Abstract
With regard to potential applications of genomic selection in small numbered breeds, we evaluated genomic models and focused on potential candidate gene annotations for weight and meat quality traits in the local Rotes Höhenvieh (RHV) breed. Traits included 6,003 birth weights (BWT), 5,719 200 d-weights (200dw), 4,594 365 d-weights (365dw), and 547 records for intramuscular fat content (IMF). A total of 581,304 SNP from 370 genotyped cattle with phenotypic records were included in genomic analyses. Model evaluations focused on single- and multiple-trait models with direct and with direct and maternal genetic effects. Genetic relationship matrices were based on pedigree (A-matrix), SNP markers (G-matrix), or both (H-matrix). Genome-wide association studies (GWASs) were carried out using linear mixed models to identify potential candidate genes for the traits of interest. De-regressed proofs (DRP) for direct and maternal genetic components were used as pseudo-phenotypes in the GWAS. Accuracies of direct breeding values were higher from models based on G or on H compared to A. Highest accuracies (> 0.89) were obtained for IMF with multiple-trait models using the G-matrix. Direct heritabilities with maternal genetic effects ranged from 0.62 to 0.66 for BWT, from 0.45 to 0.55 for 200dW, from 0.40 to 0.44 for 365dW, and from 0.48 to 0.75 for IMF. Maternal heritabilities for BWT, 200dW, and 365dW were in a narrow range from 0.21 to 0.24, 0.24 to 0.27, and 0.21 to 0.25, respectively, and from 0.25 to 0.65 for IMF. Direct genetic correlations among body weight traits were positive and favorable, and very similar from different models but showed a stronger variation with 0.31 (A), −0.13 (G), and 0.45 (H) between BWT and IMF. In gene annotations, we identified 6, 3, 1, and 6 potential candidate genes for direct genetic effect on BWT, 200dW, 365dW, and IMF traits, respectively. Regarding maternal genetic effects, four (SHROOM3, ZNF609, PECAM1, and TEX2) and two (TMEM182 and SEC11A) genes were detected as potential candidate genes for BWT and 365dW, respectively. Potential candidate genes for maternal effect on IMF were GRHL2, FGA, FGB, and CTNNA3. As the most important finding from a practical breeding perspective, a small number of genotyped RHV cattle enabled accurate breeding values for high heritability IMF.
Keywords: dual-purpose cattle, genetic parameters, meat quality, SNP marker, weight
INTRODUCTION
“Rotes Höhenvieh” (RHV) is a dual-purpose medium-sized cattle breed with a uniform red-brown hair color, originated from the Central Uplands of Germany. RHV cattle are mainly kept for quality beef production in low input grazing systems, and selection has focused on adaptation to harsh environments (Halli et al., 2020). In comparison to the mainstream beef cattle breeds, the small RHV population size implies a continuously decreasing selection intensity, with impact on the breed competitiveness. Hence, especially for such local endangered breeds, it is imperative to focus on niche markets, e.g., markets offering special meat products and aiming on meat quality improvements (Biermann et al, 2015). Furthermore, in beef breeds with small population size, it is imperative to apply all available modern technologies such as reproduction techniques or genomic selection to achieve at least moderate genetic gain per year (Pimentel and König, 2012).
Incorporation of body weight and meat quality traits into a genetic evaluation scheme requires accurate genetic parameter estimates. Moderate to high direct pedigree-based heritabilities were reported for body weight traits of several beef cattle breeds (Gutiérrez et al., 2007; Regatieri et al., 2012; Caetano et al., 2013), indicating the potential for genetic improvement. The most studied meat quality traits in beef cattle are intramuscular fat content (IMF) and shear force. In their review, Utrera and VanVleck (2004) presented direct heritabilities for both traits in the range from 0.40 to 0.50. Even higher heritabilities up to 0.88 for carcass traits in American Shorthorn cattle were reported by Pariacote et al. (1998). Such moderate to high heritability estimates when modeling pedigree relationships (A-matrix) were confirmed in more recent studies (MacNeil et al., 2010; Mateescu et al., 2015). Using commercially available genetic marker panels (i.e., two markers per panel), significant associations of the thyroglobulin-, calpastatin-, and the µ-calpain-marker with meat quality traits were observed for meat quality traits in beef cattle (Van Eenennaam et al., 2007). As shown in deterministic predictions for high heritability traits, training sets comprising 700 to 1,000 animals with genomic marker data and phenotypes enable precise estimates of genomic breeding values (Daetwyler et al., 2008). The very dense 777K SNP chip allowed quite accurate estimates for production traits in beef cattle, even when considering different breeds in the same reference set (Rolf et al., 2010). In a genome-wide association study (GWAS), Lu et al. (2013) used a sample size including 747 crossbred beef cattle and identified 34 SNP markers significantly associated with IMF and meat marbling score.
In addition, especially for weight or weight gain traits in ruminants, a strong maternal genetic component has been identified, even for traits recorded late in life (Mandal et al., 2006). Saatci et al. (1999) and Meyer (1992) indicated biased estimates when neglecting maternal genetic effects in genetic evaluations for sheep and beef cattle, respectively. For weights at different ages recorded in Holstein Friesian calves and heifers, Yin and König (2019) identified a moderate genetic maternal component up to the age of 1 yr. They considered 4,246 animals with genotypes and inferred a large number of SNP markers either contributing to the direct or maternal genetic effects on weights at the age of 3 mo.
The combination of pedigree and genomic information in so-called single-step approaches (e.g., Aguilar et al., 2010) especially offers potential to improve genetic evaluations in small-sized breeds with poorly recorded pedigrees. Single-step methods have been developed for the estimation of genetic parameters and breeding values (Misztal et al., 2018), as well as for GWASs (Wang et al., 2012). Recently, Shabalina et al. (2021) evaluated genetic parameters from pure genomic, pedigree-based, and single-step approaches considering a small sample size of only 758 genotyped cows with phenotypes for low heritability functional traits. Heritabilities and genetic variances from the single-step approach were in-between the estimates from the pure pedigree and genomic models, and associated with small standard errors.
A further modeling question addresses the application of single-trait genomic (STGM) or multiple-trait genomic models (MTGM), with generally higher accuracies of genetic parameter estimates from MTGM (Jia and Jannink, 2012; Ismael et al., 2017; Srivastava et al., 2019). In beef cattle, body weight and meat quality traits are genetically closely correlated (Gordo et al., 2016), and the benefits from a MTGM over a STGM may be smaller compared to genetic evaluations for low heritability traits (Daetwyler et al., 2012; Guo et al., 2014).
In beef cattle, there is a gap of scientific studies comparing pedigree-based and genomic models, especially for novel meat quality traits from local breeds kept in alternative low input production systems. Furthermore, to the best of our knowledge, no previous GWAS has been conducted to estimate maternal genetic effects on meat quality traits. Consequently, the objectives of this study were to 1) estimate direct and maternal genetic (co)variance components for body weight and IMF traits using pedigree-based, pure genomic, and combined pedigree-genomic models in RHV dual-purpose cattle, 2) compare estimates from single-trait and multiple-trait models with and without maternal genetic effects, and 3) perform GWAS in order to identify genomic regions that are associated with direct and maternal genetic effects on weight traits and on IMF.
MATERIALS AND METHODS
The research did not involve any direct physical contact to the animals. No experimental studies were conducted for this project. Hence, no additional statement of institutional animal care and use committee is required.
Animals and Traits
For weight traits, phenotypic data of 9,184 RHV cattle from 269 herds were provided by ‘Vereinigte Informationssysteme Tierhaltung w. V.’ (VIT) (Verden, Germany), with permission of all RHV breeder societies involved. All herds reflect a pasture-based production system. Data recording for weight traits spanned a period of 20 yr (2000 to 2019) and included birth weight (BWT), 200 d-weight (200dW), and 365 d-weight (365dW).
For intra-muscular fat content (IMF) analyses, a subset including 33 RHV herds provided meat samples from 547 RHV animals from the slaughtering years 2018 to 2020. The meat samples with a thickness of 3.5 cm were taken from the musculus longissimus dorsi between the 12th and 13th ribs. All samples were temporarily frozen and stored until analyses in the meat laboratory at Kassel University. IMF was determined by applying near-infrared (NIR) spectroscopy.
Records from herds with less than 10 phenotyped cattle were excluded. All traits were corrected for outliers by excluding values lower or higher the mean ± 3 SD. After data editing, the dataset for weight traits included 6,003 observations for BWT, 5,719 observations for 200dw, 4,594 observations for 365dw, and 547 records for IMF for ongoing genetic-statistical analyses. Means were 36.7 kg (± 5.5 kg) for BWT, 231.4 kg (± 60.4 kg) for 200dW, 342.9 kg (± 74.7 kg) for 365dW, and 2.45 % (± 2.6 %) for IMF. The pedigree dataset included 9,989 animals with 481 sires and 2,910 dams.
Genomic Data
The genomic dataset consisted of 777,963 SNP markers from 380 genotyped animals. SNP genotypes quality control was performed using the PLINK software (Purcell et al., 2007). SNPs with a call rate lower than 0.90 (2,581 SNPs), a minor allele frequency lower than 0.05 (146,469 SNPs) and a significant deviation (P-value < 10−6) from Hardy–Weinberg equilibrium (4,939 SNPs) were discarded. Furthermore, 42,670 SNPs located on the sex chromosomes, and 10 cattle due to an individual call rate lower than 0.90, were excluded. Finally, 370 genotyped cattle with 581,304 SNP markers were considered in the ongoing genomic analyses. All of the genotyped animals were phenotyped for weight traits, and a subset of 343 animals had phenotypic records for IMF.
Genetic-Statistical Models
The genetic models reflected different genetic relationship matrices. The basic model was a single-trait animal model using the pedigree relationship matrix (A) with direct genetic effects (STPM_d, model 1), which was expanded to a model with direct and maternal genetic effects (STPM_dm, model 2). The STPM_d and STPM_dm were defined as follows:
| (1) |
| (2) |
where y is the observation vector for BWT, 200dW, 365dW or IMF; b is a vector for fixed effects including herd-year-season, sex, and age at recording as a covariate for 200dW, 365dW, or IMF; a is a vector for random direct genetic effects; m is a vector for random maternal genetic effects; and e is a vector for random residual effects; X, Za, and Zm are incidence matrices relating the records to the fixed, direct genetic, and maternal genetic effects, respectively.
The (co)variance structure for random effects for the STPM_dm was (and correspondingly reduced in STPM_d without maternal genetic effect)
where , and are direct genetic, maternal genetic, and residual variances, respectively; σam is the covariance between direct and maternal genetic effects; A is the numerator relationship matrix among individuals; and I is an identity matrix for residual effects.
The direct heritability maternal heritability , and the correlation between direct and maternal genetic effects (ram) were calculated as follows: , and where , and denote direct genetic, maternal genetic, and phenotypic variances, respectively, and σam was the direct-maternal genetic covariance. The phenotypic variance was calculated as the sum of the direct genetic variance, the maternal genetic variance, the residual variance, and the covariance between direct and maternal genetic effects.
Multiple-trait pedigree-based models including the four traits simultaneously with only direct genetic effects (MTPM_d, model 3) and with direct and maternal genetic effects (MTPM_dm, model 4) were defined as follows:
| (3) |
| (4) |
where subscripts 1 to 4 refer to four traits (i.e., BWT, 200dW, 365dW, and IMF). The (co)variance structure for the MTPM_dm was
where and are direct and maternal genetic variances, respectively, for trait i (i = 1 to 4); is the covariance between the direct genetic effect for trait i and the maternal genetic effect for trait j (j = 1 to 4); and are residual variances and covariances, respectively; A is the numerator relationship matrix among animals; and I is an identity matrix.
In the multiple-trait analysis, direct genetic correlations between traits were calculated as where is the direct genetic covariance between traits i and j; and are the direct genetic variances for traits i and j, respectively. Genetic correlations between direct genetic effect for trait i and maternal genetic effect for trait j, or vice versa, were calculated as where is the covariance between direct genetic effects for trait i and maternal genetic effects for trait j; and are the direct genetic variance for trait i and the maternal genetic variance for trait j, respectively.
In genomic best linear unbiased predictions (GBLUP), matrix A was replaced with the genomic relationship matrix (G). In this regard, single- and multiple-trait genomic models with direct genetic effects (i.e., STGM_d and MTGM_d, respectively), and with both direct and maternal genetic effects (i.e., STGM_dm and MTGM_dm, respectively), were defined. Matrix G was set up for genotyped animals with as proposed by VanRaden (2008), where m denote the number of SNPs, pi is the allele frequency at locus i, and M is a centered matrix of SNP genotypes.
