Abstract
Objective:
To evaluate sex differences in microRNA (miRNA) expression, anthropometric measures and cardiometabolic risk factors in Hispanic adolescents with obesity.
Methods:
Cross-sectional study of 68 (60% male) Hispanic adolescents with obesity, aged 13–17 years, recruited from a pediatric weight management clinic. We used small RNA sequencing to identify differentially expressed circulating miRNAs. We used Ingenuity Pathway Analysis and David bioinformatic resource tools to identify target genes for these miRNAs and enriched pathways. We used standard procedures to measure anthropometric and cardiometabolic factors.
Results:
We identified five miRNAs (miR-24–3p, miR-361–3p, miR-3605–5p, miR-486–5p and miR-199b-3p) that differed between females and males. miRNA targets-enriched pathways included PI3K-AKT, AMPK, insulin resistance, sphingolipid, TGF-beta, adipocyte lipolysis regulation and oxytocin signaling pathways. In addition, there were sex differences in blood pressure, skeletal muscle mass, lean body mass and percent body fat.
Conclusion:
We have identified sex differences in miRNA expression in Hispanic adolescents relevant to cardiometabolic health. Future studies should focus on sex-specific mechanistic roles of miRNAs on gene pathways associated with obesity pathophysiology to support development of precision cardiometabolic interventions.
INTRODUCTION
The prevalence of pediatric obesity and associated metabolic syndrome has steadily increased in the United States and worldwide over the past three decades, especially in adolescents. The trend in obesity prevalence has increased more dramatically in adolescents who are Hispanic compared to non-Hispanic 1. Obesity is a risk factor for associated comorbidities, including glucose intolerance, insulin resistance, type 2 diabetes mellitus, non-alcoholic fatty liver disease, and hypertension, particularly in adolescents 2, 3. Importantly, obesity during childhood may persist into adulthood increasing the risk of these comorbidities and overt cardiovascular disease in adulthood, compared to adults who did not have obesity 4, 5. All of these considerations increase the burden on healthcare systems in the short and long term, necessitating a mechanism to identify at-risk individuals to inform a targeted approach for early intervention.
Individual characteristics such as age, race, ethnicity, socioeconomic factors, genetic and environmental factors, including lifestyle and dietary habits, may contribute to the development of obesity. In addition, several studies have shown that microRNAs (miRNAs) are associated with pediatric obesity 6, 7. miRNAs are well known biomarkers and post-transcription regulators of gene expression. 8–11. Importantly, sex differences have been associated with the development of obesity and associated comorbidities, especially in adults 12–16. However, knowledge of sex differences in miRNAs expression relative to cardiometabolic risk factors in children is lacking, especially in the Hispanic population.
The goal of our cross-sectional study was to identify sex differences in miRNA expression and cardiometabolic risk factors in Hispanic adolescents with obesity in the Child Obesity Study at Children’s Hospital of San Antonio. We hypothesized that males and females will differ in their miRNA expression and cardiometabolic risk factors.
METHODS
The Baylor College of Medicine Institutional Review Board approved the study (IRB-H-40940). We obtained signed informed consent from the parents or guardians and assent from the participants. The work described herein has been carried out in accordance with The Code of Ethics of the World Medical Association (Declaration of Helsinki) for experiments involving humans.
This was a cross-sectional analysis of baseline data collected from participants enrolled in the Child Obesity Study. Baseline data were collected from the initial visit at a pediatric weight management clinic for Hispanic adolescents with obesity at Children’s Hospital of San Antonio. Criteria for inclusion in the study were age 13–17 years, self-reported Hispanic ethnicity and a body mass index ≥ 95th percentile for age and sex. Exclusion criteria included a diagnosis of type 2 diabetes mellitus, use of neurohormonal medications, concomitant chronic or acute illnesses by self-report and unavailability of blood samples for miRNA quantification.
A trained medical assistant measured anthropometrics at the pediatric weight management clinic. Height was measured using a digital stadiometer (BSM170; InBody, Cerritos, CA). We used electric impedance (Scale 570; InBody, Cerritos, CA) to measure weight, percent body fat, skeletal muscle mass, body fat mass and lean body mass. We used a Dinamap oscillometer (GE Healthcare, Milwaukee, WI) to measure blood pressure (BP) with an appropriately sized cuff on the participant’s right upper extremity. If the initial measurement was high, the BP measurement was repeated twice manually and we recorded the average.
Blood samples (3 ml) were drawn in red-top tubes after fasting for 12 hrs. Samples were centrifuged at 2500 rpm for 20 min to obtain serum. We measured serum total cholesterol, fasting blood glucose, high-density lipoprotein cholesterol and triglycerides using standard enzymatic methods with a fully automated analyzer. We used Friewald’s equation 17 to measure low-density lipoprotein cholesterol and high-performance liquid chromatography to measure glycosylated hemoglobin A1C. Additional measurements included blood insulin, fasting blood glucose, aspartate aminotransferase, alanine amino transferase and gamma-glutamyl transferase.
Whole blood (1–3 ml) was collected in Tempus Blood RNA Tube (Thermo Fisher Scientific, Waltham, MA), containing 6 ml of RNA stabilizer according to manufacturer’s protocol. The blood sample was mixed with the stabilizer by agitating the collection tube 8 times. The samples were stored in −80°C freeze.
We used small RNA sequencing to identify all miRNAs expressed in the blood samples. Briefly, total RNA was isolated from whole blood using Direct-zol RNA Kit (Zymo Research, Irvine, CA) according to the manufacturer’s protocol with some modifications. An appropriate amount of phosphate-buffered saline was added to the sample for a final 1:1 ratio of sample and stabilization reagent. The diluted sample was vortexed vigorously at for 30 sec. We then followed the manufacturer’s protocol. RNA was quantified using Qubit Fluorimeter (DeNovix, Wilmington, DE), and the quality assessed using TapeStation RNA ScreenTape and HS reagents (Agilent, Santa Clara, CA). The RNA samples were stored at −80°C.
