Abstract
As an important component of the COVID-19 mRNA vaccines, liposomes play a key role in the efficient protection and delivery of mRNA to cells. Herein, due to the controllable release amplification strategy of liposomes, a reliable and robust single-particle collision electrochemical (SPCE) biosensor was constructed for H9N2 avian influenza virus (H9N2 AIV) detection by combining liposome encapsulation-release strategy with immunomagnetic separation. The liposomes modified with biotin and loaded with platinum nanoparticles (Pt NPs) were used as signal probes for the first time. Biotin facilitated the coupling of biomolecules (DNA or antibodies) through the specific reaction of biotin-streptavidin. Each liposome can encapsulate multiple Pt NPs, which were ruptured under the presence of 1 × PBST (phosphate buffer saline with 0.05% Tween-20) within 2 min, and the encapsulated Pt NPs were released for SPCE experiment. The combination of immunomagnetic separation not only improved the anti-interference capabilities but also avoided the agglomeration of Pt NPs, enabling the SPCE biosensor to realize ultrasensitive detection of 18.1 fg/mL H9N2 AIV. Furthermore, the reliable SPCE biosensor was successfully applied in specific detection of H9N2 AIV in complex samples (chicken serum, chicken liver and chicken lung), which promoted the universality of SPCE biosensor and its application prospect in early diagnosis of diseases.
Keywords: Single-particle collision electrochemical biosensor, Ultramicroelectrode, Liposome, Magnetic beads, H9N2 AIV
1. Introduction
Single-particle collision electrochemistry (SPCE) is a method developed in recent years with the advantages of simple operation, fast response, high sensitivity and can distinguish the response signal of a single-particle from the whole signal (Peng et al., 2017; Zhou et al., 2021). Based on random Brownian motion, single nanoparticles collide with a microelectrode in a stochastically manner and each collision generates individual step or peak-shaped transient currents (Bard et al., 2010; Patrice et al., 2018; Xiang et al., 2018). The electrocatalytic amplification (ECA) method developed by Bard's group in 2007 was considered as a powerful analytical method in which nanoparticles can catalyze a specific electrochemical reaction collide with an inert microelectrode, opening the possibility of exploring the relationship between structure, agglomeration, and activity of catalytic particles unambiguously (Xiao and Bard, 2007). Long et al. applied ECA to accurately characterize the intrinsic catalytic activity of single Au NPs as glucose-like oxidases and single Ag–Au nanohybrids as peroxidase-like enzymes (Hafez et al., 2019). Compton et al. revealed the electrocatalytic oxidation mechanism and kinetics of hydrazine (N2H4) by using graphene oxide platelets randomly decorated with palladium nanoparticles (Pd/GO) as collision objects (Miao et al., 2021). These researches showed that SPCE had more application advantages than traditional analytical methods in characterizing the intrinsic properties of single nanoparticles and collision kinetics.
How to deepen the theoretical researches of SPCE into practical applications is also a problem that to be solved. Bard et al. used the direct collision of biological macromolecules with microelectrodes to hinder the electrochemical oxidation of [Fe(CN)6]4−, and realized the detection of several biomolecules through the quantitative relationship between the collision frequency and target concentrations (Dick et al., 2015). However, the electrochemical process is complicated and various factors such as the dielectric environment and ionic strength of the electrolyte solution may affect the stability and reproducibility of electrochemical signals. Moreover, non-negligible heterogeneity of single nanoparticles may also make the current fluctuation more obvious (Jiang et al., 2021). Zhang et al. had reported that the current difference between single enzyme molecules at the same concentration was as high as 10 times when the enzyme concentration was diluted to a single molecule level(Wu et al., 2016). The commonly used quantitative method based on the standard curve between target concentrations and signal intensity was no longer applicable. Therefore, based on SPCE, collision frequency instead of current intensity was used for analytical detection, which was expected to avoid the problem of electrochemical signal fluctuations, demonstrating the application advantages of SPCE in analytical detection (Castaneda et al., 2017; Defnet and Zhang, 2021; Wang et al., 2021a).
The application of SPCE in biochemical analysis has attracted wide attention credited to its ultrasensitivity up to single-particle level and reliable quantitative model. Which can be mainly divided into two categories: 1) The direct collision of biological macromolecules or insulating particles with a microelectrode, catalyzes or hinders oxidation-reduction active molecules (Dick et al., 2015, 2016; Zhang et al., 2021). This method is simple, fast, and label-free, but lacks selectivity and specificity, in addition to the interference of complex system has non-negligible influence. Due to its good magnetic responsiveness and strong maneuverability (Fabiani et al., 2022; Sui et al., 2020; Wang et al., 2021c), the introduction of magnetic beads (MBs) is expected to achieve efficient target capture and separation in complex systems without sample pre-processing, effectively simplifying operations, saving time, and eliminating interference. 2) Indirect detection is achieved by using marking and recognition components. Collision particles are introduced for labeling, as signal markers, the target is used to trigger the release of particles, and the released particles collide with the microelectrode to perform SPCE. Crooks et al. used Pt NPs modified with capture probes to achieve specific detection of microRNA with a detection limit of 100 pM (Castaneda et al., 2017). Bai et al. applied the similar method and combined duplex-specific nuclease-mediated cycle amplification to achieve more sensitive detection of miRNA-21 (Bai et al., 2020). Signal amplification strategies such as hybridization chain reaction, chain replacement, enzyme-catalyzed reaction have been very mature for DNA/RNA detection (Castaneda et al., 2017; Wang et al., 2021a, Wang et al., 2021b). Therefore, it is interesting to combine these signal amplification methods with SPCE to increase collision frequency and detection sensitivity. However, unlike nucleic acid detection, it lacks suitable amplification strategy for other biological particles detection, such as proteins, virus, cells, etc.
