Abstract
A novel series of benzofuran-isoxazole hybrid heterocyclic unit has been synthesized and their structures characterized by 1H and 13C NMR, and mass spectral data. The synthesized products have been evaluated for their in vitro antibacterial and antifungal activity using Gentamycin sulphate and Nystatin as standard drugs, respectively. Four synthesized products have been determined as highly active against all tested bacterial and fungal strains. Structure–antimicrobial activity relationship has been supported by docking studies of the active compounds against glucosamine-6-phosphate synthase and aspartic proteinase. According to the docking studies, all derivatives exhibit good theoretical affinity with Autodock 4.2 software score in the range of –9.37 and –11.63 kcal/mol against the main protease of COVID-19.
Keywords: benzofuran-isoxazole hybrids, antimicrobial activity, docking studies, COVID-19, Autodock 4.2
INTRODUCTION
Naturally occurring and synthetically prepared compounds containing 2-benzylbenzofuran moiety exhibit a wide range of pharmacological and biological activities [1]. Among those are antihyperglycemic [2], analgesic, antibacterial [3], anti-inflammatory [4], antifungal [5], and antitumor [6] agents. Attachment of other heterocyclic rings to benzofuran may lead to compounds of even higher activity against bacteria resistant to other drugs [7]. A number of natural and synthetic isoxazole based analogues such as ibotenic acid [8–10], muscimol [11] and some more exhibited valuable biological activities. Motivated by the above information on benzofuran and isoxazole derivatives, we have synthesized conjugated benzofuran-isoxazole derivatives targeting the potential pharmacophores.
RESULTS AND DISCUSSION
Synthetic approach to the new benzofuran-isoxazole hybrids 8a–8p (Scheme 1) has not been presented in literature up to now.
Scheme 1.
Synthesis of benzofuran-isoxazoles 8a–8p.
Resorcinol (1) upon condensation with acetic acid in the presence of freshly fused ZnCl2 led to 1-(2,4-dihydroxyphenyl)ethanone (2). The following selective O-alkylation of the semiproduct 2 by propargyl bromide in presence of K2CO3 afforded para alkylated compound 3 with 90% yield. 1-[2-Hydroxy-4-(prop-2-yn-1-yloxy)phenyl]ethanone (3) was subjected to condensation - cyclisation process with phenacyl bromide (5) in the presence of potassium carbonate with formation of [3-methyl-6-(prop-2-yn-1-yloxy)benzofuran-2-yl](phenyl)methanone (6) [12].Some aliphatic and aromatic oxime intermediates 7a–7p were synthesized according to the presented earlier methods [13]. Alkyl oximes were synthesized by heating their precursors, alkyl aldehyde, with hydroxyl amine hydrochloride in presence of sodium acetate in MeOH at room temperature. Aromatic oximes were prepared from different substituted benzaldehydes using sodium acetate. Finally, the in situ generated various substituted oximes 7a–7p were subjected to 1,3-dipolar cycloaddition with terminal alkyne 6 in presence of copper sulphate to give the corresponding 3,5-disubstituted isoxazoles 8a–8p [14]. 1H and 13C NMR, FTIR and mass spectral data were used to characterize structures of the newly synthesized target compounds.
Antibactericidal activity. Antibacterial tests of all synthesized products 8a–8p exhibited activity (Table 1) on all species except Salmonella typhi. Compounds 8n, 8j, 8o, 8i, 8c, and 8k were determined to be highly active. The compounds with strong electron donating group like methoxy and weak electron withdrawing groups like fluoro and chloro on phenyl ring of oxazole enhanced antibacterial activity in comparison with aliphatic aryl substituted oxazoles. However, strong electron withdrawing groups supported the activity as well. Compounds 8m, 8g, 8a, 8h, and 8f also exhibited good antibacterial profile against the tested bacterial strains.
Table 1.
Antibactericidal activity of the synthesized benzofuran based isoxazole analogues 8a–8p
| Tested compounds | Concentration (10–6 g/mL) | Inhibition zone (in 10–3 m)a | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| gram +ve bacteria | gram –ve bacteria | ||||||||||
