Abstract
Background
A decline in the level of genetic diversity in livestock can result in reduced response to selection, greater incidence of genetic defects, and inbreeding depression. In this context, various metrics have been proposed to assess the level of genetic diversity in selected populations. Therefore, the main goals of this study were to: 1) investigate the population structure of 16 cattle populations from 15 different pure breeds or composite populations, which have been selected for different breeds goals; and, 2) identify and compare runs of homozygosity (ROH) and heterozygosity-enriched regions (HER) based on different single nucleotide polymorphism (SNP) panels and whole-genome sequence data (WGS), followed by functional genomic analyses.
Results
A total of 24,187 ROH were found across all cattle populations, with 55% classified in the 2-4 Mb size group. Fourteen homozygosity islands were found in five populations, where four ROH islands located on BTA1, BTA5, BTA16, and BTA19 overlapped between the Brahman (BRM) and Gyr (GIR) breeds. A functional analysis of the genes found in these islands revealed candidate genes known to play a role in the melanogenesis, prolactin signaling, and calcium signaling pathways. The correlations between inbreeding metrics ranged from 0.02 to 0.95, where the methods based on homozygous genotypes (FHOM), uniting of gametes (FUNI), and genotype additive variance (FGRM) showed strong correlations among them. All methods yielded low to moderate correlations with the inbreeding coefficients based on runs of homozygosity (FROH). For the HER, 3576 runs and 26 islands, distributed across all autosomal chromosomes, were found in regions containing genes mainly related to the immune system, indicating potential balancing selection. Although the analyses with WGS did not enable detection of the same island patterns, it unraveled novel regions not captured when using SNP panel data.
Conclusions
The cattle populations that showed the largest amount of ROH and HER were Senepol (SEN) and Montana (MON), respectively. Overlapping ROH islands were identified between GIR and BRM breeds, indicating a possible historical connection between the populations. The distribution and pattern of ROH and HER are population specific, indicating that different breeds have experienced divergent selection processes or different genetic processes.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-022-08384-0.
Keywords: Autozygosity, Runs of heterozygosity, Inbreeding coefficient, Signature of selection
Background
Since cattle domestication, which occurred around 10,000 years ago [1], over 1000 breeds [2, 3] have been developed through selection for different traits and breeding goals. Therefore, cattle is a valuable animal model for studying genomic changes in response to processes such as selection, crossbreeding, and domestication [4]. Genetic selection for specific traits can result in signatures of selection (also known as selective sweeps), which are characterized by genomic regions with reduced genetic variability, i.e., greater concentration of homozygous alleles [5]. In this context, various studies have revealed that one of the consequences of intensive selection is the increase of homozygosity [6], resulting in a loss of genetic diversity within populations [7, 8]. Furthermore, high levels of inbreeding are directly related to a higher incidence of ROH, which, if not controlled, could result in other issues such as congenital anomalies [7] and inbreeding depression [9].
One effect of these concentrations of homozygous alleles is the emergence of ROH. A ROH is defined as the continuous length of homozygous genotypes that are present in an individual genome due to the progenitors transmitting identical haplotypes to their descendants [8]. The identification of ROH enables the estimation of parameters regarding the genetic structure and history of the population, including autozygosity and inbreeding coefficients [10]. Given the random nature of recombination, the occurrence of ROH is highly heterogeneous across the genome, where regions with a high incidence of ROH in a large number of samples is indicative of the selection pressure suffered by that population [11]. Previous studies have been carried out to evaluate the incidence of ROH islands in cattle populations [e.g. 6-8] as well as in many other livestock species [e.g. 9].
Heterozygosity-enriched regions, also known as runs of heterozygosity, consists on the identification of genomic regions with high variability, and can provide information about the population diversity and evolutionary history [12]. For instance, Biscarini et al. [13], Marras et al. [14], and Bizarria dos Santos et al. [15] have identified HER in semi-feral cattle, poultry, and equine, respectively. This method aims to identify genomic regions with high genetic variability, providing information about the population genetic diversity level and evolutionary history [12], as well as identify specific segments in the genome where maintaining greater genetic diversity might be more beneficial [13].
Genetic diversity is not static, being a continuous process of creation and loss according to evolutionary and selection changes in a population. The maintenance of sufficient genetic diversity is important for the long-term sustainability of livestock populations [7, 8, 16]. The large majority of genetic diversity studies has been carried out based on SNP panel data, which are commonly used in genomic-based breeding programs [17]. Alternatively, WGS provides an opportunity for more accurately assessing genetic diversity in cattle breeds [18]. This is possible due to the superior genomic coverage when using WGS data. Furthermore, the use of WGS data may reveal multiple rare and common variants in ROH/HER that have not been identified when using only SNP panel data. Therefore, new possibilities to better understand the genomic structure involved in ROH/HER regions can be revealed for new populations or traits [19].
The main objectives of this study were to: 1) characterize the population structure of 16 cattle populations from 15 pure or composite breeds selected for different breeding goals; 2) quantify and classify ROH and HER in each population based on their length and alternative detection parameters; 3) perform functional annotation analyses to identify candidate genes and pathways involved in the genomic regions with higher concentration of ROH and HER; 4) estimate inbreeding coefficients and effective population size based on genomic information; and, 5) compare the results obtained from the analyzes of SNP and WGS.
Results
Runs of homozygosity
The group that presented the highest number of ROH is the SEN breed (Fig. 1) with 4198 ROHs distributed among all the autosomal chromosomes. The population with the smallest amount of ROH was the Nellore group genotyped with the 35 K SNP panel (NEL35), which also had the smallest percentage (63.16%) of individuals with at least one ROH per animal. All animals from all populations had at least one ROH, except for some animals of Angus x Simmental crossbred (ANGSIM), MON, NEL35, and SEN breeds.
Fig. 1.
Classification of runs of homozygosity (ROH) according to length size and bovine breeds
In total, 24,187 ROHs were identified, distributed throughout the autosomal chromosomes. The majority of ROHs were classified as 2-4 Mb, representing 55% of all ROHs found. In summary, only 14% of the ROHs were larger than 8 Mb; of these, 24% were larger than 16 Mb. The distribution and classification of ROHs by chromosome and population can be visualized in Additional file 1: Fig. S1. Each group showed a specific ROH concentration by chromosome. In general, BTA1, BTA6, and BTA7 showed the highest ROH concentrations across populations.
Genomic inbreeding coefficients and effective population size
The ANGSIM population showed the lowest inbreeding rate (− 0.026), except for the FROH method (Table 1). For FROH, the NEL35 population showed the lowest genomic inbreeding rate (0.001). The Hereford (HFD) population showed the highest inbreeding rate for the methodologies FHOM1 (0.086), FGRM (0.087), FHOM2 (0.087), and FUNI (0.087), while the SEN population showed the highest inbreeding rate (0.075) when FROH was used.
Table 1.
Average of inbreeding coefficients estimated for the five inbreeding calculation methodologies
| FHOM1 | FGRM | FHOM2 | FUNI | FROH | < 2 Mb | 2-4 Mb | 4-8 Mb | 8-16 Mb | > 16 Mb | < 8 Mb | > 8 Mb | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ANG | −0.003 | − 0.003 | − 0.003 | −0.003 | 0.043 | 0.000 | 0.014 | 0.015 | 0.010 | 0.004 | 0.029 | 0.014 |
| ANGSIM | −0.026 | −0.024 | −0.024 | − 0.024 | 0.001 | 0.000 | 0.001 | 0.000 | 0.000 | 0.000 | 0.001 | 0.000 |
| BOR | 0.002 | 0.002 | 0.002 | 0.002 | 0.012 | 0.001 | 0.006 | 0.001 | 0.001 | 0.002 | 0.008 | 0.004 |
| BRM | 0.014 | 0.011 | 0.011 | 0.011 | 0.036 | 0.001 | 0.020 | 0.007 | 0.006 | 0.002 | 0.028 | 0.008 |
| CGU | 0.024 | 0.024 | 0.024 | 0.024 | 0.018 | 0.000 | 0.005 | 0.003 | 0.004 | 0.006 | 0.009 | 0.009 |
| CHL | −0.004 | −0.004 | −0.004 | −0.004 | 0.018 | 0.000 | 0.005 | 0.005 | 0.004 | 0.004 | 0.011 | 0.008 |
| GIR | −0.011 | −0.011 | − 0.011 | −0.011 | 0.043 | 0.002 | 0.027 | 0.007 | 0.005 | 0.003 | 0.036 | 0.008 |
| HFD | 0.086 | 0.087 | 0.087 | 0.087 | 0.061 | 0.001 | 0.016 | 0.019 | 0.016 | 0.009 | 0.037 | 0.024 |
| HOL | −0.012 | −0.012 | −0.012 | − 0.012 | 0.026 | 0.000 | 0.006 | 0.008 | 0.008 | 0.004 | 0.014 | 0.012 |
| JER | 0.023 | 0.030 | 0.030 | 0.030 | 0.047 | 0.001 | 0.015 | 0.017 | 0.010 | 0.004 | 0.033 | 0.015 |
| LMS | −0.007 | −0.006 | −0.006 | −0.006 | 0.015 | 0.000 | 0.005 | 0.004 | 0.004 | 0.002 | 0.009 | 0.006 |
| MON | −0.017 | −0.018 | −0.018 | − 0.018 | 0.006 | 0.000 | 0.002 | 0.002 | 0.001 | 0.001 | 0.004 | 0.002 |
| NEL35 | −0.019 | −0.019 | − 0.019 | −0.019 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.001 | 0.000 | 0.000 |
| NEL50 | −0.014 | −0.013 | −0.013 | − 0.013 | 0.030 | 0.001 | 0.007 | 0.010 | 0.008 | 0.004 | 0.018 | 0.012 |
| SEN | 0.003 | 0.003 | 0.003 | 0.003 | 0.075 | 0.001 | 0.013 | 0.019 | 0.021 | 0.021 | 0.033 | 0.043 |
| SGT | −0.010 | −0.010 | −0.010 | −0.010 | 0.030 | 0.001 | 0.010 | 0.010 | 0.007 | 0.003 | 0.021 | 0.009 |
All breed abbreviations are defined in Table 5
FHOM1 Inbreeding coefficient based on the number of observed and expected homozygous genotypes, FGRM Inbreeding coefficient based on additive genotypic variance, FHOM2 Inbreeding coefficient based on homozygosity of genotypes (Similar to F1), FUNI Inbreeding coefficient based on the correlation between uniting gametes, FROH Inbreeding coefficient based on the length of the ROH’s and the total length of the autosomal genome
A strong correlation (> 0.75) was found between FHOM1 and FHOM2, FHOM1 and FUNI, and FGRM and FUNI (Fig. 2). All the methods showed weak to moderate correlations with FROH and the smallest correlation was found with the FGRM method.
