Abstract
Natural killer (NK) cells reject major histocompatibility complex class I (MHC-I)–deficient bone marrow through direct cytotoxicity but not solid organ transplants devoid of MHC-I. Here, we demonstrate an immediate switch in NK cell function upon exit from the circulation, characterized by a shift from direct cytotoxicity to chemokine/cytokine production. In the skin transplant paradigm, combining an NK cell–specific activating ligand, m157, with missing self MHC-I resulted in complete graft rejection, which was dependent on NK cells as potential helpers and T cells as effectors. Extracellular matrix proteins, collagen I, collagen III, and elastin, blocked NK cell cytotoxicity and promoted their chemokine/cytokine production. NK cell cytotoxicity against MHC-I–deficient melanoma in the skin was markedly increased by blocking tumor collagen deposition. MHC-I down-regulation occurred in solid human cancers but not leukemias, which could be directly targeted by circulating cytotoxic NK cells. Our findings uncover a fundamental mechanism that restricts direct NK cell cytotoxicity in peripheral tissues.
Collagens and elastin modulate NK cell effector function from direct cytotoxicity to helper phenotype in peripheral tissues.
INTRODUCTION
It has long been appreciated that down-regulation of major histocompatibility complex class I (MHC-I) is a primary mechanism for immune evasion used by cancer cells (1). In the era of cancer immunotherapeutics, MHC-I down-regulation has emerged as a major resistance mechanism against immunotherapy, as malignant cells escape from CD8+ T cell detection and elimination (1). However, the pervasive selection of this immune evasion strategy by cancer cells is perplexing in that MHC-I down-regulation is expected to render cells vulnerable to natural killer (NK) cell killing (2–4). The dilemma of NK cells’ role in controlling cells that fail to express self MHC-I in vivo was originally encountered in the transplantation field. In the context of hybrid resistance, parental skin grafts are accepted, but bone marrow (BM) cells are rejected by F1 hybrids in an NK cell–dependent manner (5, 6). Subsequent studies have shown that NK cells in wild-type (WT) syngeneic recipients reject donor MHC-I–deficient BM cells, while MHC-I–deficient solid organ transplants are not rejected (7, 8). Elucidating the mechanism that underlies tissue-specific NK cell responses will have major implications in cancer immunology, transplant biology, and virology.
NK cells belong to the innate lymphoid cell (ILC) family that reside in most tissues and form a swift-acting innate barrier against viral infections and cells undergoing malignant transformation (9). Conventional NK (cNK) cells, defined by expression of NK1.1, NKp46, and CD49b in mice, are found in circulation, secondary lymphoid organs, and most other tissues (9). Another member of the ILC family that is closely related in phenotype and function is the tissue-resident NK (trNK) or ILC1 cell that lacks CD49b but expresses CD49a (9). These cells are a nonmigrating, NK cell–like population (10) that are thought to develop from two distinct pathways: (i) differentiation from the ILC progenitor (ILCP) (11) or (ii) differentiation from cNK cells in the presence of transforming growth factor–β (TGFβ) (12, 13) in a nonmutually exclusive manner.
Full NK cell activation against a target cell is accomplished by the integration of multiple inhibitory and activation signals, a process that limits indiscriminate killing of healthy host cells. Self MHC-I molecules function as the dominant inhibitory signal by binding NK cell–expressed Ly49 receptors in mice, or killer inhibitory receptors in humans, to prevent degranulation and cytokine release. Multiple activation signals, together with down-regulation or loss of inhibitory signals, are required to enable complete NK cell cytotoxicity (3, 14, 15). Following their licensing to activate their cytotoxic program (16–18), NK cells destroy the target cells by production and release of granzymes and perforin in the immunological synapse (19). Fas-FasL and TNF-related apoptosis-inducing ligand (TRAIL) are also used by NK cells for target cell elimination (20).
Although in vitro models of NK cell activation have provided insights into signaling requirements that initiate NK cell killing of target cells, the conditions in vivo are much more complex in that non-MHC ligands, and potentially unknown inhibitory and activation pathways of varying strengths, acting together with cytokines, set the NK cell activation threshold (21). Most in vivo NK cell activation models have focused on natural killer group 2D (NKG2D) and other activating ligands and cytokines that also activate CD8+ T cells (22), complicating the interpretation of NK versus CD8+ T cell effects. To investigate the mechanism of NK cell activation in peripheral tissues, we used the NK cell–specific m157-Ly49H activation axis in a skin transplantation model. m157 is murine cytomegalovirus (MCMV)–encoded glycoprotein expressed on the surface of infected cells. This MHC-I–like molecule serves as an NK cell–specific activation signal, directing the targeted killing of MCMV-infected cells through recognition by the Ly49H receptor, which is only present on the surface of NK cells on the C57BL/6 (B6) background (23). As a result, B6 mice are resistant to MCMV infection, while BALB/c and other strains are susceptible (24). Thus, the m157-Ly49H axis can be used to specifically study NK cell activation and its downstream function in vivo.
Here, we demonstrate that, upon exit from the circulation into the skin graft in response to m157, NK cells are exposed to extracellular matrix (ECM) proteins, which can block NK cell cytotoxicity while promoting its helper function. NK cell activation induced by the combination of m157 and missing self MHC-I (25) resulted in skin graft rejection in an NK- and T cell–dependent manner. RNA sequencing (RNA-seq) analysis revealed substantial down-regulation in cytotoxic mediators together with up-regulation in inflammatory cytokines and chemokines, when comparing cNK cells that entered the skin with circulating cNK cells. We found that in vitro exposure to ECM components, collagens and elastin, transformed the function and signaling within cNK cells. Furthermore, control of circulating melanoma cells occurred in an NK cell–dependent manner, yet the growth of these same melanoma cells subcutaneously was not affected by NK cells embedded in ECM. Blocking collagen deposition in subcutaneous melanomas led to an NK cell–mediated tumor suppression. Human solid cancers, but not leukemias, could afford to down-regulate MHC-I to escape CD8+ T cell–mediated elimination, which suggested that NK cells within peripheral tissues, unlike those in circulation, lacked a direct cytotoxic function. Our study reveals a fundamental aspect of NK cell biology, which governs the interplay of NK cells with cancer, organ transplantation, and viral infection.
