Abstract
The sequence of an approximately 1.1-kb DNA fragment of the pbp2x gene, which encodes the transpeptidase domain, was determined for 35 clinical isolates of Streptococcus pneumoniae for which the cefotaxime (CTX) MICs varied. Strains with substitutions within a conserved amino acid motif changing STMK to SAFK and a Leu-to-Val change just before the KSG motif were highly resistant to CTX (MIC, ≧2 μg/ml). Strains with substitutions adjacent to SSN or KSG motifs had low-level resistance. The amino acid substitutions were plotted on the three-dimensional crystallographic structure of the transpeptidase domain of PBP2X. Transformants containing pbp2x from strains with high-level CTX resistance increased the CTX MIC from 0.016 μg/ml to 0.5 to 1.0 μg/ml.
Clinical isolates of penicillin (PC)-resistant Streptococcus pneumoniae (PRSP) for which cefotaxime (CTX) MICs are ≧2 μg/ml have been reported in recent years (3, 5, 8, 24, 30). In Japan, PRSP isolates for which CTX MICs ranged from 2 to 8 μg/ml have been recently isolated.
The high-molecular-weight PC-binding proteins (PBPs) 1A, 2X, and 2B, usually detected in S. pneumoniae, are involved in transpeptidase activity and contain conserved amino acid motifs of SXXK, including the active-site serine residue as a target of β-lactams, SXN, and KT(S)G. The decreased affinity of PBP 1A, 2B, and 2X for β-lactams has been shown to play an important role in the development of their resistance (2, 12, 17, 20, 29). In particular, alterations in PBP 2B mediate low-level resistance to PCs (25), while those in PBP 2X mediate low-level resistance to cephalosporins (7, 10, 13). Additional alterations in PBP 1A increased PC MICs to ≧1 μg/ml and CTX MICs to ≧0.5 μg/ml (20, 22, 29). The evidence that PBP 2A and PBP 1B with low affinity also affect β-lactam resistance was presented by Hakenbeck et al. (11).
Genetic analyses of pbp1a (1, 18), pbp2x (15, 16), and pbp2b (6) in PC-susceptible S. pneumoniae (PSSP) and PRSP have already been conducted. As for PBP 1A, of the many amino acid substitutions in the transpeptidase domain, substitution of Ala or Ser for Thr-371 in the conserved STMK motif has been most important for the development of PC resistance (1, 26). As for PBP 2B, substitutions of Ala or Ser for Thr just after the SSN motif, and of Gly for conserved Glu, were important in developing PC resistance (6, 25, 31). On the other hand, substitution of Ala for Thr just after the KSG motif in PBP 2X involved low-level resistance of cephalosporins (10, 23). Recently, structural evidence linking resistance to multiple β-lactams to amino acid substitutions for Thr-338 and/or Ser-571 within a buried cavity near the Ser-337 of a catalytic site in PBP 2X has been presented by Mouz et al. (19).
We determined the nucleotide sequence of a 1.1-kb region encoding transpeptidase activity, from bp 1018 to 2080, in the pbp2x gene sequence of S. pneumoniae strains (n = 35) isolated in Japan between 1993 and 1997. Amplification of DNA fragments and the sequencing reaction were carried out as described previously (1) with the following PCR primers: 5′-T958ATGAAAAGGATCGTCTGGG977 and 5′-A2105GAGAGTCTTTCATAGCTGAAGC2083. The correlation between amino acid substitutions in PBP 2X and the development of cephalosporin resistance was then examined.
The amino acid sequence of PBP 2X in the strains for which the CTX MICs were ≧0.125 μg/ml exhibited a variety of amino acid substitutions different from those of CTX-susceptible strains. On the basis of the substitution patterns within or adjacent to the three conserved amino acid motifs of STMK, SSN, and KSG, resistant strains were classified into five groups (Table 1). Table 1 also shows (i) the serotypes of the strains, (ii) the susceptibilities of the strains to PC, CTX, cefpodoxime, cefditoren, cefdinir, and cefaclor, and (iii) mutations in the pbp1a and pbp2b genes. Figure 1 shows the predicted amino acid sequence from residues 271 to 610 of a representative strain from each group. The nucleotide sequences of five PSSP strains were determined for comparison. They were identical to those of the strain R6.
