Abstract
Wastewater surveillance has been a useful tool complementing clinical testing during the COVID-19 pandemic. However, transitioning surveillance approaches to small populations, such as dormitories and assisted living facilities poses challenges including difficulties with sample collection and processing. Recently, the need for reliable and timely data has coincided with the need for precise local forecasting of the trajectory of COVID-19. This study compared wastewater and clinical data from the University of Delaware (Fall 2020 and Spring 2021 semesters), and evaluated wastewater collection practices for enhanced virus detection sensitivity. Fecal shedding of SARS-CoV-2 is known to occur in infected individuals. However, shedding concentrations and duration has been shown to vary. Therefore, three shedding periods (14, 21, and 30 days) were presumed and included for analysis of wastewater data. SARS-CoV-2 levels detected in wastewater correlated with clinical virus detection when a positive clinical test result was preceded by fecal shedding of 21 days (p< 0.05) and 30 days (p < 0.05), but not with new cases (p = 0.09) or 14 days of shedding (p = 0.17).
Discretely collected wastewater samples were compared with 24-hour composite samples collected at the same site. The discrete samples (n = 99) were composited examining the influence of sampling duration and time of day on SARS-CoV-2 detection. SARS-CoV-2 detection varied among dormitory complexes and sampling durations of 3-hour, 12-hour, and 24-hour (controls). Collection times frequently showing high detection values were between the hours of 03:00 to 05:00 and 23:00 to 08:00. In each of these times of day 33% of samples (3/9) were significantly higher (p < 0.05) than the control sample. The remainder (6/9) of the collection times (3-hour and 12-hour) were not different (p > 0.05) from the control. This study provides additional framework for continued methodology development for microbiological wastewater surveillance as the COVID-19 pandemic progresses and in preparation for future epidemiological efforts.
Keywords: SARS-CoV-2, COVID-19, Wastewater-based epidemiology, Virus recovery, Method development
Graphical abstract
1. Introduction
Wastewater-based epidemiology (WBE) has been widely employed for pandemic surveillance during the COVID-19 pandemic as a complement to clinical testing efforts. SARS-CoV-2, the virus causing COVID-19, is often shed in the feces and urine of both asymptomatic and symptomatic infected individuals (Park et al., 2020; Schmitz et al., 2021), allowing for the concentrations of virus to be monitored in wastewaters. In large populations, changes in wastewater concentrations of SARS-CoV-2 have been shown to correlate and precede changes in the number of COVID-19 cases. It has been demonstrated that increases of SARS-CoV-2 concentrations in municipal wastewater preceded hospital admissions by three days and positive cases of COVID-19 by 7 days (Peccia et al., 2020). Epidemic models have also shown increases in wastewater concentrations to precede rises in hospitalization by three to five days (Kaplan et al., 2020). Overall, wastewater surveillance data can be paired with clinical testing data for tracking and minimizing the spread of COVID-19. Moreover, wastewater monitoring overcomes several challenges in pandemic surveillance. Wastewater testing is cost-effective and unbiased in monitoring entire communities and it overcomes clinical testing disparities such as reluctancy or hesitancy to seek testing and the lack of general healthcare in underserved communities (Thompson et al., 2020).
The complexities and challenges of wastewater testing have become apparent as the COVID-19 pandemic has progressed. For example, poor understanding of variability observed in fecal shedding rates, concentration, and duration of SARS-CoV-2 in individuals with COVID-19 and vaccinated individuals has fueled doubt over the relationship between wastewater detection and clinical testing data. As these complications arise, alterations to wastewater testing protocols are performed to improve the overall quality of the data being produced. Detection of fecal indicators, such as pepper mild mottle virus (PMMoV), Bacteroides 16S rRNA, and human 18S rRNA, has been implemented for normalizing SARS-CoV-2 wastewater detection data and establishing correlations with COVID-19 cases (D'Aoust et al., 2020). However, use and implementation of such naturally occurring internal standards has not been widely adopted (Garcia-Aljaro et al., 2018). Attempts at normalization by per capita contribution, process control (e.g., BCoV), and fecal indicator recovery have produced variable results for locations even within the same study (Feng et al., 2021).
Challenges of wastewater monitoring are further exacerbated in small populations by limited and sporadic collection opportunities. Wastewater flow decreases in smaller populations and consequently, virus detection can be more influenced by collection location than by the contribution of human waste alone. For example, dormitories and assisted living communities have varying levels of amenities including shared restrooms and communal laundry facilities. Wastewater surveillance programs from twenty-five universities reported challenges in sample collection and processing efforts such as intermittent flow, pipe or collection instrument clogging, and heterogeneous wastewater composition (Harris-Lovett et al., 2021). A study of wastewater discharge from two private homes found significant fluctuations between the two sites in both hourly and daily discharge rates (Lucas et al., 2017). One location showed increased wastewater discharge on Friday and Saturday, while the other showed an increase on Monday. Diurnal fluctuations were also observed with peak wastewater discharge occurring in the morning and at night in both locations.
Fluctuations and inconsistencies in wastewater discharge compound difficulties in comparing viral concentrations within or between locations over time, even when utilizing twenty-four-hour composite samples. However, as the SARS-CoV-2 pandemic progresses, the importance of detecting small numbers of COVID-19 cases within both communities and small populations will increase with anticipated increases in vaccination rates and decreases in clinical testing.
This study was novel in that it included both methodological and epidemiological investigations of wastewater surveillance for SARS-CoV-2 in college dormitory populations. The influence of wastewater sampling strategies, including times of day, length of time, and location type for small populations were evaluated. A unique sampling approach was implemented to evaluate the influence of building style (traditional, suite, or apartment) on the production of wastewater. SARS-CoV-2 detection was performed for these locations over multiple times of day and time frames including 3-, 12-, and 24-hour composites. Additionally, the relationship between wastewater detection and clinical tests results was examined. While the focus of this study was SARS-CoV-2 detection, these findings should be broadly applicable for future studies monitoring infectious agents, narcotics, or other chemicals within wastewater.