We used NextFlex Small RNA-Seq Kit v3 (PerkinElmer, Waltham, MA) and 500 ng of total RNA to generate cDNA libraries. Following library amplification and the post-PCR cleanup, we assessed the quality of the libraries using TapeStation DNA ScreenTape and HS D1000 reagents (Agilent), and the quantity using KAPA® Library Quantification Kit (Roche Diagnostics Corp., Indianapolis, IN), following the manufacturer’s protocols. After normalization, we generated library two pools, 10 nM/L each, for sequencing.
We used Illumina’s reagents and instruments, including HiSeq Rapid Duo cBot Sample Loading Kit for loading sample pools in the Rapid Flow Cell, with duo lanes, for template hybridization and first extension on the cBot 2. Subsequently, we used HiSeq Rapid Cluster v2 and HiSeq Rapid SBS Kit v2 (50 cycles) for cluster generation and sequencing on a HiSeq 2500 instrument. Sequence clusters containing base calls and quality scores were streamed into Illumina’s BaseSpace Sequence Hub, where they were demultiplexed and converted to fastq files using bcl2fastq2 software.
We used the miRDeep2 pipeline 18, utilizing default settings; and miRbase v22 to analyze the fastq-formatted sequence reads and to identify all (known and novel) expressed miRNAs and their read counts. Read counts were normalized holistically using reads per million mapped to miRNA.
We used Partek Genomic Suite (PGS; Partek, Inc.) together with Prism 8 (GraphPad) for statistical analysis. We used analysis of variance tool embedded in PGS to compare differences in variable distribution between males and females. We set our two-tailed alpha at <0.05. P values were corrected using false discovery rate (15%). To identify definitive outliers in the data, we used robust outlier tool in Prism 8.
We used miRNA Target Filter Tool implemented in Ingenuity Pathway Analysis v01–16 (Qiagen, Germantown, MD) to identify putative miRNA targets. Then, we used the output, including miRNA targets predicated with high confidence and experimentally validated targets, to identify enriched pathways using David Bioinformatic Resources 6.8 19. P values were adjusted using Benjamini-Hochberg correction factor.
RESULTS
Sixty-eight participants (60% males) were included in the study. Mean age was 15 ±1.3 years with the mean body mass index percent of the 95th percentile being 137% (Table 1). Males had significantly higher mean skeletal muscle mass, lean body mass, systolic BP and diastolic BP and lower percent body fat compared to females (Figure 1). There were no other between-sex differences in the other cardiometabolic risk factors, demographics and anthropometric measures.
Table 1:
Factors showing gender differences in adolescents with obesity
| Factors | P-value | q-value* | Fold-Change (F vs. M) |
|---|---|---|---|
| hsa-miR-24–3p | 0.01840 | 0.03600 | −1.80 |
