Abstract
C-reactive protein (CRP) is an important biomarker of infection and inflammation, as CRP is one of the most prominent acute-phase proteins. CRP is usually detected using anti-CRP antibodies (Abs), where the intermolecular interactions between CRP and the anti-CRP Ab are largely affected by the pH and ionic strength of environmental solutions. Therefore, it is important to understand the environmental effects of CRP–anti-CRP Ab interactions when designing highly sensitive biosensors. Here, we investigated the efficiency of fluorescently labeled CRP–anti-CRP monoclonal antibody (mAb) interactions at different pHs and ionic strengths. Our results indicate that the affinity was insensitive to pH changes in the range of 5.9 to 8.1, while it was significantly sensitive to ionic strength changes. The binding affinity decreased by 55% at an ionic strength of 1.6 mM, when compared to that under a physiological condition (~150 mM). Based on the isoelectric focusing results, both the labeled CRP and anti-CRP mAb were negatively charged in the studied pH range, which rendered the system insensitive to pH changes, but sensitive to ionic strength changes. The decreased ionic strength led to a significant enhancement of the repulsive force between CRP and the anti-CRP mAb. Although the versality of the findings is not fully studied yet, the results provide insights into designing highly sensitive CRP sensors, especially field-effect transistor-based sensors.
Keywords: ionic strength dependence, pH dependence, fluorescence, binding affinity, surface charge
Significance
As C-reactive protein (CRP) is an important biomarker of infection and inflammation, demand exists for highly sensitive, rapid, and low-cost immunosensors. When designing biosensors, understanding the environmental effects such as pH and ionic strength change is important, because they significantly affect the binding affinity. The results of this study are useful for developing CRP sensors.
Introduction
C-reactive protein (CRP) is regarded as a universal biomarker for numerous diseases (including cardiovascular diseases and disorders) and an early indicator of infectious or inflammatory conditions [1]. In normal healthy individuals, CRP is present in the plasma with an average concentration of 0.8 μg/mL (6.8 nM) [2]. CRP levels have been categorized as being (i) low (<1.0 μg/mL), (ii) middle range (1.0 to 3.0 μg/mL), and (iii) high (>3.0 μg/mL) [2]. The CRP concentration sometimes increases to 500 μg/mL in the acute phase of the diseases [2]. Thus, CRP has emerged as an informative biomarker in humans for the early detection and accurate implementation of therapeutic interventions.
Conventional CRP-detection assays are based on turbidimetry [3], where latex particles that are covalently coated with F(ab’)2 fragments of anti-CRP antibody (Ab) aggregate if CRP is present, resulting in increased turbidity. Subsequently, lateral flow assays [4,5], enzyme-linked immunosorbent assays [6], and fluorescent assays [7] have been developed as novel methods. Recently, label-free methods using field-effect transistors (FETs) [8], surface plasmon resonance [9], and quartz crystal microbalance [10] have been used to detect CRP. In particular, great progress has been made in designing and fabricating FETs using nanomaterials, including Si nanowires [8,11,12], carbon nanotubes [13], and graphene [14–16], leading to a sensitive and rapid analysis with miniaturized and integrated sensor platforms [17,18].
However, the performance of FET-based biosensors largely depends on environmental factors, such as the pH and ionic strength. The pH of a solution drastically changes the signals of FET-based sensors [12] because the sensors respond to the molecular charges of analytes that are determined by the solution pH and the isoelectric point (pI) of target molecules. Ionic strength also affects the signal-to-noise ratio. Under physiological salt conditions, where the ionic strength is approximately 150 mM, the Debye length λD, (the distance at which a unit charge is screened by 63%) is near 0.8 nm. Therefore, physiological salt conditions may be suboptimal for detecting target molecules with FET-based sensors, as the λD is shorter than the size of an Ab (~15 nm). Indeed, some reports showed that molecules bound to Abs were not detected under physiological conditions [19,20], although they were detected at a lower ionic strength where the λD was elongated [19]. Therefore, the pH and ionic strength should be fine-tuned to improve the signal-to-noise ratios of biosensors, especially FET-based biosensors.
Environmental factors also affect the binding affinity between antigens and Abs. Such binding is mainly driven by electrostatic forces, which depend on molecular charges and is thus governed by the pH and ionic strength of environmental solutions. Some reports have shown that the binding affinity depends on the pH and ionic strength [21,22]. Previous data indicated that molecular interactions are multifactorial, as many factors may modify the binding properties. Therefore, it is necessary to further define the environmental effects of each antigen–Ab system.