2. Materials and methods
2.1. SARS-CoV-2 wastewater surveillance
2.1.1. Selection of sampling locations
Selection of populations to be monitored and collection locations was based on a variety of influential criteria to provide a dataset representative of the overall university. These criteria included selecting dormitory populations of more than 200 students to ensure sufficient wastewater flow was generated throughout the collection period and represented the three dormitory styles available to students: suite, traditional, and apartment. Also, the ease and reliability of sample collection and the proximity of the sampling location to the building complex (groups of two to eight individual buildings) were considered to reduce external influence on sample quality, potentially including dilution by stormwater infiltration. Wastewater channels were evaluated to ensure that the apparatus tubing remained in the designated position and was not susceptible to extensive clogging.
2.1.2. Housing and population dynamics
Wastewater was monitored at three housing complexes within the University of Delaware campus during the Fall 2020 (early October to mid-November) and Spring 2021 (mid-February through mid-May) semesters. These dormitories each had three distinct styles and independent populations: suite style, with two-room groups sharing a restroom (locations A and B); apartment style, with a restroom in each unit (location C); and traditional style, with common restrooms on each floor (location D). Locations A, B, and D housed undergraduate students with each complex having between 250 and 350 residents. Location C contained undergraduates including those relocated for quarantine or isolation based on positive clinical test results. Population size fluctuated in this location as quarantine and isolation needs changed throughout the semesters. After receiving a positive clinical test result, students were moved to Location C for 10-14 days after which they were considered “recovered” students in this study.
2.1.3. Wastewater production and collection
The University of Delaware water supply and wastewater collection, transmission, and treatment is provided by the City of Newark, Department of Public Works and Water Resources. Wastewater production is not monitored directly, rather calculated by the water consumption fed into dormitory buildings within the University. Water distributed to grounds keeping and other external demands is subtracted from the water consumed to calculate the wastewater produced for each building, or group of buildings, using Eq. (1). The volumes (gallons) of wastewater produced are compiled, by the University of Delaware Facilities Financial Services team, for each dormitory complex, and the monthly totals are determined using the Qlik BI tool for utility data storing and analyzing. Due to the differences in population sizes between the complexes, volumes of the total wastewater production and wastewater production per person were evaluated for the locations.
| (1) |
The method described above was selected as it is more reliable for these types of locations with smaller populations and limited flow than a direct measurement of the wastewater through a metering unit within the sewer or attached to the sampling apparatus. Limited flow and particulate accumulation surrounding the sampling apparatus can alter readings and produce inaccurate data. The complications of small-population monitoring have been previously reviewed (Harris-Lovett et al., 2021), further supporting the considerations made for selection of sampling locations in this study.
Wastewater samples were collected hourly throughout a twenty-four-hour period from each of the four locations using ISCO 3700 Portable Samplers (Teledyne Isco, Lincoln, NE; Cat. no. 68-3700-064). Samples were transported on ice to the Center for Environmental and Wastewater-based Epidemiological Research (CEWER) laboratory for processing. Thirty-eight samples were collected from Locations A, B, and C, between October 2, 2020, and November 19, 2020, for the Fall 2020 semester. Thirty-four samples were collected from the same locations between February 16, 2021, and May 13, 2021, for the Spring 2021 semester. Samples included in this study were those considered to be complete compositions with more than fifty-percent of hourly samples collected during each twenty-four-hour composite.
2.2. Wastewater composition study
Wastewater samples of approximately 100 mL were collected in 1 L bottles, hourly throughout a twenty-four-hour period. Twenty-four-hour (09:00-08:00), twelve-hour (09:00-20:00 and 21:00-08:00), and three-hour (09:00-11:00, 12:00-14:00, 15:00-17:00, 18:00-20:00, 21:00-23:00, 24:00-02:00, 03:00-05:00, 06:00-08:00) composited wastewater samples were collected. Locations A, C, and D were selected for this study and collection was repeated on three dates referred to as days i, ii, and iii. Samples were collected and processed in triplicate (n = 297) with an additional duplication during RT-qPCR detection. SARS-CoV-2 from all wastewater and clinical samples were tested using the CDC-recommended detection assay.
2.3. SARS-CoV-2 clinical testing
University of Delaware Student Health Services conducted student clinical testing, and sample analysis was performed by University of Delaware Poultry Health System technicians, part of the Department of Animal and Food Sciences. Saliva samples were collected from asymptomatic and symptomatic individuals at testing locations across the university campus. Individual saliva samples were heat inactivated before being placed on 96 well plates (200 μL per well) for RNA extraction using the MagMAX Viral/Pathogen II Nucleic Acid Isolation Kit (ThermoFisher Scientific, Waltham, MA) on a Kingfisher Flex (ThermoFisher Scientific). The approved real-time RT-PCR clinical test for the detection of the SARS-CoV-2 virus was performed using the TaqPath COVID-19 Combo Kit chemistry (ThermoFisher Scientific) on an ABI 7500 Fast Real-Time PCR System (ThermoFisher Scientific). Test results were typically provided within a 24-hour-period. Aggregate data were presented on the University of Delaware COVID-19 Dashboard available at: www.udel.edu/home/coronavirus/dashboard/.
During the Fall 2020 semester, 14,718 tests of asymptomatic (14,114 tests) and symptomatic (604 tests) students were performed from September 6, 2020 to November 21, 2020. During the Spring 2021 semester, 51,751 tests of asymptomatic (50,972 tests) and symptomatic (779 tests) students were performed from February 7, 2020 to May 22, 2021. In both time periods clinical sampling began before and ended after the wastewater sampling period.
Clinical test data for each dormitory complex was obtained separately and de-identified thus protecting the anonymity of individual students within the smaller populations. Therefore, the dormitory complex collection locations are simply referred to as A-D and the overall University of Delaware.
2.4. Virus recovery and detection
SARS-CoV-2 was recovered and detected as previously described (Anderson-Coughlin et al., 2021). The method was modified to include a multiplex RT-qPCR probe-based assay (Table 1 ) which decreased the overall time for obtaining results; all other details remained the same. Briefly, 45 mL wastewater samples were incubated for 60 min. at 60 °C immediately after arrival in the laboratory. Samples were cooled to room temperature and passed through a 0.22 μm, 13.6 cm2, PES membrane filter (Corning, Corning, NY). The filtrate was concentrated via centrifugal ultrafiltration using two 10 kDa Amicon centrifugal filters (Millipore Sigma, St. Louis, MO) in 15 mL volumes, obtaining 400 μL total concentrate. Concentrates were pooled, and extractions were performed using QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany). Detection was performed using RT-qPCR probe-based assay with target specific primer and probe sets from IDTDNA (Integrated DNA Technologies, Coralville, IA) (Table 1).
