Abstract
Nuclear pore complexes (NPCs) are 110-megadalton assemblies that mediate nucleocytoplasmic transport. NPCs are built from multiple copies of ~30 different nucleoporins, and understanding how these nucleoporins assemble into the NPC scaffold imposes a formidable challenge. Recently, it has been shown how the Ycomplex, a prominent NPC module, forms the outer rings of the nuclear pore. However, the organization of the inner ring has remained unknown until now. We used molecular modeling combined with cross-linking mass spectrometry and cryo-electron tomography to obtain a composite structure of the inner ring. This architectural map explains the vast majority of the electron density of the scaffold. We conclude that despite obvious differences in morphology and composition, the higher-order structure of the inner and outer rings is unexpectedly similar.
Nuclear pore complexes (NPCs) are the major mediators of nucleocytoplasmic exchange and are of fundamental importance to all eukaryotic cells. Alterations of the nucleocytoplasmic transport system have been linked to various types of cancer, autoimmune diseases, and aging (1, 2). Experimentally testable structural models of the vertebrate NPC could provide a mechanistic basis for understanding clinically relevant mutations. Generating predictive structural models, however, is extremely challenging because of the size and intricate composition of the NPC (3). Nucleoporins (Nups) pre-assemble into several distinct subcomplexes, which oligomerize to form an eightfold rotationally symmetric structure. Many intra-subcomplex interactions have been biochemically and structurally characterized. In contrast, the weaker inter-subcomplex interactions have remained poorly understood, mainly because of their instability ex situ. To overcome this hurdle, it has been essential to combine biochemical and classical structure determination methods with in situ structural analysis and integrative molecular modeling (4).
Previous studies revealed that 16 copies of the Y complex (also called the Nup107 complex or Nup84 complex in fungi) form a reticulated doublering arrangement within both the nuclear and cytoplasmic rings (4). However, our understanding of how the inner ring (IR) scaffold is assembled in situ has been sparse. The Nup93 and Nup62 complexes—composed of Nup62, Nup58, and Nup54 and Nup205, Nup188, Nup155, Nup93, and Nup53, respectively—have been shown to localize to the IR (Nic96 and Nsp1 complexes in fungi; fig. S1) (5). More recently, it has been shown that Nup205 binds to the IR complexes mutually exclusively with respect to its homolog Nup188 (6) and also interacts with Nup98 (7, 8). Most domains of Nup93 complex members have been analyzed by x-ray crystallography (4), and some of these show homology to Y complex members. A tomographic study (9) revealed possible locations of 32 copies each of Nup155 and the large N-terminal domain (NTD) of Nup205 or Nup188 (fig. S2) (10). However, various other components—such as Nup93, the Nup62 complex, and the C-terminal domains (CTDs) of Nup205 and Nup188—remained unassigned and left large patches of the observed electron optical density unexplained.
Recent studies structurally and biochemically analyzed the Nup62-Nup58-Nup54 heterotrimer (8, 11), the CTD of Nup205 (8), and interactions between the Nup62 and Nup93 complexes (7, 8). In this study, we fitted all of the available structures into the tomographic map of the IR (9), in consensus with the well-established domain connectivity, interactions, and symmetry that have been observed by means of cryo-electron microscopy (cryo-EM) (details are provided in fig. S2, database S2, movies S1 to S3, and the supplementary materials). We found that 32 copies each of Nup205 or Nup188 (10), Nup93, Nup155, and the Nup62 complex fit seamlessly into the IR (Fig. 1, A and B, and fig. S2); another 16 copies of Nup155 connect it to the outer rings (9). The asymmetric unit of the IR is composed of four core modules. Each core module contains one copy of the aforementioned Nups. The inner and outer copies of the core modules are rotationally shifted with respect to each other and have slightly different conformations, but they engage in identical intra-subcomplex interactions (Fig. 1C). They form heterotypic inter-subcomplex interactions and oligomerize into four horizontally stacked rings. Each ring contains eight core modules. The inner and outer pairs of the core module are symmetric with respect to the nuclear envelope plane (Fig. 1C). These architectural principles are unexpectedly similar to the Y complex of the outer rings (9, 12, 13), highlighting a potential evolutionary relationship. An obvious difference is that the inner copies of the IR core modules form a C2 symmetric interface with each other, whereas the outer rings are spatially separated.
Fig. 1. Composite structure of the human NPC.