Fig. 2.
Correlation among inbreeding estimation methods
The effective population size (Ne) for each population from the 54th to 13th generation ago is shown in Fig. 3, and as expected, the Ne of each population decreased over time. The population with the highest Ne, at the most recent generation, was the Creole from Guadalupe (CGU - 443), and the smallest Ne was found in HFD (101). On average, the Ne estimates decreased around 59.6% in the last generations, with the GIR, Limousin (LMS), Charolais (CHL), BRM, Holstein (HOL), and NEL populations presenting the highest reduction in Ne.
Fig. 3.
Behavior of effective population size (Ne) over the last generations
Homozygosity islands
The longest ROH island was found on BTA16 of the BRM breed (Table 2), with an approximate length size of 14 Mb, including 146 SNPs and 229 genes (Additional file 5: Table S1). The smallest ROH island (3 Mb and 63 SNPs) was identified in the ANGSIM population and was present in more than 75% of the ANGSIM animals included in the analyses. This region is known to code for 35 genes (Additional file 6: Table S2).
Table 2.
Homozygosity Islands found in different chromosomes and groups of individuals
| Breed | n animal | n ROH | % | CHR | Start | End | Length | n SNP |
|---|---|---|---|---|---|---|---|---|
| ANGSIM | 487 | 369 | 75.77 | 16 | 62,578,656 | 66,253,552 | 3,674,896 | 62 |
| BRM | 70 | 36 | 51.43 | 1 | 78,237,770 | 84,586,062 | 6,348,292 | 58 |
| BRM | 70 | 36 | 51.43 | 5 | 105,576,062 | 117,735,828 | 12,159,766 | 171 |
| BRM | 70 | 39 | 55.71 | 8 | 56,051,150 | 63,444,254 | 7,393,104 | 58 |
| BRM | 70 | 36 | 51.43 | 16 | 44,071,454 | 58,289,347 | 14,217,893 | 146 |
| BRM | 70 | 39 | 55.71 | 19 | 42,110,400 | 46,627,006 | 4,516,606 | 74 |
| GIR | 50 | 31 | 62.00 | 1 | 80,333,027 | 84,911,107 | 4,578,080 | 80 |
| GIR | 50 | 46 | 92.00 | 5 | 110,192,579 | 116,240,339 | 6,047,760 | 110 |
| GIR | 50 | 30 | 60.00 | 14 | 21,914,329 | 26,212,648 | 4,298,319 | 56 |
| GIR | 50 | 26 | 52.00 | 16 | 45,727,235 | 52,149,496 | 6,422,261 | 111 |
| GIR | 50 | 31 | 62.00 | 19 | 42,054,880 | 46,678,246 | 4,623,366 | 82 |
| JER | 84 | 43 | 51.19 | 6 | 100,066,570 | 110,600,517 | 10,533,947 | 180 |
| SEN | 153 | 99 | 64.71 | 1 | 776,231 | 10,605,227 | 9,828,996 | 160 |
| SEN | 153 | 82 | 53.59 | 20 | 36,135,896 | 42,174,483 | 6,038,587 | 78 |
All breed abbreviations are defined in Table 5
n animal Number of animals evaluated, n ROH Number of ROHs found in position, % Percentage of the population that presented this island, CHR Chromosome, start Start of ROH, end End of ROH, length ROH length, n SNP Number of SNPs that ROH covers
The ROH island present in the highest proportion (92%) of the population was found in the BTA5 for the GIR breed. This island was also present in the BRM breed, in a lower proportion of animals (51.43%). An overlap of ROH islands were observed in the BTA1, BTA16, and BTA19 in both GIR and BRM breeds. These breeds also showed the highest number of ROH islands (5 islands in each breed). In total, the ROH islands in these breeds contain 556 and 863 genes in the GIR and BRM breeds, respectively, which are involved in key biological processes, cellular components, and molecular functions, as detailed in Additional file 5: Tables S1 and Additional file 7: Table S3.
Heterozygous-enriched regions
Two methods were used to detect HER: the windows method, and the consecutive-SNP method [20]. Despite of being used, the windows method did not detect any runs like the consecutive SNPs method. Therefore, only the results obtained for the consecutive method are presented (Fig. 4).
Fig. 4.
Number of heterozygous-enriched regions (HER) per animal breed studied
In total, 3576 HERs were found for all populations and the highest number of HERs was found in the MON population (1702 runs). The smallest number of HER was found in the CGU population (2 HERs). In general, the highest percentage of HER was classified as having a length of 0.5-1 Mb (45.13%) and 1-1.5 Mb (41.11%). The percentage of HER higher than 2 Mb were only detected in 1% of HER. The HER distribution by chromosome in each population indicated that the BTA5 had the highest number of HER, with 12% of all HER (Additional file 2: Fig. S2).
The regions in HER islands present in at least 10% of the animals within each population can be observed in Table 3. The largest HER was found in 20.3% of the MON population with a length of 4.25 Mb (Table 3). This HER was found in BTA5, and this same region was considered as a ROH island in both BRM and GIR breeds. This region harbors 114 genes, where 71 of these are protein coding genes (Additional file 8: Table S4). The smallest region with HER concentration was found in the BTA23 of the Nellore population genotyped with 50 K panel (NEL50), with a length of approximately 0.7 Mb. This region harbors only one gene: KHDRBS2.
Table 3.
Heterozygous-enriched regions (HER), in the different populations, which appear in at least 10% of individuals
| Breed | n animal | n HER | % | CHR | Start | End | Length | n SNP |
|---|---|---|---|---|---|---|---|---|
| ANG | 99 | 20 | 20.20 | 13 | 40,318,645 | 41,433,750 | 1,115,105 | 21 |
| HFD | 61 | 8 | 13.11 | 1 | 32,091,115 | 33,034,361 | 943,246 | 23 |
| HFD | 61 | 9 | 14.75 | 1 | 101,195,925 | 104,830,589 | 3,634,664 | 47 |
| HFD | 61 | 9 | 14.75 | 3 | 75,829,200 | 76,986,902 | 1,157,702 | 27 |
| HFD | 61 | 9 | 14.75 | 3 | 89,652,649 | 90,628,822 | 976,173 | 25 |
| HFD | 61 | 13 | 21.31 | 6 | 78,147,926 | 79,620,230 | 1,472,304 | 25 |
| HFD | 61 | 12 | 19.67 | 14 | 50,316,790 | 52,240,902 | 1,924,112 | 25 |
| HFD | 61 | 7 | 11.48 | 20 | 46,124,910 | 46,912,182 | 787,272 | 20 |
| HFD | 61 | 7 | 11.48 | 24 | 28,353,262 | 29,204,162 | 850,900 | 20 |
| MON | 271 | 67 | 24.72 | 5 | 48,569,574 | 50,434,637 | 1,865,063 | 37 |
| MON | 271 | 62 | 22.88 | 5 | 95,248,428 | 96,123,569 | 875,141 | 22 |
| MON | 271 | 55 | 20.30 | 5 | 110,220,384 | 114,471,702 | 4,251,318 | 36 |
| MON | 271 | 52 | 19.19 | 12 | 44,975,037 | 46,232,946 | 1,257,909 | 21 |
| MON | 271 | 50 | 18.45 | 16 | 58,037,089 | 59,263,750 | 1,226,661 | 24 |
| MON | 271 | 35 | 12.92 | 19 | 60,384,670 | 61,177,248 | 792,578 | 23 |
| MON | 271 | 38 | 14.02 | 27 | 42,506,836 | 43,575,359 | 1,068,523 | 30 |
| NEL35 | 209 | 38 | 18.18 | 23 | 103,505 | 1,712,734 | 1,609,229 | 22 |
| NEL50 | 192 | 83 | 43.23 | 1 | 63,642,322 | 65,584,086 | 1,941,764 | 37 |
| NEL50 | 192 | 24 | 12.50 | 2 | 36,202,336 | 37,386,478 | 1,184,142 | 23 |
| NEL50 | 192 | 30 | 15.63 | 5 | 75,447,962 | 76,805,229 | 1,357,267 | 29 |
| NEL50 | 192 | 26 | 13.54 | 6 | 77,309,507 | 78,206,076 | 896,569 | 24 |
| NEL50 | 192 | 33 | 17.19 | 11 | 16,880,546 | 18,643,939 | 1,763,393 | 24 |
| NEL50 | 192 | 23 | 11.98 | 18 | 23,656,888 | 24,920,756 | 1,263,868 | 24 |
| NEL50 | 192 | 47 | 24.48 | 23 | 588,741 | 1,284,183 | 695,442 | 22 |
| SEN | 153 | 26 | 16.99 | 8 | 49,323,954 | 50,336,594 | 1,012,640 | 27 |
| SGT | 55 | 14 | 25.45 | 5 | 76,209,127 | 76,942,872 | 733,745 | 22 |
All breed abbreviations are defined in Table 5
n animal Number of animals evaluated, n HER Number of HER found in position, % Percentage of the population that presented this island, CHR Chromosome, start Start of HER, end End of HER, length HER length, n SNP Number of SNPs that HER covers
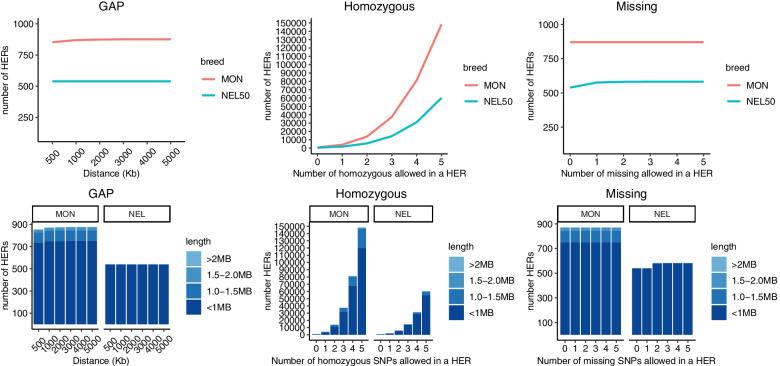
The HER with the highest proportion (43.23%) was found on BTA1 in NEL50. This region harbors 28 genes (Additional file 9: Table S5). HER islands overlapping between groups were observed on BTA5 for NEL50 and Santa Gertrudis (SGT) populations (next to the 76 Mb region) and for HFD and NEL50 (78 Mb region). We also evaluated the impact of different parameters on HER detection, as shown in Fig. 5. The main parameter affecting the amount of HER was the number of homozygous SNPs allowed in an HER specific region.
