Abstract
Pharmacovigilance is a science that involves the ongoing monitoring of adverse drug reactions to existing medicines. Traditional approaches in this field can be expensive and time-consuming. The application of natural language processing (NLP) to analyze user-generated content is hypothesized as an effective supplemental source of evidence. In this systematic review, a broad and multi-disciplinary literature search was conducted involving four databases. A total of 5318 publications were initially found. Studies were considered relevant if they reported on the application of NLP to understand user-generated text for pharmacovigilance. A total of 16 relevant publications were included in this systematic review. All studies were evaluated to have medium reliability and validity. For all types of drugs, 14 publications reported positive findings with respect to the identification of adverse drug reactions, providing consistent evidence that natural language processing can be used effectively and accurately on user-generated textual content that was published to the Internet to identify adverse drug reactions for the purpose of pharmacovigilance. The evidence presented in this review suggest that the analysis of textual data has the potential to complement the traditional system of pharmacovigilance.
Keywords: pharmacovigilance, adverse drug reactions, ADRs, computational linguistics, machine learning, public health, user-generated content
1. Introduction
In drug development, there exists a strong tension between accessibility and safety. While drugs can effectively cure diseases and improve life [1], the required process of research and development of drugs is expensive, and pharmaceutical companies have a high stake in yielding a profit on their investment [2]. This increases the urgency to make effective drugs available to the public. In contrast, medicines can also induce adverse drug reactions (ADRs) that may result in mortality, and the identification of such reactions demands thorough and time-consuming testing of the drug’s safety, drastically increasing the time-to-market of new drugs [3]. In fact, the potential consequences of ADRs are significant. In the European Union (EU), five percent of hospital admissions and almost 200,000 deaths were caused by ADRs in 2008, and the associated societal cost totaled EUR 79 billion [4].
A system that applies tools and practices from the research field of pharmacovigilance was introduced to alleviate this tension [5]. This system performs ongoing monitoring of ADRs of existing drugs [5]. It also minimizes the time-to-market of effective drugs, and it allows their long-term safety post market authorization to be continuously examined [6]. Overall, pharmacovigilance is the cornerstone in the regulation of drugs [1]. The traditional system that applies pharmacovigilance is very expensive and often fails to monitor ADRs experienced by users if these are not reported to the authorities, pharmaceutical companies, or medical professionals [6,7]. The reporting of these ADRs is important because it may help to protect public health [1].
In today’s society, many people share personal content on social media [8,9,10]. An abundance of studies have already demonstrated that user-generated content can be used accurately for remote sensing, among others, to gauge public health [11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30]. This naturally raises the valid question whether user-generated textual content can also be analyzed for the purpose of pharmacovigilance. Such automated analysis may provide a cheap and efficient supplement to the expensive and time-consuming traditional methods for pharmacovigilance, and it may also include first-hand experiences about ADRs from users that were not reported to the authorities, pharmaceuticals, or medical professionals. Although various studies [31,32,33,34,35,36,37,38,39,40,41] were conducted that investigated the suitability of natural language processing (NLP) for pharmacovigilance, to our awareness, no systematic review has yet been conducted that aggregated the reported evidence or assessed the quality of those studies.
To address this research gap, the purpose of this study is to review the existing evidence on, and the effectiveness of natural language processing to understand user-generated content for the purpose of pharmacovigilance. According to our review, it is worthwhile to analyze user-generated content that has already been published to the Internet, to proactively and automatically identify ADRs, without relying on users to actively report those cases to the authorities, pharmaceutical companies, or medical professionals.
2. Background
A severe limitation in the process of bringing new drugs to market is the potential of drugs to cause ADRs. While pre-clinical and clinical studies include testing drug safety and potential ADRs, only a total of a few hundreds or thousands of participants are included in these studies [1]. In addition, these studies are performed under controlled clinical conditions that may not represent every real-world situation or circumstance [1]. Therefore, not all ADRs may have been identified prior to making the drug generally available [3]. As long as the benefits outweigh potential costs, it is generally considered unethical to withhold the general public from using an effective drug at this stage, thus accepting that some people may develop ADRs in the future.
2.1. Traditional Approaches
To counteract the limitations of pre-clinical and clinical testing, existing drugs on the market are constantly being monitored for safety and ADRs [3]. The long-term monitoring of existing drugs is crucial, because potential ADRs, interactions, and other risk factors, may only emerge many years or even decades after the drug initially received market authorization [3].
The long-term monitoring of drug safety beyond market authorization is named pharmacovigilance [5], which is defined by the WHO as “the science and activities relating to the detection, assessment, understanding and prevention of adverse effects or any other medicine-related problem” [3]. As such, the application of tools and practices from pharmacovigilance by public health authorities results in a pro-active system that is intended to promote and protect public health [1]. It involves a wide array of activities, including data collection about drug safety, obligating pharmaceuticals and medical professionals to report ADRs, inviting patients to report experiences with drugs, and the detection of signals that may indicate drug safety issues [6]. There are, however, significant costs associated with the processing and administration of the reported cases of ADRs [7]. In addition, the current system of collecting data to monitor drug safety is suboptimal because end-users are not obliged to report cases of ADRs [6].
2.2. Improving Pharmacovigilance Using Natural Language Processing
In the preceding 15 years, many technological innovations have enabled the storage, processing, and analysis of big data [42,43,44]. In particular, with the emergence of Web 2.0 and social media platforms, there has been a significant increase of user-generated content that is published to the Internet [8,9,10]. Among others, vast amounts of textual data are generated on blogs, forums, and social media [45]. Similarly, there have been significant developments in artificial intelligence that resulted in powerful methods and algorithms for NLP [46], which enabled the processing and understanding of human-generated text [45,47]. This opened new opportunities for mining social media and analyzing texts [48]. In recent years, these fields experienced significant innovations [49].
Text mining is frequently defined as the analysis of textual data, such as unstructured or semi-structured text, with the purpose to extract hidden patterns and information [45]. As such, it combines data mining with NLP [43]. Text mining has emerged from a need to analyze large amounts of text containing human language, which can be mined for insights that facilitate data-driven decision-making [45]. However, many standard data mining techniques cannot be applied to unstructured textual data. Therefore, text mining is applied as pre-processing for unstructured data [50,51], e.g.,:
tokenization: the separation of text into smaller units, like words, characters, or sub-words (n-grams);
transformation of cases, such as uniform lowercasing or uppercasing;
stop word removal: the removal of words carrying very little meaning; and such as pronouns;
reducing inflected words to their word stem (stemming).
Once text mining has been applied to extract structured data from a semi-structured or unstructured source, conventional data mining algorithms can subsequently be used to process and analyze these structured data further to yield the valued insights [47]. The complexity that is involved with analyzing unstructured textual data and in particular its irregularities, makes the process of text mining a difficult area in artificial intelligence [52].
The applications of text mining are numerous, and include:
assigning affective states to text (sentiment analysis) [43];
the discovery of associations between words or other tokens [53];
the summarization of documents [43];
clustering texts according to some similarity measurement [54,55];
predicting words or other lexical units (as part of a word processor or chatbot) [58,59]; and
the extraction of concepts, entities, and the relationships between them [43].
Among others, NLP was used to monitor public health, such as surveilling allergies [20,24,26], depressions [21,22,30], suicide-related thoughts and conversations [11,16,27], obesity [13,17], marijuana and drug abuse [12,25,28,29], tobacco and e-cigarettes [14,18,19,23], and to gauge public health concerns [15].
3. Materials and Methods
This systematic review was guided by the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines [60,61]. However, most of the reviewed papers do not contain controlled trials, comparable statistical analysis, or methodology, making it impossible to apply the complete PRISMA 2020 checklist to this review. Therefore, we only applied items on the checklist if they were applicable, and thus our review does not conform completely to the guideline. The quality of this systematic review was evaluated using the PRISMA Checklist in Appendix A.
3.1. Search Strategy
To cover all related disciplines, a broad selection of databases was made that included PubMed, Web of Science, IEEE Xplore, and ACM Digital Library. These databases were selected because they index studies in a wide range of fields. Specifically, PubMed was included because it predominantly indexes research in the field of public health, healthcare, and medicine. IEEE Xplore and ACM Digital Library were searched because these databases index publications in information technology and information management. Web of Science was included because it is a very large database that indexes studies in various disciplines, and also because of its multidisciplinary nature, there exists a consensus among researchers that it is good practice to include this database in systematic reviews. We recognize that Google Scholar is increasingly used as a source for systematic reviews, but that there exists a debate among scientists about its appropriateness [62]. A common argument against Google Scholar is that its algorithm for ranking the relevance of publications is updated frequently, thereby making the search results unreliable for reproduction [62]. Therefore, we have excluded Google Scholar as an information source in this review. Furthermore, because it is a commonality in information technology and computational linguistics that materials are not always published in peer-reviewed journals, but instead it is frequently published only in conference proceedings or conference papers, both journal articles and conference proceedings were included in this systematic review. It was not expected that this would have a significant effect on the reliability of studies, because conference proceedings and conference papers are also subject to a peer-review process.
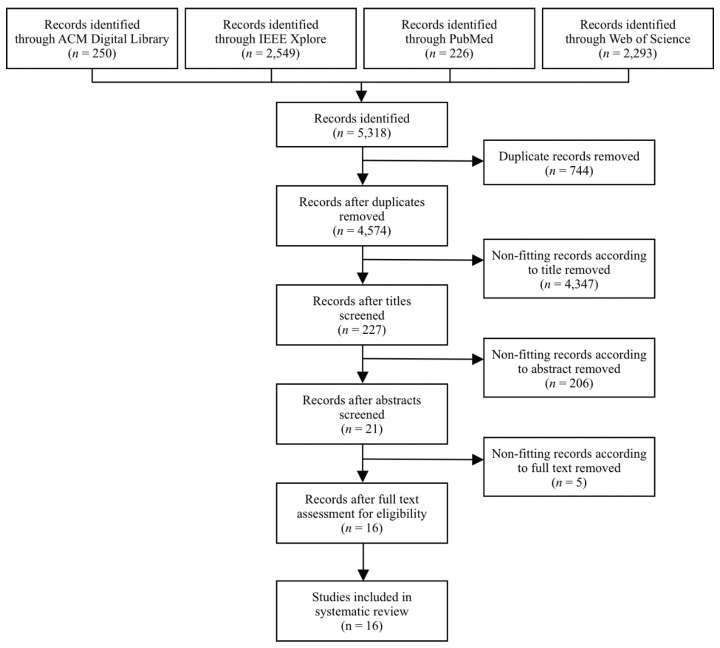
For each of the included databases, an optimized search strategy was formulated (see Appendix B). The search query was constructed from two blocks. The first block addresses the concept of NLP, and the second block includes search terms related to health surveillance. The systematic literature search was performed on 25 March 2020 for all databases. All publications appearing up to this point were considered in the search. After the databases were searched, the method for de-duplication by Bramer et al. [63] was performed to identify and remove duplicate studies. Studies eligible for this systematic review were selected in three subsequent phases and visualized in Figure 1.