In the single-step GBLUP (ssGBLUP) approach as developed by Aguilar et al. (2010), the combined pedigree-genomic relationship matrix H was used instead of matrix A. Accordingly, single- and multiple-trait models with matrix H were fitted (i.e., STHM_d and STHM_dm for single-trait models, and MTHM_d and MTHM_dm for multiple-trait models). The inverse of matrix H was defined as where GW = 0.95G + 0.05A22 and A22 is the submatrix of A for the genotyped animals. The matrix GW was constructed as proposed by VanRaden (2008), in order to render it invertible.
The analyses were conducted using GIBBS2F90 (Misztal et al., 2002). For each analysis, a single chain length of 300,000 samples was generated. After discarding the first 50,000 samples as a burn-in period, posterior means and standard deviations were calculated from every 100th samples. Convergence of the Gibbs chains was checked by visual inspection of the sample trace plots. Furthermore, the lower and upper bounds of the 95% highest posterior density (HPD) were obtained from the derived marginal densities in order to verify convergence of genetic analyses. Attempts to include maternal permanent environmental effects in the multiple-trait analyses implied convergence problems. Therefore, we focused on single- and multiple-trait models without maternal permanent environmental effects.
Model Comparisons
For both single- and multiple-trait models, comparisons considered the three different relationship matrices A, G, and H. The deviance information criterion (DIC) was used to assess goodness of fit (Spiegelhalter et al., 2002) as where is the posterior expectation of Bayesian deviance and pD is the effective number of model parameters. A smaller DIC indicates model superiority. For random effects, a priori normal distribution was assumed. For covariances, we assumed a scaled inverted chi-square distribution for the single-trait models and an inverted Wishart distribution for the multiple-trait models.
Additionally, models were evaluated based on the accuracies for direct breeding values (dEBV) and maternal breeding values (mEBV). For each trait, the accuracy of the breeding value of the ith genotyped animal (ri) was
where PEVi is the prediction error variance of the breeding value for the ith animal, and is the direct or maternal genetic variance from the respective model. The prediction error variance for a breeding value was the square of the posterior standard deviation of the respective breeding value. The EBV from the models based on A-matrix were considered as estimates being close to the true breeding value (Daetwyler et al., 2013; Badke et al., 2014). Thus, the deviation of regression coefficients of EBV on GEBV from 1.0 was a further evaluation criterion for the biasedness of genomic predictions. In the results, breeding value accuracies and the biasedness of genomic predictions are presented for the genotyped animals with phenotypes.
Genome-Wide Associations
De-regressed proofs (DRP) based on pedigree-based EBV are commonly used as pseudo-phenotypes in GWASs (Song et al., 2018; Yin and König, 2019). According to Garrick et al. (2009), the direct or maternal DRP for the ith animal was calculated as where EBVi is the pedigree-based direct or maternal breeding value for the ith genotyped animal from the MTPM_dm, and is the accuracy of the respective breeding value. The GWAS was carried out using the linear mixed model association method as implemented in the GCTA software (Yang et al., 2011). The model 5 was defined as follows:
| (5) |
where y is the vector of direct or maternal DRP for BWT, 200dW, 365dW, or IMF; 1 is the vector of ones; µ is the overall mean effect; xi is the vector of genotypes coded as 0, 1, or 2; si was the effect of the ith SNP; is the vector of random polygenic effects, with G representing the genomic relationship matrix among animals, and is the polygenic variance; Z is an incidence matrix for u, and e is the residual random effect.
Manhattan plots for the −log10P-values of the tested SNP for direct and maternal genetic effects were created using the ggplot2 package in R (Wickham, 2009). In order to evaluate the model quality, we calculated the genomic inflation factor as the median of the observed chi-squared test statistics divided by the expected median chi-squared distribution (0.4549).
The Wellcome Trust Case Control Consortium (2007) reported that the traditional Bonferroni correction based on the total number of SNP tends to produce many false-negative results. Consequently, an adjusted Bonferroni correction was applied to account for multiple testing considering an effective number of 127,730 independent SNP. Accordingly, the genome-wide significance P-value threshold was set as 0.05/n, where n was the effective number of independent SNP. The effective number of independent SNP was computed using PLINK (Purcell et al., 2007) defining a window size of 50 SNP, a step of 5 SNP and a linkage disequilibrium threshold of 0.5. In addition, a chromosome-wide significance P-value threshold was defined as 0.05/nc, where nc was the effective number of independent SNP on the respective chromosome. Chromosome-wide significance thresholds ranged from 6.71 × 10−6 for BTA1 to 1.89 × 10−5 for BTA25.
A window frame of ±250 kb around each significant SNP (i.e., according to the Bonferroni-corrected thresholds) was considered to annotate potential candidate genes. The significant SNP were mapped to corresponding genes from the Bos taurus ARS-UCD1.2 annotation release 96 assembly from the Ensembl database (http://www.ensembl.org/biomart/martview), using the R package biomaRt (Durinck et al., 2009).
RESULTS
Model Comparison
The evaluation of DIC indicated superiority of a single-trait model with direct and maternal genetic effects over a respective model ignoring the maternal genetic component (Table 1). Accordingly, DIC decreased by 2.63% for BWT, 0.75% for 200dW, 0.56% for 365dW, and 4.06% for IMF when applying STPM_dm instead of STPM_d. The modeling superiority when taking maternal genetic effects into account was very obvious for the meat quality trait IMF and consideration of SNP marker data. The DIC substantially decreased by 45.39% when using STGM_dm instead of STGM_d and by 21.37% when using STHM_dm instead of STHM_d. Also in single-step multiple-trait analyses (H-matrix), DIC were smaller when including the maternal genetic effect. However, in multiple-trait analyses based on A- or G-matrices, the simpler models ignoring the maternal genetic component indicated slight superiority according to DIC.
Table 1.
Deviance information criterion (DIC) for birth weight (BWT), 200 d-weight (200dW), 365 d-weight (365dW), and intra-muscular fat content (IMF) traits using single- and multiple-trait models considering different genetic relationship matrices
| Model1 | Trait | |||
|---|---|---|---|---|
| BWT | 200dW | 365dW | IMF | |
| STPM_d | 33550.22 | 55997.89 | 48460.38 | 1764.71 |
| STPM_dm | 32668.94 | 55579.83 | 48188.37 | 1693.14 |
| STGM_d | 33546.25 | 55939.54 | 48455.03 | 1295.16 |
| STGM_dm | 32756.95 | 55386.76 | 48141.21 | 707.25 |
| STHM_d | 33532.48 | 55988.20 | 48450.43 | 2143.64 |
| STHM_dm | 32637.26 | 55555.44 | 48167.01 | 1685.45 |
| MTPM_d | 102071.62 | |||
| MTPM_dm | 180085.57 | |||
| MTGM_d | 39480.21 | |||
| MTGM_dm | 95307.52 | |||
| MTHM_d | 196621.48 | |||
| MTHM_dm | 105188.21 | |||
ST∗M_d and ST∗M_dm denote single-trait models, and MT∗M_d and MT∗M_dm denote multiple-trait models with only direct (d) and with direct and maternal genetic (dm) effects, where ∗ refers to the pedigree-based (A), genomic (G), or combined pedigree-genomic (H) relationship matrix.
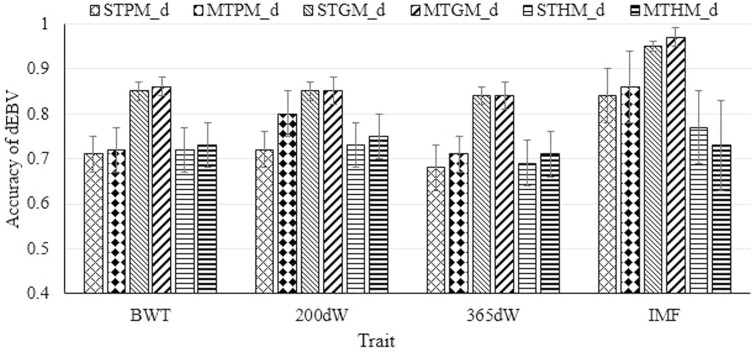
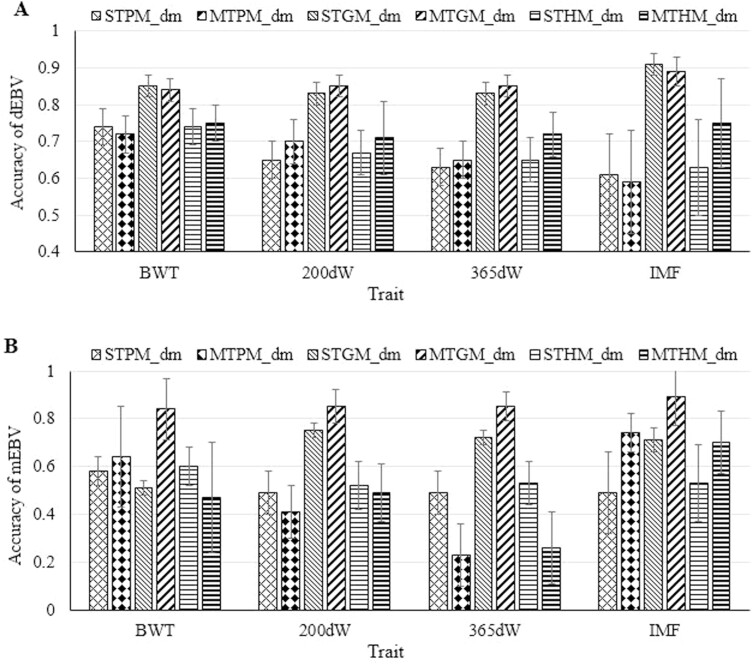
Accuracies of dEBV of genotyped cattle with phenotypes from models with only direct genetic effects are presented in Figure 1. For all traits, genomic models (i.e., STGM_d and MTGM_d) displayed the highest accuracies from 0.84 for 365dW to 0.95 for IMF. For body weight traits, a slight increase in dEBV accuracies was observed when using multiple-trait instead of single-trait models. Accuracies of dEBV of genotyped cattle from models with direct and maternal genetic effects are presented in Figure 2A. For body weight traits, dEBV accuracies were very similar when comparing to the accuracies from the respective model without maternal genetic effect as displayed in Figure 1. For IMF, dEBV accuracies were lower when using STHM_dm instead of STHM_d (0.63 ± 0.13 vs. 0.77 ± 0.08, respectively). However, with regard to the multiple-trait modeling, the MTHM_dm revealed a slightly higher accuracy than the MTHM_d (0.75 ± 0.12 vs. 0.73 ± 0.10, respectively). For all traits, the highest accuracies of mEBV were obtained from the MTGM_dm (Figure 2B). Among all models with maternal genetic effects, the accuracy of mEBV was lowest (0.26) for the trait 365dW and when considering the A-matrix in MTPM_dm.
Figure 1.
Accuracy of direct breeding values (dEBV) for birth weight (BWT), 200-d weight (200dW), 365-d weight (365dW), and intra-muscular fat content (IMF) traits. ST∗M_d and MT∗M_d denote single trait and multiple trait models with direct genetic effects, respectively, where ∗ refers to pedigree-based (A), genomic (G), or combined pedigree-genomic (H) relationship matrices.
Figure 2.
(A) Accuracy of direct breeding values (dEBV) and (B) maternal breeding values (mEBV) for birth weight (BWT), 200-d weight (200dW), 365-d weight (365dW), and intra-muscular fat content (IMF) traits. ST∗M_dm and MT∗M_dm denote single trait and multiple trait models with direct and maternal genetic effects, respectively, where ∗ refers to pedigree-based (A), genomic (G), or combined pedigree-genomic (H) relationship matrices.
The biasedness of genomic predictions decreased when considering maternal genetic effects (Table 2). The regression coefficients of EBV on GEBV from the STHM_dm were 0.97, 0.97, 1.04, and 0.93 for dEBV of BWT, 200dW, 365dW, and IMF, respectively. The biasedness increased when using the STGM_dm instead of the STHM_dm. With regard to the STGM_dm, regression coefficients were 0.62, 0.68, 0.66, and 1.24 for dEBV of BWT, 200dW, 365dW, and IMF, respectively. A significant decrease in biasedness was observed for IMF when applying multiple-trait models with the G-matrix. For instance, for dEBV of IMF, regression coefficients of 1.12 and 1.06 were observed when applying MTGM_d and MTGM_dm, respectively.
Table 2.