TABLE 1.
Classification based on PBP 2X sequence differences and properties of S. pneumoniae strains
| Group | Strain | Serotype | MIC (μg/ml)a
|
Amino acid motifb
|
PCR resultc
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PC | CTX | CPDX | CDTR | CFDN | CCL | S T M K | A H S S N V | L K S G T | pbp1a | pbp2b | |||
| None | 1/E9 | 15 | 0.031 | 0.031 | 0.125 | 0.016 | 0.125 | 1 | –––– | –––––– | ––––– | − | − |
| 2/HSB1 | 15 | 0.031 | 0.031 | 0.125 | 0.031 | 0.125 | 2 | –––– | V––––– | ––––– | − | − | |
| 3/YO42 | 3 | 0.031 | 0.063 | 0.125 | 0.031 | 0.125 | 1 | –––– | –––––– | ––––– | − | − | |
| 4/HSC21 | 6 | 0.031 | 0.063 | 0.125 | 0.031 | 0.25 | 1 | –––– | –––––– | ––––– | − | − | |
| 5/H48 | 19 | 0.031 | 0.063 | 0.125 | 0.031 | 0.25 | 1 | –––– | –––––– | ––––– | − | − | |
| I | 6/HSB5 | 3 | 0.008 | 0.125 | 0.5 | 0.031 | 0.25 | 2 | –––– | –––––– | ––––A | − | − |
| 7/T37 | 6 | 0.008 | 0.125 | 0.25 | 0.031 | 0.25 | 2 | –––– | –––––– | ––––A | − | − | |
| 8/TJ24 | 9 | 0.016 | 0.125 | 0.25 | 0.031 | 0.25 | 1 | –––– | –––––– | ––––A | − | − | |
| 9/E13 | 9 | 0.016 | 0.25 | 0.5 | 0.031 | 0.25 | 2 | –––– | –––––– | ––––A | − | − | |
| II | 14/TO22 | 19 | 0.031 | 0.063 | 0.25 | 0.063 | 0.125 | 0.5 | –––– | –L–––– | V–––– | − | − |
| 17/SU1 | 19 | 0.063 | 0.125 | 0.25 | 0.016 | 0.25 | 1 | –––– | –L–––– | V–––– | − | − | |
| 21/H3 | 10 | 0.031 | 0.125 | 0.25 | 0.063 | 0.125 | 1 | –––– | –L–––– | ––––– | − | − | |
| 16/MA37 | 23 | 0.063 | 0.125 | 0.25 | 0.125 | 0.25 | 1 | –––– | VL–––– | ––––– | − | − | |
| 22/H23 | 14 | 0.031 | 0.063 | 0.25 | 0.063 | 0.25 | 1 | –––– | VL–––– | ––––– | − | − | |
| III | 11/KM99 | 3 | 0.031 | 0.125 | 0.5 | 0.063 | 0.25 | 0.5 | –A–– | –––––– | ––––– | − | − |
| 15/HSC9 | 3 | 0.031 | 0.125 | 0.5 | 0.125 | 0.25 | 1 | –A–– | –––––– | ––––– | − | − | |
| 18/TJ41 | 3 | 0.031 | 0.125 | 0.5 | 0.063 | 0.25 | 2 | –A–– | –––––– | ––––– | − | − | |
| 23/H69 | 3 | 0.031 | 0.25 | 0.25 | 0.063 | 0.25 | 1 | –A–– | –––––– | ––––– | − | − | |
| 25/S19 | 3 | 0.031 | 0.125 | 0.5 | 0.125 | 0.25 | 1 | –A–– | –––––– | ––––– | − | − | |
| IV | 34/H31 | 6 | 1 | 0.5 | 1 | 0.5 | 4 | 32 | –A–– | –––––– | V–––– | + | + |
| 36/Z21 | 6 | 2 | 0.5 | 2 | 0.5 | 8 | 64 | –A–– | –––––– | V–––– | + | + | |
| 37/H28 | 19 | 1 | 0.5 | 2 | 0.5 | 2 | 64 | –A–– | –––––– | V–––– | + | + | |
| 38/KM90 | 19 | 2 | 0.5 | 2 | 0.5 | 4 | 64 | –A–– | –––––– | V–––– | + | + | |
| 39/Z13 | 19 | 2 | 0.5 | 1 | 0.5 | 4 | 64 | –A–– | –––––– | V–––– | + | + | |
| 40/H29 | 23 | 1 | 0.5 | 2 | 0.5 | 4 | 64 | –A–– | –––––– | V–––– | + | + | |
| 41/B99 | 23 | 1 | 0.5 | 2 | 0.5 | 4 | 64 | –A–– | –––––– | V–––– | + | + | |
| 42/H0 | 14 | 4 | 1 | 4 | 1 | 8 | 64 | –A–– | –––––– | V–––– | + | + | |
| 27/S46 | 23 | 0.5 | 0.5 | 1 | 0.25 | 2 | 32 | –A–– | –––––– | V–––– | − | + | |
| 1A22/HA5 | 6 | 0.25 | 0.5 | 1 | 0.25 | 2 | 16 | –A–– | –––––– | V–––– | − | + | |
| V | 20/SHA3 | 19 | 2 | 4 | 16 | 4 | 32 | 64 | –A–– | V––––– | V–––– | + | + |