In the case of CRP, it was previously reported that CRP pH-dependently bound to proteins in a buffer with an ionic strength of approximately 150 mM [23]. However, CRP–anti-CRP Ab interactions have not been fully studied; thus, more fundamental studies are required to design highly sensitive assay systems. Here, we investigated the effects of pH and ionic strength on the binding affinity between CRP and an anti-CRP monoclonal Ab (mAb). CRP was fluorescently labeled and incubated at various pHs and ionic strengths with an anti-CRP mAb immobilized onto 96-well polystyrene (PS) plates. After a wash step, the remaining fluorescence intensity was measured to evaluate the binding affinity. The results indicate that the affinity was insensitive to pH, but sensitive to ionic strength. Our study provides insights that are important for designing highly sensitive biosensors for CRP detection.
Materials and Methods
CRP, Anti-CRP Ab, and Solution Preparation
Human recombinant C-reactive protein (FUJIFILM Wako) and an anti-human CRP mAb (304-51283, clone 12D-2C-36, FUJIFILM Wako) were used in this study. Both CRP and anti-CRP mAb were commercially available. CRP was fluorescently labeled using Alexa Fluor 488 NHS Ester (Thermo Fisher Scientific).
Solutions at different pHs and ionic strengths were prepared as indicated in Table 1. The pH was tuned by mixing Na2HPO4·12 H2O (Wako) with KH2PO4 (Wako), and the ionic strength was tuned by adding NaCl (Wako) and KCl (Wako) in deionized water. After the preparation, the pH was measured using a pH meter (D-55, Horiba). The ionic strengths were calculated using the pH and pKa values. Phosphoric acid has three pKa values of 2.16, 7.21, and 12.32, and a pKa value of 7.21 was used for the ionic strength calculation. Solution (Sol.) 9 and 11 exhibited no buffer capacity; thus, their ionic strengths were calculated assuming that their pH values were 7. Although the pH values for Sol. 9 and Sol. 11 might be lower than 7 due to carbon dioxide, the ionic strengths do not significantly change (<10%). The calculated ionic strengths are presented in Table 1.
Table 1 .
Preparing solutions with different pHs and ionic strengths
| Sol. 1 | Sol. 2 | Sol. 3 | Sol. 4 | Sol. 5 | Sol. 6 | Sol. 7 | Sol. 8 | Sol. 9 | Sol. 10 | Sol. 11 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| [NaCl]+[KCl] (mM) | 140 | 140 | 140 | 1383 | 140 | 140 | 14 | 1.4 | 14 | 1.4 | 1.4 |
| [Na2HPO4]+[KH2PO4] (mM) | 10 | 10 | 10 | 10 | 9.6 | 5 | 5 | 10 | 1 | 5 | 0.1 |
| pH | 5.9 | 7.4 | 8.1 | 6.5 | 7.5 | 7.4 | 7.3 | 7.4 | — | 7.4 | — |
| Ionic strength (mM) | 150.7 | 159.1 | 163.2 | 1396 | 158.6 | 149.6 | 23.1 | 21.1 | 15.5 | 11.0 | 1.6 |
| λD (nm) | 0.78 | 0.76 | 0.75 | 0.26 | 0.76 | 0.79 | 2.0 | 2.1 | 2.4 | 2.9 | 7.7 |
Sol. 1–Sol. 3 were used for the study of pH-dependence, and Sol. 4–Sol. 11 were used for the study of ionic strength dependence
The λD was calculated based on equation (1) [24]:
| (1) |
where I is the ionic strength of the electrolyte (mol/m3), ε0 is the permittivity of free space, εr is the relative permittivity of water (εr=78), kB is the Boltzmann constant, T is 300 K, NA is Avogadro’s number, and e is the elementary charge. The calculated λD values are presented in Table 1.