Table 1.
Primers and probe sequences and concentrations used for detection of SARS-COV-2.
| Target | Oligo | 5′ – Sequence – 3′a |
|---|---|---|
| N1 | Forward | GACCCCAAAATCAGCGAAAT |
| Reverse | TCTGGTTACTGCCAGTTGAATCTG | |
| Probeb | FAM-ACCCCGCAT-ZEN-TACGTTTGGTGGACC | |
| N2 | Forward | TTACAAACATTGGCCGCAAA |
| Reverse | CGCCGACATTCCGAAGAA | |
| Probec | HEX-ACAATTTGC-ZEN-CCCCAGCGCTTCAG |
Sequences from the US Centers for Disease Control and Prevention (CDC) designed SARS-CoV-2 RT-PCR assay.
5′ addition of FAM fluorophore modification, absorbance and emission at 495 nm and 520 nm, respectively.
5″ addition of HEX fluorophore modification, absorbance and emission at 538 nm and 555 nm, respectively
Plasmids containing the complete nucleocapsid gene, 2019-nCoV_N Positive Control plasmid (Integrated DNA Technologies), were used to quantify SARS-CoV-2 RNA within the RT-qPCR assay. Ten-fold serial dilutions were performed to generate a standard curve to which results from the samples were compared. Undiluted plasmids were typically concentrated to 200,000 copies/μL, dilutions were performed to yield reaction concentrations (5 μL template into 22 μL reaction) 10,000, 1000, 100, 10 and 1 copies/reaction.
Amplification of target sequences, fluorescent signal above the designated threshold, for each assay were considered positive detection of SARS-CoV-2. Failure to amplify above the threshold for either target, N1 or N2, in either replicate, results in the sample being considered below the limit of detection (<10,000 copies/L). Replicate detection values for the N1 and N2 primer sets were averaged for each sample and used for analysis.
2.5. Data analyses
Microsoft Excel and JMP 16 Pro Statistical Software, were used for statistical analysis and figure and table preparation. Wastewater and clinical data were compared for potential correlations using linear regression analysis and the associated correlation value. Dunnett's Test was performed for comparison of the discrete samples with the twenty-four-hour composite as the control group. Results with a p-value of <0.05 were considered significant.
2.5.1. SARS-CoV-2 wastewater data
SARS-CoV-2 detection and quantification were performed within the Rotor-Gene Q apparatus and associated software. Data were presented as viral copies per reaction and cycle threshold (CT) values within the Rotor-Gene Q and transferred to statistical software for further analysis. Viral copies per reaction were transformed into viral copies per liter of wastewater by accounting for the dilution and concentration factors of the sample processing method. CT values were transformed into delta CT (dCT values) using the following equation:
| (2) |
where the terminal CT value was 40 cycles for all samples processed.
2.5.2. SARS-CoV-2 clinical test data
SARS-CoV-2 clinical testing data was obtained from the University Dashboard or Student Health Services as described above (Section 2.2). The data were presented as number of positive tests per day. Data were converted to weekly results and percent positivity (%) to account for fluctuations in the total number of tests performed.
3. Results
3.1. Wastewater production
Volumes of wastewater produced varied by location (dormitory complex) and month throughout the study. Wastewater production, total monthly volumes, in the fall (September-November 2020) and spring (February-May 2021) semesters were significantly different (p < 0.05) in locations C and D. This can be attributed to the increase of residents from the fall to spring semester, 113% at location C and 208% at location D. Locations A and B did not have significant differences in wastewater production (p = 0.82, p = 0.82) between the fall and spring semesters and had only minimal population increases of 12% and 2%, respectively. Total wastewater production was significantly higher (p < 0.05) at location C, apartment-style building with quarantine and isolation populations, compared to locations A, B, and D. No significant differences (p > 0.05) were observed between wastewater production of Locations A, B, and D.
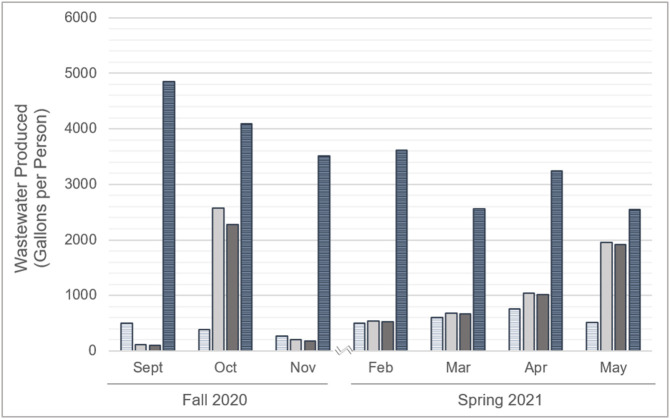
Due to the fluctuations in population sizes and differences across sampling locations, data were normalized to monthly per person wastewater production values (Fig. 1 ). Monthly wastewater production per person did not have significant differences between the fall and spring semesters at locations A (p = 0.92), B (p = 0.83), C (p = 0.07), or D (p = 0.07). However, the monthly wastewater production per person was significantly higher (p < 0.05) at location C, compared to locations A, B, and D as observed with the total wastewater production. Monthly, wastewater production ranged from 101 to 2571 gal in the suite-style dormitory complexes (locations A and B), averaging 985 gal. Monthly wastewater production in the apartment- and traditional-style complexes (locations C and D) ranged from 2550 to 4856 and 273 to 762 gal, respectively, averaging 3490 and 505 gal monthly.
Fig. 1.
Wastewater production for dormitory complexes A (suite-style, light solid), B (suite-style, dark solid), C (apartment-style, dark striped), and D (traditional style, light striped) data by month during 2020-2021 academic year. Data are presented as the total wastewater produced for each complex throughout the month (gallons).