| hsa-miR-486–5p | 0.02370 | 0.04200 | 1.06 |
| hsa-miR-199b-3p | 0.02680 | 0.04800 | 1.65 |
| hsa-miR-3605–5p | 0.03820 | 0.05400 | −1.61 |
| hsa-miR-361–3p | 0.04330 | 0.06000 | −1.45 |
| Age | 0.65596 | 0.09141 | |
| SMM (Kg) | 0.00002 | 0.00600 | −1.25 |
| LEAN MASS (Kg) | 0.00002 | 0.01200 | −1.23 |
| Diastolic BP (mmHg) | 0.00016 | 0.01800 | −1.11 |
| Percent BF | 0.00018 | 0.02400 | 1.15 |
| Systolic BP (mmHg) | 0.00020 | 0.03000 | −1.08 |
| FBG | 0.07093 | 0.06600 | 2.81 |
| TG | 0.19698 | 0.07200 | −2.07 |
| LDL-C | 0.22645 | 0.07800 | −2.05 |
| TC | 0.24412 | 0.08400 | −1.65 |
| VO2MAX | 0.27275 | 0.09000 | 1.49 |
| HDL | 0.28104 | 0.09600 | −3.29 |
| Ins | 0.35796 | 0.10200 | −2.30 |
| GGT | 0.49709 | 0.10800 | −1.82 |
| CRP | 0.60658 | 0.11400 | 1.50 |
| nHDL | 0.74726 | 0.12600 | −1.18 |
| A1C | 0.74938 | 0.13200 | 1.52 |
| ALT | 0.75742 | 0.13800 | 1.27 |
| AST | 0.79870 | 0.14400 | −1.28 |
| BMI PERCENTILE | 0.86143 | 0.15000 | 1.01 |
FDR=0.15
Figure 1:
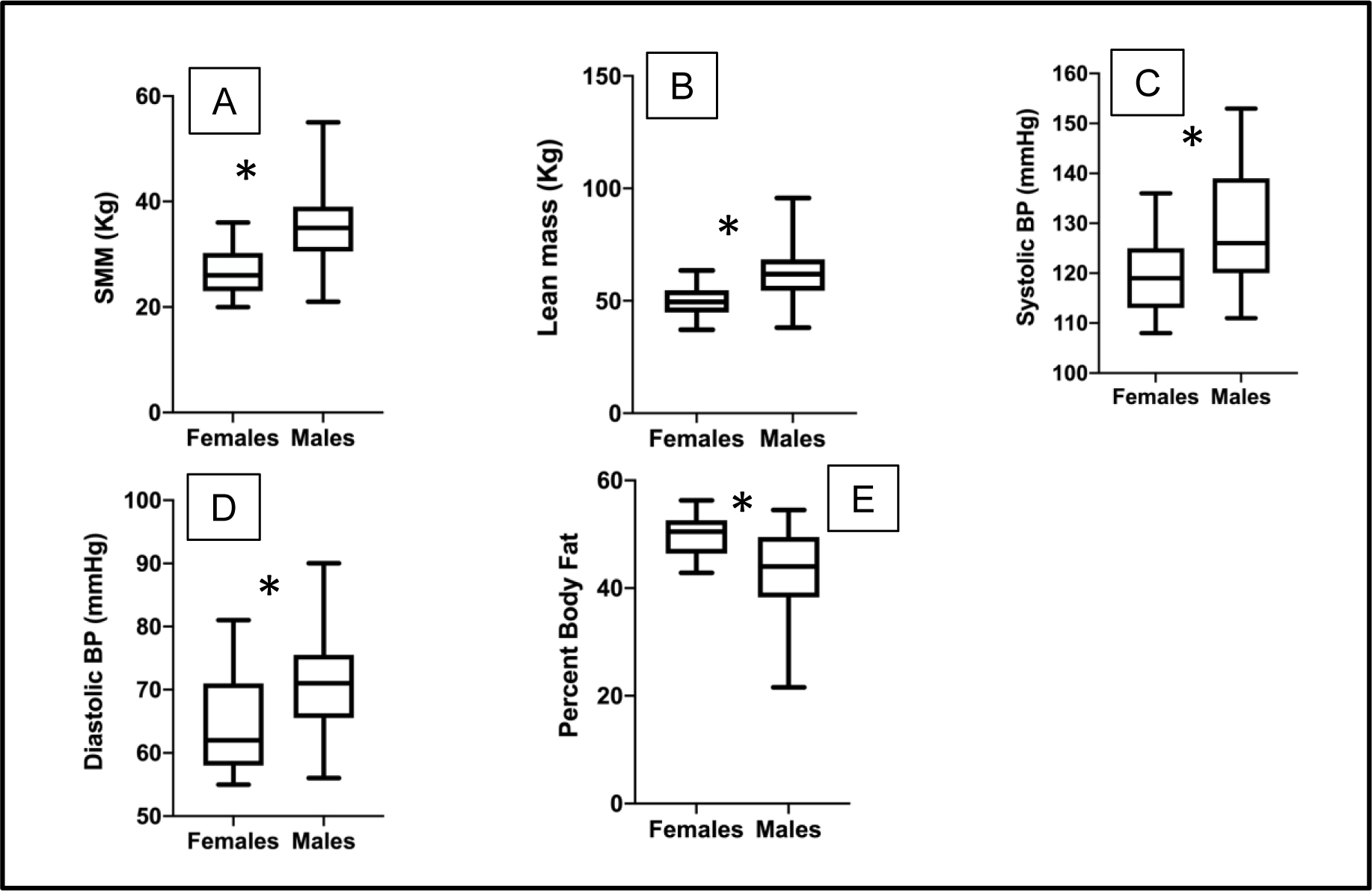
Box plots showing the distribution and mean of A. SMM (skeletal muscle mass), B. lean body mass, C. systolic BP, D. diastolic BP, and E. percent body fat for female compared to male adolescents with obesity. The boxes represent interquartile range and whiskers represent minimum and maximum values. * represents p values < 0.05.
Using small RNA sequencing, we identified 203 miRNAs expressed in blood with a minimum of 250 read counts. There was disparity in expression of five miRNAs between males and females (Table 1). The expression of miR-24–3p, miR-361–3p and miR-3605–5p was downregulated in females and upregulated in males while miR-486–5p and miR-199b-3p was upregulated in females and downregulated in males. After p value correction, the expression of miR-24–3p, miR-486–5p and miR-199b-3p remained significantly different while miR-361–3p and miR-3605–5p were statistically not significance. According to miRbase version 22, miR-361–3p is transcribed on the X-chromosome, miR-486–5p and miR-24–3p on chromosome 8 and miR-199b-3p on chromosome 2.
We identified a number of putative targets of the miRNAs that were differentially expressed between females and males (Table 2). Interestingly, we observed that there were no experimentally validated targets for two of the miRNAs (miR-3605–5p and miR-361–3p). Table 3 (online only) shows the gene symbols of the miRNA targets.
Table 2:
miRNA targets
| Candidate MicroRNAs | All targets (with low and high prediction confidence and with experimental evidence) | Targets with high prediction confidence and with experimental evidence | Targets with experimental evidence alone |
|---|---|---|---|
| miR-24–3p | 1,118 | 104 | 22 |
| miR-199b-3p | 623 | 66 | 11 |
| miR-486–5p | 423 | 37 | 4 |
| miR-3605–5p | 866 | 46 | 0 |
| miR-361–3p | 1,471 | 158 | 0 |
Table 3.