Regression coefficients of estimated breeding values (EBV) from models with pedigree-based relationship matrix on EBV from corresponding models with genomic (G) or combined pedigree-genomic (H) relationship matrix for birth weight (BWT), 200-d weight (200dW), 365-d weight (365dW), and intra-muscular fat content (IMF)
| Model1 | EBV | BWT | 200dW | 365dW | IMF | ||||
|---|---|---|---|---|---|---|---|---|---|
| H | G | H | G | H | G | H | G | ||
| STPM_d | Direct | 1.00 | 0.66 | 0.98 | 0.68 | 1.01 | 0.67 | 0.70 | 1.11 |
| STPM_dm | Direct | 0.97 | 0.62 | 0.97 | 0.68 | 1.04 | 0.66 | 0.93 | 1.24 |
| Maternal | 0.97 | 0.58 | 1.06 | 0.60 | 0.81 | 0.43 | 0.95 | 0.34 | |
| MTPM_d | Direct | 1.02 | 0.64 | 0.59 | 0.51 | 0.91 | 0.65 | 0.38 | 1.12 |
| MTPM_dm | Direct | 0.93 | 0.58 | 0.60 | 0.43 | 0.92 | 0.66 | 1.06 | 1.06 |
| Maternal | 0.88 | 0.53 | 0.67 | 0.44 | 0.85 | 0.66 | 0.63 | 0.30 | |
STPM_d and STPM_dm denote single-trait pedigree-based models, and MTPM_d and MTPM_dm denote multiple-trait pedigree-based models with only direct (d) and with direct and maternal genetic (dm) effects.
Genetic Parameters
The estimated variance components and heritabilities for weight traits and IMF from single- and multiple-trait models with only direct genetic effect are given in Table 3. For all traits, larger additive genetic variances were estimated when applying models with the pure genomic (G) relationship matrix in comparison to the models with pedigree-based (A) and with combined pedigree-genomic (H) matrices. Weight traits displayed moderate heritabilities in the range from 0.29 ± 0.03 (for 365dW; MTPM_d) to 0.42 ± 0.03 (for 200dW; STGM_d). Heritabilities for weight traits were quite similar from the models with the pedigree-based relationship matrix (i.e., STPM_d and MTPM_d) and the corresponding models with the combined pedigree-genomic relationship matrix (i.e., STHM_d and MTHM_d). Due to the increased additive genetic variances, the G-modeling approach via STGM_d and MTGM_d displayed the highest heritabilities. Heritabilities from the STGM_d were 0.39 ± 0.03 for BWT, 0.42 ± 0.03 for 200dW and 0.34 ± 0.03 for 365dW. Corresponding estimates from the MTGM_d were 0.38 ± 0.03 for BWT, 0.41 ± 0.04 for 200dW, and 0.32 ± 0.03 for 365dW. For IMF, heritabilities ranged from 0.38 ± 0.02 using the MTHM_d to 0.92 ± 0.05 using the STGM_d. In comparison to the models with A- and H-matrices, heritability estimates for IMF from the models with G-matrix (STGM_d and MTGM_d) showed significantly lower standard deviations and smaller 95% HPD intervals.
Table 3.
Genetic parameters ( = phenotypic variance, = additive genetic variance, = residual variance, = direct heritability) and the 95% highest posterior density interval (95% HPD) for heritability estimates for birth weight (BWT), 200 d-weight (200dW), 365 d-weight (365dW), and intra-muscular fat content (IMF) traits estimated via single- and multiple-trait trait models with only direct genetic effects considering different genetic relationship matrices
| Trait | Model1 | 95% HPD | ||||
|---|---|---|---|---|---|---|
| STPM_d | 18.52 (0.39) | 6.54 (0.56) | 11.68 (0.41) | 0.37 (0.03) | 0.32−0.42 | |
| STGM_d | 18.88 (0.41) | 7.31 (0.59) | 11.57 (0.41) | 0.39 (0.03) | 0.34−0.44 | |
| STHM_d | 18.55 (0.39) | 6.95 (0.58) | 11.57 (0.41) | 0.37 (0.03) | 0.32−0.43 | |
| BWT | MTPM_d | 18.53 (0.39) | 6.73 (0.56) | 11.80 (0.42) | 0.36 (0.03) | 0.31−0.41 |
| MTGM_d | 18.92 (0.42) | 7.12(0.59) | 11.89 (0.44) | 0.38 (0.03) | 0.33−0.43 | |
| MTHM_d | 18.62 (0.40) | 6.99(0.58) | 11.64 (0.41) | 0.37 (0.03) | 0.32−0.43 | |
| STPM_d | 1253.70 (33.20) | 491.35 (46.40) | 762.35 (33.20) | 0.39 (0.03) | 0.33−0.45 | |
| STGM_d | 1281.30 (30.25) | 539.06 (48.70) | 742.25 (33.29) | 0.42 (0.03) | 0.36−0.48 | |
| STHM_d | 1256.40 (29.05) | 496.37 (47.20) | 760.00 (33.50) | 0.39 (0.03) | 0.33−0.46 | |
| 200dW | MTPM_d | 1393.60 (115.54) | 529.10 (44.68) | 864.53 (109.30) | 0.38 (0.04) | 0.31−0.43 |
| MTGM_d | 1394.00 (94.01) | 570.21(51.91) | 823.82 (95.87) | 0.41 (0.04) | 0.33−0.48 | |
| MTHM_d | 1289.00 (55.07) | 506.49(44.81) | 782.49 (56.02) | 0.39 (0.03) | 0.33−0.45 | |
| STPM_d | 2497.30 (60.24) | 780.11 (91.56) | 1717.20 (70.23) | 0.31 (0.03) | 0.25−0.38 | |
| STGM_d | 2552.60 (63.77) | 857.42 (94.01) | 1695.20 (69.82) | 0.34 (0.03) | 0.27−0.40 | |
| STHM_d | 2500.50 (61.16) | 791.07 (90.33) | 1709.50 (69.83) | 0.32 (0.03) | 0.25−0.38 | |
| 365dW | MTPM_d | 2625.20 (87.32) | 767.50 (84.12) | 1857.70 (75.09) | 0.29 (0.03) | 0.24−0.34 |
| MTGM_d | 2802.20 (138.56) | 888.98(98.23) | 1913.20 (109.34) | 0.32 (0.03) | 0.26−0.37 | |
| MTHM_d | 2567.70 (70.69) | 759.16(82.39) | 1808.50 (69.70) | 0.30 (0.03) | 0.24−0.35 | |
| STPM_d | 4.91 (0.41) | 3.52 (1.11) | 1.38 (0.81) | 0.71 (0.18) | 0.34−1.05 | |
| STGM_d | 7.18 (0.69) | 6.67 (0.92) | 0.51 (0.31) | 0.92 (0.05) | 0.86−1.01 | |
| STHM_d | 4.84 (0.44) | 2.73 (1.10) | 2.10 (0.78) | 0.55 (0.18) | 0.18−0.92 | |
| IMF | MTPM_d | 5.13 (0.28) | 3.15 (0.59) | 1.98 (0.49) | 0.61 (0.10) | 0.47−0.86 |
| MTGM_d | 8.20 (0.55) | 7.24(0.52) | 0.96 (0.03) | 0.88 (0.02) | 0.85−0.94 | |
| MTHM_d | 4.82 (0.47) | 1.89(1.30) | 2.92 (0.96) | 0.38 (0.22) | 0.11−0.80 |
Posterior SD of estimates are given in brackets.
STX∗_d and MT∗M_d denote single-trait and multiple-trait models with direct genetic effect, respectively, where ∗ refers to pedigree-based (A), genomic (G), or combined pedigree-genomic (H) relationship matrix.
Variance components and genetic parameters for BWT, 200dW, 365dW, and IMF estimated from single- and multiple-trait models with direct and maternal genetic effects are presented in Table 4. For all traits, generally higher direct heritabilities were estimated when fitting the maternal genetic effect in comparison to the respective models ignoring the maternal component. The direct heritabilities for weight traits from the different models with maternal genetic effects were similar and ranged from 0.62 to 0.64 for BWT, from 0.45 to 0.55 for 200dW, and from 0.40 to 0.44 for 365dW. Again, highest heritabilities (0.48 to 0.76) were estimated for IMF, but values were generally lower than from models ignoring the maternal genetic component. Maternal heritabilities for BWT, 200dW, and 365dW from the different models with maternal genetic effects were in a narrow range from 0.21 to 0.24, 0.24 to 0.27, and 0.21 to 0.25, respectively (Table 4). However, for IMF, a wider range of maternal heritabilities was estimated from 0.25 (STGM_dm) to 0.65 (MTPM_dm). For IMF, the 95% HPD intervals for direct and maternal genetic variances overlapped when applying single-trait models with A- and H-matrices.
Table 4.
Genetic parameters ( = phenotypic variance, = additive genetic variance, = maternal genetic variance, = residual variance, = direct heritability, = maternal heritability) and 95% highest posterior density interval (95% HPD) for heritability estimates for birth weight (BWT), 200 d-weight (200dW), 365 d-weight (365dW), and intra-muscular fat content (IMF) traits estimated via single- and multiple-trait trait models with direct and maternal genetic effects considering different genetic relationship matrices
| Trait | Model1 | 95% HPD | 95% HPD | ||||||
|---|---|---|---|---|---|---|---|---|---|
| BWT | STPM_dm | 19.11 (0.45) | 12.23 (1.22) | 4.09 (0.61) | 8.41 (0.66) | 0.64 (0.05) | 0.53−0.75 | 0.21 (0.03) | 0.15−0.27 |
| STGM_dm | 19.41 (0.45) | 12.15 (1.16) | 4.12 (0.58) | 8.61 (0.61) | 0.63 (0.05) | 0.52−0.72 | 0.21 (0.03) | 0.16−0.27 | |
| STHM_dm | 19.14 (0.45) | 12.33 (1.24) | 4.10 (0.58) | 8.33 (0.66) | 0.64 (0.03) | 0.54−0.75 | 0.21 (0.02) | 0.16−0.27 | |
| MTPM_dm | 19.34 (0.44) | 12.70 (1.14) | 4.69 (0.44) | 8.25 (0.62) | 0.66 (0.05) | 0.55−0.75 | 0.24 (0.02) | 0.20−0.28 | |
| MTGM_dm | 19.64 (0.45) | 12.31 (1.11) | 4.48 (0.56) | 8.64 (0.59) | 0.62 (0.05) | 0.54−0.72 | 0.23 (0.03) | 0.18−0.28 | |
| MTHM_dm | 19.37 (0.45) | 12.32 (1.17) | 4.36 (0.58) | 8.38 (0.56) | 0.63 (0.05) | 0.53−0.72 | 0.23 (0.03) | 0.16−0.28 | |
| 200dW | STPM_dm | 1276.80 (31.75) | 573.32 (82.29) | 307.68 (55.90) | 651.00 (47.80) | 0.45 (0.06) | 0.32−0.55 | 0.24 (0.04) | 0.16−0.32 |
| STGM_dm | 1314.70 (33.95) | 663.41 (86.56) | 350.93 (59.91) | 604.77 (47.63) | 0.50 (0.06) | 0.39−0.61 | 0.27 (0.04) | 0.19−0.35 | |
| STHM_dm | 1279.90 (32.07) | 582.45 (82.78) | 311.83 (57.22) | 644.08 (47.00) | 0.45 (0.06) | 0.35−0.57 | 0.24 (0.04) | 0.16−0.32 | |
| MTPM_dm | 1295.09 (26.61) | 597.06 (59.02) | 335.44 (43.01) | 644.47 (35.44) | 0.46 (0.04) | 0.39−0.55 | 0.26 (0.03) | 0.20−0.32 | |
| MTGM_dm | 1343.10 (47.73) | 740.18 (85.77) | 369.45 (52.84) | 594.89 (53.56) | 0.55 (0.06) | 0.44−0.66 | 0.27 (0.03) | 0.21−0.34 | |
| MTHM_dm | 1326.60 (50.65) | 628.63 (89.09) | 325.19 (51.83) | 657.96 (47.68) | 0.47 (0.06) | 0.37−0.58 | 0.24 (0.03) | 0.19−0.32 | |
| 365dW | STPM_dm | 2523.10 (64.59) | 1027.70 (153.02) | 523.98 (99.45) | 1478.40 (91.12) | 0.40 (0.05) | 0.30−0.51 | 0.21 (0.04) | 0.14−0.28 |
| STGM_dm | 2589.30 (67.13) | 1026.30 (159.70) | 496.68 (105.22) | 1476.21 (90.40) | 0.44 (0.06) | 0.33−0.54 | 0.23 (0.04) | 0.15−0.29 | |
| STHM_dm | 2528.90 (64.15) | 1052.30 (154.53) | 527.30 (99.43) | 1463.30 (90.09) | 0.41 (0.06) | 0.32−0.53 | 0.21 (0.04) | 0.14−0.28 | |
| MTPM_dm | 2638.10 (64.42) | 1155.41 (132.15) | 664.34 (103.82) | 1451.91 (81.23) | 0.44 (0.04) | 0.35−0.52 | 0.25 (0.04) | 0.17−0.32 | |
| MTGM_dm | 2680.10 (69.36) | 1192.61 (159.17) | 605.22 (78.75) | 1491.12 (87.23) | 0.44 (0.05) | 0.35−0.55 | 0.23 (0.03) | 0.18−0.27 | |
| MTHM_dm | 2605.70 (60.51) | 1065.42 (144.40) | 568.90 (80.98) | 1532.00 (80.08) | 0.41 (0.05) | 0.31−0.50 | 0.22 (0.03) | 0.15−0.28 | |
| IMF | STPM_dm | 5.03 (0.40) | 2.78 (1.14) | 3.26 (1.33) | 1.04 (0.63) | 0.55 (0.21) | 0.13−0.96 | 0.44 (0.25) | 0.12−1.04 |
| STGM_dm | 7.23 (0.94) | 5.38 (0.90) | 1.81 (0.71) | 0.22 (0.19) | 0.75 (0.11) | 0.51−0.98 | 0.25 (0.10) | 0.05−0.45 | |
| STHM_dm | 5.15 (0.43) | 2.64 (1.07) | 3.18 (1.36) | 1.04 (0.66) | 0.51 (0.20) | 0.13−0.89 | 0.61 (0.26) | 0.04−0.99 | |
| MTPM_dm | 7.76 (0.33) | 3.71 (0.24) | 5.04 (0.40) | 1.14 (0.15) | 0.48 (0.04) | 0.43−0.56 | 0.65 (0.03) | 0.56−0.69 | |
| MTGM_dm | 9.81 (1.45) | 7.49 (1.44) | 2.80 (0.49) | 0.43 (0.12) | 0.76 (0.09) | 0.56−0.92 | 0.29 (0.05) | 0.21−0.37 | |
| MTHM_dm | 6.93 (0.68) | 4.04 (0.38) | 3.23 (0.48) | 0.71 (0.14) | 0.59 (0.07) | 0.47−0.70 | 0.47 (0.09) | 0.36−0.69 |
Posterior SD of estimates are given in brackets.