3.2. Fall 2020 – university clinical testing and wastewater surveillance
University of Delaware residence hall occupancy in the Fall 2020 semester was 18% of full occupancy (1246 of 6927 available spaces filled). A total of 14,718 SARS-CoV-2 clinical PCR tests were performed during the fall semester between August 30 and November 21, 2020, with an average of 1338 tests per week. Weekly positive tests ranged from 17 to 129 cases, and positivity rates ranged from 1.2% to 10.7%. Wastewater surveillance sampled effluent from approximately 70% of the student population residing on-campus in Fall 2020. Thirty-eight wastewater samples were collected from the three locations throughout the semester. Weekly averages of SARS-CoV-2 in wastewater for all locations surveilled ranged from below detection limit (< 4.00 log viral copies/L) to a maximum of 6.06 log viral copies/L.
3.3. Spring 2021 – university clinical testing and wastewater surveillance
University of Delaware residence hall occupancy in the Spring 2021 semester was 55% of full occupancy (3841 of 6927 available spaces filled). A total of 51,751 SARS-CoV-2 clinical tests were performed during the spring semester between February 7 and May 23, with an average of 3450 tests per week. Weekly positive tests ranged from 2 to 322 cases detected and positivity rates ranged from 0.1% to 7.1%. Wastewater surveillance sampled effluent from approximately 40% of the student population residing on-campus in Spring 2021. Fifty-eight wastewater samples were collected from the three locations throughout the semester, on twenty separate occasions. Weekly averages of SARS-CoV-2 levels in wastewater for all locations surveilled ranged from below detection limit to a maximum of 6.80 log viral copies/L.
3.4. Dormitory complex monitoring
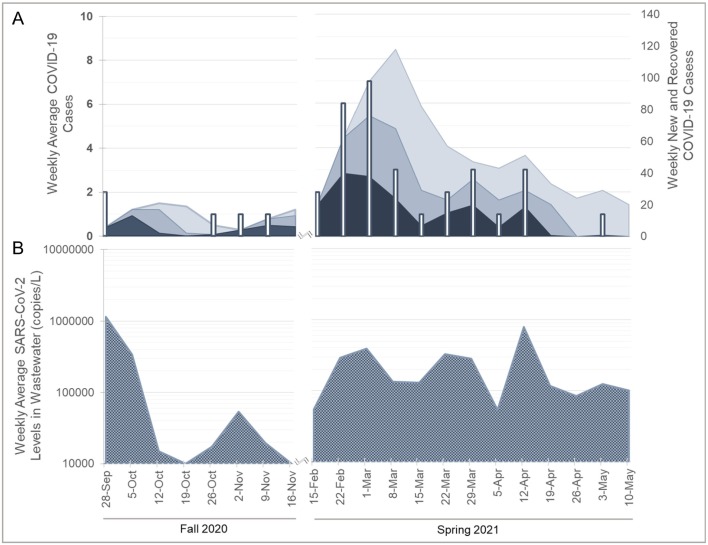
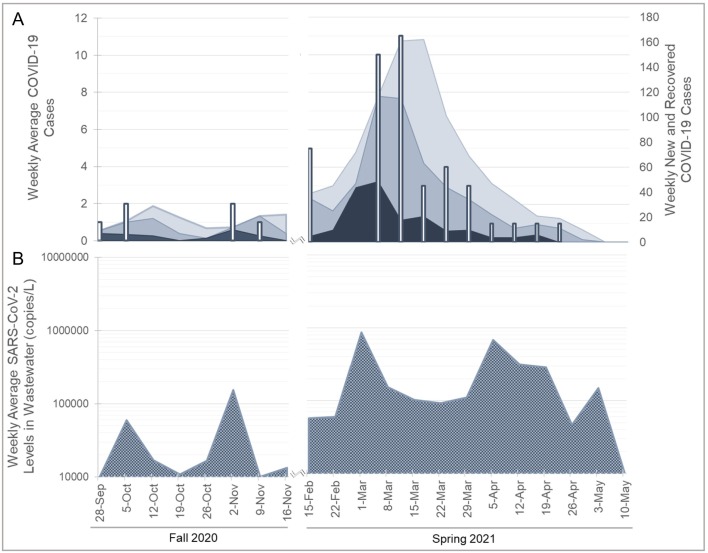
Compilation of wastewater and clinical data for locations A and B during the Fall and Spring semester (Fig. 2, Fig. 3 ) yielded significant correlations between the data sets. Significant positive correlations (p < 0.05) were observed between wastewater levels and combinations of new and recovered cases with fecal shedding for both the 21- and 30-day time periods. However, correlations were not observed for new daily clinical cases or fecal shedding during a 14-day time period (Table 2 ). Wastewater surveillance of location C is presented (Fig. 4 ) for qualitative comparison of SARS-CoV-2 levels detection in wastewater from location C, which housed the quarantine and isolation populations, to the wastewater surveillance of locations A and B. SARS-CoV-2 levels were significantly higher (p < 0.05) in wastewater collected from location C during the fall and spring semester than locations A and B. SARS-CoV-2 levels in wastewater collection from locations A and B were not significantly different (p = 0.97) from each other during the fall and spring semesters.
Fig. 2.
Dormitory complex A, SARS-CoV-2 clinical (A) and SARS-CoV-2 wastewater surveillance (B) data during the 2020-2021 academic year. Vertical bars show weekly total of positive clinical cases (left vertical axis). Shaded areas show clinical cases with potential shedding including both new and recovering case patients (right vertical axis): dark (14-days); medium (21-days); and light (30-days) of SARS-CoV-2 fecal shedding after an initial positive clinical test. Wastewater data are presented as SARS-CoV-2 viral copies per liter of wastewater.
Fig. 3.
Dormitory complex B, SARS-CoV-2 clinical (A) and SARS-CoV-2 wastewater surveillance (B) data during the 2020-2021 academic year. Vertical bars show weekly total of positive clinical cases (left vertical axis). Shaded areas show clinical cases with potential shedding including both new and recovering case patients (right vertical axis): dark (14-days); medium (21-days); and light (30-days) of SARS-CoV-2 fecal shedding after an initial positive clinical test. Wastewater data are presented as SARS-CoV-2 viral copies per liter of wastewater.
Table 2.
