Rohaim and colleagues present a strategy to generate synthetic recombinant antibody fragments (sFabs) for structural biology and studies of K+ channels. These sFabs perform comparably to traditional Fabs but are easier to produce.
Abstract
Engineered antibody fragments (Fabs) have made major impacts on structural biology research, particularly to aid structural determination of membrane proteins. Nonetheless, Fabs generated by traditional monoclonal technology suffer from challenges of routine production and storage. Starting from the known IgG paratopes of an antibody that binds to the “turret loop” of the KcsA K+ channel, we engineered a synthetic Fab (sFab) based upon the highly stable Herceptin Fab scaffold, which can be recombinantly expressed in Escherichia coli and purified with single-step affinity chromatography. This synthetic Fab was used as a crystallization chaperone to obtain crystals of the KcsA channel that diffracted to a resolution comparable to that from the parent Fab. Furthermore, we show that the turret loop can be grafted into the unrelated voltage-gated Kv1.2–Kv2.1 channel and still strongly bind the engineered sFab, in support of the loop grafting strategy. Macroscopic electrophysiology recordings show that the sFab affects the activation and conductance of the chimeric voltage-gated channel. These results suggest that straightforward engineering of antibodies using recombinant formats can facilitate the rapid and scalable production of Fabs as structural biology tools and functional probes. The impact of this approach is expanded significantly based on the potential portability of the turret loop to a myriad of other K+ channels.
Introduction
A highly successful strategy to elucidate the structures of a number of high-value classes of biomolecular molecules and complexes, especially in the case of membrane proteins, has been the use of “crystallization chaperones” (Iwata et al., 1995; Kovari et al., 1995; Ostermeier et al., 1995; Li et al., 1997; Hunte and Michel, 2002). Chaperones promote crystallization by reducing conformational heterogeneity, by masking hydrophobic surfaces, increasing solubility, and by providing primary contact points between molecules in the crystal lattice. These chaperones can take a variety of forms (reviewed in Koide [2009]), but the most prevalent type is antibody fragments (Fabs), which initially had been produced by traditional hybridoma technology (Zhou et al., 2001). However, producing them by traditional hybridoma-based technology has major drawbacks—the process is inherently slow and expensive. The resulting Fab chaperones can be unstable and moreover, there is no way to improve the chaperone’s binding or the solubility properties. More recently, Fab-based chaperones have been produced by molecular display technology (phage, yeast, or RNA display; reviewed in Koide [2009]). These methods exploit the modular form of the Fab module, which combines a rigid and conserved fold with a set of hypervariable loops (complementary-determining regions or CDRs) that confer the antibody’s specificity and binding affinity. The display methods employ large libraries that introduce diversity into the CDR loops of the Fab and rely on screening to isolate high-affinity binders from a large pool of nonbinders (Bradbury and Marks, 2004; Sidhu and Fellouse, 2006; Fellouse et al., 2007; Koenig and Fuh, 2014). The advantages of the display methods are that they are inexpensive, the selection process can be accomplished relatively quickly and importantly, they result in a recombinant protein that can be easily expressed and further engineered, for instance, to trap a desired conformation or stabilize a protein complex (Paduch and Kossiakoff, 2017).
While the Fab generation process is quickly evolving to favor recombinant approaches, there are a number of mAb-derived Fabs that remain important reagents for structure and function studies of particular systems. A case in point is the mAb (mouse IgG) against KcsA expressed from mouse hybridoma cells that were originally used to determine the x-ray crystallographic structure of the KcsA–Fab complex at high resolution (Zhou et al., 2001). Despite the inherent drawbacks, structural studies of KcsA (WT and mutants) have largely continued to rely on the original monoclonal Fab obtained from hybridoma cells (Lenaeus et al., 2005; Cordero-Morales et al., 2006; Valiyaveetil et al., 2006; Lockless et al., 2007; Cuello et al., 2010; Cuello et al., 2017; Rohaim et al., 2020). Although it remains a workhorse for this system, the production of the KcsA–Fab has remained expensive and time-consuming, with additional issues associated with a multistep purification and long-term storage. The Fab fragment of the IgG is obtained via papain proteolysis following a series of ion-exchange chromatography steps. To circumvent these steps, a recombinant synthetic Fab that binds KcsA with high specificity was designed using a CDR grafting strategy. The objective was to convert the Fab binding to KcsA to a recombinant format to facilitate the process of expression and purification without the loss of function.
Upon examination of the KcsA–Fab complex (Rohaim et al., 2020), the CDRs involved in the specific interaction were transferred to the Herceptin Fab scaffold, which has been highly characterized and has a known structure (Eigenbrot et al., 1993). The Herceptin scaffold is a modified Human Kappa isotype. Changes in the Fab light chain were previously introduced to improve its expression and stability to enhance its crystallization capacity (Slezak et al., 2020). The 3D conformation of the design was then verified using ROSETTA. We show that the newly designed synthetic Fab can readily be expressed in Escherichia coli and purified with a single-step affinity purification. As a demonstration of the suitability of the strategy for crystallographic studies, the crystal structure of sFab–KcsA complex was solved at 2.7 Å resolution. Based on this, we believe that our engineering strategy can easily be expanded in a general way to convert other monoclonal Fabs into recombinant formats, thereby endowing them with improved properties for structural and functional studies. Further, we show that the engineered KcsA sFab tightly binds to the KcsA turret loop sequence that was grafted into a related voltage-gated K+ channel, effectively inducing a significant functional response. This suggests that the turret loop can function as a portable binding site for the sFab–KcsA, which can be exploited for structure–function studies across a wide range of channels that share the same general tertiary characteristics as KcsA. Thus, the engineered sFab described here can be used as a powerful tool for both structural and functional studies.
Materials and methods
Synthetic Fab design
The wtFab conformation was obtained from the crystal structures in PDB accession nos. 1K4C and 6W0I. Structure-based homology modeling was performed with the ROSETTA algorithm using the webserver gui (robetta.bakerlab.org; Song et al., 2013). Comparative homology modeling function was selected using PDB accession no. 5UEA as the template. The UNIPROT alignment algorithm was used for sequence alignment and analysis (https://uniprot.org/align/). The final homology model was visualized, and the rmsd was estimated in PYMOL. The DNA sequence encoding the designed sFab was synthesized by Integrated DNA Technologies. The synthesized gene was cloned into the bacterial expression vector pSFV4 for recombinant antibody bacterial expression in periplasm (from Structural Genomics Consortium, Toronto), harboring an N-terminal PelB signaling peptide for periplasmic expression (Denoncin and Collet, 2013).