| M. Tub | M. lut | MRSA | B. sub | B. cer | P. aer | K. pne | E. col | P. vul | S. typ | ||
| 8a | 75 | 20 | 20 | 20 | 19 | 20 | 16 | 20 | 20 | 18 | 06 |
| 100 | 21 | 23 | 21 | 23 | 25 | 20 | 23 | 25 | 21 | 09 | |
| 8b | 75 | 20 | 16 | 22 | 19 | NA | 15 | NA | 20 | 17 | NA |
| 100 | 23 | 15 | 25 | 21 | NA | 17 | NA | 28 | 19 | NA | |
| 8c | 75 | 20 | 22 | 23 | 23 | 24 | 22 | 21 | 26 | 23 | NA |
| 100 | 22 | 25 | 25 | 27 | 29 | 28 | 24 | 29 | 27 | NA | |
| 8d | 75 | 17 | 15 | 14 | 14 | 13 | 13 | 13 | 16 | 15 | NA |
| 100 | 19 | 17 | 19 | 17 | 23 | 18 | 19 | 22 | 19 | NA | |
| 8e | 75 | 17 | 19 | 13 | 12 | 13 | 12 | 14 | 15 | 14 | NA |
| 100 | 18 | 19 | 17 | 15 | 19 | 17 | 18 | 22 | 18 | NA | |
| 8f | 75 | 20 | 19 | 18 | 13 | 15 | 17 | 14 | 20 | 13 | NA |
| 100 | 23 | 22 | 23 | 17 | 26 | 19 | 18 | 28 | 24 | NA | |
| 8g | 75 | 23 | 23 | 22 | 20 | 21 | 16 | 20 | 20 | 21 | NA |
| 100 | 25 | 25 | 26 | 27 | 28 | 20 | 24 | 28 | 24 | NA | |
| 8h | 75 | 14 | 17 | 21 | 16 | 20 | 16 | 16 | 20 | 18 | NA |
| 100 | 25 | 23 | 27 | 18 | 25 | 18 | 20 | 23 | 21 | NA | |
| 8i | 75 | 24 | 23 | 26 | 24 | 23 | 22 | 22 | 26 | 24 | NA |
| 100 | 29 | 23 | 27 | 28 | 32 | 28 | 28 | 32 | 28 | NA | |
| 8j | 75 | 28 | 23 | 25 | 23 | 26 | 22 | 23 | 28 | 24 | NA |
| 100 | 33 | 25 | 31 | 31 | 32 | 29 | 28 | 32 | 28 | NA | |
| 8k | 75 | 21 | 24 | 23 | 22 | 24 | 20 | 20 | 23 | 21 | NA |
| 100 | 23 | 26 | 24 | 25 | 28 | 23 | 23 | 28 | 25 | NA | |
| 8l | 75 | 17 | 15 | 16 | 10 | 12 | 11 | 14 | 16 | 14 | NA |
| 100 | 17 | 14 | 17 | 14 | 17 | 15 | 18 | 19 | 17 | NA | |
| 8m | 75 | 22 | 21 | 23 | 21 | 24 | 21 | 21 | 23 | 20 | NA |
| 100 | 25 | 23 | 28 | 26 | 28 | 24 | 25 | 28 | 24 | NA | |
| 8n | 75 | 28 | 26 | 27 | 27 | 32 | 28 | 28 | 33 | 29 | NA |
| 100 | 31 | 27 | 30 | 30 | 33 | 29 | 26 | 31 | 32 | NA | |
| 8m | 75 | 28 | 24 | 26 | 26 | 28 | 25 | 23 | 28 | 25 | NA |
| 100 | 29 | 27 | 31 | 30 | 31 | 27 | 27 | 31 | 29 | NA | |
| 8p | 75 | 17 | 16 | 17 | 14 | 22 | 16 | 20 | 19 | 18 | NA |
| 100 | 20 | 21 | 25 | 21 | 25 | 25 | 25 | 23 | 25 | NA | |
| Zentamycin sulphate | 75 | 29 | 27 | 31 | 30 | 31 | 28 | 27 | 31 | 29 | NA |
| 100 | 32 | 30 | 33 | 33 | 34 | 31 | 30 | 33 | 31 | NA | |
a (NA) No activity.
Antifungal activity. According to the results of tests presented in Table 2, the highest anti-fungal activity was determined for the products 8n, 8j, 8o, 8i, 8p, and 8c as compared with the standard Nystatin. Trichophyton rubrum and Trichophyton interdigitale exhibited resistance to all the products.
Table 2.
Antifungal activity of the synthesized compounds 8a–8p
| Tested compounds | Concentration (10–6 g/mL) | Inhibition zone (in 10–3 m)a | ||||
|---|---|---|---|---|---|---|
| M. canis | M. gypseum | T. rubrum | T. interdigitale | E. floccosum | ||
| 8a | 75 | 12 | 12 | NA | NA | 10 |
| 100 | 15 | 15 | NA | NA | 16 | |
| 8b | 75 | 11 | 11 | NA | NA | 08 |
| 100 | 14 | 14 | NA | NA | 12 | |
| 8c | 75 | 17 | 19 | NA | NA | 13 |
| 100 | 21 | 24 | NA | NA | 18 | |
| 8d | 75 | 10 | 10 | NA | NA | 07 |
| 100 | 14 | 16 | NA | NA | 11 | |
| 8e | 75 | 11 | 08 | NA | NA | 08 |
| 100 | 13 | 13 | NA | NA | 09 | |
| 8f | 75 | 12 | 11 | NA | NA | 08 |
| 100 | 14 | 14 | NA | NA | 13 | |
| 8g | 75 | 14 | 12 | NA | NA | 11 |
| 100 | 17 | 16 | NA | NA | 16 | |
| 8h | 75 | 11 | 11 | NA | NA | 10 |
| 100 | 15 | 15 | NA | NA | 12 | |
| 8i | 75 | 21 | 16 | NA | NA | 16 |
| 100 | 23 | 20 | NA | NA | 17 | |
| 8j | 75 | 20 | 17 | NA | NA | 16 |
| 100 | 26 | 22 | NA | NA | 18 | |
| 8k | 75 | 16 | 15 | NA | NA | 14 |
| 100 | 21 | 19 | NA | NA | 18 | |
| 8l | 75 | 08 | 07 | NA | NA | 05 |
| 100 | 12 | 09 | NA | NA | 08 | |
| 8m | 75 | 16 | 16 | NA | NA | 13 |
| 100 | 18 | 18 | NA | NA | 15 | |
| 8n | 75 | 20 | 18 | NA | NA | 18 |
| 100 | 28 | 21 | NA | NA | 20 | |
| 8o | 75 | 20 | 17 | NA | NA | 16 |
| 100 | 22 | 19 | NA | NA | 19 | |
| 8p | 75 | 21 | 20 | NA | NA | 18 |
| 100 | 23 | 24 | NA | NA | 16 | |
| Nystatin (std) | 75 | 24 | 21 | NA | NA | 21 |
| 100 | 27 | 25 | NA | NA | 24 | |
a (NA) No activity.