Analysis of Fall 2020 and Spring 2021 wastewater surveillance and clinical testing data using linear regression with variable fecal shedding timeframes for university dormitories.
| Shedding timeframea | Correlation value | Significance probability |
|---|---|---|
| 0 Days | 0.27 | 0.09 |
| 14 Days | 0.22 | 0.17 |
| 21 Days | 0.37 | <0.05 |
| 30 Days | 0.39 | <0.05 |
Bolded values indicate statistically significant results.
Viral shedding parameters of 0 days, representing new cases reported, along with 14, 21, and 30 days of viral shedding after positive clinical test results were reported, were combined and evaluated against SARS-CoV-2 levels (log viral copies/L) in wastewater.
Fig. 4.
SARS-CoV-2 wastewater surveillance data for dormitory complex C, during the 2020-2021 academic year.
3.5. Quarantine and isolation dormitory complex monitoring
Dormitory complex C contained both the general undergraduate student population and the quarantine and isolation populations. The general population consisted of more than 300 students, while the quarantine and isolation populations fluctuated throughout the study.
University clinical testing identified 701 positive cases in the Fall 2020 semester, averaging 64 per week, and ranging from 0 to 129 cases. Clinical testing in the Spring 2021 semester identified a total of 1276 positive cases, averaging 85 per week, and ranging from 0 to 322. SARS-CoV-2 detected through clinical tests performed on campus, as shown in Fig. 4.
3.6. Wastewater composition study
Three-hour wastewater composite samples were compared by both location (A, C, and D) and collection day (i, ii, and iii). SARS-CoV-2 detection in dormitory complex A was not significantly different (p > 0.05) among the 3 sampling days. Detection in complex C was significantly lower (p < 0.05) on day ii than days i and iii. Detection in complex D significantly different (p < 0.05) between all days with day i having the greatest detection and day iii having the least. All locations had significantly different detection levels on days ii and iii; on day I, locations C and D were not significantly different from one another (p = 0.13) (Table 3 ).
Table 3.
SARS-CoV-2 detected (dCT values) from 24-hour composited wastewater samples collected from three dormitory complexes (A, C, and D) on three separate occasions (days i, ii, and iii).
| Collection day | Sampling location |
||
|---|---|---|---|
| Complex A | Complex C | Complex D | |
| i | 1.4 ± 2.3 | 10.3 ± 0.9 | 9.1 ± 0.4 |
| ii | 0.5 ± 1.1 | 10.7 ± 0.9 | 4.9 ± 1.1 |
| iii | 1.9 ± 2.1 | 7.3 ± 3.4 | 3.1 ± 1.0 |
Twenty-four-hour composite samples were used as control groups for analyses of SARS-CoV-2 detection levels in the three-hour composite samples (Fig. 5 ). SARS-CoV-2 levels of the three-hour composites were compared to the 24-hour composites at each location, on each collection day, with a significance level of p < 0.05.
Fig. 5.
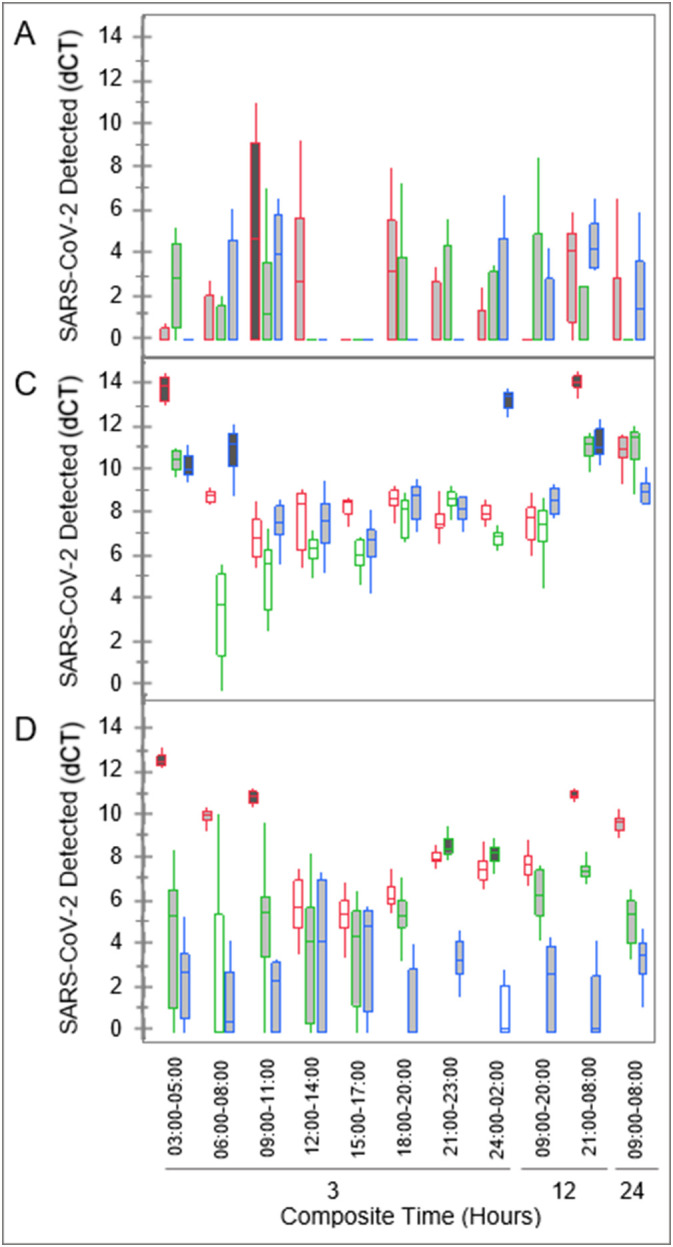
Levels of SARS-CoV-2 detected in wastewater from 3 dormitory complexes (A, C, and D), collected in 3-, 12-, and 24-hour composite samples. Detection levels displayed are dCT values, obtained from RT-qPCR performed in which greater dCT values represent greater levels of virus, SARS-CoV-2. Twenty-four-hour composite samples were used as controls. Shading represents significance (p < 0.05) of detection levels: light gray (no significant difference), dark gray (significantly higher), and white (significantly lower). Outlines represent the three collection dates: red (left), green (middle), and blue (right) in each time period.