ST∗M_dm and MT∗M_dm denote single trait and multiple trait models with direct and maternal genetic effects, respectively, where ∗ refers to pedigree-based (A), genomic (G), or combined pedigree-genomic (H) relationship matrix.
Direct Genetic and Maternal Genetic Correlations
Genetic covariances and genetic correlations among BWT, 200dW, 365dW, and IMF from the multiple-trait models with maternal genetic effects and considering the different genetic relationship matrices are displayed in Table 5. Estimates were quite similar from the different modeling approaches. The direct genetic correlations among the body weight traits BWT, 200dW, and 365dW were throughout positive and favorable. In this regard, the lowest direct genetic correlations were estimated between BWT and 365dW (0.16, 0.13, and 0.12 from the MTPM_dm, MTGM_dm, and MTHM_dm, respectively). The highest direct genetic correlations were estimated between 200dW and 365dW (0.57, 0.61, and 0.63 from the MTPM_dm, MTGM_dm, and MTHM_dm, respectively). The direct genetic correlations between BWT and IMF showed a stronger variation with 0.31, −0.13, and 0.45 from the MTPM_dm, MTGM_dm, and MTHM_dm, respectively. Slightly antagonistic direct genetic relationships were identified between IMF with 200dW and 365dW. The genetic correlations between IMF and 200dW were −0.23, −0.15, and −0.02, and between IMF and 356dW 0.12, 0.01, and 0.05 (from the MTPM_dm, MTGM_dm, and MTHM_dm, respectively).
Table 5.
Genetic covariances (above the diagonal) and genetic correlations (below the diagonal) among birth weight (BWT), 200-d weight (200dW), 365-d weight (365dW), and intra-muscular fat content (IMF) traits estimated via multiple-trait models with direct and maternal genetic effects
| Model1 | Effect2 | Trait | d | m | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| BWT | 200dW | 365dW | IMF | BWT | 200dW | 365dW | IMF | |||
| d | BWT | 23.27 (6.78) | 19.76 (8.82) | 2.16 (0.30) | −6.32 (0.59) | −2.61 (6.15) | −3.19 (7.72) | −1.61 (0.44) | ||
| 200dW | 0.27 (0.07) | 475.77 (87.07) | −10.61 (3.09) | −3.17 (3.88) | −281.10 (47.72) | −150.88 (68.30) | −23.64 (3.60) | |||
| 365dW | 0.16 (0.07) | 0.57 (0.06) | 7.85 (3.19) | −11.56 (5.99) | −154.33 (76.06) | −633.45 (114.18) | 4.84 (5.59) | |||
| MTPM_dm | IMF | 0.31 (0.05) | −0.23 (0.06) | 0.12 (0.05) | −2.81 (0.30) | 5.26 (2.35) | −9.33 (2.59) | −2.14 (0.25) | ||
| m | BWT | −0.82 (0.02) | −0.06 (0.07) | −0.16 (0.08) | −0.67 (0.06) | 7.37 (3.88) | 13.41 (5.09) | 1.84 (0.28) | ||
| 200dW | −0.04 (0.09) | −0.63 (0.05) | −0.25 (0.11) | 0.15 (0.06) | 0.19 (0.09) | 237.09 (58.22) | 18.61 (3.43) | |||
| 365dW | −0.04 (0.08) | −0.24 (0.10) | −0.72 (0.06) | −0.19 (0.06) | 0.24 (0.09) | 0.50 (0.08) | 4.54 (6.45) | |||
| IMF | −0.20 (0.06) | −0.43 (0.06) | 0.06 (0.07) | −0.49 (0.05) | 0.38 (0.07) | 0.45 (0.08) | 0.08 (0.11) | |||
| d | BWT | 24.99 (7.76) | 15.80 (9.38) | −1.31 (0.81) | −5.79 (0.70) | −1.39 (5.56) | 8.17 (7.58) | 0.87 (1.07) | ||
| 200dW | 0.26 (0.07) | 572.62 (90.63) | 12.01 (9.80) | −5.41 (6.00) | −361.42 (58.24) | −206.35 (65.07) | −1.05 (7.82) | |||
| 365dW | 0.13 (0.08) | 0.61 (0.07) | 1.12 (7.99) | −2.65 (7.10) | −196.35 (62.73) | −608.76 (99.54) | −1.73 (12.87) | |||
| MTGM_dm | IMF | −0.13 (0.09) | −0.15 (0.12) | 0.01 (0.09) | 0.42 (0.66) | −23.89 (5.85) | −32.53 (7.03) | −0.90 (0.98) | ||
| m | BWT | −0.77 (0.04) | −0.10 (0.10) | −0.04 (0.09) | −0.07 (0.11) | 6.44 (3.98) | 5.39 (4.80) | 0.20 (0.61) | ||
| 200dW | −0.02 (0.08) | −0.68 (0.04) | −0.29 (0.08) | −0.46 (0.09) | 0.16 (0.09) | 246.42 (54.11) | 7.07 (4.67) | |||
| 365dW | 0.10 (0.08) | −0.31 (0.08) | −0.71 (0.05) | −0.48 (0.08) | 0.10 (0.08) | 0.52 (0.09) | 3.72 (6.20) | |||
| IMF | 0.15 (0.18) | −0.03 (0.17) | −0.28 (0.22) | −0.18 (0.09) | 0.05 (0.17) | 0.21 (0.13) | −0.09 (0.15) | |||
| d | BWT | 17.96 (7.06) | 13.52 (9.33) | 3.14 (0.52) | −5.69 (0.69) | 0.59 (5.22) | 7.04 (8.74) | −0.34 (1.16) | ||
| 200dW | 0.20 (0.07) | 512.02 (85.72) | −1.10 (5.26) | −1.61 (4.66) | −285.18 (60.83) | −144.01 (61.71) | −2.77 (6.73) | |||
| 365dW | 0.12 (0.08) | 0.63 (0.05) | 3.45 (4.75) | −3.25 (6.89) | −178.70 (54.32) | −560.56 (84.91) | 17.94 (10.01) | |||
| MTHM_dm | IMF | 0.45 (0.06) | −0.02 (0.10) | 0.05 (0.07) | −1.71 (0.39) | −18.13 (3.44) | −24.19 (4.50) | −1.05 (0.66) | ||
| m | BWT | −0.78 (0.04) | −0.03 (0.08) | −0.05 (0.10) | −0.41 (0.08) | 4.70 (3.75) | 2.99 (5.72) | 0.55 (0.600) | ||
| 200dW | 0.01 (0.08) | −0.63 (0.06) | −0.30 (0.07) | −0.50 (0.07) | 0.12 (0.10) | 236.27 (47.06) | 8.49 (4.82) | |||
| 365dW | 0.08 (0.10) | −0.24 (0.10) | −0.72 (0.04) | −0.50 (0.07) | 0.06 (0.11) | 0.55 (0.07) | −9.60 (7.55) | |||
| IMF | −0.05 (0.18) | −0.06 (0.14) | 0.31 (0.16) | −0.29 (0.17) | 0.15 (0.15) | 0.26 (0.13) | −0.22 (0.17) | |||
Posterior SD of estimates are given in brackets.
MT∗M_dm denotes multiple trait models with direct and maternal genetic effect, where ∗ refers to pedigree-based (A), genomic (G), or combined pedigree-genomic (H) relationship matrices.
d and m denote estimates for direct and maternal genetic effects, respectively.
Low- to moderate-maternal genetic correlations were estimated among the different traits (Table 5). The highest correlations were 0.55 ± 0.07 (MTHM_dm) between 200dW and 365dW, and 0.45 ± 0.08 (MTPM_dm) between 200dW and IMF. The lowest maternal genetic correlations of 0.08 ± 0.11, −0.09 ± 0.15, and -0.22 ± 0.17 were estimated between 365dW and IMF using MTPM_dm, MTGM_dm, and MTHM_dm, respectively.
Within the same traits, quite strong negative direct-maternal genetic correlations were estimated, ranging from −0.77 to −0.82 for BWT, from −0.63 to −0.68 for 200dW, from −0.71 to −0.72 for 365dW, and from −0.18 to −0.49 for IMF (Table 5). Posterior SD were largest in all analyses including IMF.
Genome-Wide Associations
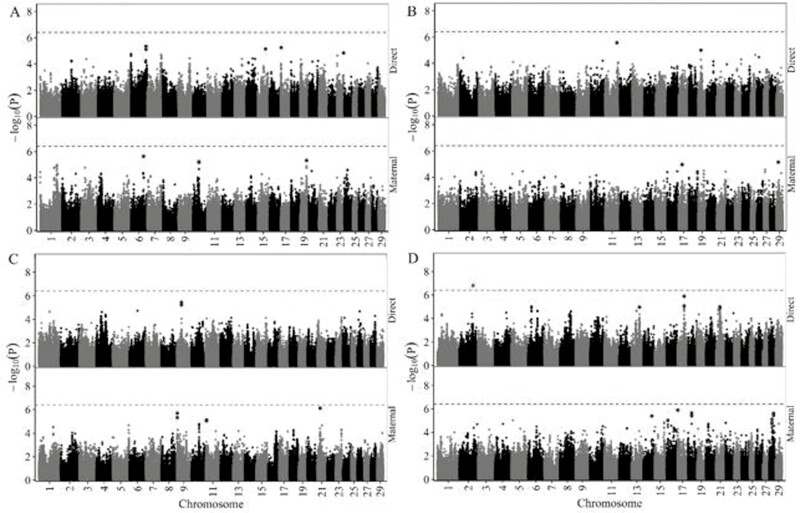
Manhattan plots from the GWAS for direct and maternal genetic effects on BWT, 200dW, 365dW, and IMF are given in Figure 3. The genomic inflation factors for additive genetic effects ranged from 0.99 for BWT to 1.01 for 200-dW. For maternal genetic effects, the genomic inflation factors ranged from 0.98 for BWT to 1.00 for 200-dW. One SNP (rs137226468) on BTA2 surpassed the genome-wide significance threshold (P = 3.91 × 10−7), which was associated with the direct genetic effect on IMF only. According to the chromosome-wide significance threshold, 5, 7, 2, and 7 SNP were significantly associated with direct genetic effects on BWT, 200dW, 365dW, and IMF, respectively. For the same significance threshold, 10, 8, 7, and 13 significant SNP were significantly associated with maternal genetic effects on BWT, 200dW, 365dW, and IMF, respectively.
Figure 3.
Manhattan plots for direct and maternal effects on birth weight (A), 200 d-weight (B), 365 d-weight (C), and intra-muscular fat content (D). The red line displays the genome-wide significance threshold according to the adjusted Bonferroni correction. The red and the blue dots represent significant SNP according to the genome-wide and the chromosome-wide significance thresholds, respectively. The genome-wide significance threshold was 3.91 × 10−7 and the chromosome-wide significance thresholds ranged from 6.71 × 10−6 for BTA1 to 1.89 × 10−5 for BTA25.