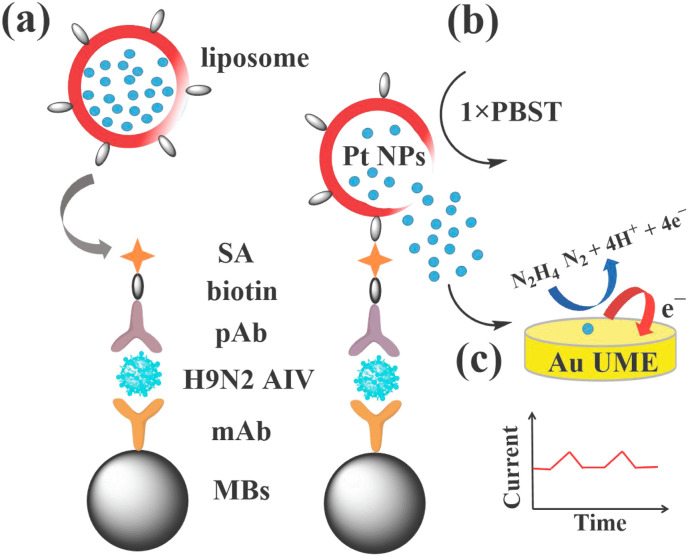
Infectious diseases have always been a serious threat to human health and property safety (Brotzakis et al., 2021), and rapid and sensitive detection methods can realize early diagnosis and treatment of diseases and control the infection and spread of pathogens. Herein, liposome encapsulation-release amplification strategy was integrated with high-efficiency separation and capture capabilities of MBs to construct a novel SPCE biosensor for H9N2 AIV detection without enzyme amplification. The liposomes modified with biotin and loaded with Pt NPs were used as signal probes for the first time. As shown in Scheme 1 , multiple Pt NPs can be encapsulated in one liposome (Tenchov et al., 2021), hence one target can correspond to multiple markers (one-to-many). Unlike the previous one or more targets corresponding to one marker (one-to-one or many-to-one), the one-to-many detection mode greatly improved the detection sensitivity. The biotin modified on liposomes facilitated the coupling of antibodies based on the specific reaction of biotin-streptavidin. Utilizing the immunosandwich reaction, magnetic immunocomplexes can be formed in the presence of H9N2 AIV. The liposomes can be ruptured to release the encapsulated Pt NPs under the presence of 1 × PBST. The process was facile, mild and the process can be completed within 2 min, which provided a universal liposome encapsulation-release strategy by changing the encapsulated substances. After magnetic separation, the Pt NPs released in the supernatant were used to collide with a gold ultramicroelectrode (Au UME) to catalyze the oxidation of N2H4. The relationship between collision frequency (the number of transient currents per unit time) and the concentrations of H9N2 AIV was used to achieve quantitative detection by recording the current-time curves. The ability of MBs to specifically capture and separate the target can not only improve the selectivity and anti-interference capability of SPCE, but also prevent the agglomeration of particles leaded by complex matrix.
Scheme 1.
Illustration diagram of the SPCE biosensor: (a) Sandwich immunoreaction. (b) The rupture of liposomes and the release of Pt NPs. (c) SPCE experiment.
2. Materials and methods
2.1. Chemicals and reagents
L-α-phosphotidylcholine (PC), cholesterol, and phosphoethanolamine-conjugated biotin (PE-PEG2000-biotin) was purchased from Aladdin Industrial Corporation. Sulfo–NHS–LC-biotin was purchased from Pierce Biotechnology (Rockford, IL). Anti-influenza A H9N2 hemagglutinin rabbit polyclonal antibody (pAb) and mouse monoclonal antibody (mAb) were purchased from Yiqiaoshenzhou Technology Co. Ltd. (Beijing, China). H1N1 AIV, H7N9 AIV, H9N2 AIV, pseudorabies virus (PRV) and newcastle disease virus (NDV) were obtained from Wuhan Institute of Virology, Chinese Academy of Sciences. Magnetic beads (MBs) were obtained from Ademtech SA (Pessac, France). Dylight 488-labeled goat anti-mouse IgG was purchased from Abbkine. N-hydroxysuccinimide (NHS) was purchased from thermo. Bovine serum albumin (BSA) and N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC) were purchased from Sigma-Aldrich. Streptavidin (SA) was purchased from Promega Corporation. A dialysis bag for dialysis (G2, 3500 MWCO, Thermo Scientific Inc.). All other chemical reagents were purchased from Aladdin Industrial Corporation.
2.2. Apparatus
All electrochemical assays were performed on a CHI 660E electrochemical workstation and a CHI Faraday cage (CH Instruments, Inc. Shanghai, China). Fluorescence spectra were recorded by an RF-5301 PC fluorometry (Shimadzu, Japan). Transmission electron microscopy (TEM, FEI Tecnai G2 20 TWIN) was applied to obtain the shape, dispersity, and size of nanomaterials. UV-2550 (Shimadzu, Japan) and dynamic light scattering (DLS, Malvern) were applied to characterize the concentration and surface potential of nanoparticles.
2.3. Synthesis of Pt NPs-loaded and biotin-modified liposome (Pt-Liposome-Biotin)
Uniform Pt NPs with a diameter of 4.4 nm were synthesized based on citric acid reduction method. Details were reported in S1 in the supporting information (SI). Pt-Liposome-Biotin were synthesized by rotary evaporation method. First, 5 mg mixtures of L-α-phosphotidylcholine (PC), cholesterol, and phosphoethanolamine-conjugated biotin (PE-PEG2000-biotin) with a molar ratio of 70:10:20 was dissolved in 1 mL chloroform. The solution was evaporated within 2–3 min by using a rotary evaporator at 40 °C under reduced pressure, and a thin film of phospholipid/cholesterol (PC) was formed on the inside wall of the pear-shaped flask. Then 5 mL 1 μM Pt NPs solution was added to peel off the lipid film and Pt-Liposome-Biotin was formed. The solution continued to become turbidity, and a milky white suspension was obtained. Subsequently, the solution was then passed through a 400 and 100 nm polycarbonate filters (Avanti Inc, USA) respectively by a micro extruder to produce liposome with a uniform diameter of 100 nm. Finally, a dialysis bag for dialysis was used for at least 1.5 h. The obtained Pt-Liposome-Biotin solution was stored at 4 °C for later use.
2.4. Preparation processes of Pt NPs with different agglomeration degree
Pt NPs with different agglomeration degree were prepared as follows: (1) The synthesized 4.4 ± 0.5 nm Pt NPs were ultrasonically dispersed before SPCE test. (2) The Pt NPs were not ultrasonically dispersed before SPCE test. (3) The Pt NPs were placed at room temperature (25 °C) for one day, and they were used for SPCE test after vortex. (4) The Pt NPs were placed at −20 °C for 1 h, then they were taken out for SPCE test after melt and vortex.
2.5. Preparation of MBs-mAb
The carboxylated MBs reacted with the amino group of the antibody, and the antibody was modified on MBs as capture probes. First, 200 μL PBS (0.01 M, pH 6.1) solution containing 50 mM EDC and 50 mM NHS was added to 15 μL (10 mg/mL) MBs, shaking and reacting at 37 °C for 30 min. After magnetic separation, the MBs was washed three times with PBS (0.01 M, pH 7.2). Next, 0.01 mg/mL mAb was added and reacted at 37 °C for 4 h. After being washed 3 times again with PBS (0.01 M, pH 7.2), 1% BSA was added and reacted at 37 °C for 30 min to block the unbinding sites. Finally, MBs-mAb was dissolved in PBS (0.01 M, pH 7.2) and stored at 4 °C for later use.