Samples from complex A had the highest variability observed in detected SARS-CoV-2 levels, which coincided with high turbidity (observational data, not shown) and a lower population overall. Only one sample (09:00-11:00) out of 30 samples was significantly different (p < 0.05) from the 24-hour-composite controls. Detection levels in samples from complexes C and D fluctuated throughout the 3-hour-composite periods, compared to the 24-hour-composite controls. Decreased detection levels were observed in complex C samples primarily between 09:00 and 23:00 with 67% of samples (n = 15) having significantly lower levels (p < 0.05) than the controls and the remaining 33% with no significant difference from the controls (p > 0.05). Similarly, decreased detection levels were primarily observed in complex D samples between 12:00 and 20:00 with 33% of samples (n = 9) having significantly lower levels (p < 0.05) than the controls and the remaining 67% with no significant difference from the controls (p > 0.05).
4. Discussion
Throughout the COVID-19 pandemic, multiple universities employed wastewater-based epidemiological wastewater sampling to assist in tracking and minimizing the spread of the disease within the on-campus populations. Wastewater surveillance data has been used on university campuses for directing clinical testing efforts and finding both pre-symptomatic and asymptomatic individuals infected with SARS-CoV-2 (Gibas et al., 2021). One such on-campus study at the University of Arizona found that of the 711 clinically identified infections in college students residing in the dormitories, ~ 80% were asymptomatic and the remaining 20% were symptomatic (Schmitz et al., 2021). Shedding of SARS-CoV-2 RNA has been detected in both stool and urine samples collected from asymptomatic individuals (Sharkey et al., 2021). University of Arizona wastewater-based epidemiology efforts also yielded an 82% positive and 89% negative predictive power which was used to direct clinical testing efforts and indicated the presence of infected individuals prior to identification through clinical testing (Betancourt et al., 2021). It has been posited that if clinical surveillance efforts are expansive, with rapid clinical test results available for symptomatic individuals and routine testing of the asymptomatic population, then wastewater sampling is of limited benefit in pandemic monitoring (Bibby et al., 2021). Due to the extensive clinical testing efforts employed by the University of Delaware, stringent social distancing and mask-wearing protocols, and limitations placed on dormitory capacities, predictive power was not observed in our study. However, the concentrations of SARS-CoV-2 detected in wastewater from the dormitories was monitored throughout the Fall 2020 and Spring 2021 semesters and the data were shared with university administration for support in the direction of clinical surveillance efforts across campus.
Students, typically aged eighteen to twenty-two years old, had mean shedding concentrations of more than 6-log genomic copies of SARS-CoV-2 per gram of feces. Additionally, SARS-CoV-2 fecal shedding durations ranged from 6 days for asymptomatic individuals to 14 days for serious infections and up to 32 days for critical COVID-19 cases after a positive clinical test detected SARS-CoV-2. Therefore, students who are considered active COVID-19 cases and those who have recovered can contribute to the SARS-CoV-2 concentrations detected in wastewater. Our study yielded correlations of SARS-CoV-2 levels in wastewater with 21 and 30 days of fecal shedding after a positive clinical test. However, no correlations were observed with new cases or 14 days of fecal shedding. Due to the routine clinical testing of students residing in the dormitories, clinical testing may have identified SARS-CoV-2 infections in the respiratory system prior to infection of the gastrointestinal tract and subsequent fecal shedding. Students were housed separately in Location C for 10 to 14 days after receiving a positive clinical test prior to re-entering their dormitories. Those individuals with serious infections may have remained in Location C through the duration of their fecal shedding period and would not have been contributing to the wastewater at Locations A or B upon their return.
In our study, no significant differences were observed in the average wastewater SARS-CoV-2 levels when comparing communal (traditional) and suite-style dormitories. This could be due to limited population densities, 18% and 55% occupancy of total capacity during the Fall 2020 and Spring 2021 semesters, at the University of Delaware during this study or the social distancing protocols employed. Additionally, increased turbidity and larger variability in SARS-CoV-2 detection levels were observed in wastewater collected from the traditional-style dormitory which may have impacted the ability to obtain significantly significant differences in detection levels between the two dormitories. In a prior study, dormitories with communal-style restrooms (traditional-style dormitory) produced greater detection rates and higher concentrations of SARS-CoV-2 than those with apartment or suite-style restrooms (Scott et al., 2021). Communal restrooms were shared by more than 10 people, suite-style dorms 5-10 people, and apartment style less than 5 people. It was hypothesized that this could be attributed to the population density in these locations as the cases significantly decreased after social distancing guidelines were implemented.
In our study, monthly wastewater production per person in the traditional- and suite-style dormitories was significantly less than wastewater production per person in the apartment-style dormitories. The increased wastewater production in apartment-style dormitories is likely attributed to the additional graywater production by use of kitchen and laundry facilities present in each of the apartments, and not an increase of wastewater production. Increased turbidity and variability in virus detection (standard deviations) were observed in the traditional-style dormitory wastewater. The variability of detected SARS-CoV-2 levels could have contributed to the lack of differences observed between the types of dormitories. Though the wastewater was potentially diluted with graywater in the apartment-stye dormitory, the levels of SARS-CoV-2 produced by the quarantine and isolation populations within that complex resulted in significantly higher detection of the virus compared to traditional- and suite-style dormitories.
The findings of our study and others support the need to investigate populations and environmental factors which may impact target detection prior to employing surveillance efforts. The style of dormitory influences the interactions between individuals within the facilities and how wastewater is diluted by graywater. When sampling locations are selected, these elements need to be considered and accounted for to determine sample collection frequency and composition periods or grab sample timepoints.
Variability in wastewater detection of SARS-CoV-2 with time of collection has been reported with fluctuations as large as >5 log genomic copies/L to below detection limit within a single hour time period (Betancourt et al., 2021). This study also demonstrated the need for careful selection of sample collection times when sampling small populations. While 24-hour composited wastewater may provide the most reliable data for monitoring of trends, this sampling strategy is costly and, in some cases, can delay the reporting of wastewater data. However, composite samples of limited timeframes or even grab samples may provide the best chance at detection if only presence-absence data is the desired end. In our study, greater detection typically occurred during the evening and early morning hours compared to mid-day and afternoon. This could likely be partially attributed to the dilution of wastewater by graywater through more active use of sink, shower, dish and clothes washing fixtures and appliances during waking hours.