Fig. 5.
Number of heterozygous-enriched regions (HER), and length size classification, based on different values of parameters in Montana (MON) and Nellore (NEL50) breeds
When zero homozygous SNP was allowed in the HER analyses, 870 and 539 HERs were found for MON and NEL50 populations, respectively. When five homozygous SNPs were allowed, 148,233 HERs were found for MON and 60,155 for NEL50. Thus, the modification from zero to five homozygous SNPs allowed in the analyses represented an increase on the number of HERs of 17,039% and 11,160%, for MON and NEL50, respectively. However, no relevant effect was observed when the maximum distance among two consecutive SNPs were increased, demonstrating that the increase on gap parameter did not affect the number of HERs obtained. Finally, another parameter tested was the number of missing SNPs permitted in HER analyses, which for MON none relevant effect was observed, but for NEL50 an increase in 8% of HER found was detected when the parameter was changed from zero to five missing SNPs allowed.
Comparison between SNP panel and WGS results
The island found in SNP panel analysis for the GIR and BRM breeds was not presented in WGS analysis (Fig. 6). This result was also repeated in the other groups evaluated (Additional file 3: Fig. S3). One interesting point was observed in BTA20, where HOL animals based on the WGS analysis had ROHs close to the island found in SEN in the analyses using SNP panel data (Fig. 7). For the HER analyses, the pattern was similar to what happens in ROH analysis where there was no repetition of the island in WGS analysis (Additional file 4: Fig. S4).
Fig. 6.
Comparison between runs of homozygosity SNP panel and WGS analyzes for chromosome 1
Fig. 7.
Comparison between runs of homozygosity SNP panel and WGS analyzes for chromosome 20
Functional genomic analyses
In summary, 1662 positional genes were found in the ROH islands and 850 genes in the HER. The genes found in ROH islands are related to 379 pathways, 389 cellular components, 400 biological processes, and 319 molecular functions. The genes found in HER islands are related to 217 pathways, 180 cellular components, 373 biological processes, and 224 molecular functions (Additional file 5: Table S1, Additional file 6: Table S2, Additional file 7: Table S3, Additional file 8: Table S4, Additional file 9: Table S5, Additional file 10: Table S6, Additional file 11: Table S7, Additional file 12: Table S8, Additional file 13: Table S9, Additional file 14: Table S10, Additional file 15: Table S11 and Additional file 16: Table S12). The ROH islands from GIR and BRM populations contain genes related to the melanogenesis pathway such as DVL3 in BTA1, EP300 in BTA5, CREB3 in BTA8 (only for the BRM animals), DVL1 in BTA16, and FZD2 and WNT3 in BTA19.
Prolactin signaling was another important pathway observed in this study. Four genes identified for BRM are involved in this pathway (CCND2, STAT5B, STAT5A, and STAT3); in the GIR breed, three genes are involved (STAT5B, STAT5A, and STAT3), while for Jersey (JER) and SEN, only the genes MPK10 and PRLR were involved, respectively. Another pathway in common between groups was the calcium signaling pathway in ANGSIM and JER, where the CACNA1E and DRD5 genes were found related to this pathway.
Regarding HER, the majority of the genes found are involved with immunity as ANXA1, INFG, KLRD1, RAC2, and NKG2C. For SEN, the identified gene ANXA1 is known for influencing biological processes like immunity response of type 2 and antibiotic response, proliferation, leucocytes migration, adaptive immunity responses, and immune system development. In the MON population, six genes known for processing and presentation of antigens pathway were identified: IFNG, ENSBTAG00000046268, NKG2C, KLRD1, ENSBTAG00000000966, and CD4. Still, in NEL50, the leukocyte transendothelial migration pathway is influenced by three genes: NCF4, RAC2, and MMP2, or by genes that are involved in leukocyte proliferation such as CD80, MARCHF7, and RAC2.
Discussion
There is a large variability in the literature on the parameters used to identify ROH, which makes it difficult to compare results from different studies, as reinforced by Peripolli et al. [16] and shown in Table 4. Besides the ROH parameters, we also evaluated the impact of the SNP panel density on ROH detection.
Table 4.
Parameters for identifying Runs of Homozygosity in different studies with different densities of genotyping panels
| Studies | SNP Panel | het | miss | trhs | gap (kb) | min (kb) | nSNP | Density | window | |
|---|---|---|---|---|---|---|---|---|---|---|
| snp | kb | |||||||||
| Our study | 35K, 50K | 1 | 1 | 0.05 | 1000 | 500 | 30 | 1 | 50 | 50 |
| Peripolli et al. (2018a) [21] | 20K, 30K, 50K, 70K E HD | 1 | 5 | – | 500 | 1000 | 100 | 1 | 50 | 50 |
| Mastrangelo et al. (2018) [4] | 50K | 1 | 1 | – | 1000 | 4000 | 30 | 1 | 100 | – |
| Zavarez et al. (2015) [11] | HD | 2 | 5 | – | 500 | 100 | 30 | 1 | 100 | – |
| Peripolli et al. (2020) [22] | 30K | 0 | 2 | – | 1000 | 500 | 15 | 1 | 120 | 40 |
| Peripolli et al. (2018b) [23] | HD | 1 | 5 | 0.05 | 500 | 1000 | 100 | 1 | 50 | 50 |
| Fonseca et al. (2016) [24] | 50K | 0 | 5 | – | 1000 | – | 15 | – | – | – |
| Zanella et al. (2018) [25] | HD | 1 | 1 | – | – | 1000 | – | – | – | 50 |
| Ventura et al. (2020) [26] | HD | 1 | 5 | 0.05 | 500 | 1000 | 100 | 1 | 50 | 50 |
All breed abbreviations are defined in Table 5
het Number of heterozygous SNPs allowed in an ROH, miss Number of missing SNPs allowed in an ROH, trhs Windows threshold, min Minimum size of an ROH, nSNP Minimum number of SNP that make up an ROH
As shown in Table 4, the diversity of parameters chosen among studies leads to difficulty in comparison of studies and/or breeds [16]. As mentioned by Biscarini et al. [13], few studies aimed to evaluate the impact of different parameters on ROH detection, with such parameters still not well established. This definition is of crucial importance, as it has a direct effect on the results obtained [27]; not only in ROH detection, but also in the secondary analyses (e.g., inbreeding coefficient based on ROH).
We evaluated 16 populations of cattle selected for different breeding goals to compare ROHs and HERs. The majority of ROHs were defined as being 2-4 Mb and taking into consideration that the recombination events that happen in each generation break the homozygous segments into smaller haploblocks, these runs were likely formed in more ancient generations. Howrigan et al. [28] estimated that the ROH lengths of 10 Mb, 5 Mb, and 2.5 Mb would be correlated with 5, 10, and 20 generations ago, respectively. Moreover, Cardoso et al. [29] estimated that the ROH length higher than 16 Mb was formed less than three generations ago and the ROH less than 8 Mb was formed more than six generations ago.
SEN showed the highest ROH number and the largest number of ROH segments greater than 16 Mb. This suggests that recent inbreeding events have occurred more frequently in this population. As expected, the crossbred or composite populations, such as ANGSIM and MON, presented the smallest amount of ROHs. The verification of ROH incidence in composite breeds and crossbreeds is an important factor, as a high ROH incidence can indicate a decrease of heterosis [22].
The populations with high selection pressure, such as ANG, NEL50, HOL, and JER, showed higher numbers of ROHs. It is well known that selection increase autozygosity, although there is still a lack of information on selection effects regarding ROH distribution along the genome [7]. The ROH prevalence is more common in regions with higher linkage disequilibrium and low recombination, or regions of low genetic diversity [30]. However, it is also known that selection can cause a substantial pressure in specific genomic regions, resulting in an increase of ROH numbers and determining an appearance pattern in the population [31, 32].
The difference in the SNP panel density affected the number of ROHs detected. For NEL35 and NEL50 (same breed but genotyped using different SNP panels), there was a considerable discrepancy between the amount of ROH detected (Fig. 1 and Additional file 1: Fig. S1). As discussed by Hillested et al. [33], denser SNP panels tend to result in the identification of a larger number of ROH segments. This happens because an increasing density of markers allows the detection of heterozygote markers that are not presented in the lower density SNP panels. However, this might not be the only reason in this context, since the SNP panel used for NEL35 was created specifically for the Bos taurus indicus breeds (GGP indicus 35 K), with the selection of SNPs with higher MAF in Zebu cattle populations and designed to optimize equidistant spacing of markers [34]. These factors might have affected the ROH detection of ROHs based on the parameters used.