Binding-Affinity Assays in 96-Well PS Plates
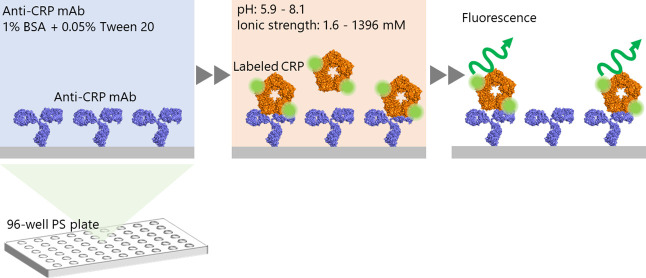
Figure 1 illustrates the experimental procedure used to analyze binding affinities. Anti-CRP mAb (768 nM) was immobilized overnight in each well of PS plates at 4°C. In addition, an anti-influenza A virus nucleoprotein mAb (Bio Matrix Research) was immobilized in different wells as a negative control. After Ab immobilization, each well was rinsed three times with 0.05% polyoxyethylene sorbitan monolaurate (Tween 20, Tokyo Chemical Industry) in Dulbecco’s phosphate-buffered saline (D-PBS(–), Nacalai Tesque). After rinsing, the wells were incubated for 1 h with a blocking buffer containing 1% bovine serum albumin (BSA, Sigma-Aldrich) and 0.05% Tween 20 in D-PBS(–) to prevent non-specific adsorption, after which the wells were rinsed again. Fluorescently labeled CRP was diluted to different concentrations (ranging from 0.5 to 100 nM) with different pHs and ionic strengths, according to Table 1. The prepared CRP solutions were incubated in each well for 1 h and then rinsed four times with 0.05% Tween 20 in D-PBS(–). Two or four test wells were used for each condition. The binding affinity was estimated by measuring the fluorescence from labeled CRP, based on the assumption that the intensity correlates linearly with the number of bound CRP molecules. The fluorescence intensity was measured using a multimode plate reader (Infinite 200 PRO M Plex, excitation wavelength: 490 nm, emission wavelength: 525 nm, Tecan).
Figure 1 .
Schematic illustration of the binding-affinity assay, performed at different pHs and ionic strengths.
Measuring pI Values by Isoelectric Focusing (IEF)
An IEF gel (Novex pH 3-10 IEF Protein Gels, Thermo Fisher Scientific) was set in an electrophoresis device (XCell SureLock Mini-Cell, Thermo Fisher Scientific), and IEF running buffers (Novex IEF pH 3-10 Cathode Buffer and Novex IEF Anode Buffer, Thermo Fisher Scientific) were loaded. The samples, including CRP, labeled CRP, anti-CRP mAb, and BSA, as well as the IEF marker (IEF marker 3-10, Thermo Fisher Scientific) were diluted with a sample buffer (Novex IEF Sample Buffer pH 3-10, Thermo Fisher Scientific) and loaded onto the gel. IEF was performed under the following sequential conditions: (i) 100 V constant for 1 h, (ii) 200 V constant for 1 h, and (iii) 500 V constant for 0.5 h. After the run, the samples were fixed for 1 h with a 12% trichloroacetic acid solution (Hayashi Pure Chemical). Subsequently, the gel was rinsed with deionized water and stained for 1 h with Coomassie Brilliant Blue (EzStain AQua, ATTO) and finally destained with deionized water.
Evaluating the Antigen–Ab Binding Kinetics Via Biolayer Interferometry (BLI)
Binding kinetics were measured by performing BLI (BLItz, Sartorius). First, an anti-CRP mAb (768 nM) was immobilized on sensor chips (Anti-Murine IgG Quantitation, AMQ, Sartorius) and then the sensor chips were dipped into CRP in D-PBS(–) with an ionic strength of 150 mM and a pH of 7.4. The binding kinetics were measured at CRP (labeled CRP) concentrations of 25, 50, and 100 nM, respectively.
Results and Discussion
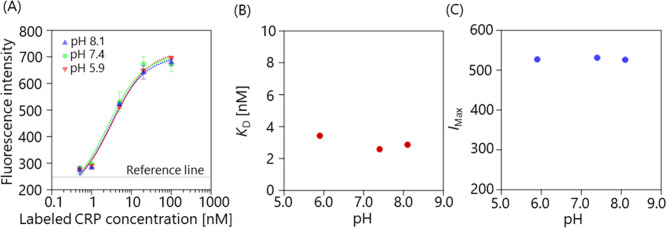
We first investigated the pH dependence of the binding affinity, while varying the pH from 5.9 to 8.1, and holding the ionic strength constant at approximately 155 mM (Sol. 1–3, Table 1. Figure 2(A) shows the fluorescence intensity as a function of the labeled CRP concentration at different pH values. The reference line (horizontal line) represents the average intensity of the negative control (100 nM CRP plus an anti-influenza mAb), showing that non-specific adsorption was suppressed. To evaluate the affinity quantitatively, the plots were fitted using equation (2) [24]:
Figure 2 .