Assessment of fluctuations in fecal concentrations can be performed using fecal markers in lieu of pathogen detection. Ammonium (NH4-N) levels can be used to estimate fecal concentration and dilution and were monitored by Been et al. (2014) to track fluctuations within their populations. Peaks in ammonium levels were observed daily in the morning hours, with the time shifting slightly on weekend days to later morning hours. Fecal content can also be monitored by concentrations of microbial fecal indicators such as human-specific bacteriophage e.g., HF183, or viruses e.g., pepper mild mottle virus (Feng et al., 2021; Greaves et al., 2020; Scott et al., 2021). Employing detection of fecal indicators alongside detection of SARS-CoV-2 could provide a more complete depiction of the wastewater composition and of ideal sample collection periods as well as providing a normalization element for use with wastewater-based epidemiology studies.
As the COVID-19 pandemic has progressed, wastewater-based epidemiology as a field has grown considerably. The lessons learned over this time have generated numerous datasets and resulted in the development of the National Wastewater Surveillance System (NWSS) as a collaboration among The Centers for Disease Control, Department of Health and Human Services as well as multiple agencies, public and private partnerships. Universal methods for sample collection, SARS-CoV-2 recovery and detection, and data reporting are now available and gain further value as the pandemic progresses. Reduction in clinical testing efforts, public aversion to seeking testing, or “testing fatigue,” is becoming more common and decreases the probability of SARS-CoV-2 detection within the community. These factors support the continued use of employing wastewater surveillance as an impartial and affordable means for monitoring the COVID-19 pandemic as well as potential future pandemics as they emerge.
The existing infrastructure can be readily adapted for use with other microbial organisms of concern. The potential for molecular methods to be used for detection of SARS-CoV-2 and additional organisms has been explored (Spurbeck et al., 2021). Sequencing of viral RNA isolated from wastewater is costly compared to the RT-qPCR method and targeted detection performed in this study. Identification of genetic sequences for microbial targets will still be needed for future wastewater-based epidemiology studies; however, standardized methods of recovery and detection are now available which will expedite the process of detection and data reporting. Additionally, the routes of SARS-CoV-2 transmission through various environments has been reviewed along with the potential for artificial intelligence to be used for adaptation of transmission models to emerging infectious diseases (Abdeldayem et al., 2021). Continued wastewater testing could potentially serve a valuable role in ongoing efforts to monitor and support public health even as crises subside. The inclusion of emerging technologies with existing wastewater surveillance infrastructure could help to identify potential organisms and pathways of transmission prior to a disease becoming epidemic or pandemic.
Identification of the factors influencing detection of SARS-CoV-2 within community subsets, such as the dormitories monitored in this study, is crucial to detection of limited cases as the COVID-19 pandemic progresses as well as the adaptation of methods for other microbial organisms of concern. As we now know, detection rates vary by location and housing type, thus individual sampling locations will need to be assessed to select the appropriate collection periods. Emerging organisms may be shed in different concentrations, particularly if the digestive tract is primarily infected, which will undoubtably require additional investigation. However, the factors influencing detection of microbiological organisms in wastewater have been identified, creating a framework for future WBE applications.
CRediT authorship contribution statement
Brienna L. Anderson-Coughlin: Conceptualization, Methodology, Investigation, Formal Analysis, Writing – Original Draft. Adrienne E.H. Shearer: Methodology, Investigation, Resources. Alexis N. Omar: Investigation, Resources. Pushpinder K. Litt: Investigation. Erin Bernberg: Investigation. Marcella Murphy: Investigation. Amy Anderson: Investigation. Lauren Sauble: Investigation. Bri Ames: Investigation. Oscar Damminger. Jr.: Investigation. Brian S. Ladman: Methodology, Investigation, Writing – Review & Editing. Timothy F. Dowling: Data Curation, Project Administration. K. Eric Wommack: Methodology, Resources, Supervision, Funding Acquisition, Writing – Review & Editing. Kalmia E. Kniel: Methodology, Resources, Supervision, Writing – Review & Editing, Visualization, Supervision, Funding Acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was funded in part by New Castle County, Delaware through the COVID-19 Wastewater Surveillance Research Agreement with the University of Delaware.
We would like to thank Michael Harris and Vincent DiSciullo from the New Castle County, Department of Public Works for their assistance with project development and sample collection.
We would also like to thank the University of Delaware Facilities Plumbing services, especially Joe Williams and Jamie Shuler for their assistance in determining sampling locations, troubleshooting collection issues, and overall support during this project. The authors also thank Mary Jane Keyser and the University Facilities Financial Services team within the Facilities, Real Estate, and Auxiliary Services Department for collecting, analyzing, and providing the water consumption data to our team for use in this study.