Molecular Docking Studies
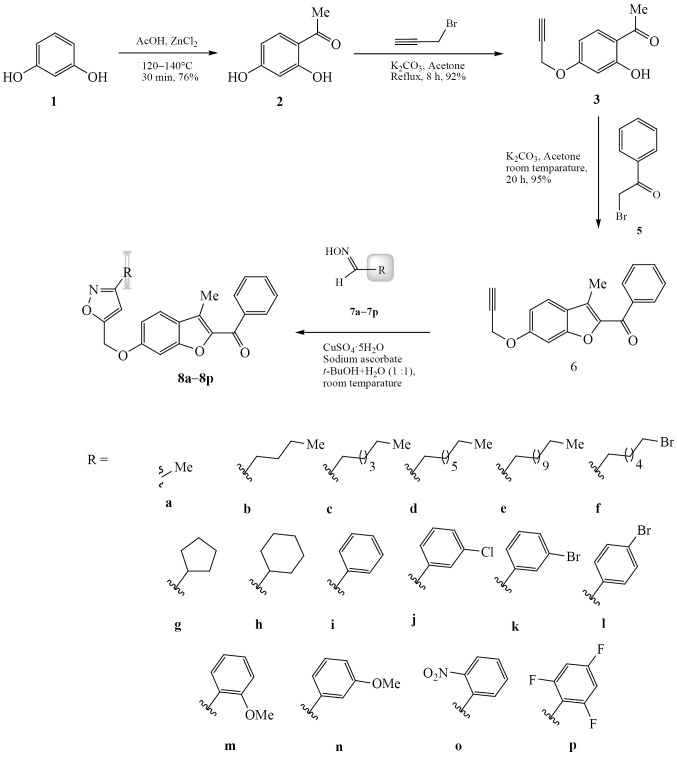
Antibacterial docking study. The compound 8m and reference compound gentamycin were docked with Glucosamine-6-phosphate synthase (PDB ID: 2VF5). The Grid box was set up with 70 : 64 : 56 Å along x, y, and z points and coordinates 30.59, 15.822, and –3.497 were assigned to 2VF5 (Fig. 1) [15]. Both ligand and a molecule were loaded into ADT tool and saved in .pdbqt format. For obtaining best docking results 10 confirmers of ligand were run in Autodock 4.2. Docking score of the best conformer of compound 8m was –9.29 kcal/mol whereas gentamycin score was –6.96 kcal/mole, and it demonstrated the higher value of binding energy than the reference compound with H-bond interactions with amino acid residues Thr302 and Ser303, and hydrophobic interactions with Leu484, Glu488, Tyr491, and Leu601 of 2VF5. Hence, the docking studies revealed that the newly synthesized compound 8m was the efficient bacterial inhibitor.
Fig. 1.
Docking pose and 2D interactions of compound 8m with 2VF5.
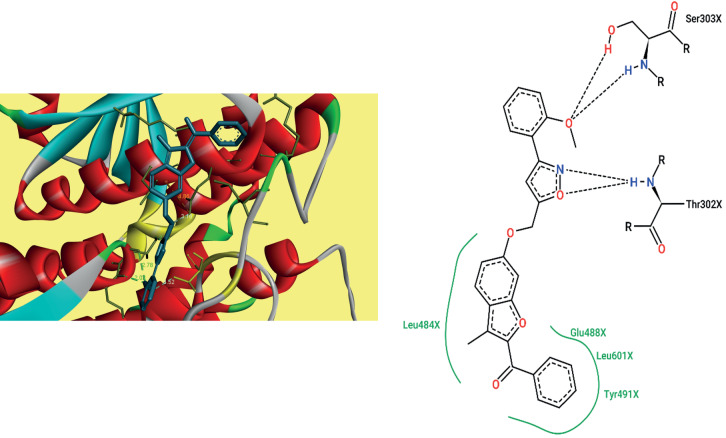
Antifungal docking study. For estimating antifungal activity of compound 8n and Nystatin these were docked into the active site of secreted aspartic proteinase (PDB ID: 2QZW). The ligand and proteins were loaded into ADT tool and saved in .pdbqt format. The Grid box for 2QZW (Fig. 2) was set up with 64 : 64 : 64 Å and coordinates –16.302, –23.24, and –16.245 were assigned [16]. The ligand was characterized by H-bond and hydrophobic interactions with the docking score of –10.03 kcal/mol on par with Nystatin whose score was –12.43 kcal/mol. The amino acid residues Asn131 and Arg192 of 2QZW were involved in H-bond interactions whereas Ile30, Ser35, Ile82, Tyr84, Gly85, Ile123, Ala335, and Asn337 were involved in hydrophobic interactions with the ligand 8n. Hydrophobic interactions were not observed for Nystatin confirming the ligand 8n to be the efficient antifungal agents.
Fig. 2.
Docking pose and 2D interactions of compound 8n with 2QZW.
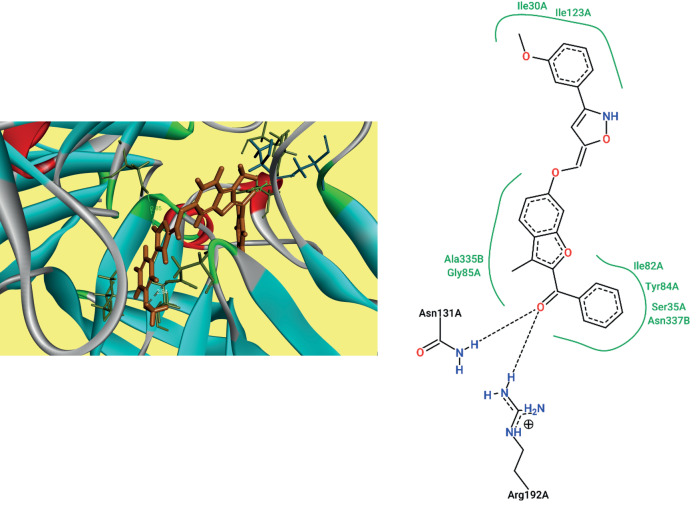
Anti-Covid19 docking study. Docking simulations were carried out with each and every ligand on to the active site of COVID-19 main protease (PDB ID: 6LU7). After loading the ligands and protein those were saved in .pdbqt format. For 6LU7 (Fig. 3) the grid box was set up with 60 : 60 : 60 Å and coordinates –11.824, 14.735, and 74.152 were assigned [17]. Docking scores of the ligands were ranging from –9.37 to –11.63 kcal/mol. Binding energies of all the ligands are presented in Table 3. All the ligands were characterized by H-bond and hydrophobic interactions with 6LU7 except compound 8l.
Fig. 3.
Docking pose and 2D interactions of compound 8j with 6LU7.
Table 3.