Expression and purification
The sFab expression vector was transformed into BL21 cells. A small-scale culture in LB medium supplemented with 0.1 mg/ml ampicillin was incubated overnight at 37°C. 5 ml of overnight culture was used to inoculate 1 liter of LB medium supplemented with 0.1 mg/ml ampicillin and then incubated at 37°C. At OD600 = 0.6, the expression was induced by adding 1 mM IPTG at 37°C for 3 h. The cells were collected by centrifugation for 40 min at 6,000 g and resuspended in lysis buffer (50 mM Tris pH 8.0, 200 mM NaCl, 1 mM PMSF, and 20 μg/ml of DNase I). The cells were lyzed by homogenization using an Avestin Emulsiflex C5. The cell lysate was clarified by centrifugation for 30 min at 30,000 g, loaded on a protein G column (Bailey et al., 2014), and incubated with agitation for 1 h at 4°C. The protein G column was washed with five column volumes of wash buffer (50 mM Tris, pH 7.5, 200 mM NaCl). The sFab was eluted with three column volumes of elution buffer (0.1 M Glycine, pH 2.6) and neutralized by adding 1 M Tris pH 7.5 directly to the sample to a final concentration of 0.1 M Tris pH 7.5. The yield of sFab was 4 mg/liter. KcsA in pQE32 vector (Rohaim et al., 2020) was transformed into XL-1 Blue cells in LB media supplemented with 0.1 mg/ml ampicillin and grown at 37°C until reaching an OD600 of 0.8–1.0, after which expression was induced by adding 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG). After 3 h, cell pellets were collected and resuspended in lysis buffer (20 mM Tris, pH 8.0, 0.2 M NaCl) supplemented with 1 mM PMSF and 20 μg/ml of DNase I. The resuspended cells were lysed using an Avestin Emulsiflex C5, and the membranes were collected by centrifugation at 100,000 g at 4°C for 1 h. The membrane pellet was resuspended in 20 mM Tris, pH 7.5, 0.15 M KCl and solubilized by incubation with 40 mM DM (n-decyl-β-D-maltoside) overnight at 4°C. The sample was centrifuged at 100,00 g for 1 h, and the supernatant was loaded on a HisTrap FF 5 ml nickel metal affinity column previously equilibrated with buffer A (20 mM Tris, pH 7.5, 0.15 M KCl, 5 mM DM, and 20 mM imidazole) and eluted with buffer B (buffer A + 300 mM imidazole). The C-terminal tail was removed by incubation with 1 μg/ml chymotrypsin for 1 h at room temperature. The chymotrypsin-treated protein was further purified by size exclusion chromatography (SEC) using a Superdex 200 HR 10/30 column with an AKTA FPLC system (GE Healthcare) in buffer C (50 mM Tris, pH 7.5, 0.15 M KCl, and 5 mM DM).
Crystallization and structure determination
The KcsA peak fraction was incubated with sFab at 20% molar excess of sFab to ensure saturated complex formation. SEC was repeated to remove excess Fabs using Superdex 200 HR 10/30 in buffer C. The peak fractions were analyzed on 4–20% SDS-PAGE and then the sample was concentrated to 14 mg/ml prior to crystallization in a hanging drop vapor diffusion format. Crystals appeared after 24 h at room temperature in 50 mM sodium acetate (or ammonium acetate) pH 5.5, 50 mM magnesium acetate, and 25–27% PEG 400. The crystals were flash-frozen in 40% PEG solution. X-ray diffraction datasets were collected at the NECAT 24-ID-E beamline at the Advanced Photon Source. Crystal structures were determined by molecular replacement method using the structures of the sFab homology model and one monomer of KcsA as a search model using PHASER (McCoy et al., 2007). The structure refinements were performed using REFMAC (Murshudov et al., 1997). The electron density map and sFab–KcsA models were inspected using COOT (Emsley and Cowtan, 2004). Mapping of the amino acid interaction was performed using CONTACT (Winn et al., 2011). The 3-D structure prediction of KvKcsA chimera was performed using AlphaFold (Jumper et al., 2021) with the amino acid sequence ...AE(RGAPGAQLIS)IP..., where the KcsA turret is in parentheses.
Electrophysiology
The full-length, chimeric Kv1.2/2.1 (KvWT) cDNA from Rattus norvegicus, obtained from the MacKinnon lab at Rockefeller University, was cloned into the pMax vector. The cDNA of the turret loop (ADERDSQFP) was deleted from KvWT chimera and replaced with cDNA of the KcsA turret loop (RGAPGAQLIS) using site-directed mutagenesis to produce the KvKcsA chimera. Mutations were confirmed by Sanger sequencing. Mutations were introduced via site-directed mutagenesis and confirmed by Sanger sequencing to produce the KvKcsA chimera. cRNA was synthesized using a T7 RNA expression Kit (Ambion, Invitrogen). Approximately 24 h after surgical removal from adult frogs, in accordance with animal usage protocol 71475 of the University of Chicago Institutional Animal Care and Use Committee, 50–100 ng cRNA in 50 nl RNase-free water was injected into enzymatically defolliculated oocytes. Oocytes were maintained at 18°C in a standard oocyte solution (SOS), a solution containing 10 mM HEPES, pH 7.5, 100 mM NaCl, 5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, and 50 μg ml−1 gentamycin. Macroscopic currents were recorded 24–36 h postinjection on a two-electrode voltage clamp (TEVC) setup, comprising an OC-720C amplifier (Warner Instruments), Digidata 1322A 16-bit digitizer (Axon Instruments), and a Windows XP PC running Clampex10.3. Oocytes were impaled with two 3 M KCl-filled Ag/AgCl electrodes with resistances in the range 0.2–1.0 MΩ in a bath containing SOS. More than five recordings were obtained for both KvWT and KvKcsA, each from a different oocyte and some from different injection sessions to test for the effect of sFab on each construct’s conductance. K+ currents were evoked by voltage steps of 2.5 s, going from −60 to +70 mV in 10-mV steps. The holding potential was set at −90 mV, but the data acquisition protocol varied slightly between KvWT and KvKcsA. For KvWT, holding of −90 mV for 0.35 s was followed by variable pulse for 0.8 s, recovery at −140 mV for 0.5 s, and then back to −90 mV for 0.85 s for a total of 2.5 s. For KvKcsA, holding of −90 mV for 0.2 s was followed by a variable pulse for 1.6 s, and then back to −90 mV for 0.7 s for a total of 2.5 s. The isochronal tail currents were measured in isopotential condition after the decay of the oocyte linear capacitive response for the I–V curve.
