The elements that mediate VanA glycopeptide resistance in enterococci are heterogeneous. Twenty-four distinct VanA elements were identified among a collection of enterococci isolated from hospital patients and nonhuman sources in the United Kingdom (9). Despite their diversity, all these elements showed extensive homology with the prototype VanA transposon, Tn1546, from Enterococcus faecium BM4147 (2); specifically, the vanRSHAX genes, which are critical for the expression of glycopeptide resistance, were conserved in all 24 elements (8).
We recently identified a blood culture isolate of E. faecium (designated ARMRL 26) in which the vanRSHAX gene cluster was distinct from that observed in all the other United Kingdom strains examined. The strain yielded a long PCR (L-PCR) amplicon larger than the expected size of 4.4 kb (8), and digestion of this amplicon with DdeI indicated loss of the 418-bp fragment present in Tn1546 digests and the appearance of a new fragment of ca. 1 kb (Fig. 1). As the 418-bp fragment results from digestion of Tn1546 at nucleotide positions 5382 and 5800, the loss of this fragment suggested an intragenic insertion located towards the 3′ end of vanS. Sequencing detected a copy of the insertion sequence IS1216V (1) flanked by 8-bp direct repeats of GCTTCCAG (corresponding to Tn1546 nucleotides 5761 to 5768) and consistent with target site duplication following a transposition event. Insertion at this position would be predicted to cause the loss of 11 amino acids (AMPDLVDKRRS) from the C terminus of the VanS peptide and their possible replacement by 10 amino acids (GFCCKVLBKE) resulting from read-through of the inserted IS1216V sequence. It is unlikely that this change would affect the function of the VanS sensor peptide, because the critical residues remain intact (7). Even if the insertion did cause a functional change, VanS is not essential for expression of glycopeptide resistance (3). Strain ARMRL 26 had a normal VanA phenotype (vancomycin and teicoplanin MICs of >32 μg/ml).
FIG. 1.
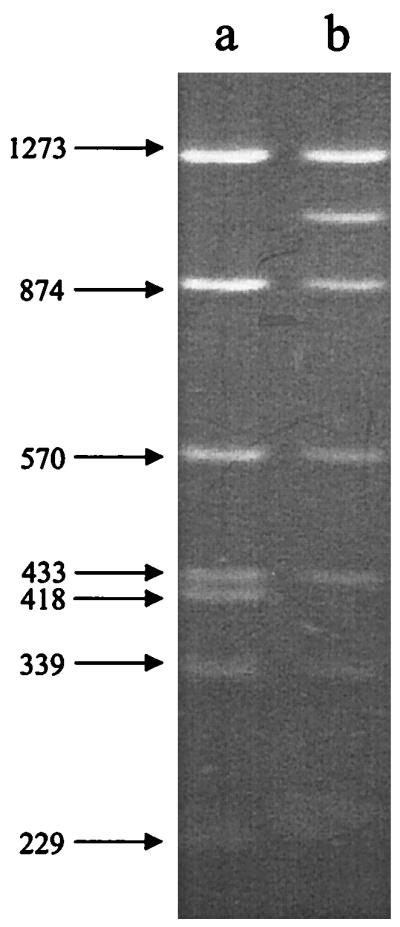
Digestion with DdeI of vanRSHAX L-PCR amplicons derived from E. faecium BM4147 containing Tn1546 (lane a) or E. faecium ARMRL 26 (lane b). Sizes are shown in base pairs.
To our knowledge, E. faecium ARMRL 26 is only the second glycopeptide-resistant enterococcus in which an insertion has been identified within a van gene; disruption of vanY by IS1476 has been reported previously (6). L-PCR has been stated previously to be a useful method for fingerprinting and comparing glycopeptide resistance elements for epidemiological and evolutionary purposes (4, 8). This report further emphasizes the ability of L-PCR to identify rapidly enterococci that have insertions within the vanRSHAX gene cluster. It would therefore detect readily strains that carry copies of IS1251 in the vanS-vanH intergenic region which have been documented in the United States (5). Furthermore, restriction fragment length polymorphism analysis of the resulting amplicons can be used to pinpoint the likely position of the insertion and so permit relevant sequencing, allowing cost-effective analysis of VanA elements to be undertaken.
Acknowledgments
A. Darini was funded by a grant from the Fundacao de Amparo a Pesquisa do Estado de Sao Paulo, Brazil.
REFERENCES
- 1.Arthur M, Depardieu F, Gerbaud G, Galimand M, Leclercq R, Courvalin P. The VanS sensor negatively controls VanR-mediated transcriptional activation of glycopeptide resistance genes in Tn1546 and related elements in the absence of induction. J Bacteriol. 1997;179:97–106. doi: 10.1128/jb.179.1.97-106.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arthur M, Molinas C, Depardieu F, Courvalin P. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol. 1993;175:117–127. doi: 10.1128/jb.175.1.117-127.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arthur M, Molinas C, Courvalin P. The VanS-VanR two-component regulatory system controls synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol. 1992;174:2582–2591. doi: 10.1128/jb.174.8.2582-2591.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haaheim H, Dahl K H, Simonsen G S, Olsvik O, Sundsfjord A. Long PCRs of transposons in the structural analysis of genes encoding acquired glycopeptide resistance in enterococci. BioTechniques. 1998;24:432–437. doi: 10.2144/98243st02. [DOI] [PubMed] [Google Scholar]
- 5.Handwerger S, Skoble J, Discotto L F, Pucci M J. Heterogeneity of the vanA gene cluster in clinical isolates of enterococci from the northeastern United States. Antimicrob Agents Chemother. 1995;39:362–368. doi: 10.1128/aac.39.2.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.MacKinnon M G, Drebot M A, Tyrell G J. Identification and characterization of IS1476, an insertion sequence-like element that disrupts VanY function in a vancomycin-resistant Enterococcus faecium strain. Antimicrob Agents Chemother. 1997;41:1805–1807. doi: 10.1128/aac.41.8.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morel-Deville F, Fauvel F, More P. Two-component signal-transducing systems involved in stress responses and vancomycin susceptibility in Lactobacillus sakei. Microbiology. 1998;144:2873–2883. doi: 10.1099/00221287-144-10-2873. [DOI] [PubMed] [Google Scholar]
- 8.Palepou M-F I, Adebiyi A-M A, Tremlett C H, Jensen L B, Woodford N. Molecular analysis of diverse elements mediating VanA glycopeptide resistance in enterococci. J Antimicrob Chemother. 1998;42:605–612. doi: 10.1093/jac/42.5.605. [DOI] [PubMed] [Google Scholar]
- 9.Woodford N, Adebiyi A-M A, Palepou M-F I, Cookson B D. Diversity of VanA glycopeptide resistance elements in enterococci from humans and nonhuman sources. Antimicrob Agents Chemother. 1998;42:502–508. doi: 10.1128/aac.42.3.502. [DOI] [PMC free article] [PubMed] [Google Scholar]