Figure 1.
Flow diagram for literature search and study selection.
3.2. Study Selection
First, the titles were screened for the presence of subjects related to public health monitoring or public health surveillance. The screening was very global to prevent the unnecessary exclusion of studies. Therefore, not only terms such as “adverse drug reactions” were considered relevant, but titles containing more indirect terms such as “medication outcomes” were also included. In addition, if it was ambiguous whether a study was relevant or not, it was still included for further screening in the next phase. Studies that were not relevant were omitted from the library.
Second, the abstracts were screened for information related to NLP, public health monitoring, public health surveillance, and pharmacovigilance. The keywords provided with the manuscript for indexing purposes were also screened for these concepts. This phase was also intended to be broad. For example, abstracts were considered relevant if they contained terms directly related to pharmacovigilance, such as “adverse effects of drug treatment”, but also indirectly related terms such as “drug reviews”. Drug reviews involve an extensive process where experimental drugs are assessed on safety (e.g., toxicity and side effects) and effectiveness using various clinical trials [1]. Drug reviews are mainly performed by pharmaceutical companies which document their tests for review by the European Medicines Agency (EMA) or the U.S. Food and Drug Administration (FDA) [1]. Post market authorization, existing drugs and their side effects are continuously being monitored by medical doctors, laboratories, pharmaceutical organizations, and health authorities [1]. Publications were still included if their relevance was considered ambiguous, for further screening in the next phase. Irrelevant manuscripts were removed.
Third, the full text was downloaded and read. Studies were considered relevant if they investigated the application of NLP to understand text with the purpose of public health monitoring or public health surveillance within the discipline of pharmacovigilance. Eligible studies reported on the application and results of using computational linguistics to identify adverse drug reactions from textual sources, such as forums, patient records, and social media.
3.3. Inclusion and Exclusion Criteria
Overall, studies were only eligible for inclusion in this systematic review if they aimed to identify adverse drug reactions using computational linguistics. Both journal articles and conference proceedings were included. In addition, we only included studies if written in the English language, irrespective of the language of the dataset of user-generated content that these studies utilized. There were no limitations regarding the publication date, institutional affiliation, or the journal that these studies were published in.
Publications were excluded if they only reported on a framework instead of the actual application. For example, authors may suggest a process to investigate adverse drug reactions using computational linguistics without actually applying it and evaluating the results. Likewise, studies were excluded if they were published in a language other than English. We only excluded studies if the manuscript was not written in English, irrespective of the language that its dataset was written in. Furthermore, if the same publication was published in different formats, for example as both a conference proceeding and a journal article, only one format of the publication, namely the journal article, was retained.
3.4. Reliability and Validity
The included publications were evaluated on quality by assessing their reliability and validity. This assessment was performed using the strategy of Kampmeijer et al. [64]. A publication was evaluated as reliable if it reported a thorough and repeatable description of the performed process, methods, data collection, and data analysis [64]. A reliable study provides a well-defined, transparent, and consistent protocol for the collection, processing, and analysis of data. It facilitates researchers to establish its consistency and identify potential flaws in the research design. In addition, a reliable study provides sufficient details such that it can be reproduced. Under the same conditions, if repeated, a reliable study will produce similar findings.
A publication was evaluated as valid if the reported findings are logically the result of the described process, methods, data, and analyses that were used to find that result [64]. The validity of a study refers to its accuracy; the study indeed measures what it intended to measure. This evaluation requires that researchers are transparent about their protocol. Assessing the validity of a study involves identifying that the reported results and conclusions in a study are consistent with the study hypotheses and research design. In addition, it involves the verification that the reported findings from one study are comparable to other studies utilizing a comparable research protocol. The identified consistency within one paper, or consistency in reported findings among comparable papers, are indications that a paper was evaluated as valid.
The reliability and validity of studies were assessed qualitatively and discussed among researchers until consensus was achieved. Studies with “low” reliability did not provide a well-defined, transparent, and consistent protocol or this information provided insufficient details. Instead, studies with “high” reliability provided this information and this information was thorough. In all intermediate cases, these studies were marked as “medium” reliability. Similarly, studies with “low” validity had either limited consistency between the hypotheses and research design on the one hand, with the results and conclusions on the other hand, or their findings were not consistent with studies that utilized a comparable research design. Instead, if the consistency was high and their findings were comparable to similar studies, the validity of these studies was marked as “high”. In all intermediate cases, the validity was considered “medium”.
Although the quality assessment was rigorous and based on scientific standards, all identified publications were included in the systematic review.
3.5. Data Analysis
Thematic analysis was used to analyze the included publications [65]. The themes were defined by the objectives of the present systematic review. The following themes were extracted from the full text: authors, year of publication, type of drugs, data source, sample size, users, unique users, origin of users, average number of followers, years of data collection, horizon of data collection, software used, techniques and classifiers used, outcome, drugs studied, result, and a description of the result.
For each publication, the extracted themes were processed into an extraction matrix. This matrix was used to synthesize and narratively present the extracted information by theme. The results are summarized and presented using tables.
4. Results
The procedure that was followed for the selection of studies is presented in Figure 1. The 5318 initial records, which were identified through an inclusive search strategy, were assessed for the presence of duplicate publications. Consequently, 744 duplicate results were identified and omitted. Therefore, the literature search yielded 4574 unique studies. According to the thorough study selection strategy described in Section 3.2, the first selection phase identified 4347 irrelevant studies to be excluded. In the second phase, the remaining 227 results were screened by reading the abstract; 206 irrelevant studies were omitted. For example, studies were excluded when not mentioning “adverse effects of drug treatment” or other related but rather general terms such as “drug reviews” in the abstract. In the third phase, the full text of the remaining 21 publications was read. Five studies were considered irrelevant because they did not investigate the application of computational linguistics to understand text, with the purpose of public health monitoring or public health surveillance within the discipline of pharmacovigilance.
Overall, this yielded 16 publications that were considered relevant and were included in this systematic review. All studies were published between 2009 and 2019. Most of the studies (69%) were published in the last five years (2015–2019) [31,32,33,34,35,36,37,38,39,40,41]. A summary is provided in Table 1, and it will be further elaborated in the rest of this section.
Table 1.
Summary of characteristics of publications included in the analysis.
| Authors | Data Source | Sample Size | Horizon of Data Collection | Software Used | Techniques and Classifiers Used | Outcome | Result | Description of Result |
|---|---|---|---|---|---|---|---|---|
| [31] | Social media | Twitter.com: 1642 tweets | 3 years | Toolkit for Multivariate Analysis | Artificial Neural Networks (ANN), Boosted Decision Trees with AdaBoost (BDT), Boosted Decision Trees with Bagging (BDTG), Sentiment Analysis, Support Vector Machines (SVM) | Reported ADRs for HIV treatment | Positive | Reported adverse effects are consistent with well-recognized toxicities. |
| [32] | Forums | DepressionForums.org: 7726 posts | 10 years | General Architecture for Text Engineering (GATE), NLTK Toolkit within MATLAB, RapidMiner | Hyperlink-Induced Topic Search (HITS), k-Means Clustering, Network Analysis, Term-Frequency-Inverse Document Frequency (TF-IDF) | User sentiment on depression drugs | Positive | Natural language processing is suitable to extract information on ADRs concerning depression. |
| [66] | Social media | Twitter.com: 2,102,176,189 tweets | 1 year | Apache Lucene | MetaMap, Support Vector Machines (SVM) | Reported ADRs for cancer | Neutral | Classification models had limited performance. Adverse events related to cancer drugs can potentially be extracted from tweets. |
| [33] | Social media | Twitter.com: 6528 tweets | Unknown | GENIA tagger, Hunspell, Snowball stemmer, Stanford Topic Modelling Toolbox, Twokenizer | Backward/Forward Sequential Feature Selection (BSFS/FSFS) Algorithm, k-Means Clustering, Sentiment Analysis, Support Vector Machines (SVM) | Reported ADRs | Positive | ADRs were identified reasonably well. |
| [34] | Social media | Twitter.com: 32,670 tweets |
Unknown | Hunspell, Twitter tokenizer | Term Frequency-Inverse Document Frequency (TF-IDF) | Reported ADRs | Neutral | ADRs were not identified very well. |
| [67] | Social media | Twitter.com: 10,822 tweets |
Unknown | Unknown | Naive Bayes (NB), Natural Language Processing (NLP), Support Vector Machines (SVM) | Reported ADRs | Positive | ADRs were identified well. |
| [35] | Drug reviews | Drugs.com, Drugslib.com: 218,614 reviews |
Unknown | BeautifulSoup | Logistic Regression, Sentiment Analysis |
Patient satisfaction with drugs, Reported ADRs, Reported effectiveness of drugs | Positive | Classification results were very good. |
| [36] | Social media | Twitter.com: 172,800 tweets |
1 year | Twitter4J | Decision Trees, Medical Profile Graph, Natural Language Processing (NLP) | Reported ADRs | Positive | Building a medical profile of users enables the accurate detection of adverse drug events. |
| [37] | Social media | Twitter.com: 1245 tweets |
Unknown | CRF++ Toolkit, GENIA tagger, Hunspell, Twitter REST API, Twokenizer | Natural Language Processing (NLP) | Reported ADRs | Positive | ADRs were identified reasonably well. |
| [38] | Drug reviews | WebMD.com: Unknown | Unknown | SentiWordNet, WordNet | Sentiment Analysis, Support Vector Machines (SVM), Term document Matrix (TDM) | User sentiment on cancer drugs | Positive | Sentiment on ADRs was identified reasonably well. |
| [39] | Drug reviews, Social media | DailyStrength.org: 6279 reviews, Twitter.com: 1784 tweets |
Unknown | Unknown | ARDMine, Lexicon-based, MetaMap, Support Vector Machines (SVM) | Reported ADRs | Positive | ADRs were identified very well. |
| [40] | Drug reviews, Social media | PatientsLikeMe.com: 796 reviews, Twitter.com: 39,127 tweets, WebMD.com: 2567 reviews, YouTube.com: 42,544 comments |
Not applicable | Deeply Moving | Unknown | Patient-reported medication outcomes | Positive | Social media serves as a new data source to extract patient-reported medication outcomes. |
| [68] | Forums | Medications.com: 8065 posts, SteadyHealth.com: 11,878 |
Not applicable | Java Hidden Markov Model library, jsoup | Hidden Markov Model (HMM), Natural Language Processing (NLP) | Reported ADRs | Positive | Reported adverse effects are consistent with well-recognized side-effects. |
| [69] | Electronic Health Record (EHR) | 25,074 discharge summaries | Not applicable | MedLEE | Unknown | Reported ADRs | Positive | Reported adverse effects are consistent with well-recognized toxicities (recall: 75%; precision: 31%). |
| [41] | Social media | Twitter.com: 3251 tweets |
Not applicable | AFINN, Bing Liu sentiment words, Multi-Perspective Question Answering (MPQA), SentiWordNet, TextBlob, Tweepy, WEKA | MetaMap, Naive Bayes (NB), Natural Language Processing (NLP), Sentiment Analysis, Support Vector Machines (SVM) | Reported ADRs | Positive | Several well-known ADRs were identified. |
| [70] | Forums | MedHelp.org: 6244 discussion threads | Unknown | Unknown | Association Mining | Reported ADRs | Positive | ADRs were identified. |
The reliability and validity of all studies were assessed as medium. While all studies were performed reasonably well, they failed to be entirely transparent about their process, methodology, used software, and the used technologies and classifiers. As is presented in the detailed overview of the characteristics of the included studies in Appendix C, all studies failed to disclose a complete overview of crucial information.