| 29/KK133 | 23 | 2 | 8 | 16 | 4 | 32 | 64 | –AF– | –––––– | V–––– | + | + | |
| 30/NG44 | 23 | 2 | 2 | 16 | 2 | 16 | 64 | –AF– | –––––– | V–––– | + | + | |
| 31/KU5 | 23 | 1 | 2 | 8 | 2 | 8 | 64 | –AF– | –––––– | V–––– | + | + | |
| 32/KU81 | 19 | 4 | 4 | 16 | 2 | 16 | 64 | –AF– | –––––– | V–––– | + | + | |
| 33/AK5 | 14 | 0.125 | 4 | 16 | 2 | 2 | 8 | –AF– | –––––– | V–––– | + | − | |
Susceptibilities to PC, CTX, cefpodoxime (CPDX), cefditoren (CDTR), cefdinir (CFDN), and cefaclor (CCL) were determined by previously described methods (1).
Only amino acid residues differing from PBP 2X conserved motif sequences of the PSSP strain R6 are shown. Conserved amino acid motifs are underlined.
Altered PBP genes were detected by PCR as described previously (29). +, altered; −, not altered.
FIG. 1.
Deduced amino acid sequences of part of PBP 2X from representative strains from each group. The sequence of PSSP R6 is shown on the top line. Numbering is based on published data on the R6 strain (15). Only amino acids differing from the R6 sequence are shown. Boxes represent conserved amino acid motifs.
Four strains were classified into group I. In this group, Thr-550 just after the KSG motif was replaced with Ala. From the X-ray crystallographic structure of a complex of a homologous dd-peptidase and CTX, Kuzin et al. (14) showed that the loss of a hydrogen bond between the Thr and CTX by the change to Ala can account for the higher CTX MICs. Other than this Thr, only one or two amino acid substitutions were confirmed. The CTX MIC for these strains was 0.125 μg/ml, which was four to eight times higher than that for PSSP.
Eight strains were classified into group II. Ala-393 and His-394 just before the SSN motif were replaced with Val and Leu, respectively, or His-394 and Leu-546 just before the SSN and KSG motifs were replaced with Leu and Val, respectively. The CTX MICs for these strains were also about 0.125 μg/ml.
Five strains that were classified into group III showed a substitution of Ala for Thr-338 in the STMK motif. The CTX MICs for these strains ranged from 0.125 to 0.25 μg/ml. The homology of amino acid sequences between these strains and the R6 strain was 90.7%.
PRSP strains classified into group IV were isolated predominantly in Japan. These strains had altered pbp1a, pbp2x, and pbp2b genes and two amino acid substitutions, Ala for Thr-338 in STMK and Val for Leu-546 adjacent to KSG, in the pbp2x gene product.
CTX MICs for strains that were classified into group V ranged from 2 to 8 μg/ml, which is four to eight times higher than those for group IV. The general amino acid substitutions in group V strains were virtually the same as those in the group IV strains, but Thr-Met in STMK was replaced with Ala-Phe. The amino acid sequence of these strains was highly homologous to that of the high-level CTX resistant strain CS111 isolated in the United States in 1991 (homology ranging from 99.7 to 100%) (5). The serotypes of the strains with high-level resistance were 14, 19, and 23.
