Abstract
A clinical isolate of Pseudomonas aeruginosa RP-1 produced the extended-spectrum β-lactamase (ESBL) SHV-2a. Its gene was expressed from a composite promoter made of the −35 region derived from the left inverted repeat of IS26 and the −10 region from the blaSHV-2a promoter itself. The DNA sequences immediately surrounding blaSHV-2a were homologous to plasmid pMPA2a from Klebsiella pneumoniae KpZU-3, while further away and 3′ to the blaSHV-2a gene, a sequence corresponding to the left end of Tn1721 was detected, thus indicating a likely enterobacterial origin of this ESBL gene.
The so-called extended-spectrum β-lactamases (ESBLs) hydrolyze extended-spectrum cephalosporins such as ceftriaxone, cefotaxime, and ceftazidime and monobactams such as aztreonam, while their activity is inhibited by clavulanic acid. Most of them are penicillinases (Ambler class A β-lactamases) (2), which are members of the 2be group of the Bush functional classification (3, 10), being mainly point mutation derivatives of TEM-1/TEM-2 or SHV-1 (22). They have been extensively described worldwide and are mostly plasmid mediated in members of the family Enterobacteriaceae (22).
Pseudomonas aeruginosa possesses inducible, naturally occurring cephalosporinases (3) which confer low-level resistance to aminopenicillins, narrow-spectrum cephalosporins such as cephalothin, and cephamycins such as cefoxitin. These Ambler class C β-lactamases are not inhibited by clavulanic acid (3). The most common mechanism for increased resistance to ceftazidime and other extended-spectrum cephalosporins in P. aeruginosa is derepression of the chromosomal class C enzyme, resulting in its overproduction (4, 5, 26).
However, within the last 4 years, three clavulanic acid-inhibitable ESBLs were found in P. aeruginosa. Among the 2be Bush group enzymes, two ESBLs are known, PER-1 and TEM-42 (16, 19). PER-1 was originally identified as chromosomally located in a P. aeruginosa isolate from the urinary tract of a Turkish patient hospitalized in Paris in 1992 (16). The PER-1 gene was later also identified as plasmid mediated (6). Recently, a Turkish study shows that PER-1 is found in 11% of P. aeruginosa hospital isolates and in 43% of Acinetobacter sp. strains, underlining its wide spread in this country (27). PER-1 is weakly related to the ESBLs of TEM or SHV derivatives (18). TEM-42 is the second ESBL found in a P. aeruginosa isolate in Paris in 1992 and is so far limited to just one isolate (16). It is also plasmid mediated. The only oxacillin-hydrolyzing β-lactamase (Ambler class D) with clavulanic acid-inhibited extended-spectrum properties is OXA-18, identified in a P. aeruginosa strain from an Italian patient hospitalized in Paris in 1995 (23). blaOXA-18 was chromosomally located.
In this study, we analyzed the β-lactamase content of a P. aeruginosa clinical strain for which a slight synergy between the ceftazidime and clavulanic acid discs was found in the double-disc diffusion test on a routine antibiogram.
(This work was presented in part at the 37th Interscience Conference on Antimicrobial Agents and Chemotherapy, Toronto, Ontario, Canada, on 26 September 1997.)
P. aeruginosa RP-1 was isolated in 1995 from a bronchoalveolar brush of a 52-year-old patient hospitalized in the intensive care unit at the Raymond Poincaré hospital (Garches, France). This French patient had recently returned from a trip to Tunisia, where he was hospitalized. This laboratory specimen was collected because the patient suffered from pneumonia. The strain was identified by using an API 20NE system (bioMérieux, Marcy l’Etoile, France). According to routine antibiogram results, it was additionally resistant to fluoroquinolones (ciprofloxacin, ofloxacin, and pefloxacin), aminoglycosides (amikacin, isepamicin, netilmicin, and tobramycin), chloramphenicol, and rifampin. The isolated strain showed a slight synergy between ceftazidime and clavulanate discs, which was best evidenced when the discs were put 1 cm from one another, suggesting the presence of an ESBL. Such a synergy test was performed as routine screening for all P. aeruginosa isolates in order to detect any ESBL-possessing strains. To search for any other gastrointestinal carriers of P. aeruginosa strains with the same unusual β-lactam resistance profile, rectal swab samples were collected from patients in the same hospitalization unit over the same period of time. The negative results ruled out any cross-contamination or any outbreak in this hospitalization unit.