Docking scores of the compounds 8a–8p with COVID-19 main protease (PDB ID: 6LU7)
| Compound | Binding score, kcal/mol | Interacting amino acid residues |
|---|---|---|
| 8a | –9.37 | Thr25, Cys145, His163 |
| 8b | –10.45 | Phe140, Leu141, Gly143, Glu166, Arg188 |
| 8c | –9.68 | Met49, His41, Leu141, Gly143, Cys145, Met165 |
| 8d | –9.83 | Thr25, Cys145, Met165, Glu166, Arg188 |
| 8e | –9.82 | Met165, Glu166, Gln189 |
| 8f | –10.69 | His41, Gly143, Cys145, Met165, Asp187 |
| 8g | –10.59 | His41, Met49, Leu141, Gly143, Cys145, Met165 |
| 8h | –10.95 | His41, Met49, Leu141, Gly143, Cys145, Met165, Leu167, Pro168, Gln192 |
| 8i | –10.73 | Thr25, Leu27, His41, Ser144, Cys145, Met165, Asp187 |
| 8j | –11.63 | His41, Cys145, Met165, Glu166, Asp187 |
| 8k | –11.38 | His41, Met49, Leu141, Gly143, Cys145, Met165, Leu167 |
| 8l | –10.97 | No interactions |
| 8m | –10.91 | His41, Met49, Gly143, Cys145, Met165, Glu166, Asp187, Arg188, Gln189 |
| 8n | –10.94 | His41, Leu141, Gly143, Cys145, Met165, Leu167, Pro168 |
| 8o | –11.29 | Thr25, Leu27, His41, Met49, Gly143, Ser144, Cys145, Met165, Asp187, Gln189 |
| 8p | –10.62 | His41, Met49, Leu141, Gly143, His163, Met165 |
Compound 8j demonstrated the highest docking score of –11.63 kcal/mol, and compounds 8k, 8n and 8m exhibited binding energies of –11.38, –10.94 and –10.91 kcal/mol, respectively. These compounds were characterized by H-bond and hydrophobic interactions with COVID-19 main protease at active site amino residues His41, Met49, Leu141, Gly143, Cys145, Met165, Glu166, Leu167, Pro168, Asp187, Arg188, and Gln189. These results revealed that the ligands were potent inhibitors of COVID-19 main protease.
EXPERIMENTAL
Melting points were measured in open capillaries and are uncorrected. TLC was carried out on silica gel-G, and the spots were visualized under UV light at 254 nm. Column chromatography was performed on a Merck silica gel 60A (100–200 mesh). IR spectra (KBr discs) were recorded on a Perkin-Elmer 1000 spectrophotometer. NMR spectra were measured on a Bruker AV-400 and AV-300 spectrometers using CDCl3 as a solvent and TMS as an internal standard. Mass spectra were measured on an Agilent LC-MS instrument.
Synthesis of 1-(2,4-dihydroxyphenyl) ethanone (2). A mixture of freshly fused ZnCl2 (6.8 g, 0.05mol) with acetic acid (3 g, 0.05mol) was boiled at 120°C for 30 min, then resorcinol (1) (5.5 g, 0.05mol) was added to it. The mixture was boiled for 30 min at 140°C upon monitoring of the process by TLC. The mixture was cooled down to room temperature, mixed with cold H2O (100 mL) and extracted with ethyl acetate (3×50 mL). The mixture of organic layers was washed by 20% hydrochloric acid (50 mL), saturated NaHCO3 (25 mL) and brine solution (2×25 mL) then dried by anhydrous sodium sulphate, filtered and concentrated under vacuum. The crude product was purified by column chromatography with silica gel 100–200 mesh using 10% CH3COOC2H5–hexane. The pure product 2 was isolated in the form of reddish brown needles.
Synthesis of 1-[2-hydroxy-4-(prop-2-yn-1-yloxy)phenyl]ethanone (3). Propargyl bromide was added dropwise to a well agitated solution of 1-(2,4-dihydroxyphenyl) ethanone (2) (3.0 g, 0.019 mol) in dry acetone and K2CO3 (2.72 g, 0.019 mol) followed by refluxing the mixture for 8 h (TLC). The mixture was cooled down to room temperature, and acetone was evaporated under reduced pressure. The residue was extracted with water (50 mL) and ethyl acetate (3×50 mL). The mixture of organic layers was washed with salt solution (2× 25 mL). Organic layer was dried over anhydrous Na2SO4, then filtered and concentrated under vacuum. The crude product was purified by column chromatography using 5% ethyl acetate in hexane to obtain compound 3 as white solid.
Synthesis of [3-methyl-6-(prop-2-yn-1-yloxy) benzofuran-2-yl]phenylmethanone (6). A mixture of (2 g, 0.010 mol) phenacylbromide (5) with compound 3 (2.09 g, 0.010 mol) and K2CO3 (2.90 g, 0.02mol) was heated in acetone (20 mL) for 20 h [12] (TLC). The residue was filtered off and washed with acetone (2 × 15 mL), then it was purified by column chromatography using 10% ethyl acetate in petroleum ether as an eluent, yield 95%.
General procedure for the synthesis of oxime mediates 7a–7p. To the aldehyde (1 eq.) solution in methanol were added hydroxylamine hydrochloride (1 eq.) followed by sodium acetate (1.5 eq.). The mixture was agitated for ca 3 h (TLC). Upon completion of the reaction, the mixture was quenched by ice. The resulting precipiate was filtered and extracted, then washed by hexane and dried.
General procedure for the synthesis of isoxazoles 8a–8p. The intermediate 5 (200 mg, 0.375 mmol) was dissolved in 10 mL of aqueous t-BuOH (50%) and mixed with CuSO4·5H2O (5 mol %), sodium ascorbate (10 mol %) and a desired oxime 7a–7p (0.45 mmol). The reaction mixture was stirred for 1 h at room temperature (TLC). Upon completion of the process, the reaction mixture was diluted with H2O (25 mL) and extracted with C2H5OAc (3×25 mL). The combined organic layers were washed with brine (2×25 mL), dried over anhydrous sodium sulphate, filtered, and concentrated in vacuum. The crude product was purified by column chromatography using ethyl acetate in hexane as an eluent to afford the corresponding product 8a–8p.
