Abstract
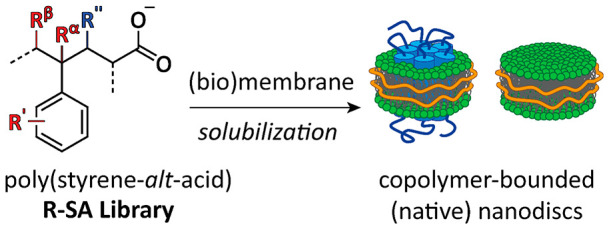
Amphipathic copolymers such as poly(styrene-maleic acid) (SMA) are promising tools for the facile extraction of membrane proteins (MPs) into native nanodiscs. Here, we designed and synthesized a library of well-defined alternating copolymers of SMA analogues in order to elucidate polymer properties that are important for MP solubilization and stability. MP extraction efficiency was determined using KcsA from E. coli membranes, and general solubilization efficiency was investigated via turbidimetry experiments on membranes of E. coli, yeast mitochondria, and synthetic lipids. Remarkably, halogenation of SMA copolymers dramatically improved solubilization efficiency in all systems, while substituents on the copolymer backbone improved resistance to Ca2+. Relevant polymer properties were found to include hydrophobic balance, size and positioning of substituents, rigidity, and electronic effects. The library thus contributes to the rational design of copolymers for the study of MPs.
Introduction
Membrane proteins (MPs) have a prominent biological and pharmacological importance. Nevertheless, their structures remain highly underrepresented as compared to those of water-soluble proteins,1,2 mainly because MPs tend to destabilize when taken out of their native lipid environment. The use of styrene-maleic acid (SMA) copolymers, as first described in 2009,3 has given a substantial impetus to the field of MP research, as these amphipathic polymers can solubilize MPs together with an annulus of native lipids, forming so-called native nanodiscs.4−7 This preservation of the endogenous lipidic environment confers high stability to the MPs and allows for the study of (native) lipid–protein and protein–protein interactions.7−11 The MPs in the nanodiscs furthermore are amenable to functional and structural studies with an array of biophysical techniques,12 including mass spectrometry,13 mass photometry,14 NMR spectroscopy,15,16 and cryo-electron microscopy.5,17,18
The efficiency of membrane solubilization by SMA copolymers is determined by many factors, including environmental conditions such as ionic strength and pH,19,20 physicochemical properties of the target membrane,21−23 and properties of SMA, such as length and chemical composition.20,24−26 Copolymers with relatively short chains24,27 and with a ratio of styrene-to-maleic acid of ∼2:1 and ∼3:120,28 generally are efficient at solubilization, while either more hydrophobic (∼4:1) or more hydrophilic (∼1:1) copolymers are not.20,25,26
Unfortunately, SMA copolymers that are efficient solubilizers (i.e., with a ∼ 2:1 or 3:1 styrene-to-maleic acid ratio) tend to be very heterogeneous in size27,29 and composition20,24 with a highly irregular distribution of comonomers along the copolymer backbone.24 This is because during copolymerization styrene and maleic anhydride prefer to form alternating (1:1) copolymers and because the polymers are synthesized in a free-radical copolymerization reaction, which is a random process.29−31 To facilitate studies on MP solubilization by SMA, much work has been performed on preparing copolymers with more uniform size dispersity and/or with well-defined comonomer sequence distributions.24,27,32−35 Furthermore, by introducing various substitutions,36−38 different types of copolymers have been developed to overcome some of the limitations of SMA, e.g., enabling use in a different pH range or in the presence of divalent cations.39−43 However, these copolymers are not always effective solubilizers, and ultimately, a clearer understanding that allows for a more comprehensive predictive and rational design has remained elusive.
Here, we present a library of well-defined, alternating (1:1) amphipathic copolymers with systematic substitutions to allow elucidation of polymer parameters that are important for biologically relevant properties, such as solubilization efficiency and divalent cation resistance. The new library expands the toolkit available for the isolation and characterization of MPs.
Materials and Methods
Chemicals and Reagents
The commercially available SMAnh (∼2:1) with the product name XiranSZ30010 was a kind gift from Polyscope Polymers (Geleen, Netherlands). Phospholipids were obtained from Avanti Polar Lipids (Alabaster, United States), namely, 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC), 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC), and 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC). 4-Iodostyrene was purchased from Apollo Scientific (Stockport, United Kingdom). All other chemicals were sourced from Sigma-Aldrich (Darmstadt, Germany). Demineralized water that has run through a Milli-Q water purification system is simply referred to as water. Styrene and acrylic acid were freshly distilled before use.
SMAnh/SAA Synthesis: Reversible–Addition–Fragmentation Transfer (RAFT)-Mediated Copolymerizations
The general procedure for RAFT copolymerizations was adapted from the literature.24,33−35,44−46 For the RAFT polymerization reaction, a round-bottom flask was charged with the hydrophilic monomer, either maleic anhydride or acrylic acid (5 mmol, 1 equiv), RAFT agent, 2-(dodecylthiocarbonothioylthio)-2-methylpropionic acid (DDMAT; 2 or 4 mol % equiv), and free-radical initiator, azobis(isobutyronitrile) (AIBN; [RAFT]/[initiator] 1:0.2 equiv). The hydrophobic monomer, styrene, or an analogue thereof (5 mmol, 1 equiv) was diluted in dioxane (anhydrous, [1.42 M]) and passed through a column of aluminum oxide (basic) to remove the inhibitor (i.e., 4-tert-butylcatechol) from the commercial monomer products. All chemicals were brought together, along with a Teflon coated magnetic stirring bar. The flask was connected to a Schlenk line, and to remove all oxygen from the solution, the mixture was put through freeze–pump–thaw cycles (4×) and then kept in an inert atmosphere of nitrogen. The reaction mixture was stirred and heated (80 °C) overnight. The yellow-colored solution was then cooled before being added dropwise to diethyl ether to precipitate out the product. In some cases, precipitation was not possible in ether in which case either water or n-hexane was used. The suspension was then filtered under vacuum over a glass frit and washed several times with the appropriate solvent. The vacuum was left on for ∼15 min to dry the solids. Finally, the solid material was dried in an oven (∼40 °C) until the mass stabilized. This yielded the copolymers as (bright) yellow powders.
SMA-PEA Synthesis
The following synthesis was modified from literature procedures.16,39,42 The maleic anhydride starting material, SMAnh (1:1 copolymer) (0.65 g, ∼3.32 mmol MAnh, 1 equiv), was loaded in a round-bottom flask together with a Teflon coated stirring bar and dissolved in DMF (dry, 5 mL). Phenethylamine (0.51 mL, 4 mmol, ∼1.2 equiv) was added to the bright yellow solution, followed by triethylamine (1.12 mL, 8 mmol, 2.4 equiv). The flask was flushed with nitrogen gas. The reaction mixture was continuously stirred and heated (∼60 °C) for 3 days. The resulting dark red solution was added dropwise to diethyl ether (∼250 mL) under stirring. The formed precipitate was filtered under vacuum with a glass frit, and the solids were washed first with acetone (3×, ∼30 mL) and then with diethyl ether (3×, ∼30 mL). The material was left to dry on the filter under vacuum for ∼15 min, followed by air drying for several days, yielding the amide product (SMA-PEA) as a cream colored solid (0.9 g, 86% yield).
Gel Permeation Chromatography (GPC)
Polymers were dissolved in THF at a concentration of 1 mg/mL, and the solution was filtered using a pore diameter of 0.45 μm. Then, 20 μL of the polymer solution was injected onto a Shimadzu gel permeation chromatography (GPC) system, using THF as the eluent and an isocratic flow with a flow rate of 1 mL/min for a total run time of 14 min. The oven temperature was set to 30 °C. Dual detection was done with a RI detector and a PDA detector with a wavelength range of 200–400 nm, looking specifically at 254 and 211 nm with a bandwidth of 4 nm. A calibration curve consisting of 13 polystyrene standards in the range of 266–300 000 Da was used to determine copolymer size and dispersity.
SMAnh Hydrolysis
The maleic anhydride (R/X-SMAnh) copolymers were suspended in an aqueous alkaline solution of NaOH (0.6 M) at a concentration of 10% (w/v). The reaction mixtures were heated (∼42 °C) in a water bath with mild shaking overnight. When the solids had dissolved to make a bright yellow solution, the mixtures were left on a roller (42 rpm) overnight at ambient temperature. The hydrolyzed products were isolated by the addition of HCl (3 M) to precipitate out the maleic acid copolymers, which were pelleted down by centrifugation at 3000g for 10 min, and the supernatant was discarded. The pellets were washed by the addition of HCl (1 M) and resuspended thoroughly, after which they were pelleted down again. The washing steps were repeated in succession with 0.1 M HCl and finally 0.01 M HCl. After the final washing step, the pellets were dried in an oven (∼50 °C).
Preparation of Polymer Stock Solutions
The free acid copolymers were suspended in NaOH (0.4 M, aq) at a concentration of 10% (w/v). The mixture was left on the roller at 42 rpm overnight to allow the complete dissolution of the solids. Once dissolved, the pH of the solutions was carefully titrated with NaOH/HCl to obtain pH 8 ± 0.5. The solutions were then diluted with water to a concentration of 5% (w/w), and finally, the stocks were diluted further with buffer (50 mM Tris-HCl, pH 8, 150 mM NaCl) to a final concentration of 1% (w/w).
Fourier-Transform Infrared (FT-IR) Spectroscopy
To characterize the copolymers and assess ring-opening from the anhydrides (C=O anhydride stretching 1775 cm–1) to the acids (C=O acid stretching 1705 cm–1),47 they were analyzed using Fourier-transform infrared (FT-IR) spectroscopy. Data were obtained using a PerkinElmer Spectrum One FT-IR spectrometer with an UATR accessory; ATR correction was not performed. Analysis was performed on solid samples. The spectra were recorded in the range of 650–4000 cm–1 with a resolution of 1 cm–1 and taken as an average of 4 scans.
Ultraviolet–Visible (UV–vis) Spectroscopy
Ultraviolet–visible (UV–vis) spectra were obtained for diluted copolymer solutions (final concentration 0.025% (w/v)) in a total volume of 1 mL of water. A calibration curve using DDMAT was prepared using a concentration range of 0–0.003% (w/v) in water. Measurements were performed using quartz cuvettes (10 mm path length) equilibrated at 21 °C using a Peltier system. Scans were recorded in the wavelength range of 200–500 nm with a slit width of 0.5 nm and a speed of 100 nm/min with data points measured every 0.1 nm. Measurements were performed using a PerkinElmer Lambda 18 Spectrophotometer.
Nuclear Magnetic Resonance (NMR) Spectroscopy
The copolymers (anhydrides and acids) were characterized by nuclear magnetic resonance (NMR) spectroscopy. To this end, solid polymer samples (20–60 mg) were dissolved in 0.5 mL of deuterated solvent (DMSO-d6). 1H and 13C NMR spectra were recorded at 298 K using a Bruker Avance Neo (600 MHz) spectrometer equipped with a cryogenic probe. Chemical shifts are reported with respect to residual solvent peaks. Data was processed using MestReNova software (Mestrelab Research S.L.).
Preparation of Multilamellar Vesicles (MLVs)
For the preparation of multilamellar vesicles (MLVs), the phospholipids were first dried from a chloroform stock under a stream of nitrogen gas in a heated (∼45 °C) water bath. To remove traces of organic solvent, the films were further placed in a desiccator under a high vacuum for at least 1 h. The thin lipid films were then hydrated in buffer (50 mM Tris-HCl, pH 8, 150 mM NaCl) at a lipid concentration of 20 mM. The hydration process was performed above the lipid phase transition temperature in a water bath with gentle shaking for ∼1 h. Afterward, the MLVs stock suspensions were subjected to 10 freeze–thaw cycles and finally stored in the freezer (−20 °C). The phosphate concentration was determined using a Rouser assay.48
Dynamic Light Scattering (DLS)
Nanoparticle sizes and distributions were analyzed using dynamic light scattering (DLS) with a Malvern Zetasizer Nano-ZS machine equipped with a red laser (633 nm) detecting backscattering at an angle of 173°. All measurements were performed at 25 °C with an equilibration time of 60 s before the experiment. Data are the average from 7 measurements, consisting of at least 10 subruns of 10 s each. The analysis model applied was general purpose (normal resolution) with the assumption of spherical particles. All samples were diluted in buffer consisting of 50 mM Tris-HCl, pH 8, 150 mM NaCl (viscosity 0.96066 cP, RI 1.332), and the material was approximated as polystyrene latex (RI 1.59, abs 0.01). Nanodisc samples contained ∼2 mM phospholipid (DMPC) and ∼0.4% (w/v) polymer and were allowed to reach equilibrium by incubation at 30 °C for 1 h with shaking at 650 rpm, followed by incubation in the fridge (4 °C) for at least 1 day. Polymer only samples had a concentration of ∼0.4% (w/v) polymer and were allowed to equilibrate by the same treatment as performed on the nanodiscs.