Plasmid DNA extractions from P. aeruginosa RP-1 failed, despite repeated attempts using four different extraction methods (23). Conjugation assays performed as previously described (23), by using as the recipient strain P. aeruginosa PU21 or in vitro-obtained ciprofloxacin-resistant Escherichia coli JM109, also failed. Genomic DNA from P. aeruginosa RP-1 was then prepared as described previously (23). Preliminary dot blot hybridizations were performed with probes consisting of several class A or D β-lactamase genes, i.e., blaPER-1, blaSHV-3, blaTEM-1, and blaOXA-18 (23). Only the blaSHV-3 probe gave a positive signal with genomic DNA of P. aeruginosa RP-1. Partially Sau3AI-digested genomic DNA from P. aeruginosa RP-1 was then ligated into the BamHI site of a pBK-CMV cloning vector as previously described (23). Ten recombinant plasmids were obtained after selection of the E. coli JM109 electroporants on trypticase soy plates containing amoxicillin (100 μg/ml). The recombinant strains had decreased susceptibility to extended-spectrum cephalosporins such as ceftazidime compared to E. coli JM109 (data not shown). Analysis of their plasmid content revealed insert sizes ranging from 2.9 to 10 kb. One of them, pPL20 harboring the 2.9-kb insert, was retained for further analysis (Fig. 1).
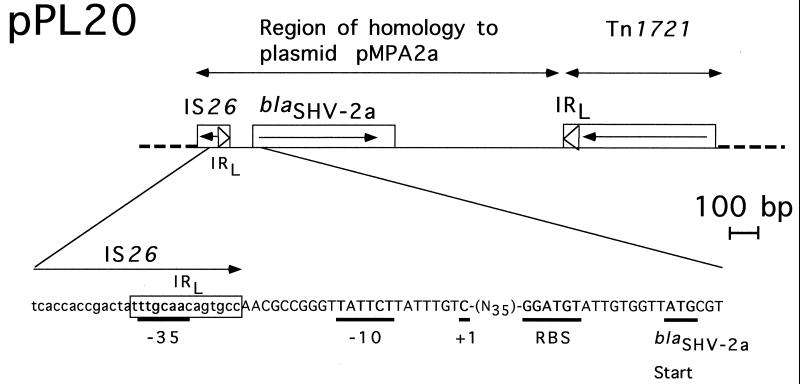
FIG. 1.
Schematic map of recombinant plasmid pPL20 which possesses blaSHV-2a. The thin, solid line represents the cloned insert from P. aeruginosa RP-1, while the dotted lines indicate the pBK-CMV cloning vector. The open boxes represent genes, and the arrow indicates their translational orientation. pMPA2a (20) and Tn1721 (1) homology regions are indicated. Details of the nucleotide sequence of the blaSHV-2a promoter region are shown below. The boxed sequence corresponds to the left inverted repeat (IRL) of IS26, the −35, −10, and +1 promoter sequences of blaSHV2-a are those described by Podbielski et al. (24). RBS, ribosome-binding site.
Susceptibility testing of E. coli JM109 harboring pPL20 and of P. aeruginosa RP-1 was performed by an agar dilution method as previously described (17, 23). Additionally, MICs were determined for reference strain P. aeruginosa ATCC 27853 and its in vitro-obtained, stably derepressed, cephalosporinase-producing mutant. This mutant, obtained after selection on ceftazidime-containing plates, produced an 85-fold increase of cephalosporinase activity determined as described for an Enterobacter cloacae isolate (14). This β-lactamase level, as well as the β-lactam MICs (Table 1), corresponded to a P. aeruginosa strain with a high basal level of constitutive cephalosporinase production (4). As shown in Table 1, P. aeruginosa RP-1 had decreased susceptibility to all of the β-lactams tested except imipenem. The MIC of ceftazidime (32 μg/ml) was reduced to 8 μg/ml in the presence of clavulanic acid. E. coli JM109(pPL20) had decreased susceptibility to all of the β-lactams tested except cefoxitin and imipenem; the MICs of the extended-spectrum cephalosporins and aztreonam were markedly reduced in the presence of clavulanic acid, indicating the presence of an ESBL (Table 1). Crude extracts from P. aeruginosa RP-1 and from E. coli JM109(pPL20) were analyzed by isoelectric focusing (23) and revealed a β-lactamase with a pI of 7.6 in both cases and an additional β-lactamase with a pI of 8.2 (likely corresponding to an AmpC cephalosporinase) found only in P. aeruginosa RP-1 extracts (data not shown).