4.1. General Characteristics
A general description of the publications included in the analysis is provided in Table 2. To establish differences between them, various characteristics of these publications were compared and the observed differences are presented in Table 3.
Table 2.
General description of publications included in the analysis.
| Category | Sub-Categories | n (%) | References |
|---|---|---|---|
| Year of publication | 2009 | 1 (6) | [69] |
| 2010 | 0 (0) | - | |
| 2011 | 0 (0) | - | |
| 2012 | 2 (13) | [66,70] | |
| 2013 | 0 (0) | - | |
| 2014 | 2 (13) | [67,68] | |
| 2015 | 7 (43) | [31,36,37,38,39,40,41] | |
| 2016 | 2 (13) | [32,33] | |
| 2017 | 0 (0) | - | |
| 2018 | 1 (6) | [35] | |
| 2019 | 1 (6) | [34] | |
| Type of drugs | Asthma | 1 (5) | [40] |
| Cancer | 2 (11) | [38,66] | |
| Cystic fibrosis | 1 (5) | [40] | |
| Depression | 1 (5) | [32] | |
| HIV | 1 (5) | [31] | |
| Rheumatoid arthritis | 1 (5) | [40] | |
| Type 2 diabetes | 1 (5) | [40] | |
| Unknown | 11 (58) | [33,34,35,36,37,39,41,67,68,69,70] | |
| Data source | Drug reviews | 4 (22) | [35,38,39,40] |
| Electronic Health Records (EHR) | 1 (6) | [69] | |
| Forums | 3 (17) | [32,68,70] | |
| Social media | 10 (56) | [31,33,34,36,37,39,40,41,66,67] | |
| Sample size | Less than 5000 | 3 (19) | [31,37,41] |
| 5000 to 9999 | 4 (25) | [32,33,39,70] | |
| 10,000 to 14,999 | 1 (6) | [67] | |
| 15,000 to 19,999 | 1 (6) | [68] | |
| 20,000 or more | 6 (38) | [34,35,36,40,66,69] | |
| Unknown | 1 (6) | [38] | |
| Users | HIV-infected persons undergoing drug treatment | 1 (6) | [31] |
| Unknown | 15 (94) | [32,33,34,35,36,37,38,39,40,41,66,67,68,69,70] | |
| Unique users | Less than 5000 | 2 (13) | [31,36] |
| 5000 to 9999 | 0 (0) | - | |
| 10,000 to 14,999 | 0 (0) | - | |
| 15,000 to 19,999 | 0 (0) | - | |
| 20,000 or more | 0 (0) | - | |
| Unknown | 14 (88) | [32,33,34,35,37,38,39,40,41,66,67,68,69,70] | |
| Origin of users | Canada | 1 (5) | [31] |
| South Africa | 1 (5) | [31] | |
| United Kingdom | 1 (5) | [31] | |
| United States | 1 (5) | [31] | |
| Unknown | 15 (79) | [32,33,34,35,36,37,38,39,40,41,66,67,68,69,70] | |
| Average number of followers | Less than 5000 | 1 (6) | [31] |
| 5000 to 9999 | 0 (0) | - | |
| 10,000 to 14,999 | 0 (0) | - | |
| 15,000 to 19,999 | 0 (0) | - | |
| 20,000 or more | 0 (0) | - | |
| Unknown | 15 (94) | [32,33,34,35,36,37,38,39,40,41,66,67,68,69,70] | |
| Years of data collection | 2004 | 2 (6) | [32,69] |
| 2005 | 1 (3) | [32] | |
| 2006 | 1 (3) | [32] | |
| 2007 | 1 (3) | [32] | |
| 2008 | 1 (3) | [32] | |
| 2009 | 2 (6) | [32,66] | |
| 2010 | 3 (10) | [31,32,66] | |
| 2011 | 2 (6) | [31,32] | |
| 2012 | 3 (10) | [31,32,68] | |
| 2013 | 2 (6) | [31,32] | |
| 2014 | 3 (10) | [32,36,40] | |
| 2015 | 2 (6) | [36,41] | |
| Unknown | 8 (26) | [33,34,35,37,38,39,67,70] | |
| Horizon of data collection | 1 year | 2 (13) | [36,66] |
| 2 to 5 years | 1 (6) | [31] | |
| 6 to 10 years | 1 (6) | [32] | |
| Not applicable | 4 (25) | [40,41,68,69] | |
| Unknown | 8 (50) | [33,34,35,37,38,39,67,70] | |
| Software used | AFINN | 1 (3) | [41] |
| Apache Lucene | 1 (3) | [66] | |
| BeautifulSoup | 1 (3) | [35] | |
| Bing Liu sentiment words | 1 (3) | [41] | |
| CRF++ toolkit | 1 (3) | [37] | |
| Deeply Moving | 1 (3) | [40] | |
| General Architecture for Text Engineering (GATE) | 1 (3) | [32] | |
| GENIA tagger | 2 (6) | [33,37] | |
| Hunspell | 3 (9) | [33,34,37] | |
| Java Hidden Markov Model library | 1 (3) | [68] | |
| jsoup | 1 (3) | [68] | |
| MedLEE | 1 (3) | [69] | |
| Multi-Perspective Question Answering (MPQA) | 1 (3) | [41] | |
| NLTK toolkit within MATLAB | 1 (3) | [32] | |
| RapidMiner | 1 (3) | [32] | |
| SentiWordNet | 2 (6) | [38,41] | |
| Snowball stemmer | 1 (3) | [33] | |
| Stanford Topic Modelling Toolbox | 1 (3) | [33] | |
| TextBlob | 1 (3) | [41] | |
| Toolkit for Multivariate Analysis | 1 (3) | [31] | |
| Tweepy | 1 (3) | [41] | |
| Twitter REST API | 1 (3) | [37] | |
| Twitter tokenizer | 1 (3) | [34] | |
| Twitter4J | 1 (3) | [36] | |
| Twokenizer | 2 (6) | [33,37] | |
| Unknown | 3 (9) | [39,67,70] | |
| WEKA | 1 (3) | [41] | |
| WordNet | 1 (3) | [38] | |
| Techniques and classifiers used | ARDMine | 1 (2) | [39] |
| Artificial Neural Networks (ANN) | 1 (2) | [31] | |
| Association Mining | 1 (2) | [70] | |
| Backward/forward sequential feature selection (BSFS/FSFS) algorithm | 1 (2) | [33] | |
| Boosted Decision Trees with AdaBoost (BDT) | 1 (2) | [31] | |
| Boosted Decision Trees with Bagging (BDTG) | 1 (2) | [31] | |
| Decision Trees | 1 (2) | [36] | |
| Hidden Markov Model (HMM) | 1 (2) | [68] | |
| Hyperlink-Induced Topic Search (HITS) | 1 (2) | [32] | |
| k-Means Clustering | 2 (5) | [32,33] | |
| Lexicon-based | 1 (2) | [39] | |
| Logistic Regression | 1 (2) | [35] | |
| Medical Profile Graph | 1 (2) | [36] | |
| MetaMap | 3 (7) | [39,41,66] | |
| Naive Bayes (NB) | 2 (5) | [41,67] | |
| Natural Language Processing (NLP) | 5 (12) | [36,37,41,67,68] | |
| Network Analysis | 1 (2) | [32] | |
| Sentiment Analysis | 5 (12) | [31,33,35,38,41] | |
| Support Vector Machines (SVM) | 7 (17) | [31,33,38,39,41,66,67] | |
| Term Document Matrix (TDM) | 1 (2) | [38] | |
| Term-Frequency-Inverse Document Frequency (TF-IDF) | 2 (5) | [32,34] | |
| Unknown | 2 (5) | [40,69] | |
| Outcome | Patient satisfaction with drugs | 1 (6) | [35] |
| Patient-reported medication outcomes | 1 (6) | [40] | |
| Reported ADRs | 11 (61) | [33,34,35,36,37,39,41,67,68,69,70] | |
| Reported ADRs for cancer | 1 (6) | [66] | |
| Reported ADRs for HIV treatment | 1 (6) | [31] | |
| Reported effectiveness of drugs | 1 (6) | [35] | |
| User sentiment on depression drugs | 1 (6) | [32] | |
| User sentiment on cancer drugs | 1 (6) | [38] | |
| Drugs studied | Less than 5 | 1 (6) | [31] |
| 5 to 9 | 4 (25) | [32,41,66,69] | |
| 10 to 14 | 2 (13) | [40,70] | |
| 15 to 19 | 0 (0) | - | |
| 20 or more | 5 (31) | [36,38,39,67,68] | |
| Unknown | 4 (25) | [33,34,35,37] | |
| Result | Positive | 14 (88) | [31,32,33,35,36,37,38,39,40,41,67,68,69,70] |
| Neutral | 2 (13) | [34,66] | |
| Negative | 0 (0) | - | |
| Reliability | Low | 0 (0) | - |
| Medium | 16 (100) | [31,32,33,34,35,36,37,38,39,40,41,66,67,68,69,70] | |
| High | 0 (0) | - | |
| Validity | Low | 0 (0) | - |
| Medium | 16 (100) | [31,32,33,34,35,36,37,38,39,40,41,66,67,68,69,70] | |
| High | 0 (0) | - |
Table 3.