Many strains in groups IV and V showed simultaneous alterations in PBP 1A, 2X, and 2B, while strains of other groups showed resistance mediated by an alteration in PBP 2X only. To clarify the effect of substitutions in the conserved amino acid motifs of PBP 2X on CTX MICs, the amplified pbp2x genes of strains in groups IV and V were used to transform a PSSP strain, KK97 (Table 2). Transformation of pbp2x genes was monitored by procedures previously described (27, 28). Transformants were selected on blood agar containing CTX and were confirmed by sequencing to contain the pbp2x gene of donor DNA. CTX MICs for transformants containing pbp2x DNA of the 34/H31 and 40/H29 strains (group IV) increased from 0.016 to 0.5 μg/ml. In contrast, CTX MICs for transformants from the 29/KK133 and 32/KU81 strains (group V) increased to 1.0 μg/ml. Although the pbp2x gene from group V strains could not transform a susceptible recipient strain to donor-level CTX resistance by itself, it increased the resistance to a slightly higher level than that of the others.
TABLE 2.
Susceptibilities to β-lactam antibiotics of S. pneumoniae transformants obtained with pbp2x DNA
| Straina | MIC (μg/ml)b
|
|||||
|---|---|---|---|---|---|---|
| PC | CTX | CPDX | CDTR | CFDN | CCL | |
| Recipient, KK97 | 0.031 | 0.016 | 0.031 | 0.016 | 0.063 | 0.5 |
| Transformant | ||||||
| KK972X34/H31 | 0.063 | 0.5 | 2.0 | 0.5 | 4.0 | 2.0 |
| KK972X40/H29 | 0.063 | 0.5 | 2.0 | 0.5 | 4.0 | 2.0 |
| KK972X29/KK133 | 0.063 | 1.0 | 2.0 | 0.5 | 8.0 | 2.0 |
| KK972X32/KU81 | 0.063 | 1.0 | 4.0 | 0.5 | 8.0 | 2.0 |
Nonencapsulated strain KK97 was transformed with PCR-amplified pbp2x from group IV and V strains as the donor DNA. Transformants were selected on plates containing 0.2 or 0.5 μg of CTX per ml. Example of transformant nomenclature, KK972X34/H31 corresponds to KK97 strain containing the pbp2x gene from strain 34/H31.
For abbreviations of β-lactams, see Table 1, footnote a.
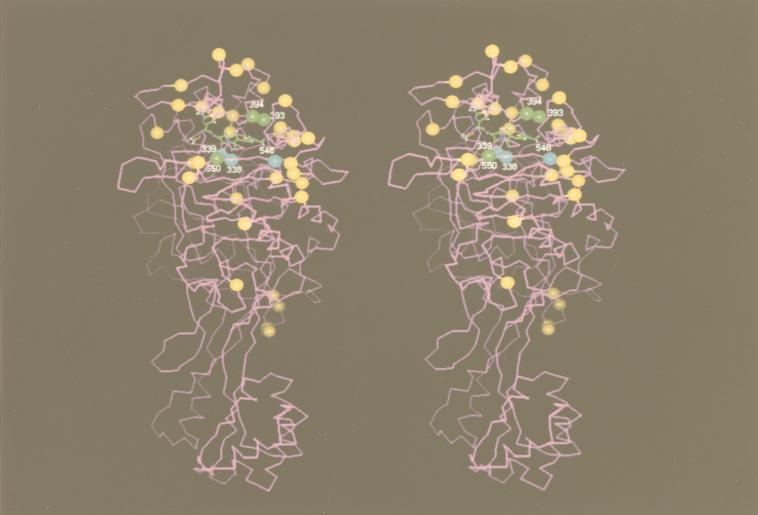
Charlier et al. (4) crystallized PBP 2X, and Pares et al. (21) determined its three-dimensional structure by X-ray crystallography. According to their observations, PBP 2X consisted of three domains. Its central domain was the transpeptidase domain, with a long groove surrounded with conserved motifs of STMK, SSN, and KSG. The active-site serine to which β-lactam binds was located in the STMK motif at the center of this groove. Figure 2 shows a stereoview of the PBP 2X transpeptidase domain of the PRSP strain 31/KU5, in which amino acids differing from the R6 sequence are marked with yellow, green, and blue circles.