RESULTS
NK cell–activating ligand together with loss of MHC-I is insufficient to induce rejection of syngeneic skin graft
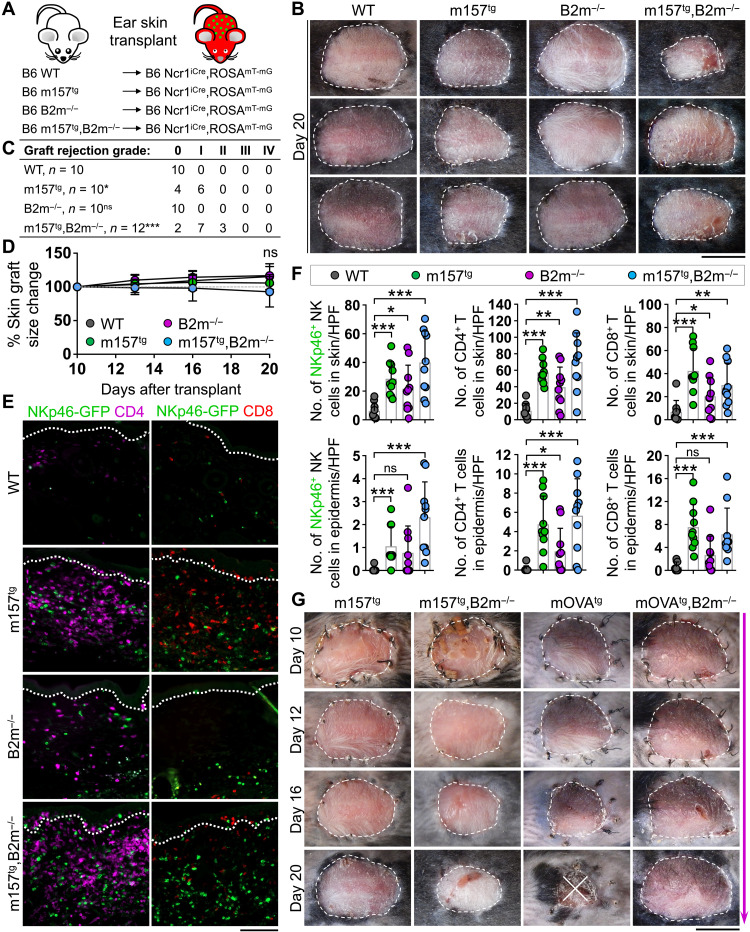
To assess the requirements for NK cell cytotoxicity in solid organs, we developed a syngeneic skin transplant model. Ear skin from donor mice on an albino B6 background was transplanted onto the back of B6 recipients. WT and m157tg mice with or without the deletion of beta-2 microglobulin (B2m) gene were used as donors. Ncr1iCre,ROSAmT-mG reporter mice expressing green fluorescent protein (GFP) in NKp46+ NK cells and TdTomato in all other cells were used as recipients (Fig. 1A) (26). This approach enabled us to examine the impact of an NK cell–specific activating ligand, loss of MHC-I (through B2m deletion), or the combination of both on skin transplant outcomes while precisely distinguishing donor from recipient skin by skin graft color (albino ear skin) and histologically by immunofluorescence (IF) imaging (fig. S1A). WT, B2m−/−, m157tg, and m157tg,B2m−/− skin grafts were monitored for signs of inflammation and rejection over time (fig. S1B). At 20 days after transplant, B2m−/− skin grafts showed no signs of rejection (Fig. 1, B and C). In contrast, B2m−/− BM transplants were readily rejected in WT B6 recipients (fig. S1C). m157tg and m157tg,B2m−/− skin grafts displayed minor signs of inflammation, which was most apparent in the latter (Fig. 1, B and C), yet no significant change in the size of the skin grafts was observed between the four groups from day 10 to day 20 after transplant (Fig. 1D). Monitoring m157tg,B2m−/− skin grafts out to day 60 after transplant revealed no additional changes (fig. S1D). Massive numbers of NKp46-GFP+ NK, CD4+ T, and CD8+ T cells infiltrated the skin and the epidermis of m157tg and m157tg,B2m−/− compared with WT grafts (Fig. 1, E and F). B2m−/− skin grafts also showed higher NK and T cell infiltrates compared with WT grafts (Fig. 1, E and F). No NK or T cell infiltrate was detected in the recipient skin adjacent to the skin grafts across the four groups (fig. S1, E and F). These findings establish that the expression of NK cell–specific activating ligand, m157, in the skin grafts results in markedly enhanced recruitment of NK cells and an associated increase in T cell numbers in the grafts but does not lead to graft rejection.
Fig. 1. Presence of NK cell–activating m157 ligand and loss of inhibitory B2m are insufficient to induce skin graft rejection in syngeneic recipients.
(A) Schematic diagram of ear skin transplantation from B6 albino WT, B2m−/−, m157tg, and m157tg,B2m−/− donor mice to B6 Ncr1iCre,ROSAmT-mG recipients. (B) Representative images of skin grafts at day 20 after transplant. (C) Quantification of graft rejection grades assessed at day 20 after transplant of B6 albino WT, B2m−/−, m157tg, and m157tg,B2m−/− skin grafts transplanted onto B6 Ncr1iCre,ROSAmT-mG recipients. (D) The percentage change in skin graft size from day 10 to day 20 after transplant. (E) Representative IF images of NKp46-GFP+ (green), CD4+ (purple), and CD8+ (red) cells in skin grafts at day 20 after transplant. Dotted lines indicate the epidermal basement membrane. (F) Quantification of NKp46-GFP+ NK, CD4+ T, and CD8+ T cells in the skin and epidermis of skin grafts at day 20 after transplant. (B to F) n = 9 to 12 mice per group from nine independent experiments. HPF, high-power field. (G) Time course of skin graft maintenance from day 10 to day 20 after transplant comparing m157tg, m157tg,B2m−/−, mOVAtg, and mOVAtg,B2m−/− skin grafts. m157tg and m157tg,B2m−/− images from the same experiment as (B). mOVAtg and mOVAtg,B2m−/−, n = 2 to 5 from one to two independent experiments. (C) Fisher’s exact test. ns, not significant; *P < 0.05 and ***P < 0.001 compared with WT group. (D and F) Graphs show means ± SD. Mann-Whitney U test; *P < 0.05, **P < 0.01, and ***P < 0.001. (B and G) Scale bars, 1 cm. (E) Scale bar, 100 μm. Photo Credit: Mark D. Bunting, Massachusetts General Hospital.
Next, we examined the strength of m157-Ly49H interaction in NK cells that infiltrated the skin grafts. Recipient-derived CD49b+ cNK and CD49a+ trNK cells were found in m157-expressing skin grafts, which expressed Ly49H and exhibited m157-induced Ly49H down-regulation compared with Ly49H− and Ly49H+ cNK and Ly49H− trNK cells in the recipient’s liver (fig. S1G) (27). No significant alteration in NKG2D, CD48, DNAM1 (DNAX accessory molecule-1), NKG2A, Ly49A, and Ly49C/I expression on the surface of CD49b+ cNK and CD49b+CD49a+ double-positive NK (dpNK) cells in m157tg,B2m−/− skin graft was detected (fig. S1, H to M). Although a large number of NKp46-GFP+ cNK and trNK cells infiltrated m157-expressing skin grafts, these cells did not up-regulate interferon-γ (IFNγ), tumor necrosis factor–α (TNFα), or granzyme B expression (fig. S2A). NK cells isolated from the blood, spleen, and lymph nodes of WT recipients transplanted with m157tg skin did not down-regulate Ly49H expression in contrast to those of m157tg mice, ruling out systemic NK cell hyporesponsiveness in recipient animals (fig. S2B) (27). In addition, WT NK cells were fully responsive to m157 stimulation in the presence of serum from m157tg mice in vitro, ruling out the possibility that a soluble form of m157 blocked NK cell activation in m157-expressing skin grafts (fig. S2C). NK cells can become hyporesponsive upon chronic exposure to the loss of MHC-I (28, 29). To investigate this concept in our transplant system, we tested the ability of m157tg,B2m−/− skin graft-infiltrating NK cells at day 10 after transplantation to degranulate and produce IFNγ in response to ex vivo stimulation. Compared with splenic cNK cells, skin graft–derived cNK cells had comparable expression levels of MHC-I–specific inhibitory receptors and were not found to be degranulating at baseline (fig. S3, A to D). However, upon ex vivo coculture, Ly49H+ cNK cells from m157tg,B2m−/− skin graft showed robust degranulation and IFNγ secretion compared with splenic Ly49H+ cNK cells in control immunoglobulin G (IgG)–coated, anti-NK1.1 antibody–coated plates as well as upon phorbol myristate acetate and ionomycin (PMA/Ion) stimulation (fig. S3E). We did not observe any increase in degranulation and IFNγ secretion by Ly49H− cNK cells in the skin graft versus spleen except for a minor increase in degranulation of skin Ly49H− NK cells in an IgG-coated plate (fig. S3F). These findings demonstrate that Ly49H+ cNK cells from m157tg,B2m−/− skin graft are not hyporesponsive and are fully capable of activation and degranulation outside of the skin.