TABLE 1.
MICs of β-lactams for P. aeruginosa RP-1, E. coli JM109 harboring recombinant plasmid pPL20, reference strain E. coli JM109, reference strain P. aeruginosa ATCC 27853, and its stably derepressed cephalosporinase-producing mutant, Mut
| Antibiotic(s) | MIC (μg/ml) against:
|
||||
|---|---|---|---|---|---|
| P. aeruginosa RP-1 | E. coli JM109 (pPL20) | E. coli JM109 | P. aeruginosa ATCC 27853 | P. aeruginosa Mut | |
| Amoxicillin | >512 | 512 | 2 | >512 | >512 |
| Amoxicillin + CLAa | >512 | 4 | 2 | >512 | >512 |
| Ticarcillin | >512 | >512 | 2 | 2 | 32 |
| Ticarcillin + CLA | 64 | 4 | 1 | 2 | 32 |
| Piperacillin | 256 | 256 | 1 | 1 | 16 |
| Piperacillin + CLA | 32 | 2 | 1 | 1 | 16 |
| Cephalothin | >512 | >512 | 4 | >512 | >512 |
| Cephalothin + CLA | >512 | 4 | 2 | >512 | >512 |
| Cefoxitin | >512 | 8 | 8 | >512 | >512 |
| Ceftazidime | 32 | 8 | 0.25 | 0.25 | 8 |
| Ceftazidime + CLA | 8 | 0.25 | 0.25 | 0.25 | 8 |
| Cefotaxime | >512 | 32 | 0.06 | 2 | >512 |
| Cefotaxime + CLA | 256 | 0.12 | 0.06 | 2 | >512 |
| Imipenem | 2 | 0.12 | 0.06 | 0.5 | 0.5 |
| Aztreonam | 32 | 8 | 0.12 | 0.5 | 2 |
| Aztreonam + CLA | 16 | 0.12 | 0.12 | 0.5 | 2 |
CLA, clavulanic acid at a fixed concentration of 2 μg/ml (MIC for E. coli JM109, 16 μg/ml).
The 2.9-kb cloned fragment from pPL20 was sequenced on both strands with an Applied Biosystems ABI 377 sequencer. Analysis of the sequenced DNA and the deduced protein revealed a sufficiently large open reading frame of 861 bp encoding a 286-amino-acid protein identified as SHV-2a. First described from a Klebsiella pneumoniae isolate in Germany (25), SHV-2a is a point mutation derivative of SHV-2 differing only by a leucine-to-glutamine replacement at the unusual position 35 of the Ambler numbering. Compared to the consensus sequence for SHV-1 (21), SHV-2 and SHV-2a possess a G238S change, explaining their extended-spectrum catalytic properties. Although initially considered insignificant (25), the position 35 mutation in SHV-2a compared to SHV-2 increased its resistance to ceftazidime but reduced the MICs of all other cephalosporins (20). The prevalence of SHV-2a in Enterobacteriaceae in Western Europe or in the United States is not known, although it was recently reported as being widespread among K. pneumoniae strains in Korea (11).
The −35 (5′-TTGCAA-3′) and −10 (5′-TATTCT-3′) promoter sequences of blaSHV-2a present on the 2.9-kb insert of pPL20 (Fig. 1) corresponded exactly to those previously identified for this β-lactamase gene in a K. pneumoniae isolate (25). No promoter sequence specific for P. aeruginosa genes was found upstream the blaSHV-2a structural gene. The −35 and −10 boxes showed higher homology to E. coli promoter sequences described by Hawley and McClure (9) than does the promoter belonging to blaSHV-2, thus explaining the blaSHV-2a promoter strength (24). In fact, a detailed analysis of blaSHV2a revealed that the immediate upstream and downstream sequences retained 100% DNA identity with parts of plasmid pMPA2a from K. pneumoniae KpZU-3 (20). Analysis of the promoter sequences of blaSHV2a indicated that they resulted from a fusion of the −35 sequence from IS26 (15) to the −10 sequence of the native blaSHV-2a promoter (Fig. 1). IS26 was, in fact, previously reported as being the promoter for the expression of an aminoglycoside resistance gene within a multidrug resistance operon (12). A similar hybrid promoter was identified for ESBL gene blaTEM-6, for which an IS1-like element provided the −35 sequence, thus allowing high-level expression of TEM-6 (8). From a general point of view, it is known that insertion sequences may act as mobile promoters on prokaryotic gene expression (7). Their inverted repeats contain −35 sequences that, upon insertion next to −10 sequences, may boost gene expression (7).