Some studies reported that the SNP panel density affect ROH detection [8, 19, 34]. According to Ceballos et al. [19], the accurate detection of ROH is affected by the density of markers and their distribution along the genome. Rebelato et al. [10] reported that ROHs are better identified in SNP panels with more than 50,000 SNPs. In the present study, the 35 K SNP panel was less appropriate to detect ROHs compared to the higher density SNP panels. One possible alternative would be the imputation to higher-density SNP panels once the accuracy of such procedure can be high Nellore cattle [34].
Genomic inbreeding coefficients and effective population size
In general, the inbreeding estimates of the first four methodologies showed similar results. Each one of the methodologies used in the present study has their particularities regarding inbreeding estimation and are dependent on some factors. For example, the first three methods (FHOM1, FHOM2, FGRM) are dependent on genotype allele frequency different to the methodology by uniting gamete [35]. These particularities must be taken into account when defining inbreeding concepts, for example, the UNI method that takes into account the Wright [36] and Malécot [37] definition or the HOM methods that take into consideration the heterozygosity reduction. The HOM and ROH methodologies weigh every allele equally, while the UNI and GRM methods give more weight to rare alleles [38].
The majority of molecular information measured on inbreeding coefficients estimation is based on marker allele identity and does not directly separate the regions that are identical-by-descendent and those that are identical-by-state [39]. The advantage of the ROH inbreeding estimate, besides not being dependent on allele frequency, is the ability to differentiate recent and ancient inbreeding [40]. However, the ROH estimate method is highly dependent on the detection parameters used in the analyses.
Low to moderate correlations were observed between the first four methods based on the ROHs analyses (Fig. 2). The correlation among the four methodology and the ROH length groups decreases for shorter ROH segments, reinforcing that some inbreeding estimation methods have lower power of detection of more ancient inbreeding [41]. These results corroborate with previous findings in cattle studies such as Gurgul et al. [42] that evaluated the correlation of GRM and ROH methodologies, or Zhang et al. [43] which evaluated the correlation of the HOM and UNI methodologies with the ROH method.
Regarding Ne, there was a reduction, over the generations, for all populations. However, the highest reduction of Ne was detected for GIR and LMS populations. The probable effect of a Ne decrease is the genetic diversity reduction, affecting the homozygous and heterozygous rates in these populations. This Ne reduction might be a consequence of intense selection practices and use of the reproductive technologies [8, 10, 16]. Attention is required in the management of genetic diversity of populations with Ne lower than 100 [44]. Reduction in Ne values can result in increases of the linkage disequilibrium and rates of fixed alleles, causing a reduction in the variability of certain genomic regions. All populations in the present study had recent Ne values higher than 100, which demonstrates a reasonable control of inbreeding. However, it is recommended, especially for GIR, HFD, SGT, HOL, and JER that the Ne rates should be constantly monitored to avoid a loss of diversity in the next generations. Edea et al. [45] working with some of the same breeds (ANG, CHL, HFD, HOL, and JER) found similar Ne pattern.
Homozygosity islands
The homozygosity islands were defined as the regions with the incidence of ROHs in at least 50% plus one individual of the population. This occurs due to increase of allele frequency in certain regions as a response to adaptive processes or intense selection of traits with high economic interest [10]. Many islands were present in both GIR and BRM animals. Interestingly, GIR contributed to the formation of BRM [46], suggesting that these islands might have been maintained over generations and were kept in BRM. One of the metabolic pathways found with the genes into these islands is the melanogenesis pathway, which is responsible for determining the coat color pattern in each breed [47], sustaining the balance between the brown-black eumelanin and reddish-yellow phaeomelanin [48], and is also associated with thermoregulation, resulting in better adaptation to certain environmental conditions [49, 50].
Another pathway found to be in common in four populations that presented the homozygosity islands was the prolactin signaling pathway, which is responsible for many biological processes. Various genes in this pathway were previously associated with important traits in cattle. For example, the STAT gene family (signal transducer and transcription activator) is responsible for the development and differentiation of mammary gland cells in different life stages [51]. The MAPK10 gene (mitogen-activated protein kinase 10) is linked with the inflammatory response and immunity of epithelial cells in the mammary gland. According to Silva et al. [52], MAPK10 is a candidate gene to somatic cell score (SCS). The PRLR gene (prolactin receptor), also known as the SLICK gene, was associated with heat tolerance [53], because results in short and slick hair that result in better adaptation to high temperatures [54].
The calcium signaling pathway found in the common islands was observed in ANGSIM and JER. In the ANGSIM breed, the gene related to this pathway is CACNA1E (calcium voltage-gated channel subunit alpha1 E). Hay and Roberts [55] reported CACNA1E as a candidate gene for growth and carcass-related traits. In JER breed, the DRD5 gene (dopamine receptor D5) was associated with the calcium signaling pathway, which has been associated with feed intake regulation and energy homeostasis [56]. Other pathways and genes associated with the homozygosity island found in this study can be accessed in the Supplementary Materials.
Heterozygosity-enriched regions
Different from ROHs, it is expected that the HER occur in regions under balancing selection or with high recombination rate, as low linkage disequilibrium leads to high region diversity. Normally, HER are concentrated in genomic regions associated with disease resistance, where higher levels of diversity can help the population to deal with novel (and changing) potential infirmity issues [13]. Despite being associated with interesting regions, HER are not as widely studied as ROHs [15].
The population that showed the highest amount of HER was MON (a composite population), which represented more than 47% of HER found in this study. This result was expected as the MON population is a composite breed that combines different biological groups, including breeds from Bos taurus indicus and Bos taurus taurus [22].
It is also important to point out the difference found between NEL35 and NEL50 groups for HER, mostly because the difference on the density among SNP panels. The length classification of HER was different between SNP panels: the NEL50 group presented a smaller length of HER compared to the NEL35 group. As the same regions were detected based on both SNP panels, this result suggests that many of the long HER regions are composed of small HER, broken by regions with homozygous SNPs.
Only one gene was found in the smaller HER – KHDRBS2 (KH RNA Binding Domain Containing, Signal Transduction Associated 2). This gene has been associated with fertility and reproductive performance in Sanmartinero cattle [57]. However, most genes found in the HER islands showed some relation with animal immunity, demonstrating a relation of the biological responses to environmental challenges. Another important gene is IFNG (interferon-gamma), which was found in the HER island for MON breed. IFNG has been previously associated with tick resistance in taurine and zebu cattle [58]. The ANXA1 gene (Annexin A1), a protein regulated by glucocorticoids, which was identified for SEN, plays an inhibitory role in the synthesis of arachidonic acid, a precursor of inflammatory processes.
These high heterozygosity concentrations in the region of immune response control are an interesting point to be investigated in these populations, as they can contribute to disease resilience [12]. Additionally, the genomic regions where there is higher variability in key traits is an important point to check in studies estimating the genetic diversity of the populations [13], as sustaining diversity can be an advantage for adaptive traits and selective breeding [59].
Although only few studies have focused on identifying HER islands, it seems that the use of the consecutive approach is preferred, as the case of Biscarini et al .[13], Marras et al. [14], and Santos et al. [15]. In our work, the use of sliding window did not capture any HER run. Otherwise, considering the consecutive approach, the analyses demonstrated a good power of detection of HER. In this context, the number of homozygous and missing allowed, and the gap between two consecutive SNPs are directly correlated to the flexibility in the criteriums to identify HER. When we tested different parameters of HER identification, the modification on gap and missing parameters did not seem to interfere in HER detection. However, the quantity of homozygous permitted within a HER is an important parameter affecting the number of HER detected, as also observed by Biscarini et al. [13]. Even with an impact on the number of HER detection, is important to state that the majority of HER identified in our study continued to be small (< 1 MB), regardless of the number of homozygous allowed in the analyses (from 0 to 5). Therefore, we recommend future studies allowing a small number of homozygous alleles when detecting HER.
SNP panels and WGS comparison
Currently, some WGS studies have been used to detect population structure and identify the regions influencing economically-important traits in livestock [60]. The present study did not find similarities between SNP panels and WGS results in any of the evaluated populations. The majority of ROHs with long-homozygous sequences are actually many short ROHs distributed side by side. In addition, as in the SNP panels, the loci between two homozygous SNPs are assumed to also be in homozygosity, as the ROH tends to be long, overestimating the lengths of some ROHs. In the other hand, the commonly used of low-density SNPs in the detection of ROH may be sufficient in some cases, whereas defining ROH at the WGS level is not easy due to obstacles such as the high values of the technique, occasional sequencing errors, and new individual mutations. The results of ROH with commercial SNP panels can be consistent with WGS analyses when an apparent long haplotype is present in certain genomic regions.
One interesting observation in the comparison between breeds and SNP panels that was not previously identified with the analysis only with SNP panels, was a small ROH concentration observed in the WGS analysis for Holstein breed close to an island region found in SNP panel analysis of the SEN population. This region contains the SLICK gene, already discussed previously and originally found in SEN animals. Recent studies have already shown that the SLICK gene has been introduced into some HOL populations in a selection process to control heat stress [54].
As found in our study, the number of ROH and HER regions differ among populations and provide insights on their differences in selective breeding and evolutionary histories. These differences are expected once the events that acted in each population are different or show different intensity [45]. Some particularities of this study must be taken into account, as the unbalanced number of animals among populations. This could affect the total number of ROH and HER identified for each population and the comparison of the results obtained. Furthermore, the WGS datasets were not obtained in the same animals genotyped for SNP panels and hence, the lack of common regions cannot be attributed solely to the genotyping platform.