(Online only): MicroRNAs predicted targets
| hsa-miR-199b-3p | hsa-miR-24–3p | hsa-miR-3605–5p | hsa-miR-361–3p | hsa-miR-486–5p |
|---|---|---|---|---|
| ADAM22 | ABCB9 | ABLIM1 | AATK | ARID4B |
| ADAMTS3 | ACVR1B | ADORA1 | ABI1 | BCAS2 |
| AK4 | ADD2 | ADRB3 | ADGRG3 | CD247 |
| AKAP9 | ADORA1 | AK4 | AJUBA | CDKN2B |
| BCAR3 | AGPAT1 | ALOXE3 | AP2A1 | CRLS1 |
| CBLB | AGPAT3 | AMHR2 | APLN | CSPG5 |
| CD2AP | ARHGEF15 | ATP5F1C | APOL3 | CSTF1 |
| CD44 | ATG4A | BHMT2 | APOL5 | CTSK |
| CDK17 | ATP6V0E2 | CACNG3 | AQP7 | DHFR2 |
| CDK7 | AURKB | CHMP1A | ARF3 | DNAJC19 |
| CELSR2 | BAX | CHRM1 | ARHGEF1 | EHHADH |
| CFL2 | BBC3 | CTRC | ARPC5L | FGF7 |
| CHKA | BCL2L11 | CX3CL1 | ASH2L | FOXO1 |
| CHRM3 | BRCA1 | CXCL9 | ATP2A3 | GPR78 |
| CNOT7 | CALCR | CYGB | BAK1 | GPX8 |
| COL4A5 | CCNA2 | CYP19A1 | CACNG6 | KLK2 |
| COPS2 | CDK1 | CYP3A4 | CACNG8 | LILRB4 |
| CREB1 | CDK4 | DDT | CAMK2B | LMTK2 |
| CREBZF | CDKN1B | DHRS3 | CAMKK1 | MDH1 |
| CXCL11 | CDKN2A | ECHS1 | CAVIN1 | MRAS |
| EIF3M | CHST4 | EIF2AK1 | CBX7 | NEK2 |
| FCGR3A/FCGR3B | CISH | ENTPD3 | CCL18 | PARP2 |
| FGF7 | COX6B2 | FAS | CCN2 | PDGFC |
| FN1 | CRH | FFAR3 | CD40 | PIM1 |
| GIP | CTSD | GJB3 | CD79A | PLA1A |
| GNA12 | CXCR2 | GNG2 | CDKN2D | PTEN |
| GPAT3 | CYP46A1 | GSTA4 | CEBPA | PTGDR |
| GREM1 | DHFR | HACD2 | CELSR3 | PYCR2 |
| GRK3 | DNAJB12 | HCRTR1 | CEMIP2 | RCOR3 |
| ITGB8 | DNAJB2 | HIBADH | CHD4 | RFC2 |
| ITPK1 | DNAJC16 | HMGA1 | CHMP1A | SELENOT |
| KTN1 | DOT1L | IGBP1 | CLDN1 | SLC38A1 |
| LPAR4 | DUSP1 | KLK5 | CXCR5 | SNRPD1 |
| LPAR5 | E2F2 | NDRG1 | CYP19A1 | TOB1 |
| MAP3K4 | EIF3I | NRG3 | DDR1 | TWF1 |
| MET | ENTPD6 | PARD6G | DLL3 | VTI1A |
| MTOR | FASLG | PPM1A | DNAJB2 | ZNF331 |
| NLK | FBLIM1 | PRKACG | DNAJB5 | |
| PAK4 | FEN1 | RHOA | DTX4 | |
| PAWR | FGF11 | RPS16 | DUSP15 | |
| PIK3CB | FMO1 | SRR | DUSP2 | |
| PLEKHA3 | FST | STXBP1 | EEF1AKMT3 | |
| PNRC1 | FURIN | TEAD1 | EFNA4 | |
| PPP1R1C | GJD2 | UCKL1 | EFNA5 | |
| PPP2R2A | GNE | UGT2B11 | ELK1 | |
| PPP2R5E | GPAT3 | ZWILCH | ENO2 | |
| PRDX6 | GSTM5 | ENSA | ||
| PTGS2 | H2AX | EPHA10 | ||
| RNGTT | HIP1R | ETV4 | ||
| RORB | HNF1B | EXTL3 | ||
| RPS6KA6 | HNRNPA1 | FADS6 | ||
| RUNX1 | IFNG | FGF4 | ||
| SCD | IL10RB | FKBP4 | ||
| SDC2 | IL1A | FOXH1 | ||
| SERPINE2 | INMT | FRAT1 | ||
| SMAD1 | IRF1 | GAB2 | ||
| SUZ12 | LAMB3 | GGCT | ||
| SYT16 | LILRA6 | GGT1 | ||
| TFAM | LIPT2 | GGT5 | ||
| TMSB10/TMSB4X | LPAR6 | GJC2 | ||
| UBE2W | MAP2K4 | GJD2 | ||
| UBQLN1 | MAPK14 | GLI1 | ||
| UPRT | MOGAT3 | GNA12 | ||
| VAMP3 | MPI | GPR37L1 | ||
| VCAN | MT-ND4L | GPR89A/GPR89B | ||
| YES1 | MT1E | GUCA1A | ||
| MT1M | H3–3A/H3–3B | |||
| MYC | HAP1 | |||
| NDST1 | HBB | |||
| NEFM | HEXD | |||
| NFKBIE | HOXA10 | |||
| NOTCH1 | HOXB1 | |||
| PCYOX1 | IHH | |||
| PDGFRB | IL13 | |||
| PER2 | IL1RL2 | |||
| PIM2 | IL21R | |||
| PMAIP1 | IL31 | |||
| POLR3D | IL36RN | |||
| PPARG | IP6K2 | |||
| PPCS | ISY1-RAB43 | |||
| PRKCH | ITGB6 | |||
| PRSS8 | ITPR2 | |||
| PSMB8 | IVD | |||
| RAB4B | KCNJ4 | |||
| RAP1A | KCNJ6 | |||
| RAP1B | KHK | |||
| RASD2 | KLK2 | |||
| RNF2 | KRT2 | |||
| RPL36 | LCN6 | |||
| SCP2 | LIMK2 | |||
| SIRPA | LPIN3 | |||
| SMAD3 | MAFK | |||
| SMAD4 | MAP3K10 | |||
| SMAD5 | MEF2D | |||
| SPN | MEX3B | |||
| SSR1 | MKNK2 | |||
| TNFSF9 | MMP24 | |||
| TPSAB1/TPSB2 | MPRIP | |||
| TPSD1 | MRM2 | |||
| TRIB3 | MTNR1B | |||
| UBE2D4 | NCOA2 | |||
| UCK1 | NDST1 | |||
| USP18 | NDUFS7 | |||
| WNT8B | NFIC | |||
| NLRC5 | ||||
| P2RX6 | ||||
| P2RY2 | ||||
| PADI1 | ||||
| PALM2AKAP2 | ||||
| PAX5 | ||||
| PEBP1 | ||||
| PHF1 | ||||
| PIAS4 | ||||
| PIM2 | ||||
| PIP4K2C | ||||
| PLA2G2C | ||||
| PLK5 | ||||
| PODXL | ||||
| POLR2D | ||||
| PPP2R5B | ||||
| PRSS33 | ||||
| PSAP | ||||
| PSMD2 | ||||
| PSPN | ||||
| PTAFR | ||||