To investigate the timing of inflammation in m157-expressing skin grafts, we compared m157tg grafts to skin grafts expressing a model T cell antigen, ovalbumin (OVA). WT B6 mice were transplanted with m157tg, m157tg,B2m−/−, mOVAtg, and mOVAtg,B2m−/− skin grafts and monitored. As expected, mOVAtg donor skin grafts were mostly rejected by day 20 after transplant, while the absence of MHC-I inhibited the rejection of mOVAtg,B2m−/− grafts (Fig. 1G). The paradoxical increase in inflammation in m157tg,B2m−/− compared with m157tg grafts supported the dominant role of NK rather than T cells in initiating the immune response in m157-expressing grafts. However, we noticed that inflammation in m157tg,B2m−/− skin grafts developed after day 12 after transplant, along a similar adaptive immune response timeline to mOVAtg grafts (Fig. 1G). These data suggest that early NK cell activation following m157tg skin transplantation may initiate a delayed T cell response leading to skin inflammation. Nonetheless, NK cell activation against syngeneic skin expressing m157 and lacking MHC-I is insufficient to cause skin graft rejection.
cNK cells recruited from the circulation into skin grafts give rise to trNK cells and fail to reject the graft upon additional cytokine stimulation
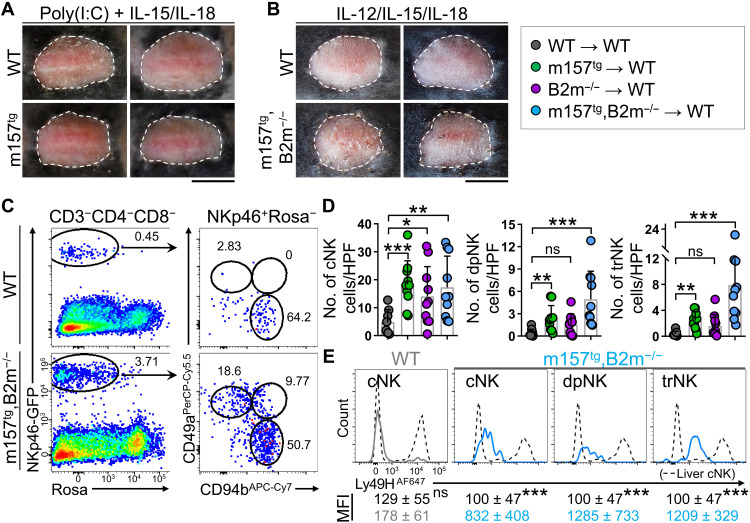
To overcome the apparent inhibition of cytotoxicity experienced by NK cells in m157-expressing skin grafts, we treated the recipient mice with interleukin-15 (IL-15), IL-18 and a Toll-like receptor 3 agonist, polyinosinic:polycytidylic acid [poly(I:C)], or IL-12, IL-15, and IL-18 (fig. S4A). These NK cell survival and activating factors are commonly used to enhance NK cell cytotoxicity (30, 31). However, m157tg and m157tg,B2m−/− skin grafts treated with these agents showed no sign of rejection at day 21 after transplant (Fig. 2, A and B). Although combination of IL-12, IL-15, and IL-18 had no effect on NK cell recruitment and their activation and maturation profile, these cytokines appeared to enhance the conversion of cNK cells into trNK cells in the skin graft, which may be mediated by IL-15 (fig. S4, B to E) (32, 33). To elucidate the basis for this profound lack of NK cell cytotoxicity, we investigated the phenotype of CD49b+ cNK and CD49a+ trNK cells, which were highly enriched in m157tg,B2m−/− skin grafts (Fig. 2C). Recipient liver cNK cells were largely negative for CD69, CD103, and TRAIL, while the large majority of liver trNK cells expressed CD69 and TRAIL (fig. S4F). cNK cells in m157tg,B2m−/− skin grafts expressed CD69 with minimal CD103 and TRAIL expression (fig. S4G). In contrast, two-thirds of trNK cells in the skin graft coexpressed CD69 and CD103 (fig. S4G), supporting their tissue-resident status. Unexpectedly, trNK cells in the skin graft expressed low levels of TRAIL (fig. S4G). Combining the ratio of NK cell subtypes from flow cytometry data with absolute NK cell counts in histological sections showed that CD49b+ cNK cells were significantly increased in B2m−/−, m157tg, and m157tg,B2m−/− skin grafts compared with WT grafts (Fig. 2D). In addition, CD49a+ trNK cells and CD49b+CD49a+ dpNK cells were markedly increased in m157tg and m157tg,B2m−/− skin grafts compared with WT grafts, which largely lacked trNK and dpNK cells (Fig. 2D). In contrast to cNK cells in WT skin grafts, all three NK cell subsets in m157tg,B2m−/− skin grafts expressed Ly49H at levels indicative of Ly49H down-regulation upon binding to m157 on the donor skin cells (Fig. 2E). Labeling the circulating immune cells with intravenous injection of anti-CD45 antibody further demonstrated that most of the cNK cells in WT and m157tg skin grafts were CD45 negative and had exited circulation similar to trNK cells in the skin (fig. S4H). Considering that trNK cells do not migrate (34–36) and ILCP-derived trNK cells are Ly49H negative (36), the large population of NKp46-GFP+Ly49H+ trNK cells in m157-expressing skin grafts indicates that circulating cNK cells give rise to trNK cells in the donor skin.
Fig. 2. NK cells fail to induce skin graft rejection upon cytokine stimulation.