The comparison between the 35 K and 50 K SNP panels in the Nellore breed evidenced a divergence in the identification and classification of ROH and HER (Additional file 1: Fig. S1 and Additional file 2: Fig. S2). As previously reported [8, 19, 34], the use of at least a 50 K panel in the analysis of ROH and HER is recommended. The use of lower-density panels can underestimate the number of ROH/HER and the classification, where higher length segments are less identified. This situation occurs mainly due to the higher distance between markers and less markers distributed along the genome, resulting in less information available for the analyses.
It is noteworthy that our work reports substantial information about the genetic diversity in different cattle breeds, presenting new genomic regions with homozygosity islands and heterozygous-enriched regions, where a great number of genes are located (Additional file 5: Table S1, Additional file 6: Table S2, Additional file 7: Table S3, Additional file 8: Table S4, Additional file 9: Table S5, Additional file 10: Table S6, Additional file 11: Table S7, Additional file 12: Table S8, Additional file 13: Table S9, Additional file 14: Table S10, Additional file 15: Table S11 and Additional file 16: Table S12). One of the main challenges of managing genetic diversity of livestock populations is to know which genomic regions are in homozygosity/heterozygosity, since they are highly heterogeneous across the genome. This management and characterization of the genetic structure of a population is essential to access the diversity and help to understand the time action over specific breeds. With the advancement of molecular technologies, novel insights into the animal genome can be accessed and populations compared, to verify which regions are in high or low diversity in each population and thus better manage future generations.
Conclusions
The ROH and HER numbers differ for each population suggesting that different events acted in the distinct populations over time. The population with the highest number of ROH was the SEN and for HER it was the MON population. Overlapping islands were identified between GIR and BRM suggesting that these regions may have been shared during their formation. The different methodologies of inbreeding estimates presented low to moderate correlation with the ROH method, mainly with a smaller ROH length, suggesting that ancient inbreeding was not well captured for these populations. HER islands were identified in regions related to animals’ immunity response. Lastly, in the comparison between SNP panels and WGS, it was observed that long ROH and HER identified on SNP panels are shorter runs side by side. It was observed an incidence of ROH in WGS analyses for HOL animals in a region containing a ROH island, on SNP panels analyses, for SEN animals that are related to heat tolerance indicating that a possible selection for such trait has been applied in this population.
Methods
Genotypes
Genomic datasets (n = 2415) from 16 worldwide cattle populations selected for different purposes were used in this study. The sample size of each breed and the density of the SNP panels used are presented in Table 5. These datasets were provided by: Purdue University (West Lafayette, IN, USA) – data of Angus x Simmental crossbreed (ANGSIM - F1 population); University of São Paulo (Pirassununga, SP, Brazil) – provided the data of the Montana Tropical Composite population (MON); Katayama Ltd. Livestock Company (Guararapes, SP, Brazil) – provided the data of the Nellore breed with two SNP panels: 35 K (NEL35) and 50 K (NEL50); the WIDDE database ([61] - http://widde.toulouse.inra.fr/widde/) provided the data of Angus (ANG), Borgou (BOR), Brahman (BRM), Creolo from Guadalupe (CGU), Charolais (CHL), Gyr (GIR), Hereford (HFD), Holstein (HOL), Jersey (JER), Limousin (LMS), Senepol (SEN), and Santa Gertrudis (SGT). To increase the sample size for some breeds, datasets from high-density and 50 K SNP panels were merged for the ANG, BRM, GIR, HFD, JER, and LIM breeds. This was done on the WIDDE platform prior to downloading the data. All SNP coordinates were updated to the ARS-UCD 1.2 [62] reference genome prior to performing further analyses.
Table 5.
Herds used in the study with the respective samplings (N), acronym referring to the herd, density of the genotyping chip and the database to which the genotypes are found
| Population | N | Abbreviation | Density | Database |
|---|---|---|---|---|
| Angus | 99 | ANG | 46,989 | WIDDE |
| Angus x Simental | 487 | ANGSIM | 52,597 | Purdue University |
| Borgou | 158 | BOR | 52,497 | WIDDE |
| Brahman | 70 | BRM | 46,989 | WIDDE |
| Criolo de Guadalupe | 140 | CGU | 52,497 | WIDDE |
| Charolais | 62 | CHL | 46,989 | WIDDE |
| GIR | 50 | GIR | 46,989 | WIDDE |
| Hereford | 61 | HFD | 46,989 | WIDDE |
| Holstein | 137 | HOL | 46,989 | WIDDE |
| Jersey | 84 | JER | 46,989 | WIDDE |
| Limousin | 87 | LMS | 46,989 | WIDDE |
| Montana | 271 | MON | 51,084 | USP |
| Nellore 35K | 209 | NEL35 | 35,237 | Katayama |
| Nellore 50K | 192 | NEL50 | 54,791 | Katayama |
| Senepol | 153 | SEN | 52,497 | WIDDE |
| Santa Gertrudis | 55 | SGT | 46,989 | WIDDE |
Quality control
The genotype quality control (QC) was done separately for each analysis. For ROH and HER, we removed SNPs with call rate lower than 0.90, duplicated position, non-autosomes, or without a defined position. For the other analyses, the minor frequency allele (MAF < 0.05) and Hardy-Weinberg equilibrium (HWE < 10− 6) parameters were also used to filter out SNPs. The density of genotype panels and the amount of discarded SNP in each QC are shown in Table 6.
Table 6.
Descriptive statistics of the genomic datasets after the genotype quality control
| Breeds | N | SNP | Call rate | Duplicates | NA/SDP | Total 1 | MAF | HWE | Total 2 |
|---|---|---|---|---|---|---|---|---|---|
| ANG | 99 | 46,989 | 260 | 0 | 0 | 46,729 | 10,399 | 19 | 36,311 |
| ANGSIM | 487 | 52,597 | 0 | 0 | 4014 | 48,583 | 9633 | 31 | 38,919 |
| BOR | 158 | 52,497 | 1315 | 0 | 0 | 51,182 | 16,525 | 161 | 34,496 |
| BRM | 70 | 46,989 | 1047 | 0 | 0 | 45,942 | 19,456 | 44 | 26,442 |
| CGU | 140 | 52,497 | 3435 | 0 | 0 | 49,062 | 10,428 | 130 | 38,504 |
| CHL | 62 | 46,989 | 401 | 0 | 0 | 46,588 | 9352 | 18 | 37,218 |
| GIR | 50 | 46,989 | 1260 | 0 | 0 | 45,729 | 24,289 | 33 | 21,407 |
| HFD | 61 | 46,989 | 252 | 0 | 0 | 46,737 | 8816 | 105 | 37,816 |
| HOL | 137 | 46,989 | 1350 | 0 | 0 | 45,639 | 9416 | 16 | 36,207 |
| JER | 84 | 46,989 | 1377 | 0 | 0 | 45,612 | 13,163 | 56 | 32,393 |
| LMS | 87 | 46,989 | 243 | 0 | 0 | 46,746 | 9925 | 37 | 36,784 |
| MON | 271 | 51,084 | 0 | 0 | 0 | 51,084 | 142 | 127 | 50,815 |
| NEL 35 | 209 | 35,237 | 780 | 16 | 1624 | 32,817 | 2529 | 1385 | 28,903 |
| NEL 50 | 192 | 54,791 | 942 | 9 | 3647 | 50,193 | 9895 | 1804 | 38,494 |
| SEN | 153 | 52,497 | 1633 | 0 | 0 | 50,864 | 12,635 | 91 | 38,138 |
| SGT | 55 | 46,989 | 619 | 0 | 0 | 46,370 | 9768 | 28 | 36,574 |
All breed abbreviations are defined in Table 5
N Number of animals used in the analyses, NA / SDP Non-autosomal SNPs or SNPs without defined positions, Total 1 Number of SNPs used for ROH and HER analyses, Total 2 Number of SNPs used in the inbreeding and effective population size analyses performed, MAF Lower allele frequency, HWE Hardy-Weinberg equilibrium
Runs of homozygosity
The PLINK v1.9 software [63] was used for the ROH identification based on the following criteria:
1 heterozygous and 1 missing SNP were allowed;
The window of threshold used was 0.05;
The gap between consecutive SNPs could not be higher than 1000 kb;
The minimum length of a ROH was 500 kb;
The minimum number of consecutive SNPs that create an ROH must be equal or greater than 30;
The density of 1 SNP used in at least 50 kb;
A sliding genomic window was used with 50 SNPs.
ROHs were classified in the following classes: < 2 Mb, 2-4 Mb, 4-8 Mb, 4-16 Mb, and > 16 Mb. The genomic regions that showed at least 50% plus one of the individuals with ROH were considered as ROH islands, which were used for the subsequent functional analyses.
Heterozygosity-enriched regions
The detectRUNS package [20] was used for the detection of HER following the two methods proposed by the package authors – the methodology based on SNP windows and the methodology based on consecutive SNPs. The method with windows is used to scan the genome and the window parameters selected to determine if the SNP is included or not in a HER. The methodology based on consecutive SNPs checks SNP by SNP in the genome. For the SNP window analyses, the following parameters were considered:
A window of 50 SNPs;
A minimum of 20 consecutive SNPs constitute an ROH;
A minimum length of 500 kb;
The density of 1 SNP at 100 kb;
Allowing the minimum number of two homozygous and one missing SNP; and,
The maximum gap between consecutive SNPs could not be larger than 1000 kb.
For the SNPs’ consecutive analysis, the following parameters were considered:
A minimum number of 20 consecutive SNPs constitutes a HER;
A minimum length of 500 kb;
A minimum of two homozygous and one missing SNP is allowed; and,
The maximum gap between consecutive SNPs could not be higher than 1000 kb.
The genomic regions that showed at least 10% of the population with HER were included in the subsequent functional analyses.
To better understand the impact of parameters modifications on HER identification, additional analyses using different threshold metrics were made for the main parameters utilized to determine HER, including: the gap between consecutive SNPs (500 to 5000 kb), number of homozygous SNPs allowed in a HER (0 to 5), and number of missing SNPs allowed in a HER (0 to 5). In this case, the two population with the highest amount of HER were used to test the impact of parameters alterations on HER results (MON and NEL50).