KcsA Containing Membranes from E. coli
Biological membranes with overexpressed KcsA protein were obtained from E. coli, as previously described.7,49 Briefly, E. coli strain BL21(λDE3) cells were transformed via a heat shock protocol with a plasmid for KcsA containing N-terminal His-tag from pT7-KcsA.50,51 Protein expression was induced with IPTG, and the bacteria were cultured in LB medium until an OD600 of ∼0.8 was reached. Cells were harvested by centrifugation and lysed by mechanical disruption through a French press. Soluble proteins were removed, and whole membranes (mixture of both inner and outer membrane fragments) were obtained via ultracentrifugation (100 000g). These membrane pellets were resuspended in buffer (50 mM Tris-HCl, pH 8, 300 mM NaCl, 15 mM KCl) to an OD600 of ∼4. A small sample was taken, and the lipids were isolated following the extraction protocol of Bligh and Dyer,52 and the phosphate concentration was determined to be ∼7 mM on the basis of the protocol of Rouser et al.48 The concentrated whole membrane stocks were stored at −20 °C before being used for subsequent solubilization experiments.
Densitometry: Membrane Protein Solubilization
To assess membrane protein extraction efficiency, E. coli membranes overexpressing KcsA were used. The biomembranes were diluted to a phosphate (lipid) concentration of 1.5 mM in buffer (50 mM Tris-HCl, pH 8, 300 mM NaCl, 15 mM KCl). The copolymers were added to a final concentration of 0.25% (w/v) and incubated for 2 h in a shaking block set at 650 rpm and 25 °C. Next, the samples were centrifuged at 21 000g at 4 °C for 1 h. The supernatant (soluble) material was carefully removed from the pellet and subsequently taken up in solubilization buffer containing 1% (w/v) SDS. Laemilli sample buffer (4×, without reducing agent) was added to all samples before loading on a gel of 13% acrylamide. SDS-PAGE was run initially at 75 V for 15 min and then at 175 V for ∼55 min. The gels were stained using Coomassie Brilliant Blue R-250 for 1 h followed by destaining overnight. Gels were scanned in grayscale at 1200 dpi for quantification. Densitometric analysis was performed, and the percentage of KcsA extracted was determined by comparing the density of the supernatant relative to the sum of the densities for the supernatant and pellet for each copolymer sample. The whole solubilization experiment was repeated in triplicate for all copolymers.
Isolation of Yeast Mitochondrial Membranes
A yeast strain expressing the chromosomally encoded GFP-tagged voltage-dependent anion channel VDAC (Por1, encoded by ORF YNL055C) in the BY4741 strain background (MATazhis3Δ1 leu2Δ0 met15Δ0 ura3Δ0)53 was purchased from Thermo Fisher and maintained on YPD (1% w/v yeast extract, 2% w/v peptone, 2% w/v dextrose).
Yeast cells precultured overnight in 5 mL of YPD at 30 °C were transferred to 20 mL of semisynthetic lactate medium (SSL) containing 0.3% (w/v) yeast extract, 0.1% (w/v) glucose, 2% (w/v) lactate, and various salts,54 adjusted to pH 5.5 by the addition of KOH, and incubated for 4 h at 30 °C. An aliquot corresponding to 2 OD600 units was used to inoculate 800 mL of SSL and cultured overnight under aerobic conditions at 30 °C. Cells were harvested at an OD600 value of 2 and washed with 1 mM EDTA, pH 7.4.
The isolation of mitochondrial membranes was based on Daum et al.54 Briefly, after incubating the cells in 0.1 M Tris-HCl, pH 9.4, 10 mM DTT for 10 min at 30 °C, spheroplasts were prepared by treatment with Zymolyase 100T (Seikagaku Corporation, Tokyo, Japan) at 0.5 mg/g cells (wet weight) in 1.2 M sorbitol and 20 mM KH2PO4, pH 7.4, for 30 min at 30 °C while shaking. After a wash step in 1.2 M sorbitol and 20 mM KH2PO4, pH 7.4, the spheroplasts were resuspended in ice cold 0.6 M sorbitol, 10 mM Tris-HCl, pH 7.4, and 1 mM PMSF at a concentration corresponding to 0.075 g cells (wet weight)/mL and broken by 20 strokes in a 40 mL glass Dounce homogenizer with a tight-fitting pestle. The homogenate was centrifuged for 5 min at 1400g to remove unbroken spheroplasts and nuclei. Mitochondria were pelleted from the supernatant by centrifugation for 15 min at 20 000g, resuspended in 10 mM Tris-HCl, pH 8.2, 300 mM NaCl, and 0.6 M sorbitol at a phosphate concentration of ∼20 mM (based on the protocol of Rouser et al.48) and stored at −80 °C.
Turbidimetry: Kinetics of Solubilization of (Bio)membranes
Turbidimetry experiments were conducted to study the kinetics of lipid membrane solubilization, as previously described by Scheidelaar et al.,22 except that a wavelength of 430 nm was used instead of 350 nm to avoid interference from the RAFT end-groups (see Figure S3). A PerkinElmer Lambda 18 UV/vis Spectrophotometer was used, equipped with a Julabo PC pump and a PTP-1+1 Peltier System to ensure constant stirring and temperature control. In all experiments, a total volume of 700 μL was used. The concentration of lipid membranes was adjusted to an absorbance lower than 1, and the polymer concentration was adjusted accordingly using a 1% (w/w) stock solution (see Table 1 for an overview of the experimental conditions). The membrane suspensions were temperature-equilibrated for 5 min before starting the measurement. At t = 1 min, the polymer was added using a micropipette; all was mixed with a 200 μL pipet, and the measurement was left to run to a total time of 15 min unless stated otherwise. All turbidity experiments were performed in duplicate.
Table 1. Experimental Conditions of Turbidimetry Experiments.
| membrane | buffer | temperature (°C) | phosphate [Pi] (mM) | [polymer] (final) (%) |
|---|---|---|---|---|
| DMPC | 50 mM Tris-HCl pH 8 | 15 | 0.5 | 0.1 |
| 150 mM NaCl | 30 | 0.5 | 0.1 | |
| DSPC | 50 mM Tris-HCl pH 8 | 30 | 0.5 | 0.1 |
| 150 mM NaCl | 60 | 0.5 | 0.1 | |
| E. coli | 50 mM Tris-HCl pH 8 | 25 | 0.25 | 0.125 |
| 300 mM NaCl | ||||
| 15 mM KCl | ||||
| yeast mitochondria | 50 mM Tris-HCl pH 8 | 25 | 0.125 | 0.0625 |
| 300 mM NaCl |
Affinity Purification of KcsA Native Nanodiscs
Eight polymers were chosen (4-BrSMA, 4-ClSMA, 4-CF3SMA, 4-tBuSMA, β-NMA, and SAA with SMA and SMA (2:1) as a control/comparison) for purification and characterization of native nanodiscs from E. coli membranes. These polymers were selected because of their ability to solubilize membranes effectively. One mL of 1% polymer samples was added to 1 mL of a 1.5 mM (phosphate) E. coli membrane suspension so that a final polymer concentration of 0.5% was achieved. This was left to incubate in a heating block at 25 °C with 600 rpm shaking for 2.5 h. The samples were then transferred to a spinning disc for gentle agitation at 4 °C overnight. The next morning, the samples were centrifuged at 21 000g for 45 min to separate any undissolved membrane fragments. His-Pur Ni-NTA agarose beads (Thermo Scientific, 8 mL suspension) were transferred to buffer (50 mM Tris-HCl, 150 mM NaCl, 10 mM KCl, 10 mM imidazole, pH 8). One mL of a bead suspension was mixed with 1 mL of supernatant, and the samples were placed in a spinning disc for gentle agitation at 4° overnight. The samples were centrifuged (700g, 2 min, 4 °C); the supernatant was collected, and the beads were resuspended in buffer containing 10 mM imidazole. This was repeated with 10 and 50 mM imidazole, and finally, the nanodiscs were separated from the beads using a buffer with 300 mM imidazole. Buffer exchange was done using Amicon Ultra, 2 mL 3 kDa filter Eppendorf tubes (Merck Millipore) with 10 mM Tris-HCl, 100 mM NaCl, and 5 mM KCl, pH 8.
Ca2+ Stability
To quantify the stability of the polymers in the presence of Ca2+, the minimum calcium concentration at which each polymer precipitates was determined. A 1 M CaCl2 solution was made with buffer (50 mM Tris-HCl and 150 mM NaCl, pH 8) and titrated in a flat-bottom 96-well plate with concentrations of 0–200 mM Ca2+. Buffer was used to bring all samples to volume; samples were then mixed well. The polymer was added to a final concentration of 0.5%, and all samples were mixed using a 200 μL 8-channel pipet. A Ca2+ titration was done without any polymers as a negative control. The well plates were covered with a lid, and the edges were sealed off using parafilm to prevent possible evaporation, and subsequently, the plates were left to incubate at 30 °C overnight. Directly before measurements, parafilm and the lid were removed and any bubbles in the samples were popped using a clean pipet tip, after which the absorbance in the plates was measured at 430 nm using a BMT Labtech CLARIOstar PLUS microplate reader. The plates were temperature-equilibrated to 30 °C for 10 min, followed by a 300 rpm double-orbital shake for 30 s and subsequent absorbance measurement adjusted for focal height. All experiments were done in duplicate.
Transmission Electron Microscopy (TEM)
Nanodiscs were prepared by incubating MLVs composed of DPPC with various copolymers at a polymer-to-lipid mass ratio of 1.125:1 on the basis of a protocol by Dominguez Pardo et al.55 The final concentration of the samples was ∼0.8 mg/mL (polymer + lipid) in a buffer of 50 mM Tris-HCl, pH 8, and 150 mM NaCl. The samples were incubated at 41 °C for 1 h before being centrifuged at 115 000g for 1 h at 4 °C. The supernatant was isolated and stored at 4 °C until analysis.
For transmission electron microscopy (TEM), samples were prepared by the negative stain-mica-carbon flotation technique (MFT), Valentine procedure.56 Briefly, samples were absorbed to the clean side of a carbon film on mica, stained, and transferred to a 400-mesh copper grid. Samples were stained using sodium silico tungstate (SST) H4Na4O40SiW12 at 2% in distilled water (pH 7–7.5). The images were taken under low dose conditions (<10 e–/Å2) with defocus values between 1.2 and 2.5 μm on a Tecnai 12 LaB6 electron microscope at 120 kV accelerating voltage using a CCD Camera Gatan Orius 1000. Micrographs were taken at 13 000× and 30 000× magnification. The average size of the nanodiscs was estimated from at least 120 particles per sample and determined using the software MeasureIT (Olympus).
Results and Discussion
Synthesis and Characterization of a Library of Amphipathic Copolymers
A library of amphipathic copolymers was synthesized according to the general procedure in Scheme 1. The aim was to make alternating SMA copolymer analogues that are more lipophilic by systematically varying their chemical composition. This was done either by using styrene analogues with a more hydrophobic nonpolar moiety or by making the polar maleic acid moiety less hydrophilic.
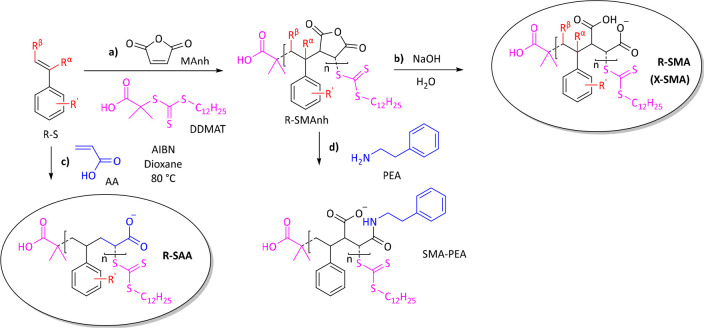
Scheme 1. General Synthetic Scheme Showing the Synthesis of Different Sets of SMA Analogues.