Although blaSHV2a was not plasmid located, our report indicates that the ESBL SHV derivatives may be identified in P. aeruginosa. The sequences surrounding blaSHV-2a had strong homology with K. pneumoniae plasmid sequences, and the more distantly related downstream sequences from blaSHV2a had homology with Tn1721, a transposon often encountered in Enterobacteriaceae (1) (Fig. 1). Pulsed-field gel electrophoresis of XbaI-restricted genomic DNA of P. aeruginosa RP-1 (13), followed by blaSHV-2a-specific hybridization, revealed that the fragment containing blaSHV-2a was larger than 300 kb, indicating its likely chromosomal origin (data not shown). We therefore believe that a putative plasmid derived from K. pneumoniae (containing blaSHV-2a) became integrated into P. aeruginosa RP-1 chromosomal DNA either by homologous recombination or by insertion sequence- or transposon-mediated specific cointegration (7).
Our report indicates that the ESBL genes are no longer limited to Enterobacteriaceae, from which they may have originated. From a clinical point of view, detection of these ESBLs based on the double-disc synergy test remains difficult in P. aeruginosa. This bacterial species may therefore become a hidden reservoir for such ESBLs, as is the case for oxacillinase extended-spectrum derivatives.
Nucleotide sequence accession number.
The nucleotide sequence reported in this work will appear in the GenBank nucleotide sequence database under accession no. AF074950.
Acknowledgments
This work was financed in part by a grant from the Ministère de l’Education Nationale et de la Recherche, Faculté de Médecine Paris-Sud (UPRES, JE 2227), Université Paris XI, France.
REFERENCES
- 1.Allmeier H, Gesnar B, Greck M, Schmitt R. Complete nucleotide sequence of Tn1721: gene organization and a novel gene product with features of chemotaxis protein. Gene. 1992;111:11–20. doi: 10.1016/0378-1119(92)90597-i. [DOI] [PubMed] [Google Scholar]
- 2.Ambler R P. The structure of β-lactamases. Philos Trans R Soc Lond B Biol Sci. 1980;289:321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- 3.Bush K, Jacoby G A, Medeiros A A. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell J I A, Ciofu O, Hoiby N. Pseudomonas aeruginosa isolates from patients with cystic fibrosis have different β-lactamase expression phenotypes but are homogeneous in the ampC-ampR genetic region. Antimicrob Agents Chemother. 1997;41:1380–1384. doi: 10.1128/aac.41.6.1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen H Y, Yuan M, Livermore D M. Mechanisms of resistance to β-lactam antibiotics amongst Pseudomonas aeruginosa isolates collected in the UK in 1993. J Med Microbiol. 1995;43:300–309. doi: 10.1099/00222615-43-4-300. [DOI] [PubMed] [Google Scholar]
- 6.Danel F, Hall L M C, Gur D, Akalin H E, Livermore D M. Transferable production of PER-1 β-lactamase in Pseudomonas aeruginosa. J Antimicrob Chemother. 1995;35:281–294. doi: 10.1093/jac/35.2.281. [DOI] [PubMed] [Google Scholar]
- 7.Galas D J, Chandler M. Bacterial insertion sequences. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C: American Society for Microbiology; 1989. pp. 109–162. [Google Scholar]
- 8.Goussard S, Sougakoff W, Mabilat C, Bauernfeind A, Courvalin P. An IS1-like element is responsible for high-level synthesis of extended-spectrum β-lactamase TEM-6 in Enterobacteriaceae. J Gen Microbiol. 1991;137:2681–2687. doi: 10.1099/00221287-137-12-2681. [DOI] [PubMed] [Google Scholar]
- 9.Hawley D K, McClure W R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983;11:2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacoby G A, Medeiros A A. More extended-spectrum β-lactamases. Antimicrob Agents Chemother. 1991;35:1697–1704. doi: 10.1128/aac.35.9.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim J, Kwon Y, Pai H, Kim J W, Cho D T. Survey of Klebsiella pneumoniae strains producing extended-spectrum β-lactamases: prevalence of SHV-12 and SHV-2a in Korea. Antimicrob Agents Chemother. 1998;36:1446–1449. doi: 10.1128/jcm.36.5.1446-1449.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee K Y, Hopkins J D, Syvanen M. Direct involvement of IS26 in an antibiotic resistance operon. J Bacteriol. 1990;172:3229–3236. doi: 10.1128/jb.172.6.3229-3236.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maslow J N, Slutsky A M, Arbeit R D. Application of pulse-field gel electrophoresis to molecular microbiology. In: Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology: principles and applications. Washington, D.C: American Society for Microbiology; 1993. pp. 563–572. [Google Scholar]