(A) Fluorescence intensity as a function of the labeled CRP concentration at different pH values, ranging from 5.9 to 8.1. The error bar represents the standard deviation. The plots were fitted with equation (2). The horizontal dashed line represents the reference line which was calculated from the negative-control sample (100 nM CRP plus an anti-influenza mAb. (B) pH dependence of KD. (C) pH dependence of IMax.
| (2) |
where I, [CRP], KD, IMax, and Ibase are the fluorescence intensity, concentration of labeled CRP, dissociation constant, maximum, and baseline fluorescence intensity, respectively. Figure 2(B) and (C) show the KD and IMax values as a function of pH, respectively. The results indicate that both KD and IMax were nearly constant over this range. Thus, it can be concluded that the binding affinity was unaffected by changing the pH within the studied range.
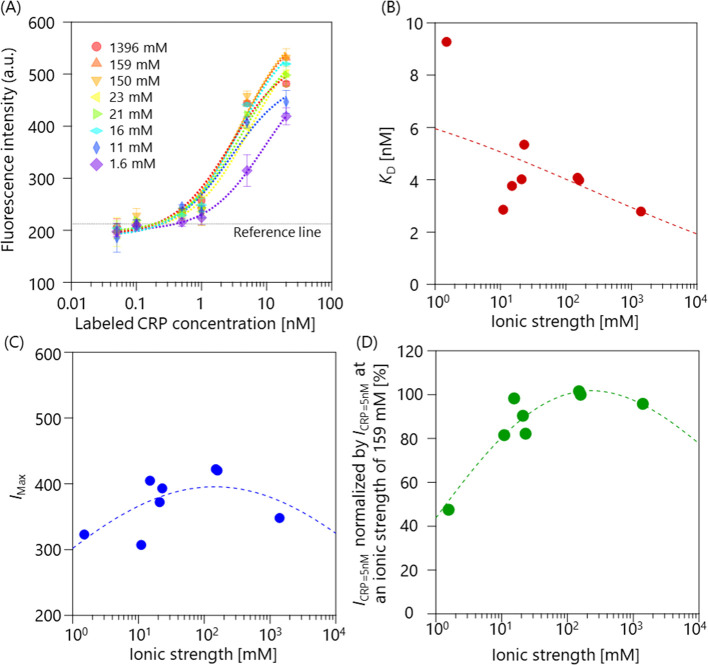
We next studied how the binding affinity depended on the ionic strength, which was varied from 1.6 to 1396 mM (Sol. 4–11, Table 1). Figure 3(A) shows the fluorescence intensity as a function of the labeled CRP concentration at different ionic strengths. The reference line (horizontal dashed line) represents the average intensity of the negative control (20 nM CRP plus an anti-influenza mAb), showing that non-specific adsorption was suppressed. The plots were fitted with equation (2) in the same manner. KD and IMax are plotted as functions of the ionic strength in Fig. 3(B) and (C), respectively. The trend was visualized by applying Gaussian process regression (GPR) [25], which is a nonlinear and nonparametric regression tool and infers a continuous function from a set of individual data points. The mean prediction of the GPR is also plotted in Fig. 3(B) and (C). In contrast to the pH dependence, the KD and IMax values varied with changes in the ionic strength. As the average CRP concentration in normal healthy individuals is 6.8 nM [2], highly sensitive CRP sensors should be designed to detect such a low concentration. Thus, we focused on the ionic strength dependence at a CRP concentration of 5 nM. Each I at a CRP concentration of 5 nM (ICRP=5nM) was normalized by ICRP=5nM at an ionic strength of 159 mM (near the physiological concentration) and plotted against the ionic strength (Fig. 3(D)).
Figure 3 .
(A) Fluorescence intensity as a function of the labeled CRP concentration at different ionic strengths, ranging from 1.6 to 1396 mM. The error bar represents the standard deviation. The plots were fitted with equation (2). The horizontal dashed line represents the average intensity of the negative control (20 nM CRP plus an anti-influenza mAb). (B) Ionic strength-dependence of KD. (C) Ionic strength-dependence of IMax. (D) The relative binding affinity plotted against the ionic strength, where I at a CRP concentration of 5 nM (ICRP=5nM) was normalized by ICRP=5nM at an ionic strength of 159 mM. GPR was applied for the plots shown in (B)-(D). In each case, the mean GPR prediction is shown with a dashed line.
The trend was also visualized by applying GPR in the same manner as shown in Fig. 3(B) and (C). The plot clearly indicates that the binding affinity was strong under the physiological condition, while became weaker at lower and higher ionic strengths. The relative binding affinity at 1.6 mM decreased by 55%, compared with that under the physiological condition (~150 mM).