Publications by classification category and result.
| Category | Sub-Categories | Positive (n %) | Neutral (n %) | Negative (n %) | References |
|---|---|---|---|---|---|
| Type of drugs | Asthma | 1 (5) | 0 (0) | 0 (0) | [40] |
| Cancer | 1 (5) | 1 (5) | 0 (0) | [38,66] | |
| Cystic fibrosis | 1 (5) | 0 (0) | 0 (0) | [40] | |
| Depression | 1 (5) | 0 (0) | 0 (0) | [32] | |
| HIV | 1 (5) | 0 (0) | 0 (0) | [31] | |
| Rheumatoid arthritis | 1 (5) | 0 (0) | 0 (0) | [40] | |
| Type 2 diabetes | 1 (5) | 0 (0) | 0 (0) | [40] | |
| Unknown | 10 (53) | 1 (5) | 0 (0) | [33,34,35,36,37,39,41,67,68,69,70] | |
| Data source | Drug reviews | 4 (22) | 0 (0) | 0 (0) | [35,38,39,40] |
| Electronic Health Records (EHR) | 1 (6) | 0 (0) | 0 (0) | [69] | |
| Forums | 3 (17) | 0 (0) | 0 (0) | [32,68,70] | |
| Social media | 8 (44) | 2 (11) | 0 (0) | [31,33,34,36,37,39,40,41,66,67] | |
| Origin of users | Canada | 1 (5) | 0 (0) | 0 (0) | [31] |
| South Africa | 1 (5) | 0 (0) | 0 (0) | [31] | |
| United Kingdom | 1 (5) | 0 (0) | 0 (0) | [31] | |
| United States | 1 (5) | 0 (0) | 0 (0) | [31] | |
| Unknown | 13 (68) | 2 (11) | 0 (0) | [32,33,34,35,36,37,38,39,40,41,66,67,68,69,70] | |
| Horizon of data collection | 1 year | 1 (6) | 1 (6) | 0 (0) | [36,66] |
| 2 to 5 years | 1 (6) | 0 (0) | 0 (0) | [31] | |
| 6 to 10 years | 1 (6) | 0 (0) | 0 (0) | [32] | |
| Not applicable | 4 (25) | 0 (0) | 0 (0) | [40,41,68,69] | |
| Unknown | 7 (44) | 1 (6) | 0 (0) | [33,34,35,37,38,39,67,70] | |
| Outcome | Patient satisfaction with drugs | 1 (6) | 0 (0) | 0 (0) | [35] |
| Patient-reported medication outcomes | 1 (6) | 0 (0) | 0 (0) | [40] | |
| Reported ADRs | 10 (56) | 1 (6) | 0 (0) | [33,34,35,36,37,39,41,67,68,69,70] | |
| Reported ADRs for cancer | 0 (0) | 1 (6) | 0 (0) | [66] | |
| Reported ADRs for HIV treatment | 1 (6) | 0 (0) | 0 (0) | [31] | |
| Reported effectiveness of drugs | 1 (6) | 0 (0) | 0 (0) | [35] | |
| User sentiment on depression drugs | 1 (6) | 0 (0) | 0 (0) | [32] | |
| User sentiment on cancer drugs | 1 (6) | 0 (0) | 0 (0) | [38] | |
| Drugs studied | Less than 5 | 1 (6) | 0 (0) | 0 (0) | [31] |
| 5 to 9 | 3 (19) | 1 (6) | 0 (0) | [32,41,66,69] | |
| 10 to 14 | 2 (13) | 0 (0) | 0 (0) | [40,70] | |
| 15 to 19 | 0 (0) | 0 (0) | 0 (0) | - | |
| 20 or more | 5 (31) | 0 (0) | 0 (0) | [36,38,39,67,68] | |
| Unknown | 3 (19) | 1 (6) | 0 (0) | [33,34,35,37] | |
| Reliability | Low | 0 (0) | 0 (0) | 0 (0) | - |
| Medium | 14 (88) | 2 (13) | 0 (0) | [31,32,33,34,35,36,37,38,39,40,41,66,67,68,69,70] | |
| High | 0 (0) | 0 (0) | 0 (0) | - | |
| Validity | Low | 0 (0) | 0 (0) | 0 (0) | - |
| Medium | 14 (88) | 2 (13) | 0 (0) | [31,32,33,34,35,36,37,38,39,40,41,66,67,68,69,70] | |
| High | 0 (0) | 0 (0) | 0 (0) | - |
Only one study by Adrover et al. [31] discussed the geographical location of users that published the included posts. They report that the users were from Canada, South Africa, the United Kingdom, or the United States [31]. The remaining 15 studies did not disclose the geographical location of users.
Studies disclosing the date of publication of the textual samples (74%) were published between 2004 and 2015 [31,32,36,40,41,66,68,69]. Content published since 2010 was included in more studies compared to content published before 2010. The remaining 26% of studies did not discuss when the posts were published [33,34,35,37,38,39,67,70].
Studies that reported the date of publication of the included content (50%) were used to compute the time horizon of the collected data [31,32,36,40,41,66,68,69]. In 13% of studies, this horizon was one calendar year [36,66]. In 6% of the studies, this horizon was between two and five years [31]. In another 6%, the horizon ranged between 6 and 10 years [32]. In four studies (25%), the time horizon could not be computed because the data were published within the same calendar year [40,41,68,69]. The remaining studies (50%) did not present the date on which the included data were published [33,34,35,37,38,39,67,70]. Therefore, the horizon of data collection could not be computed.
Discounting the studies that did not present the type of drugs that were studied, drugs to treat asthma (5%) [40], cancer (11%) [38,66], cystic fibrosis (5%) [40], depression (5%) [32], HIV (5%) [31], rheumatoid arthritis (5%) [40], and type 2 diabetes (5%) [40] were investigated. In a majority of studies (58%), the type of drugs was not specified [33,34,35,36,37,39,41,67,68,69,70].
The studies also differed with respect to the number of drugs for which ADRs were investigated. Most studies (31%) included posts concerning 20 or more drugs [36,38,39,67,68], followed by 25% that studied between five and nine drugs [32,41,66,69]. Two studies (13%) included between ten and fourteen drugs [40,70], while only one study (6%) addressed less than five drugs [31]. No studies included between 15 and 19 drugs. The remaining 25% of studies did not disclose the number of drugs that were investigated [33,34,35,37].
4.2. Input Sources
Publications used data from four sources. A majority of studies (56%) used textual information from social media to extract ADRs [31,33,34,36,37,39,40,41,66,67]. Drug reviews were also a popular source of unstructured data (22%) [35,38,39,40]. Forums (17%) [32,68,70] and electronic health records (6%) [69] were used less often.
There was a wide diversity in the sample size of the posts used, which ranged from 1245 [37] to more than two billion [66] tweets. Three studies (19%) included less than 5000 posts [31,37,41], six publications (38%) used at least 5000 but less than 20,000 posts [32,33,39,67,68,70], and six studies (38%) were performed using more than 20,000 posts [34,35,36,40,66,69]. The sample size was not reported in one study (6%) [38].
Only one study (6%) by Adrover et al. [31] provided contextual information about the background of the publishers of the included posts. In the remaining studies (94%), the background of these users was not disclosed [32,33,34,35,36,37,38,39,40,41,66,67,68,69,70].
The vast majority of studies (88%) did not provide information about the unique number of users that published the analyzed content [32,33,34,35,37,38,39,40,41,66,67,68,69,70]. In only two studies (13%), it was disclosed that less than 5000 users had published the posts [31,36]. Similarly, only one study (6%) discussed that a user had an average of fewer than 5000 followers [31]. The remaining studies (94%) provided no information about this theme [32,33,34,35,36,37,38,39,40,41,66,67,68,69,70].
Most studies (56%) included at least unstructured data from social media [31,33,34,36,37,39,40,41,66,67]. In particular, all of these studies included data from Twitter, while one study additionally used content from YouTube [40]. This study analyzed 42,544 comments from YouTube to identify patient-reported medication outcomes [40]. The majority of these studies reported that ADRs could indeed be extracted from textual content from social media. In addition, drug reviews (22%) [35,38,39,40] and content from forums (17%) [32,68,70] were studied less frequently. For both data sources, ADRs were identified correctly. The least studied source involved electronic health records, for which also positive results were reported [69].
4.3. Employed Methods
The studies reported a vast difference in software that was used (see Table 3). In total, 27 software products were discussed. Often, studies also used alternatives for the same type of software. For example, although some studies used Tweepy (3%) [41], Twitter REST API (3%) [37], or Twitter4J (3%) [36] to retrieve data from Twitter using a different programming language, these software products can be aggregated in the type of Twitter API. By frequency, the spelling checker Hunspell was used most often (9%) [33,34,37]. This tool can reduce the dimensionality of NLP tasks by considering various spellings of a single word. For example, misspellings (e.g., “organiezation”) or inconsistent types of English such as “organization” and “organisation”, would then be transformed into the U.S. word “organization”. Notably, three studies (9%) did not present the software that was used [39,67,70].
Likewise, a vast number of 21 different techniques and classifiers were reported. The more generic term “NLP” [36,37,41,67,68] and the more particular task of sentiment analysis [31,33,35,38,41] were mentioned in five studies (12%). In terms of particular techniques, two studies (5%) and one study (2%) reported the use of term-frequency-inverse document frequency (TF-IDF) [32,34] and term document matrix (TDM) [38], respectively. In terms of models, support vector machines (SVM) was used most often in seven studies (17%) to analyze the quantitative features that were extracted from unstructured data [31,33,38,39,41,66,67]. The remaining 5% of the studies did not disclose the techniques and classifiers that were used [40,69].
4.4. Study Effectiveness
Although all included studies investigated how NLP can be applied to understand text for pharmacovigilance, and thus all studies investigated reported ADRs, some studies were more explicit than others in discussing their outcome. For example, several studies explicitly disclosed that the outcome was reported ADRs for cancer (6%) [66], reported ADRs for HIV treatment (6%) [31], user sentiment on depression drugs (6%) [32], and user sentiment on cancer drugs (6%) [38]. The remaining studies were rather inconsistent by presenting the outcome as patient satisfaction with drugs (6%) [35], patient-reported medication outcomes (6%) [40], reported ADRs (61%) [33,34,35,36,37,39,41,67,68,69,70], and reported the effectiveness of drugs (6%) [35].
The studies reported consistent evidence that NLP can be successfully used to understand text for the purpose of pharmacovigilance. A vast majority of studies (88%) presented positive results [31,32,33,35,36,37,38,39,40,41,67,68,69,70]. These studies claimed that ADRs could indeed be extracted accurately and reliably from content published by patients. These studies often compared the accuracy of the adverse effects that were extracted from posts against a list of known ADRs, for example, from the medical package insert or from other reliable sources. Only 13% of the studies reported neutral findings [34,66]. No studies reported a negative result.
For example, Nikfarjam et al. [39] addressed the challenges of patients who use informal language and express medical concepts in lay terms, which may obstruct utilizing patients’ digital content for public health monitoring for pharmacovigilance. They developed a system for extracting ADRs from highly informal and unstructured content from both Twitter and a website for drug reviews. They find that it is possible, with reasonably high performance, to extract complex medical concepts from these platforms. In addition, Sampathkumar et al. [68] aimed to identify mentions about ADRs from user-generated content that were published by drug users to healthcare-related forums and to use those mentions for the purpose of pharmacovigilance. They find that it is possible to extract those mentions about ADRs with good performance and that the mentions are consistent with known ADRs. Likewise, Wu et al. [41] developed a pipeline “for collecting, processing, and analyzing tweets to find signals” about ADRs. They were able to identify several well-known ADRs. Furthermore, Yang et al. [70] mined the associations between drugs and the ADRs that patients published to online healthcare communities. These identified associations were then compared to ADR alerts from the U.S. Food and Drug Administration. They find that association mining appears to be promising for the detection of ADRs.