2.6. Preparation of biotinylated antibody (B-pAb)
Sulfo–NHS–LC-biotin was used to react with the amino groups of pAb to prepare biotinylated antibody (B-pAb). First, 0.5 mg sulfo–NHS–LC-biotin was dissolved in 60 μL PBS (0.01 M, pH 7.2). Then 40 μL (0.1 mg/mL) pAb was added and reacted at 37 °C for 2 h. A desalting column was used to remove excess reagents, and the absorbance of sample at 280 nm was utilized to quantify the concentration of B-pAb. Finally, 1% BSA and 0.05% sodium azide were added to B-pAb and stored at 4 °C for later use.
2.7. SPCE biosensor for ultrasensitive detection of H9N2 AIV
The ultrasensitive detection of H9N2 AIV was based on an immunosandwich reaction. First, 10 μL MBs-mAb solution was mixed with 100 μL PBS solution containing different concentrations of H9N2 AIV, and reacted at 37 °C at 200 rpm for 30 min to obtain the MBs-H9N2 complexes. After being washed three times with PBS, 90 μL PBS and 10 μL B-pAb (50 μg/mL) was added, reacted at 37 °C at 200 rpm for shaking 1 h. After being washed three times with PBS on a magnetic shelf again, 90 μL 1% BSA-PBS and 10 μL SA (1.25 μg/mL) were added to react at 37 °C for 30 min. Then 90 μL PBS and 10 μL Pt-Liposome-Biotin were added and reacted at 200 rpm at 37 °C for 30 min. Finally, 96 μL PBS and 4 μL 1 × PBST were added to rupture liposomes and release Pt NPs. After magnetic separation, 100 μL supernatant containing the released Pt NPs was added into 2.5 mL solution containing 10 mM PBS (pH 7.2), and 10 mM N2H4. For SPCE experiments, the appropriate voltage was held at −0.1 V vs Ag/AgCl (3 M KCl), and the home-made Au UME were used as working electrode. By recording the current-time curves, the relationship between collision frequency and H9N2 AIV concentrations was used for quantitative detection.
3. Results and discussion
3.1. Encapsulation of Pt NPs in liposomes for signal amplification
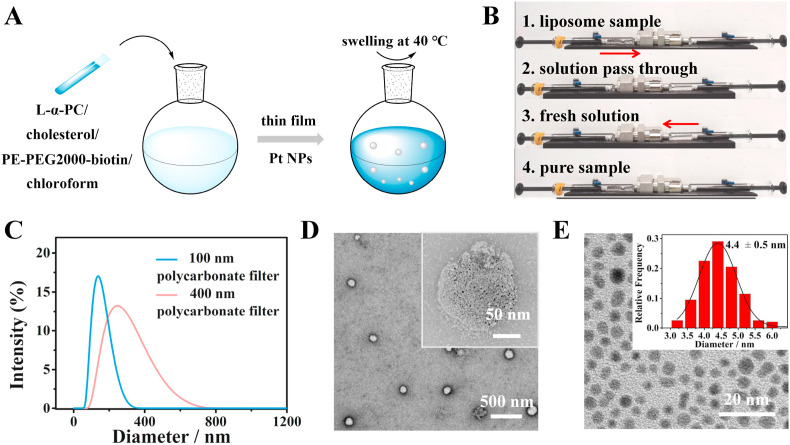
Pt NPs-loaded liposomes were prepared and first used as signal probes to improve detection sensitivity. The procedure for the preparation of Pt NPs-loaded liposomes can be divided into two main steps: (1) encapsulation of Pt NPs in liposomes by rotary evaporation method (Fig. 1 A); (2) liposome extruder purification (LEP). As shown in Fig. 1B, the liposome sample was loaded into the starting syringe, then the solution was passed through a 400 and 100 nm polycarbonate filters respectively and injected into another syringe by the micro-extruder. Due to the flexibility of liposome, large liposomes would be blocked at the entrance of the membrane pores. The fresh solution obtained was injected back into the starting syringe again to get the pure sample to complete a LEP cycle. Fig. 1C showed that uniform liposomes with a diameter of 100 nm can be obtained by LEP. Therefore, 100 nm liposomes were used because of its less steric hindrance and good flexibility. Fig. 1D revealed that the Pt NPs-loaded liposomes were spherical and the particle size was approximately 100 nm. As shown in Fig. 1E, the Pt NPs were uniform dispersed and the average particle size was 4.4 ± 0.5 nm. The concentration of Pt NPs was calculated to be about 48 μM by the ultraviolet–visible absorption method (the detailed calculation process was shown in S2 of SI). The inset TEM image in Fig. 1D showed that abundant 4.4 nm Pt NPs were successfully encapsulated inside liposomes. To improve encapsulation rate, excessive Pt NPs was added, and the encapsulation rate was calculated to be 32.63% (the detailed calculation process was shown in S3 of SI). Multiple Pt NPs can be encapsulated in one liposome, which was ruptured within 2 min in the presence of 1 × PBST. To characterize the amount of biotin on liposomes, streptavidin-modified quantum dots (SA-QDs) was used based on the specific reaction between streptavidin and biotin. The amount of biotin on each liposome was calculated to be 7.75 × 109, which enabled efficient labeling with rich binding sites (the detailed calculation process was shown in S4 of SI). To prove the feasibility and universality of the liposome encapsulation-release amplified method, a visual experiment of encapsulating cysteine molecules in liposomes was performed (Fig. S4). The use of liposomes not only increased the load of colliding nanoparticles, but also provided a universal encapsulation-release strategy.
Fig. 1.
(A) Schematic illustration for the preparation of Pt NPs-loaded liposomes. (B) Visual demonstration of the liposome extruder purification (LEP) procedure. (C) Dynamic light scattering data of liposomes after being passed through a 100 or 400 nm polycarbonate filter. (D) TEM image of Pt NPs-loaded liposomes (Inset: a higher magnification image). (E) TEM image of Pt NPs (Inset: size distribution).
All in all, Pt NPs-loaded liposomes were successfully synthesized by the rotary evaporation method, and uniform liposomes were obtained by the LEP method. In the presence of 1 × PBST, the synthesized Pt NPs-loaded liposomes were ruptured and the encapsulated Pt NPs were released for subsequent SPCE experiments. Realizing the one-to-one or many-to-one detection mode to one-to-many amplification mode, which successfully achieved signal amplification and greatly improved the detection sensitivity.