Styrenic derivatives (R-S) were copolymerized either with maleic anhydride (MAnh) to obtain R-SMAnh copolymers (a) or with acrylic acid (AA) to obtain R-SAA copolymers (c). Following copolymerization, R-SMAnh parent copolymers were hydrolyzed to the water-soluble free acid forms (R/X-SMA) under aqueous alkaline conditions (b). SMA-PEA was prepared by reacting SMAnh with phenethylamine (PEA) (d). Comonomers were added at an equimolar concentration (1:1 mol ratio). Other reaction conditions included the use of azobisisobutyronitrile (AIBN) as radical source, 2-(dodecylthiocarbonothioylthio)-2-methylpropionic acid (DDMAT) as the RAFT agent (highlighted in pink), and anhydrous dioxane as solvent at a reaction temperature of 80 °C.
Three sets of copolymers were synthesized in a living free-radical reaction via Reversible–Addition–Fragmentation Transfer (RAFT)-mediated copolymerization. R-SMA was synthesized by copolymerizing styrenic analogues (R-S) with maleic anhydride (MAnh) to obtain R-SMAnh copolymers, followed by hydrolysis (Scheme 1a,b, respectively). It has modifications of the hydrophobic moiety (highlighted in red), either on the pendant group (R′) or on the backbone (Rα/β). X-SMA is a special subset of R-SMA where the derivatives (R′) represent halogen atoms (X). R-SAA has only one carboxylic acid group (highlighted in blue) and was obtained by copolymerization with acrylic acid (AA) (Scheme 1c). A separate type of modification is SMA-PEA, in which the copolymer was made less hydrophilic by opening the maleic anhydride rings with phenethylamine (PEA; highlighted in blue) (Scheme 1d).
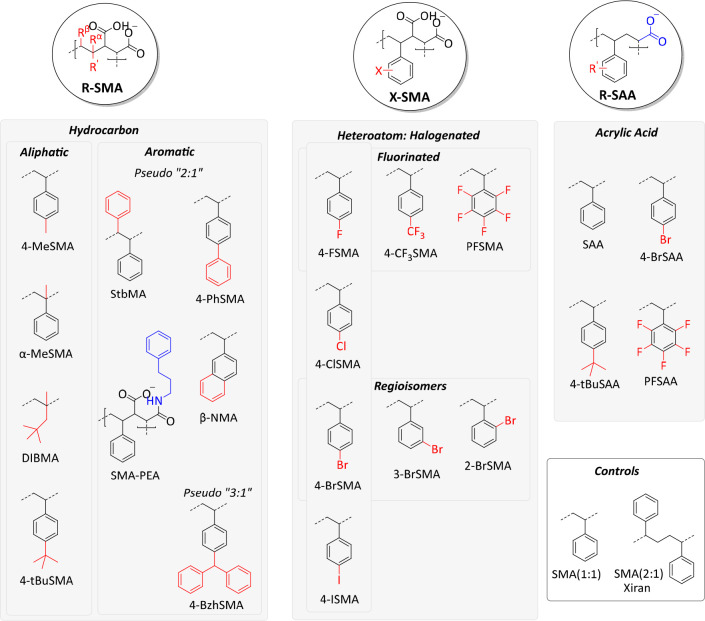
The modifications in the synthesized library are illustrated in Figure 1, and the full chemical structures are shown in Figure S1. The copolymers were fully characterized by FT-IR (Figure S2), UV–vis (Figure S3), and NMR spectroscopy (Figures S4–S8). NMR spectra reveal extremely broad peaks that are characteristic of (SMA and related) copolymeric materials.24,34,35,37,44,57 The relative purity of the copolymers is estimated to be >95%. Subsequently, copolymer size (Mn, Mw) and dispersity (Đ) were determined by gel permeation chromatography (GPC) (Figure S9). Table 2 summarizes the properties of the polymers. Most copolymers gave roughly the targeted peak sizes as well as good size distributions (Đ < 1.3, as expected for RAFT polymerization, with only α-MeSMA having a slightly larger Đ of 1.57). For stilbene (StbMA) and β-naphthalene (β-NMA), the GPC experiments showed bimodal distributions with the main peak coinciding with the anticipated smaller RAFT-copolymer size and Đ. The second peak represented a much longer polymer and more disperse fraction, which could be due to standard free-radical polymerization occurring alongside living (RAFT) polymerization. Finally, it should be noted that in all copolymer syntheses the comonomers were added together at an equimolar ratio, and it is assumed that the copolymers are alternating (i.e., 1:1). Whereas SMA analogues indeed probably have a highly alternating character, based on the reactivity ratios of styrene and maleic acid, the styrene acrylic acid analogues likely have somewhat less of an alternating character. However, this was not explicitly determined.
Figure 1.
Library of SMA copolymer analogues showing the three different classes (R-SMA, X-SMA, and R-SAA) and their subdivisions. Modifications of the styrene units are highlighted in red and those of the hydrophilic units, in blue.
Table 2. Characterization of RAFT Synthesized Amphipathic Copolymersa.
| # | name | R | Rα | Rβ | hydrophilic moiety | yield (%) | DP | Mn (kDa) | Mw (kDa) | Đ |
|---|---|---|---|---|---|---|---|---|---|---|
| R-SMA | ||||||||||
| 1 | SMA | H | H | H | MA | 93 | 42 | 4.6 | 5.3 | 1.15 |
| 2 | α-MeSMA | H | Me | H | MA | 63 | 25 | 3.1 | 4.8 | 1.57 |
| 3 | 4-MeSMA | 4-Me | H | H | MA | 100 | 46 | 5.4 | 6.3 | 1.17 |
| 4 | 4-PhSMA | 4-Ph | H | H | MA | 71 | 24 | 3.8 | 4.9 | 1.29 |
| 5 | 4-BzhSMA | 4-Bzh | H | H | MA | 90 | 35 | 6.8 | 8.8 | 1.29 |
| 6 | 4-tBuSMA | 4-tBu | H | H | MA | 100 | 51 | 6.9 | 8.3 | 1.20 |
| 7 | DIBMA | DIB | Me | H | MA | 57 | 27 | 3.2 | 3.6 | 1.12 |
| 8 | StbMA | H | H | Ph | MA | 58 | 29, 1621b | 4.5, 226b | 5.3, 457b | 1.19, 2.02b |
| 9 | β-NMA | Np | H | H | MA | 70 | 16, 3315c | 2.4, 419c | 2.9, 1520c | 1.23, 3.63c |
| 10 | SMA-PEA | H | H | H | AA-PEA | 86 | 42 | 4.7 | 5.4 | 1.15 |
| X-SMA | ||||||||||
| 11 | SMA | H | H | H | MA | 76 | 17 | 2.1 | 2.6 | 1.21 |
| 12 | 4-FSMA | 4-F | H | H | MA | 75 | 20 | 2.6 | 3.1 | 1.17 |
| 13 | 4-ClSMA | 4-Cl | H | H | MA | 68 | 15 | 2.2 | 2.6 | 1.19 |
| 14 | 4-BrSMA | 4-Br | H | H | MA | 63 | 11 | 1.9 | 2.4 | 1.24 |
| 15 | 4-ISMA | 4-I | H | H | MA | 49 | 9 | 1.8 | 2.2 | 1.24 |
| 16 | 3-BrSMA | 3-Br | H | H | MA | 61 | 11 | 1.9 | 2.3 | 1.21 |
| 17 | 2-BrSMA | 2-Br | H | H | MA | 64 | 13 | 2.2 | 2.6 | 1.16 |
| 18 | 4-CF3SMA | 4-CF3 | H | H | MA | 77 | 14 | 2.3 | 2.7 | 1.17 |
| 19 | PFSMA | 2,3,4,5,6-F | H | H | MA | 60 | 10 | 1.8 | 2.1 | 1.14 |
| R-SAA | ||||||||||
| 20 | SAA | H | H | H | AA | 46 | 11 | 1.4 | 1.6 | 1.17 |
| 21 | 4-BrSAA | 4-Br | H | H | AA | 46 | 11 | 1.7 | 2.0 | 1.18 |
| 22 | PFSAA | 2,3,4,5,6-F | H | H | AA | 60 | 18 | 2.7 | 3.2 | 1.18 |
| 23 | 4-tBuSAA | 4-tBu | H | H | AA | 59 | 23 | 3.0 | 3.8 | 1.25 |
DP, degree of polymerization; Mn, number average molecular weight; Mw, weight average molecular weight; Đ, dispersity; MA, maleic acid; AA, acrylic acid.
12% (AUC).
45% (AUC).
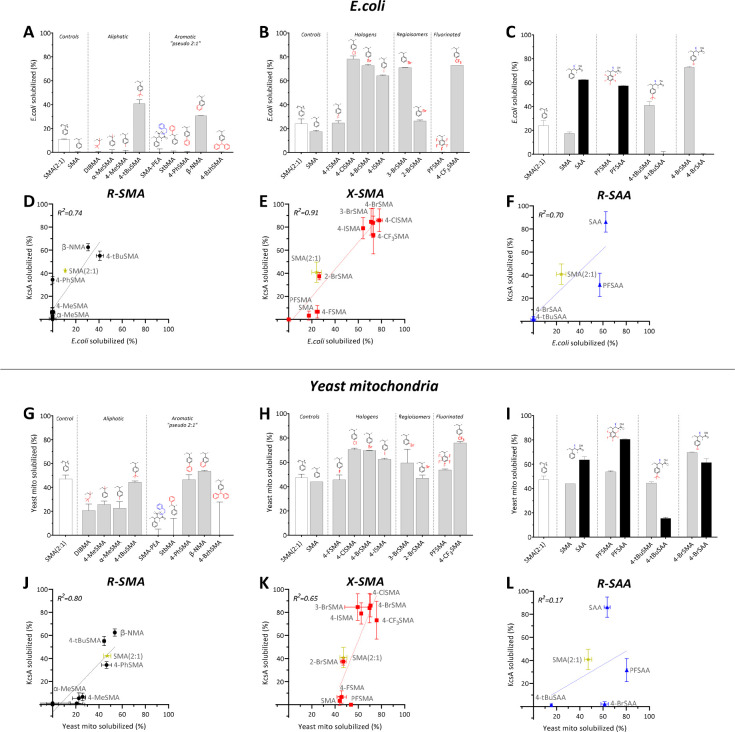
Efficiency of Membrane Protein Solubilization from E. coli
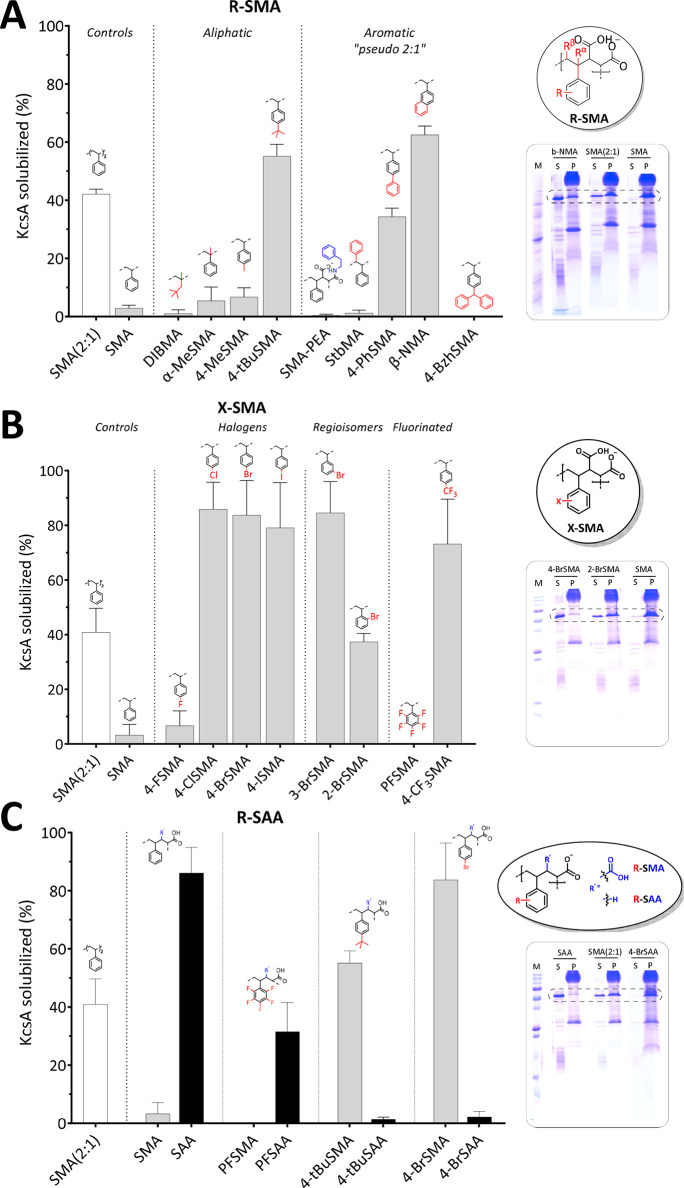
The ability of the copolymers to solubilize biomembranes was first tested using the tetrameric K+-channel KcsA as a model protein, overexpressed in Escherichia coli.7,19 The solubilization efficiency was quantified by densitometry on SDS-PAGE gels.7,19 To create an optimal window for the evaluation of the efficacy of all substitutions, also with respect to the commercially available “gold standard” Xiran SZ30010 (SMA 2:1, styrene-to-maleic acid ratio), a relatively low concentration of copolymer (0.25% w/v) was used, at which SMA (2:1) does not give complete solubilization. Another advantage of using a relatively low polymer concentration is that it reduces the chance of artifacts due to the solubility issues of the polymers.