For named diseases, only one study observed neutral effectiveness for oncological drugs [66]. Specifically, Bian et al. [66] developed an approach to identify drug users and extract ADRs concerning cancer from tweets. They used high-performance computing to analyze more than two billion tweets using NLP, and classified tweets using support vector machines. They, however, find that their classification model had limited performance.
There were no significant inconsistencies in the effectiveness of NLP to identify ADRs with respect to the outcome that was under investigation in each study. For a vast majority of the outcomes, ADRs could indeed be established. For the outcome of reported ADRs for cancer, only neutral effectiveness was reported [66]. Although most of the studies that investigated the outcome of reported ADRs observed positive findings, only one study found a neutral result [34]. There were no notable differences in the effectiveness of NLP with respect to the number of drugs that were considered in the publications.
5. Discussion
The purpose of this study was to review the existing evidence on the methods and effectiveness of natural language processing to understand user-generated textual content for the purpose of pharmacovigilance.
The first main finding of this systematic review is that the potential of applying NLP for pharmacovigilance looks very promising. Studies included in this systematic review consistently reported positive results on the effectiveness and accuracy of using NLP that is applied to user-generated digital content to identify ADRs. For all diseases investigated, a vast majority of studies reported that the identified ADRs were consistent with the information provided on the medical package insert. For example, Ru et al. [40] analyzed and compared content about patient-reported medication outcomes concerning asthma, cystic fibrosis, rheumatoid arthritis, and type 2 diabetes, published to the social media sites PatientsLikeMe.com, WebMD.com, Twitter, and YouTube. They find that, although advising that more emphasis should be placed on developing more reliable methods for NLP and text mining, social media platforms are indeed suitable and complementary sources for investigating outcomes of medication. In addition, Mishra et al. [38] assessed pharmaceutical oncological drug reviews authored by patients and published to user forums, and they compared the reported drug-related issues with official drug labels. They used support vector machines to classify sentiments about ADRs with good performance. Furthermore, Akay et al. [32] investigated how user-generated content on a depression-related forum can be used for modeling the exchange of information between users about drug-related treatments for depression. They find that it is possible to use NLP on this content to identify the ADRs of these drugs in greater detail, and they confirmed the identified ADRs using medical literature about these drugs. In another study, Androver et al. [31] studied the potential of using user-generated tweets to identify ADRs for HIV-related drugs and to gauge patient sentiments about these drug treatments. They find that the identified ADRs are consistent with well-recognized toxicities.
The second main finding of this systematic review is that some studies also correctly identified ADRs that were previously unknown. In [68], the authors identified increased suicide risk for the drug Singulair, and an increased risk of acute pancreatitis and altered kidney function for the drug Byetta. In both cases, the FDA required the manufacturing companies to conduct an investigation, and update the labels of the products with a warning indicating these risks. This result suggests that NLP may also be used to identify novel ADRs, and it may serve as a suitable tool for pro-active and real-time health surveillance using remote sensing. As such, this automated system may identify trends and periodically report novel insights to policymakers and public health professionals, and it may support and enable these professionals to initiate interventions timely to protect public health and to maintain, and perhaps even increase, the quality of healthcare further [32].
Although this systematic review finds that the application of computational linguistics may be effective for pharmacovigilance, it does not suggest that the traditional system is obsolete and should be replaced by computational linguistics. Instead, it may be worthwhile to apply computational linguistics as a complementary tool to retrieve and process adverse drug reactions that end-users share on the Internet. This information and the insights may be combined with the adverse drug reactions that are reported by medical professionals, with the purpose to achieve a more complete overview of adverse drug reactions. Similarly, computational linguistics may be a suitable tool for the real-time monitoring of adverse drug reactions.
6. Limitations
The systematic literature search and study selection were performed by only one researcher. Therefore, it was not possible to establish inter-rater reliability. However, the process of study selection and the included studies were discussed by the authors until consensus was achieved. Nevertheless, it may be possible that this has introduced selection bias, but this could not be verified.
All studies that were included in this systematic review were found to have a medium quality. Quality was operationalized using reliability and validity. The process and assessment of the quality of the included studies were discussed by the authors until consensus was achieved.
It was observed that studies often failed to report information on the themes that were used to extract relevant information (see Appendix C). Consequently, the absence of these data limited the analyses of the studies with respect to their methodology, sample characteristics, and the utilized techniques. In addition, various publications failed to disclose information on the diseases that the identified ADRs were related to. We highly recommend authors disclose this information. This information has great value, among others, to establish the quality of these studies and to enable replicability, but it may also benefit the research community if the methodology and processes are explained in greater detail.
Because it is a commonality in the field of information technology and computational linguistics that findings are not always published in peer-reviewed journals, but instead it is often only published in conference proceedings or conference papers, both types of publications were included in this systematic review. This is important, because it may be possible that the process of peer-review is more rigorous when performed by journals compared to conferences. It was also observed that a significant number of included publications were not journal articles.
A common and unavoidable limitation of studies using user-generated content from social media (including all data sources listed in this review) is the inherent noisiness and bias of these data sources. In the context of the reviewed studies, users are usually unqualified to assess their symptoms, they might exaggerate mild or unrelated symptoms, they might just follow popular trends in criticism, or be biased or even malicious (e.g., seeking to discredit competition). These factors have to be taken into account when judging the effectiveness of the proposed tools.
As we discussed in the methods section, due to the interdisciplinary nature of the reviewed studies and their limitations, it was impossible to apply the complete PRISMA 2020 checklist in this systematic review. Therefore no registration was made in PROSPERO either. We acknowledge this as a limitation of this work.
7. Conclusions and Future Outlook
Our findings suggest that the user-generated textual content that drug users share on the Internet may have the potential to augment or enhance the expensive and time-consuming traditional system of pharmacovigilance. NLP may thus be used to automate the monitoring of ADRs using content that users publish to social media and other digital platforms [40]. This novel tool may not only contribute to improving public health and the quality of healthcare, but it could potentially also reduce the costs and processing time that are associated with conducting pharmacovigilance. Therefore, this tool may be a viable solution that addresses two of the most prominent challenges of traditional pharmacovigilance, namely the reduction of the high associated costs [7] and the inclusion of ADRs as experienced by the end-users [6]. It is strongly suggested for policymakers to consider the automated analysis of user-generated textual content for the purpose of pharmacovigilance, and to employ it ethically, responsibly, and with great respect to the privacy and anonymity of these drug users.
We acknowledge that we ought to be limited in describing the architecture of such tools for pharmacovigilance in the present paper. On an abstract level, this tool would subscribe to the Twitter API and filters Tweets based on keywords related to ADRs. Relevant Tweets are then subject to sentiment analysis and processed using NLP techniques. Based on the informational needs of researchers, further processing and analysis can be performed to extract key information on ADRs for medicines of interest and the related sentiment expressed by drug users.
Appendix A. PRISMA 2020 Checklist
Table A1.
PRISMA checklist.
| Section/Topic | # | Checklist Item | Reported on Page # |
|---|---|---|---|
| TITLE | |||
| Title | 1 | Identify the report as a systematic review, meta-analysis, or both. | 1 |
| ABSTRACT | |||
| Abstract | 2 | See the PRISMA 2020 for Abstracts checklist. | 1 |
| INTRODUCTION | |||
| Rationale | 3 | Describe the rationale for the review in the context of what is already known. | 1 |
| Objectives | 4 | Provide an explicit statement of the objective(s) or question(s) the review addresses. | 2 |
| METHOD | |||
| Eligibility criteria | 5 | Specify the inclusion and exclusion criteria for the review and how studies were grouped for the syntheses. | 5 |
| Information sources | 6 | Specify all databases, registers, websites, organisations, reference lists and other sources searched or consulted to identify studies. Specify the date when each source was last searched or consulted. | 3 |
| Search strategy | 7 | Present the full search strategies for all databases, registers and websites, including any filters and limits used. | 3, 19 |
| Selection process | 8 | Specify the methods used to decide whether a study met the inclusion criteria of the review, including how many reviewers screened each record and each report retrieved, whether they worked independently, and if applicable, details of automation tools used in the process. | 4 |
| Data collection process | 9 | Specify the methods used to collect data from reports, including how many reviewers collected data from each report, whether they worked independently, any processes for obtaining or confirming data from study investigators, and if applicable, details of automation tools used in the process. | 4 |
| Data items | 10a | List and define all outcomes for which data were sought. Specify whether all results that were compatible with each outcome domain in each study were sought (e.g. for all measures, time points, analyses), and if not, the methods used to decide which results to collect. | 4 |
| 10b | List and define all other variables for which data were sought (e.g. participant and intervention characteristics, funding sources). Describe any assumptions made about any missing or unclear information. | 4, 5 | |
| Study risk of bias assessment | 11 | Specify the methods used to assess risk of bias in the included studies, including details of the tool(s) used, how many reviewers assessed each study and whether they worked independently, and if applicable, details of automation tools used in the process. | 5 |
| Effect measures | 12 | Specify for each outcome the effect measure(s) (e.g. risk ratio, mean difference) used in the synthesis or presentation of results. | n. a. |
| Synthesis methods | 13a | Describe the processes used to decide which studies were eligible for each synthesis (e.g. tabulating the study intervention characteristics and comparing against the planned groups for each synthesis (item #5)). | 5 |
| 13b | Describe any methods required to prepare the data for presentation or synthesis, such as handling of missing summary statistics, or data conversions. | 6 | |
| 13c | Describe any methods used to tabulate or visually display results of individual studies and syntheses. | 6 | |
| 13d | Describe any methods used to synthesize results and provide a rationale for the choice(s). If meta-analysis was performed, describe the model(s), method(s) to identify the presence and extent of statistical heterogeneity, and software package(s) used. | 6 | |
| 13e | Describe any methods used to explore possible causes of heterogeneity among study results (e.g. subgroup analysis, meta-regression). | n. a. | |
| 13f | Describe any sensitivity analyses conducted to assess robustness of the synthesized results. | n. a. | |
| Reporting bias assessment | 14 | Describe any methods used to assess risk of bias due to missing results in a synthesis (arising from reporting biases). | n. a. |
| Certainty assessment | 15 | Describe any methods used to assess certainty (or confidence) in the body of evidence for an outcome. | 5 |
| RESULTS | |||
| Study selection | 16a | Describe the results of the search and selection process, from the number of records identified in the search to the number of studies included in the review, ideally using a flow diagram. | 4 |
| 16b | Cite studies that might appear to meet the inclusion criteria, but which were excluded, and explain why they were excluded. | n. a. | |
| Study characteristics | 17 | Cite each included study and present its characteristics. | 7 |
| Risk of bias within studies | 18 | Present assessments of risk of bias for each included study. | 20 |
| Results of individual studies | 19 | For all outcomes, present, for each study: (a) summary statistics for each group (where appropriate) and (b) an effect estimate and its precision (e.g. confidence/credible interval), ideally using structured tables or plots. | 9 |
| Results of syntheses | 20a | For each synthesis, briefly summarise the characteristics and risk of bias among contributing studies. | n. a. |
| 20b | Present results of all statistical syntheses conducted. If meta-analysis was done, present for each the summary estimate and its precision (e.g. confidence/credible interval) and measures of statistical heterogeneity. If comparing groups, describe the direction of the effect. | n. a. | |
| 20c | Present results of all investigations of possible causes of heterogeneity among study results. | n. a. | |
| 20d | Present results of all sensitivity analyses conducted to assess the robustness of the synthesized results. | n. a. | |
| Reporting biases | 21 | Present assessments of risk of bias due to missing results (arising from reporting biases) for each synthesis assessed. | 15 |
| Certainty of evidence | 22 | Present assessments of certainty (or confidence) in the body of evidence for each outcome assessed. | n. a. |
| DISCUSSION | |||
| Discussion | 23a | Provide a general interpretation of the results in the context of other evidence. | 14 |
| 23b | Discuss any limitations of the evidence included in the review. | 15 | |
| 23c | Discuss any limitations of the review processes used. | 15 | |
| 23d | Discuss implications of the results for practice, policy, and future research. | 16 | |
| OTHER INFORMATION | |||
| Registration and protocol | 24a | Provide registration information for the review, including register name and registration number, or state that the review was not registered. | 15 |
| 24b | Indicate where the review protocol can be accessed, or state that a protocol was not prepared. | 15 | |
| 24c | Describe and explain any amendments to information provided at registration or in the protocol. | 15 | |
| Support | 25 | Describe sources of financial or non-financial support for the review, and the role of the funders or sponsors in the review. | 16 |
| Competing interests | 26 | Declare any competing interests of review authors. | 16 |
| Availability of data, code and other materials | 27 | Report which of the following are publicly available and where they can be found: template data collection forms; data extracted from included studies; data used for all analyses; analytic code; any other materials used in the review. | n. a. |
Appendix B. Search Strategies
The systematic literature search was performed on 25 March 2020 for all databases. All publications appearing up to this point were considered in the search.