To understand why the CRP–anti-CRP mAb system was insensitive to pH changes, but sensitive to ionic strength changes, the pI of CRP, labeled CRP, and anti-CRP mAb were measured using IEF. The pI of BSA was measured as a control sample. The results are shown in Table 2. The pIs of CRP and BSA agreed with previous findings [26,27], indicating that the IEF was performed properly, although the pI of the anti-CRP mAb was slightly lower than that of other IgG antibodies (pI=6.1–9.4) [28]. The pI of labeled CRP was smaller than that of native CRP, probably because of the fluorescence tag. Based on the absorption spectrum (data not shown), the average number of fluorescence tags per molecule was estimated to be 8.1. The fluorescent tag was reacted with primary amines (-NH2) on CRP; thus, the pI decrease possibly resulted from the conjugation. In addition, variations of the pI (pI=5.3–5.9) may be attributed to variations in the number of tags per molecule.
Table 2 .
pIs of an anti-CRP mAb, labeled CRP, CRP, and BSA
| Anti-CRP mAb | Labeled CRP | CRP | BSA | |
|---|---|---|---|---|
| pI | 5.3 | 5.3–5.9 | 6.2 | 4.7 |
The IEF results indicate that both labeled CRP and the anti-CRP mAb were negatively charged when the pH was higher than 5.9. Therefore, the sign of surface charge did not change during the pH change from 5.9 to 8.1, resulting in the system being insensitive to pH changes. Although the sign of the charge did not change within the pH range, the intermolecular force between CRP and anti-CRP mAb can depend significantly on the ionic strength. The electric double-layer force (which acts as a repulsive force) increases as the ionic strength decreases, that is, the λD increases [29]. Therefore, the repulsive force may be responsible for the system being markedly sensitive to changes in the ionic strength. Moreover, changes in ionic strength may also affect the stability of biomolecules. A previous report showed that the conformation of biomolecules was altered at an extreme ionic strength [30], which may also occur in our system.
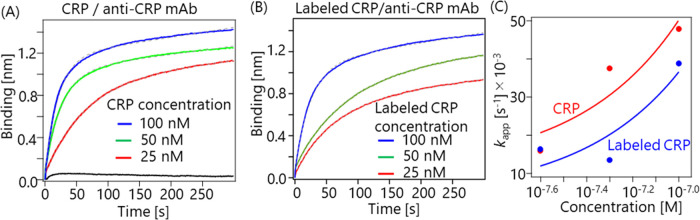
Finally, since the CRP used in our study was fluorescently labeled, we investigated whether the labeling may have affected the binding affinity. To test this possibility, KD values of native CRP and labeled CRP were compared. Figure 4(A) and (B) show the binding kinetics between an immobilized anti-CRP mAb and CRP (labeled CRP) at different concentrations. In Fig. 4(A), a negative control, which measured the binding kinetics for CRP (80 nM) in the absence of an anti-CRP mAb, is also shown as a black line. The binding kinetics were fitted with equations (3) and (4), where a linear slope component, corresponding to baseline drift, was added to the fitting function described previously [31]:
Figure 4 .
(A) Binding kinetics of immobilized anti-CRP mAb and CRP at concentrations of 25, 50, and 100 nM, with an ionic strength of 150 mM and a pH of 7.4. The black line represents the binding kinetics of negative control. (B) Binding kinetics for immobilized anti-CRP mAb and labeled CRP at concentrations of 25, 50, and 100 nM, with an ionic strength of 150 mM and a pH of 7.4. (C) kapp at different CRP concentrations. The kon and koff values were calculated, based on the fitting.
| (3) |
| (4) |
where S, S0, kapp, kon, koff, [CRP], y0, and t are the binding intensity, maximum binding intensity, apparent on-rate constant, on-rate constant, off-rate constant, concentration of CRP or labeled CRP, baseline-drift coefficient, and elapsed time, respectively. By measuring the binding kinetics at different CRP concentrations, the kon and koff were fitted, as shown in Fig. 4(C). The fitting results are summarized in Table 3. Herein, the KD values (KD=koff/kon) were calculated for CRP and labeled CRP, respectively (Table 3). Table 3 indicates that labeling affected the koff value, but not the kon value, resulting in a decrease in the KD. As the KD was changed, it was inferred that some labels may be close to the interaction sites. Although the KD was slightly different, the observed pH dependence and ionic dependence should be qualitatively consistent between labeled or unlabeled CRP and the anti-CRP mAb when the pH was above 6.3, because both CRP and labeled CRP were negatively charged in that pH range.