R-SMA Analogues
The first set of analogues, referred to as R-SMA, contains aliphatic or aromatic substituents to increase the hydrophobicity of the alternating SMA copolymers (Figure 2A). As expected, SMA (2:1) yielded only partial solubilization (∼42% KcsA extraction), while the unsubstituted SMA (alternating, 1:1) was unable to solubilize the membranes (<3%). The aliphatic copolymer DIBMA also showed negligible solubilization (∼1%) under these suboptimal conditions.
Figure 2.
Solubilization of the membrane protein, KcsA, from E. coli membranes by R-SMA (A), X-SMA (B), and R-SAA (C) copolymers. The general scaffolds are circled on the right with deviations from styrene marked in red and depicted above the bars and deviations from the maleic acid moiety marked in blue. Insets on the right show representative SDS-PAGE lanes for selected polymers (for complete overview see Figure S10) with the dashed box highlighting the band corresponding to the KcsA tetramer. M, molecular weight marker; S, supernatant; P, pellet. Solubilized KcsA (%) determined from the density of the KcsA band in S relative to the sum of S plus P. Data points indicate the mean ± SD (n ≥ 3 independent experiments). Densitometry data were obtained 2 h post-solubilization at 25 °C with a polymer concentration of 0.25% (w/v) (polymer-to-lipid ratio of ∼2.3:1 (w/w)).
Homologues of SMA containing an extra methyl group, either on the backbone in the α-position (α-MeSMA) or on the para-position of the aromatic ring (4-MeSMA), gave slightly increased yields (∼6%) compared to the underivatized SMA. Interestingly, a tert-butyl group on styrene (4-tBuSMA) showed a solubilization efficacy of ∼55%, outperforming the commercial SMA (2:1).
The aromatic substitutions yielded pseudo “2:1” SMA copolymers. The introduction of a second phenyl ring, either by opening up the maleic anhydride ring using phenethylamine (SMA-PEA) or by grafting it on the β-position of the backbone (StbMA), resulted in negligible solubilization (∼1%), in agreement with previous studies, where a StbMA copolymer was found to be ineffective in dissolving lipid vesicles.45 However, when the phenyl was grafted directly to the styrene on the para-position (4-PhSMA), the polymer had a solubilization efficacy of ∼34%. Finally, an analogue where two aromatic rings are fused into a rigid naphthalene group (β-NMA) was found to be the best solubilizer (∼63%) in the series. The addition of another aromatic ring in the 3:1 mimic benzhydryl (4-BzhSMA) did not result in any membrane activity (∼0%), possibly because the groups are either (i) too hydrophobic and readily form a hydrophobic core whereby the lipophilic monomers are shielded and unavailable for membrane insertion or (ii) too bulky for efficient insertion due to the three large phenyl rings.
X-SMA Halogenated Analogues
A well-established phenomenon in the drug discovery field is that halogenation of compounds increases their lipophilicity58 and enhances membrane binding and permeation.59 This inspired us to synthesize a subset of R-SMA copolymers bearing halogen substitutions (X-SMA). As shown in Figure 2B, upon replacement of the proton at the 4-position with fluorine (the least hydrophobic halogen), the solubilization efficiency increases slightly but remains poor (∼7%). However, substitution with chlorine, bromine, and iodine all result in a remarkable jump to near-complete solubilization (∼86%, ∼84%, and ∼79%, respectively).
The importance of the position of the substitutions was investigated by preparing regioisomers of BrSMA. While 3-BrSMA shows a similar high solubilization efficiency (∼85%) as that of 4-BrSMA, for 2-BrSMA, an ∼2-fold drop in extraction efficiency (∼37%) is observed, which still is comparable to that of SMA (2:1) (∼41%).
To further probe substitution with fluorine, two more analogues were tested. Substitution of all aromatic protons by fluorine in perfluoro (PFSMA) resulted in a complete loss of MP extraction ability (∼0%). Surprisingly, substitution by trifluoromethyl (4-CF3SMA) again resulted in a copolymer with very good solubilization capability (∼73%).
R-SAA Analogues
In the R-SAA set of polymers, the hydrophobicity of SMA is increased by replacing the dicarboxylic acid in maleic acid (MA) by a monoacid derived from acrylic acid (AA), as first reported by Appel and co-workers.46Figure 2C shows that, whereas SMA and PFSMA both are inefficient solubilizers (<3%), SAA is highly active, and also, PFSAA shows significant activity (∼86% and ∼32% extraction, respectively). In contrast, whereas 4-BrSMA and 4-tBuSMA are efficient solubilizers (∼80% and ∼55%, respectively), their acrylic acid equivalents, 4-BrSAA and 4-tBuSAA, both show negligible membrane protein extraction (<3%). Presumably, these acrylic variants are too hydrophobic, highlighting that a suitable hydrophobic balance is marked by sharp boundaries.
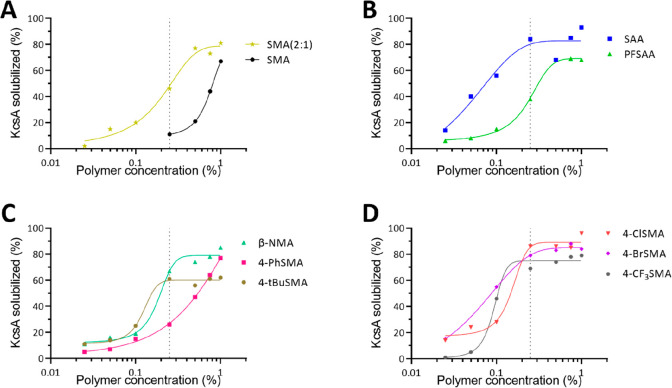
Dose–Response Curves
The efficacy of the best performing copolymers was tested via a dose–response curve (Figure 3). For all tested copolymers, the solubilization performance increased with increasing concentration. The tested copolymers all plateau between 60% and 100% solubilization. Interestingly, at 0.25% (w/v), which is the concentration used in all previous experiments, a large window is achieved in which to assess differences between the various copolymers. Having more potent copolymers, such as SAA or some of the halogenated derivatives (4-Br/Cl/CF3-SMA), will be advantageous as a lower amount of copolymer can be employed for maximal solubilization. Hence, there will be less excess copolymer, which could potentially interfere with downstream applications or analyses, including affinity purification, structural determination, or size characterization.
Figure 3.
Dose–response curves showing the (%) solubilization of KcsA for several selected copolymers, (A) controls, (B) R-SAA, (C) R-SMA, and (D) X-SMA, at various concentrations ranging from 0.025% to 1% (w/v). The dashed line highlights a concentration of 0.25% (w/v) as used in previous experiments. KcsA was extracted using whole E. coli membranes at a lipid concentration of 1.5 mM, and solubilization was performed at 25 °C for 2 h.
Efficiency of Membrane Solubilization in Different Systems as Measured by Turbidimetry
The membrane solubilizing efficacy of the copolymers thus far was based on extraction and quantification of KcsA from the E. coli inner membrane. We next explored turbidimetry as a more general approach to gain insight into the membrane solubilizing properties of the polymers. For these experiments, an excess of copolymer relative to lipid was used to allow monitoring of solubilization properties on a relatively short time scale.
Solubilization of Biological Membranes
Figure 4A–C shows the percentage decrease in optical density (OD) after incubation with E. coli membranes (mixture of inner and outer) for the three polymer sets. Importantly, for all three polymer sets, good correlations (R2 of 0.7–0.9) are found when the percentage of KcsA extracted is plotted against the percentage decrease in OD (Figure 4D–F), indicating that turbidimetry is a valuable tool to analyze solubilization efficiency. Notably, solubilization of E. coli membranes generally appears to be less efficient than KcsA extraction, likely because of the shorter incubation times and because the turbidimetry measurements also include outer E. coli membranes, which are more difficult to solubilize.
Figure 4.
Whole membrane solubilization as determined by turbidimetry for E. coli (A–C) and yeast mitochondria (G–I) and the correlation with membrane protein (KcsA) extraction as determined by SDS-PAGE densitometry (D–F and J–L). The percentage of membrane solubilized is based on the relative decrease in optical density after 14 min (data are the average of 2 independent experiments ± range). Solubilization data are shown for analogues of R-SMA (left panels), X-SMA (middle panels), and R-SAA (right panels). Dashed lines indicate the line of linear fit, and the corresponding correlation coefficients (R2) are given in the left upper corner. For the complete turbidimetry traces, see Figure S11.
Additional turbidimetry measurements on mitochondrial membranes from Saccharomyces cerevisiae showed that most of the polymers are able to also solubilize these yeast membranes (Figure 4G–I). The polymers generally appear more efficient than with E. coli membranes, likely due to differences in membrane composition. For R-SMA and X-SMA, again, good correlations (R2 of 0.8 and 0.65, respectively) are found with E. coli solubilization, as based on KcsA extraction, while for R-SAA, rather surprisingly, the variation (R2 of 0.17) is much larger (Figure 4J–L). Overall, these data suggest that yeast membranes tolerate a broader hydrophobic balance range than E. coli membranes.
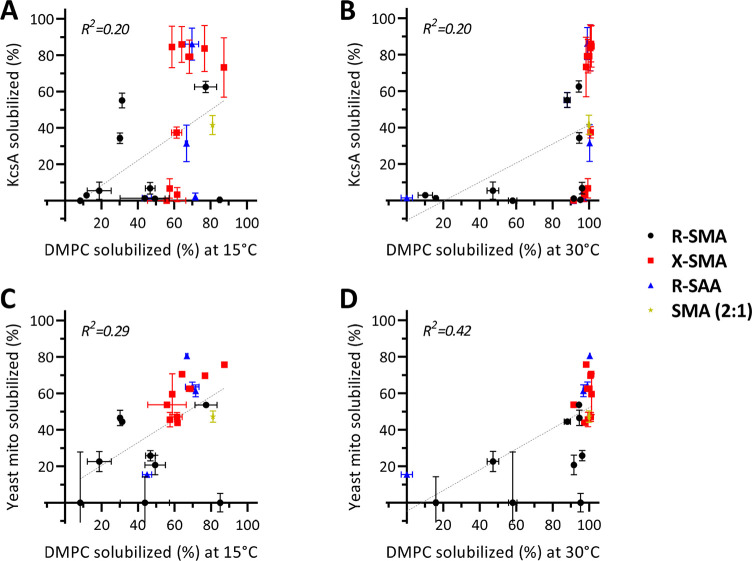
Solubilization of Model Lipid Membranes
Model membranes are frequently used test systems to investigate the solubilization efficiency of amphipathic copolymers.16,22,60 Here, we used dimyristoylphosphatidylcholine (DMPC) vesicles at different temperatures (see Figure S12 for traces and Figure S13 for bar graphs) and plotted the solubilization efficiency against that observed for E. coli inner membranes (based on KcsA extraction) and yeast mitochondrial membranes.
Figure 5A,B shows that both in the gel phase at 15 °C and in the fluid phase at 30 °C there is a poor correlation (R2 of 0.2) between KcsA extraction and DMPC vesicle solubilization. For yeast membranes (Figure 5C), which contain a substantial amount of PC lipids,61,62 the correlation is somewhat better (R2 of 0.29), particularly at 30 °C (R2 of 0.42, Figure 5D). Correlations are further improved when using distearoylphosphatidylcholine (DSPC) (Figure S14), in particular when compared to yeast mitochondrial membranes (R2 of 0.60–0.67), most likely because these longer PC lipids better represent the lipid composition of yeast membranes. These results thus highlight the importance of membrane properties for solubilization efficiency.21,22
Figure 5.