(A and B) Representative images of WT or m157tg skin grafts treated with poly(I:C)/IL-15/IL-18 (A) and m157tg,B2m−/− skin grafts treated with IL-12/IL-15/IL-18 (B) on WT B6 recipients at day 21 after transplant. n = 2 to 5 mice per group from one to three independent experiments. (C) Representative flow cytometry dot plots of NK cells gated as NKp46-GFP+ROSA-TdTomato (ROSA)−CD3−CD4−CD8− in WT and m157tg,B2m−/− skin grafts. cNK, dpNK, and trNK were distinguished on the basis of CD49a and CD49b expression. Numbers on the dot plots represent the percent cells within each gate. APC, allophycocyanin; PerCP, peridinin chlorophyll protein. (D) Enumeration of cNK, dpNK, and trNK populations in the skin by combining flow cytometry frequency data with NKp46-GFP+ cell counts from Fig. 1. n = 9 to 10 mice per group from nine independent experiments. (E) Ly49H expression by cNK, dpNK, and trNK cells from m157tg,B2m−/− and cNK cells from WT skin grafts at day 20 after transplant compared with cNK cells from the liver. Note that WT skin grafts contain Ly49H+ and Ly49H− cNK cells, while all NK subsets in m157tg,B2m−/− skin grafts express low levels of Ly49H. Average ± SD of mean fluorescence intensity (MFI) of Ly49H on NK cell subsets in m157tg,B2m−/− skin grafts, and Ly49H− cNK cells in WT skin grafts at day 20 after transplant are compared with Ly49H− cNK cells in the liver of the recipient mice. n = 4 to 10 mice per group from three independent experiments. (D and E) Graphs are shown as means ± SD. Mann-Whitney U test; *P < 0.05, **P < 0.01, and ***P < 0.001. (A and B) Scale bars, 1 cm. Photo Credit: Mark D. Bunting, Massachusetts General Hospital.
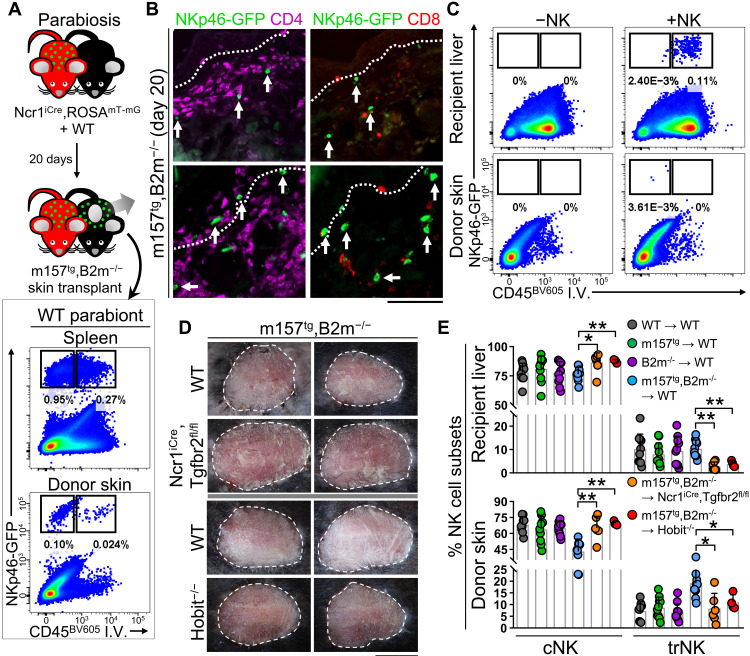
To determine whether NKp46-GFP+ NK cells in m157tg,B2m−/− skin grafts were recruited from circulation or the surrounding recipient skin, we performed a parabiosis experiment. Ncr1iCre,ROSAmT-mG mice were partnered with WT mice, and their circulatory systems were allowed to conjoin for 20 days before the m157tg,B2m−/− skin was transplanted onto the WT parabiont (Fig. 3A). At 20 days following skin transplantation, NKp46-GFP+ NK cells were found in the spleen and the skin graft, most of which were negative for anti-CD45 antibody injected intravenously (Fig. 3A). NKp46-GFP+ NK cells migrated into the dermis and epidermis of m157tg,B2m−/− skin followed by a large T cell infiltrate into the skin grafts (Fig. 3B). To directly test recruitment from circulation, we sorted splenic NKp46-GFP+CD49b+CD49a− cNK cells from Ncr1iCre,ROSAmT-mG mice and injected them intravenously into WT mice 1 day following m157tg,B2m−/− skin transplantation. After 20 days, NKp46-GFP+ NK cells were readily identified circulating in the recipient liver and a few NKp46-GFP+ NK cells were detected in the skin grafts, which had exited the circulation (Fig. 3C and fig. S4I). Together, these data demonstrate that NK cells in the donor graft are recruited from the circulation and migrate into the dermis and epidermis where they likely adopt tissue-resident cell properties.
Fig. 3. Circulating cNK cells enter the skin grafts and give rise to trNK cells.
(A) Schematic of parabiosis between WT and Ncr1iCre,ROSAmT-mG B6 mice followed by m157tg,B2m−/− skin transplantation onto the WT parabiont. Representative dot plots of the spleen and donor skin from the WT parabiont gating on CD3−CD4−CD8− cells and showing CD45 intravenously (I.V.) and NKp46-GFP at day 20 after skin transplant. (B) Representative IF images of m157tg,B2m−/− skin graft from the WT parabiont showing the presence of NKp46-GFP+ (green, white arrows), CD4+ (purple), and CD8+ (red) cells. Dotted lines indicate the epidermal basement membrane. n = 5 parabiosis pairs. (C) Sorted splenic cNK cells from Ncr1iCre,ROSAmT-mG mice were adoptively transferred into WT mice 1 day after skin transplantation with m157tg,B2m−/− skin grafts. Representative dot plots of transferred NK cells (defined as CD3−CD4−CD8−NKp46-GFP+) in liver and skin grafts of mice that did not (left) and did (right) receive NKp46-GFP+ NK cells. n = 2 mice per +NK group from two independent experiments. (D) Representative images of m157tg,B2m−/− skin grafts transplanted onto WT, Ncr1iCre,Tgfbr2fl/fl, and Hobit−/− recipient mice at day 20 after transplant. n = 4 to 7 mice per group. (E) Frequencies of cNK and trNK cells in the recipient liver (top) and donor skin (bottom) at day 20 after transplant. Ncr1iCre,ROSAmT-mG mice transplanted with B6 albino WT, m157tg, B2m−/−, and m157tg,B2m−/− donor skin from Fig. 1 (n = 9 to 10 mice per group) and Ncr1iCre,Tgfbr2fl/fl and Hobit−/− recipients transplanted with m157tg,B2m−/− donor skin at day 20 after transplant (n = 4 to 7 mice per group). (E) Graphs are shown as means ± SD. Mann-Whitney U test; *P < 0.05 and **P < 0.01. (B) Scale bar, 100 μm. (D) Scale bar, 1 cm. Photo Credit: Mark D. Bunting, Massachusetts General Hospital.