- 14.Mimoz O, Jacolot A, Léotard S, Hidri N, Samii K, Nordmann P, Petitjean O. Efficacies of cefepime, ceftazidime, and imipenem alone or in combination with amikacin in rats with experimental pneumonia due to ceftazidime-susceptible or -resistant Enterobacter cloacae strains. Antimicrob Agents Chemother. 1998;42:3304–3308. doi: 10.1128/aac.42.12.3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mollet B, Iida S, Shepherd J, Arber W. Nucleotide sequence of IS26, a new prokaryotic mobile genetic element. Nucleic Acids Res. 1983;11:6319–6330. doi: 10.1093/nar/11.18.6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mugnier P, Dubrous S, Arlet G, Collatz E. A TEM-derived extended-spectrum β-lactamase in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1996;40:2488–2493. doi: 10.1128/aac.40.11.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A3. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1993. [Google Scholar]
- 18.Nordmann P, Naas T. Sequence analysis of PER-1 extended-spectrum β-lactamase from Pseudomonas aeruginosa and comparison with class A β-lactamases. Antimicrob Agents Chemother. 1994;38:104–114. doi: 10.1128/aac.38.1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nordmann P, Ronco E, Naas T, Duport C, Michel-Briand Y, Labia R. Characterization of PER-1, a novel extended-spectrum β-lactamase from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1993;37:962–969. doi: 10.1128/aac.37.5.962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nüesch-Inderbinen M T, Hächler H, Kayser F H. New system based on site-directed mutagenesis for highly accurate comparison of resistance levels conferred by SHV β-lactamases. Antimicrob Agents Chemother. 1995;39:1726–1730. doi: 10.1128/aac.39.8.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nüesch-Inderbinen M T, Kayser F H, Hächler H. Survey and molecular genetics of SHV β-lactamases in Enterobacteriaceae in Switzerland: two novel enzymes, SHV-11 and SHV-12. Antimicrob Agents Chemother. 1997;41:943–949. doi: 10.1128/aac.41.5.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Philippon A, Arlet G, Lagrange P H. Origin and impact of plasmid-mediated extended-spectrum beta-lactamases. Eur J Clin Microbiol Infect Dis. 1994;13(Suppl. 1):17–29. doi: 10.1007/BF02390681. [DOI] [PubMed] [Google Scholar]
- 23.Philippon L N, Naas T, Bouthors A T, Barakett V, Nordmann P. OXA-18, a class D clavulanic-acid inhibited extended-spectrum β-lactamase from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1997;41:2188–2195. doi: 10.1128/aac.41.10.2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Podbielski A, Schönling J, Melzer B, Haase G. Different promoters of SHV-2 and SHV-2a β-lactamase lead to diverse levels of cefotaxime resistance in their bacterial producers. J Gen Microbiol. 1991;137:1667–1675. doi: 10.1099/00221287-137-7-1667. [DOI] [PubMed] [Google Scholar]
- 25.Podbielski A, Schönling J, Melzer B, Warnatz K, Leusch H G. Molecular characterization of a new plasmid-encoded SHV-type β-lactamase (SHV-2 variant) conferring high level cefotaxime resistance upon K. pneumoniae. J Gen Microbiol. 1991;137:569–578. doi: 10.1099/00221287-137-3-569. [DOI] [PubMed] [Google Scholar]
- 26.Sanders C C, Gates M L, Sanders W E., Jr Heterogeneity of class I β-lactamase expression in clinical isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1988;32:1893–1895. doi: 10.1128/aac.32.12.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vahaboglu H, Ozturk R, Aygun G, Coskunkan F, Yaman A, Kaygusuz A, Leblebicioglu H, Balik I, Aydin K, Otkun M. Widespread detection of PER-type extended-spectrum β-lactamases among nosocomial Acinetobacter and Pseudomonas aeruginosa isolates in Turkey: a nationwide multicenter study. Antimicrob Agents Chemother. 1997;41:2265–2269. doi: 10.1128/aac.41.10.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]