For the direct genetic effect on BWT, we identified six potential candidate genes. Two of these genes (ENSBTAG00000048983 and FBXW7) are located on BTA17 (Table 6). Three genes (NOL10, ODC1, and SMG6) and one gene (FRK) were also annotated as potential candidate genes for 200dW and for 365-dW, respectively. Six potential candidate genes (i.e., ENSBTAG00000052399, VWC2L, BMP7, TMEM132D, UBE2Q2, and CHRNB4) were annotated for the direct genetic effects on IMF. Regarding maternal genetic effects on BWT, four potential candidate genes (SHROOM3 on BTA6, ZNF609 on BTA10, and PECAM1 and TEX2 on BTA19) were identified. The ZNF609 gene included the largest number of significant SNP (seven SNP), located within or in close distance. Two genes (TMEM182 and SEC11A) and four genes (GRHL2, FGA, FGB, and CTNNA3) were inferred as potential candidate genes for the maternal genetic effect on 365-dW and on IMF, respectively.
Table 6.
Potential candidate genes related to the identified single-nucleotide polymorphisms (SNP) associated with direct (d) and maternal (m) genetic effects on birth weight (BWT), 200-d weight (200dW), 365-d weight (365dW), and intra-muscular fat content (IMF) traits
| Trait | Effect | BTA1 | Gene name | Gene position | No. of significant SNP within/close to gene2 |
Most significant SNP | |||
|---|---|---|---|---|---|---|---|---|---|
| rs number | Position | in gene | P-value | ||||||
| BWT | d | 6 | HS3ST1 | 106504062-106540229 | 0/2 | rs136251364 | 106314217 | No | 4.50E-06 |
| 15 | BBOX1 | 57758972-57839046 | 1/0 | rs137689845 | 57781671 | Yes | 7.08E-06 | ||
| 17 | ENSBTAG00000048983 | 5007893-5022532 | 0/1 | rs109750506 | 5131401 | No | 5.58E-06 | ||
| 17 | FBXW7 | 5256463-5351434 | 0/1 | rs109750506 | 5131401 | No | 5.58E-06 | ||
| 23 | TUBB2A | 50525012-50528874 | 0/1 | rs109760348 | 50501825 | No | 1.43E-05 | ||
| 23 | TUBB2B | 50489737-50494257 | 0/1 | rs109760348 | 50501825 | No | 1.43E-05 | ||
| m | 6 | SHROOM3 | 91317426-91647743 | 1/0 | rs110666415 | 91494841 | Yes | 2.27E-06 | |
| 10 | ZNF609 | 45385273-45582446 | 7/0 | rs135585847 | 45410963 | Yes | 5.94E-06 | ||
| 19 | PECAM1 | 48541625-48619840 | 1/0 | rs41919301 | 48557862 | Yes | 4.69E-06 | ||
| 19 | TEX2 | 48368410-48482779 | 0/1 | rs41919273 | 48538199 | No | 4.69E-06 | ||
| 200-dW | d | 11 | NOL10 | 86971480-87061426 | 0/6 | rs110940186 | 87095463 | No | 2.69E-06 |
| 11 | ODC1 | 87197374-87204771 | 0/6 | rs110940186 | 87095463 | No | 2.69E-06 | ||
| 19 | SMG6 | 23080158-23280691 | 1/0 | rs41904211 | 23107053 | Yes | 9.76E-06 | ||
| 365-dW | d | 9 | FRK | 34593961-34691721 | 0/2 | rs43339651 | 34929448 | No | 3.61E-06 |
| m | 11 | TMEM182 | 7459624-7502937 | 0/2 | rs43661127 | 7695135 | No | 7.48E-06 | |
| 21 | SEC11A | 22328984-22362069 | 2/0 | rs134746982 | 22359151 | Yes | 7.37E-07 | ||
| IMF | d | 2 | ENSBTAG00000052399 | 101838614-102121965 | 0/1 | rs137226468 | 102157462 | No | 1.54E-07 |
| 2 | VWC2L | 102407028-102619802 | 0/1 | rs137226468 | 102157462 | No | 1.54E-07 | ||
| 13 | BMP7 | 58889622-58975046 | 0/1 | rs108949054 | 59057952 | No | 1.07E-05 | ||
| 17 | TMEM132D | 47426664-48084896 | 3/0 | rs109093435 | 47665130 | Yes | 1.29E-06 | ||
| 21 | UBE2Q2 | 31126645-31188035 | 1/0 | rs42815709 | 31132302 | Yes | 9.87E-06 | ||
| 21 | CHRNB4 | 30995468-31015323 | 0/1 | rs43136852 | 31093734 | No | 1.25E-05 | ||
| m | 14 | GRHL2 | 62721044-62888891 | 1/0 | rs136368193 | 62727223 | Yes | 4.01E-06 | |
| 17 | FGA | 2859932-2868867 | 0/1 | rs134206866 | 2888493 | No | 1.25E-06 | ||
| 17 | FGB | 2894488-2932611 | 1/0 | rs136514361 | 2895691 | Yes | 1.25E-06 | ||
| 28 | CTNNA3 | 22282909-24121400 | 4/0 | rs136938446 | 22852412 | Yes | 7.61E-06 | ||
BTA, Bos taurus chromosome.
Number of SNP that reached the Bonferroni-corrected genome-wide (in bold) or chromosome-wide significance threshold.
DISCUSSION
Direct Heritabilities for Weight and Meat Quality Traits
In the present study, direct heritabilities for weight traits ranged from 0.41 to 0.66 when applying the enhanced models considering the direct and the maternal genetic effect. For IMF, heritabilities ranged from 0.48 to 0.76. Generally, already published heritabilities for weight and meat quality traits as well as the corresponding variance components display a broad range of estimates (Marshall, 1994; Utrera and Van Vleck, 2004). Possible explanations address breed particularities, applied statistical methods and considered respective model effects, the number of phenotypic records, pedigree inconsistencies as well as specific slaughtering effects for meat quality traits (Utrera and Van Vleck, 2004). Reported heritabilities for BWT were 0.22 (Müllenhoff, 2008), 0.23 (Brandt et al., 2010), 0.32 (Chud et al., 2014), and 0.61 (Mackinnon et al., 1991). Heritabilities for weaning weight were 0.12 (Brandt et al., 2010), 0.18 to 0.21 (Jeyaruban et al., 2009), 0.20 (Mackinnon et al., 1991), 0.20 to 0.23 (Meyer, 1995), 0.21 (Santana et al., 2013), and 0.26 to 0.28 (Williams et al., 2012). Heritabilities for yearling weight were 0.26 to 0.29 (Jeyaruban et al., 2009), 0.25 (Mackinnon et al., 1991), 0.28 to 0.31 (Meyer, 1995), 0.29 (Zuin et al., 2012), and 0.38 to 0.64 (Guidolin et al., 2012). Heritabilities for IMF in cattle were 0.27 (Torres-Vázquez and Spangler, 2016), 0.31 (MacNeil et al., 2010), 0.38 (Seroba et al., 2011), 0.47 to 0.52 (Crews et al., 2003), and 0.93 (Shackelford et al., 1994). For marbling score, which is genetically highly correlated with IMF (Devitt and Wilton, 2001; MacNeil et al., 2010), Srivastava et al. (2019) estimated a heritability of 0.35 and 0.38 in Korean Hanwoo cattle from a single-trait pedigree-based model and a multiple-trait pedigree-based model, respectively. Hence, heritabilities for weight traits and IMF in the dual-purpose RHV population reflect the range of estimates as published for different commercial beef cattle breeds.
In comparison to the models with only direct genetic effects, the models with both direct and maternal genetic effects implied higher heritabilities as direct-maternal genetic covariances were negative. For body weight traits, quite similar heritabilities were estimated when using models with different relationship matrices. However, for IMF, we estimated larger direct heritabilities and smaller 95% HPD intervals when using the models with G-matrix compared to the models with A- and H-matrices. Differences in genetic parameter estimates might be due to the differences when constructing the relationship matrices. In GBLUP, unrelated animals are also connected through common genetic markers. In our study, only a subset of animals from the pedigree-based and the combined pedigree-genomic analyses were considered when constructing the G-matrix for the pure genomic models. Most obvious differences in parameter estimates from the different relationship matrices were observed for IMF, especially when considering the large 95% HPD intervals. Such large 95% HPD intervals for IMF reflect the uncertainty of the heritabilities estimates, which might be related to the limited number of phenotypic records. Heritabilities for IMF from single and multiple trait models with the same relationship matrices were very similar. For instance, direct heritabilities for IMF from both models STGM_dm and MTGM_dm were 0.75 and 0.76, respectively. The quite high IMF heritability and genetic variance for IMF in the local RHV dual purpose cattle population suggest a breeding focus on meat quality and RHV cattle genotyping, enabling a leadership in meat quality traits which have been neglected in German beef cattle breeding goals during the past decades.
Maternal Genetic Impact on Weight and Meat Quality Traits
The inclusion of the maternal genetic effect into genetic evaluation models is imperative; otherwise, the direct heritability will be overestimated (Meyer, 1992). Recently, even from an extended time-lagged and across-generation perspective in dairy cattle, consideration of maternal genetic effects improved the statistical modeling quality (Kipp et al., 2021). For BWT, weaning weight, and yearling weight in beef cattle, the maternal effects explained a large percentage of the phenotypic variation (Waldron et al., 1993). Accordingly, in our study, maternal heritabilities were quite high in the range from 0.21 to 0.24 for BWT, from 0.24 to 0.27 for 200dW, and from 0.21 to 0.25 for 365dW. These estimates for the maternal genetic component in dual-purpose RHV cattle correspond with maternal heritabilities for BWT, weaning weight, and yearling weight in specialized beef cattle breeds as reported by Brandt et al. (2010), Eler et al. (1995), Meyer (1992), Meyer (1994), Robinson (1996), and Tosh et al. (1999). Nevertheless, the maternal genetic impact especially for trait recorded late in life was larger in the RHV outdoor population than in the commercial beef breeds. Accordingly, in the RHV breed, cows and their offspring are raised on pasture without feeding supplements. Hence, the dam’s milk is the most important energy supply for offspring of all ages and explains the respective strong maternal genetic component. Dam milk quality may also affect meat quality traits recorded in offspring, explaining the quite high maternal heritabilities for IMF in the range from 0.29 to 0.65 in the present study. In Duroc pigs with a breeding focus on meat quality and which are predominantly used in low input systems, Hoque et al. (2008) estimated maternal heritabilities between 0.13 and 0.15 for IMF. In contrast, for meat quality traits of beef cattle kept in feedlots, the maternal heritability was close to zero, e.g., 0.03 for rib eye area (Abdel-Aziz et al., 2005) and 0.03 for marbling score in Japanese Black cattle (Shimada et al., 1998).
Correlations Between Direct and Maternal Genetic Effects for Weight and Meat Quality Traits
Correlations between the direct genetic effect and the maternal genetic effect for weight traits in the RHV breed, estimated via multiple-trait models, were unfavorable and ranged between −0.77 and −0.82 for BWT, between −0.63 and −0.68 for 200dW, and between −0.71 and −0.72 for 365dW. Similarly, pronounced negative direct-maternal correlations for BWT were reported by Müllenhoff (2008) and by Eler et al. (1995). Contrarily, Brandt et al. (2010), Meyer et al. (1992), Müllenhoff (2008), and Tosh et al. (1999) reported positive direct-maternal correlations for BWT in the range from 0.04 to 0.32.
For weight or weight gain traits recorded later in life, direct-maternal correlations as estimated in commercial beef cattle displayed a huge variation, i.e., from −0.57 to 0.16 for weaning weight in the study by Müllenhoff (2008) and in the range from −0.13 to −0.32 in the study by Eler et al. (1995). For yearling weight, the direct-maternal correlations were −0.48 to 0.49 (Meyer et al., 1992), −0.22 (Swalve, 1993), and −0.17 (Eler et al., 1995).
The correlations between the direct and the maternal genetic effects for IMF in the present study from the different models ranged from −0.18 to −0.49. Accordingly, regarding meat quality in beef cattle, Johnson et al. (2002) reported antagonistic relationships between direct and maternal genetic effects for rib eye area with a correlation estimate of −0.67. As stated by Lee (2001), a clear biological explanation for the antagonistic relationships between maternal and direct genetic effects for meat quality traits is not available. Nevertheless, regarding conservation programs in local small sized populations, a negative covariance between direct and maternal genetic effects might favorably contribute to genetic variation (Maniatis, et al., 2013). Hence, especially in the RHV outdoor system, it may be the challenge to improve the economically important meat quality trait IMF without neglecting the also very important maternal abilities.