{3-Methyl-6-[(3-methylisoxazol-5-yl)methoxy}benzofuran-2-yl}phenylmethanone (8a). Yellow solid, yield 70%, mp 167–169°C. IR spectrum, ν, cm–1: 1646 (C=O), 1560, 1250, 1100. 1H NMR spectrum, δ, ppm: 8.14–7.99 m (2H), 7.69–7.44 m (5H), 7.14 s (1H), 7.07–6.95 m (1H), 5.27 s (2H), 4.37 t (2H, J = 6.47 Hz), 2.60 s (3H), 1.98–1.82 m (2H), 1.43–1.30 m (2H), 1.03–0.87 m (3H). 13C NMR spectrum, δ, ppm: 185.3, 159.6, 155.5, 148.3, 143.5, 138.0, 132.3, 129.6 (2C), 128.2 (2C), 127.6, 123.1, 122.6, 121.9, 114.0, 96.7, 62.5, 50.4, 31.0, 30.1, 22.4, 10.2. MS: m/z: 390.2 [M + H]+.
{3-Methyl-6-[(3-methylisoxazol-5-yl)methoxy]benzofuran-2yl}phenylmethanone (8b). White solid, yield 93%, mp 196–198°C. IR spectrum, ν, cm–1: 1720 (C=O), 1545, 1260, 1015, 980. 1H NMR spectrum, δ, ppm: 8.08 d (2H, J = 7.42 Hz), 7.66–7.51 m (5H), 7.16 d (1H, J = 1.72 Hz), 7.06–7.02 m (1H), 5.30 s (2H), 4.37 t (2H, J = 7.27 Hz,), 2.62 s (3H), 1.98–1.88 m (2H), 1.31 d (6H, J = 10.93 Hz,), 0.88 t (3H, J = 6.86 Hz). 13C NMR spectrum, δ, ppm: 185.3, 159.6, 155.5, 148.3, 143.5, 138.1, 132.3, 129.6 (2C), 128.2 (2C), 127.5, 123.2, 122.5, 121.9, 114.0, 96. MS: m/z: 348.1 [M + H]+.
{6-[(3-Hexylisoxazol-5-yl)methoxy]-3-methylbenzofuran-2-yl}phenylmethanone (8c). White solid, yield 93%, mp 103–105°C. IR spectrum, ν, cm–1:1720 (C=O), 1635, 1452, 1250, 1150. 1H NMR spectrum, δ, ppm: 8.08 d (2H, J = 7.42 Hz), 7.66–7.51 m (5H), 7.16 d (1H, J = 1.72 Hz), 7.06–7.02 m (1H), 5.30 s (2H), 4.37 t (2H, J = 7.27 Hz), 2.62 s (3H), 1.98–1.88 m (2H), 1.31 d (6H, J = 10.93 Hz), 0.88 t (3H, J = 6.86 Hz). 13C NMR spectrum, δ, ppm: 185.3, 159.6, 155.5, 148.3, 143.5, 138.1, 132.3, 129.6, 128.2, 127.5, 123.2, 122.5, 121.9, 114.0, 96.8, 62.6, 50.5, 31.1, 30.2, 26.1, 22.3, 13.9, 10.1. MS: m/z: 418 [M + H]+.
{3-Methyl-6-[(3-octylisoxazol-5-yl)methoxy]benzofuran-2-yl}phenylmethanone (8d). Yellow solid, yield 94%, mp 165–167°C. IR spectrum, ν, cm–1: 1760 (C=O), 1645, 1287, 1150, 960. 1H NMR spectrum, δ, ppm: 8.06 d (2H , J = 6.72 Hz), 7.69–7.44 m (5H), 7.14 s (1H), 7.03 d (1H, J = 7.90 Hz), 5.28 s (2H), 4.36 t (2H, J = 6.47 Hz), 2.61 s (3H), 1.99–1.82 m (2H), 1.38–1.17 m (10H), 0.94–0.79 m (3H). 13C NMR spectrum, δ, ppm: 185.3, 159.6, 155.5, 148.3, 143.5, 138.0, 132.3, 129.6, 128.3, 127.6, 123.1, 122.6, 121.9, 114.0, 96.7, 62.5, 50.5, 31.7, 30.2, 29.0, 28.9, 26.5, 22.6, 14.0, 10.2. MS: m/z: 446.5 [M + H]+.
{6-[(3-Dodecylisoxazol-5-yl)methoxy]-3-methylbenzofuran-2-yl}phenylmethanone (8e). White solid, yield 94%, mp 169–171°C. IR spectrum, ν, cm–1: 1646 (C=O), 1620, 1263, 1075. 1H NMR spectrum, δ, ppm: 8.08 d (2H, J = 7.68 Hz), 7.65 s (1H), 7.63–7.49 m (4H), 7.16 s (1H), 7.04 d (1H, J = 8.66 Hz), 5.29 s (2H), 4.37 t (2H, J = 7.19 Hz), 2.62 s (3H), 2.03–1.84 m (2H), 1.38–1.24 m (18H), 0.89 t (3H, J = 6.56 Hz). 13C NMR spectrum, δ, ppm: 185.3, 159.6, 155.5, 148.3, 143.5, 138.0, 132.3, 129.5, 128.2, 127.6, 123.0, 122.6, 121.9, 114.0, 96.6, 62.5, 51.1, 33.7, 32.2, 30.6, 28.9, 26.9, 26.5, 24.5, 22.6, 20.2, 17.3, 14.0, 10.2. MS: m/z: 502.4 [M + H]+.
(6-{[3-(6-Bromohexyl)isoxazol-5-yl]methoxy}-3-methylbenzofuran-2-yl)phenylmethanone (8f). Yellow solid, yield 73%, mp 168–170°C. IR spectrum, ν, cm–1: 1754, 1612, 1478, 1150, 980. 1H NMR spectrum, δ, ppm: 8.06 d (2H, J = 7.10 Hz), 7.67–7.49 m (5H), 7.14 s (1H), 7.07–6.99 m (1H), 5.28 s (2H), 4.38 t (2H, J = 6.36 Hz), 3.38 t (2H, J = 6.06 Hz), 2.61 s (3H), 1.98–1.77 m (4H), 1.49–1.28 m (4H). 13C NMR spectrum, δ, ppm: 185.3, 159.5, 155.4, 148.2, 143.5, 138.0, 132.3, 129.5, 128.2, 127.5, 123.1, 122.6, 121.9, 114.0, 96.6, 62.5, 50.2, 33.5, 32.3, 30.0, 27.4, 25.6, 10.2. MS: m/z: 496.2 [M + H]+.