3.2. Electrochemical property of the home-made Au UME
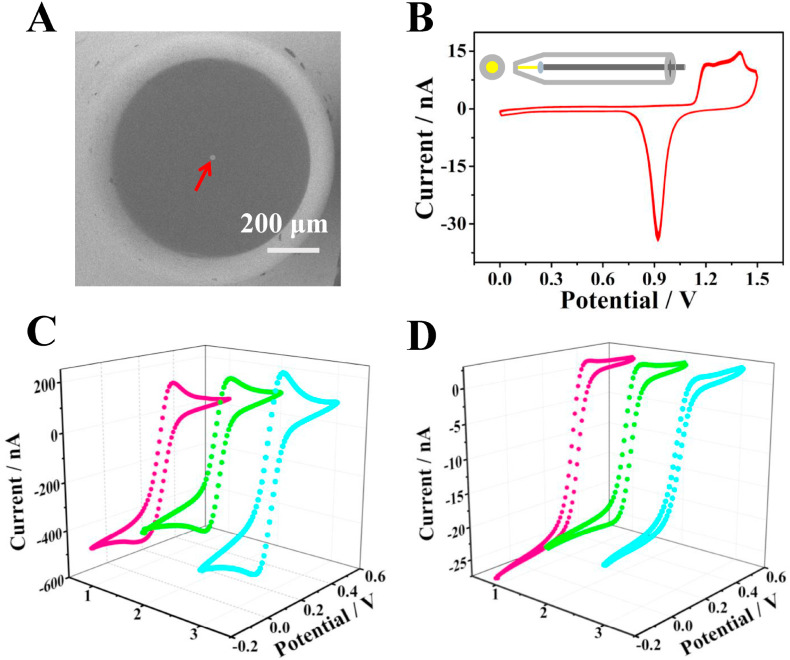
The preparation of UME with excellent electrochemical response is a key factor for the collision process of SPCE. Based on fire-sealing and grinding method, a disk-shaped Au UME was fabricated as the working electrode (Fig. 2 A), the insert of Fig. 2B showed a schematic drawing of the Au UME. Cyclic voltammogram (CV) of Au UME in 0.5 M H2SO4 (Fig. 2B) depicted that obvious adsorption oxidation peaks appeared between 1.2 and 1.5 V vs Ag/AgCl, and a corresponding reduction peak appeared at 0.9 V, indicating the home-made Au UME was successful prepared. Considering that the charge required for the reduction of adsorbed oxygen on Au electrode is 386 μC cm−2 (Woods and Bard, 1976), then the microscopic area of the prepared Au UME was estimated as 1270 μm2 according to the desorption peak area. The roughness factor (ρ) was calculated as follows:
| (1) |
where A m is the microscopic area of Au UME, and A g is the geometric area of Au UME. The calculated result was consistent with the reported roughness of the polished metal surface (ρ = 2–3) (Lee and Park, 2010). Before polishing, the CV of the microelectrode was not S-shaped, and the currents were significantly different from each other (Fig. 2C). After being polished, the Au UME demonstrated good electrochemical performance (Fig. 2D) (Ortiz-Ledon and Zoski, 2018). From the CV diagram in Fig. 2D, the radius of the homemade Au UME can be calculated to be 13.6 μm by the following diffusion-limited steady-state current equation:
| (2) |
where i ss is the steady-state current for the Au UME, F is Faraday's constant. D and C are the diffusion coefficient (D = 7.6 × 10−6 cm2/s) (Karfa et al., 2018) and bulk concentration of ferrocyanide (5 mM), and n is the number of electrons transferred per molecule (the detailed calculation process was shown in S6). However, it could be seen that there were slight differences in electrochemical responses among microelectrodes prepared in the same batch, which might cause current fluctuations. Especially for single entities, their heterogeneity may make the current fluctuation more obvious. Therefore, the use of collision frequency instead of current intensity for analytical detection in SPCE was expected to solve the problem and provide a reliable quantitative model. In general, the disk-shaped Au UME with a radius of 13.6 μm was successfully prepared and showed excellent electrochemical property, providing the possibility for SPCE experiments.
Fig. 2.
(A) Scanning electron microscopy image of the Au UME (As indicated by the red arrow). (B) Cyclic voltammogram of the Au UME in 0.5 M H2SO4. (Inset: schematic drawing of an Au UME). Cyclic voltammograms of three different Au UMEs before (C) and after (D) being polished in 5 mM K3Fe(CN)6/0.1 M KCl (scan rate: 10 mV/s).
3.3. Single-particle collision electrochemical: theory vs experiment
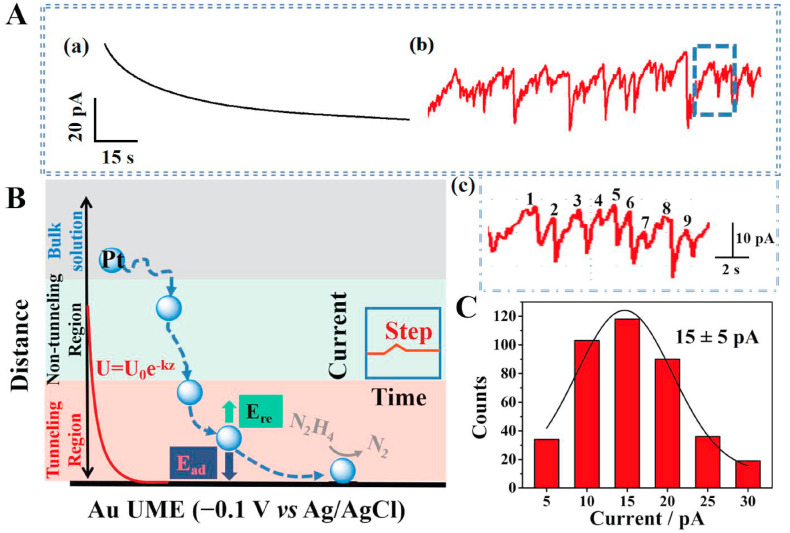
Electrocatalytic amplified SPCE was realized by the collision of Pt NPs on an Au UME to catalyze the oxidation of N2H4. In the experiment, PBS was used as a supporting electrolyte, whose concentration was found to have a great influence on the stability of Pt NPs (Zhang et al., 2019). By comparing the hydrodynamic diameter of Pt NPs in 100 mM and 10 mM PBS, 10 mM PBS concentration was selected since the Pt NPs was more uniformly dispersed (Fig. S6). In SPCE, the electrode potential was optimized to −0.1 V vs Ag/AgCl, so that only when Pt NPs were present and collided with an Au UME could catalyze the oxidation of N2H4. By comparing the difference of current signals under different voltages (Fig. S7A), it can be found that when the applied voltage becoming more and more negative, the peak signal intensity became smaller and even disappeared. However, as the applied voltage becoming positive, the background current increased accordingly. It can be seen from CV of Au UME in the presence and absence of 10 mM N2H4 solution that the limit current of N2H4 was small (Fig. S7B). Moreover, the home-made Au UME can be reused after polishing (Fig. S8), which greatly saved time and energy. Therefore, collision tests of single Pt NPs in a 2.5 mL solution containing 10 mM PBS (pH 7.2) and 10 mM N2H4 were performed. The movement behavior of particles on the electrode surface had a significant impact on the shape of collision signals. As shown in Fig. 3 B, as the size of NPs increased, electrostatic adsorption energy (E ad) and repulsion energy (E re) between Pt NPs and Au UME decreased and increased respectively (Ma et al., 2017). Since the Pt NPs of the citric acid ligand was negatively charged in the solution, there was an electrostatic attraction between the Pt NPs and the Au UME. After Pt NPs collided with the UME, the initial charge of the oxidation of N2H4 was obviously negatively shifted, indicating that Pt NPs were adhered to the surface of Au UME after collision (Fig. S9). Therefore, the transient currents observed was step−shaped. The current−time curves in Fig. 3A (a and b) showed that obvious step-shaped transient currents could be observed only when 4.4 nm Pt NPs were present. The average current intensity was 15 ± 5 pA by randomly counting 200 transient currents (Fig. 3C), confirming the feasibility of SPCE. It was worth noting that current fluctuation appeared in the current distribution of Fig. 3C, which might be explained by the heterogeneity between single particles and the difference between microelectrodes (Fig. 2D). Therefore, based on SPCE, collision frequency instead of current intensity was used for quantification, which was expected to avoid the problem of current fluctuation, demonstrating the application advantages of SPCE in analytical detection. The experimental collision frequency was calculated as follows:
| (3) |
Fig. 3.