Population genomic structure
Genomic inbreeding metrics
Five models of inbreeding coefficient estimates were analyzed. The first method was based on the homozygous genotypes observed and expected (FHOM1), calculated as [63]:
where, Hexp is the expected value for homozygous genotypes and Hobs is the observed value for the homozygous genotypes.
The second method was based on genotype additive variance (FGRM), using the following model [64]:
where, xi is the number of reference allele copies of ith SNP, pi is the reference allele frequency in the population. Similar to the first method, the methodology FHOM2 was based on homozygous genotype following the model:
The above models are all dependents of genotype allele frequency, for this reason, a fourth model was a test based on the correlation between uniting gametes (FUNI) using the following model [65]:
The last method was based on the sum of ROH individual length divided by the total length of the autosomal genome (FROH) using the following eq. [66]:
where f(ROHi) is the ROH length of individual ith, n is the total intact homozygous genomic regions of each individual, h(j) is the length of chromosome jth, and A is the number of autosomal chromosomes (A = 29). Still, for each class of ROH (< 2 Mb, 2-4 Mb, 4-8 Mb, 4-16 Mb, > 16 Mb, < 8 Mb, and > 8 Mb), inbreeding estimates were made dividing the total sum of ROH segments by the total length of the cattle autosomal genome covered by SNPs. All the genomic inbreeding coefficients were calculated using the PLINK v1.9 software [63]. The PROC CORR option of the SAS statistical software [67] was used to correlate the inbreeding coefficient estimates. A heatmap was created for better visualization of the results through the plotly package [68].
Effective population size
The effective population size (Ne) was investigated with the relationship method between linkage disequilibrium variances and the effective population size using the SNeP software [69] and the following formula [70]:
where Ne is the effective population size at the Tth generation, ct is the recombination rate for the physical distance between the markers, α is the probability for the occurrence of mutation, and radj is the linkage disequilibrium value calculated by the correlation between two alleles in separate loci, assuming the following model:
where pa is the frequency of haplotype-a, pb is the frequency of haplotype-b, and pab is the haplotype frequency with allele a on the first locus and allele b on the second locus.
Functional analyses
The genomic regions considered as ROH and HER islands were used for genomic annotations. The GALLO package [71] was used for the annotation of genes in these regions, with the annotated data for Bos taurus from the Ensembl database (https://www.ensembl.org/Bos_taurus/Info/Index), version ARS-UCD1.2 [62]. Subsequently, the WebGestaltR package [72] was used for the Gene Ontology (GO) analyses to identify biological processes, molecular functions, and cellular components in which the positional candidate genes are involved in.
Comparison between SNP and WGS-based regions
The results of the SNP panel analysis, for both ROH and HER analyses were also carried out using WGS data. The WGS data was obtained from the “The 1000 Bull Genomes Project – Run 8” database [73]. A total of 1842 animals of 138 breeds between Bos taurus taurus and Bos taurus indicus and their crosses. In our analyses, we only considered breeds in common for the analyses in the SNP panel, this configured a sum of 914 animals. We removed SNPs with call-rate lower than 0.90, duplicated positions, non-autosomes, or without a defined position for the quality control. The analyses were made separately for each chromosome. The parameters used to identify both ROHs and HER in the WGS data was basically the same than those used in the SNP panel analyses, except the number of heterozygous/homozygous SNPs (3) and missing SNPs (5). These parameters were adapted from Ceballos et al. [19].
Supplementary Information
Additional file 1: Figure S1. Classification of runs of homozygosity according to length size by chromosome in the different breeds.
Additional file 2: Figure S2. Classification of heterozygous-enriched regions according to length size by chromosome in the different breeds.
Additional file 3: Figure S3. Comparison between runs of homozygosity SNP panel and whole-genome sequence (WGS) analyzes.
Additional file 4: Figure S4. Comparison between heterozygous-enriched regions SNP panel and whole-genome sequence (WGS) analyzes.
Additional file 5: Table S1. Genes found in Brahman’s runs homozygosity islands and the processes that are involved.
Additional file 6: Table S2. Genes found in AngusxSimmental crossbreed’s runs homozygosity islands and the processes that are involved.
Additional file 7: Table S3. Genes found in Gyr’s runs of homozygosity islands and the processes that are involved.
Additional file 8: Table S4. Genes found in Montana’s heterozygous-enriched regions and the processes that are involved.
Additional file 9: Table S5. Genes found in Nellore 50 K’s heterozygous-enriched regions and the processes that are involved.
Additional file 10: Table S6. Genes found in Jersey’s runs of homozygosity islands and the processes that are involved.
Additional file 11: Table S7. Genes found in Angus’ heterozygous-enriched regions and the processes that are involved.
Additional file 12: Table S8. Genes found in Hereford’s heterozygous-enriched regions and the processes that are involved.
Additional file 13: Table S9. Genes found in Nellore 35 K’s heterozygous-enriched regions and the processes that are involved.
Additional file 14: Table S10. Genes found in Senepol’s heterozygous-enriched regions and the processes that are involved.
Additional file 15: Table S11. Genes found in Senepol’s runs of homozygosity islands and the processes that are involved.
Additional file 16: Table S12. Genes found in Santa Gertrudis’ heterozygous-enriched regions and the processes that are involved.
Acknowledgments
The authors thank the Purdue University, University of São Paulo, and Katayama Ltd. livestock company for providing the genotype data. The 1,000 Bull Genomes Project are acknowledged for providing whole genome sequence genotypes. Support from the Foundation of the State of Bahia (FAPESB) are acknowledged for granting the scholarships and the funds to develop this research.
Abbreviations
- ANG
Angus population
- ANGSIM
Angus x Simmental crossbreed population
- BOR
Borgou population
- BRM
Brahman population
- CGU
Creolo from Guadalupe population
- CHL
Charolais population
- FGRM
Inbreeding coefficient estimate based on genotype additive variance
- FHOM
Inbreeding coefficient estimate based on the homozygous genotypes
- FROH
Inbreeding coefficient estimate based on ROH
- FUNI
Inbreeding coefficient estimate based on the correlation between uniting gametes
- GIR
Gyr population
- HER
Heterozygosity-enriched regions
- HFD
Hereford population
- HOL
Holstein population
- JER
Jersey population
- LMS
Limousin population
- MON
Montana Tropical Composite population
- Ne
Effective population size
- NEL35
Nellore population genotype with 35 K panel
- NEL50
Nellore population genotype with 35 K panel
- QC
Quality control
- ROH
Runs of homozygosity
- SEN
Senepol population
- SGT
Santa Gertrudis population
- SNP
Single nucleotide polymorphism
- WGS
Whole-genome sequence
Authors’ contributions
HAM, LFB, and VBP conceived, designed, and conducted the data analyses. MRS, LFB, JBSF, LG, and VBP contributed to the data acquisition. HAM, LFB, LFBP, and VBP wrote and edited the manuscript. All authors reviewed and contributed to editing of the manuscript and approved of its final publication.
Funding
Not applicable.
Availability of data and materials
The 1000 Bull Genomes Project data is available at http://www.1000bullgenomes.com/. Genotypes of WIDDE databases are available at http://widde.toulouse.inra.fr/widde/. Genotypes from databases of Purdue University, University of São Paulo, and Katayama are not publicly available, but can be obtained through reasonable request via the corresponding author.