| RAD9A | ||||
| RAP2B | ||||
| RELA | ||||
| RGS21 | ||||
| RHO | ||||
| RORC | ||||
| RPS21 | ||||
| RPS6KB2 | ||||
| SDR9C7 | ||||
| SH3GL3 | ||||
| SNRPD3 | ||||
| SOST | ||||
| SRF | ||||
| SSTR5 | ||||
| SYMPK | ||||
| TFE3 | ||||
| TFR2 | ||||
| TFRC | ||||
| THRA | ||||
| TNF | ||||
| TNNC1 | ||||
| TNNI1 | ||||
| TOMM20 | ||||
| TRAF3 | ||||
| TSPAN1 | ||||
| U2AF2 | ||||
| UBASH3B | ||||
| UBE2V1 | ||||
| VASP | ||||
| WASF2 | ||||
| ZBP1 | ||||
| ZBTB16 | ||||
| ZFP36 | ||||
For each miRNA, we identified pathways, annotated in Kyoto Encyclopedia of Genes and Genomes that were enriched in the list of miRNA targets that were predicted with high confidence and experimentally validated (Tables 4, 5 and 6; all online only). For miR-199b-3p, enriched pathways for phosphatidylinositol 3-kinase-protein kinase B (PI3K-AKT) signaling pathway and pathways associated with cancer had the greatest statistical significance and remained significant after p-value correction. Other enriched pathways for miR-199b-3p targets included the 5’ AMP-activated protein kinase (AMPK), insulin resistance and sphingolipid signaling pathways. For miR-24–3p targets, we observed the following enriched pathways: cell cycle, cancer, hepatitis B and TGF-beta signaling pathways. Enriched pathways for miR-3605–5p and miR-361–3p targets included adipocyte lipolysis regulation and oxytocin signaling pathways, respectively.
Table 4.
(online only): Enriched KEGG pathways associated with miR-199b-3p
| miR-199b-3p | ||||
|---|---|---|---|---|
| KEGG pathways | # Genes involved | Fold Enrichment | P-Value | Benjamini correction |
| PI3K-Akt signaling pathway | 12 | 5.20 | 0.00001 | 0.00139 |
| Pathways in cancer | 12 | 4.57 | 0.00003 | 0.00236 |
| Regulation of actin cytoskeleton | 8 | 5.70 | 0.00038 | 0.01848 |
| AMPK signaling pathway | 6 | 7.29 | 0.00116 | 0.04209 |
| Bacterial invasion of epithelial cells | 5 | 9.59 | 0.00160 | 0.04635 |
| Proteoglycans in cancer | 7 | 5.23 | 0.00177 | 0.04281 |
| Focal adhesion | 6 | 4.36 | 0.01059 | 0.20159 |
| Small cell lung cancer | 4 | 7.04 | 0.01779 | 0.28258 |
| ECM-receptor interaction | 4 | 6.88 | 0.01892 | 0.26957 |
| ErbB signaling pathway | 4 | 6.88 | 0.01892 | 0.26957 |
| Insulin resistance | 4 | 5.54 | 0.03316 | 0.39289 |
| Sphingolipid signaling pathway | 4 | 4.98 | 0.04322 | 0.44816 |
Table 5.
(online only): Enriched KEGG pathways associated with miR-24–3p
| miR-24–3p | ||||
|---|---|---|---|---|
| KEGG pathway | # genes involved | Fold Enrichment | P-Value | Benjamini correction |
| Cell cycle | 9 | 5.61 | 0.0002 | 0.0290 |
| Pathways in cancer | 15 | 2.95 | 0.0004 | 0.0351 |
| Hepatitis B | 9 | 4.80 | 0.0005 | 0.0281 |
| TGF-beta signaling pathway | 7 | 6.44 | 0.0007 | 0.0286 |
| p53 signaling pathway | 6 | 6.92 | 0.0016 | 0.0530 |
| Chronic myeloid leukemia | 6 | 6.44 | 0.0022 | 0.0607 |
| Epstein-Barr virus infection | 7 | 4.43 | 0.0046 | 0.1063 |
| MAPK signaling pathway | 10 | 3.06 | 0.0048 | 0.0988 |
| HTLV-I infection | 10 | 3.04 | 0.0050 | 0.0905 |
| Osteoclast differentiation | 7 | 4.13 | 0.0064 | 0.1052 |
| Signaling pathways regulating pluripotency of stem cells | 7 | 3.86 | 0.0088 | 0.1294 |
| Pancreatic cancer | 5 | 5.95 | 0.0094 | 0.1260 |
| Melanoma | 5 | 5.44 | 0.0127 | 0.1553 |
| Bladder cancer | 4 | 7.54 | 0.0152 | 0.1715 |
| Neurotrophin signaling pathway | 6 | 3.86 | 0.0184 | 0.1922 |
| Small cell lung cancer | 5 | 4.55 | 0.0231 | 0.2221 |
| Measles | 6 | 3.49 | 0.0274 | 0.2448 |
| FoxO signaling pathway | 6 | 3.46 | 0.0282 | 0.2388 |
| PI3K-Akt signaling pathway | 10 | 2.24 | 0.0315 | 0.2517 |
| Glycerolipid metabolism | 4 | 5.33 | 0.0377 | 0.2814 |
| Chagas disease (American trypanosomiasis) | 5 | 3.72 | 0.0438 | 0.3069 |
| Colorectal cancer | 4 | 4.99 | 0.0446 | 0.2998 |
Table 6.