Next, we took genetic approaches to block trNK cell development and thereby augment the cytotoxic function of cNK cells in the skin. In a cancer setting, enhanced NK cell cytotoxicity is accomplished through the elimination of the immunosuppressive signal from TGFβ (37), which converts cNK to trNK cells (13). WT and Ncr1iCre,Tgfbr2fl/fl mice were transplanted with m157tg,B2m−/− skin grafts; however, no rejection was observed at day 20 after transplant (Fig. 3D). Likewise, the deletion of transcription factor Hobit, which is required for the development of liver trNK cells (38), in recipient mice did not affect m157tg,B2m−/− skin grafts after 20 days (Fig. 3D). Quantification of the frequency of cNK and trNK cells in the recipient liver and skin grafts revealed increased cNK and decreased trNK cells in m157tg,B2m−/− skin grafts in Ncr1iCre,Tgfbr2fl/fl and Hobit−/− compared with WT recipients (Fig. 3E). Therefore, the signal(s) in the skin that restrict NK cell cytotoxicity are not superseded by exogenous NK cell–stimulating factors, restriction of immunosuppressive TGFβ signaling, or prevention of cNK-to-trNK cell conversion.
m157 in the absence of self MHC-I induces robust skin graft rejection
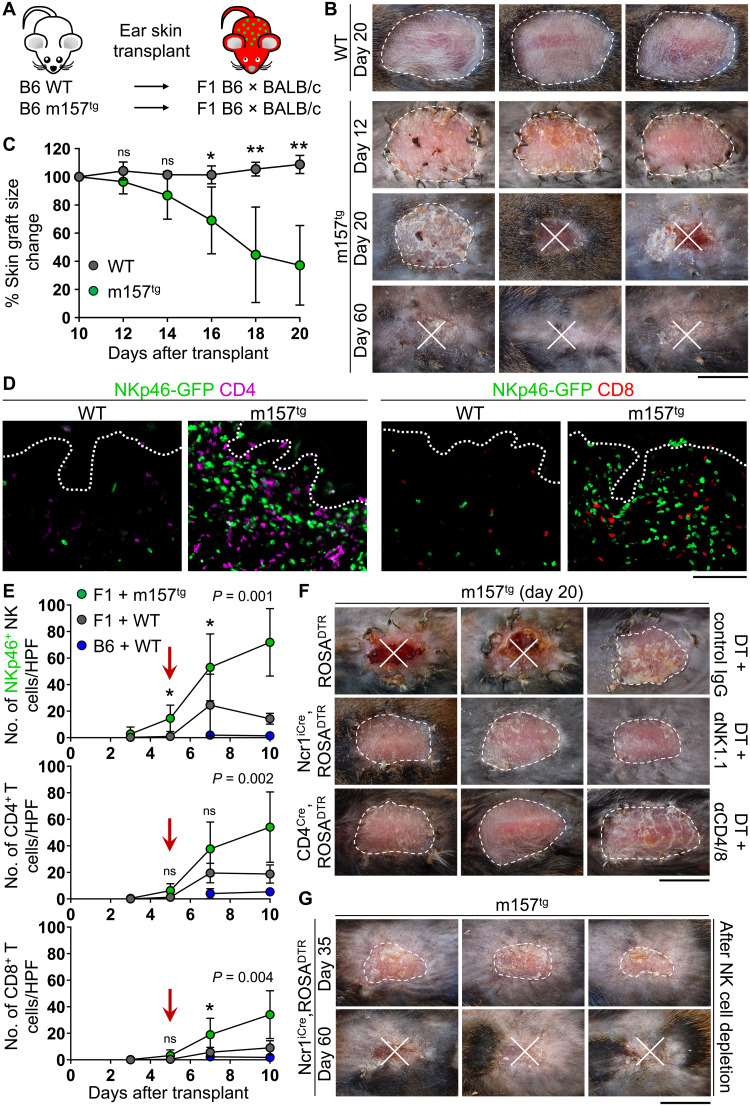
Our previous attempts to induce rejection of skin grafts through activation of the Ly49H pathway together with loss of MHC-I failed in a syngeneic transplant system. To preserve CD8+ T cell functionality while removing NK cell inhibition through the absence of self MHC-I, we generated Ncr1iCre,ROSAmT-mG recipient mice that were first-generation (F1) progeny of B6 × BALB/c parents and transplanted them with WT and m157tg B6 skin grafts (Fig. 4A). In this setting, CD8+ T cells were still functional through matched MHC-I signaling, while NK cells licensed by BALB/c MHC-I lost MHC-I–mediated inhibition against B6 donor skin cells. Although WT skin grafts remained intact in F1 recipients, most m157tg skin grafts were rejected by day 20 (Fig. 4B). The remainder of m157tg grafts were fully rejected by day 60 after transplant (Fig. 4B). m157tg skin graft size was significantly reduced compared with WT grafts starting from day 16 after transplant, similar to the timeline of mOVAtg skin graft rejection in WT B6 mice (Fig. 4C and fig. S5A). Thus, an NK cell–specific activating ligand in the context of missing-self in F1 recipients induces skin graft rejection; however, this rejection does not initiate within a 48- to 72-hour window expected of NK cell direct cytotoxicity. Because BALB/c mice do not express Ly49H, the proportion of Ly49H-expressing cNK cells in the B6 × BALB/c F1 mice was reduced compared with B6 mice (fig. S5B). However, similar to B6 recipient mice (Fig. 2E), most of the cNK cells that migrated into the m157tg skin graft in B6 × BALB/c F1 mice were Ly49H+ that down-regulated Ly49H receptor expression upon m157 engagement in the donor skin (fig. S5C). These data indicate that, despite fewer circulating Ly49H+ cNK cells in B6 × BALB/c F1 mice, these cells were fully active against m157-expressing B6 skin graft.
Fig. 4. Presence of activating m157 and loss of self-MHC induces robust skin graft rejection in allogeneic recipients.
(A) Schematic of ear skin transplant from B6 albino WT and m157tg donor mice to F1 B6 × BALB/c Ncr1iCre,ROSAmT-mG recipients. (B) Representative images of skin grafts at days 12, 20, and 60 after transplant. (C) The percentage change in skin graft size from day 10 to day 20 after transplant. n = 4 to 8 mice per group from two independent experiments. (D) Representative IF images of NKp46-GFP+ (green), CD4+ (purple), and CD8+ (red) cells in skin grafts at day 10 after transplant. Dotted lines indicate the epidermal basement membrane. (E) Quantification of NKp46-GFP+ NK, CD4+ T, and CD8+ T cells in the skin grafts at days 3, 5, 7, and 10 after transplant. Red arrows highlight NK cells infiltrating the skin before T cells. n = 3 to 9 mice per group from one to three independent experiments. (F) Comparison of m157tg skin graft rejection in control (ROSADTR + DT/control IgG), NK cell–depleted [Ncr1iCre,ROSADTR + DT/anti-NK1.1 (αNK1.1) antibody], and T cell–depleted [CD4Cre,ROSADTR + DT/anti-CD4/8 (αCD4/8) antibodies] F1 mice at day 20 after transplant. (G) Representative images of m157tg skin graft rejection in NK cell–depleted mice at days 35 and 60 after transplant following cessation of NK cell depletion at day 17 after transplant. (B, D, F, and G) n = 4 to 8 mice per group from two independent experiments. (C and E) Graphs show means ± SD, Mann-Whitney U test; *P < 0.05 and ***P < 0.001. (B, F, and G) Scale bars, 1 cm. (D) Scale bar, 100 μm. Photo Credit: Mark D. Bunting and Marta Requesens, Massachusetts General Hospital.