Comparison of KcsA solubilization from E. coli (top) and yeast mitochondria solubilization (bottom) to model lipid-only vesicle solubilization. Data are shown for DMPC at 15 °C (A, C) and 30 °C (B, D). The different classes are depicted as follows: R-SMA, black circles; X-SMA, red squares; R-SAA, blue triangles; SMA (2:1), tan star. For systems with T < TM, the vesicle solubilization rate was derived after 14 min, and for T > TM, time points were taken at 4 min (see Figure S7 for turbidity traces and Figure S8 for the corresponding bar graphs). In all cases, biomembranes (yeast, E. coli) were solubilized at 25 °C. Dashed lines indicate the line of linear fit, and the corresponding correlation coefficients (R2) are given in the left upper corner.
Strikingly, for solubilization of both DMPC and DSPC in the fluid phase, a sharp transition is observed between poor solubilization and complete solubilization when plotted against KcsA solubilization (Figures 5B and S14B). The transition is less sharp for yeast membranes, but here, the correlation between solubilization efficiencies in the two model systems tends to be better (Figures 5D and S14D). Together, these results suggest that polymers that are not able to efficiently solubilize DMPC or DSPC vesicles in the fluid phase can be considered poor biomembrane solubilizers. Hence, this may serve as a convenient screening assay to test new copolymers.
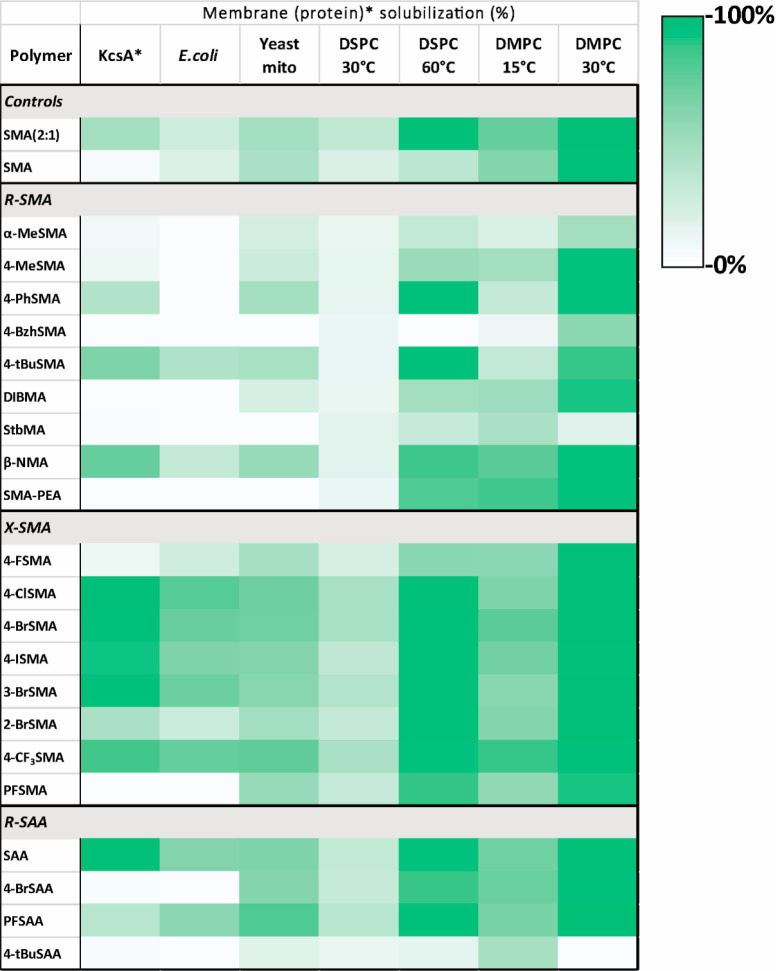
Summary of Membrane Solubilization Properties of the Different Polymers
Figure 6 summarizes the solubilization data and gives an overview of the structure–activity relationships for the various copolymers and the different membrane systems tested, biological as well as synthetic. The map illustrates the huge variation in solubilization efficiency of the different copolymers as well as the variation for each polymer with respect to solubilization of different lipid systems.
Figure 6.
Heatmap summarizing membrane solubilization properties of the polymers in the copolymer library in different membrane systems. KcsA membrane protein solubilization is based on densitometric analysis of protein bands on SDS-PAGE, i.e., the percentage of KcsA extracted. Solubilization of all other (whole) membranes is based on turbidimetric analysis with the percentage of solubilization given as a relative decrease in OD.
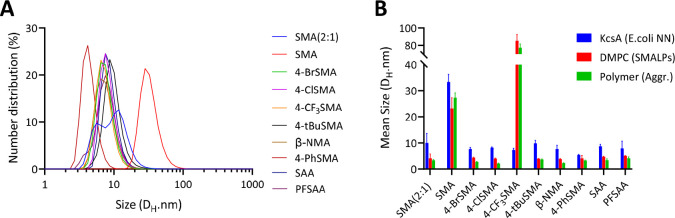
Nanoparticle Sizes
From each set of copolymers, we selected efficient solubilizers to compare the sizes of the purified KcsA nanodiscs by dynamic light scattering (DLS). As shown in Figure 7A, most of the nanodiscs have a homogeneous size distribution and a small particle size of d ∼ 8–10 nm. Exceptions were the controls of the nanodiscs prepared from SMA, which were significantly larger (d ∼ 30 nm), 4-PhSMA which was smaller (d ∼ 5 nm), and nanodiscs from SMA (2:1), which showed a less homogeneous size distribution.
Figure 7.
DLS analysis of KcsA native nanodiscs (A) as well as DMPC nanodiscs and polymer aggregates (B). The values are the average from 7 measurements with error bars reflecting the SD. Sizes are reported as the hydrodynamic diameters based on the peak maximum from the number distributions. For the full size distribution plots, please see Figure S16. All samples contain ∼0.4% (w/v) polymer; nanodisc samples also contain ∼2 mM lipid (polymer-to-lipid ratio of ∼3:1 (w/w)).
When sizes of the KcsA nanodiscs are compared with those of the nanoparticles solubilized from the DMPC vesicles and copolymers only, the KcsA nanodiscs are larger (except for 4-CF3SMA) and the latter two appear to be rather similar (Figure 7B). This was also observed for the other copolymers in the library (Figures S15A and S16). Likely, the excess copolymer contributes to the scattering, influencing the apparent size of the particles, even though a relatively low polymer concentration was used. This is supported by the large size difference for 4-CF3SMA between lipid-only nanodiscs and affinity purified KcsA-containing nanodiscs (Figure 7B). Furthermore, while it is not clear what determines the size of the polymer aggregates or the nanodiscs, we do note that polymers that form small aggregates in aqueous solution are the most efficient membrane solubilizers with 4-CF3SMA and 3-BrSMA being the only exceptions (Figure S15B).
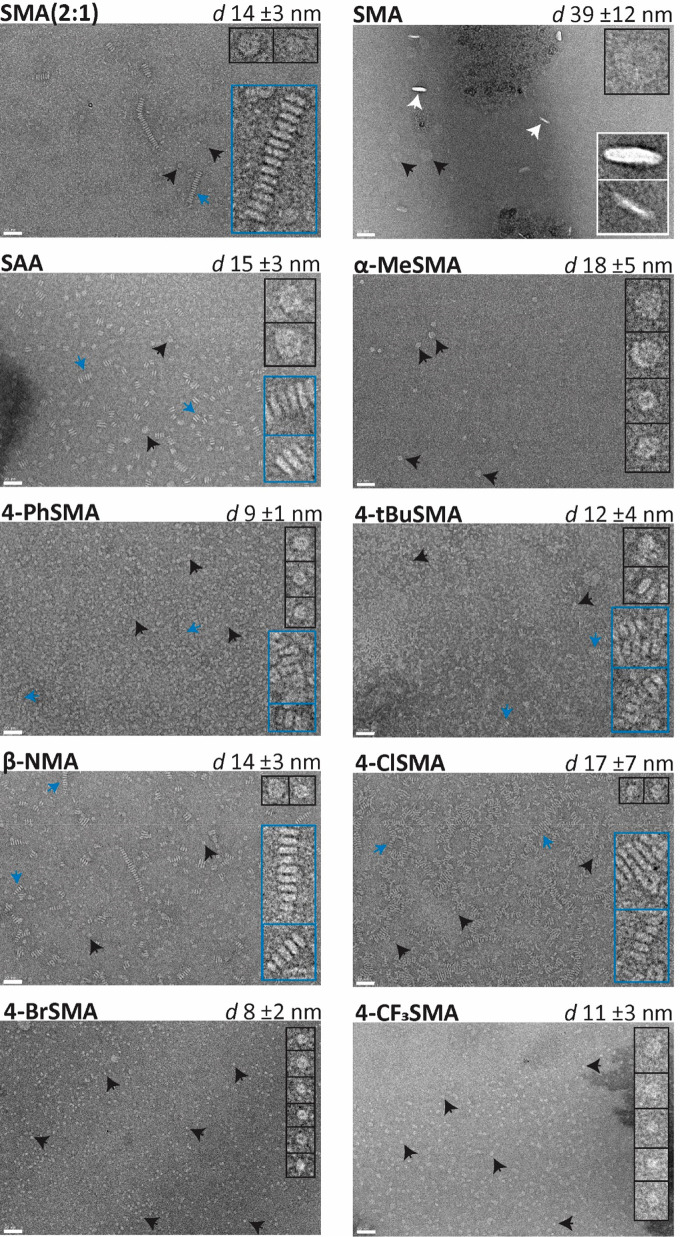
The morphology of the particles was investigated using negative-stain transmission electron microscopy (TEM) (Figure 8). The copolymers together with lipids form nanodiscs as can be seen by the discoidal particles, both top views and side views (see SMA), as well as the formation of stacks of nanodiscs into rouleaux. This phenomenon is an artifact of the negative stain interacting with the phosphate head groups and has previously been observed for nanodiscs under specific conditions (e.g., certain polymer-to-lipid ratios, salt concentration, and the use of tungsten based stains with TEM).55,63,64 Importantly, the formation of these rouleaux suggests that the polymers surround the acyl chains of the nanodiscs but do not strongly interact with the phosphate headgroup region. Finally, it should be noted that the sizes of the nanoparticles are somewhat larger than determined by DLS. As mentioned above, this may be explained by the fact that DLS data are skewed toward smaller sizes due to interference from excess unbound copolymers. Furthermore, a lower copolymer-to-lipid (1.125:1 m/m) ratio was used for TEM, which can result in both having less free copolymer around as well as generally having larger particle sizes.39,65
Figure 8.
Negative-stain transmission electron micrographs of nanodiscs composed of dipalmitoylphosphatidylcholine (DPPC) lipids and various amphipathic copolymers, images taken at 30 000× magnification. Solubilization was performed at a polymer-to-lipid ratio of 1.125:1 (m/m). Nanodisc sizes are reported as the mean diameter ± standard deviation of at least 100 particles. Insets show enlarged (4×) views of several representative particles, i.e., rouleaux stacks of nanodiscs (blue), top view (black), and side view (white). Scale bars represent 50 nm.
Resistance against Ca2+-Induced Aggregation
For studies on membrane proteins, it can be useful when the copolymers tolerate the presence of divalent cations. Therefore, aggregation induced by titration with calcium ions was analyzed by visual inspection and quantified by OD measurements (see Figure S17). Table 3 shows that most of the copolymers precipitated in the low millimolar range (≤10 mM). In agreement with the literature,43,66 DIBMA had a high resistance, remaining up to ∼40 mM Ca2+ in solution. Of the SMA analogues, only StbMA and α-MeSMA showed high resistance to precipitation up to calcium concentrations of ∼45 and ∼100 mM, respectively. Interestingly, these are the only three polymers with substitutions along the backbone: StbMA has a phenyl group on the β-position, while DIBMA and α-MeSMA have a methyl group on the α-position.
Table 3. Maximum Tolerated Concentration of Calcium Ions Where No Polymer Precipitation Was Observeda,b.
| R-SMA | [Ca2+] (mM) | X-SMA | [Ca2+] (mM) | R-SAA | [Ca2+] (mM) |
|---|---|---|---|---|---|
| 4-BzhSMA | <1 | PFSMA | ∼2 | 4-BrSAA | ∼1 |
| SMA-PEA | ∼2 | 4-ISMA | ∼3 | 4-tBuSAA | ∼1 |
| β-NMA | ∼5 | 3-BrSMA | ∼3 | PFSAA | ∼3 |
| 4-PhSMA | ∼5 | 4-CF3SMA | ∼3 | SAA | ∼5 |
| 4-MeSMA | ∼6 | 4-BrSMA | ∼4 | ||
| 4-tBuSMA | ∼6 | 2-BrSMA | ∼5 | ||
| DIBMA | ∼40 | 4-ClSMA | ∼5 | ||
| StbMA | ∼45 | 4-FSMA | ∼9 | ||
| α-MeSMA | ∼100 |
See Figure S17.