(A) Current-time curves in 10 mM N2H4 solution with (a) or without (b) Pt NPs. (c) A larger version of Fig. (b). (B) Schematic diagram of the motion trajectories of Pt NPs on an Au UME. (C) Current distribution (n = 200) of individual Pt NPs collision events.
As shown in Fig. S10, the collision frequency increased with Pt NPs concentrations and a linear relationship appeared from 1 pM to 10 pM, indicating the possibility of SPCE in analytical application. Since the collision process is diffusion-controlled, the theoretical collision frequency can be expressed by equation (4):
| (4) |
where D NP (5 × 10−11 m2/s) and C NP are the diffusion coefficient and concentration of NPs, a is the radius of the UME, and N A is Avogadro's constant. Among them, D NP can be calculated according to the Stokes−Einstein equation (5):
| (5) |
where k B is Boltzmann's constant, T is temperature, η is the viscosity of the solvent at 298.15 K (0.89 × 10−3 kg m−1s−1), and r NP is the radius of the NP.
When C NP and radius of the UME were 10 pM and 12.5 μm respectively, the collision frequency value was calculated to be about 15.1 Hz by equations (4), (5), while the experimental collision frequency calculation result was 0.75 Hz by equation (3). The experimental collision frequency was lower than the theoretical values. The possible reasons might be that only a fraction of Pt NPs in the solution can move to the UME surface and conduct effective collision due to the randomness of the Brownian movement. Therefore, collision frequency is important for the detection sensitivity and great efforts are still needed to improve the collision frequency.
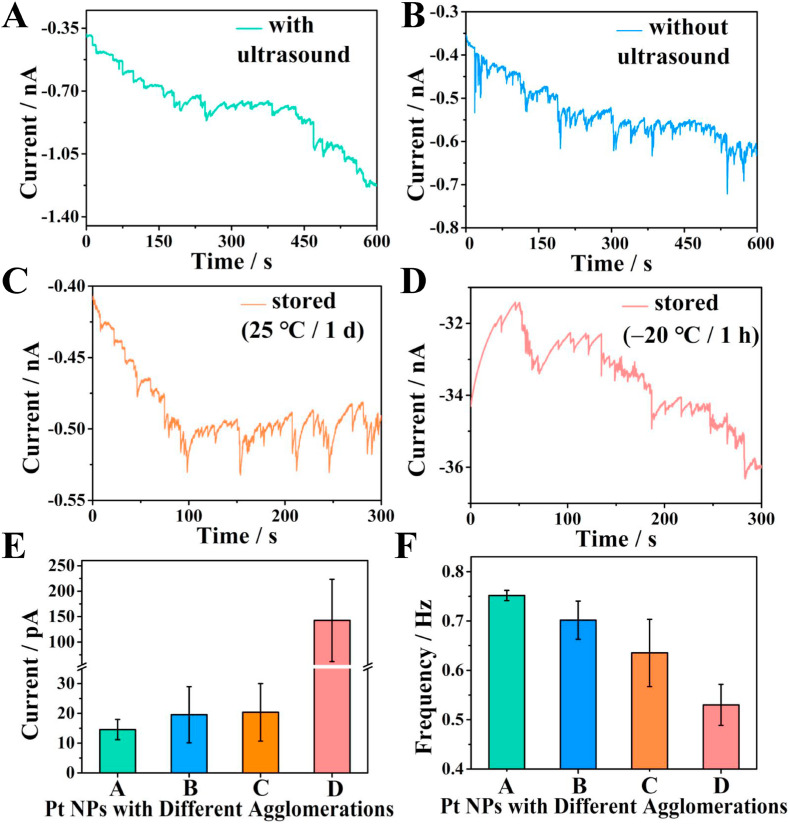
3.4. Investigation the collision of Pt NPs with different agglomeration degree
The stability of NPs is necessary to the success of SPCE experiment. Particle agglomeration can be divided into two types: reversible agglomeration and irreversible agglomeration (Ortiz-Ledón and Zoski, 2017; Sokolov et al., 2015). Reversible agglomeration refers to the weak physical interaction between particles. Through external disturbance, such as ultrasound, the agglomerated particles can be re-dispersed. When the particles are bound together through strong chemical bonds, they cannot be dispersed again. The agglomeration of NPs also has a great impact on its catalytic performance (Zhou et al., 2018; Wei et al., 2010). Therefore, it is very important to develop an efficient and convenient method for in-situ and real-time monitoring of the agglomeration status of NPs in solution. SPCE, as a promising technique with high sensitivity and resolution, has triggered researcher's interest in monitoring the agglomeration degree of NPs in solution in recent years.