Appendix B.1. Search Strategy for PubMed
Block 1: Computational Linguistics
artificial intelligence[Title/Abstract] OR Artificial Intelligence[MeSH Terms] OR machine learning[Title/Abstract] OR Machine Learning[MeSH Terms] OR text mining[Title/ Abstract] OR computational linguistics[Title/Abstract] OR natural language processing[Title/ Abstract] OR Natural Language Processing[MeSH Terms] OR nlp[Title/Abstract] OR sentiment analysis[Title/Abstract] OR word embedding*[Title/Abstract] OR natural language toolkit[Title/Abstract] OR nltk[Title/Abstract]
Block 2: Health Surveillance
public health surveillance[Title/Abstract] OR Public Health Surveillance[MeSH Terms] OR health surveillance[Title/Abstract] OR public health monitoring[Title/Abstract] OR health monitoring[Title/Abstract]
Filters:
Article types: Journal Article
Languages: English
Appendix B.2. Search Strategy for Web of Science
Block 1: Computational Linguistics
TS=(artificial intelligence) OR TS=(machine learning) OR TS=(text mining) OR TS=(computational linguistics) OR TS=(natural language processing) OR TS=(nlp) OR TS=(sentiment analysis) OR TS=(word embedding*) OR TS=(natural language toolkit) OR TS=(nltk)
Block 2: Health Surveillance
TS=(public health surveillance) OR TS=(health surveillance) OR TS=(public health monitoring) OR TS=(health monitoring)
Filters:
Document types: Article, Proceedings Paper
Languages: English
Appendix B.3. Search Strategy for IEEE Xplore
Block 1: Computational Linguistics
“All Metadata”:artificial intelligence OR “All Metadata”:machine learning OR “All Metadata”:text mining OR “All Metadata”:computational linguistics OR “All Metadata”: natural language processing OR “All Metadata”:nlp OR “All Metadata”:sentiment analysis OR “All Metadata”:word embedding* OR “All Metadata”:natural language toolkit OR “All Metadata”:nltk
Block 2: Health Surveillance
“All Metadata”:public health surveillance OR “All Metadata”:health surveillance OR “All Metadata”:public health monitoring OR “All Metadata”:health monitoring
Filters:
Document types: Conferences, Journals
Appendix B.4. Search Strategy for ACM Digital Library
Block 1: Computational Linguistics
artificial intelligence OR machine learning OR text mining OR computational linguistics OR natural language processing OR nlp OR sentiment analysis OR word embedding* OR natural language toolkit OR nltk
Block 2: Health Surveillance
public health surveillance OR health surveillance OR public health monitoring OR health monitoring
Filters:
Searched within: Abstract, Author Keyword, Title All publications: Proceedings, Research Article
Appendix C. Characteristics of Publications
Table A2.
Characteristics of publications included in the analysis.
| Authors | Type of Drugs | Drugs Studied | Data Source | Sample Size | Users | Unique Users | Origin of Users | Average Number of Followers | Years of Data Collection | Horizon of Data Collection | Software Used | Techniques and Classifiers Used | Outcome | Result | Description of Result | Reliability | Validity |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| [31] | HIV | Atripla, Sustiva, Truvada |
Social media | Twitter.com: 1642 tweets |
HIV-infected persons undergoing drug treatment | 512 (male: 247; female: 83; unknown: 182) |
Canada, South Africa, United Kingdom, United States (New York City, San Fransisco) |
2300 | 2010, 2011, 2012, 2013 | 3 years | Toolkit for Multivariate Analysis | Artificial Neural Networks (ANN), Boosted Decision Trees with AdaBoost (BDT), Boosted Decision Trees with Bagging (BDTG), Sentiment Analysis, Support Vector Machines (SVM) |
Reported ADRs for HIV treatment | Positive | Reported adverse effects are consistent with well-recognized toxicities. | Medium | Medium |
| [32] | Depression | Citalopram, Chlorpromazine, Cyclobenzaprine, Dulexetine, Promethazine |
Forums | Depression Forums.org: 7726 posts |
Unknown | Unknown | Unknown | Unknown | 2004–2014 | 10 years | General Architecture for Text Engineering (GATE), NLTK Toolkit within MATLAB, RapidMiner |
Hyperlink-Induced Topic Search (HITS), k-Means Clustering, Network Analysis, Term-Frequency-Inverse Document Frequency (TF-IDF) |
User sentiment on depression drugs | Positive | Natural language processing is suitable to extract information on ADRs concerning depression. | Medium | Medium |
| [66] | Cancer | Avastin, Melphalan, Rupatadin, Tamoxifen, Taxotere |
Social media | Twitter.com: 2,102,176,189 tweets |
Unknown | Unknown | Unknown | Unknown | 2009, 2010 |
1 year | Apache Lucene | MetaMap, Support Vector Machines (SVM) |
Reported ADRs for cancer | Neutral | Classification models had limited performance. Adverse events related to cancer drugs can potentially be extracted from tweets. | Medium | Medium |
| [33] | Unknown | Unknown | Social media | Twitter.com: 6528 tweets |
Unknown | Unknown | Unknown | Unknown | Unknown | Unknown | GENIA tagger, Hunspell, Snowball stemmer, Stanford Topic Modelling Toolbox, Twokenizer |
Backward/forward sequential feature selection (BSFS/FSFS) algorithm, k-Means Clustering, Sentiment Analysis, Support Vector Machines (SVM) |
Reported ADRs | Positive | ADRs were identified reasonably well. | Medium | Medium |
| [34] | Unknown | Unknown | Social media | Twitter.com: 32,670 tweets |
Unknown | Unknown | Unknown | Unknown | Unknown | Unknown | Hunspell, Twitter tokenizer |
- Term Frequency-Inverse Document Frequency (TF-IDF) | Reported ADRs | Neutral | ADRs were not very well identified. | Medium | Medium |
| [67] | Unknown | 65 drugs | Social media | Twitter.com: 10,822 tweets |
Unknown | Unknown | Unknown | Unknown | Unknown | Unknown | Unknown | Naive Bayes (NB), Natural Language Processing (NLP), Support Vector Machines (SVM) |
Reported ADRs | Positive | ADRs were identified good. | Medium | Medium |
| [35] | Unknown | Unknown | Drug reviews | Drugs.com, Drugslib.com: 218,614 reviews |
Unknown | Unknown | Unknown | Unknown | Unknown | Unknown | BeautifulSoup | Logistic Regression, Sentiment Analysis |
Patient satisfaction with drugs, Reported ADRs, Reported effectiveness of drugs |
Positive | Classification results were very good. | Medium | Medium |
| [36] | Unknown | 103 drugs | Social media | Twitter.com: 172,800 tweets |
Unknown | 864 | Unknown | Unknown | 2014, 2015 |
1 year | Twitter4J | Decision Trees, Medical Profile Graph, Natural Language Processing (NLP) |
Reported ADRs | Positive | Building a medical profile of users enables the accurate detection of adverse drug events. | Medium | Medium |
| [37] | Unknown | Unknown | Social media | Twitter.com 1245 tweets |
Unknown | Unknown | Unknown | Unknown | Unknown | Unknown | CRF++ Toolkit, GENIA tagger, Hunspell, Twitter REST API, Twokenizer |
Natural Language Processing (NLP) | Reported ADRs | Positive | ADRs were identified reasonably well. | Medium | Medium |
| [38] | Cancer | 146 drugs | Drug reviews | WebMD.com: Unknown |
Unknown | Unknown | Unknown | Unknown | Unknown | Unknown | SentiWordNet, WordNet |
Sentiment Analysis, Support Vector Machines (SVM), Term document Matrix (TDM) |
User sentiment on cancer drugs | Positive | Sentiment on ADRs was identified reasonably well. | Medium | Medium |
| [39] | Unknown | 81 drugs | Drug reviews, Social media |
DailyStrength.org: 6279 reviews, Twitter.com: 1784 tweets |
Unknown | Unknown | Unknown | Unknown | Unknown | Unknown | Unknown | ARDMine, Lexicon-based, MetaMap, Support Vector Machines (SVM) |
Reported ADRs | Positive | ADRs were identified very well. | Medium | Medium |
| [40] | Asthma, Cystic fibrosis, Rheumatoid arthritis, Type 2 diabetes |
Albuterol, Azithromycin, Bromocriptine, Insulin, Ipratropium, Ivacaftor, Meloxicam, Metformin, Prednisone, Sulfasalazine |
Drug reviews, Social media |
PatientsLikeMe.com: 796 reviews, Twitter.com: 39,127 tweets, WebMD.com: 2567 reviews, YouTube.com: 42,544 comments |
Unknown | Unknown | Unknown | Unknown | 2014 | Not applicable | Deeply Moving | Unknown | Patient-reported medication outcomes | Positive | Social media serves as a new data source to extract patient-reported medication outcomes. | Medium | Medium |
| [68] | Unknown | Medications.com: 168 drugs, SteadyHealth.com: 316 drugs |
Forums | Medications.com: 8065 posts, SteadyHealth.com: 11,878 |
Unknown | Unknown | Unknown | Unknown | 2012 | Not applicable | Java Hidden Markov Model library, jsoup |
Hidden Markov Model (HMM), Natural Language Processing (NLP) |
Reported ADRs | Positive | Reported adverse effects are consistent with well-recognized side-effects. | Medium | Medium |
| [69] | Unknown | ACE inhibitors, Bupropion, Ibuprofen, Morphine, Paroxetine, Rosiglitazone, Warfarin |
Electronic Health Record (EHR) | 25,074 discharge summaries | Unknown | Unknown | Unknown | Unknown | 2004 | Not applicable | MedLEE | Unknown | Reported ADRs | Positive | Reported adverse effects are consistent with well-recognized toxicities (recall: 75%; precision: 31%). | Medium | Medium |
| [41] | Unknown | Baclofen, Duloxetine, Gabapentin, Glatiramer, Pregabalin |
Social media | Twitter.com: 3251 tweets |
Unknown | Unknown | Unknown | Unknown | 2015 | Not applicable | AFINN, Bing Liu sentiment words, Multi-Perspective Question Answering (MPQA), SentiWordNet, TextBlob, Tweepy, WEKA |
MetaMap, Naive Bayes (NB), Natural Language Processing (NLP), Sentiment Analysis, Support Vector Machines (SVM) |
Reported ADRs | Positive | Several well-known ADRs were identified. | Medium | Medium |
| [70] | Unknown | Adenosine, Biaxin, Cialis, Elidel, Lansoprazole, Lantus, Luvox, Prozac, Tacrolimus, Vyvanse |
Forums | MedHelp.com: 6244 discussion threads |
Unknown | Unknown | Unknown | Unknown | Unknown | Unknown | Unknown | Association Mining | Reported ADRs | Positive | ADRs were identified. | Medium | Medium |
Author Contributions
Study design: P.P. and M.L.; implementation: P.P.; analysis: P.P.; writing and editing: P.P. and A.B. All authors have read and agreed to the published version of the manuscript.