Declarations
Ethics approval and consent to participate
No ethics approval was obtained for this study because no new animals were handled in this experiment.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Pitt D, Sevane N, Nicolazzi EL, MacHugh DE, Park SDE, Colli L, et al. Domestication of cattle: two or three events? Evol Appl. 2019;12:123–136. doi: 10.1111/eva.12674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Felius M. Cattle breeds: an encyclopedia. 1995. [Google Scholar]
- 3.Felius M, Koolmees PA, Theunissen B, Lenstra JA, Baumung R, Manatrinon S, et al. On the breeds of cattle-historic and current classifications. Diversity. 2011;3:660–692. doi: 10.3390/d3040660. [DOI] [Google Scholar]
- 4.Mastrangelo S, Sardina MT, Tolone M, Di Gerlando R, Sutera AM, Fontanesi L, et al. Genome-wide identification of runs of homozygosity islands and associated genes in local dairy cattle breeds. Animal. 2018;12:2480–2488. doi: 10.1017/S1751731118000629. [DOI] [PubMed] [Google Scholar]
- 5.Marchesi JAP, Buzanskas ME, Cantão ME, Ibelli AMG, Peixoto JO, Joaquim LB, et al. Relationship of runs of homozygosity with adaptive and production traits in a paternal broiler line. Animal. 2018;12:1126–1134. doi: 10.1017/S1751731117002671. [DOI] [PubMed] [Google Scholar]
- 6.Baes CF, Makanjuola BO, Miglior F, Marras G, Howard JT, Fleming A, et al. Symposium review: the genomic architecture of inbreeding: how homozygosity affects health and performance. J Dairy Sci. 2019;102:2807–2817. doi: 10.3168/jds.2018-15520. [DOI] [PubMed] [Google Scholar]
- 7.Forutan M, Ansari Mahyari S, Baes C, Melzer N, Schenkel FS, Sargolzaei M. Inbreeding and runs of homozygosity before and after genomic selection in north American Holstein cattle. BMC Genomics. 2018;19:98. doi: 10.1186/s12864-018-4453-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Purfield DC, Berry DP, McParland S, Bradley DG. Runs of homozygosity and population history in cattle. BMC Genet. 2012;13:70. doi: 10.1186/1471-2156-13-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Curik I, Ferenčaković M, Sölkner J. Inbreeding and runs of homozygosity: a possible solution to an old problem. Livest Sci. 2014;166:26–34. doi: 10.1016/j.livsci.2014.05.034. [DOI] [Google Scholar]
- 10.Rebelato AB, Caetano AR, Rebelato AB, Caetano AR. Runs of homozygosity for autozygosity estimation and genomic analysis in production animals. Pesqui Agropecuária Bras. 2018;53:975–984. doi: 10.1590/s0100-204x2018000900001. [DOI] [Google Scholar]
- 11.Zavarez LB, Utsunomiya YT, Carmo AS, Neves HHR, Carvalheiro R, Ferencakovic M, et al. Assessment of autozygosity in Nellore cows (Bos indicus) through high-density SNP genotypes. Front Genet. 2015;5 JAN:5. doi: 10.3389/fgene.2015.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Samuels DC, Wang J, Ye F, He J, Levinson RT, Sheng Q, et al. Heterozygosity ratio, a robust global genomic measure of autozygosity and its association with height and disease risk. Genetics. 2016;204:893–904. doi: 10.1534/genetics.116.189936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Biscarini F, Mastrangelo S, Catillo G, Senczuk G, Ciampolini R. Insights into genetic diversity, runs of homozygosity and heterozygosity-rich regions in Maremmana semi-feral cattle using pedigree and genomic data. Animals. 2020;10:2285. doi: 10.3390/ani10122285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marras G, Wood BJ, Makanjuola B, Malchiodi F, Peeters K, Van As P, et al. 11th World Congr Genet Appl to Livest Prod. 2018;11 Szwaczkowski. 2017. Characterization of runs of homozygosity and heterozygosity-rich regions in a commercial turkey (Meleagris gallopavo) population; p. 763. [Google Scholar]
- 15.Bizarria dos Santos W, Pimenta Schettini G, Fonseca MG, Pereira GL, Loyola Chardulo LA, Rodrigues Machado Neto O, et al. Fine-scale estimation of inbreeding rates, runs of homozygosity and genome-wide heterozygosity levels in the Mangalarga Marchador horse breed. J Anim Breed Genet. 2021;138:161–173. doi: 10.1111/jbg.12508. [DOI] [PubMed] [Google Scholar]
- 16.Peripolli E, Munari DP, Silva MVGB, Lima ALF, Irgang R, Baldi F. Runs of homozygosity: current knowledge and applications in livestock. Anim Genet. 2017;48:255–271. doi: 10.1111/age.12526. [DOI] [PubMed] [Google Scholar]
- 17.Szmatoła T, Gurgul A, Ropka-Molik K, Jasielczuk I, Zabek T, Bugno-Poniewierska M. Characteristics of runs of homozygosity in selected cattle breeds maintained in Poland. Livest Sci. 2016;188:72–80. doi: 10.1016/j.livsci.2016.04.006. [DOI] [Google Scholar]
- 18.Weldenegodguad M, Popov R, Pokharel K, Ammosov I, Ming Y, Ivanova Z, et al. Whole-genome sequencing of three native cattle breeds originating from the northernmost cattle farming regions. Front Genet. 2019;9 JAN:728. doi: 10.3389/fgene.2018.00728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ceballos FC, Hazelhurst S, Ramsay M. Assessing runs of homozygosity: a comparison of SNP array and whole genome sequence low coverage data. BMC Genomics. 2018;19:106. doi: 10.1186/s12864-018-4489-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Biscarini F, Cozzi P, Gaspa G, Marras G. detectRUNS: an R package to detect runs of homozygosity and heterozygosity in diploid genomes. CRAN.R. 2019. [Google Scholar]
- 21.Peripolli E, Stafuzza NB, Munari DP, Lima ALF, Irgang R, Machado MA, et al. Assessment of runs of homozygosity islands and estimates of genomic inbreeding in Gyr (Bos indicus) dairy cattle. BMC Genomics. 2018;19:34. doi: 10.1186/s12864-017-4365-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peripolli E, Stafuzza NB, Amorim ST, de Lemos MVA, Grigoletto L, Kluska S, et al. Genome-wide scan for runs of homozygosity in the composite Montana Tropical® beef cattle. J Anim Breed Genet. 2020;137:155–165. doi: 10.1111/jbg.12428. [DOI] [PubMed] [Google Scholar]
- 23.Peripolli E, Metzger J, de Lemos MVA, Stafuzza NB, Kluska S, Olivieri BF, et al. Autozygosity islands and ROH patterns in Nellore lineages: evidence of selection for functionally important traits. BMC Genomics. 2018;19:680. doi: 10.1186/s12864-018-5060-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Souza Fonseca PA, dos Santos FC, Rosse IC, Ventura RV, Brunelli FÂT, Penna VM, et al. Retelling the recent evolution of genetic diversity for Guzerá: inferences from LD decay, runs of homozygosity and ne over the generations. Livest Sci. 2016;193:110–117. doi: 10.1016/j.livsci.2016.10.006. [DOI] [Google Scholar]
- 25.Zanella R, Lago LV, da Silva AN, Pértille F, de Carvalho NS, do Carmo Panetto JC, et al. Genetic characterization of indubrasil cattle breed population. Vet Sci. 2018;5:98. doi: 10.3390/vetsci5040098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ventura RV, Brito LF, Oliveira GA, Daetwyler HD, Schenkel FS, Sargolzaei M, et al. A comprehensive comparison of high-density SNP panels and an alternative ultra-high-density panel for genomic analyses in Nellore cattle. Anim Prod Sci. 2020;60:333. doi: 10.1071/AN18305. [DOI] [Google Scholar]
- 27.Macciotta NPP, Colli L, Cesarani A, Ajmone-Marsan P, Low WY, Tearle R, et al. The distribution of runs of homozygosity in the genome of river and swamp buffaloes reveals a history of adaptation, migration and crossbred events. Genet Sel Evol. 2021;53:20. doi: 10.1186/s12711-021-00616-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Howrigan DP, Simonson MA, Keller MC. Detecting autozygosity through runs of homozygosity: a comparison of three autozygosity detection algorithms. BMC Genomics. 2011;12:460. doi: 10.1186/1471-2164-12-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cardoso DF, Fernandes Júnior GA, Scalez DCB, Alves AAC, Magalhães AFB, Bresolin T, et al. Uncovering sub-structure and genomic profiles in across-countries subpopulations of Angus cattle. Sci Rep. 2020;10:8770. doi: 10.1038/s41598-020-65565-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ceballos FC, Joshi PK, Clark DW, Ramsay M, Wilson JF. Runs of homozygosity: windows into population history and trait architecture. Nat Rev Genet. 2018;19:220–234. doi: 10.1038/nrg.2017.109. [DOI] [PubMed] [Google Scholar]
- 31.Fleming A, Abdalla EA, Maltecca C, Baes CF. Invited review: reproductive and genomic technologies to optimize breeding strategies for genetic progress in dairy cattle. Arch Anim Breed. 2018;61:43–57. doi: 10.5194/aab-61-43-2018. [DOI] [Google Scholar]
- 32.Nandolo W, Utsunomiya YT, Mészáros G, Wurzinger M, Khayadzadeh N, Torrecilha RBP, et al. Misidentification of runs of homozygosity islands in cattle caused by interference with copy number variation or large intermarker distances. Genet Sel Evol. 2018;50:43. doi: 10.1186/s12711-018-0414-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hillestad B, Woolliams JA, Boison SA, Grove H, Meuwissen T, Våge DI, et al. Detection of runs of homozygosity in Norwegian red: density, criteria and genotyping quality control. Acta Agric Scand Sect A Anim Sci. 2017;67:107–116. doi: 10.1080/09064702.2018.1501088. [DOI] [Google Scholar]
- 34.Aliloo H, Mrode R, Okeyo AM, Ni G, Goddard ME, Gibson JP. The feasibility of using low-density marker panels for genotype imputation and genomic prediction of crossbred dairy cattle of East Africa. J Dairy Sci. 2018;101:9108–9127. doi: 10.3168/jds.2018-14621. [DOI] [PubMed] [Google Scholar]
- 35.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wright S. Coefficients of inbreeding and relationship. Am Nat. 1922;56:330–338. doi: 10.1086/279872. [DOI] [Google Scholar]
- 37.Malécot G. Les mathématiques de l’hérédité: Masson; 1948. https://books.google.com.br/books/about/Les_mathématiques_de_l_hérédité.html?id=QOIfAAAAIAAJ&redir_esc=y. Accessed 22 Apr 2021