In order to demonstrate the probability of SPCE in monitoring the agglomeration process of NPs, four control experiments (Pt NPs with ultrasonic dispersion, Pt NPs without ultrasonic dispersion, Pt NPs stored at room temperature for one day, and Pt NPs stored at −20 °C for 1 h) were carried out. From Fig. 4 A, it could be found that the collision current intensity of Pt NPs after ultrasonic dispersion was relatively uniform, indicating that the Pt NPs were well dispersed. On the contrary, the current intensity of the Pt NPs without ultrasonic dispersion (Fig. 4B), the Pt NPs stored at room temperature (25 °C) (Fig. 4C) or at −20 °C (Fig. 4D) were obviously uneven. By randomly counting 200 transient currents, a histogram of the average current intensity was obtained. It could be seen that different degrees of agglomeration have occurred in B, C, D (Fig. 4E). The result in Fig. 4F showed that only the Pt NPs with ultrasonic dispersion had the highest collision frequency, and the other three process methods would cause the agglomeration of Pt NPs and the collision frequency to decrease. In our work, the released Pt NPs from liposomes were tested immediately after ultrasonic dispersion.
Fig. 4.
Current-time curves of Pt NPs with different agglomeration degree in 10 mM N2H4 solution: Pt NPs with (A) or without (B) ultrasonic dispersion. (C) Store of Pt NPs at room temperature for one day. (D) Store of Pt NPs at −20 °C for 1 h. Collision frequency (E) and the average current intensity (F) of Pt NPs with different agglomeration degree.
In short, SPCE can be regarded as a useful tool for in-situ and real-time monitoring of the agglomeration degree of nanoparticles, which provides some guidance for optimizing experimental conditions to reduce or avoid agglomeration.
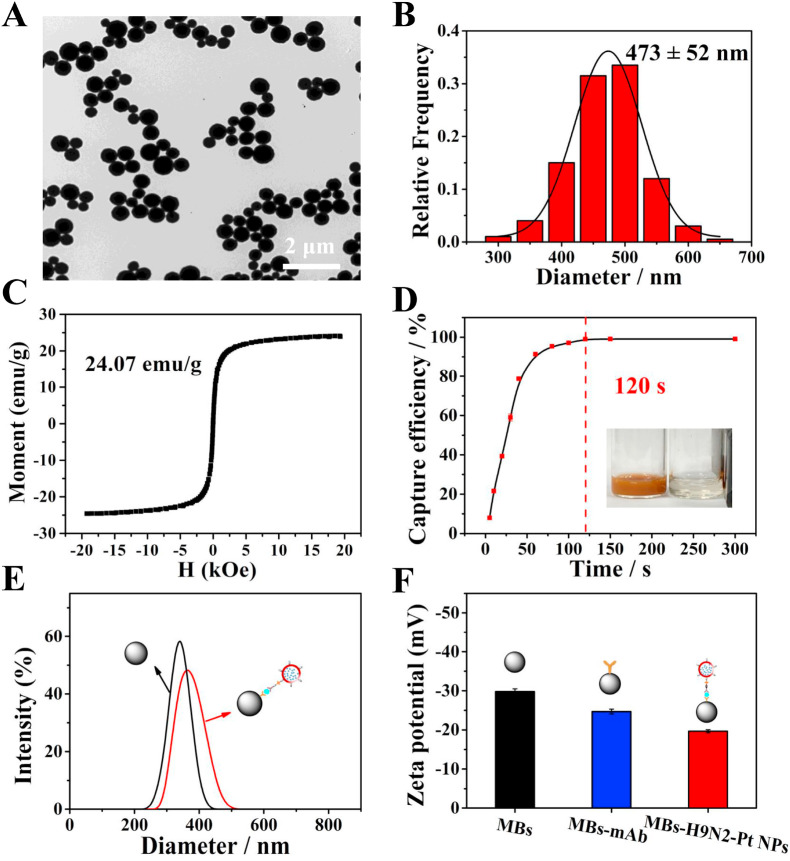
3.5. MBs-enabled effective separation of H9N2 AIV from complex samples
In order to achieve fast and efficient capture and separation of targets from complex systems, MBs was introduced because of its superparamagnetism, fast magnetic response and large specific surface area (Zhan et al., 2021). MBs had good dispersion and uniform size (Fig. 5 A) with an average particle size of about 473 nm (Fig. 5B). It could be seen from its hysteresis loop (Fig. 5C) that the saturation magnetization was as high as 24.07 emu/g, which made MBs exhibit strong magnetism in a magnetic field. After removing the magnetic field, MBs maintained good dispersion quickly. With the presence of a magnetic shelf, the capture efficiency of MBs within 120 s was as high as 100% (Fig. 5D), which can achieve rapid separation and efficient enrichment of the target. Due to its large surface area and abundant –COOH groups, multiple antibodies and Pt NPs-loaded liposomes could be modified to MBs surface. The hydrodynamic size of MBs changed from 310.1 nm to 339.2 nm (Fig. 5E), and the zeta (ζ) potential of MBs changed from −29.8 mV to −24.7 mV and further to −19.6 mV due to changes in surface functional groups (Fig. 5F), which indicated the successful fabrication of magnetic complexes. During reaction and washing process, it is difficult to avoid the loss of MBs. We have tried to reduce the loss of MBs by pretreating the tube and extending the adsorption time of MBs on the magnetic shelf. The remaining concentration of MBs was calculated to be 221.69 μg/mL after mAb modification, which was enough to realize efficient capture of H9N2 AIV (the detailed calculation process was shown in S12). And the number of mAb coupled to each MBs was calculated to be 452 (the detailed calculation process was in S13), which indicated that MBs can achieve efficient capture of virus from complex samples.
Fig. 5.
(A) TEM image of MBs. (B) Histogram of the size distribution of MBs (n = 200). (C) Hysteresis loop of MBs. (D) Capture efficiency of MBs on a magnetic shelf (Inset: camera picture of MBs on a magnet). Hydrodynamic diameter (E) and ζ potential (F) of MBs and MBs-mAb-pAb-Pt NPs complexes.