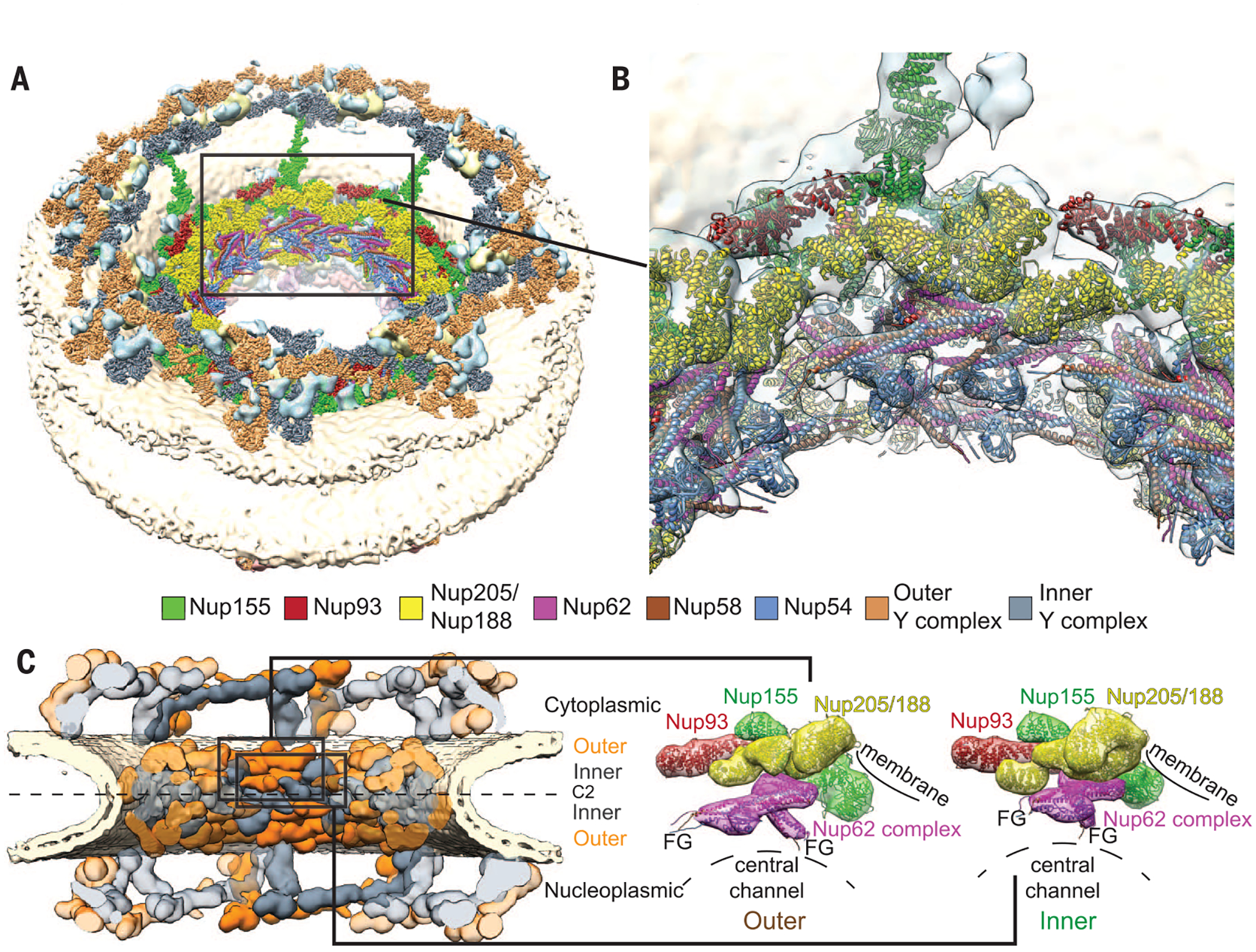
(A) Overview of the composite structure of the entire NPC, in which previous structural assignments in the outer rings (9, 13) are represented together with the assignments in the IR that were undertaken in this study (details are shown in fig. S2). Unassigned density is shown in cyan; the nuclear ring is facing up. (B) Zoomed-in view of the IR region framed in (A). High-resolution structures (colored ribbons) are shown in the context of the tomographic map (transparent isosurface). (C) Conceptual outline of the NPC architecture. Inner (gray) and outer (orange) copies of the Ycomplex (top and bottom) and the IR core module (middle) are shown in comparison.The configurations of the outer and inner copies of the IR core module are shown enlarged on the right. FG repeat domains can readily reach out into the central channel (F, phenylalanine; G, glycine).