The control values for SMA (2:1) and SM were ∼5 and ∼11 mM, respectively.
Summary of Relevant Copolymer Parameters
Here, we will discuss how properties of the copolymer backbone and of the pendant chains may influence membrane solubilization and how this may help explain the results obtained from the three sets of copolymers.
Amphiphilicity
Acting at the interface between the hydrophobic fatty acid tails and the aqueous environment, the copolymers require a high degree of amphipathicity. They need to be sufficiently polar for solubility in water and sufficiently hydrophobic to drive insertion into the membrane and to allow the formation of stable nanodiscs. The balance between these two opposing parameters can be delicate, as clearly illustrated for example for the SAA analogues.
Flexibility of the Backbone and Pendant Chains
In addition to amphipathicity, the copolymer must have sufficient flexibility to expose its hydrophilic groups to the aqueous phase and the hydrophobic groups to the lipid acyl chains without too many conformational constraints. This may be the reason hydrophobic substituents on the pendant chains seem much more effective in increasing solubilization efficiency than hydrophobic substituents on the backbone, such as in the “pseudo 2:1 R-SMA polymers” SMA-PEA and StbMA, where the substitutions may interfere with backbone flexibility.
In contrast, for the pendant chains, an increased rigidity may be advantageous for efficient solubilization, as it will reduce the loss of entropy upon membrane insertion. This can be achieved by having fewer rotatable bonds or more symmetrical substitutions, as in the β-NMA analogue or in the para-substituted halogen analogues.
Size of the Polymers and the Pendant Chains
Previously, it was shown that copolymer sizes of <10 kDa are optimal for solubilization,25,26 likely due to steric hindrance in longer polymers and a tendency to form aggregates in the aqueous phase. This is in line with our present observation that copolymers that form larger aggregates in solution generally are less efficient solubilizers. Importantly, the polymers in this study vary somewhat in length. To gain information on the influence of this variation for solubilization efficiency, we synthesized a series of 4-BrSMA copolymers with varying lengths (for full characterization and evaluation, see Figure S18). When a 4-BrSMA copolymer was prepared by standard free-radical polymerization, a large and disperse batch was obtained (Mn = 35 kDa, Đ = 2), which as expected, was inefficient at solubilizing KcsA (∼6%) (see Figure S18L). However, when 4-BrSMA copolymers are prepared by RAFT synthesis in the size range of 1–5 kDa, they are effective solubilizers (∼70%) and there is only a small variation (less than 15%) in membrane protein solubilization efficiency. This suggests that the size variation of the copolymers in the present library has only a minor influence and that the chemical composition is the dominant factor in determining solubilization efficiency. One caveat is that the RAFT synthesized copolymers still contain the alkyl tail terminus group, and it is not known if or to what extent this affects solubilization. Further research would be required in this regard.
Also, for the pendant groups, size plays an important role. To allow insertion into the membrane, a smaller size might be beneficial, combined with sufficient hydrophobicity. However, the formation of nanodiscs should be more favorable than insertion at the interface, and therefore, the polymer should also be able to destabilize the bilayer by disrupting lipid packing. This may be promoted by a larger size of the pendant chain or deeper penetration into the bilayer. Possibly, the halogen analogues as studied here strike an optimal balance, as they can introduce significant lipophilicity with an intermediate bulk and size and can efficiently solubilize a range of target membranes with different lipid packing properties.
Positioning of Substitutions on Backbone and Pendant Chains
Additional (hydrophobic) substituents on the backbone (i.e., DIBMA, α-MeSMA, and StbMA) do not result in efficient solubilization under the suboptimal environmental conditions used here. The main reason for this relatively low polymer concentration was to get an optimal window to compare the relative solubilization efficiencies of the copolymers with those of the traditional SMA (2:1) (Xiran). However, the dose–response curves in this study show that polymers that are not efficient at these suboptimal concentrations may still be good solubilizers at higher concentrations. Indeed, as demonstrated for DIBMA,43,66 at higher concentrations, such backbone-substituted analogues may be useful for biological systems, in particular, since they tolerate divalent cations. The reason for this tolerance is likely that steric hindrance prevents chelation of the cations to the carboxyl groups, either directly by physically obstructing access or by changing the backbone conformation and flexibility.
Also, for the pendant groups, the positioning of the substituents is important. The attachment of the derivative on the 3- and 4-position of the phenyl ring is superior to the 2-position, as shown for the brominated SMA analogues. Possible reasons are that the closeness of the 2-position to the backbone results in steric hindrance, reducing the conformational freedom of the polymer backbone and/or that a deeper penetration into the hydrophobic core for the 3- and 4-positions facilitates membrane disruption.
Electronic Effects
Phenyl groups have an electronic surface potential that is negative above and below the ring and positive in the plane of the ring.67 This may play a role in membrane insertion of styrene-containing polymers, in particular since the membrane core has a positive dipole potential.68,69 Halogens generally have high electronegativities and are electron-withdrawing, thereby modifying the electrostatic surface potential of the aromatic rings.70 It is not clear how this would affect insertion, but we do note that substitution with F, which is the most electronegative element in the periodic table, does not result in efficient solubilization. Another property of the heavier halogens (Cl, Br, I) is that they have an electrostatically positive region (σ-hole) that can act as a Lewis acid and undergo halogen bonding with nucleophiles,71,72 either within the copolymer molecule(s) themselves or with lipids and proteins.
Copolymer Purity and the Potential Role of Contaminants
The copolymers were used as crude products; i.e., they were used directly after precipitation from the reaction mixtures without further purification. This is standard practice in the use of commercial copolymer products. Nevertheless, at this point, it cannot be ruled out that the activities are due in part to the impurities present, and the same holds true for the commercial SMA copolymer products. Further research is required to investigate exactly to what extent and how impurities may be playing a part in the process.
Implications for Polymer Design
The solubilization efficiency of any copolymer obviously will depend on the membrane environment and on environmental parameters. However, for maximum solubilization efficiency in a wide range of target membranes under the conditions used here, the halogen substitutions seem most promising, together with the naphthalene variant and SAA polymers. Substitutions on the backbone, on the other hand, may improve activity in the presence of divalent cations. This raises the question of whether the different modifications can be combined to make a copolymer that is both highly effective at solubilization and resistant to divalent cations. As a first trial, we synthesized and tested an α-MeSAA copolymer via conventional free-radical copolymerization (for full characterization and evaluation, see Figure S19). Depending on the polymer concentration, the α-MeSAA copolymer showed moderate solubilization (35–57% KcsA extracted) with reasonable divalent cation resistance (∼20 mM Ca2+), demonstrating overall properties indeed lying in between those of α-MeSMA and SAA. An even better approach may be to prepare halogenated (e.g., 4-Cl/Br/CF3) α-MeSMA copolymers.
The new library of copolymers offers several advantages over commercially available copolymers. First, the copolymers in the present study are well-defined in terms of sequence distribution and size, which is useful for systematic studies to understand their mode of action. For example, it will help improve the accuracy of molecular dynamics simulations as it allows a better representation of the polymers. Furthermore, through the size control of RAFT polymerization, copolymers can be employed without interference (band smearing) of longer copolymers on SDS gels. In addition, RAFT synthesized copolymers have the potential to be selectively modified on the end groups, allowing the incorporation of a single label such as a fluorophore or affinity-tag per copolymer molecule. Last but not least, halogenated copolymers may be useful for dedicated biophysical techniques, e.g., the use of (i) fluorinated copolymers, such as 4-CF3SMA, for 19F-NMR studies, (ii) brominated copolymers for MS experiments due to their convenient isotopic signature, and (iii) copolymers with heavier halogens (i.e., 4-BrSMA or 4-ISMA) for EM microscopy where the polymers/particles could potentially be visualized more easily due to the scattering of the dense halogen atoms. Although beyond the scope of this work, it should be noted that varying copolymer properties may affect the functionality of membrane proteins.73
Conclusion
We have introduced a library of poly(styrene-alt-maleic/acrylic acid) analogues with well-defined composition and length. The library contains several promising new analogues with equivalent or better membrane protein solubilization when compared to the benchmark of SMA (2:1) (Xiran30010). By systematic variation of the nature and positioning of different substituents, we obtained new insights into the parameters that govern efficient solubilization and tolerance of divalent cations. This knowledge can be utilized for the targeted and rational design of future copolymer generations for membrane protein solubilization. In addition, the library expands the toolbox for the study of membrane proteins, allowing improvement of yields and stability of precious membrane protein samples.
Acknowledgments
We are grateful to Anton I. P. M. de Kroon for assistance with editing parts of the manuscript, Helene Jahn for preparing the yeast mitochondrial membranes, and Bonny W. M. Kuipers for support in DLS analysis as well as Polyscope Polymers (NL) for gifting the SMAnh(2:1) copolymer (Xiran SZ30010). This work used the EM facilities at the Grenoble Instruct-ERIC Center (ISBG; UAR 3518 CNRS CEA-UGA-EMBL) with support from the French Infrastructure for Integrated Structural Biology (FRISBI; ANR-10-INSB-05-02) and GRAL, a project of the University Grenoble Alpes graduate school (Ecoles Universitaires de Recherche) CBH-EUR-GS (ANR-17-EURE-0003) within the Grenoble Partnership for Structural Biology. The IBS Electron Microscope facility is supported by the Auvergne Rhône-Alpes Region, the Fonds Feder, the Fondation pour la Recherche Médicale, and GIS-IBiSA. We thank Daphna Fenel, Christine Moriscot, and Guy Schoehn, all from the Electron Microscopy platform of the Institut de Biologie Structurale, CEA, CNRS, Université Grenoble Alpes Grenoble France for performing the TEM imaging. This work was supported financially by the Division of Chemical Sciences (CW) of the Dutch Research Council (Nederlandse Organisatie voor Wetenschappelijk Onderzoek, NWO) via ECHO grant No. 711-017-006 (A.H.K.).
Glossary
Abbreviations
- MP
membrane protein
- SMAnh
styrene-maleic anhydride
- SMA
styrene-maleic acid
- SA
styrene acid
- Đ
dispersity
- RAFT
reversible-addition–fragmentation transfer
- AA
acrylic acid
- AIBN
azobis(isobutyronitrile)
- DMPC
1,2-dimyristoyl-sn-glycero-3-phosphocholine
- DPPC
1,2-dipalmitoyl-sn-glycero-3-phosphocholine
- DSPC
1,2-distearoyl-sn-glycero-3-phosphocholine
- PEA
phenethylamine
- MLV
multilamellar vesicle
- Mn
number-average molecular weight
- SAA
styrene-acrylic acid
- Tm
(gel-to-fluid crystalline) phase transition temperature
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.biomac.1c01166.