Genetic Correlations Among Weight and Meat Quality Traits
Genetic correlations between the direct genetic effect among BWT and weight traits from later ages (200dW and 365dW) from all multiple-trait models were throughout positive. Genetic correlations between measurements within a closer time interval (i.e., between BWT and 200dW) were slightly higher than between BWT and 365dW. Different gene activities for different body weight traits recorded at birth and recorded more than 1 yr later have been reported by Yin and König (2019) in cattle and by Aikins-Wilson et al. (2021) in pigs. In beef cattle, Brandt et al. (2010) and Eler et al. (1995) reported a genetic correlation between BWT and weaning weight of 0.20 and of 0.23, respectively, reflecting our estimate between BWT and 200dW. Higher genetic correlations between these two traits were estimated by Chud et al. (2014) with 0.36, Mackinnon et al. (1991) with 0.57, and by Tosh et al. (1999) with 0.73.
In the present study, we found differing genetic correlations between IMF and BWT, depending on the considered relationship matrix. The correlation was 0.31 on the basis of the A-matrix (MTPM_dm) and 0.45 on the basis of the H-matrix (MTHM_dm), but close to zero with −0.13 when modeling the pure genomic G-matrix (MTGM_dm). We assume that the small number of genotyped animals explains the similarities of estimates from A- and H-matrices, but differencing results with higher accuracies of breeding values (Figure 1) from the G-matrix. Such aspects were outlined by Shabalina et al. (2021) in the context of genetic evaluations in an organic dairy cattle production system, including a very small number of genotyped cows from organic herds.
In the present study, genetic correlations between IMF and 200dW were of antagonistic nature (−0.02 to −0.23), but slightly positive between IMF and 365dW (0.01 to 0.12). However, when taking the posterior SD into account, all correlations were very close to zero, and reflecting estimates from the literature between weight at later ages and meat quality and carcass traits in cattle. For example, a genetic correlation of 0.04 between 210-d weight and back fat thickness, and of −0.09 between 210-d weight and rump fat, was reported by Zuin et al. (2012). Similar genetic correlations of −0.10 and −0.09, respectively, were estimated by Yokoo et al. (2010). With regard to 365dW and carcass traits, Yokoo et al. (2010) and Zuin et al. (2012) estimated genetic correlations of 0.04 and 0.15 with back fat thickness, respectively, and of −0.01 and 0.03 with rump fat thickness, respectively. In Japanese Black cattle, the genetic correlation between body weight at the beginning and at the end of the fattening period with marbling score was 0.04 and 0.17, respectively (Abdel-Aziz et al., 2005). Nephawe et al. (2004) correlated mature weight of steers with meat quality traits. The genetic correlation with marbling score was −0.15, and 0.15 with Warner–Bratzler shear force. In contrast, a moderate genetic correlation of 0.34 between body weight and IMF was reported in pigs (Solanes et al., 2009), supporting the results of Suzuki et al. (2005) and Lo et al. (1992), who also estimated positive genetic correlations between daily gain and IMF of 0.23 and 0.27, respectively. A possible physiological explanation for the differences between the species addresses the growth rate of subcutaneous fat and IMF relative to total fat which differs between cattle and pigs (Kempster et al., 1976; Kempster and Evans, 1979).
Accuracies of Breeding Values
Cesarani et al. (2019), Lourenco et al. (2015), and Wei et al. (2020) suggested genetic-statistical models using the combined H-matrix. Accordingly, in the present study and with focus on the enhanced models with direct and maternal genetic effects, accuracies of dEBV on the basis of G- or H-matrices were higher than from the pure pedigree-based approach. However, for the moderate- to high-heritability weight traits and IMF, dEBV accuracies from MTPM_dm and MTHM_dm only differed slightly, probably due to the small number of cattle with genotypes. This was also reflected by the regression coefficients of EBV on GEBV close to 1.0. In the present study, especially on the basis of pure genomic data, STGM_dm and MTGM_dm indicated model superiority according to DIC, the quite small 95% HPD intervals for direct heritabilities and the highest accuracies of dEBV.
Regarding the most enhanced models with maternal genetic effects, accuracies were lowest when considering only the pedigree relationships. In beef cattle, Nwogwugwu et al. (2019) reported a strong impact of pedigree errors on EBV accuracies, genetic gain, and heritability estimates in classical pedigree-based modeling approaches. The problem in this regard of missing or wrong pedigree data was carefully outlined by Harder et al. (2005). Especially in local small-sized beef, dual-purpose, and pig populations in Germany, there is often a lack of deep and complete pedigrees, encouraging genetic studies on the basis of animal genotypes (e.g., Biermann et al., 2015). Especially for meat quality traits displaying a high heritability, consideration of dense SNP markers (Pimentel and König, 2012) greatly improved accuracies of overall breeding goals. In contrast, for traits displaying low heritabilities such as health or female fertility, it is imperative to consider training sets with more than 10,000 genotyped animals. However, also in such large-scale genomic studies as mostly conducted in dairy cattle with Bayesian, machine learning, or GBLUP approaches (Naderi et al., 2018; Bohlouli et al, 2019), consideration of genomic marker data contributed to the best prediction accuracies.
Genome-Wide Associations for Weight and Meat Quality Traits
The genomic inflation factors for all traits were close to 1.0, indicating that there was no need to correction for population stratification. Many of the potential candidate genes detected in the present study support previous reports for the same or correlated traits in beef cattle populations. The gene HS3ST1 (a potential candidate gene for direct genetic effects on BWT) and the genes ZNF609, PECAM1, and TEX2 genes (potential candidate genes for maternal genetic effects on BWT) were associated with carcass and body weight traits in beef cattle (Kim, et al., 2011; Naserkheil et al., 2020; Bruscadin et al., 2021). In a selection signature segment on BTA10, Boitard et al. (2021) also detected ZNF609 as a candidate gene for carcass traits in a beef cattle population. SMG6, a potential candidate gene for the maternal genetic effect on 200dW, was significantly associated with rib eye area in beef cattle (Wang et al., 2020). The results from the present study support the associations reported by Wang et al. (2020), because Caetano et al. (2013) estimated a moderate positive correlation between rib eye area and weight gain from 210 to 365 d. For 365-dW, the FRK and TMEM182 genes were identified for direct and maternal genetic effects, respectively. FRK is a protein coding gene which is associated with feed efficiency and feed intake in beef cattle (Sherman et al., 2010; Higgins et al., 2018). The TMEM182 gene is known to be involved in the development of muscle tissues (Wu et al., 2008; Freebern et al., 2020).
For direct genetic effects on IMF, the most significant SNP (rs137226468) is located in close distance to the ENSBTAG00000052399 and VWC2L genes on BTA2. ENSBTAG00000052399 is a protein coding gene, but the function of this gene is unknown. VWC2L and BMP7, another potential candidate gene for direct genetic effects on IMF, are reported as candidate genes for feed efficiency in pigs (Wang et al., 2015) and in beef cattle (Zhang et al., 2020). The VWC2L gene regulates the osteoclast activation and matrix mineralization (Ohyama, et al., 2012), and the BMP7 gene is a lipid-related candidate gene. In several studies conducted in beef cattle (Srikanth et al., 2020; Wang et al., 2020; Zhang et al., 2020), BMP7 was also found to be associated with carcass traits (e.g., backfat thickness, rib eye area, lean meat yield, and carcass marbling score) and average daily gain.
The estimated genetic parameters for weight traits and IMF in the local RHV dual-purpose cattle population kept in outdoor production systems indicate trait improvement possibilities via breeding approaches. Especially for the meat quality trait IMF, quite high heritabilities were estimated. Especially from the background that meat quality traits do not play a role in breeding goals for commercial beef cattle breeds in Germany, we strongly suggest to develop an RHV breeding goal including IMF. Probably due to the intensive calf–cow relationship in the outdoor suckler system, we identified a quite strong maternal genetic component on weight traits at different ages as well as on IMF. Hence, it may be imperative to apply models with maternal genetic effects in genetic evaluations. Furthermore, we suggest to focus on RHV cattle genotyping, especially when aiming on breeding goal traits with high heritabilities. In this regard, a comparatively small number including less than 1,000 genotyped animals implied reliable genetic parameter estimates with acceptable standard errors. Furthermore, on the basis of the G-matrix, we identified breeding values with highest accuracies.
Footnotes
This work was financially supported by the German Federal Ministry of Food and Agriculture (BMEL) through the Federal Office for Agriculture and Food (BLE), grant number 2816BM010. We gratefully thank for the support.
Conflict of Interest Statement
None declared.
LITERATURE CITED
- Abdel-Aziz, M., Nishida S., Suzuki K., and Nishida A.. . 2005. Estimation of direct and maternal genetic parameters for growth and carcass traits in a herd of Japanese Black cattle in Miyagi prefecture, using a multitrait animal model. Anim. Sci. J. 76:187–193. doi: 10.1111/j.1740-0929.2005.00255.x. [DOI] [Google Scholar]
- Aguilar, I., Misztal I., Johnson D. L., Legarra A., Tsuruta S., and Lawlor T. J.. . 2010. Hot topic: a unified approach to use phenotypic, full pedigree and genomic information for genetic evaluation of Holstein for final score. J. Dairy Sci. 93:743–752. doi: 10.3168/jds.2009-2730. [DOI] [PubMed] [Google Scholar]
- Aikins-Wilson, S., Bohlouli M., and König S.. . 2021. Maternal and direct genetic parameters for tail length, tail lesions, and growth traits in pigs. J. Anim. Sci. 99:1–11. doi: 10.1093/jas/skaa398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badke, Y. M., Bates R. O., Ernst C. W., Fix J., and Steibel J. P.. . 2014. Accuracy of estimation of genomic breeding values in pigs using low-density genotypes and imputation. G3. 4:623–631. doi: 10.1534/g3.114.010504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biermann, A. D., Yin T., König von Borstel U. U., Rübesam K., Kuhn B., and König S.. . 2015. From phenotyping towards breeding strategies: using in vivo indicator traits and genetic markers to improve meat quality in an endangered pig breed. Animal. 9:919–927. doi: 10.1017/S1751731115000166. [DOI] [PubMed] [Google Scholar]
- Bohlouli, M., Alijani S., Naderi S., Yin T., and König S.. . 2019. Prediction accuracies and genetic parameters for test-day traits from genomic and pedigree-based random regression models with or without heat stress interactions. J. Dairy Sci. 102:488–502. doi: 10.3168/jds.2018-15329. [DOI] [PubMed] [Google Scholar]
- Boitard, S., Paris C., Sevane N., Servin B., Bazi-Kabbaj K., and Dunner S.. . 2021. Gene banks as reservoirs to detect recent selection: the example of the Asturiana de los Valles Bovine Breed. Front. Genet. 12:575405. doi: 10.3389/fgene.2021.575405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt, H., Müllenhoff A., Lambertz C., Erhardt G., and Gauly M.. . 2010. Estimation of genetic and crossbreeding parameters for preweaning traits in German Angus and Simmental beef cattle and reciprocal crosses. J. Anim. Sci. 88:80–86. doi: 10.2527/jas.2008-1742. [DOI] [PubMed] [Google Scholar]