Online supplemental material
In Fig. S1, the full-length sequences of sFab and wtFab are described. In addition, Table S1 summarizes the x-ray diffraction and crystal structure statistics.
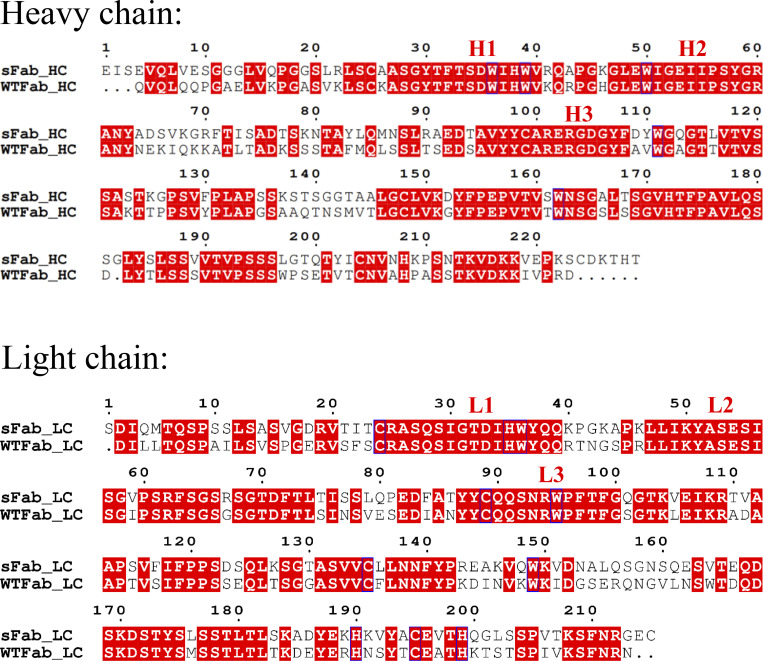
Figure S1.
Full-length alignment of sFab and wtFab for the heavy and light chain. The regions highlighted in red indicates similarity between the sFab and the wtFab. The complementarity-determining regions (CDRs) are indicated as H1, H2, and H3 for the heavy chain, and L1, L2, and L3 for the light chain.
Results
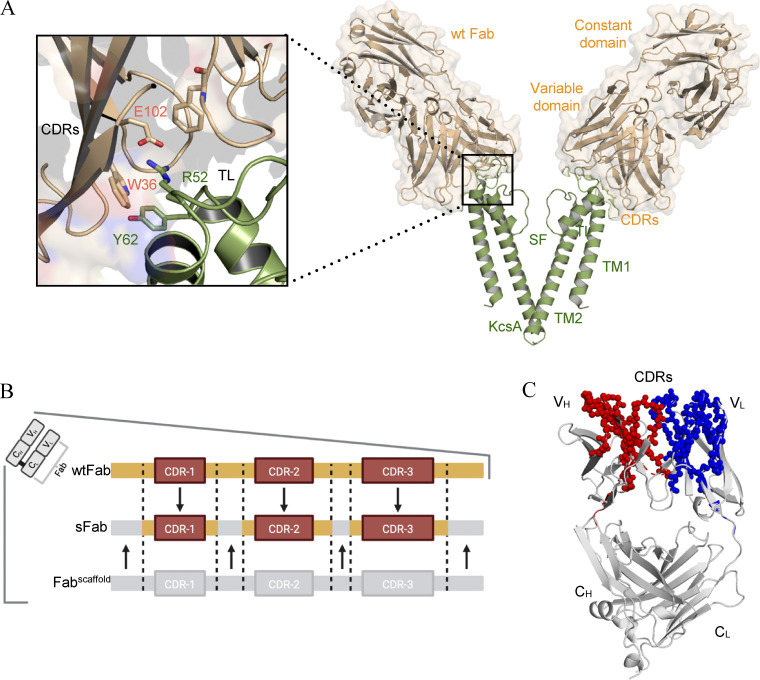
The Fab fragment of the monoclonal mouse Ig-γ (IgG) developed against KcsA is obtained by papain proteolysis following a series of ion-exchange chromatography steps. In a first attempt to circumvent the onerous expression in hybridoma cells, the mouse Fab heavy and light chains were cloned into a recombinant bacterial expression vector. However, this approach was not viable for structural studies, as this construct expressed poorly and was unstable during purification. Fig. 1 A shows the interaction of the CDRs of IgG-Fab (here after called wtFab) with the turret loop of KcsA (residues R52-Y62). Based on this analysis, a synthetic Fab with the wtFab paratope was designed by a computer-assisted loop grafting strategy. Sequences which matched the CDRs amino acid composition (see Fig. S1), length, and flanking regions of the interaction interface of wtFab were grafted into a human Herceptin Fab scaffold (Slezak et al., 2020). Other parts of the sFab, not harboring the CDRs, were slightly modified to enhance stability and the crystallization capability of the sFab (Fig. 1 B). This yielded an overall ∼40% sequence similarity with the wtFab, with most of the similarities occurring in the Fab–KcsA interface of the variable domain (Fig. 1 C). To validate the positions of the CDRs and the overall fold of the variable and constant regions, the final model was compared with wtFab using comparative ROSETTA modeling. The structure-based homology model of sFab is in good agreement with the wtFab with an overall backbone rmsd of 0.4 Å (Fig. 1 C).
Figure 1.
