: a population-based, exposure science approach to health disparities research. Int. J. Environ. Res. Public Health. 2014;11:12866–12895. doi: 10.3390/ijerph111212866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S., Peters T.J., Yang P., Luu L.D.W., Vuong J., Krycer J.R., O’Donoghue S.I. Temporal ordering of omics and multiomic events inferred from time-series data. NPJ Syst. Biol. Appl. 2020;6:1–7. doi: 10.1038/s41540-020-0141-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohane I.S., Masys D.R., Altman R.B. The incidentalome. JAMA. 2006;296:212. doi: 10.1001/jama.296.2.212. [DOI] [PubMed] [Google Scholar]
- Lenart P., Kuruczova D., Kukla L., Scheringer M., Vasku J.B. Putting the dynamic pathosome in practice: a novel way of analyzing longitudinal data. arXiv. 2021 doi: 10.48550/arXiv.2103.12822. [DOI] [Google Scholar]
- Lenart P., Scheringer M., Bienertova-Vasku J. The pathosome: a dynamic three-dimensional view of disease–environment interaction. Bioessays. 2019;41:1–6. doi: 10.1002/bies.201900014. [DOI] [PubMed] [Google Scholar]
- Li J., Li X., Zhang S., Snyder M. Gene-environment interaction in the era of precision medicine. Cell. 2019;177:38–44. doi: 10.1016/j.cell.2019.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Lu Q., Wen Y. Multi-kernel linear mixed model with adaptive lasso for prediction analysis on high-dimensional multi-omics data. Bioinformatics. 2020;36:1785–1794. doi: 10.1093/bioinformatics/btz822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Cirillo P., Hu X., Tran V., Krigbaum N., Yu S., Jones D.P., Cohn B. Understanding mixed environmental exposures using metabolomics via a hierarchical community network model in a cohort of California women in 1960’s. Reprod. Toxicol. 2019;92:57–65. doi: 10.1016/j.reprotox.2019.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan A.C., Prescott S.L., Haahtela T., Katz D.L. The importance of the exposome and allostatic load in the planetary health paradigm. J. Physiol. Anthropol. 2018;37:15. doi: 10.1186/s40101-018-0176-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahner M., Kary M. What exactly are genomes, genotypes and phenotypes? and what about phenomes? J. Theor. Biol. 1997;186:55–63. doi: 10.1006/jtbi.1996.0335. [DOI] [PubMed] [Google Scholar]
- Maitre L., Bustamante M., Hernandez-Ferrer C., Thiel D., Lau C.-H.E., Siskos A.P., Vives-Usano M., Robinson O., Wright J., Cadiou S., et al. Multi-omics signatures of the human early life exposome. medRxiv. 2021 doi: 10.1101/2021.05.04.21256605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason J.W. A historical view of the stress field. J. Hum. Stress. 1975;1:6–12. doi: 10.1080/0097840X.1975.9940399. [DOI] [PubMed] [Google Scholar]
- Mayeux R. Biomarkers: potential uses and limitations. NeuroRx. 2004;1:182–188. doi: 10.1602/neurorx.1.2.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen B.S., Wingfield J.C. The concept of allostasis in biology and biomedicine. Horm. Behav. 2003;43:2–15. doi: 10.1016/S0018-506X(02)00024-7. [DOI] [PubMed] [Google Scholar]
- Miller G.W. Integrating the exposome into a multi-omic research framework. Exposome. 2021;1:2–4. doi: 10.1093/exposome/osab002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G.W., Jones D.P. The nature of nurture: refining the definition of the exposome. Toxicol. Sci. 2014;137:1–2. doi: 10.1093/toxsci/kft251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers S.S., Gaffikin L., Golden C.D., Ostfeld R.S., Redford K.H., Ricketts T.H., Turner W.R., Osofsky S.A. Human health impacts of ecosystem alteration. Proc. Natl. Acad. Sci. U S A. 2013;110:18753–18760. doi: 10.1073/pnas.1218656110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura J., Mutlu E., Sharma V., Collins L., Bodnar W., Yu R., Lai Y., Moeller B., Lu K., Swenberg J. The endogenous exposome. DNA Repair (Amst.) 2014;19:3–13. doi: 10.1016/j.dnarep.2014.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson J.K., Wilson I.D. Understanding “global” systems biology: metabonomics and the continuum of metabolism. Nat. Rev. Drug Discov. 2003;2:668–676. doi: 10.1038/nrd1157. [DOI] [PubMed] [Google Scholar]