- Bruscadin, J. J., de Souza M. M., de Oliveira K. S., Rocha M. I. P., Afonso J., Cardoso T. F., Zerlotini A., Coutinho L. L., Niciura S. C. M., and de Almeida Regitano L. C.. . 2021. Muscle allele-specific expression QTLs may affect meat quality traits in Bos indicus. Sci. Rep. 11:7321. doi: 10.1038/s41598-021-86782-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caetano, S. L., Savegnago R. P., Boligon A. A., Ramos S. B., Chud T. C. S., Lôbo R. B., and Munari D. P.. . 2013. Estimates of genetic parameters for carcass, growth and reproductive traits in Nellore cattle. Livest. Sci. 155:1–7. doi: 10.1016/j.livsci.2013.04.004. [DOI] [Google Scholar]
- Cesarani, A., Gaspa G., Correddu F., Cellesi M., Dimauro C., and Macciotta N. P. P.. . 2019. Genomic selection of milk fatty acid composition in Sarda dairy sheep: effect of different phenotypes and relationship matrices on heritability and breeding value accuracy. J. Dairy Sci. 102:3189–3203. doi: 10.3168/jds.2018-15333. [DOI] [PubMed] [Google Scholar]
- Chud, T. C. S., Caetano S. L., Buzanskas M. E., Grossi D. A., Guidolin D. G. F., Nascimento G. B., Rosa J. O., Lôbo R. B., and Munari D. P.. . 2014. Genetic analysis for gestation length, birth weight, weaning weight, and accumulated productivity in Nellore beef cattle. Livest. Sci. 170:16–21. doi: 10.1016/j.livsci.2014.09.024. [DOI] [Google Scholar]
- Crews, D. H., Jr, Pollak E. J., Weaber R. L., Quaas R. L., and Lipsey R. J.. . 2003. Genetic parameters for carcass traits and their live animal indicators in Simmental cattle. J. Anim. Sci. 81:1427–1433. doi: 10.2527/2003.8161427x. [DOI] [PubMed] [Google Scholar]
- Daetwyler, H. D., Calus M. P., Pong-Wong R., de Los Campos G., and Hickey J. M.. . 2013. Genomic prediction in animals and plants: simulation of data, validation, reporting, and benchmarking. Genetics. 193:347–365. doi: 10.1534/genetics.112.147983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daetwyler, H. D., Swan A. A., van der Werf J. H., and Hayes B. J.. . 2012. Accuracy of pedigree and genomic predictions of carcass and novel meat quality traits in multi-breed sheep data assessed by cross-validation. Genet. Sel. Evol. 44:33. doi: 10.1186/1297-9686-44-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daetwyler, H. D., Villanueva B., and Woolliams J. A.. . 2008. Accuracy of predicting the genetic risk of disease using a genome-wide approach. PLoS One. 3:e3395. doi: 10.1371/journal.pone.0003395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devitt, C. J., and Wilton J. W.. . 2001. Genetic correlation estimates between ultrasound measurements on yearling bulls and carcass measurements on finished steers. J. Anim. Sci. 79:2790–2797. doi: 10.2527/2001.79112790x. [DOI] [PubMed] [Google Scholar]
- Durinck, S., Spellman P. T., Birney E., and Huber W.. . 2009. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 4:1184–1191. doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eler, J. P., Van Vleck L. D., Ferraz J. B., and Lôbo R. B.. . 1995. Estimation of variances due to direct and maternal effects for growth traits of Nelore cattle. J. Anim. Sci. 73:3253–3258. doi: 10.2527/1995.73113253x. [DOI] [PubMed] [Google Scholar]
- Freebern, E., Santos D. J. A., Fang L., Jiang J., Parker Gaddis K. L., Liu G. E., VanRaden P. M., Maltecca C., Cole J. B., and Ma L.. . 2020. GWAS and fine-mapping of livability and six disease traits in Holstein cattle. Bmc Genomics. 21:41. doi: 10.1186/s12864-020-6461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrick, D. J., Taylor J. F., and Fernando R. L.. . 2009. Deregressing estimated breeding values and weighting information for genomic regression analyses. Genet. Sel. Evol. 41:55. doi: 10.1186/1297-9686-41-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordo, D. G., Espigolan R., Tonussi R. L., Júnior G. A., Bresolin T., Magalhães A. F., Feitosa F. L., Baldi F., Carvalheiro R., Tonhati H., . et al. 2016. Genetic parameter estimates for carcass traits and visual scores including or not genomic information. J. Anim. Sci. 94:1821–1826. doi: 10.2527/jas.2015-0134. [DOI] [PubMed] [Google Scholar]
- Guidolin, D. G. F., Buzanskas M. E., Ramos S. B., Venturini G. C., Lôbo R. B., Paz C. C. P., Munari D. P., and Oliveira J. A.. . 2012. Genotype-environment interaction for post-weaning traits in Nellore beef cattle. Anim. Prod. Sci. 52:975–980. doi: 10.1071/AN11037. [DOI] [Google Scholar]
- Guo, G., Zhao F., Wang Y., Zhang Y., Du L., and Su G.. . 2014. Comparison of single-trait and multiple-trait genomic prediction models. Bmc Genet. 15:30. doi: 10.1186/1471-2156-15-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez, J. P., Goyache F., Fernández I., Alvarez I., and Royo L. J.. . 2007. Genetic relationships among calving ease, calving interval, birth weight, and weaning weight in the Asturiana de los Valles beef cattle breed. J. Anim. Sci. 85:69–75. doi: 10.2527/jas.2006-168. [DOI] [PubMed] [Google Scholar]
- Halli, K., Brügemann K., Bohlouli M., and König S.. . 2020. Time-lagged and acute impact of heat stress on production and fertility traits in the local dual-purpose cattle breed “Rotes Höhenvieh” under pasture-based conditions. Transl. Anim. Sci. 4:txaa148. doi: 10.1093/tas/txaa148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder, B., Bennewitz J., Reinsch N., Mayer M., and Kalm E.. . 2005. Effect of missing sire information on genetic evaluation. Arch. Anim. Breed. 48:219–232. doi: 10.5194/aab-48-219-2005. [DOI] [Google Scholar]
- Higgins, M. G., Fitzsimons C., McClure M. C., McKenna C., Conroy S., Kenny D. A., McGee M., Waters S. M., and Morris D. W.. . 2018. GWAS and eQTL analysis identifies a SNP associated with both residual feed intake and GFRA2 expression in beef cattle. Sci. Rep. 8:14301. doi: 10.1038/s41598-018-32374-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoque, M. A., Kadowaki H., Shibata T., and Suzuki K.. . 2008. Maternal and direct genetic parameters for production traits and maternal correlations among production and feed efficiency traits in Duroc pigs. Asian-Aust. J. Anim. Sci. 21:961–966. doi: 10.5713/ajas.2008.70641. [DOI] [Google Scholar]
- Ismael, A., Løvendahl P., Fogh A., Lund M. S., and Su G.. . 2017. Improving genetic evaluation using a multitrait single-step genomic model for ability to resume cycling after calving, measured by activity tags in Holstein cows. J. Dairy Sci. 100:8188–8196. doi: 10.3168/jds.2017-13122. [DOI] [PubMed] [Google Scholar]
- Jeyaruban, M. G., Johnston D. J., and Graser H.-U.. . 2009. Estimation of genotype x environment interactions for growth, fatness and reproductive traits in Australian Angus cattle. Anim. Prod. Sci. 49:1–8. doi: 10.1071/EA08098. [DOI] [Google Scholar]
- Jia, Y., and Jannink J. L.. . 2012. Multiple-trait genomic selection methods increase genetic value prediction accuracy. Genetics. 192:1513–1522. doi: 10.1534/genetics.112.144246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, Z. B., Chewning J. J., and R. A.Nugent, 3rd. 2002. Maternal effects on traits measured during postweaning performance test of swine from four breeds. J. Anim. Sci. 80:1470–1477. doi: 10.2527/2002.8061470x. [DOI] [PubMed] [Google Scholar]
- Kempster, A. J., Cuthbertson A., and Harrington G.. . 1976. Fat distribution in steer carcasses of different breeds and crosses. 1. Distribution between depots. Anim. Sci. 23:25–34. doi: 10.1017/S0003356100031044. [DOI] [Google Scholar]
- Kempster, A. J., and Evans D. G.. . 1979. The effects of genotype, sex and feeding regimen on pig carcass development. 2. Tissue weight distribution and fat partition between depots. J. Agric. Sci. 93:349–358. doi: 10.1017/S0021859600038028. [DOI] [Google Scholar]
- Kim, Y., Ryu J., Woo J., Kim J. B., Kim C. Y., and Lee C.. . 2011. Genome-wide association study reveals five nucleotide sequence variants for carcass traits in beef cattle. Anim. Genet. 42:361–365. doi: 10.1111/j.1365-2052.2010.02156.x. [DOI] [PubMed] [Google Scholar]
- Kipp, C., Brügemann K., Yin T., Halli K., and König S.. . 2021. Genotype by heat stress interactions for production and functional traits in dairy cows from an across-generation perspective. J. Dairy Sci. 104:10029–10039. doi: 10.3168/jds.2021-20241. [DOI] [PubMed] [Google Scholar]
- Lee, C. 2001. On the negative estimates of direct and maternal genetic correlation—a review. Asian-Australas. J. Anim. Sci. 15:1222–1226. [Google Scholar]
- Lo, L. L., McLaren D. G., McKeith F. K., Fernando R. L., and Novakofski J.. . 1992. Genetic analyses of growth, real-time ultrasound, carcass, and pork quality traits in Duroc and Landrace pigs. 2. Heritabilities and correlations. J. Anim. Sci. 70:2387–2396. doi: 10.2527/1992.7082387x. [DOI] [PubMed] [Google Scholar]
- Lourenco, D. A., Tsuruta S., Fragomeni B. O., Masuda Y., Aguilar I., Legarra A., Bertrand J. K., Amen T. S., Wang L., Moser D. W., . et al. 2015. Genetic evaluation using single-step genomic best linear unbiased predictor in American Angus. J. Anim. Sci. 93:2653–2662. doi: 10.2527/jas.2014-8836. [DOI] [PubMed] [Google Scholar]
- Lu, D., Sargolzaei M., Kelly M., Vander Voort G., Wang Z., Mandell I., Moore S., Plastow G., and Miller S. P.. . 2013. Genome-wide association analyses for carcass quality in crossbred beef cattle. Bmc Genet. 14:80. doi: 10.1186/1471-2156-14-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackinnon, M. J., Meyer K., and Hetzel D. J. S.. . 1991. Genetic variation and covariation for growth, parasite resistance and heat tolerance in tropical cattle. Livest. Prod. Sci. 27:105–122. doi: 10.1016/0301-6226(91)90090-D. [DOI] [Google Scholar]
- MacNeil, M. D., Nkrumah J. D., Woodward B. W., and Northcutt S. L.. . 2010. Genetic evaluation of Angus cattle for carcass marbling using ultrasound and genomic indicators. J. Anim. Sci. 88:517–522. doi: 10.2527/jas.2009-2022. [DOI] [PubMed] [Google Scholar]
- Mandal, A., Neser F. W. C., Rout P. K., Roy R., and Notter D. R.. . 2006. Estimation of direct and maternal (co) variance components for pre-weaning growth traits in Muzaffarnagari sheep. Livest. Sci. 99:79–89. doi: 10.1016/j.livprodsci.2005.06.001. [DOI] [Google Scholar]
- Maniatis, G., Demiris N., Kranis A., Banos G., and Kominakis A.. . 2013. Model comparison and estimation of genetic parameters for body weight in commercial broilers. Can. J. Anim. Sci. 93:67–77. doi: 10.4141/cjas2012-070. [DOI] [Google Scholar]
- Marshall, D. M. 1994. Breed differences and genetic parameters for body composition traits in beef cattle. J. Anim. Sci. 72:2745–2755. doi: 10.2527/1994.72102745x. [DOI] [PubMed] [Google Scholar]
- Mateescu, R. G., Garrick D. J., Garmyn A. J., VanOverbeke D. L., Mafi G. G., and Reecy J. M.. . 2015. Genetic parameters for sensory traits in longissimus muscle and their associations with tenderness, marbling score, and intramuscular fat in Angus cattle. J. Anim. Sci. 93:21–27. doi: 10.2527/jas.2014-8405. [DOI] [PubMed] [Google Scholar]
- Meyer, K. 1992. Variance components due to direct and maternal effects for growth traits of Australian beef cattle. Livest. Prod. Sci. 31:179–204. doi: 10.1016/0301-6226(92)90017-X. [DOI] [Google Scholar]
- Meyer, K. 1994. Estimates of direct and maternal correlations among growth traits in Australian beef cattle. Livest. Prod. Sci. 38:91–105. doi: 10.1016/0301-6226(94)90053-1. [DOI] [Google Scholar]
- Meyer, K. 1995. Estimates of genetic parameters and breeding values for New Zealand and Australian Angus cattle. Aust. J. Agric. Res. 46:1219–1229. doi: 10.1071/AR9951219. [DOI] [Google Scholar]
- Misztal, I., Tsuruta S., Lourenco D. A. L., Masuda Y., Aguilar I., Legarra A., and Vitezica Z.. . 2018. Manual for BLUPF90 Family Programs.University of Georgia,Athens, GA: —[accessed April 21, 2021]. http://nce.ads.uga.edu/wiki/doku.php?id=documentation [Google Scholar]
- Misztal, I., Tsuruta S., Strabel T., Auvray B., Druet T., and Lee D. H.. . 2002. BLUPF90 and related programs (BGF90). Proceedings of the 7th World Congress on Genetics Applied to Livestock Production. 33; p. 743–744. [Google Scholar]