Structural analysis of the wtFab–KcsA crystal structure. (A) Side view (right) of only two diagonally opposing monomers of KcsA for clarity (green), showing the transmembrane helices (TM1, TM2), selectivity filter (SF), and the wtFab (gold) interacting near the turret loop (TL). A closeup view of the interaction interface (left) showing the interaction of the CDRs with residues in the TL; the crystal structure is obtained from PDB accession no. 1K4C. (B) Design of the sFab based on the human Herceptin Fab scaffold (Fabscaffold) showing the heavy and/or light CDRs (orange) necessary for restoring binding to KcsA. (C) ROSETTA homology modeling of the sFab (gray). Amino acid residues of high similarity with wtFab are shown as spheres in red and blue for variable heavy (VH) and light (VL) domains, respectively.
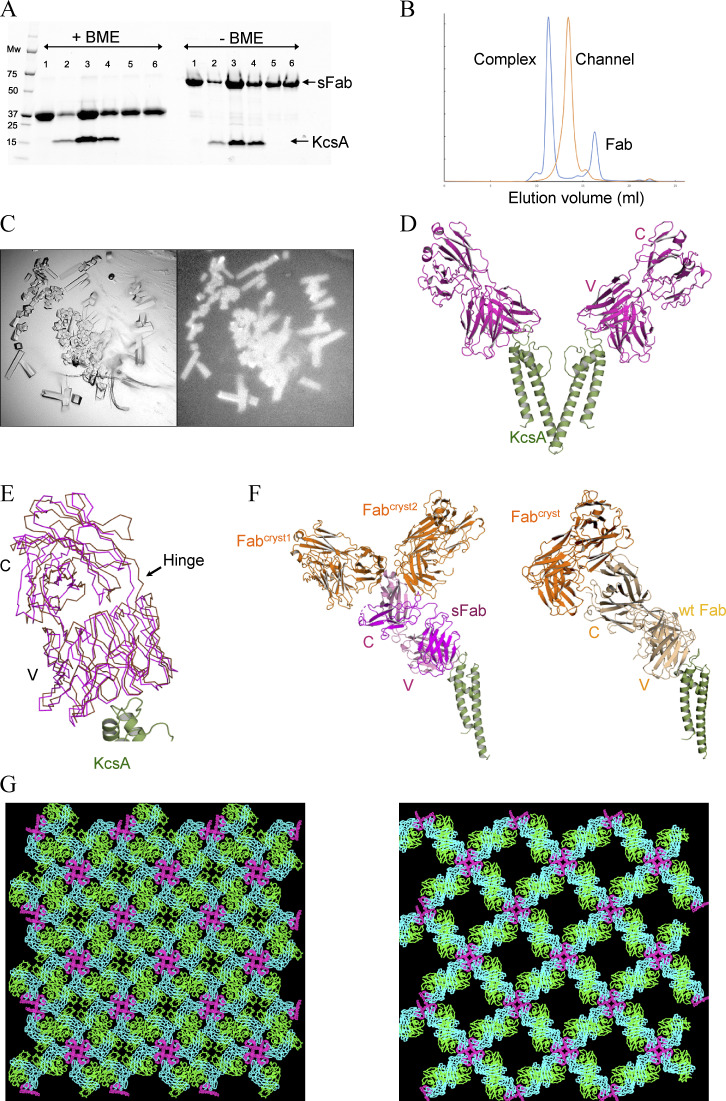
DNA encoding the engineered sFab was synthesized and subsequently cloned in pSFV4 for recombinant expression in E. coli. To ensure proper native-like folding, the expressed sFab was targeted to the periplasmic space (see Materials and methods). The expressed sFab was purified by a single step protein-G affinity column (Bailey et al., 2014) after which the final eluted sample was neutralized to pH ∼7.5. The yield was 4 mg/liter. The purity of the sFab was assessed by SDS-PAGE showing a single band at the expected mol wt (Fig. 2 A). To investigate the binding properties, the KcsA channel and sFab were mixed at equimolar ratios, and the protein–Fab complex formation was analyzed by analytical SEC (Fig. 2 B). A decrease in the retention volume of SEC was observed, indicative of a stably formed KcsA–sFab complex. The binary complex was analyzed by SDS-PAGE, showing single bands at the expected molecular weights for KcsA and sFab, respectively (Fig. 2 A). To test the ability of the sFab to promote crystallization, the purified protein complex was crystallized using similar conditions as for the wtFab complex. Protein crystals appeared after 24 h at room temperature (faster than with the original wtFab) and were confirmed by tryptophan fluorescence (Fig. 2 C). The crystal structure was solved at 2.7 Å resolution by molecular replacement method with an ensemble of sFab and KcsA monomers used as a search model (Fig. 2 D). Maps having well-defined electron density for the KcsA and sFab monomers were observed covering most of the regions of the models of the binary complex. Model building and refinement improved the overall quality of the electron density maps but lacked density at the distal loop areas of the constant light (CL) and heavy (CH) chain regions of the sFab, which is a common feature of Fab–antigen crystal structures. Notably, the KcsA–sFab complex crystallized in the same space group (I4) as reported for its wtFab counterpart and reflect the channel’s inherent fourfold symmetry. However, the unit cell dimensions of the KcsA–sFab crystal form are significantly shorter in two dimensions, which leads to a reduction of ∼20% in the unit cell volume. This results in a tighter packing arrangement with a significant lower solvent content than the wtFab crystal form (56% versus 67%; Table S1). Structural superposition revealed that the sFab deviates from the wtFab at the constant domain with a slight counterclockwise rotation about the hinge regions (Fig. 2 E). Examining the crystal lattice packing, the CL domain of the sFab in one asymmetric unit interacts with two sFab (Fabcryst1,2) molecules from nearby symmetry mates. This is in contrast with wtFab where its CL interacts with only one molecule, Fabcryst (Fig. 2 F). The interaction explains the crystallization of the two complexes in the same space group, but with different unit cells and lattice packing properties (Fig. 2 G). It is probable that the differences in some of the surface residues in the CL domains of wtFab versus sFab lead to the altered lattice packing that differentiates into the two crystal forms.
Figure 2.