Table 3 .
kon, koff, and KD values for CRP/Anti-CRP mAb and labeled CRP/Anti-CRP
| CRP/Anti-CRP mAb | Labeled CRP/Anti-CRP mAb | |
|---|---|---|
| kon [s–1·M–1] | 3.9×105 | 3.3×105 |
| koff [s–1] | 0.011 | 0.0037 |
| KD [nM] | 27 | 11 |
Conclusion
In conclusion, we investigated pH and ionic strength dependences of the binding affinity between fluorescently labeled CRP and an anti-CRP mAb. Our results clearly demonstrate that the affinity was insensitive to pH changes, but was sensitive to ionic strength changes. The binding affinity at an ionic strength of 1.6 mM significantly decreased by 55%, when compared to that under physiological conditions. The IFE results indicate that both labeled CRP and anti-CRP mAb were negatively charged in the pH range of 5.9 to 8.1. The surface charge was likely responsible for the ionic strength dependence, as the repulsive force between CRP and anti-CRP mAb was enhanced at lower ionic strengths. Our experiments were conducted in phosphate-based solutions, and the results might be different if buffers contain different ion species, as some reports have shown specific protein-ion interactions [32,33]. Although lowering the ionic strength is a common strategy to overcome the Debye screening in FET-based biosensors, our results imply that diluting the buffer may not be an effective solution for enhancing the signal-to-noise ratio in the case of CRP detection, because there is a tradeoff between the binding affinity and λD. Although the versality of the findings is not fully studied yet, which will be addressed in the future, the results provide insights into designing highly sensitive CRP sensors, especially FET-based sensors. Furthermore, since the pH and ionic dependence may be different for each antigen–Ab system, our approach should pave the way for developing other antigen–Ab systems.
Conflict of Interest
The authors declare that they have no conflict of interest.
Author Contributions
Y.O. conceived the study. All authors discussed the results and wrote the paper. All authors gave final approval of the version to be published.
Acknowledgments
This work was also supported by JST-PRESTO (JPMJPR19G3), JST-CREST (JPMJCR15F4), and JST-Mirai (JPMJMI19D4).
References
- [1].Vashist, S. K., Venkatesh, A. G., Schneider, E. M., Beaudoin, C., Luppa, P. B., Luong, J. H. T.. Bioanalytical advances in assays for C-reactive protein. Biotechnol. Adv. 34, 272–290 (2016). https://doi.org/10.1016/j.biotechadv.2015.12.010 [DOI] [PubMed] [Google Scholar]
- [2].Dhara, K., Mahapatra, D. R.. Review on electrochemical sensing strategies for C-reactive protein and cardiac troponin I detection. Microchem. J. 156, 104857 (2020). https://doi.org/10.1016/j.microc.2020.104857 [Google Scholar]
- [3].Collet-Cassart, D., Abbeele, E. van den, Poncelet, S.. A quantitative C-reactive protein assay using latex agglutination in microtiter plates. J. lmmunol. Methods 125, 137–141 (1989). https://doi.org/10.1016/0022-1759(89)90086-0 [DOI] [PubMed] [Google Scholar]
- [4].Oh, Y. K., Joung, H., Han, H. S., Suk, H., Kim, M.. A three-line lateral flow assay strip for the measurement of C-reactive protein covering a broad physiological concentration range in human sera. Biosens. Bioelectron. 61, 285–289 (2014). https://doi.org/10.1016/j.bios.2014.04.032 [DOI] [PubMed] [Google Scholar]
- [5].Oyama, Y., Osaki, T., Kamiya, K., Kawano, R., Honjoh, T., Shibata, H., et al. A glass fiber sheet-based electroosmotic lateral flow immunoassay for point-of-care testing. Lab. Chip 12, 5155–5159 (2012). https://doi.org/10.1039/c2lc40994a [DOI] [PubMed] [Google Scholar]