FIG. 2.
Stereoview of the transpeptidase domain of PBP 2X of the high-level CTX-resistant PRSP strain 31/KU5. The structure was constructed by using the crystallographic coordinates of PBP 2X of Pares et al. (PDB entry code 1 PMD) (21). Cefditoren of oral cephalosporin was shown in the long groove as a β-lactam model. Amino acids differing from the R6 sequence are indicated by colored circles (blue, substitutions within or adjacent to conserved motifs of STMK and KTG; green, substitutions adjacent to SSN and KSG in strains classified into groups I and II; yellow, general substitutions).
Distinct amino acid changes, as well as those near the conserved motif, are probably important but are more difficult to understand. An extended structural analysis, while far beyond the scope of this paper, would be worthwhile.
Zhao et al. (32) reported that the kinetics of the transpeptidase activity of PBP 2X differed significantly between penicillin-resistant and -susceptible strains of S. pneumoniae. The resistant strain used in their study was classified into group IV in the present study. Furthermore, Garcia-Bustos and Tomasz (9) used whole cells and documented that the products of pentaglycine bridge reactions differed between susceptible and resistant strains. It was speculated that these changes were attributable to modification of the three-dimensional structure of the active domain of high-molecular-mass PBPs in the resistant strains.
We have not yet isolated S. pneumoniae with high-level CTX resistance with substitutions such as Pro for Ser-571 in PBP 2X. Nevertheless, since many oral cephalosporins are currently in use in Japan, we fear that the strains with high-level cephalosporin resistance with the PBP 2X alterations so far described will predominate among strains isolated in the future.
Nucleotide sequence accession numbers.
The nucleotide sequences determined in this study will appear in the DDBJ/EMBL/Gen Bank nucleotide sequence databases under the following accession numbers: AB011198 to AB011210 and AB015846 to AB015852.
REFERENCES
- 1.Asahi Y, Ubukata K. Association of a Thr-371 substitution in a conserved amino acid motif of penicillin-binding protein 1A with penicillin resistance of Streptococcus pneumoniae. Antimicrob Agents Chemother. 1998;42:2267–2273. doi: 10.1128/aac.42.9.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barcus V A, Ghanekar K, Yeo M, Coffey T J, Dowson C G. Genetics of high level penicillin resistance in clinical isolates of Streptococcus pneumoniae. FEMS Microbiol Lett. 1995;126:299–303. doi: 10.1111/j.1574-6968.1995.tb07433.x. [DOI] [PubMed] [Google Scholar]
- 3.Bradley J S, Connor J D. Ceftriaxone failure in meningitis caused by Streptococcus pneumoniae with reduced susceptibility to beta-lactam antibiotics. Pediatr Infect Dis J. 1991;10:871–873. [PubMed] [Google Scholar]
- 4.Charlier P, Buisson G, Dideberg O, Wierenga J, Keck W, Laible G, Hakenbeck R. Crystallization of a genetically engineered water-soluble primary penicillin target enzyme. The high molecular mass PBP2x of Streptococcus pneumoniae. J Mol Biol. 1993;232:1007–1009. doi: 10.1006/jmbi.1993.1452. [DOI] [PubMed] [Google Scholar]
- 5.Coffey T J, Daniels M, McDougal L K, Dowson C G, Tenover F C, Spratt B G. Genetic analysis of clinical isolates of Streptococcus pneumoniae with high-level resistance to expanded-spectrum cephalosporins. Antimicrob Agents Chemother. 1995;39:1306–1313. doi: 10.1128/aac.39.6.1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dowson C G, Hutchison A, Spratt B G. Extensive re-modelling of the transpeptidase domain of penicillin-binding protein 2B of a penicillin-resistant South African isolate of Streptococcus pneumoniae. Mol Microbiol. 1989;3:95–102. doi: 10.1111/j.1365-2958.1989.tb00108.x. [DOI] [PubMed] [Google Scholar]