Expression, purification, and structural determination of the sFab–KcsA complex. (A) An affinity pulldown SDS-PAGE showing the association between KcsA and sFab with (left) and without (right) β-mercaptoethanol (BME). Lane 1: sFab, lane 2–6: sFab–KcsA complex; excess fab is shown in lane 5 and 6 from SEC. (B) Comparison of the analytical size-exclusion chromatograms for the apo (orange) and complex form of KcsA with sFab (blue), with ∼2 ml shift of the elution volume to the left upon complex formation. (C) Crystals of the sFab–KcsA complex visualized under visible (left) and UV (right) light microscope obtained at room temperature after 24 h. (D) The crystal structure of sFab–KcsA where the sFab (magenta) binds at the turret loop of KcsA (green), showing two diagonally opposing monomers for clarity (PDB accession no. 7MDJ). (E) Crystal lattice packing of the sFab–KcsA, structural analysis of the sFab (magenta), and wtFab (gold) showing the positional deviation around the constant domain. (F) Comparison of the interaction between Fabs in crystallographic symmetry mates (Fabcryst, in orange) showing the asymmetric unit of sFab (left) and wtFab (right). (G) Comparison of the crystal lattice from the new sFab–KcsA on the left with that from wtFab–KcsA (PDB accession no. 1K4C) on the right.
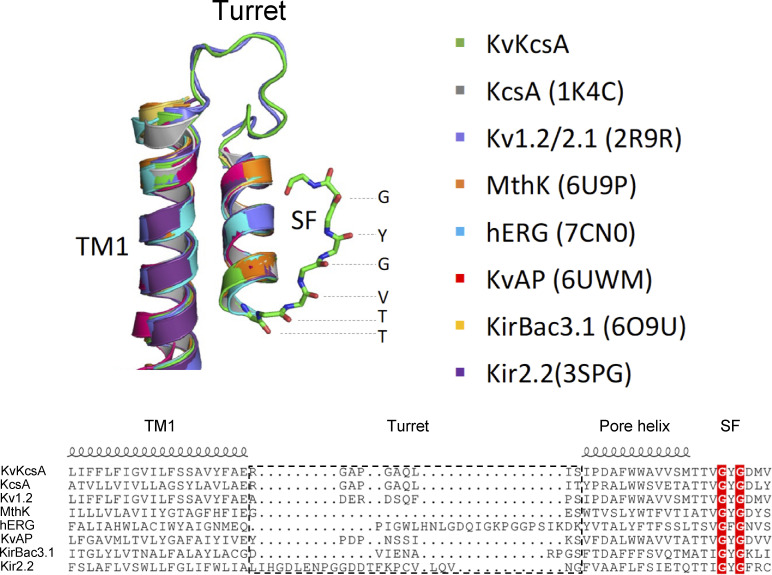
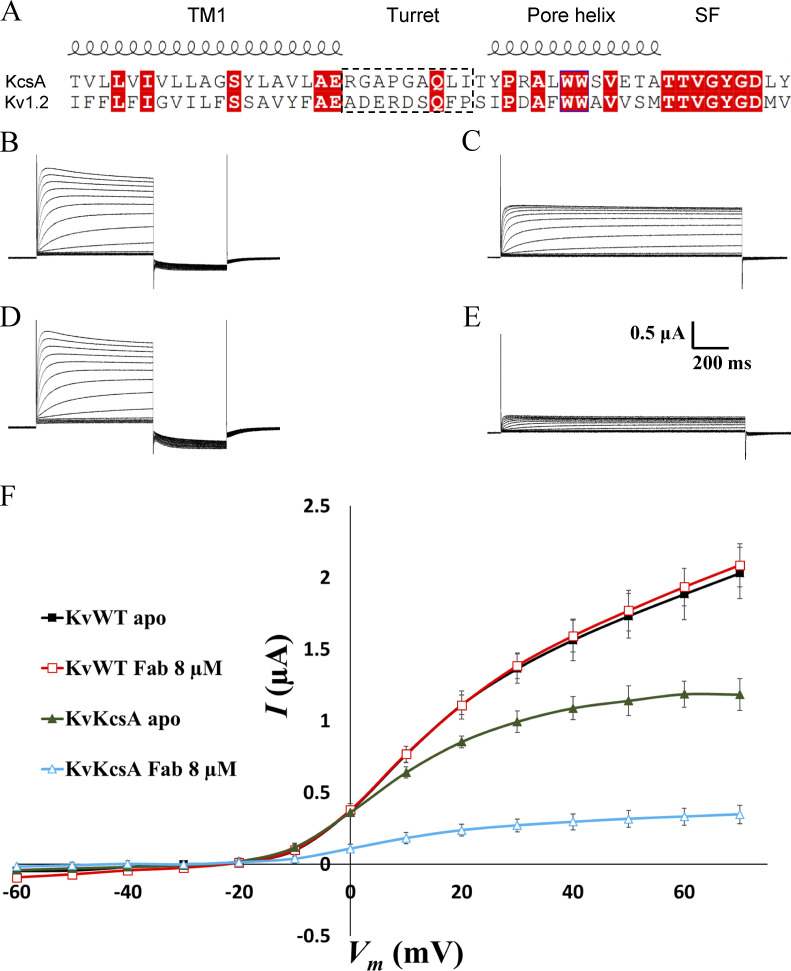
We sought to determine whether the sFab could serve as a useful tool in functional and structural studies of other K+ channels. To address this question, we superimposed the structures of KcsA and seven familiar K+ channels. As depicted in Fig. 3, there is a remarkable structural similarity among K+ channels, suggesting that porting the KcsA turret loop may be an effective strategy in most cases. Specifically, the turret is located between the outer helix (TM1) and the pore helix in the pore domain. Since the relative orientation of the outer and pore helices is highly conserved among K+ channels, one may imagine that the substitution of the KcsA turret (RGAPGAQLI) could preserve its conformation and its ability to bind the sFab with high affinity. Based on the high structural similarity, it is reasonable to envision that such simple protein engineering could yield an effective strategy to study a variety of K+ channels. To test this hypothesis, a voltage-gated K+ channel chimera was designed in which the turret loop of the mammalian Kv1.2/2.1 channel was swapped with that of the KcsA channel and referred to as KvKcsA. Using the amino acid sequence, the 3-D structure prediction of KvKcsA was determined using the AlphaFold structure prediction engine (Jumper et al., 2021; Fig. 3). Subsequently, the predicted structure of KvKcsA was superimposed to the seven homologous K+ channels as shown in Fig. 3. Overall, the predicted tertiary structure of the KvKcsA channel was identical to the the homologous K+ channels with the turret loop adopting a conformation similar to the KcsA turret loop (Fig. 3). We chose to use electrophysiological measurements in Xenopus laevis oocytes to examine the interaction of the sFab with the KvKcsA channel, a convenient technique allowing for a rapid functional assay. For clarity, the sequence alignment of the pore domain of the KvKcsA channel with that of the KcsA channel is shown in Fig. 4 A. As a control, electrophysiological measurements were also performed on the unmodified Kv1.2/2.1 chimera (referred to as the KvWT). Macroscopic currents were recorded in Xenopus oocytes using pulses from −60 to +70 mV at 10 mV increments (Fig. 4, B–F). It was observed that the addition of sFab strongly affects the voltage-dependent activation of the KvKcsA construct, but not of the KvWT channel. The presence of sFab had virtually no effect on the voltage-dependent activation and conductance of KvWT (Fig. 4, B and C). Conversely, binding of sFab dramatically reduced the voltage-dependent activation of the KvKcsA construct (Fig. 4, D and E), reducing its conductance to ∼26% (Fig. 4 F). Washout of the sFab for up to 30 min resulted in a conductance recovery up to only ∼37% of the original apo state (data not shown), indicating that sFab binds tightly to the turret loop of the KvKcsA construct. The functional assay shows that the sFab is able to bind to the KcsA turret substituted in another K+ channel. From a functional perspective, these observations are intriguing because the Fab fragment from the original mAb (mouse IgG) is known to reduce the amount of inactivation observed in macroscopic recordings of the WT KcsA channel (Cordero-Morales et al., 2006). At this point, one cannot determine whether sFab promotes inactivation or some other nonconductive state of the KvKcsA construct. Further studies will be required to draw more definitive conclusions about the functional impact of the sFab on K+ channels.
Figure 3.