- [6].Vashist, S. K., Schneider, E. M., Lam, E., Hrapovic, S., Luong, J. H. T.. One-step antibody immobilization-based rapid and highly-sensitive sandwich ELISA procedure for potential in vitro diagnostics. Sci. Rep. 4, 4407 (2014). https://doi.org/10.1038/srep04407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Baldini, F., Carloni, A., Giannetti, A., Porro, G., Trono, C.. An optical PMMA biochip based on fluorescence anisotropy: Application to C-reactive protein assay. Sens. Actuators B Chem. 139, 64–68 (2009). https://doi.org/10.1016/j.snb.2008.08.027 [Google Scholar]
- [8].Lee, M. H., Lee, D. H., Jung, S. W., Lee, K. N., Park, Y. S., Seong, W. K.. Measurements of serum C-reactive protein levels in patients with gastric cancer and quantification using silicon nanowire arrays. Nanomedicine 6, 78–83 (2010). https://doi.org/10.1016/j.nano.2009.04.004 [DOI] [PubMed] [Google Scholar]
- [9].Hu, W. P., Hsu, H., Chiou, A., Tseng, K. Y., Lin, H., Chang, G. L., et al. Immunodetection of pentamer and modified C-reactive protein using surface plasmon resonance biosensing. Biosens. Bioelectron. 21, 1631–1637 (2006). https://doi.org/10.1016/j.bios.2005.11.001 [DOI] [PubMed] [Google Scholar]
- [10].Balevicius, Z., Ramanaviciene, A., Baleviciute, I., Makaraviciute, A., Mikoliunaite, L., Ramanavicius, A.. Evaluation of intact- and fragmented-antibody based immunosensors by total internal reflection ellipsometry. Sens. Actuators B Chem. 160, 555–562 (2011). https://doi.org/10.1016/j.snb.2011.08.029 [Google Scholar]
- [11].Patolsky, F., Zheng, G., Hayden, O., Lakadamyali, M., Zhuang, X., Lieber, C. M.. Electrical detection of single viruses. Proc. Natl. Acad. Sci. U.S.A. 101, 14017–14022 (2004). https://doi.org/10.1073/pnas.0406159101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kim, A., Ah, C. S., Yu, H. Y., Yang, J., Baek, I., Ahn, C., et al. Ultrasensitive, label-free, and real-time immunodetection using silicon field-effect transistors. Appl. Phys. Lett. 91, 103901 (2007). https://doi.org/10.1063/1.2779965 [Google Scholar]
- [13].Son, M., Kim, D., Ko, H. J., Hong, S., Park, T. H.. A portable and multiplexed bioelectronic sensor using human olfactory and taste receptors. Biosens. Bioelectron. 87, 901–907 (2017). https://doi.org/10.1016/j.bios.2016.09.040 [DOI] [PubMed] [Google Scholar]
- [14].Kanai, Y., Ohmuro-Matsuyama, Y., Tanioku, M., Ushiba, S., Ono, T., Inoue, K., et al. Graphene field effect transistor-based immunosensor for ultrasensitive noncompetitive detection of small antigens. ACS Sens. 5, 24–28 (2020). https://doi.org/10.1021/acssensors.9b02137 [DOI] [PubMed] [Google Scholar]
- [15].Ushiba, S., Okino, T., Miyakawa, N., Ono, T., Shinagawa, A., Kanai, Y., et al. State-space modeling for dynamic response of graphene FET biosensors. Jpn. J. Appl. Phys. 59, SGGH04 (2020). https://doi.org/10.7567/1347-4065/ab65ac [Google Scholar]
- [16].Miyakawa, N., Shinagawa, A., Kajiwara, Y., Ushiba, S., Ono, T. Kanai, Y., et al. Drift suppression of solution-gated graphene field-effect transistors by cation doping for sensing platforms. Sensors (Basel) 21, 7455 (2021). https://doi.org/10.3390/s21227455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Syedmoradi, L., Ahmadi, A., Norton, M. L., Omidfar, K.. A review on nanomaterial-based field effect transistor technology for biomarker detection. Microchim. Acta 186, 739 (2019). https://doi.org/10.1007/s00604-019-3850-6 [DOI] [PubMed] [Google Scholar]
- [18].Ushiba, S., Miyakawa, N., Ito, N., Shinagawa, A., Nakano, T., Okino, T., et al. Deep-learning-based semantic image segmentation of graphene field-effect transistors. Appl. Phys. Express 14, 036504 (2021). https://doi.org/10.35848/1882-0786/abe3db [Google Scholar]
- [19].Elnathan, R., Kwiat, M., Pevzner, A., Engel, Y., Burstein, L., Khatchtourints, A., et al. Biorecognition layer engineering: Overcoming screening limitations of nanowire-based FET devices. Nano Lett. 12, 5245–5254 (2012). https://doi.org/10.1021/nl302434w [DOI] [PubMed] [Google Scholar]