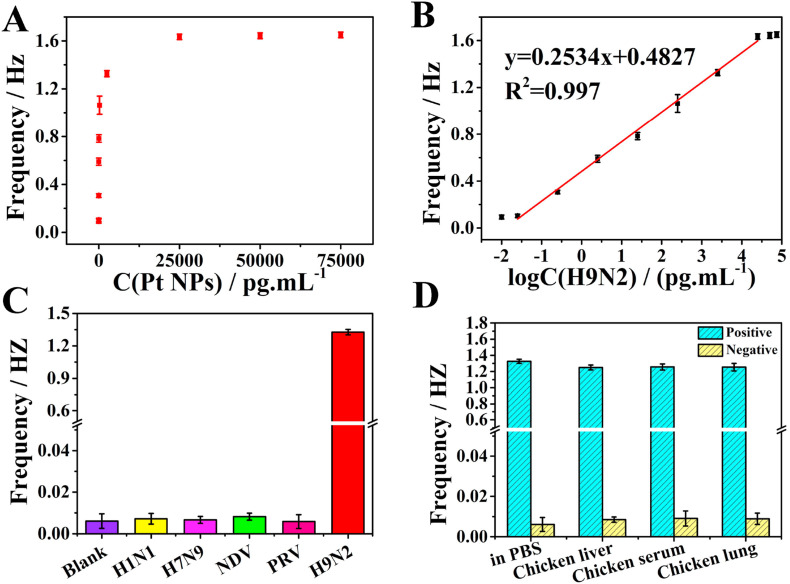
3.6. Reliable and robust SPCE biosensor for ultrasensitive H9N2 AIV detection
By using the prepared Pt NPs-loaded liposomes, MBs-mAb and Au UME were used as signal probes, capture probes and working electrodes, a novel SPCE biosensor was developed for H9N2 AIV detection. Under the condition of adding 10 μL of Pt NPs-loaded liposomes, the collision frequency was the highest (Fig. S12). To explore the feasibility of the SPCE biosensor, four control experiments were carried out, including without mAb, without H9N2, without pAb and all existed. Significant transient currents occurred only when all reagents were present, while no transient currents appeared in the other three control groups (Fig. S13). The results showed that the collision was effective between the released Pt NPs and Au UME, and proved the reliability of the SPCE biosensor. The collision frequency increased with H9N2 AIV concentration and then reached a platform (Fig. 6 A), resulting from the limited number of antibodies on MBs surface and the binding constant (K a) of the antibody. K a was calculated to be 4.36 × 108 L/mol and the detailed calculation process was shown in S15, which was consistent with the result reported in the literature (Chen et al., 2011). A linear relationship between collision frequencies and the logarithm of H9N2 AIV concentrations was appeared from 25 fg/mL to 25 ng/mL with a correlation coefficient (R 2) of 0.997 (Fig. 6B). The limit of detection (LOD) was as low as 18.1 fg/mL (S/N = 3), which was more sensitive than other H9N2 AIV detection methods with signal amplification (Table S1). Possible explanations were as follows: (1) Multiple Pt NPs were encapsulated in one liposome to promote collision frequency and detection sensitivity, ∼1.57 × 109 Pt NPs by 250 fg/mL H9N2 AIV (The detailed calculation process was shown in S16). (2) Antibody modified MBs could achieve efficient capture and separation of targets from complex systems. (3) Unlike conventional one-to-one or many-to-one detection mode, a one-to-many amplification mode was achieved.
Fig. 6.
(A) Relationship between collision frequencies and H9N2 AIV concentrations. (B) Linear curve between collision frequencies and the logarithm of H9N2 AIV concentrations (from 25 fg/mL to 25 ng/mL). (C) Histogram for the specificity of this method by using H9N2 AIV samples and other viruses (H1N1, H7N9, PRV, and NDV). (D) Collision frequencies responses for H9N2 AIV in complex systems.
In practical applications, the detection method needs high selectivity and specificity to ensure the accuracy of the results. Other viruses, H1N1 AIV (2.5 ng/mL), H7N9 AIV (5 ng/mL), PRV (10 ng/mL), NDV (25 ng/mL) as control groups, and H9N2 AIV (2.5 ng/mL) as the experimental group, were conducted the same immune process and SPCE test. Only the experimental group showed significant collision behavior, as shown in Fig. 6C, however the collision frequency of control groups was almost negligible, proving that the method had good specificity.
In clinical diagnosis, the practical samples usually existed in a complex environment, and the detection method was needed to have good anti-interference ability. The chicken lung and chicken liver were grinded and the solid insoluble matter was removed by centrifugation to prepare a positive sample. Then 2.5 ng/mL of H9N2 AIV was added to fresh chicken serum, chicken lung, and chicken liver respectively. Without adding H9N2 AIV, the same process was used to prepare negative samples. As shown in Fig. 6D, only positive samples showed obvious current response, and the collision frequencies obtained in the complex system of chicken serum, chicken lung, and chicken liver were comparable to that of in PBS, indicating that the SPCE biosensor had good anti-interference ability and could be directly used in complex systems. The practical samples were directly tested without sample pre-processing, which had great practical value in clinical diagnosis.
4. Conclusions
In summary, a reliable and robust SPCE biosensor was developed by the combination of liposome encapsulation-release amplification with MBs, realizing the efficient capture and detection of H9N2 AIV from complex samples. Collision frequency instead of current intensity was used for quantification, which solved the current fluctuations of individual Pt NPs and provided a reliable quantitative method. The reliable SPCE biosensor presented a universal encapsulation-release strategy for other disease markers by changing the encapsulated substances, specially, ∼1.57 × 109 Pt NPs can be released by 250 fg/mL H9N2 AIV. Unlike conventional one-to-one or many-to-one detection mode, the one-to-many amplification mode with high signal conversion efficiency was achieved. Importantly, highly sensitive SPCE biosensor had good performances in complex system by virtue of MBs, and the detection limit was as low as 18.1 fg/mL, making the assay promising for early clinical diagnosis and disease treatment.