{6-[(3-Cyclopentylisoxazol-5-yl)methoxy]-3-methylbenzofuran-2-yl-phenylmethanone (8g). White solid, yield 85%, mp 168–170°C. IR spectrum, ν, cm–1: 1647 (C=O), 1450, 1230, 1122, 1015, 860. 1H NMR spectrum, δ, ppm: 8.06 d (2H, J = 7.29 Hz), 7.70 s (1H), 7.64–7.48 m (4H), 7.15 s (1H), 7.03 d.d (1H, J = 8.65, 1.70 Hz), 5.26 s (2H), 4.99-4.89 m (1H), 2.63 s (3H), 2.32–2.21 m (2H), 2.12–2.01 m (2H), 1.97–1.86 m (2H), 1.82–1.70 m (2H). 13C NMR spectrum, δ, ppm: 185.3, 159.7, 155.5, 148.3, 143.2, 138.0, 132.3, 129.6, 128.2, 127.5, 123.1, 121.9, 121.3, 114.0, 96.7, 62.6, 62.0, 33.3, 24.0, 10.1. MS: m/z: 402.3 [M + H]+.
{6-[(3-Cyclohexylisoxazol-5-yl)methoxy]-3-methylbenzofuran-2-yl}phenylmethanone (8h). Pale yellow solid, yield 81%, mp 157–159°C. IR spectrum, ν, cm–1: 1670 (C=O), 1600, 1450, 1240, 630. 1H NMR spectrum, δ, ppm: 8.05 d (2H, J = 6.40 Hz), 7.65 s (1H), 7.61–7.42 m (4H), 7.13 s (1H), 7.01 d (1H, J = 7.75 Hz), 5.24 s (2H), 4.43 s (1H), 2.58 s (3H), 2.20 d (2H, J = 9.62 Hz), 1.90 d (2H, J = 11.15 Hz), 1.73 d (3H, J = 10.99 Hz), 1.43 d (2H, J = 12.43 Hz), 1.25 d (1H, J = 11.51 Hz). 13C NMR spectrum, δ, ppm: 185.3, 159.7, 155.5, 148.2, 142.9, 138.0, 132.3, 129.5, 128.2, 127.6, 123.0, 121.9, 120.6, 114.0, 96.5, 62.5, 60.2, 33.5, 25.1, 25.0, 10.2. MS: m/z: 416.5 [M + H]+.
{3-Methyl-6-[(3-phenylisoxazol-5-yl)methoxy]benzofuran-2-yl}phenylmethanone (8i). White solid, yield 88%, mp 127–130°C. IR spectrum, ν, cm–1: 1720 (C=O), 1650, 1578, 1240, 1100, 680. 1H NMR spectrum, δ, ppm: 8.12–8.07 m (3H), 7.77–7.75 m (2H), 7.62–7.59 m (2H), 7.57–7.52 m (4H), 7.48–7.45 m (1H), 7.20 d (1H, J = 2.12 Hz), 7.08 d.d (1H, J = 8.69, 2.20 Hz), 5.39 s (2H), 2.63 s (3H). 13C NMR spectrum, δ, ppm: 185.3, 159.5, 155.5, 148.4, 144.4, 138.0, 136.9, 132.3, 129.8, 129.6, 128.9, 128.2, 127.5, 123.3, 122.0, 121.0, 120.6, 114.0, 96.9, 62.5, 10.1. MS: m/z: 410.4 [M + H]+.
(6-{[3-(3-Chlorophenyl)isoxazol-5-yl]methoxy}-3-methylbenzofuran-2-yl)phenylmethanone (8j). Pale yellow solid, yield 78%, mp 168–170°C. IR spectrum, ν, cm–1: 1664, 1498, 1232, 1100, 1098, 680. 1H NMR spectrum, δ, ppm: 8.15–7.99 m (3H), 7.77 s (1H), 7.67–7.37 m (7H), 7.14 s (1H), 7.03 d (1H, J = 8.28 Hz), 5.33 s (2H), 2.59 s (3H). 13C NMR spectrum, δ, ppm: 185.3, 159.4, 155.4, 148.3, 137.9, 137.6, 135.6, 132.4, 130.8, 129.6, 129.5, 129.0, 128.2, 127.5, 123.2, 122.0, 121.0, 120.7, 118.4, 113.9, 96.6, 62.2, 10.2. MS: m/z: 444 [M + H]+.
(6-{[3-(3-Bromophenyl)isoxazol-5-yl]methoxy}-3-methylbenzofuran-2-yl)phenylmethanone (8k). Light yellow solid, yield 81%, mp 154–156°C. IR spectrum, ν, cm–1: 1663 (C=O), 1600, 1548, 1230, 1100, 1005, 980, 750. 1H NMR spectrum, δ, ppm: 8.11 s (1H), 8.07 d (2H, J = 7.16 Hz), 7.66–7.43 m (8H), 7.19 d (1H, J = 2.01 Hz), 7.07 d.d (1H, J = 8.69, 2.11 Hz), 5.38 s (2H), 2.61 s (3H). 13C NMR spectrum, δ, ppm: 185.4, 159.6, 155.5, 148.3, 143.4, 138.0, 134.7, 132.4, 130.9, 130.8, 129.6, 128.5, 128.3, 128.0, 127.8, 127.6, 125.0, 123.3, 122.0, 114.1, 96.8, 62.4, 10.2. MS: m/z: 444.2 [M + H]+.
(6-{[3-(4-Bromophenyl)isoxazol-5-yl]methoxy}-3-methylbenzofuran-2-yl)phenylmethanone (8l). Brick red solid, yield 91%, mp 138–140°C. IR spectrum, ν, cm–1: 1666 (C=O), 1546, 1456, 1234, 1100, 1005, 965, 650. 1H NMR spectrum, δ, ppm: 8.13–8.04 m (3H), 7.70 d (2H, J = 8.81 Hz), 7.63–7.57 m (2H), 7.56–7.48 m (4H), 7.17 d (1H, J = 1.99 Hz), 7.07–7.03 m (1H), 5.38 s (2H), 2.61 s (3H). 13C NMR spectrum, δ, ppm: 185.3, 159.5, 155.5, 148.3, 144.6, 138.0, 135.4, 134.8, 132.4, 130.0, 129.6, 128.3, 127.5, 123.3, 122.0, 121.7, 121.0, 113.9, 96.8, 62.4, 10.2. MS: m/z: 444.2 [M + H]+.