Funding
Parts of this work have been funded by the Applied AI Digital Innovation Hub North project, funded by the European Regional Development Fund.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The resources used in this systematic review are available from the first author.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Santoro A., Genov G., Spooner A., Raine J., Arlett P. Promoting and Protecting Public Health: How the European Union Pharmacovigilance System Works. Drug Saf. 2017;40:855–869. doi: 10.1007/s40264-017-0572-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barlow J. Managing Innovation in Healthcare. World Scientific; London, UK: 2017. [Google Scholar]
- 3.WHO Policy Perspectives on Medicines—Pharmacovigilance: Ensuring the Safe Use of Medicines. WHO Policy Perspectives on Medicines, World Health Organization; Geneva, Switzerland: 2004. [Google Scholar]
- 4.Strengthening Pharmacovigilance to Reduce Adverse Effects of Medicines. European Commission; Brussels, Belgium: 2008. [Google Scholar]
- 5.Beninger P. Pharmacovigilance: An Overview. Clin. Ther. 2018;30:1991–2004. doi: 10.1016/j.clinthera.2018.07.012. [DOI] [PubMed] [Google Scholar]
- 6.Pharmacovigilance. Volume 2020 European Commission; Brussels, Belgium: 2020. [Google Scholar]
- 7.Schmider J., Kumar K., LaForest C., Swankoski B., Naim K., Caubel P.M. Innovation in Pharmacovigilance: Use of Artificial Intelligence in Adverse Event Case Processing. Clin. Pharmacol. Ther. 2019;105:954–961. doi: 10.1002/cpt.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaplan A.M., Haenlein M. Users of the world, unite! The challenges and opportunities of Social Media. Bus. Horiz. 2010;53:59–68. doi: 10.1016/j.bushor.2009.09.003. [DOI] [Google Scholar]
- 9.Monkman G.F., Kaiser M.J., Hyder K. Text and data mining of social media to map wildlife recreation activity. Biol. Conserv. 2018;228:89–99. doi: 10.1016/j.biocon.2018.10.010. [DOI] [Google Scholar]
- 10.Moro S., Rita P., Vala B. Predicting social media performance metrics and evaluation of the impact on brand building: A data mining approach. J. Bus. Res. 2016;69:3341–3351. doi: 10.1016/j.jbusres.2016.02.010. [DOI] [Google Scholar]
- 11.Brown R.C., Bendig E., Fischer T., Goldwich A.D., Baumeister H., Plener P.L. Can acute suicidality be predicted by Instagram data? Results from qualitative and quantitative language analyses. PLoS ONE. 2019;14:e0220623. doi: 10.1371/journal.pone.0220623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cavazos-Rehg P.A., Zewdie K., Krauss M.J., Sowles S.J. “No High Like a Brownie High”: A Content Analysis of Edible Marijuana Tweets. Am. J. Health Promot. 2018;32:880–886. doi: 10.1177/0890117116686574. [DOI] [PubMed] [Google Scholar]
- 13.Cesare N., Dwivedi P., Nguyen Q.C., Nsoesie E.O. Use of social media, search queries, and demographic data to assess obesity prevalence in the United States. Palgrave Commun. 2019;5:106. doi: 10.1057/s41599-019-0314-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cole-Lewis H., Pugatch J., Sanders A., Varghese A., Posada S., Yun C., Schwarz M., Augustson E. Social Listening: A Content Analysis of E-Cigarette Discussions on Twitter. J. Med. Internet Res. 2015;17:e243. doi: 10.2196/jmir.4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ji X., Chun S.A., Wei Z., Geller J. Twitter sentiment classification for measuring public health concerns. Soc. Netw. Anal. Min. 2015;5:13. doi: 10.1007/s13278-015-0253-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kavuluru R., Ramos-Morales M., Holaday T., Williams A.G., Haye L., Cerel J., ACM Classification of Helpful Comments on Online Suicide Watch Forums; Proceedings of the 7th Acm International Conference on Bioinformatics, Computational Biology, and Health Informatics; Seattle, WA, USA. 2–5 October 2016; New York, NY, USA: Association for Computing Machinery; 2016. pp. 32–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kent E.E., Prestin A., Gaysynsky A., Galica K., Rinker R., Graff K., Chou W.Y.S. “Obesity is the New Major Cause of Cancer”: Connections Between Obesity and Cancer on Facebook and Twitter. J. Cancer Educ. 2016;31:453–459. doi: 10.1007/s13187-015-0824-1. [DOI] [PubMed] [Google Scholar]
- 18.Kim A., Miano T., Chew R., Eggers M., Nonnemaker J. Classification of Twitter Users Who Tweet About E-Cigarettes. JMIR Public Health Surveill. 2017;3:e63. doi: 10.2196/publichealth.8060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kostygina G., Tran H., Shi Y., Kim Y., Emery S. “Sweeter Than a Swisher”: Amount and themes of little cigar and cigarillo content on Twitter. Tob. Control. 2016;25:i75–i82. doi: 10.1136/tobaccocontrol-2016-053094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee K., Agrawal A., Choudhary A. Mining social media streams to improve public health allergy surveillance; Proceedings of the 2015 IEEE/ACM International Conference on Advances in Social Networks Analysis and Mining (ASONAM); Paris, France. 25–28 August 2015; pp. 815–822. [DOI] [Google Scholar]
- 21.Leis A., Ronzano F., Mayer M.A., Furlong L.I., Sanz F. Detecting Signs of Depression in Tweets in Spanish: Behavioral and Linguistic Analysis. J. Med. Internet Res. 2019;21:16. doi: 10.2196/14199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mowery D., Smith H., Cheney T., Stoddard G., Coppersmith G., Bryan C., Conway M. Understanding Depressive Symptoms and Psychosocial Stressors on Twitter: A Corpus-Based Study. J. Med. Internet Res. 2017;19:e48. doi: 10.2196/jmir.6895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Myslin M., Zhu S.H., Chapman W., Conway M. Using twitter to examine smoking behavior and perceptions of emerging tobacco products. J. Med. Internet Res. 2013;15:e174. doi: 10.2196/jmir.2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nargund K., Natarajan S. Public health allergy surveillance using micro-blogs; Proceedings of the 2016 International Conference on Advances in Computing, Communications and Informatics (ICACCI); Jaipur, India. 21–24 September 2016; pp. 1429–1433. [DOI] [Google Scholar]
- 25.Phan N., Chun S.A., Bhole M., Geller J. Enabling Real-Time Drug Abuse Detection in Tweets; Proceedings of the 2017 IEEE 33rd International Conference on Data Engineering; San Diego, CA, USA. 19–22 April 2017; New York, NY, USA: IEEE; 2017. pp. 1510–1514. [DOI] [Google Scholar]
- 26.Rong J., Michalska S., Subramani S., Du J., Wang H. Deep learning for pollen allergy surveillance from twitter in Australia. BMC Med. Inform. Decis. Mak. 2019;19:208. doi: 10.1186/s12911-019-0921-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song J.Y., Song T.M., Seo D.C., Jin J.H. Data Mining of Web-Based Documents on Social Networking Sites That Included Suicide-Related Words Among Korean Adolescents. J. Adolesc. Health. 2016;59:668–673. doi: 10.1016/j.jadohealth.2016.07.025. [DOI] [PubMed] [Google Scholar]
- 28.Tran T., Nguyen D., Nguyen A., Golen E. Sentiment Analysis of Marijuana Content via Facebook Emoji-Based Reactions; Proceedings of the 2018 IEEE International Conference on Communications (ICC); Kansas City, MO, USA. 20–24 May 2018; pp. 1–6. [DOI] [Google Scholar]
- 29.Ward P.J., Rock P.J., Slavova S., Young A.M., Bunn T.L., Kavuluru R. Enhancing timeliness of drug overdose mortality surveillance: A machine learning approach. PLoS ONE. 2019;14:e0223318. doi: 10.1371/journal.pone.0223318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zucco C., Calabrese B., Cannataro M. Sentiment analysis and affective computing for depression monitoring; Proceedings of the 2017 IEEE International Conference on Bioinformatics and Biomedicine (BIBM); Kansas City, MO, USA. 13–16 November 2017; pp. 1988–1995. [DOI] [Google Scholar]
- 31.Adrover C., Bodnar T., Huang Z., Telenti A., Salathé M. Identifying Adverse Effects of HIV Drug Treatment and Associated Sentiments Using Twitter. JMIR Public Health Surveill. 2015;1:10. doi: 10.2196/publichealth.4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Akay A., Dragomir A., Erlandsson B.E., Akay M. Assessing anti-depressants using intelligent data monitoring and mining of online fora. IEEE J. Biomed. Health Inform. 2016;20:977–986. doi: 10.1109/JBHI.2016.2539972. [DOI] [PubMed] [Google Scholar]
- 33.Dai H.J., Touray M., Jonnagaddala J., Syed-Abdul S. Feature Engineering for Recognizing Adverse Drug Reactions from Twitter Posts. Information. 2016;7:27. doi: 10.3390/info7020027. [DOI] [Google Scholar]
- 34.Dai H.J., Wang C.K. Classifying Adverse Drug Reactions from Imbalanced Twitter Data. Int. J. Med. Inform. 2019;129:122–132. doi: 10.1016/j.ijmedinf.2019.05.017. [DOI] [PubMed] [Google Scholar]
- 35.Gräßer F., Kallumadi S., Malberg H., Zaunseder S. Aspect-Based Sentiment Analysis of Drug Reviews Applying Cross-Domain and Cross-Data Learning; Proceedings of the 2018 International Conference on Digital Health, DH ’18; Lyon, France. 23–26 April 2018; New York, NY, USA: Association for Computing Machinery; 2018. pp. 121–125. [DOI] [Google Scholar]