Editor: Warish Ahmed
References
- Abdeldayem O.M., Dabbish A.M., Habashy M.M., Mostafa M.K., Elhefnawy M., Amin L., Al-Sakkari E.G., Ragab A., Rene E.R. Viral outbreaks detection and surveillance using wastewater-based epidemiology, viral air sampling, and machine learning techniques: a comprehensive review and outlook. Sci. Total Environ. 2021;803 doi: 10.1016/j.scitotenv.2021.149834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson-Coughlin B.L., Shearer A.E.H., Omar A.N., Wommack K.E., Kniel K.E. Recovery of SARS-CoV-2 from wastewater using centrifugal ultrafiltration. Methods Protoc. 2021;4(32):1–9. doi: 10.3390/mps4020032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Been F., Rossi L., Ort C., Rudaz S., Delemont O., Esseiva P. Population normalization with ammonium in wastewater-based epidemiology: application to illicit drug monitoring. Environ. Sci. Technol. 2014;48(14):8162–8169. doi: 10.1021/es5008388. [DOI] [PubMed] [Google Scholar]
- Betancourt W.Q., Schmitz B.W., Innes G.K., Prasek S.M., Brown K.M.P., Stark E.R., Foster A.R., Sprissler R.S., Harris D.T., Sherchan S.P., Gerba C.P., Pepper I.L. COVID-19 containment on a college campus via wastewater-based epidemiology, targeted clinical testing and an intervention. Sci. Total Environ. 2021;779(146408):1–8. doi: 10.1016/j.scitotenv.2021.146408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibby K., Bivins A., Wu Z., North D. Making waves: plausible Lead time for wastewater based epidemiology as an early warning system for COVID-19. Water Res. 2021 doi: 10.1016/j.watres.2021.117438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Aoust P.M., Mercier E., Montpetit D., Jia J., Alexandrov I., Neault N., Baig A.T., Mayne J., Zhang X., Alain T., Langlois M., Servos M.R., MacKenzie M., Figeys D., MacKenzie A.E., Graber T.E., Delatolla R. Quantitative analysis of SARS-CoV-2 RNA from wastewater solids in communities with low COVID-19 incidence and prevalence. Water Res. 2020;188(116560) doi: 10.1016/j.watres.2020.116560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Roguet A., McClary-Gutierrez J.S., Newton R.J., Kloczko N., Meiman J.G., McLellan S.L. Evaluation of sampling, analysis, and normalization methods for SARS-CoV-2 concentrations in wastewater to assess COVID-19 burdens in Wisconsin communities. ACS EST Water. 2021;1(8):1955–1965. doi: 10.1021/acsestwater.1c00160. [DOI] [Google Scholar]
- Garcia-Aljaro C., Blanch A.R., Campos C., Jofre J., Lucena F. Pathogens, faecal indicators and human-specific microbial source-tracking markers in sewage. J. Appl. Microbiol. 2018;126(3):701–717. doi: 10.1111/jam.14112. [DOI] [PubMed] [Google Scholar]
- Gibas C., Lambirth K., Mittal N., Juel M.A.I., Barua V.B., Brazell L.R., Hinton K., Lontai J., Stark N., Young I., Quach C., Russ M., Kauer J., Nicolosi B., Chen D., Akella S., Tang W., Schlueter J., Munir M. Implementing building-level SARS-CoV-2 wastewater surveillance on a university campus. Sci. Total Environ. 2021;782(146749):1–9. doi: 10.1016/j.scitotenv.2021.146749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves J., Stone D., Wu Z., Bibby K. Persistence of emerging viral fecal indicators in large-scale freshwater mesocosms. Water Res. X. 2020;9(100067):1–10. doi: 10.1016/j.wroa.2020.100067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris-Lovett S., Nelson K.L., Beamer P., Bischel H.N., Bivins A., Bruder A., Butler C., Camenisch T.D., De Long S.K., Karthikeyan S., Larsen D.A., Meierdiercks K., Mouser P.J., Pagsuyoin S., Prasek S.M., Radniecki T.S., Ram J.L., Roper D.K., Safford H., Sherchan S.P., Shuster W., Stalder T., Wheeler R.T., Korfmacher K.S. Wastewater surveillance for SARS-CoV-2 on college campuses: initial efforts, lessons learned, and research needs. Int. J. Environ. Res. Public Health. 2021;18(4455):1–20. doi: 10.3390/ijerph18094455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan E.H., Wang D., Wang M., Malik A.A., Zulli A., Peccia J. Aligning SARS-CoV-2 indicators via an epidemic model: application to hospital admissions and RNA detection in sewage sludge. Health Care Manag. Sci. 2020;24:320–329. doi: 10.1038/s4187-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas S.A., Coombes P.J., Geary P.M., Horn K. On-site wastewater systems: investigating dynamics and diurnal patterns impacting on the performance of mound systems. J. Environ. Anal. Toxicol. 2017;7(498):1–10. doi: 10.4172/2161-0525.1000498. [DOI] [Google Scholar]
- Park S., Lee C., Park D., Woo H., Cheong H.S., Shin H.C., Ahn K., Kwon M., Joo E. Detection of SARS-CoV-2 in fecal samples from patients with asymptomatic and mild COVID-19 in Korea. Clin. Gastroenterol. Hepatol. 2020;19(7):1387–1394. doi: 10.1016/j.cgh.2020.06.005. e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., Ko A.I., Malik A.A., Wang D., Wang M., Warren J.L., Weinberger D.M., Arnold W., Omer S.B. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nature Biotech. 2020;38:1164–1167. doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz B.W., Innes G.K., Prasek S.M., Betancourt W.Q., Stark E.R., Foster A.R., Abraham A.G., Gerba C.P., Pepper I.L. Enumerating asymptomatic COVID-19 cases and estimating SARS-CoV-2 fecal shedding rates via wastewater-based epidemiology. Sci. Total Environ. 2021;801(149794):1–7. doi: 10.1016/j.scitotenv.2021.149794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott L.C., Aubee A., Babahaji L., Vigil K., Tims S., Aw T.G. Targeted wastewater surveillance of SARS-CoV-2 on a university campus for COVID-19 outbreak detection and mitigation. Environ. Res. 2021;200(111374):1–10. doi: 10.1016/j.envres.2021.111374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharkey M.E., Kumar N., Mantero A.M.A., Babler K.M., Boone M.M., Cardentey Y., Cortizas E.M., Grills G.S., Herrin J., Kemper J.M., Kenney R., Kobetz E., Laine J., Lamar W.E., Mason C.E., Quintero A.Z., Reding B.D., Roca M.A., Ryon K., Schuurer S.C., Shukla B., Solle N., Stevenson M., Stone T., Tallon J.J., Venkatapuram S.S., Vidovic D., Williams S.L., Young B., Solo-Gabriele H.M. Lessons learned from SARS-CoV-2 measurements in wastewater. Sci. Total Environ. 2021;798(149177):1–13. doi: 10.1016/j.scitotenv.2021.149177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurbeck R.R., Minard-Smith A., Catlin L. Feasibility of neighborhood and building scale wastewater-based genomic epidemiology for pathogen surveillance. Sci. Total Environ. 2021;789(147829):1–8. doi: 10.1016/j.scitotenv.2021.147829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.R., Nancharaiah Y.V., Gu X., Lee W.L., Rajal V.B., Haines M.B., Girones R., Ng L.C., Alm E.J., Wuertz S. Making waves: wastewater surveillance of SARS-CoV-2 for population-based health management. Water Res. 2020;184(116181):1–6. doi: 10.1016/j.watres.2020.116181. [DOI] [PMC free article] [PubMed] [Google Scholar]