Structural alignment and modeling illustrating the high structural similarity of different K+ channels. The conformation of the pore and outer helices (TM1) is highly conserved among different K+ channels. The upper panel illustrates the superposition of the TM1-pore helix motif for eight homologous K+ channels with their PDB accession nos. in parentheses except for the predicted structure of KvKcsA (in green). For clarity, the turret loops of KvKcsA and KcsA only are shown in ribbon representation. The lower panel shows the structure-based sequence alignment for the same channels illustrating the modularity of the respective domains: first transmembrane helix (TM1), P-loop (or pore helix), and selectivity filter (SF). The turret loop is framed in an orange box.
Figure 4.
Functional effect of the sFab on the voltage-gated Kv1.2–Kv2.1 chimera channel (KvWT) and on a construct in which the Kv1.2–Kv2.1 turret was substituted by that of KcsA (KvKcsA). (A) Sequence alignment of the pore domain of the Kv1.2–Kv2.1 channel with that of the KcsA channel. The turret interacting with the sFab is indicated by a dashed box. (B–E) Macroscopic currents of the full-length KvWT channel and of the KvKcsA construct recorded in Xenopus oocytes using pulses from −60 to +70 mV at 10 mV increments to test the functional impact of the sFab (n = 10). (B and C) KvWT current without sFab (B) and with 8 µM sFab (C). (D and E) KvKcsA construct current without sFab (D) and with 8 µM sFab (E). (F) I–V curve for the different experiments showing the effect of sFab on the conductance of the KvWT and KvKcsA channels.
Discussion
With the availability of synchrotron sources and advanced structure determination software, the success of determining macromolecule structures by x-ray diffraction is often limited to challenges related to obtaining ordered crystals diffracting at high resolution. Numerous early studies showed the promise of using antibody-based reagents as effective crystallization chaperones (Ostermeier et al., 1995; Zhou et al., 2001; Hunte and Michel, 2002), and building on these successes, the approach has transformed the field of membrane structural biology (Uysal et al., 2009; Li et al., 2014; Borowska et al., 2015; Mateja et al., 2015; Stuwe et al., 2015). Analysis of these structures indicates that Fabs improve crystal structures via stabilization of certain protein conformations or by forming crystal lattice contacts that permit more productive crystal packing opportunities. More recently, Fabs have also been employed in myriad single particle cryo-EM studies to stabilize certain conformations and increase the overall size of small protein molecules (Wu et al., 2012; Dominik et al., 2016; Paduch and Kossiakoff, 2017). Nonetheless, Fabs produced by monoclonal technology remain a challenge to produce since they are obtained from animal immunizations and require proteolytic digestion of full Igs. To circumvent these problems, we developed a simple computational methodology for converting a Fab from monoclonal to recombinant form.
An sFab that targets the turret loop of the KcsA channel was designed based on grafting the CDR loops from the parent monoclonal onto the Herceptin Fab scaffold modified for structural studies. The overall conformation and the CDRs of the sFab were then verified using ROSETTA. The sFab can be expressed in E. coli in a form that can be directly used for structural and functional studies, thus, eliminating the time-consuming and costly steps required for the generation and purification of Fabs from monoclonal antibodies. From a practical point of view, the described methodology can guide the rapid production of recombinant Fabs needed for structural studies in a cost-effective manner. The sFab could be used to successfully obtain ordered crystals of the KcsA channel diffracting to a good resolution. Importantly, the sFab is clearly not a simple copy of the original wtFab, as reflected by the significant changes in the water content and unit cell dimensions of the crystals (Fig. 2).
An important adjunct for the development of sFab for structural studies of KcsA is the finding that the turret loop, which serves as the sFab’s docking site, is structurally conserved across a number of homologous potassium channels (Fig. 3). Thus, it can potentially be exploited as a docking module that can be coupled with sFab to facilitate structure determination of a number of related K+ channels by x-ray crystallography as a crystallization chaperone. Additionally, because K+ channels are generally too small for cryo-EM, the turret loop sFab could be employed as a fiducial marker to increase the size of the particle. Recently, a similar approach of using Fabs targeting portable structure motifs have been successfully utilized as crystallization chaperones and/or a fiducial markers for small membrane proteins (Dutka et al., 2019; Mukherjee et al., 2020; Bloch et al., 2021; McIlwain et al., 2021) Furthermore, the observation that the sFab inhibits a voltage-gated channel construct, in which the binding KcsA turret loop was substituted (Fig. 4), is very encouraging. While these results were obtained with only one system, they suggest that the combination of the sFab and turret loop engineering could provide a useful platform for rapid functional and structural studies of a variety of K+ channels.