- 38.Alemu SW, Kadri NK, Harland C, Faux P, Charlier C, Caballero A, et al. An evaluation of inbreeding measures using a whole-genome sequenced cattle pedigree. Heredity (Edinb) 2021;126:410–423. doi: 10.1038/s41437-020-00383-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meuwissen THE, Sonesson AK, Gebregiwergis G, Woolliams JA. Management of genetic diversity in the era of genomics. Front Genet. 2020;11:880. doi: 10.3389/fgene.2020.00880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ghoreishifar SM, Moradi-Shahrbabak H, Fallahi MH, Jalil Sarghale A, Moradi-Shahrbabak M, Abdollahi-Arpanahi R, et al. Genomic measures of inbreeding coefficients and genome-wide scan for runs of homozygosity islands in Iranian river buffalo, Bubalus bubalis. BMC Genet. 2020;21:16. doi: 10.1186/s12863-020-0824-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doekes HP, Veerkamp RF, Bijma P, de Jong G, Hiemstra SJ, Windig JJ. Inbreeding depression due to recent and ancient inbreeding in Dutch Holstein–Friesian dairy cattle. Genet Sel Evol. 2019;51:54. doi: 10.1186/s12711-019-0497-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gurgul A, Szmatoła T, Topolski P, Jasielczuk I, Żukowski K, Bugno-Poniewierska M. The use of runs of homozygosity for estimation of recent inbreeding in Holstein cattle. J Appl Genet. 2016;57:527–530. doi: 10.1007/s13353-016-0337-6. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Q, Calus MP, Guldbrandtsen B, Lund MS, Sahana G. Estimation of inbreeding using pedigree, 50k SNP chip genotypes and full sequence data in three cattle breeds. BMC Genet. 2015;16:88. doi: 10.1186/s12863-015-0227-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meuwissen TH. Accuracy of breeding values of “unrelated” individuals predicted by dense SNP genotyping. Genet Sel Evol. 2009;41:35. doi: 10.1186/1297-9686-41-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edea Z, Dadi H, Dessie T, Uzzaman MR, Rothschild MF, Kim ES, et al. Genome-wide scan reveals divergent selection among taurine and zebu cattle populations from different regions. Anim Genet. 2018;49:550–563. doi: 10.1111/age.12724. [DOI] [PubMed] [Google Scholar]
- 46.Koufariotis L, Hayes BJ, Kelly M, Burns BM, Lyons R, Stothard P, et al. Sequencing the mosaic genome of Brahman cattle identifies historic and recent introgression including polled. Sci Rep. 2018;8:17761. doi: 10.1038/s41598-018-35698-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Randhawa IAS, Khatkar MS, Thomson PC, Raadsma HW. A Meta-assembly of selection signatures in cattle. PLoS One. 2016;11:e0153013. doi: 10.1371/journal.pone.0153013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim SH, Hwang SY, Yoon JT. Microarray-based analysis of the differential expression of melanin synthesis genes in dark and light-muzzle Korean cattle. PLoS One. 2014;9:e96453. doi: 10.1371/journal.pone.0096453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.D’Mello S, Finlay G, Baguley B, Askarian-Amiri M. Signaling pathways in melanogenesis. Int J Mol Sci. 2016;17:1144. doi: 10.3390/ijms17071144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Senczuk G, Guerra L, Mastrangelo S, Campobasso C, Zoubeyda K, Imane M, et al. Fifteen Shades of Grey: combined analysis of genome-wide SNP data in steppe and Mediterranean Grey cattle sheds new light on the molecular basis of coat color. Genes (Basel) 2020;11:932. doi: 10.3390/genes11080932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Otto PI, Guimarães SEF, Calus MPL, Vandenplas J, Machado MA, Panetto JCC, et al. Single-step genome-wide association studies (GWAS) and post-GWAS analyses to identify genomic regions and candidate genes for milk yield in Brazilian Girolando cattle. J Dairy Sci. 2020;103:10347–10360. doi: 10.3168/jds.2019-17890. [DOI] [PubMed] [Google Scholar]
- 52.Silva AA, Silva DA, Silva FF, Costa CN, Silva HT, Lopes PS, et al. GWAS and gene networks for milk-related traits from test-day multiple lactations in Portuguese Holstein cattle. J Appl Genet. 2020;61:465–476. doi: 10.1007/s13353-020-00567-3. [DOI] [PubMed] [Google Scholar]
- 53.Hansen PJ. Prospects for gene introgression or gene editing as a strategy for reduction of the impact of heat stress on production and reproduction in cattle. Theriogenology. 2020;154:190–202. doi: 10.1016/j.theriogenology.2020.05.010. [DOI] [PubMed] [Google Scholar]
- 54.Dikmen S, Khan FA, Huson HJ, Sonstegard TS, Moss JI, Dahl GE, et al. The SLICK hair locus derived from Senepol cattle confers thermotolerance to intensively managed lactating Holstein cows. J Dairy Sci. 2014;97:5508–5520. doi: 10.3168/jds.2014-8087. [DOI] [PubMed] [Google Scholar]
- 55.Hay EH, Roberts A. Genotype × prenatal and post-weaning nutritional environment interaction in a composite beef cattle breed using reaction norms and a multi-trait model. J Anim Sci. 2018;96:444–453. doi: 10.1093/jas/skx057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Msalya G, Kim ES, Laisser ELK, Kipanyula MJ, Karimuribo ED, Kusiluka LJM, et al. Determination of genetic structure and signatures of selection in three strains of Tanzania Shorthorn Zebu, Boran and Friesian cattle by genome-wide SNP analyses. PLoS One. 2017;12(1):e0171088. doi: 10.1371/journal.pone.0171088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.De León C, Manrique C, Martínez R, Rocha JF. Research article genomic association study for adaptability traits in four Colombian cattle breeds. Genet Mol Res. 2019;18. 10.4238/gmr18373.
- 58.Maryam J, Babar ME, Nadeem A, Hussain T. Genetic variants in interferon gamma (IFN-γ) gene are associated with resistance against ticks in Bos taurus and Bos indicus. Mol Biol Rep. 2012;39:4565–4570. doi: 10.1007/s11033-011-1246-8. [DOI] [PubMed] [Google Scholar]
- 59.Williams JL, Hall SJG, Del Corvo M, Ballingall KT, Colli L, Ajmone Marsan P, et al. Inbreeding and purging at the genomic level: the Chillingham cattle reveal extensive, non-random SNP heterozygosity. Anim Genet. 2016;47:19–27. doi: 10.1111/age.12376. [DOI] [PubMed] [Google Scholar]
- 60.Xia X, Zhang S, Zhang H, Zhang Z, Chen N, Li Z, et al. Assessing genomic diversity and signatures of selection in Jiaxian red cattle using whole-genome sequencing data. BMC Genomics 2021;22:43. doi:10.1186/s12864-020-07340-0. [DOI] [PMC free article] [PubMed]
- 61.Sempéré G, Moazami-Goudarzi K, Eggen A, Laloë D, Gautier M, Flori L. WIDDE: a web-interfaced next generation database for genetic diversity exploration, with a first application in cattle. BMC Genomics. 2015;16:1–8. doi: 10.1186/s12864-015-2181-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rosen BD, Bickhart DM, Schnabel RD, Koren S, Elsik CG, Tseng E, et al. De novo assembly of the cattle reference genome with single-molecule sequencing. Gigascience. 2020;9:1–9. doi: 10.1093/gigascience/giaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.VanRaden PM. Efficient methods to compute genomic predictions. J Dairy Sci. 2008;91:4414–4423. doi: 10.3168/jds.2007-0980. [DOI] [PubMed] [Google Scholar]
- 65.Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McQuillan R, Leutenegger A-L, Abdel-Rahman R, Franklin CS, Pericic M, Barac-Lauc L, et al. Runs of homozygosity in European populations. Am J Hum Genet. 2008;83:359–372. doi: 10.1016/j.ajhg.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.SAS Institute Inc . SAS 9.1.3 help and documentation. Cary: ADABAS; 2013. [Google Scholar]
- 68.Sievert C. Interactive web-based data visualization with R, plotly, and shiny. 2020. [Google Scholar]
- 69.Barbato M, Orozco-terWengel P, Tapio M, Bruford MW. SNeP: a tool to estimate trends in recent effective population size trajectories using genome-wide SNP data. Front Genet. 2015;6. 10.3389/fgene.2015.00109. [DOI] [PMC free article] [PubMed]
- 70.Corbin LJ, Liu AYH, Bishop SC, Woolliams JA. Estimation of historical effective population size using linkage disequilibria with marker data. J Anim Breed Genet. 2012;129:257–270. doi: 10.1111/j.1439-0388.2012.01003.x. [DOI] [PubMed] [Google Scholar]
- 71.Fonseca PAS, Suárez-Vega A, Marras G, Cánovas Á. GALLO: an R package for genomic annotation and integration of multiple data sources in livestock for positional candidate loci. Gigascience. 2020;9:1–9. doi: 10.1093/gigascience/giaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang J, Liao Y, Jaehnig E, Shi Z, Sheng Q. Gene set analysis toolkit WebGestaltR. 2020. [Google Scholar]
- 73.Hayes BJ, Daetwyler HD. 1000 bull genomes project to map simple and complex genetic traits in cattle: applications and outcomes. Annu Rev Anim Biosci. 2019;7:89–102. doi: 10.1146/annurev-animal-020518-115024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Classification of runs of homozygosity according to length size by chromosome in the different breeds.
Additional file 2: Figure S2. Classification of heterozygous-enriched regions according to length size by chromosome in the different breeds.
Additional file 3: Figure S3. Comparison between runs of homozygosity SNP panel and whole-genome sequence (WGS) analyzes.
Additional file 4: Figure S4. Comparison between heterozygous-enriched regions SNP panel and whole-genome sequence (WGS) analyzes.
Additional file 5: Table S1. Genes found in Brahman’s runs homozygosity islands and the processes that are involved.
Additional file 6: Table S2. Genes found in AngusxSimmental crossbreed’s runs homozygosity islands and the processes that are involved.
Additional file 7: Table S3. Genes found in Gyr’s runs of homozygosity islands and the processes that are involved.
Additional file 8: Table S4. Genes found in Montana’s heterozygous-enriched regions and the processes that are involved.
Additional file 9: Table S5. Genes found in Nellore 50 K’s heterozygous-enriched regions and the processes that are involved.
Additional file 10: Table S6. Genes found in Jersey’s runs of homozygosity islands and the processes that are involved.
Additional file 11: Table S7. Genes found in Angus’ heterozygous-enriched regions and the processes that are involved.
Additional file 12: Table S8. Genes found in Hereford’s heterozygous-enriched regions and the processes that are involved.
Additional file 13: Table S9. Genes found in Nellore 35 K’s heterozygous-enriched regions and the processes that are involved.
Additional file 14: Table S10. Genes found in Senepol’s heterozygous-enriched regions and the processes that are involved.
Additional file 15: Table S11. Genes found in Senepol’s runs of homozygosity islands and the processes that are involved.
Additional file 16: Table S12. Genes found in Santa Gertrudis’ heterozygous-enriched regions and the processes that are involved.
Data Availability Statement
The 1000 Bull Genomes Project data is available at http://www.1000bullgenomes.com/. Genotypes of WIDDE databases are available at http://widde.toulouse.inra.fr/widde/. Genotypes from databases of Purdue University, University of São Paulo, and Katayama are not publicly available, but can be obtained through reasonable request via the corresponding author.