- Niedzwiecki M.M., Walker D.I., Vermeulen R., Chadeau-Hyam M., Jones D.P., Miller G.W. The exposome: molecules to populations. Annu. Rev. Pharmacol. Toxicol. 2019;59:107–127. doi: 10.1146/annurev-pharmtox-010818-021315. [DOI] [PubMed] [Google Scholar]
- Pan Y., Lei X., Zhang Y. Association predictions of genomics, proteinomics, transcriptomics, microbiome, metabolomics, pathomics, radiomics, drug, symptoms, environment factor, and disease networks: a comprehensive approach. Med. Res. Rev. 2022;42:441–461. doi: 10.1002/med.21847. [DOI] [PubMed] [Google Scholar]
- Peters A., Hoek G., Katsouyanni K. Understanding the link between environmental exposures and health : does the exposome promise too much? J. Epidemiol. Community Health. 2012;66:103–105. doi: 10.1136/jech-2011-200643. [DOI] [PubMed] [Google Scholar]
- Peters A., Nawrot T.S., Baccarelli A.A. ll Hallmarks of environmental insults. Cell. 2021;184:1455–1468. doi: 10.1016/j.cell.2021.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior L. Allostatic load and exposure histories of disadvantage. Int. J. Environ. Res. Public Health. 2021;18:7222. doi: 10.3390/ijerph18147222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prohaska S.J., Stadler P.F. The use and abuse of -omes. Methods Mol. Biol. 2011;719:173–196. doi: 10.1007/978-1-61779-027-0_8. [DOI] [PubMed] [Google Scholar]
- Purdy K.J., Hurd P.J., Moya-Laraño J., Trimmer M., Oakley B.B., Woodward G. Systems biology for ecology. From molecules to ecosystems. Adv. Ecol. Res. 2010;43:87–149. doi: 10.1016/B978-0-12-385005-8.00003-4. [DOI] [Google Scholar]
- Rappaport S.M. Implications of the exposome for exposure science. J. Expo. Sci. Environ. Epidemiol. 2011;21:5–9. doi: 10.1038/jes.2010.50. [DOI] [PubMed] [Google Scholar]
- Rappaport S.M., Smith M.T. Environment and disease risks. Science. 2010;330:460–461. doi: 10.1126/science.1192603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbioni G., Berset J.D., Day B.W. Is it realistic to propose determination of a lifetime internal exposome? Chem. Res. Toxicol. 2020;33:2010–2021. doi: 10.1021/acs.chemrestox.0c00092. [DOI] [PubMed] [Google Scholar]
- Scholz S., Nichols J.W., Escher B.I., Ankley G.T., Altenburger R., Blackwell B., Brack W., Burkhard L., Collette T.W., Doering J.A., et al. The eco-exposome concept: supporting an integrated assessment of mixtures of environmental chemicals. Environ. Toxicol. Chem. 2022;41:30–45. doi: 10.1002/etc.5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher B., Pothof J., Vijg J., Hoeijmakers J.H.J. The central role of DNA damage in the ageing process. Nature. 2021;592:695–703. doi: 10.1038/s41586-021-03307-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selye H. Psychopathology of Human Adaptation. Springer US; 1976. Stress without distress; pp. 137–146. [DOI] [Google Scholar]
- Selye H. Confusion and controversy in the stress field. J. Hum. Stress. 1975;1:37–44. doi: 10.1080/0097840X.1975.9940406. [DOI] [PubMed] [Google Scholar]
- Senier L., Brown P., Shostak S., Hanna B. The socio-exposome: advancing exposure science and environmental justice in a postgenomic era. Environ. Sociol. 2017;3:107–121. doi: 10.1080/23251042.2016.1220848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sher K.J., Dick D.M., Crabbe J.C., Hutchison K.E., O’Malley S.S., Heath A.C. Consilient research approaches in studying gene × environment interactions in alcohol research. Addict. Biol. 2010;15:200–216. doi: 10.1111/j.1369-1600.2009.00189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillé F. The exposome – a new approach for risk assessment. ALTEX. 2020;37:3–23. doi: 10.14573/altex.2001051. [DOI] [PubMed] [Google Scholar]