Supplementary Material
summarizes data collection and refinement statistics for sFab–KcsA.
Acknowledgments
Crina M. Nimigean served as editor.
The authors are grateful to Eduardo Perozo for his insight and support.
This research was supported by the National Institutes of Health (NIH) through grant R0-GM062342 (B. Roux) and R0-GM117372 (A.A. Kossiakoff). This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences from the National Institutes of Health (P30 GM124165). The Eiger 16M detector on 24-ID-E is funded by a NIH-ORIP (Office of Research Infrastructure Programs) HEI grant (S10OD021527). This research used resources of the Advanced Photon Source; a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. An allocation on the Blue Waters computer at NCSA from the National Science Foundation (NSF) was supported through grant PRAC-1640888, and the Beagle computer was supported in part by NIH through resources provided by the Computation Institute and the Biological Sciences Division of the University of Chicago and Argonne National Laboratory, under grant 1S10OD018495-01.
The authors declare no competing financial interests.
Authors contributions: A. Rohaim and T. Slezak designed the sFab, solved the crystal structure, and carried out homology modeling. L. Blachowicz and A. Rohaim performed cloning and purification. Y.H. Koh performed electrophysiology. A.A. Kossiakoff and B. Roux designed and supervised the research.
Data availability
All data that support the method described in this study is provided in the manuscript and by the corresponding author upon request. All the structures are deposited and found in the PDB under accession code 7MDJ.
References
- Bailey, L.J., Sheehy K.M., Hoey R.J., Schaefer Z.P., Ura M., and Kossiakoff A.A.. 2014. Applications for an engineered protein-G variant with a pH controllable affinity to antibody fragments. J. Immunol. Methods. 415:24–30. 10.1016/j.jim.2014.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch, J.S., Mukherjee S., Kowal J., Filippova E.V., Niederer M., Pardon E., Steyaert J., Kossiakoff A.A., and Locher K.P.. 2021. Development of a universal nanobody-binding Fab module for fiducial-assisted cryo-EM studies of membrane proteins. Proc. Natl. Acad. Sci. USA. 118:e2115435118. 10.1073/pnas.2115435118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowska, M.T., Dominik P.K., Anghel S.A., Kossiakoff A.A., and Keenan R.J.. 2015. A YidC-like protein in the archaeal plasma membrane. Structure. 23:1715–1724. 10.1016/j.str.2015.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury, A.R., and Marks J.D.. 2004. Antibodies from phage antibody libraries. J. Immunol. Methods. 290:2910–2949. 10.1016/j.jim.2004.04.007 [DOI] [PubMed] [Google Scholar]
- Cordero-Morales, J.F., Cuello L.G., Zhao Y.X., Jogini V., Cortes D.M., Roux B., and Perozo E.. 2006. Molecular determinants of gating at the potassium-channel selectivity filter. Nat. Struct. Mol. Biol. 13:311–318. 10.1038/nsmb1069 [DOI] [PubMed] [Google Scholar]
- Cuello, L.G., Cortes D.M., and Perozo E.. 2017. The gating cycle of a K+ channel at atomic resolution. eLife. 6:e28032. 10.7554/eLife.28032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuello, L.G., Jogini V., Cortes D.M., and Perozo E.. 2010. Structural mechanism of C-type inactivation in K+ channels. Nature. 466:203–208. 10.1038/nature09153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denoncin, K., and Collet J.F.. 2013. Disulfide bond formation in the bacterial periplasm: major achievements and challenges ahead. Antioxid. Redox Signal. 19:63–71. 10.1089/ars.2012.4864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominik, P.K., Borowska M.T., Dalmas O., Kim S.S., Perozo E., Keenan R.J., and Kossiakoff A.A.. 2016. Conformational chaperones for structural studies of membrane proteins using antibody phage display with nanodiscs. Structure. 24:300–309. 10.1016/j.str.2015.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutka, P., Mukherjee S., Gao X., Kang Y., de Waal P.W., Wang L., Zhuang Y., Melcher K., Zhang C., Xu H.E., and Kossiakoff A.A.. 2019. Development of “plug and play” fiducial marks for structural studies of GPCR signaling complexes by single-particle cryo-EM. Structure. 27:1862–1874.e7. 10.1016/j.str.2019.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eigenbrot, C., Randal M., Presta L., Carter P., and Kossiakoff A.A.. 1993. X-ray structures of the antigen-binding domains from three variants of humanized anti-p185HER2 antibody 4D5 and comparison with molecular modeling. J. Mol. Biol. 229:969–995. 10.1006/jmbi.1993.1099 [DOI] [PubMed] [Google Scholar]
- Emsley, P., and Cowtan K.. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132. 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- Fellouse, F.A., Esaki K., Birtalan S., Raptis D., Cancasci V.J., Koide A., Jhurani P., Vasser M., Wiesmann C., Kossiakoff A.A., et al. 2007. High-throughput generation of synthetic antibodies from highly functional minimalist phage-displayed libraries. J. Mol. Biol. 373:924–940. 10.1016/j.jmb.2007.08.005 [DOI] [PubMed] [Google Scholar]
- Hunte, C., and Michel H.. 2002. Crystallisation of membrane proteins mediated by antibody fragments. Curr. Opin. Struct. Biol. 12:503–508. 10.1016/s0959-440x(02)00354-8 [DOI] [PubMed] [Google Scholar]
- Iwata, S., Ostermeier C., Ludwig B., and Michel H.. 1995. Structure at 2.8 A resolution of cytochrome c oxidase from Paracoccus denitrificans. Nature. 376:660–669. 10.1038/376660a0 [DOI] [PubMed] [Google Scholar]
- Jumper, J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Zidek A., Potapenko A., et al. 2021. Highly accurate protein structure prediction with AlphaFold. Nature. 596:583–589. 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig, P., and Fuh G.. 2014. Selection and screening using antibody phage display libraries. Methods Mol. Biol. 1131:133–149. 10.1007/978-1-62703-992-5_9 [DOI] [PubMed] [Google Scholar]