- 36.Katragadda S., Karnati H., Pusala M., Raghavan V., Benton R. Detecting Adverse Drug Effects Using Link Classification on Twitter Data; Proceedings of the 2015 IEEE International Conference on Bioinformatics and Biomedicine (BIBM); Washington, DC, USA. 9–12 November 2015; pp. 675–679. [DOI] [Google Scholar]
- 37.Lin W.S., Dai H.J., Jonnagaddala J., Chang N.W., Jue T.R., Iqbal U., Shao J.Y.H., Chiang I.J., Li Y.C. Utilizing different word representation methods for twitter data in adverse drug reactions extraction; Proceedings of the 2015 Conference on Technologies and Applications of Artificial Intelligence (TAAI); Tainan, Taiwan. 20–22 November 2015; pp. 260–265. [DOI] [Google Scholar]
- 38.Mishra A., Malviya A., Aggarwal S. Towards Automatic Pharmacovigilance: Analysing Patient Reviews and Sentiment on Oncological Drugs; Proceedings of the 2015 IEEE 15th International Conference on Data Mining Workshops; Atlantic City, NJ, USA. 14–17 November 2015; pp. 1402–1409. [DOI] [Google Scholar]
- 39.Nikfarjam A., Sarker A., O’Connor K., Ginn R., Gonzalez G. Pharmacovigilance from social media: Mining adverse drug reaction mentions using sequence labeling with word embedding cluster features. J. Am. Med. Inform. Assoc. 2015;22:671–681. doi: 10.1093/jamia/ocu041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ru B., Harris K., Yao L. A Content Analysis of Patient-Reported Medication Outcomes on Social Media; Proceedings of the 2015 IEEE International Conference on Data Mining Workshop; Atlantic City, NJ, USA. 14–17 November 2015; pp. 472–479. [DOI] [Google Scholar]
- 41.Wu L., Moh T.S., Khuri N. Twitter Opinion Mining for Adverse Drug Reactions; Proceedings of the 2015 IEEE International Conference on Big Data (Big Data); Santa Clara, CA, USA. 29 October–1 November 2015; pp. 1570–1574. [DOI] [Google Scholar]
- 42.Mikalef P., Pappas I.O., Krogstie J., Pavlou P.A. Big data and business analytics: A research agenda for realizing business value. Inf. Manag. 2020;57:103237. doi: 10.1016/j.im.2019.103237. [DOI] [Google Scholar]
- 43.Tekin M., Etlioğlu M., Koyuncuoğlu Ö., Tekin E. Data Mining in Digital Marketing. In: Durakbasa N.M., Gencyilmaz M.G., editors. Proceedings of the 2018 International Symposium for Production Research; Vienna, Austria. 28–31 August 2018; Cham, Switzerland: Springer International Publishing; 2018. pp. 44–61. [Google Scholar]
- 44.Wall J., Krummel T. The digital surgeon: How big data, automation, and artificial intelligence will change surgical practice. J. Pediatr. Surg. 2020;55:47–50. doi: 10.1016/j.jpedsurg.2019.09.008. [DOI] [PubMed] [Google Scholar]
- 45.Gandomi A., Haider M. Beyond the hype: Big data concepts, methods, and analytics. Int. J. Inf. Manag. 2015;35:137–144. doi: 10.1016/j.ijinfomgt.2014.10.007. [DOI] [Google Scholar]
- 46.Zeng Z., Shi H., Wu Y., Hong Z. Survey on Natural Language Processing Techniques in Bioinformatics. Comput. Math. Methods Med. 2015;2015:674296. doi: 10.1155/2015/674296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iglesias J.A., Tiemblo A., Ledezma A., Sanchis A. Web news mining in an evolving framework. Inf. Fusion. 2016;28:90–98. doi: 10.1016/j.inffus.2015.07.004. [DOI] [Google Scholar]
- 48.Kennedy H., Elgesem D., Miguel C. On fairness: User perspectives on social media data mining. Converg. Int. J. Res. New Media Technol. 2015;23:270–288. doi: 10.1177/1354856515592507. [DOI] [Google Scholar]
- 49.Szpakowicz S., Feldman A., Kazantseva A. Editorial: Computational Linguistics and Literature. Front. Digit. Humanit. 2018;5:1–2. doi: 10.3389/fdigh.2018.00024. [DOI] [Google Scholar]
- 50.Jurafsky D., Martin J.J. Speech and Language Processing: An Introduction to Natural Language Processing, Computational Linguistics, and Speech Recognition. 3rd ed. Stanford University; Stanford, CA, USA: 2019. [Google Scholar]
- 51.Rizk A., Elragal A. Data science: Developing theoretical contributions in information systems via text analytics. J. Big Data. 2020;7:7. doi: 10.1186/s40537-019-0280-6. [DOI] [Google Scholar]
- 52.Liao S.H., Chu P.H., Hsiao P.Y. Data mining techniques and applications—A decade review from 2000 to 2011. Expert Syst. Appl. 2012;39:11303–11311. doi: 10.1016/j.eswa.2012.02.063. [DOI] [Google Scholar]
- 53.Sunikka A., Bragge J. Applying text-mining to personalization and customization research literature—Who, what and where? Expert Syst. Appl. 2012;39:10049–10058. doi: 10.1016/j.eswa.2012.02.042. [DOI] [Google Scholar]
- 54.Aggarwal C.C., Zhai C. Mining Text Data. Springer; New York, NY, USA: 2012. A Survey of Text Clustering Algorithms; pp. 77–128. [Google Scholar]
- 55.Lu Y., Zhang P., Liu J., Li J., Deng S. Health-Related Hot Topic Detection in Online Communities Using Text Clustering. PLoS ONE. 2013;8:e56221. doi: 10.1371/journal.pone.0056221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sadiq A.T., Abdullah S.M. Hybrid Intelligent Techniques for Text Categorization. Int. J. Adv. Comput. Sci. Appl. 2014;2:23–40. [Google Scholar]
- 57.Tan A. Text Mining: The State of the Art and the Challenges; Proceedings of the PAKDD 1999 Workshop on Knowledge Discovery from Advanced Databases; Beijing, China. 26–28 April 1999; pp. 65–70. [Google Scholar]
- 58.Nassirtoussi A.K., Aghabozorgi S., Wah T.Y., Ngo D.C.L. Text mining for market prediction: A systematic review. Expert Syst. Appl. 2014;41:7653–7670. doi: 10.1016/j.eswa.2014.06.009. [DOI] [Google Scholar]
- 59.Oberreuter G., Velsquez J.D. Text mining applied to plagiarism detection: The use of words for detecting deviations in the writing style. Expert Syst. Appl. 2013;40:3756–3763. doi: 10.1016/j.eswa.2012.12.082. [DOI] [Google Scholar]
- 60.Liberati A., Altman D.G., Tetzlaff J., Mulrow C., Gøtzsche P.C., Ioannidis J.P.A., Clarke M., Devereaux P.J., Kleijnen J., Moher D. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: Explanation and elaboration. BMJ. 2009;339:b2700. doi: 10.1136/bmj.b2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moher D., Liberati A., Tetzlaff J., Altman D.G., The P.G. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. PLoS Med. 2009;6:e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Giustini D., Boulos M.N.K. Google Scholar is not enough to be used alone for systematic reviews. Online J. Public Health Inform. 2013;5:214. doi: 10.5210/ojphi.v5i2.4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bramer W.M., Giustini D., de Jonge G.B., Holland L., Bekhuis T. De-duplication of database search results for systematic reviews in EndNote. J. Med. Libr. Assoc. JMLA. 2016;104:240–243. doi: 10.3163/1536-5050.104.3.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kampmeijer R., Pavlova M., Tambor M., Golinowska S., Groot W. The use of e-health and m-health tools in health promotion and primary prevention among older adults: A systematic literature review. BMC Health Serv. Res. 2016;16:290. doi: 10.1186/s12913-016-1522-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hsieh H.F., Shannon S.E. Three Approaches to Qualitative Content Analysis. Qual. Health Res. 2005;15:1277–1288. doi: 10.1177/1049732305276687. [DOI] [PubMed] [Google Scholar]
- 66.Bian J., Topaloglu U., Yu F. Towards Large-Scale Twitter Mining for Drug-Related Adverse Events; Proceedings of the 2012 International Workshop on Smart Health and Wellbeing, SHB ’12; Maui, HI, USA. 29 October 2012; New York, NY, USA: Association for Computing Machinery; 2012. pp. 25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ginn R., Pimpalkhute P., Nikfarjam A., Patki A., O’Connor K., Sarker A., Smith K., Gonzalez G. Mining Twitter for adverse drug reaction mentions: A corpus and classification benchmark; Proceedings of the Fourth Workshop on Building and Evaluating Resources for Health and Biomedical Text Processing; Reykjavik, Iceland. 31 May 2014; pp. 1–8. [Google Scholar]
- 68.Sampathkumar H., Chen X.W., Luo B. Mining Adverse Drug Reactions from online healthcare forums using Hidden Markov Model. BMC Med. Inform. Decis. Mak. 2014;14:91. doi: 10.1186/1472-6947-14-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang X., Hripcsak G., Markatou M., Friedman C. Active Computerized Pharmacovigilance Using Natural Language Processing, Statistics, and Electronic Health Records: A Feasibility Study. J. Am. Med. Inform. Assoc. 2009;16:328–337. doi: 10.1197/jamia.M3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang C.C., Yang H., Jiang L., Zhang M. Social Media Mining for Drug Safety Signal Detection; Proceedings of the 2012 International Workshop on Smart Health and Wellbeing, SHB ’12; Maui, HI, USA. 29 October 2012; New York, NY, USA: Association for Computing Machinery; 2012. pp. 33–40. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The resources used in this systematic review are available from the first author.