A further-refined tomographic map of the IR solidifies the assignments (fig. S3 and database S2). It resolves the subdomain structures of Nup205 and Nup188 and the asymmetric features of Nup93, and it facilitates an unambiguous determination of the orientation of those proteins that could not be positioned previously (9).
Next, we generated spatial restraints by using cross-linking mass spectrometry. We affinity-purified Nup205-Nup93 complex (fig. S4) and Nup62 complex from human cells and reconstituted a complex in vitro, which consisted of the fungal orthologs of Nup205, Nup155, Nup93, Nup53, and Nup98 (fig. S5), as previously described (7). Spatial restraints were combined and mapped onto human proteins where necessary (Fig. 2A and fig. S6). Our composite structure is consistent with most of the spatial restraints (table S1), and the cross-links confirm several of the key features (Fig. 2, B to D). The very few cross-links that are violated in the structure are well in line with the false-positive discovery rate of cross-linking mass spectrometry. They may also arise from conformational heterogeneity due to the ex situ cross-linking (table S1 and figs. S4B and S7). We systematically validated our model by imposing biochemical restraints that were generated by several previous studies. We extracted 89 restraints from 35 publications and found strong agreement with our composite structure (table S2). To assess whether the available data could converge into alternative configurations of the IR structure, we used the Integrative Modeling Platform software suite (14) to set up an unbiased computational modeling approach that takes into account the matching of the high-resolution structures with the tomographic map, the cross-links, and domain connectivity. A set of seven top-scoring solutions were highly similar to the composite structure (fig. S8A). We therefore conclude that our composite structure is an objective and robust representation of the IR architecture. It is also in line with independent analyses of stoichiometry (15) and spatial proximities (16).
Fig. 2. Cross-linking mass spectrometry confirms intraand inter-subcomplex interactions of the IR architecture.

(A) Primary schematic of the IR proteins, showing the cross-links obtained by mass spectrometry. Inter-protein cross-links are shown in black, intra-protein links in light purple, and homodimeric cross-links (linking the same residues of a protein) in red. For clarity, Nup53 is not drawn to scale. Colored areas within the primary structures mark domains and motifs as individually labeled (residues are indicated below). A larger version is shown in fig. S6, and table S1 provides details on the cross-link data sets. The cross-links [blue lines in (B) to (D)] confirm specific features suggested by the composite structure, such as (B) head-to-tail arrangement of the inner and outer copy of Nup93, (C) interaction between Nup93 and Nup205, and (D) positioning of the Nup54 ferredoxin-like domain within the Nup62 complex.
Nup155 is homologous to Nup133 and Nup160 of the Y complex, whereas Nup93 is homologous to Nup107 and Nup85 of the Y complex (17–19). The interface of Nup133 and Nup107 (13) also seamlessly superimposes with that of Nup160 and Nup85 (fig. S9) (20). In our blueprint of the IR architecture, Nup155 and Nup93 are similarly positioned (Fig. 3, A and B, and fig. S9). A potential direct interface between Nup155 and Nup93 would have to be much weaker than its outer ring counterparts, because it has escaped biochemical characterization. However, it would be in agreement with the high sequence conservation of the respective surfaces (figs. S9C and S10) (19, 21) and the observation that the CTD of Nup93 is necessary and sufficient for the assembly of the NPC scaffold (22). Complex formation is strengthened in the presence of Nup53, as reported by various studies (table S2). This structural signature reoccurs throughout the inner and outer rings and directly engages with the membrane through the b-propellers of Nup160, Nup133, and Nup155, thereby forming a membrane-binding NPC coat (Fig. 3C).
Fig. 3. Simple and highly similar organizational principles govern the architecture of the inner and outer rings of the NPC.

(A) Schematic representation of the branching motif formed by the different heterodimers. (B) Structural similarity of the Nup155-Nup93 dimer to the Nup107-Nup133 and Nup120-Nup85 dimers. The structures are shown in low-resolution representation within the corresponding segments of the tomographic map. Homologous Nups are colored identically. (C) Arrangement of the branching motifs within the NPC membrane-binding coat [homologous Nups are colored as in (B)]. (D) Schematic of the human NPC architecture (details are shown in figs. S9 to S11). Trailing lines indicate FG repeats.