CRediT authorship contribution statement
Jinrong Liu: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. Chong Ma: Conceptualization, Data curation, Formal analysis, Investigation, Methodology. Siwei Shi: Conceptualization, Data curation, Formal analysis, Investigation, Methodology. Heng Liu: Conceptualization, Data curation, Formal analysis, Investigation. Wei Wen: Conceptualization, Data curation, Formal analysis, Investigation. Xiuhua Zhang: Conceptualization, Data curation, Formal analysis, Investigation. Zhen Wu: Resources, Supervision, Validation, Project administration, Writing – review & editing. Shengfu Wang: Resources, Supervision, Validation, Project administration, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 21904032, 21775033 and 22076042) and the Key Laboratory of Emergency and Trauma (Hainan Medical University), Ministry of Education (Grant No. KLET-202009).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2022.114182.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Bai Y.Y., Wu Z., Xu C.M., Zhang L., Feng J., Pang D.W., Zhang Z.L. Anal. Chem. 2020;92:853–858. doi: 10.1021/acs.analchem.9b03492. [DOI] [PubMed] [Google Scholar]
- Bard A.J., Zhou H., Kwon S.J. Isr. J. Chem. 2010;50:267–276. [Google Scholar]
- Brotzakis Z.F., Lohr T., Vendruscolo M. Chem. Sci. 2021;12:9168–9175. doi: 10.1039/d1sc00244a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaneda A.D., Brenes N.J., Kondajji A., Crooks R.M. J. Am. Chem. Soc. 2017;139:7657–7664. doi: 10.1021/jacs.7b03648. [DOI] [PubMed] [Google Scholar]
- Chen Y.W., Luo W.X., Song H.J., Yin B.Y., Tang J.X., Chen Y.X., Ng M.H., Yeo A.E., Zhang J., Xia N.S. PLoS One. 2011;6 doi: 10.1371/journal.pone.0024144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defnet P.A., Zhang B. J. Am. Chem. Soc. 2021;143:16154–16162. doi: 10.1021/jacs.1c07164. [DOI] [PubMed] [Google Scholar]
- Dick J.E., Hilterbrand A.T., Strawsine L.M., Upton J.W., Bard A.J. Proc. Natl. Acad. Sci. U.S.A. 2016;113:6403–6408. doi: 10.1073/pnas.1605002113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick J.E., Renault C., Bard A.J. J. Am. Chem. Soc. 2015;137:8376–8379. doi: 10.1021/jacs.5b04545. [DOI] [PubMed] [Google Scholar]
- Fabiani L., Mazzaracchio V., Moscone D., Fillo S., De Santis R., Monte A., Amatore D., Lista F., Arduini F. Biosens. Bioelectron. 2022;200:113909. doi: 10.1016/j.bios.2021.113909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafez M.E., Ma H., Ma W., Long Y.T. Angew. Chem. Int. Ed. 2019;58:6327–6332. doi: 10.1002/anie.201901384. [DOI] [PubMed] [Google Scholar]
- Jiang W., Wei W., Yuan T., Liu S., Niu B., Wang H., Wang W. Chem. Sci. 2021;12:8556–8562. doi: 10.1039/d1sc01623g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karfa P., Majhi K.C., Madhuri R. ACS Catal. 2018;8:8830–8843. [Google Scholar]
- Lee Y.J., Park J.Y. Biosens. Bioelectron. 2010;26:1353–1358. doi: 10.1016/j.bios.2010.07.048. [DOI] [PubMed] [Google Scholar]
- Ma W., Ma H., Chen J.F., Peng Y.Y., Yang Z.Y., Wang H.F., Ying Y.L., Tian H., Long Y.T. Chem. Sci. 2017;8:1854–1861. doi: 10.1039/c6sc04582k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao R., Shao L., Compton R.G. Nano Res. 2021;14:4132–4139. [Google Scholar]
- Ortiz-Ledon C.A., Zoski C.G. Anal. Chem. 2018;90:12616–12624. doi: 10.1021/acs.analchem.8b02785. [DOI] [PubMed] [Google Scholar]
- Ortiz-Ledón C.A., Zoski C.G. Anal. Chem. 2017;89:6424–6431. doi: 10.1021/acs.analchem.7b00188. [DOI] [PubMed] [Google Scholar]
- Patrice F.T., Qiu K., Zhao L.J., Kouadio Fodjo E., Li D.W., Long Y.T. Anal. Chem. 2018;90:6059–6063. doi: 10.1021/acs.analchem.7b05405. [DOI] [PubMed] [Google Scholar]
- Peng Y.Y., Qian R.C., Hafez M.E., Long Y.T. Chemelectrochem. 2017;4:977–985. [Google Scholar]
- Sokolov S.V., Tschulik K., Batchelor-McAuley C., Jurkschat K., Compton R.G. Anal. Chem. 2015;87:10033–10039. doi: 10.1021/acs.analchem.5b02639. [DOI] [PubMed] [Google Scholar]
- Sui C., Yin H., Wang L., Zhou Y., Ai S. Biosens. Bioelectron. 2020;150:111908. doi: 10.1016/j.bios.2019.111908. [DOI] [PubMed] [Google Scholar]
- Tenchov R., Bird R., Curtze A.E., Zhou Q. ACS Nano. 2021;15:16982–17015. doi: 10.1021/acsnano.1c04996. [DOI] [PubMed] [Google Scholar]
- Wang H., Yang C., Tang H., Li Y. Anal. Chem. 2021;93:4593–4600. doi: 10.1021/acs.analchem.0c05055. [DOI] [PubMed] [Google Scholar]
- Wang H.Y., Ruan Y.F., Zhu L.B., Shi X.M., Zhao W.W., Chen H.Y., Xu J.J. Angew. Chem. Int. Ed. 2021;60:13244–13250. doi: 10.1002/anie.202014798. [DOI] [PubMed] [Google Scholar]
- Wang S., Shu J., Lyu A., Huang X., Zeng W., Jin T., Cui H. Anal. Chem. 2021;93:14238–14246. doi: 10.1021/acs.analchem.1c03208. [DOI] [PubMed] [Google Scholar]
- Wei Y.H., Han S.B., Kim J., Soh S., Grzybowski B.A. J. Am. Chem. Soc. 2010;132:11018–11020. doi: 10.1021/ja104260n. [DOI] [PubMed] [Google Scholar]
- Woods R., Bard A.J. vol. 9. Marcel Dekker; New York: 1976. (Electroanalytical Chemistry). [Google Scholar]
- Wu Z., Zhou C.H., Pan L.J., Zeng T., Zhu L., Pang D.W., Zhang Z.L. Anal. Chem. 2016;88:9166–9172. doi: 10.1021/acs.analchem.6b02284. [DOI] [PubMed] [Google Scholar]
- Xiang Z.P., Deng H.Q., Peljo P., Fu Z.Y., Wang S.L., Mandler D., Sun G.Q., Liang Z.X. Angew. Chem. Int. Ed. 2018;57:3464–3468. doi: 10.1002/anie.201712454. [DOI] [PubMed] [Google Scholar]
- Xiao X.Y., Bard A.J. J. Am. Chem. Soc. 2007;129:9610–9612. doi: 10.1021/ja072344w. [DOI] [PubMed] [Google Scholar]
- Zhan X., Yang S., Huang G., Yang L., Zhang Y., Tian H., Xie F., Lamy de la Chapelle M., Yang X., Fu W. Biosens. Bioelectron. 2021;188:113314. doi: 10.1016/j.bios.2021.113314. [DOI] [PubMed] [Google Scholar]
- Zhang J.H., Shen Q., Zhou Y.G. ACS Sens. 2021;6:2320–2329. doi: 10.1021/acssensors.1c00361. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Mao J., Ji W., Feng T., Fu Z., Zhang M., Mao L. Anal. Chem. 2019;91:5654–5659. doi: 10.1021/acs.analchem.8b05457. [DOI] [PubMed] [Google Scholar]
- Zhou Y.G., Zhang W.M., Li J., Xia X.H. Angew. Chem. Int. Ed. 2021 doi: 10.1002/anie.202115819. [DOI] [Google Scholar]
- Zhou Z., Liu X., Yue L., Willner I. ACS Nano. 2018;12:10725–10735. doi: 10.1021/acsnano.8b05452. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.