Chemical structures of the library of amphipathic copolymers; characterization of the copolymer library: FT-IR spectra and UV–vis spectra as well as 1H and 13C NMR spectra of the copolymers; GPC traces of copolymers; SDS-PAGE for the densitometric analysis of membrane protein extraction; turbidimetry traces of biomembrane solubilization; turbidity data of model membrane solubilization; comparison of biological versus synthetic membrane solubilization; nanoparticle sizes as determined by DLS; DLS size-distribution data for nanodisc and copolymer samples; spectroscopic analysis of sensitivity to divalent cations; characterization and evaluation of the 4-BrSMA copolymer size series as well as α-MeSAA copolymer (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Li F.; Egea P. F.; Vecchio A. J.; Asial I.; Gupta M.; Paulino J.; Bajaj R.; Dickinson M. S.; Ferguson-Miller S.; Monk B. C.; Stroud R. M. Highlighting Membrane Protein Structure and Function: A Celebration of the Protein Data Bank. J. Biol. Chem. 2021, 296, 100557. 10.1016/j.jbc.2021.100557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbaye M. N.; Hou Q.; Basu S.; Teheux F.; Pucci F.; Rooman M. A Comprehensive Computational Study of Amino Acid Interactions in Membrane Proteins. Sci. Rep. 2019, 9 (1), 1–14. 10.1038/s41598-019-48541-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles T. J.; Overduin M.; Finka R.; Smith C.; Lin Y.-P.; Dafforn T. Membrane Proteins Solubilized Intact in Lipid Containing Nanoparticles Bounded by Styrene Maleic Acid Copolymer. J. Am. Chem. Soc. 2009, 131 (22), 7484–7485. 10.1021/ja810046q. [DOI] [PubMed] [Google Scholar]
- Prabudiansyah I.; Kusters I.; Caforio A.; Driessen A. J. M. Characterization of the Annular Lipid Shell of the Sec Translocon. Biochim. Biophys. Acta - Biomembr. 2015, 1848 (10), 2050–2056. 10.1016/j.bbamem.2015.06.024. [DOI] [PubMed] [Google Scholar]
- Sun C.; Benlekbir S.; Venkatakrishnan P.; Wang Y.; Hong S.; Hosler J.; Tajkhorshid E.; Rubinstein J. L.; Gennis R. B. Structure of the Alternative Complex III in a Supercomplex with Cytochrome Oxidase. Nat. Lett. 2018, 557, 123–126. 10.1038/s41586-018-0061-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. C.; Knowles T. J.; Postis V. L. G.; Jamshad M.; Parslow R. A.; Lin Y. P.; Goldman A.; Sridhar P.; Overduin M.; Muench S. P.; Dafforn T. R. A Method for Detergent-Free Isolation of Membrane Proteins in Their Local Lipid Environment. Nat. Protoc. 2016, 11 (7), 1149–1162. 10.1038/nprot.2016.070. [DOI] [PubMed] [Google Scholar]
- Dörr J. M.; Koorengevel M. C.; Schäfer M.; Prokofyev A. V.; Scheidelaar S.; van der Cruijsen E. A. W.; Dafforn T. R.; Baldus M.; Killian J. A. Functional Reconstitution of a Tetrameric K + Channel: The Power of Native Nanodiscs. Proc. Natl. Acad. Sci. U.S.A. 2014, 111 (52), 18607–18612. 10.1073/pnas.1416205112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swainsbury D. J. K.; Scheidelaar S.; Van Grondelle R.; Killian J. A.; Jones M. R. Bacterial Reaction Centers Purified with Styrene Maleic Acid Copolymer Retain Native Membrane Functional Properties and Display Enhanced Stability. Angew. Chemie - Int. Ed. 2014, 53 (44), 11803–11807. 10.1002/anie.201406412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calzada E.; Avery E.; Sam P. N.; Modak A.; Wang C.; Mccaffery J. M.; Han X.; Alder N. N.; Claypool S. M. Phosphatidylethanolamine Made in the Inner Mitochondrial Membrane Is Essential for Yeast Cytochrome Bc1 Complex Function. Nat. Commun. 2019, 10, 1432. 10.1038/s41467-019-09425-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo A. C. K.; Lee S. C.; Pollock N. L.; Stroud Z.; Hall S.; Thakker A.; Pitt A. R.; Dafforn T. R.; Spickett C. M.; Roper D. I. Analysis of SMALP Co-Extracted Phospholipids Shows Distinct Membrane Environments for Three Classes of Bacterial Membrane Protein. Sci. Rep. 2019, 9 (1813), 1–10. 10.1038/s41598-018-37962-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellwig N.; Peetz O.; Ahdash Z.; Tascón I.; Booth P. J.; Mikusevic V.; Diskowski M.; Politis A.; Hellmich Y.; Hänelt I.; Reading E.; Morgner N. Native Mass Spectrometry Goes More Native: Investigation of Membrane Protein Complexes Directly from SMALPs. Chem. Commun. 2018, 54 (97), 13702–13705. 10.1039/C8CC06284F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overduin M.; Trieber C.; Prosser R. S.; Picard L. P.; Sheff J. G. Structures and Dynamics of Native-State Transmembrane Protein Targets and Bound Lipids. Membranes (Basel). 2021, 11 (6), 451. 10.3390/membranes11060451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reading E.; Hall Z.; Martens C.; Haghighi T.; Findlay H.; Ahdash Z.; Politis A.; Booth P. J. Interrogating Membrane Protein Conformational Dynamics within Native Lipid Compositions. Angew. Chemie - Int. Ed. 2017, 56 (49), 15654–15657. 10.1002/anie.201709657. [DOI] [PubMed] [Google Scholar]
- Olerinyova A.; Sonn-Segev A.; Gault J.; Eichmann C.; Schimpf J.; Kopf A. H.; Rudden L. S. P.; Ashkinadze D.; Bomba R.; Frey L.; Greenwald J.; Degiacomi M. T.; Steinhilper R.; Killian J. A.; Friedrich T.; Riek R.; Struwe W. B.; Kukura P. Mass Photometry of Membrane Proteins. Chem. 2021, 7 (1), 224–236. 10.1016/j.chempr.2020.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bersch B.; Dörr J. M.; Hessel A.; Killian J. A.; Schanda P. Proton-Detected Solid-State NMR Spectroscopy of a Zinc Diffusion Facilitator Protein in Native Nanodiscs. Angew. Chemie - Int. Ed. 2017, 56 (9), 2508–2512. 10.1002/anie.201610441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravula T.; Ramadugu S. K.; Di Mauro G.; Ramamoorthy A. Bioinspired, Size-Tunable Self-Assembly of Polymer–Lipid Bilayer Nanodiscs. Angew. Chemie - Int. Ed. 2017, 56 (38), 11466–11470. 10.1002/anie.201705569. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Parmar M.; Rawson S.; Scarff C. A.; Goldman A.; Dafforn T. R.; Muench S. P.; Postis V. L. G. Using a SMALP Platform to Determine a Sub-Nm Single Particle Cryo-EM Membrane Protein Structure. Biochim. Biophys. Acta - Biomembr. 2018, 1860 (2), 378–383. 10.1016/j.bbamem.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tascón I.; Sousa J. S.; Corey R. A.; Mills D. J.; Griwatz D.; Aumüller N.; Mikusevic V.; Stansfeld P. J.; Vonck J.; Hänelt I. Structural Basis of Proton-Coupled Potassium Transport in the KUP Family. Nat. Commun. 2020, 11 (1), 1–10. 10.1038/s41467-020-14441-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopf A. H.; Dörr J. M.; Koorengevel M. C.; Antoniciello F.; Jahn H.; Killian J. A. Factors Influencing the Solubilization of Membrane Proteins from Escherichia Coli Membranes by Styrene–Maleic Acid Copolymers. Biochim. Biophys. Acta - Biomembr. 2020, 1862 (2), 183125. 10.1016/j.bbamem.2019.183125. [DOI] [PubMed] [Google Scholar]
- Scheidelaar S.; Koorengevel M. C.; Van Walree C. A.; Dominguez J. J.; Dörr J. M.; Killian J. A. Effect of Polymer Composition and PH on Membrane Solubilization by Styrene-Maleic Acid Copolymers. Biophys. J. 2016, 111 (9), 1974–1986. 10.1016/j.bpj.2016.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez Pardo J. J.; Dörr J. M.; Iyer A.; Cox R. C.; Scheidelaar S.; Koorengevel M. C.; Subramaniam V.; Killian J. A. Solubilization of Lipids and Lipid Phases by the Styrene–Maleic Acid Copolymer. Eur. Biophys. J. 2017, 46 (1), 91–101. 10.1007/s00249-016-1181-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheidelaar S.; Koorengevel M. C.; Dominguez Pardo J. J.; Meeldijk J. D.; Breukink E.; Killian J. A. Molecular Model for the Solubilization of Membranes into Nanodisks by Styrene Maleic Acid Copolymers. Biophys. J. 2015, 108 (2), 279–290. 10.1016/j.bpj.2014.11.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janson K.; Zierath J.; Kyrilis F. L.; Semchonok D. A.; Hamdi F.; Skalidis I.; Kopf A. H.; Das M.; Kolar C.; Rasche M.; Vargas C.; Keller S.; Kastritis P. L.; Meister A. Solubilization of Artificial Mitochondrial Membranes by Amphiphilic Copolymers of Different Charge. BBA - Biomembr. 2021, 1863 (12), 183725. 10.1016/j.bbamem.2021.183725. [DOI] [PubMed] [Google Scholar]
- Cunningham R. D.; Kopf A. H.; Elenbaas B. O. W.; Staal B. B. P.; Pfukwa R.; Killian J. A.; Klumperman B. Iterative RAFT-Mediated Copolymerization of Styrene and Maleic Anhydride toward Sequence- and Length-Controlled Copolymers and Their Applications for Solubilizing Lipid Membranes. Biomacromolecules 2020, 21 (8), 3287–3300. 10.1021/acs.biomac.0c00736. [DOI] [PubMed] [Google Scholar]
- Swainsbury D. J. K.; Scheidelaar S.; Foster N.; Van Grondelle R.; Killian J. A.; Jones M. R. The Effectiveness of Styrene-Maleic Acid (SMA) Copolymers for Solubilisation of Integral Membrane Proteins from SMA-Accessible and SMA-Resistant Membranes. BBA - Biomembr. 2017, 1859 (10), 2133–2143. 10.1016/j.bbamem.2017.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison K. A.; Akram A.; Mathews A.; Khan Z. A.; Patel J. H.; Zhou C.; Hardy D. J.; Moore-Kelly C.; Patel R.; Odiba V.; Knowles T. J.; Javed M. -u.-H.; Chmel N. P.; Dafforn T. R.; Rothnie A. J. Membrane Protein Extraction and Purification Using Styrene-Maleic Acid (SMA) Copolymer: Effect of Variations in Polymer Structure. Biochem. J. 2016, 473 (23), 4349–4360. 10.1042/BCJ20160723. [DOI] [PubMed] [Google Scholar]
- Domínguez Pardo J. J.; Koorengevel M. C.; Uwugiaren N.; Weijers J.; Kopf A. H.; Jahn H.; van Walree C. A.; van Steenbergen M. J.; Killian J. A. Membrane Solubilization by Styrene-Maleic Acid Copolymers: Delineating the Role of Polymer Length. Biophys. J. 2018, 115 (1), 129–138. 10.1016/j.bpj.2018.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grethen A.; Oluwole A. O.; Danielczak B.; Vargas C.; Keller S. Thermodynamics of Nanodisc Formation Mediated by Styrene/Maleic Acid (2:1) Copolymer. Sci. Rep. 2017, 7 (1), 1–14. 10.1038/s41598-017-11616-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörr J. M.; Scheidelaar S.; Koorengevel M. C.; Dominguez J. J.; Schäfer M.; van Walree C. A.; Killian J. A. The Styrene–Maleic Acid Copolymer: A Versatile Tool in Membrane Research. Eur. Biophys. J. 2016, 45 (1), 3–21. 10.1007/s00249-015-1093-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill D. J. T.; O'Donnell J. H.; O'Sullivan P. W. Analysis of the Mechanism of Copolymerization of Styrene and Maleic Anhydride. Macromolecules 1985, 18 (1), 9–17. 10.1021/ma00143a002. [DOI] [Google Scholar]
- Klumperman B. Mechanistic Considerations on Styrene–Maleic Anhydride Copolymerization Reactions. Polym. Chem. 2010, 1 (5), 558–562. 10.1039/b9py00341j. [DOI] [Google Scholar]
- Zhu M.; Wei L.; Li M.; Jiang L.; Du F.; Li Z.; Li F. A Unique Synthesis of a Well-Defined Block Copolymer Having Alternating Segments Constituted by Maleic Anhydride and Styrene and the Self-Assembly Aggregating Behavior Thereof. Chem. Comm 2001, 365–366. 10.1039/b009815i. [DOI] [Google Scholar]