Significant increases in the number of NKp46-GFP+ NK, CD4+ T, and CD8+ T cells were found in m157tg skin grafts at day 10 after transplant (Fig. 4D). Quantifying the immune cell infiltrates over time revealed that significant numbers of NKp46-GFP+ NK cells appeared in m157tg skin grafts at day 5 after transplant, while CD4+ T and CD8+ T cells infiltrated m157tg skin grafts with a delay (Fig. 4E). Recruitment of NK and T cells into the donor epidermis was also detected at later time points (fig. S5D). To determine whether m157tg graft rejection in F1 recipients was NK cell dependent, we transplanted m157tg skin onto Ncr1iCre,ROSADTR F1 recipient mice and treated them with diphtheria toxin (DT) and anti-NK1.1 antibody over 18 days to deplete NK cells (fig. S5, E and F). ROSADTR F1 recipient mice administered with DT and control antibody were used as controls. Depletion of NK cells in F1 recipients prevented rejection of m157tg skin grafts (Fig. 4F). Depletion of T cells using CD4Cre,ROSADTR F1 mice together with DT and CD4- and CD8- depleting antibodies also prevented rejection of m157tg skin grafts (Fig. 4F and fig. S5, E and G). Tracking the survival of m157tg skin grafts in Ncr1iCre,ROSADTR mice after the termination of DT plus anti-NK1.1 antibody treatment demonstrated that recovery of NK cells in recipient mice led to complete graft rejection by day 60 after transplant (Fig. 4G). m157tg skin grafts began to recede and develop scabs approximately 20 days after NK cell depletion was stopped (Fig. 4G). These data demonstrate that NK and T cells are required for the rejection of m157tg skin grafts in the context of missing self. The NK cell–specific nature of the m157 activation signal and the timeline of NK and, later, T cell infiltration into the skin suggest that activated NK cells may have recruited T cells as effectors to reject the skin graft.
cNK cells switch their function from cytotoxicity to chemokine/cytokine production as soon as they exit the circulation
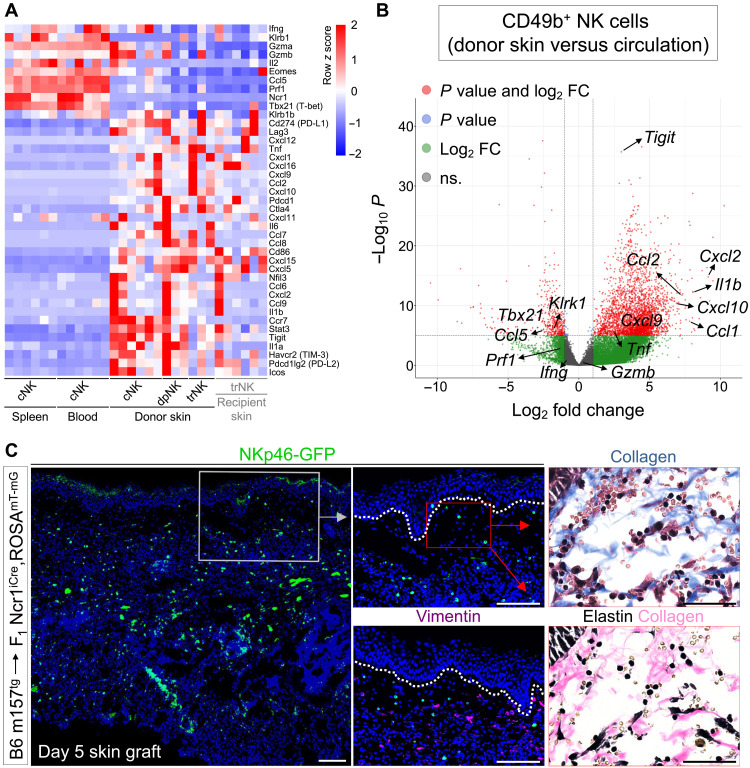
To define the function of NK cells that infiltrated m157-expressing skin grafts, we examined the transcriptional profiles of cNK cells in the circulation (spleen and blood); cNK cells recently emigrated from the circulation into m157-expressing skin grafts, dpNK and trNK cells in the skin grafts, and trNK cells in the recipient skin (fig. S6A). This strategy enabled us to identify pathways that were up- or down-regulated in cNK cells early during their entry into the skin microenvironment. Notably, we found a marked switch in cNK cells’ profile as they entered the skin grafts, which was highlighted by down-regulation of cytotoxicity-related genes and up-regulation of chemokines and inflammatory cytokines (Fig. 5A and fig. S6B). Ccl1, Ccl2, Cxcl2, Cxcl9, and Cxcl10 were significantly up-regulated in cNK cells in the skin graft compared to cNK cells in circulation (Fig. 5, A and B). Likewise, Il1a, Il1b, and Tnf were up-regulated in donor skin cNK cells (Fig. 5, A and B). No up-regulation was seen in Ifng, Gzma, or Gzmb expression, and Prf1 (perforin), Klrk1, Ccl5, Eomes, and Tbx21 (T-bet) were significantly down-regulated in donor skin cNK cells compared with their baseline expression in circulating cNK cells (Fig. 5, A and B). On the basis of RNA-seq data and confirmed by flow cytometry, we identified TGFβRII, IL-4Rα, CCR7, and TIGIT (T cell immunoreceptor with Ig and ITIM domains) as previously unknown markers to distinguish cNK cells in the skin from circulating cNK cells (fig. S6, C and D). Together, these data demonstrate that, upon entry into the skin, cNK cells undergo a drastic change in their function, which involves down-regulating their cytotoxic program while boosting their ability to provide “help” to neighboring immune cell populations through the production of chemokines and inflammatory cytokines.
Fig. 5. cNK cells’ transcriptome changes markedly as they emigrate from the circulation into ECM-rich skin grafts.
(A) Comparison of up- and down-regulated genes in circulating cNK cells, m157-expressing donor skin–derived cNK, dpNK, and trNK cells from donor and recipient skin as determined by RNA-seq analysis of sorted NK cell subpopulations. n = 6 mice for spleen, blood, cNK cell in donor skin, and trNK cells in recipient skin; n = 3 mice for donor dpNK and trNK cells in donor skin. Note that three F1 recipients with m157tg skin graft and three B6 recipients with m157tg,B2m−/− skin graft were included. NK cell sorting strategy is shown in fig. S6A. (B) Gene expression changes in donor skin–derived cNK cells compared with circulating cNK cells in the spleen and blood. FC, fold change. (C) Representative images of NKp46-GFP+ NK cells, vimentin+ fibroblasts, and collagen/elastin-stained adjacent sections of m157tg skin graft at day 5 after transplant onto F1 recipient mice. 4′,6-Diamidino-2-phenylindole (blue) marks the cell nuclei. n = 7 from two independent experiments. Dotted lines indicate the epidermal basement membrane. Scale bars, 100 μm; insets, 50 μm.