(6-{[3-(2-Methoxyphenyl)isoxazol-5-yl]methoxy}-3-methylbenzofuran-2-yl)phenylmethanone (8m). White solid, yield 86%, mp 168–170°C. IR spectrum, ν, cm–1: 1750 (C=O), 1600, 1554, 1420, 1230, 1100, 630. 1H NMR spectrum, δ, ppm: 8.23 s (1H), 8.11–8.04 m (2H), 7.80 d.d (1H, J = 7.88,1.54 Hz), 7.64–7.49 m (4H), 7.47–7.40 m (1H), 7.20 d (1H, J = 2.04 Hz), 7.15–7.04 m (3H), 5.37 s (2H), 3.88 s (3H), 2.62 s (3H). 13C NMR spectrum, δ, ppm: 185.4, 159.7, 155.5, 151.0, 148.3, 142.8, 138.0, 132.3, 130.2, 129.6, 128.3, 127.6, 126.1, 125.5, 125.2, 123.2, 121.9, 121.2, 114.2, 112.2, 96.8, 62.5, 56.0, 10.2. MS: m/z: 440.4 [M + H]+.
(6-{[3-(3-methoxyphenyl)isoxazol-5-yl]methoxy}-3-methylbenzofuran-2-yl)phenylmethanone (8n). Brick red solid, yield 87%, mp 136–138°C. IR spectrum, ν, cm–1: 1650 (C=O), 1540, 1250, 1003, 630. 1H NMR spectrum, δ, ppm: 8.41 d (2H, J = 8.84 Hz), 8.22 s (1H), 8.08–7.95 m (4H), 7.62–7.56 m (2H), 7.54–7.48 m (2H), 7.16 s (1H), 7.05 d (1H, J = 8.60 Hz), 5.38 s (2H), 2.59 s (3H). 13C NMR spectrum, δ, ppm: 185.3, 159.2, 155.4, 148.3, 147.3, 145.3, 140.9, 137.9, 132.4, 129.5, 128.2, 127.4, 125.5, 123.4, 122.1, 120.8, 120.5, 113.8, 96.7, 62.2, 10.1. MS: m/z: 455.0 [M + H]+.
(3-Methyl-6-{[3-(2-nitrophenyl)isoxazol-5-yl]methoxy}benzofuran-2-yl)phenylmethanone (8o). Yellow solid, yield, 91%, mp 148–150°C. IR spectrum, ν, cm–1: 1680 (C=O), 1520, 1423, 1122, 1010, 890. 1H NMR spectrum, δ, ppm: 8.59 s (1H), 8.32–8.23 m (2H), 8.22–8.13 m (1H), 8.04 d (2H, J = 7.43 Hz), 7.73 t (1H, J = 8.15 Hz), 7.62–7.45 m (4H), 7.14 s (1H), 7.03 d (1H, J = 8.29 Hz), 5.36 s (2H), 2.57 s (3H). 13C NMR spectrum, δ, ppm: 185.3, 159.3, 155.4, 148.8, 148.3, 145.1, 137.9, 137.5, 132.4, 131.0, 129.5, 128.2, 127.5, 125.9, 123.3, 123.3, 122.1, 121.0, 115.2, 113.9, 96.6, 62.2, 10.2. MS: m/z: 455.2 [M + H]+.
(3-Methyl-6-{[3-(2,4,6-trifluorophenyl)isoxazol-5-yl]methoxy}benzofuran-2-yl)phenylmethanone (8p). White solid, yield 85%, mp 149–147°C. IR spectrum, ν, cm–1: 1770 (C=O), 1650, 1432, 1012, 960. 1H NMR spectrum, δ, ppm: 8.17 d (1H, J = 2.72 Hz), 8.10–8.03 m (2H), 7.78–7.69 m (1H), 7.63–7.48 m (4H), 7.24–7.14 m (2H), 7.06 d.d (1H, J = 8.69, 2.15 Hz), 5.37 s (2H), 2.61 s (3H). 13C NMR spectrum, δ, ppm: 185.3, 159.4, 155.4, 148.3, 144.5, 139.3, 138.0, 132.4, 129.6, 128.3, 127.5, 123.9, 123.3, 122.6, 122.1, 118.8, 114.01, 113.0, 96.8, 62.2, 10.2. MS: m/z: 464.3 [M + H]+.
Anti-microbial assay. Solution of a tested compound (1 mg/mL) in DMSO was impregnated on sterilized standard discs of filter paper (5 mm). The discs soaked with the test compound were placed on an agar plate injected with test organism. The tests were carried out in triplicates. The zentamycin sulphate and nystatin were used as the standards. All the petri plates were incubated at 37°C for one to five days. The results were estimated by measuring the diameter of inhibition zones (Tables 1 and 2). The most active compounds were further subjected to determination of their minimum inhibitory concentrations (MICs).
Minimal inhibition concentration (MIC) measurement. To determine the MIC vulnerability tests of microorganism in nutrient and dextrose broths were employed. DMSO (1000 µg/mL) was used to prepare stock solutions of Ciproflaxin (standard antibacterial agent), Nystatin (standard antifungal agent) and test compounds. The following dilutions of the above solutions gave concentrations ranging from 25 to 250 µg/mL. The suspensions of microbial cultures were inoculated on to agar plates and then discs of test and control compounds of different concentrations were placed on agar surface (Tables 1 and 2).
Docking procedure. The open source software Autodock 4.2 was downloaded from the Scripps Research Institute (www.scripps.edu) into the computer configured with Intel(R) Core(TM) i5-8250U CPU @ 1.60GHz 1.80 GHz processor and RAM capacity of 8.00GB. The ligand molecules were drawn using the tool ChemSketch (www.acdlabs.com) in .mol format and converted to PDB file using Pymol (pymol.org) program tool.