- Harding B. D.; Dixit G.; Burridge K. M.; Sahu I. D.; Dabney-Smith C.; Edelmann R. E.; Konkolewicz D.; Lorigan G. A. Characterizing the Structure of Styrene-Maleic Acid Copolymer-Lipid Nanoparticles (SMALPs) Using RAFT Polymerization for Membrane Protein Spectroscopic Studies. Chem. Phys. Lipids 2019, 218, 65–72. 10.1016/j.chemphyslip.2018.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall S. C. L.; Tognoloni C.; Price G. J.; Klumperman B.; Edler K. J.; Dafforn T. R.; Arnold T. Influence of Poly(Styrene-Co-Maleic Acid) Copolymer Structure on the Properties and Self-Assembly of SMALP Nanodiscs. Biomacromolecules 2018, 19, 761–772. 10.1021/acs.biomac.7b01539. [DOI] [PubMed] [Google Scholar]
- Smith A. A. A.; Autzen H. E.; Laursen T.; Wu V.; Yen M.; Hall A.; Hansen S. D.; Cheng Y.; Xu T. Controlling Styrene Maleic Acid Lipid Particles through RAFT. Biomacromolecules 2017, 18 (11), 3706–3713. 10.1021/acs.biomac.7b01136. [DOI] [PubMed] [Google Scholar]
- Esmaili M.; Acevedo-Morantes C.; Wille H.; Overduin M. The Effect of Hydrophobic Alkyl Sidechains on Size and Solution Behaviors of Nanodiscs Formed by Alternating Styrene Maleamic Copolymer. Biochim. Biophys. Acta - Biomembr. 2020, 1862 (10), 183360. 10.1016/j.bbamem.2020.183360. [DOI] [PubMed] [Google Scholar]
- Brady N. G.; Workman C. E.; Cawthon B.; Bruce B. D.; Long B. K. Protein Extraction Efficiency and Selectivity of Esterified Styrene-Maleic Acid Copolymers in Thylakoid Membranes. Biomacromolecules 2021, 22 (6), 2544–2553. 10.1021/acs.biomac.1c00274. [DOI] [PubMed] [Google Scholar]
- Hardin N. Z.; Ravula T.; Di Mauro G.; Ramamoorthy A. Hydrophobic Functionalization of Polyacrylic Acid as a Versatile Platform for the Development of Polymer Lipid Nanodisks. Small 2019, 15 (9), 1804813. 10.1002/smll.201804813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravula T.; Hardin N. Z.; Ramadugu S. K.; Cox S. J.; Ramamoorthy A. Formation of PH-Resistant Monodispersed Polymer–Lipid Nanodiscs. Angew. Chemie - Int. Ed. 2018, 57 (5), 1342–1345. 10.1002/anie.201712017. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Fiori M. C.; Jiang Y.; Altenberg G. A.; Liang H. Polymer-Encased Nanodiscs with Improved Buffer Compatibility. Sci. Rep. 2017, 7 (7432), 1–10. 10.1038/s41598-017-07110-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin N. Z.; Kocman V.; Di Mauro G. M.; Ravula T.; Ramamoorthy A. Metal-Chelated Polymer Nanodiscs for NMR Studies. Angew. Chemie - Int. Ed. 2019, 58 (48), 17246–17250. 10.1002/anie.201910118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravula T.; Hardin N. Z.; Ramadugu S. K.; Ramamoorthy A. PH Tunable and Divalent Metal Ion Tolerant Polymer Lipid Nanodiscs. Langmuir 2017, 33, 10655–10662. 10.1021/acs.langmuir.7b02887. [DOI] [PubMed] [Google Scholar]
- Oluwole A. O.; Danielczak B.; Meister A.; Babalola J. O.; Vargas C.; Keller S. Solubilization of Membrane Proteins into Functional Lipid-Bilayer Nanodiscs Using a Diisobutylene/Maleic Acid Copolymer. Angew. Chemie - Int. Ed. 2017, 56 (7), 1919–1924. 10.1002/anie.201610778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge K. M.; Harding B. D.; Sahu I. D.; Kearns M. M.; Stowe R. B.; Dolan M. T.; Edelmann R. E.; Dabney-Smith C.; Page R. C.; Konkolewicz D.; Lorigan G. A. Simple Derivatization of RAFT-Synthesized Styrene-Maleic Anhydride Copolymers for Lipid Disk Formulations. Biomacromolecules 2020, 21 (3), 1274–1284. 10.1021/acs.biomac.0c00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esmaili M.; Brown C. J.; Shaykhutdinov R.; Acevedo-Morantes C.; Wang Y. L.; Wille H.; Gandour R. D.; Turner S. R.; Overduin M. Homogeneous Nanodiscs of Native Membranes Formed by Stilbene-Maleic-Acid Copolymers. Nanoscale 2020, 12 (32), 16705–16709. 10.1039/D0NR03435E. [DOI] [PubMed] [Google Scholar]
- Smith A. A. A.; Autzen H. E.; Faust B.; Mann J. L.; Muir B. W.; Howard S.; Postma A.; Spakowitz A. J.; Cheng Y.; Appel E. A. Lipid Nanodiscs via Ordered Copolymers. Chem. 2020, 6 (10), 2782–2795. 10.1016/j.chempr.2020.08.004. [DOI] [Google Scholar]
- Kopf A. H.; Koorengevel M. C.; van Walree C. A.; Dafforn T. R.; Killian J. A. A Simple and Convenient Method for the Hydrolysis of Styrene-Maleic Anhydride Copolymers to Styrene-Maleic Acid Copolymers. Chem. Phys. Lipids 2019, 218, 85–90. 10.1016/j.chemphyslip.2018.11.011. [DOI] [PubMed] [Google Scholar]
- Rouser G.; Fleischer S.; Yamamoto A. Two Dimensional Thin Layer Chromatographic Separation of Polar Lipids and Determination of Phospholipids by Phosphorus Analysis of Spots. Lipids 1970, 5 (5), 494–496. 10.1007/BF02531316. [DOI] [PubMed] [Google Scholar]
- Kopf A. H.; Dörr J. M.; Koorengevel M. C.; Antoniciello F.; Jahn H.; Killian J. A. Factors Influencing the Solubilization of Membrane Proteins from Escherichia Coli Membranes by Styrene–Maleic Acid Copolymers. Biochim. Biophys. Acta - Biomembr. 2020, 1862 (2), 183125. 10.1016/j.bbamem.2019.183125. [DOI] [PubMed] [Google Scholar]
- Van Dalen A.; Schrempf H.; Killian J. A.; De Kruijff B. Efficient Membrane Assembly of the KcsA Potassium Channel in Escherichia Coli Requires the Protonmotive Force. EMBO Rep. 2000, 1 (4), 340–346. 10.1093/embo-reports/kvd067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dalen A.; Hegger S.; Killian J. A.; De Kruijff B. Influence of Lipids on Membrane Assembly and Stability of the Potassium Channel KcsA. FEBS Lett. 2002, 525 (1–3), 33–38. 10.1016/S0014-5793(02)03061-2. [DOI] [PubMed] [Google Scholar]
- Bligh E. G.; Dyer W. J. A Rapid Method of Total Lipid Extraction and Purification. Can. J. Biochem. Physiol. 1959, 37 (8), 911–917. 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Huh W. K.; Falvo J. V.; Gerke L. C.; Carroll A. S.; Howson R. W.; Weissman J. S.; O’Shea E. K. Global Analysis of Protein Localization in Budding Yeast. Nature 2003, 425 (6959), 686–691. 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Daum G.; Böhni P. C.; Schatz G. Import of Proteins into Mitochondria. Cytochrome B2 and Cytochrome c Peroxidase Are Located in the Intermembrane Space of Yeast Mitochondria. J. Biol. Chem. 1982, 257 (21), 13028–13033. 10.1016/S0021-9258(18)33617-2. [DOI] [PubMed] [Google Scholar]
- Dominguez Pardo J. J.; Dörr J. M.; Renne M. F.; Ould-Braham T.; Koorengevel M. C.; van Steenbergen M. J.; Killian J. A. Thermotropic Properties of Phosphatidylcholine Nanodiscs Bounded by Styrene-Maleic Acid Copolymers. Chem. Phys. Lipids 2017, 208, 58–64. 10.1016/j.chemphyslip.2017.08.010. [DOI] [PubMed] [Google Scholar]
- Valentine R. C.; Shapiro B. M.; Stadtman E. R. Regulation of Glutamine Synthetase. XII. Electron Microscopy of the Enzyme from Escherichia Coli. Biochemistry 1968, 7 (6), 2143–2152. 10.1021/bi00846a017. [DOI] [PubMed] [Google Scholar]
- Ball L. E.; Riley L. J.; Hadasha W.; Pfukwa R.; Smith C. J. I.; Dafforn T. R.; Klumperman B. Influence of DIBMA Polymer Length on Lipid Nanodisc Formation and Membrane Protein Extraction. Biomacromolecules 2021, 22 (2), 763–772. 10.1021/acs.biomac.0c01538. [DOI] [PubMed] [Google Scholar]
- Priimagi A.; Cavallo G.; Metrangolo P.; Resnati G. The Halogen Bond in the Design of Functional Supramolecular Materials: Recent Advances. Acc. Chem. Res. 2013, 46 (11), 2686–2695. 10.1021/ar400103r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerebtzoff G.; Li-Blatter X.; Fischer H.; Frentzel A.; Seelig A. Halogenation of Drugs Enhances Membrane Binding and Permeation. ChemBioChem. 2004, 5, 676–684. 10.1002/cbic.200400017. [DOI] [PubMed] [Google Scholar]
- Cuevas Arenas R.; Klingler J.; Vargas C.; Keller S. Nanoscale Influence of Lipid Bilayer Properties on Nanodisc Formation Mediated by Styrene/Maleic Acid. Nanoscale 2016, 8 (32), 15016–15026. 10.1039/C6NR02089E. [DOI] [PubMed] [Google Scholar]
- Boumann H. A.; Damen M. J. A.; Versluis C.; Heck A. J. R.; De Kruijff B.; De Kroon A. I. P. M. The Two Biosynthetic Routes Leading to Phosphatidylcholine in Yeast Produce Different Sets of Molecular Species. Evidence for Lipid Remodeling. Biochemistry 2003, 42 (10), 3054–3059. 10.1021/bi026801r. [DOI] [PubMed] [Google Scholar]
- Horvath S. E.; Daum G. Lipids of Mitochondria. Prog. Lipid Res. 2013, 52 (4), 590–614. 10.1016/j.plipres.2013.07.002. [DOI] [PubMed] [Google Scholar]
- Zhang L.; Song J.; Cavigiolio G.; Ishida B. Y.; Zhang S.; Kane J. P.; Weisgraber K. H.; Oda M. N.; Rye K. A.; Pownall H. J.; Ren G. Morphology and Structure of Lipoproteins Revealed by an Optimized Negative-Staining Protocol of Electron Microscopy. J. Lipid Res. 2011, 52 (1), 175–184. 10.1194/jlr.D010959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan C. P. L.; Chiu M. H.; Wu X.; Lee S. K.; Prenner E. J.; Weers P. M. M. Apolipoprotein-Induced Conversion of Phosphatidylcholine Bilayer Vesicles into Nanodisks. Biochim. Biophys. Acta - Biomembr. 2011, 1808 (3), 606–613. 10.1016/j.bbamem.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R.; Sahu I. D.; Liu L.; Osatuke A.; Comer R. G.; Dabney-Smith C.; Lorigan G. A. Characterizing the Structure of Lipodisq Nanoparticles for Membrane Protein Spectroscopic Studies. Biochim. Biophys. Acta - Biomembr. 2015, 1848 (1), 329–333. 10.1016/j.bbamem.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielczak B.; Meister A.; Keller S. Influence of Mg2+ and Ca2+ on Nanodisc Formation by Diisobutylene/Maleic Acid (DIBMA) Copolymer. Chem. Phys. Lipids 2019, 221, 30–38. 10.1016/j.chemphyslip.2019.03.004. [DOI] [PubMed] [Google Scholar]
- Dhotel A.; Chen Z.; Delbreilh L.; Youssef B.; Saiter J.; Tan L. Molecular Motions in Functional Self-Assembled Nanostructures. Int. J. Mol. Sci. 2013, 14 (2), 2303–2333. 10.3390/ijms14022303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockman H. Dipole Potential of Lipid Membranes. Chem. Phys. Lipids 1994, 73, 57–79. 10.1016/0009-3084(94)90174-0. [DOI] [PubMed] [Google Scholar]
- Cladera J.; O’Shea P. Intramembrane Molecular Dipoles Affect the Membrane Insertion and Folding of a Model Amphiphilic Peptide. Biophys. J. 1998, 74 (5), 2434–2442. 10.1016/S0006-3495(98)77951-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y.; Liu Y.; Xu Z.; Li H.; Liu H.; Zhu W. Halogen Bonding for Rational Drug Design and New Drug Discovery. Expert Opin. Drug Discovery 2012, 7 (5), 375–383. 10.1517/17460441.2012.678829. [DOI] [PubMed] [Google Scholar]
- Auffinger P.; Hays F. A.; Westhof E.; Ho P. S. Halogen Bonds in Biological Molecules. Proc. Natl. Acad. Sci. U. S. A. 2004, 101 (48), 16789–16794. 10.1073/pnas.0407607101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcken R.; Zimmermann M. O.; Lange A.; Joerger A. C.; Boeckler F. M. Principles and Applications of Halogen Bonding in Medicinal Chemistry and Chemical Biology. J. Med. Chem. 2013, 56 (4), 1363–1388. 10.1021/jm3012068. [DOI] [PubMed] [Google Scholar]
- Grime R. L.; Logan R. T.; Nestorow S. A.; Sridhar P.; Edwards P. C.; Tate C. G.; Klumperman B.; Dafforn T. R.; Poyner D. R.; Reeves P. J.; Wheatley M. Differences in SMA-like Polymer Architecture Dictate the Conformational Changes Exhibited by the Membrane Protein Rhodopsin Encapsulated in Lipid Nano-Particles. Nanoscale 2021, 13, 13519–13528. 10.1039/D1NR02419A. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.