Considering the significant up-regulation of Tigit, Pdcd1, and Ctla4 in cNK cells entering the skin grafts (Fig. 5, A and B), we examined the impact of immune checkpoint blockade on the rejection of m157-expressing skin grafts (fig. S6E). Programmed cell death protein 1 (PD-1), TIGIT, or TIGIT/PD-1/CTLA4 (cytotoxic T-lymphocyte-associated antigen 4) triple antibody blockade did not result in rejection of m157tg or m157tg,B2m−/− skin grafts in WT B6 recipients (fig. S6, F and G). Furthermore, TIGIT/PD-1/CTLA4 triple antibody blockade did not accelerate the rejection of m157tg skin grafts in F1 recipients (fig. S6G), indicating the persistent lack of NK cell direct cytotoxicity in the skin grafts.
ECM proteins as potential modulators of cNK cell’s effector functions in the skin
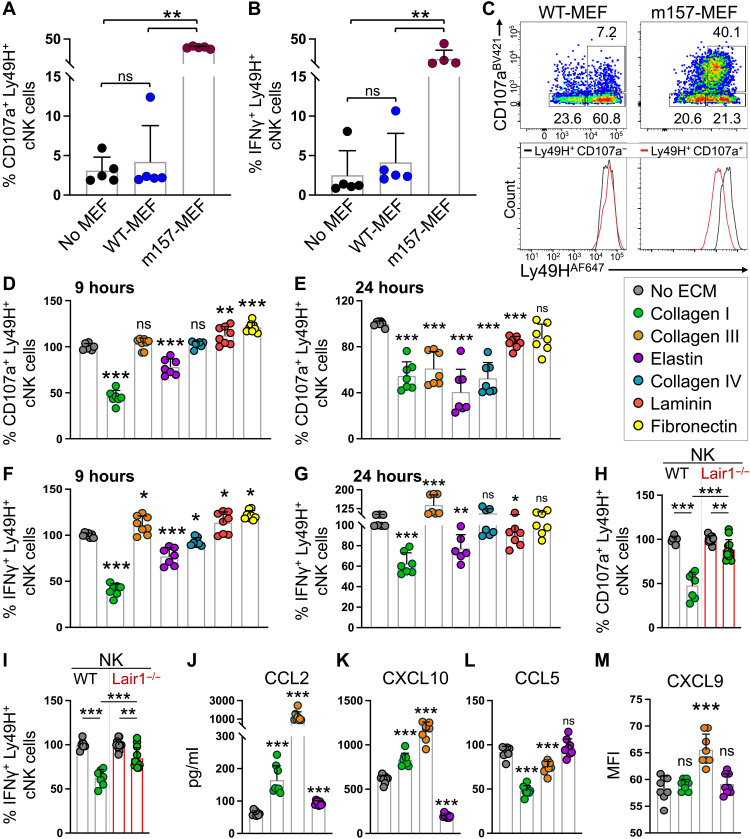
To identify the mediator(s) of NK cell functional switch in the skin, we examined the components of the skin microenvironment that interacted with NK cells as soon as they entered the skin grafts at day 5 after transplant (Fig. 4E). NK cells infiltrated broadly into m157tg dermal ECM, surrounded by collagen and, to a lesser extent, elastin (Fig. 5C and fig. S7A). In addition, occasional NK cells were detected contacting dermal fibroblasts at day 5 after transplant (Fig. 5C and fig. S7B). To elucidate the impact of ECM proteins and fibroblasts on cNK cell cytotoxicity, we examined splenic cNK cell degranulation and cytokine production in coculture assays. WT mouse embryonic fibroblasts (WT-MEFs) did not block Ly49H+ cNK cell degranulation or IFNγ production in response to IL-12 and IL-15 stimulation (Fig. 6, A and B). Furthermore, fibroblasts expressing m157 (m157-MEFs) markedly induced Ly49H+ cNK cell degranulation and IFNγ production, which was associated with Ly49H down-regulation (Fig. 6, A to C). In contrast, major dermal ECM proteins, collagen I, collagen III, and elastin, potently blocked Ly49H+ cNK cell degranulation after 9 and 24 hours of coculture with m157-MEFs (Fig. 6, D and E). Collagen I and elastin also suppressed IFNγ production by Ly49H+ cNK cells exposed to m157-MEFs (Fig. 6, F and G). On the other hand, major epidermal basement membrane proteins—collagen IV, laminin, and fibronectin—played a minor role in altering NK cell functions (Fig. 6, D to G). Leukocyte-associated Ig-like receptor 1 (LAIR1) is an inhibitory receptor expressed by NK cells that binds collagens (39, 40). Deletion of Lair1 partially rescued collagen I–induced blockade of Ly49H+ cNK cell degranulation and IFNγ production upon in vitro exposure to m157-MEFs (Fig. 6, H and I). However, global Lair1 deletion failed to induce the rejection of m157tg,B2m−/− skin graft in Lair1−/− mice (fig. S7C). This suggests that the partial LAIR1 dependency observed in our in vitro study may be compensated by complex interplay between NK cells and other ECM proteins in the skin. The close linkage between Ncr1 and Lair1 genes on chromosome 7 in mice prevented us from generating Ncr1iCre,Lair1fl/fl mice to examine the NK cell–specific role of LAIR1 in vivo. Notably, collagen I and III significantly induced the chemokines CCL2 and CXCL10 but suppressed CCL5 release by splenic cNK cells cultured with m157-MEFs (Fig. 6, J to L). Collagen III also induced CXCL9 release by cNK cells (Fig. 6M).
Fig. 6. Dermal ECM proteins modulate cNK cell effector function in vitro.
(A and B) Quantification of CD107a (A) and IFNγ (B) by Ly49H+ splenic cNK cells after 7 hours coculture with no MEFs, WT-MEFs, or m157-MEFs in the presence of IL-12 and IL-15. n = 5 per group. (C) Representative dot plots of CD107a by splenic cNK cells cocultured with WT-MEFs or m157-MEFs. Representative histograms of Ly49H expression by Ly49H+CD107a+ NK cells following coculture with m157-MEFs and WT-MEFs. Histograms of Ly49H+CD107a− cNK cells (black) are shown as controls. Numbers on the dot plots represent the percent cells within each gate. (D to G) Quantification of CD107a (D and E) and IFNγ (F and G) expression by Ly49H+ splenic cNK cells cocultured with different ECM proteins and m157-MEFs at 9 hours (D and F) and 24 hours (E and G). Percentages are normalized on the basis of the “No ECM” group. n = 7 per group from two independent experiments. (H and I) Comparison of CD107a (H) and IFNγ (I) by Ly49H+ splenic cNK cells from WT and Lair1−/− mice in the presence of m157-MEFs and collagen I. Percentages are normalized on the basis of the no ECM group in WT (n = 7) and Lair1−/− (n = 12) experiments. (J to M) Measurement of CCL2 (J), CXCL10 (K), CCL5 (L), and CXCL9 (M) in the culture medium of splenic cNK cells cocultured with m157-MEFs in the presence of no ECM, collagen I and III, or elastin at 12 hours (CXCL10 and CCL5) and 24 hours (CCL2 and CXCL9). n = 8 per group from two independent experiments. (A, B, and D to M) Graphs show means ± SD. Mann-Whitney U test; *P < 0.05, **P < 0.01, and ***P < 0.001 compared with no ECM group.