To study the binding interactions between the newly synthesized ligands and the target molecules, glucosamine 6-phosphate synthase from Escherichia Coli (PDB ID: 2VF5), the Secreted aspartic proteinase from Candida albicans (PDB ID: 2QZW) and Covid-19 main protease (PDB ID: 6LU7) were downloaded from Protein Data Bank (www.rcsb.org). The ligands and the target proteins were loaded into Autodock 4.2, the number of torsions were set to the ligands. Both ligand and target proteins were saved into .pdbqt format. The Grid box and x, y, z centres were assigned to the active site of proteins.
The Autodock 4.2 uses a Lamarckian genetic algorithm program to calculate different ligand conformers. Conformations were ranked according to the binding energy obtained from docked procedure and the confirmation with lowest binding energy was considered as the best docking score. The Autodock 4.2 results were visualized by using BIOVIA Discovery Studio Visualizer and Proteins Plus Server (https://proteins.plus/).
CONCLUSIONS
A new class of benzofuran-isoxazoles (8a–8p) has been successfully synthesized in high yields and characterized by spectroscopic methods. All newly synthesized compounds have demonstrated high antibacterial and antifungal activities. The compounds 8m, 8n, 8j, and 8k have been determined to be the most active. Molecular docking studies have been performed for all compounds into the binding cavity of protein 2VF5, 2QZW, and 6LU7. Docking scores of the best conformers of compounds are as follows: –9.29 kcal/mol for 8m against 2VF5, –10.03 kcal/mol for 8n against 2QZW, and –11.63 kcal/mol for 8j against 6LU7. All products have demonstrated docking scores ranging from –9.37 to –11.63 kcal/mol with COVID-19 Main Protease. Hence, all newly synthesized benzofuran-isoxazoles ligands need further studies as the potential therapeutic agents for COVID-19.
ACKNOWLEDGMENTS
N. Umapathi thanks the Head of the Department, Osmania University, Hyderabad, India and Research and Development central facilities, Osmania University, Hyderabad, India for providing analytical support. B.S. thanks the University Grants Commission in India for the award of a Dr. D. S. Kothari postdoctoral fellowship.
CONFLICT OF INTEREST
No conflict of interest was declared by the authors.
REFERENCES
- 1.Rawat P., Kumar M., Rahuja N., Srivastava D.S.L., Srivastava A.K., Maurya R. Bioorg. Med. Chem. Lett. 2011;21:228. doi: 10.1016/j.bmcl.2010.11.031Get. [DOI] [PubMed] [Google Scholar]
- 2.Cottineau B., Toto P., Marot C., Pipaud A., Chenault J. Bioorg. Med. Chem. Lett. 2002;12:2105. doi: 10.1016/S0960-894X(02)00380-3. [DOI] [PubMed] [Google Scholar]
- 3.Manna K., Agrawal Y.K. Bioorg. Med. Chem. Lett. 2009;19:2688. doi: 10.1016/j.bmcl.2009.03.161. [DOI] [PubMed] [Google Scholar]
- 4.Chakrabarti J.K., Eggleton R.J., Gallagher P.T., Harvey J., Hicks T.A., Kitchen E.A., Smith C.W. J. Med. Chem. 1987;30:1663. doi: 10.1021/jm00392a024. [DOI] [PubMed] [Google Scholar]
- 5.Gündoğdu-Karaburun N., Benkli K., Tunali Y., Uçucu Ü., Demirayak Ş. Eur. J. Med. Chem. 2006;41:651. doi: 10.1016/j.ejmech.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 6.Galal S.A., El-All A.S.A., Abdallah M.M., El-Diwani H.I. Bioorg. Med. Chem. Lett. 2009;19:2420. doi: 10.1016/j.bmcl.2009.03.069. [DOI] [PubMed] [Google Scholar]
- 7.Kumbhare Ravindra M. Bioorg. Med. Chem. Lett. 2012;22:5424. doi: 10.1016/j.bmcl.2012.07.041. [DOI] [PubMed] [Google Scholar]
- 8.Naeimi H., Amini A., Moradian M. Org. Chem. Front. 2014;1:415. doi: 10.1039/C4QO00031E. [DOI] [Google Scholar]
- 9.Naeimi H., Moradi L. Catal. Commun. 2006;7:1067. doi: 10.1016/j.catcom.2006.04.012. [DOI] [Google Scholar]
- 10.Sharghi, H., Hosseini-Sarvari, M., and Eskandari, R., Synthesis, 2006, p. 2047.
- 11.Kumar S., Reddy L., Shekhar C., Kumar Y., Kumar A., Singh B.K., Kumar V., Malhotra S., Pandey M.K., Jain R. Arch. Pharm. 2012;345:368. doi: 10.1002/ardp.201100279. [DOI] [PubMed] [Google Scholar]
- 12.Shankar B., Jalapathi P., Ramesh M., Kishore Kumar A., Ragavender M., Bharath G. Russ. J. Gen. Chem. 2016;86:1711. doi: 10.1134/S107036321607029X. [DOI] [Google Scholar]
- 13.Sharghi, H. and Hosseini-Sarvari, R.M., and Eskandari, Synthesis, 2006, p. 2047.
- 14.Hansen T.V., Wu P., Fokin V.V. J. Org. Chem. 2005;70:7761. doi: 10.1021/jo050163b. [DOI] [PubMed] [Google Scholar]
- 15.Akansha A., Rajesh Kumar P., Virendra K. Arab. J. Sci. Eng. 2022;47:347. doi: 10.1007/s13369-021-05377-1. [DOI] [Google Scholar]
- 16.Abhishek B., Venkataraghavan R., Vivek P., Ivo Romauld S. J. App. Pharm. Sci. 2019;9:21. doi: 10.7324/JAPS.2019.90503. [DOI] [Google Scholar]
- 17.Nagamani, M., Vishnu, T., Jalapathi, P., and Srinivas, M., J. Iran. Chem. Soc., 2021. 10.1007/s13738-021-02365-y