- Müllenhoff, A. 2008. Schätzung genetisch-statistischer Parameter bei Fleischrindern der Rassen Deutsche Angus und Deutsches Fleckvieh sowie deren Einfachkreizungen [PhD Diss]. Giessen, Germany: Justus-Liebig-Univ. [Google Scholar]
- Naderi, S., Bohlouli M., Yin T., and König S.. . 2018. Genomic breeding values, SNP effects and gene identification for disease traits in cow training sets. Anim. Genet. 49:178–192. doi: 10.1111/age.12661. [DOI] [PubMed] [Google Scholar]
- Naserkheil, M., Bahrami A., Lee D., and Mehrban H.. . 2020. Integrating single-step GWAS and bipartite networks reconstruction provides novel insights into yearling weight and carcass traits in Hanwoo beef cattle. Animals. 10:1836. doi: 10.3390/ani10101836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nephawe, K. A., Cundiff L. V., Dikeman M. E., Crouse J. D., and Van Vleck L. D.. . 2004. Genetic relationships between sex-specific traits in beef cattle: mature weight, weight adjusted for body condition score, height and body condition score of cows, and carcass traits of their steer relatives. J. Anim. Sci. 82:647–653. doi: 10.2527/2004.823647x. [DOI] [PubMed] [Google Scholar]
- Nwogwugwu, C. P., Kim Y., Chung Y. J., Jang S. B., Roh S. H., Kim S., Lee J. H., Choi T. J., and Lee S. H.. . 2019. Effect of errors in pedigree on the accuracy of estimated breeding value for carcass traits in Korean Hanwoo cattle. Asian-Australas. J. Anim. Sci. 33:1057–1067. doi: 10.5713/ajas.19.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama, Y., Katafuchi M., Almehmadi A., Venkitapathi S., Jaha H., Ehrenman J., Morcos J., Aljamaan R., and Mochida Y.. . 2012. Modulation of matrix mineralization by Vwc2-like protein and its novel splicing isoforms. Biochem. Biophys. Res. Commun. 418:12–16. doi: 10.1016/j.bbrc.2011.12.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pariacote, F., Van Vleck L. D., and Hunsley R. E.. . 1998. Genetic and phenotypic parameters for carcass traits of American Shorthorn beef cattle. J. Anim. Sci. 76:2584–2588. doi: 10.2527/1998.76102584x. [DOI] [PubMed] [Google Scholar]
- Pimentel, E. C., and König S.. . 2012. Genomic selection for the improvement of meat quality in beef. J. Anim. Sci. 90:3418–3426. doi: 10.2527/jas.2011-5005. [DOI] [PubMed] [Google Scholar]
- Purcell, S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A., Bender D., Maller J., Sklar P., de Bakker P. I., Daly M. J., . et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regatieri, I. C., Boligon A. A., Baldi F., and Albuquerque L. G.. . 2012. Genetic correlations between mature cow weight and productive and reproductive traits in Nellore cattle. Genet. Mol. Res. 11:2979–2986. doi:10.4238(2012.May.10.4. [DOI] [PubMed] [Google Scholar]
- Robinson, D. L. 1996. Estimation and interpretation of direct and maternal genetic parameters for weights of Australian Angus cattle. Livest. Prod. Sci. 45:1–11. doi: 10.1016/0301-6226(95)00083-6. [DOI] [Google Scholar]
- Rolf, M. M., McKay S. D., McClure M. C., Decker J. E., Taxis T. M., Chapple R. H., Vasco D. A., Gregg S. J., Kim J. W., Schnabel R. D., . et al. 2010. How the next generation of genetic technologies will impact beef cattle selection. Proceedings of the Beef Improvement Federations 42nd Annual Research Symposium and Annual Meeting, Columbia, MO. [Google Scholar]
- Saatci, M., Ap Dewi I., and Ulutas Z.. . 1999. Variance components due to direct and maternal effects and estimation of breeding values for 12-week weight of Welsh Mountain lambs. Anim. Sci. 69:345.– . doi: 10.1017/S1357729800050918. [DOI] [Google Scholar]
- Santana, M. L., Eler J. P., Cardoso F. F., Albuquerque L. G., and Ferraz J. B.. . 2013. Phenotypic plasticity of composite beef cattle performance using reaction norms model with unknown covariate. Animal 7:202–210. doi: 10.1017/S1751731112001711. [DOI] [PubMed] [Google Scholar]
- Seroba, M. M., Maiwashe A., Nephawe K. A., and Norris D.. . 2011. Genetic parameter estimates for live animal ultrasound measures of carcass traits in South African Angus cattle. S. Afr. J. Anim. Sci. 41:243–249. doi: 10.4314/sajas.v41i3.6. [DOI] [Google Scholar]
- Shabalina, T., Yin T., May K., and König S.. . 2021. Proofs for genotype by environment interactions considering pedigree and genomic data from organic and conventional cow reference populations. J. Dairy Sci. 104:4452–4466. doi: 10.3168/jds.2020-19384. [DOI] [PubMed] [Google Scholar]
- Shackelford, S. D., Koohmaraie M., Cundiff L. V., Gregory K. E., Rohrer G. A., and Savell J. W.. . 1994. Heritabilities and phenotypic and genetic correlations for bovine postrigor calpastatin activity, intramuscular fat content, Warner-Bratzler shear force, retail product yield, and growth rate. J. Anim. Sci. 72:857–863. doi: 10.2527/1994.724857x. [DOI] [PubMed] [Google Scholar]
- Sherman, E. L., Nkrumah J. D., and Moore S. S.. . 2010. Whole genome single nucleotide polymorphism associations with feed intake and feed efficiency in beef cattle. J. Anim. Sci. 88:16–22. doi: 10.2527/jas.2008-1759. [DOI] [PubMed] [Google Scholar]
- Shimada, Kitamura K. C., Nomura T., and Hayashi T.. . 1998. Maternal effects on carcass traits in Japanese Black steers (Wagyu). Anim. Sci. Technol. ISSN: 0918-2365. [Google Scholar]
- Solanes, F. X., Reixach J., Tor M., Tibau J., and Estany J.. . 2009. Genetic correlations and expected response for intramuscular fat content in a Duroc pig line. Livest. Sci. 123:63–69. doi: 10.1016/j.livsci.2008.10.006. [DOI] [Google Scholar]
- Song, H., Li L., Zhang Q., Zhang S., and Ding X.. . 2018. Accuracy and bias of genomic prediction with different de-regression methods. Animal. 12:1111–1117. doi: 10.1017/S175173111700307X. [DOI] [PubMed] [Google Scholar]
- Spiegelhalter, D. J., Best N. G., Carlin B. P., and Van Der Linde A.. . 2002. Bayesian measures of model complexity and fit. J. R. Stat. Soc. Series B. 64:583–639. doi: 10.1111/1467-9868.00353 [DOI] [Google Scholar]
- Srikanth, K., Lee S.-H., Chung K.-Y., Park J.-E., Jang G.-W., Park M.-R., Kim N. Y., Kim T.-H., Chai H.-H., Park W. C., . et al. 2020. A gene-set enrichment and protein-protein interaction network-based GWAS with regulatory SNPs identifies candidate genes and pathways associated with carcass traits in Hanwoo cattle. Genes. 11. doi: 10.3390/genes11030316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava, S., Lopez B. I., Heras-Saldana S. D. L., Park J.-E., Shin D.-H., Chai H.-H., Park W., Lee S.-H., and Lim D.. . 2019. Estimation of genetic parameters by single-trait and multi-trait models for carcass traits in Hanwoo cattle. Animals. 9:1061. doi: 10.3390/ani9121061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, K., Kadowaki H., Shibata T., Uchida H., and Nishida A.. . 2005. Selection for daily gain, loin-eye area, backfat thickness and intramuscular fat based on desired gains over seven generations of Duroc pigs. Livest. Prod. Sci. 97:193–202. doi: 10.1016/j.livprodsci.2005.04.007. [DOI] [Google Scholar]
- Swalve, H. H. 1993. Estimation of direct and maternal (co)variance components for growth traits in Australian Simmental beef cattle. J. Anim. Breed. Genet. 110:241–252. doi: 10.1111/j.1439-0388.1993.tb00736.x. [DOI] [PubMed] [Google Scholar]
- The Wellcome Trust Case Control Consortium. 2007. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Vázquez, J. A., and Spangler M. L.. . 2016. Genetic parameters for docility, weaning weight, yearling weight, and intramuscular fat percentage in Hereford cattle. J. Anim. Sci. 94:21–27. doi: 10.2527/jas.2015-9566. [DOI] [PubMed] [Google Scholar]
- Tosh, J. J., Kemp R. A., and Ward D. R.. . 1999. Estimates of direct and maternal genetic parameters for weight traits and backfat thickness in a multibreed population of beef cattle. Can. J. Anim. Sci. 79:433–439. doi: 10.4141/A99-014. [DOI] [Google Scholar]
- Utrera, A. R., and Van Vleck L. D.. . 2004. Heritability estimates for carcass traits of cattle: a review. Genet. Mol. Res. 3:380–394. PMID: 15614729. [PubMed] [Google Scholar]
- Van Eenennaam, A. L., Li J., Thallman R. M., Quaas R. L., Dikeman M. E., Gill C. A., Franke D. E., and Thomas M. G.. . 2007. Validation of commercial DNA tests for quantitative beef quality traits. J. Anim. Sci. 85:891–900. doi: 10.2527/jas.2006-512. [DOI] [PubMed] [Google Scholar]
- VanRaden, P. M. 2008. Efficient methods to compute genomic predictions. J. Dairy Sci. 91:4414–4423. doi: 10.3168/jds.2007-0980. [DOI] [PubMed] [Google Scholar]
- Waldron, D. F., Morris C. A., Baker R. L., and Johnson D. L.. . 1993. Maternal effects for growth traits in beef cattle. Livest. Prod. Sci. 34:57–70. doi: 10.1016/0301-6226(93)90035-G. [DOI] [Google Scholar]
- Wang, H., Misztal I., Aguilar I., Legarra A., and Muir W. M.. . 2012. Genome-wide association mapping including phenotypes from relatives without genotypes. Genet. Res. 94:73–83. doi: 10.1017/S0016672312000274. [DOI] [PubMed] [Google Scholar]
- Wang, K., Liu D., Hernandez-Sanchez J., Chen J., Liu C., Wu Z., Fang M., and Li N.. . 2015. Genome wide association analysis reveals new production trait genes in a male duroc population. PLoS One 10:e0139207. doi: 10.1371/journal.pone.0139207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Zhang F., Mukiibi R., Chen L., Vinsky M., Plastow G., Basarab J., Stothard P., and Li C.. . 2020. Genetic architecture of quantitative traits in beef cattle revealed by genome wide association studies of imputed whole genome sequence variants. II. Carcass merit traits. Bmc Genomics 21:38. doi: 10.1186/s12864-019-6273-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, C., Luo H., Zhao B., Tian K., Huang X., Wang Y., Fu X., Tian Y., Di J., Xu X., . et al. 2020. The effect of integrating genomic information into genetic evaluations of Chinese Merino sheep. Animals. 10:569. doi: 10.3390/ani10040569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham, H. 2009. ggplot2: elegant graphics for data analysis/ by Hadley Wickham. Use R!. Springer,New York, London. [Google Scholar]
- Williams, J. L., Bertrand J. K., Misztal I., and Łukaszewicz M.. . 2012. Genotype by environment interaction for growth due to altitude in United States Angus cattle. J. Anim. Sci. 90:2152–2158. doi: 10.2527/jas.2011-4365. [DOI] [PubMed] [Google Scholar]
- Wu, Y., and Smas C. M.. . 2008. Expression and regulation of transcript for the novel transmembrane protein Tmem182 in the adipocyte and muscle lineage. Bmc Res. Notes 1:85. doi: 10.1186/1756-0500-1-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J., Lee S. H., Goddard M. E., and Visscher P. M.. . 2011. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, T., and König S.. . 2019. Genome-wide associations and detection of potential candidate genes for direct genetic and maternal genetic effects influencing dairy cattle body weight at different ages. Genet. Sel. Evol. 51:4. doi: 10.1186/s12711-018-0444-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoo, M. J., Lobo R. B., Araujo F. R., Bezerra L. A., Sainz R. D., and Albuquerque L. G.. . 2010. Genetic associations between carcass traits measured by real-time ultrasound and scrotal circumference and growth traits in Nelore cattle. J. Anim. Sci. 88:52–58. doi: 10.2527/jas.2008-1028. [DOI] [PubMed] [Google Scholar]
- Zhang, F., Wang Y., Mukiibi R., Chen L., Vinsky M., Plastow G., Basarab J., Stothard P., and Li C.. . 2020. Genetic architecture of quantitative traits in beef cattle revealed by genome wide association studies of imputed whole genome sequence variants. I. Feed efficiency and component traits. Bmc Genomics 21:36. doi: 10.1186/s12864-019-6362-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuin, R. G., Buzanskas M. E., Caetano S. L., Venturini G. C., Guidolin D. G., Grossi D. A., Chud T. C., Paz C. C., Lôbo R. B., and Munari D. P.. . 2012. Genetic analysis on growth and carcass traits in Nelore cattle. Meat Sci. 91:352–357. doi: 10.1016/j.meatsci.2012.02.018. [DOI] [PubMed] [Google Scholar]