(online only): Enriched KEGG pathways associated with miR-3605–5p, miR-361–3p and miR-486–5p
| miR-3605–5p | ||||
| KEGG pathway | # genes involved Count | Fold Enrichment | P-Value | Benjamini correction |
| Chemokine signaling pathway | 5.00 | 5.78 | 0.0092 | 0.7048 |
| Drug metabolism - other enzymes | 3.00 | 14.02 | 0.0180 | 0.6982 |
| Regulation of lipolysis in adipocytes | 3.00 | 11.52 | 0.0260 | 0.6866 |
| Steroid hormone biosynthesis | 3.00 | 11.12 | 0.0278 | 0.6054 |
| Retinol metabolism | 3.00 | 10.08 | 0.0333 | 0.5913 |
| Renin secretion | 3.00 | 10.08 | 0.0333 | 0.5913 |
| Drug metabolism - cytochrome P450 | 3.00 | 9.48 | 0.0372 | 0.5661 |
| Metabolism of xenobiotics by cytochrome P450 | 3.00 | 8.71 | 0.0434 | 0.5671 |
| miR-361–3p | ||||
| KEGG pathway | # genes involved Count | Fold Enrichment | P-Value | Benjamini correction |
| Oxytocin signaling pathway | 8 | 3.67 | 0.0057 | 0.6539 |
| Fc gamma R-mediated phagocytosis | 6 | 4.91 | 0.0070 | 0.4808 |
| Acute myeloid leukemia | 5 | 6.14 | 0.0084 | 0.4060 |
| Transcriptional misregulation in cancer | 8 | 3.30 | 0.0100 | 0.3747 |
| MAPK signaling pathway | 10 | 2.72 | 0.0103 | 0.3202 |
| HIF-1 signaling pathway | 6 | 4.30 | 0.0122 | 0.3155 |
| Inflammatory bowel disease | 5 | 5.37 | 0.0133 | 0.2987 |
| Hypertrophic cardiomyopathy | 5 | 4.41 | 0.0256 | 0.4527 |
| Regulation of actin cytoskeleton | 8 | 2.62 | 0.0312 | 0.4808 |
| Dilated cardiomyopathy | 5 | 4.09 | 0.0325 | 0.4586 |
| NF-kappa B signaling pathway | 5 | 3.95 | 0.0363 | 0.4644 |
| miR-486–5p | ||||
| KEGG pathway | # genes involved Count | Fold Enrichment | P-Value | Benjamini correction |
| Melanoma | 3 | 10.77 | 0.0291 | 0.9138 |
| Prostate cancer | 3 | 8.69 | 0.0431 | 0.8394 |
DISCUSSION
We observed sex differences in miRNA expression pattern, cardiometabolic risk factors and anthropometric measures in a cohort of Hispanic adolescents with obesity. miRNA expression patterns can inform our knowledge of the diverse mechanisms involved in cellular metabolism and obesity pathophysiology as well as genetic factors that may contribute to obesity and related sex differences.
Our study identified five miRNAs (miR-199a-3p, miR-486–5p, miR-361–3p, miR-3605–5p and miR-24–3p) that were expressed differentially between Hispanic adolescent males and females with obesity. Previous studies have reported sex differences in miRNA gene regulation 20. Other studies have suggested that miRNAs transcribed on the X-chromosome may escape inactivation leading to suppression of the genes involved in lipid metabolism 21, 22. Moreover, studies have shown that the number of activated X-chromosomes in cells may explain mechanisms underlying sex differences in pathophysiologic processes 23, 24. One of the five miRNAs identified in our study (miR-361–3p) is transcribed on the X-chromosome. Future studies will explore whether this miRNA escapes inactivation and the implications for obesity in adolescents. We conclude that sex differences in expression of miRNAs observed in our study may be one mechanism underlying sex differences in cardiometabolic disorders in adolescents with obesity.
We have identified genes predicted to be targeted by the miRNAs exhibiting sex differences. Many of these predicted miRNA targets have been validated experimentally, according to miRTarBase, TarBase and miRecords databases. However, we did not find statistically significant validated targets for miR-3605–5p, miR-361–3p and miR-486–5p. Future studies will focus on validation of these targets and their mechanistic effect on sex differences in obesity. This is important because miRNAs fine-tune gene expression, and understanding their effects on cell physiology may have clinical implications that can uncover novel therapeutic targets for prevention and treatment in individuals at high risk of obesity and its cardiometabolic complications.
We identified several intriguing miRNA targets-enriched pathways relevant to obesity and development of hypertension, diabetes and cardiovascular disease, including PI3K-AKT and AMPK pathways. The PI3K-AKT pathway is enriched in miR-199b-3p targets and is involved in promotion of cell proliferation, cell survival, growth and angiogenesis in response to extracellular stimuli such as insulin. Interestingly, there is a large body of evidence linking miR-199b-3p with obesity and associated comorbidities 25–28. This miRNA was upregulated in fat-exposed hepatocytes derived from human fetal brains in females compared to males. In addition, the PI3K-AKT pathway was found to be enriched in targets of differentially expressed miRNAs 25. A previous study revealed that miR-26b modulates PI3K-AKT pathway by targeting the phosphatase and tension homolog gene to promote glucose uptake by adipocytes and insulin sensitivity 29.