- [20].Gao, N., Zhou, W., Jiang, X., Hong, G., Fu, T., Lieber, C. M.. General strategy for biodetection in high ionic strength solutions using transistor-based nanoelectronic sensors. Nano Lett. 15, 2143–2148 (2015). https://doi.org/10.1021/acs.nanolett.5b00133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Nguyen, T. H., Greinacher, A.. Effect of pH and ionic strength on the binding strength of anti-PF4/polyanion antibodies. Eur. Biophys. J. 46, 795–801 (2017). https://doi.org/10.1007/s00249-017-1240-8 [DOI] [PubMed] [Google Scholar]
- [22].Dejaegere, A., Choulier, L., Lafont, V., De Genst, E., Altschuh, D.. Variations in antigen-antibody association kinetics as a function of pH and salt concentration: A QSAR and molecular modeling study. Biochemistry 44, 14409–14418 (2005). https://doi.org/10.1021/bi050986v [DOI] [PubMed] [Google Scholar]
- [23].Hammond, D. J., Singh, S. K., Thompson, J. A., Beeler, B. W., Rusiñol, A. E., Pangburn, M. K.. Identification of acidic pH-dependent ligands of pentameric C-reactive protein. J. Biol. Chem. 285, 36235–36244 (2010). https://doi.org/10.1074/jbc.M110.142026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lloret, N., Frederiksen, R. S., Møller, T. C., Rieben, N. I., Upadhyay, S., Vico, L. D., et al. Effects of buffer composition and dilution on nanowire field-effect biosensors. Nanotechnology 24, 035501 (2013). https://doi.org/10.1088/0957-4484/24/3/035501 [DOI] [PubMed] [Google Scholar]
- [25].Deringer, V. L., Bart, A. P., Bernstein, N., Wilkins, D. M., Ceriotti, M., Csanyi, G.. Gaussian process regression for materials and molecules. Chem. Rev. 121, 10073–10141 (2021). https://doi.org/10.1021/acs.chemrev.1c00022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Laurent, P., Potempa, L. A., Gewurz, H., Fiedel, B. A., Allen, R. C.. The titration curve of native C reactive protein. Electrophoresis 4, 316–317 (1983). https://doi.org/10.1002/elps.1150040414 [Google Scholar]
- [27].Salis, A., Bostrom, M., Medda, L., Cugia, F., Barse, B., Parsons, D. F., et al. Measurements and theoretical interpretation of points of zero charge/potential of BSA protein. Langmuir 27, 11597–11604 (2011). https://doi.org/10.1021/la2024605 [DOI] [PubMed] [Google Scholar]
- [28].Goyon, A., Exco, M., Janin-bussat, M., Bobaly, B., Fekete, S., Guillarme, D., et al. Determination of isoelectric points and relative charge variants of 23 therapeutic monoclonal antibodies. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 1066, 119–128 (2017). https://doi.org/10.1016/j.jchromb.2017.09.033 [DOI] [PubMed] [Google Scholar]
- [29].Popa, I., Sinha, P., Finessi, M., Maroni, P., Papastavrou, G., Borkovec, M.. Importance of charge regulation in attractive double-layer forces between dissimilar surfaces. Phys. Rev. Lett. 104, 228301 (2010). https://doi.org/10.1103/PhysRevLett.104.228301 [DOI] [PubMed] [Google Scholar]
- [30].Dzubiella, J. Salt-specific stability and denaturation of a short salt-bridge-forming α-helix. J. Am. Chem. Soc. 130, 14000–14007 (2008). https://doi.org/10.1021/ja805562g [DOI] [PubMed] [Google Scholar]
- [31].Zeilinger, M., Pichler, F., Nics, L., Wadsak, W., Spreitzer, H., Hacker, M.. New approaches for the reliable in vitro assessment of binding affinity based on high-resolution real-time data acquisition of radioligand-receptor binding kinetics. EJNMMI Res. 7, 22 (2017). https://doi.org/10.1186/s13550-016-0249-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Rembert, K. B., Paterova, J., Heyda, J., Hilty, C., Jungwirth, P., Cremer, P. S.. Molecular mechanisms of ion-specific effects on proteins. J. Am. Chem. Soc. 134, 10039–10046 (2012). https://doi.org/10.1021/ja301297g [DOI] [PubMed] [Google Scholar]
- [33].Batoulis, H., Schmidt, T. H., Weber, P., Schloetel, J., Kandt, C., Lang, T.. Concentration dependent ion-protein interaction patterns underlying protein oligomerization behaviours. Sci. Rep. 6, 24131 (2016). https://doi.org/10.1038/srep24131 [DOI] [PMC free article] [PubMed] [Google Scholar]