- Koide, S. 2009. Engineering of recombinant crystallization chaperones. Curr. Opin. Struct. Biol. 19:449–457. 10.1016/j.sbi.2009.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovari, L.C., Momany C., and Rossmann M.G.. 1995. The use of antibody fragments for crystallization and structure determinations. Structure. 3:1291–1293. 10.1016/s0969-2126(01)00266-0 [DOI] [PubMed] [Google Scholar]
- Lenaeus, M.J., Vamvouka M., Focia P.J., and Gross A.. 2005. Structural basis of TEA blockade in a model potassium channel. Nat. Struct. Mol. Biol. 12:454–459. 10.1038/nsmb929 [DOI] [PubMed] [Google Scholar]
- Li, H., Dunn J.J., Luft B.J., and Lawson C.L.. 1997. Crystal structure of Lyme disease antigen outer surface protein A complexed with an Fab. Proc. Natl. Acad. Sci. USA. 94:3584–3589. 10.1073/pnas.94.8.3584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q., Wanderling S., Sompornpisut P., and Perozo E.. 2014. Structural basis of lipid-driven conformational transitions in the KvAP voltage-sensing domain. Nat. Struct. Mol. Biol. 21:160–166. 10.1038/nsmb.2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockless, S.W., Zhou M., and MacKinnon R.. 2007. Structural and thermodynamic properties of selective ion binding in a K+ channel. PLoS Biol. 5:e121. 10.1371/journal.pbio.0050121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateja, A., Paduch M., Chang H.Y., Szydlowska A., Kossiakoff A.A., Hegde R.S., and Keenan R.J.. 2015. Protein targeting. Structure of the Get3 targeting factor in complex with its membrane protein cargo. Science. 347:1152–1155. 10.1126/science.1261671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy, A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., and Read R.J.. 2007. Phaser crystallographic software. J. Appl. Crystallogr. 40:658–674. 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlwain, B.C., Erwin A.L., Davis A.R., Ben Koff B., Chang L., Bylund T., Chuang G.Y., Kwong P.D., Ohi M.D., Lai Y.T., and Stockbridge R.B.. 2021. N-terminal transmembrane-helix epitope tag for X-ray crystallography and electron microscopy of small membrane proteins. J. Mol. Biol. 433:166909. 10.1016/j.jmb.2021.166909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee, S., Erramilli S.K., Ammirati M., Alvarez F.J.D., Fennell K.F., Purdy M.D., Skrobek B.M., Radziwon K., Coukos J., Kang Y., et al. 2020. Synthetic antibodies against BRIL as universal fiducial marks for single-particle cryoEM structure determination of membrane proteins. Nat. Commun. 11:1598. 10.1038/s41467-020-15363-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov, G.N., Vagin A.A., and Dodson E.J.. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240–255. 10.1107/S0907444996012255 [DOI] [PubMed] [Google Scholar]
- Ostermeier, C., Iwata S., Ludwig B., and Michel H.. 1995. Fv fragment-mediated crystallization of the membrane protein bacterial cytochrome c oxidase. Nat. Struct. Biol. 2:842–846. 10.1038/nsb1095-842 [DOI] [PubMed] [Google Scholar]
- Paduch, M., and Kossiakoff A.A.. 2017. Generating conformation and complex-specific synthetic antibodies. Methods Mol. Biol. 1575:93–119. 10.1007/978-1-4939-6857-2_6 [DOI] [PubMed] [Google Scholar]
- Rohaim, A., Gong L., Li J., Rui H., Blachowicz L., and Roux B.. 2020. Open and closed structures of a barium-blocked potassium channel. J. Mol. Biol. 432:4783–4798. 10.1016/j.jmb.2020.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhu, S.S., and Fellouse F.A.. 2006. Synthetic therapeutic antibodies. Nat. Chem. Biol. 2:682–688. 10.1038/nchembio843 [DOI] [PubMed] [Google Scholar]
- Slezak, T., Bailey L.J., Jaskolowski M., Nahotko D.A., Filippova E.V., Davydova E.K., and Kossiakoff A.A.. 2020. An engineered ultra-high affinity Fab-Protein G pair enables a modular antibody platform with multifunctional capability. Protein Sci. 29:141–156. 10.1002/pro.3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, Y., DiMaio F., Wang R.Y., Kim D., Miles C., Brunette T., Thompson J., and Baker D.. 2013. High-resolution comparative modeling with RosettaCM. Structure. 21:1735–1742. 10.1016/j.str.2013.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuwe, T., Bley C.J., Thierbach K., Petrovic S., Schilbach S., Mayo D.J., Perriches T., Rundlet E.J., Jeon Y.E., Collins L.N., et al. 2015. Architecture of the fungal nuclear pore inner ring complex. Science. 350:56–64. 10.1126/science.aac9176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uysal, S., Vasquez V., Tereshko V., Esaki K., Fellouse F.A., Sidhu S.S., Koide S., Perozo E., and Kossiakoff A.. 2009. Crystal structure of full-length KcsA in its closed conformation. Proc. Natl. Acad. Sci. USA. 106:6644–6649. 10.1073/pnas.0810663106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valiyaveetil, F.I., Leonetti M., Muir T.W., and Mackinnon R.. 2006. Ion selectivity in a semisynthetic K+ channel locked in the conductive conformation. Science. 314:1004–1007. 10.1126/science.1133415 [DOI] [PubMed] [Google Scholar]
- Winn, M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A., et al. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67:235–242. 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, S., Avila-Sakar A., Kim J., Booth D.S., Greenberg C.H., Rossi A., Liao M., Li X., Alian A., Griner S.L., et al. 2012. Fabs enable single particle cryoEM studies of small proteins. Structure. 20:582–592. 10.1016/j.str.2012.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y., Morais-Cabral J.H., Kaufman A., and MacKinnon R.. 2001. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution. Nature. 414:43–48. 10.1038/35102009 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
summarizes data collection and refinement statistics for sFab–KcsA.
Data Availability Statement
All data that support the method described in this study is provided in the manuscript and by the corresponding author upon request. All the structures are deposited and found in the PDB under accession code 7MDJ.