Enriched pathways associated with miR-361–3p included the oxytocin pathway. Previous studies have reported that plasma oxytocin concentration is associated with obesity and diabetes 30, 31, and animal and cellular studies have unraveled the protective effect of oxytocin on metabolic outcomes 32. However, estrogen influences the effects of oxytocin-mediated reduction in chow diet intake to the extent that the effect is more pronounced in male than female rats 33.
Previous studies in animal models have investigated the mechanisms underlying the influence of sex differences on body fat. It is well documented that sex hormones and their receptors, including estrogen progesterone and androgens have profound effects on adipose tissue and exhibit sex differences 13. A previous study described the mechanistic role of miR-22 in modulating sex-specific lipid metabolism and body fat 34. MiR-22 suppressed expression of estrogen receptor alpha leading to decreased lipid metabolism and fatty acid oxidation and ultimately visceral white fat accumulation in male mice. In females, estrogen receptor alpha promotes self-activation by binding to the miR-22 precursor to inhibit mature miRNA processing. This observation indicates that miRNAs have the ability to modulate sex-specific pathophysiology processes.
There are limitations of the present study that should be considered. First, we did not investigate the association of sex hormones with the observed sex differences, in order to confirm their role in this relationship. Second, our findings are based only on Hispanic adolescents with obesity and we did not recruit adolescents without obesity, younger children, or children from other racial and ethnic groups for comparison, which limits our finding’s generalizability. Third, our sample size is small and may have limited our power to detect differences in other traits. Even though multiple testing correction was performed, type-I error is plausible. Finally, we did not fully account for several sources of bias, including confounding bias. There is a need to validate our findings in a longitudinal study of a larger multiethnic cohort of adolescents with obesity with a comparator group without obesity and using analytic methods to account for bias. In conclusion we identified sex differences in miRNA expression that are relevant to key cardiometabolic pathways in this study of Hispanic adolescents with obesity.
ACKNOWLEDGEMENT
We acknowledge the following persons for their contribution to the study: Jeremy Glen and Clint Christensen of the Texas Biomedical Research Institute for RNA isolation and cDNA library generation. Julee Carlton of the Children’s Hospital San Antonio for clinical data collection.
Funding source:
This study was supported by the Texas Biomedical Research Institute Pilot grant (17-04625) to GMK and SC, Texas Biomedical Research Institute Healthy Babies Project to ACB and LAC, and the National Institutes of Health (K01 HL130697) to GMK.
Footnotes
Conflict of interest: None
REFERENCES
- [1].Ogden CL, Fryar CD, Martin CB, Freedman DS, Carroll MD, Gu Q, Hales CM: Trends in Obesity Prevalence by Race and Hispanic Origin-1999–2000 to 2017–2018. JAMA 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Anderson EL, Howe LD, Fraser A, Callaway MP, Sattar N, Day C, Tilling K, Lawlor DA: Weight trajectories through infancy and childhood and risk of non-alcoholic fatty liver disease in adolescence: the ALSPAC study. J Hepatol 2014, 61:626–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Sinha R, Fisch G, Teague B, Tamborlane WV, Banyas B, Allen K, Savoye M, Rieger V, Taksali S, Barbetta G, Sherwin RS, Caprio S: Prevalence of impaired glucose tolerance among children and adolescents with marked obesity. N Engl J Med 2002, 346:802–10. [DOI] [PubMed] [Google Scholar]
- [4].Malhotra S, Sivasubramanian R, Singhal V: Adult obesity and its complications: a pediatric disease? Curr Opin Endocrinol Diabetes Obes 2021, 28:46–54. [DOI] [PubMed] [Google Scholar]
- [5].Weihe P, Spielmann J, Kielstein H, Henning-Klusmann J, Weihrauch-Bluher S: Childhood Obesity and Cancer Risk in Adulthood. Curr Obes Rep 2020, 9:204–12. [DOI] [PubMed] [Google Scholar]
- [6].Oses M, Margareto Sanchez J, Portillo MP, Aguilera CM, Labayen I: Circulating miRNAs as Biomarkers of Obesity and Obesity-Associated Comorbidities in Children and Adolescents: A Systematic Review. Nutrients 2019, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Prats-Puig A, Grau-Cabrera P, Riera-Perez E, Cortes-Marina R, Fortea E, Soriano-Rodriguez P, de Zegher F, Ibanez L, Bassols J, Lopez-Bermejo A: Variations in the obesity genes FTO, TMEM18 and NRXN3 influence the vulnerability of children to weight gain induced by short sleep duration. Int J Obes (Lond) 2013, 37:182–7. [DOI] [PubMed] [Google Scholar]
- [8].Karere G, Glenn J, Galindo S, Garcia R, Chevalier F, Dick E, Cox L: A Six-Microrna Panel Identified as a Potential Biomarker for Early-Stage Atherosclerotic Lesions. Arterioscl Throm Vas 2017, 37. [Google Scholar]
- [9].Karere GM, Glenn JP, Birnbaum S, Garcia R, VandeBerg JL, Cox LA: Identification of coordinately regulated microRNA-gene networks that differ in baboons discordant for LDL-cholesterol. PloS one 2019, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Karere GM, Glenn JP, Birnbaum S, Kuhn N, Lange K, Christensen C, Rice K, Mahaney M, Havill L, VandeBerg JL, Cox LA: A Potential MicroRNA Biomarker for Atherosclerotic Lesions in Baboons. Circ Res 2013, 113.23048070 [Google Scholar]