It has been previously assumed that the Y and Nup93 complexes are spatially separated into the morphological features observed with classical EM—namely, the outer and inner rings, respectively. Our analysis, taken together with previous work on the outer rings (9), suggests that multiple copies of Nup205 or Nup188 independently bind to the coat of the NPC in both the IR and the outer rings (Fig. 3D and fig. S11). The Nup62 complex binds to this scaffold (Fig. 3D), whereby short linear motifs in Nup53, Nup98, and Nup93 are important for the interconnection of subcomplexes (6–8). Our composite structure of the IR explains both the bulk of the electron optical density observed in situ and the biochemical data. In the future, cryo-EM maps at subnanometer resolution will be required to enable flexible fitting of x-ray structures and to precisely determine the orientations of Nup53, Ndc1, Nup98, and the linker regions of Nup53 and Nup93. Inter-subcomplex interfaces must be targeted by x-ray crystallography. Our model is suggestive of various such interfaces and will remain a cornerstone for further crystallographic analysis. It also highlights the anchoring positions of the intrinsically disordered Nup domains that facilitate the translocation of cargo, and it lays the foundation for understanding their spatial organization and functional role in healthy and diseased tissues.
Supplementary Material
ACKNOWLEDGMENTS
We thank A. Andres-Pons for technical support and critical reading of the manuscript. We acknowledge support from EMBL’s Electron Microscopy and Proteomics Core Facilities. W.W. and J.A.G.B. thank N. Grigorieff for sharing data before publication. J.K. was supported by the EMBL Interdisciplinary Postdoc Programme under Marie Curie COFUND actions. A.L.D. was supported by the Robert Crooks Stanley Fellowship at the Stevens Institute of Technology and National Institute on Aging (NIA) grant 1R21AG047433-01. J.S.G. was supported by an Ignition Grant Initiative from Stevens Institute of Technology and NIA grant 1R21AG047433-01. M.B. acknowledges funding by EMBL and the European Research Council (grant 309271-NPCAtlas). The density maps have been deposited in the Electron Microscopy Data Bank with accession numbers 8085, 8086, and 8087. The composite structure and the top-scoring automated models have been deposited in the Protein Data Bank with accession numbers 5IJN and 5IJO.
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencemag.org/content/352/6283/363/suppl/DC1
Materials and Methods
Figs. S1 to S11
Tables S1 to S4
References (23–34)
Movies S1 to S3
Databases S1 and S2
REFERENCES AND NOTES
- 1.Raices M, D’Angelo MA, Nat. Rev. Mol. Cell Biol 13, 687–699 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Mor A, White MA, Fontoura BM, Curr. Opin. Cell Biol 28, 28–35 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoelz A, Debler EW, Blobel G, Annu. Rev. Biochem 80, 613–643 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Hurt E, Beck M, Curr. Opin. Cell Biol 34, 31–38 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Vollmer B, Antonin W, Biol. Chem 395, 515–528 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Amlacher S et al. , Cell 146, 277–289 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Fischer J, Teimer R, Amlacher S, Kunze R, Hurt E, Nat. Struct. Mol. Biol 22, 774–781 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Stuwe T et al. , Science 350, 56–64 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.von Appen A et al. , Nature 526, 140–143 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.As noted by von Appen et al. (9), the structural homologs Nup205 and Nup188 cannot be distinguished at the given resolution of the tomographic map, although biochemical data point to an association of Nup188 with the outer rings.
- 11.Chug H, Trakhanov S, Hülsmann BB, Pleiner T, Görlich D, Science 350, 106–110 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Bui KH et al. , Cell 155, 1233–1243 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Stuwe T et al. , Science 347, 1148–1152 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Russel D et al. , PLOS Biol 10, e1001244 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ori A et al. , Mol. Syst. Biol 9, 648 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim DI et al. , Proc. Natl. Acad. Sci. U.S.A 111, E2453–E2461 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Devos D et al. , PLOS Biol 2, e380 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brohawn SG, Leksa NC, Spear ED, Rajashankar KR, Schwartz TU, Science 322, 1369–1373 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whittle JR, Schwartz TU, J. Biol. Chem 284, 28442–28452 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boehmer T, Jeudy S, Berke IC, Schwartz TU, Mol. Cell 30, 721–731 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeudy S, Schwartz TU, J. Biol. Chem 282, 34904–34912 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Sachdev R, Sieverding C, Flötenmeyer M, Antonin W, Mol. Biol. Cell 23, 740–749 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