- 7.Dowson C G, Johnson A P, Cercenado E, George R C. Genetics of oxacillin resistance in clinical isolates of Streptococcus pneumoniae that are oxacillin resistant and penicillin susceptible. Antimicrob Agents Chemother. 1994;38:49–53. doi: 10.1128/aac.38.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Figueiredo A M S, Connor J D, Severin A, Vaz Pato M V, Tomasz A. A pneumococcal clinical isolate with high-level resistance to cefotaxime and ceftriaxone. Antimicrob Agents Chemother. 1992;36:886–889. doi: 10.1128/aac.36.4.886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia-Bustos J, Tomasz A. A biological price of antibiotic resistance: major changes in the peptidoglycan structure of penicillin-resistant pneumococci. Proc Natl Acad Sci USA. 1990;87:5415–5419. doi: 10.1073/pnas.87.14.5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grebe T, Hakenbeck R. Penicillin-binding proteins 2b and 2x of Streptococcus pneumoniae are primary resistance determinants for different classes of β-lactam antibiotics. Antimicrob Agents Chemother. 1996;40:829–834. doi: 10.1128/aac.40.4.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hakenbeck R, König A, Kern I, van der Linden M, Keck W, Billot-Klein D, Legrand R, Schoot B, Gutmann L. Acquisition of five high-Mr penicillin-binding protein variants during transfer of high-level β-lactam resistance from Streptococcus mitis to Streptococcus pneumoniae. J Bacteriol. 1998;180:1831–1840. doi: 10.1128/jb.180.7.1831-1840.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kell C M, Sharma U K, Dowson C G, Town C, Balganesh T S, Spratt B G. Deletion analysis of the essentiality of penicillin-binding proteins 1A, 2B, 2X of Streptococcus pneumoniae. FEMS Microbiol Lett. 1993;106:171–175. doi: 10.1111/j.1574-6968.1993.tb05954.x. [DOI] [PubMed] [Google Scholar]
- 13.Krauss J, van der Linden M, Grebe T, Hakenbeck R. Penicillin-binding protein 2x and 2b as primary PBP targets in Streptococcus pneumoniae. Microb Drug Resist. 1996;2:183–186. doi: 10.1089/mdr.1996.2.183. [DOI] [PubMed] [Google Scholar]
- 14.Kuzin A P, Liu H, Kelly J A, Knox J R. Binding of cephalothin and cefotaxime to d-Ala-d-Ala-peptidase reveals a functional basis of a natural mutation in a low-affinity penicillin-binding protein and in extended-spectrum β-lactamases. Biochemistry. 1995;34:9532–9540. doi: 10.1021/bi00029a030. [DOI] [PubMed] [Google Scholar]
- 15.Laible G, Hakenbeck R, Sicard M A, Joris B, Ghuysen J M. Nucleotide sequences of the pbpX genes encoding the penicillin-binding proteins 2X from Streptococcus pneumoniae R6 and a cefotaxime-resistant mutant, C506. Mol Microbiol. 1989;3:1337–1348. doi: 10.1111/j.1365-2958.1989.tb00115.x. [DOI] [PubMed] [Google Scholar]
- 16.Laible G, Spratt B G, Hakenbeck R. Interspecies recombinational events during the evolution of altered PBP 2x genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Mol Microbiol. 1991;5:1993–2002. doi: 10.1111/j.1365-2958.1991.tb00821.x. [DOI] [PubMed] [Google Scholar]
- 17.Markiewicz Z, Tomasz A. Variation in penicillin-binding protein patterns of penicillin-resistant clinical isolates of pneumococci. J Clin Microbiol. 1989;27:405–410. doi: 10.1128/jcm.27.3.405-410.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin C, Briese T, Hakenbeck R. Nucleotide sequences of genes encoding penicillin-binding proteins from Streptococcus pneumoniae and Streptococcus oralis with high homology to Escherichia coli penicillin-binding proteins 1A and 1B. J Bacteriol. 1992;174:4517–4523. doi: 10.1128/jb.174.13.4517-4523.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mouz N, Gordon E, Di Guilmi A M, Petit I, Petillot Y, Dupont Y, Hakenbeck R, Vernet T, Dideberg O. Identification of a structural determinant for resistance to β-lactam antibiotics in gram-positive bacteria. Proc Natl Acad Sci USA. 1998;95:13403–13406. doi: 10.1073/pnas.95.23.13403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muñoz R, Dowson C G, Daniels M, Coffey T J, Martin C, Hakenbeck R, Spratt B G. Genetics of resistance to third-generation cephalosporins in clinical isolates of Streptococcus pneumoniae. Mol Microbiol. 1992;6:2461–2465. doi: 10.1111/j.1365-2958.1992.tb01422.x. [DOI] [PubMed] [Google Scholar]