- [11].Karere GM, Glenn JP, VandeBerg JL, Cox LA: Differential microRNA response to a high-cholesterol, high-fat diet in livers of low and high LDL-C baboons. Bmc Genomics 2012, 13:320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Calcaterra V, Larizza D, De Silvestri A, Albertini R, Vinci F, Regalbuto C, Dobbiani G, Montalbano C, Pelizzo G, Cena H: Gender-based differences in the clustering of metabolic syndrome factors in children and adolescents. J Pediatr Endocrinol Metab 2020, 33:279–88. [DOI] [PubMed] [Google Scholar]
- [13].Chang E, Varghese M, Singer K: Gender and Sex Differences in Adipose Tissue. Curr Diab Rep 2018, 18:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Sabbatini AR, Kararigas G: Estrogen-related mechanisms in sex differences of hypertension and target organ damage. Biol Sex Differ 2020, 11:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].van Mil SR, Biter LU, van de Geijn GJM, Birnie E, Dunkelgrun M, Ijzermans JNM, van der Meulen N, Mannaerts GHH, Castro Cabezas M: The effect of sex and menopause on carotid intima-media thickness and pulse wave velocity in morbid obesity. Eur J Clin Invest 2019, 49:e13118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Varghese M, Griffin C, Singer K: The Role of Sex and Sex Hormones in Regulating Obesity-Induced Inflammation. Adv Exp Med Biol 2017, 1043:65–86. [DOI] [PubMed] [Google Scholar]
- [17].Friedewald WT, Levy RI, Fredrickson DS: Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem 1972, 18:499–502. [PubMed] [Google Scholar]
- [18].Friedlander MR, Mackowiak SD, Li N, Chen W, Rajewsky N: miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic acids research 2012, 40:37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Huang da W, Sherman BT, Lempicki RA: Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009, 4:44–57. [DOI] [PubMed] [Google Scholar]
- [20].Morgan CP, Bale TL: Sex differences in microRNA regulation of gene expression: no smoke, just miRs. Biol Sex Differ 2012, 3:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Berletch JB, Yang F, Disteche CM: Escape from X inactivation in mice and humans. Genome Biol 2010, 11:213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Yang F, Babak T, Shendure J, Disteche CM: Global survey of escape from X inactivation by RNA-sequencing in mouse. Genome Res 2010, 20:614–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Itoh Y, Melamed E, Yang X, Kampf K, Wang S, Yehya N, Van Nas A, Replogle K, Band MR, Clayton DF, Schadt EE, Lusis AJ, Arnold AP: Dosage compensation is less effective in birds than in mammals. J Biol 2007, 6:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lopes GB, Matos CM, Leite EB, Martins MT, Martins MS, Silva LF, Robinson BM, Port FK, James SA, Lopes AA: Depression as a potential explanation for gender differences in health-related quality of life among patients on maintenance hemodialysis. Nephron Clin Pract 2010, 115:c35–40. [DOI] [PubMed] [Google Scholar]
- [25].Joshi A, Azuma R, Akumuo R, Goetzl L, Pinney SE: Gestational diabetes and maternal obesity are associated with sex-specific changes in miRNA and target gene expression in the fetus. Int J Obes (Lond) 2020, 44:1497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Nesca V, Guay C, Jacovetti C, Menoud V, Peyot ML, Laybutt DR, Prentki M, Regazzi R: Identification of particular groups of microRNAs that positively or negatively impact on beta cell function in obese models of type 2 diabetes. Diabetologia 2013, 56:2203–12. [DOI] [PubMed] [Google Scholar]
- [27].Shi C, Zhang M, Tong M, Yang L, Pang L, Chen L, Xu G, Chi X, Hong Q, Ni Y, Ji C, Guo X: miR-148a is Associated with Obesity and Modulates Adipocyte Differentiation of Mesenchymal Stem Cells through Wnt Signaling. Sci Rep 2015, 5:9930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wang J, Yang X: The function of miRNA in cardiac hypertrophy. Cell Mol Life Sci 2012, 69:3561–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Xu G, Ji C, Song G, Zhao C, Shi C, Song L, Chen L, Yang L, Huang F, Pang L, Zhang N, Zhao Y, Guo X: MiR-26b modulates insulin sensitivity in adipocytes by interrupting the PTEN/PI3K/AKT pathway. Int J Obes (Lond) 2015, 39:1523–30. [DOI] [PubMed] [Google Scholar]
- [30].Drusco A, Bottoni A, Lagana A, Acunzo M, Fassan M, Cascione L, Antenucci A, Kumchala P, Vicentini C, Gardiman MP, Alder H, Carosi MA, Ammirati M, Gherardi S, Luscri M, Carapella C, Zanesi N, Croce CM: A differentially expressed set of microRNAs in cerebro-spinal fluid (CSF) can diagnose CNS malignancies. Oncotarget 2015, 6:20829–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Weingarten MFJ, Scholz M, Wohland T, Horn K, Stumvoll M, Kovacs P, Tonjes A: Circulating Oxytocin Is Genetically Determined and Associated With Obesity and Impaired Glucose Tolerance. J Clin Endocrinol Metab 2019, 104:5621–32. [DOI] [PubMed] [Google Scholar]
- [32].Jankowski M, Broderick TL, Gutkowska J: Oxytocin and cardioprotection in diabetes and obesity. BMC Endocr Disord 2016, 16:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Liu CM, Davis EA, Suarez AN, Wood RI, Noble EE, Kanoski SE: Sex Differences and Estrous Influences on Oxytocin Control of Food Intake. Neuroscience 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Schweisgut J, Schutt C, Wust S, Wietelmann A, Ghesquiere B, Carmeliet P, Drose S, Korach KS, Braun T, Boettger T: Sex-specific, reciprocal regulation of ERalpha and miR-22 controls muscle lipid metabolism in male mice. The EMBO journal 2017, 36:1199–214. [DOI] [PMC free article] [PubMed] [Google Scholar]