- Stringhini S., Carmeli C., Jokela M., Avendaño M., Muennig P., Guida F., Ricceri F., D’Errico A., Barros H., Bochud M., et al. Socioeconomic status and the 25 × 25 risk factors as determinants of premature mortality: a multicohort study and meta-analysis of 1·7 million men and women. Lancet. 2017;389:1229–1237. doi: 10.1016/S0140-6736(16)32380-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira A.P., Dias J.M.L., Carinhas N., Sousa M., Clemente J.J., Cunha A.E., von Stosch M., Alves P.M., Carrondo M.J.T., Oliveira R. Cell functional enviromics: unravelling the function of environmental factors. BMC Syst. Biol. 2011;5:92. doi: 10.1186/1752-0509-5-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipton M. A case for combined environmental stressor studies. Extrem. Physiol. Med. 2012;1:1–2. doi: 10.1186/2046-7648-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toscano W.A., Oehlke K.P. Systems biology: new approaches to old environmental health problems. Int. J. Environ. Res. Public Health. 2005;2:4–9. doi: 10.3390/ijerph2005010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. Food and Drug Administration (FDA) - National Institutes of Health (NIH), Biomarker Working Group BEST (biomarkers, EndpointS, and other tools) resource. 2016. https://www.ncbi.nlm.nih.gov/books/NBK326791/pdf/Bookshelf_NBK326791.pdf [PubMed]
- United States Environmental Protection Agency Terminology servies glossary. 2009. https://sor.epa.gov/sor_internet/registry/termreg/searchandretrieve/glossariesandkeywordlists/
- VanderWeele T.J. Outcome-wide epidemiology. Epidemiology. 2017;28:399–402. doi: 10.1097/EDE.0000000000000641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanderWeele T.J., Mathur M.B., Chen Y. Outcome-wide longitudinal designs for causal inference: a new template for empirical studies. Stat. Sci. 2020;35:437–466. doi: 10.1214/19-sts728. [DOI] [Google Scholar]
- Vermeulen R., Schymanski E.L., Barabási A.L., Miller G.W. The exposome and health: where chemistry meets biology. Science. 2020;367:392–396. doi: 10.1126/science.aay3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijg J., Montagna C. Genome instability and aging: cause or effect? Transl. Med. Aging. 2017;1:5–11. doi: 10.1016/j.tma.2017.09.003. [DOI] [Google Scholar]
- Wagner M., Grodstein F., Leffondre K., Samieri C., Proust-Lima C. Time-varying associations between an exposure history and a subsequent health outcome: a landmark approach to identify critical windows. BMC Med. Res. Methodol. 2021;21:1–15. doi: 10.1186/s12874-021-01403-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild C.P. The exposome: from concept to utility. Int. J. Epidemiol. 2012;41:24–32. doi: 10.1093/ije/dyr236. [DOI] [PubMed] [Google Scholar]
- Wild C.P. Complementing the genome with an “exposome”: the outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol. Biomarkers Prev. 2005;14:1847–1850. doi: 10.1158/1055-9965.EPI-05-0456. [DOI] [PubMed] [Google Scholar]
- World Health Organization & International Programme on Chemical Safety Assessing human health risks of chemicals: derivation of guidance values for health-based exposure limits. 1994. https://apps.who.int/iris/handle/10665/40675 Environ. Heal. Criteria 170.
- World Health Organization & International Programme on Chemical Safety Biomarkers and risk assessment: concepts and principles. 1993. https://apps.who.int/iris/handle/10665/39037 Environ. Heal. Criteria 155.
- Xu Y. Envirotyping for deciphering environmental impacts on crop plants. Theor. Appl. Genet. 2016;129:653–673. doi: 10.1007/s00122-016-2691-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue B.K., Sartori P., Leibler S. Environment-to-phenotype mapping and adaptation strategies in varying environments. Proc. Natl. Acad. Sci. U S A. 2019;116:13847–13855. doi: 10.1073/pnas.1903232116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaehringer J., Jennen-Steinmetz C., Schmahl C., Ende G., Paret C. Psychophysiological effects of downregulating negative emotions: insights from a meta-analysis of healthy adults. Front. Psychol. 2020;11 doi: 10.3389/fpsyg.2020.00470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zartarian V., Bahadori T., McKone T. Adoption of an official ISEA glossary. J. Expo. Anal. Environ. Epidemiol. 2005;15:1–5. doi: 10.1038/sj.jea.7500411. [DOI] [PubMed] [Google Scholar]