- 21.Pares S, Mouz N, Petillot Y, Hakenbeck R, Dideberg O. X-ray structure of Streptococcus pneumoniae PBP2x, a primary target enzyme. Nat Struct Biol. 1996;3:284–289. doi: 10.1038/nsb0396-284. [DOI] [PubMed] [Google Scholar]
- 22.Reichmann P, König A, Marton A, Hakenbeck R. Penicillin-binding proteins as resistance determinants in clinical isolates of Streptococcus pneumoniae. Microb Drug Resist. 1996;2:177–181. doi: 10.1089/mdr.1996.2.177. [DOI] [PubMed] [Google Scholar]
- 23.Sifaoui F, Kitzis M D, Gutmann L. In vitro selection of one-step mutants of Streptococcus pneumoniae resistant to different oral β-lactam antibiotics is associated with alterations of PBP2x. Antimicrob Agents Chemother. 1996;40:152–156. doi: 10.1128/aac.40.1.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sloas M M, Barrett F F, Chesney P J, English B K, Hill B C, Tenover F C, Leggiadro R J. Cephalosporin treatment failure in penicillin- and cephalosporin-resistant Streptococcus pneumoniae meningitis. Pediatr Infect Dis J. 1992;11:662–666. [PubMed] [Google Scholar]
- 25.Smith A M, Klugman K P. Alterations in penicillin-binding protein 2B from penicillin-resistant wild-type strains of Streptococcus pneumoniae. Antimicrob Agents Chemother. 1995;39:859–867. doi: 10.1128/aac.39.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith A M, Klugman K P. Alterations in PBP 1A essential for high-level penicillin resistance in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1998;42:1329–1333. doi: 10.1128/aac.42.6.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomasz A, Hotchkiss R D. Regulation of the transformability of pneumococcal cultures by macromolecular cell products. Proc Natl Acad Sci USA. 1964;51:480–487. doi: 10.1073/pnas.51.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tomasz A, Mosser J L. On the nature of the pneumococcal activator substance. Proc Natl Acad Sci USA. 1966;55:58–66. doi: 10.1073/pnas.55.1.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ubukata K, Muraki T, Igarashi A, Asahi Y, Konno M. Identification of penicillin and other beta-lactam resistance in Streptococcus pneumoniae by polymerase chain reaction. J Infect Chemother. 1997;3:190–197. doi: 10.1007/BF02490033. [DOI] [PubMed] [Google Scholar]
- 30.Viladrich P F, Gudiol F, Linares J, Pallares R, Sabate I, Rufi G, Ariza J. Evaluation of vancomycin for therapy of adult pneumococcal meningitis. Antimicrob Agents Chemother. 1991;35:2467–2472. doi: 10.1128/aac.35.12.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamane A, Nakano H, Asahi Y, Ubukata K, Konno M. Directly repeated insertion of 9-nucleotide sequence detected in penicillin-binding protein 2B gene of penicillin-resistant Streptococcus pneumoniae. Antimicrob Agents Chemother. 1996;40:1257–1259. doi: 10.1128/aac.40.5.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao G, Yeh W K, Carnahan R H, Flokowitsch J, Meier T I, Alborn W E, Jr, Becker G W, Jaskunas S R. Biochemical characterization of penicillin-resistant and -sensitive penicillin-binding protein 2x transpeptidase activities of Streptococcus pneumoniae and mechanistic implications in bacterial resistance to β-lactam antibiotics. J Bacteriol. 1997;179:4901–4908. doi: 10.1128/jb.179.15.4901-4908.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]