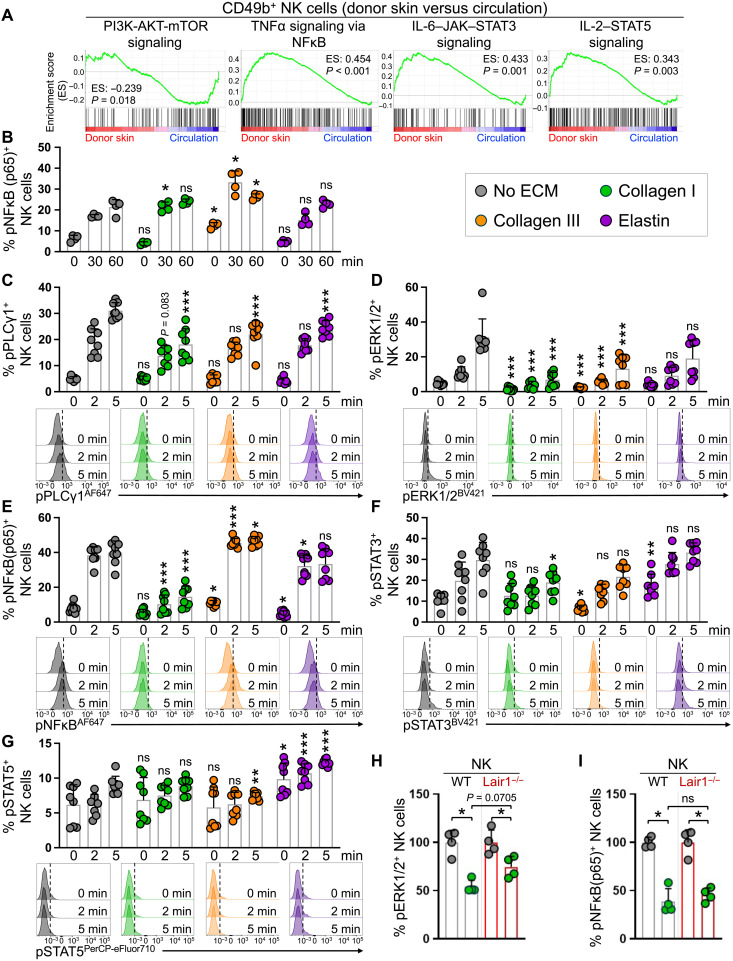
To gain a greater insight into the underlying molecular mechanisms linking the ECM to changed NK cell functionalities, we investigated the signaling pathways that were associated with the switch in cNK cells’ profile from cytotoxicity to an inflammatory response (fig. S6B). cNK cells entering the skin down-regulated phosphatidylinositol 3-kinase (PI3K)–AKT pathway while up-regulating nuclear factor κB (NFκB), signal transducer and activator of transcription 3 (STAT3), and STAT5 signaling pathways compared with circulating cNK cells (Fig. 7A). We found that collagen III significantly induced phosphorylated NFκB (pNFκB) (p65) in splenic cNK cells at baseline, which was highly amplified after 30- and 60-min exposure to m157-MEFs (Fig. 7B). To examine the impact of ECM proteins on NK cell signaling upon activation in a time-controlled setting that excluded the possibility for ECM proteins to physically block activating ligand-receptor interactions, we activated NK cells with H2O2 in the presence of collagens and elastin (41). Notably, collagens and elastin markedly reduced cytotoxicity-associated phosphorylated phospholipase C–γ1 (pPLCγ1) and phosphorylated extracellular signal–regulated kinase 1/2 (pERK1/2) levels in the NK cells (Fig. 7, C and D). In contrast, collagen III increased pNFκB in NK cells at baseline and after 2- and 5-min exposure to H2O2 (Fig. 7E). Elastin increased phosphorylated STAT3 (pSTAT3) at baseline and highly induced pSTAT5 at baseline and after 2- and 5-min exposure to H2O2 (Fig. 7, F and G). Collagen I showed a suppressive effect on pNFκB and pSTAT3 (Fig. 7, E and F). Loss of Lair1 in NK cells partially reversed pERK1/2 suppression by collagen I but had no impact on NFκB phosphorylation after 2-min exposure to H2O2 (Fig. 7, H and I). Consistent with their critical role in regulating cNK cell function, we found the expression of multiple ECM protein receptors on cNK cells entering the skin microenvironment (fig. S8). These findings indicate that ECM proteins including collagen I, collagen III, and elastin are potent regulators of cNK cell function, which collectively may lead to reduced cytotoxicity and increased chemokine and cytokine production by NK cells as they enter the peripheral tissues.
Fig. 7. Collagens and elastin modulate cytotoxicity and inflammation-associated signaling pathways in NK cells.
(A) Gene set enrichment analysis (GSEA) enrichment plots of PI3K-AKT-mTOR, TNFα-NFκB, IL-6–JAK–STAT3, and IL-2–STAT5 signaling in CD49b+ cNK cells in the donor skin compared with the circulation. The enrichment scores (ES) and P values are indicated in each plot. (B) Quantification of phospho-NFκB (pNFκB) (p65) in splenic cNK cells at 30 and 60 min after coculture with m157-MEFs in the presence of different ECM proteins. n = 4 per group, data representative of two independent experiments. (C to G) Quantification of phospho-PLCγ1+ (pPLCγ1+) (C), pERK1/2+ (D), pNFκB (p65) + (E), pSTAT3+ (F), and pSTAT5+ (G) in splenic cNK cells at 2 and 5 min after stimulation with H2O2 in the presence of different ECM proteins. n = 8 per group from two independent experiments. Representative flow cytometry histograms comparing the phosphorylation of key signaling proteins at 2 and 5 min after stimulation with H2O2 in response to no ECM (gray), collagen I (green), collagen III (orange), and elastin (purple). Black dotted line corresponds to the gating on the basis of the “no ECM – 0 min” histogram for each signaling protein. (H and I) Comparison of pERK1/2 (H) and pNFκB (p65) (I) in splenic NK cells from WT and Lair1−/− mice in response to 2-min H2O2 stimulation in the presence of collagen I. Percentages are normalized on the basis of the no ECM group in WT and Lair1−/− experiments. n = 4 per group; data are representative of two independent experiments. (B to I) Graphs show means ± SD. Mann-Whitney U test; *P < 0.05, **P < 0.01, and ***P < 0.001. (C to G) Each time point is compared with the corresponding time point in no ECM group.
ECM proteins suppress NK cell antitumor response in the skin
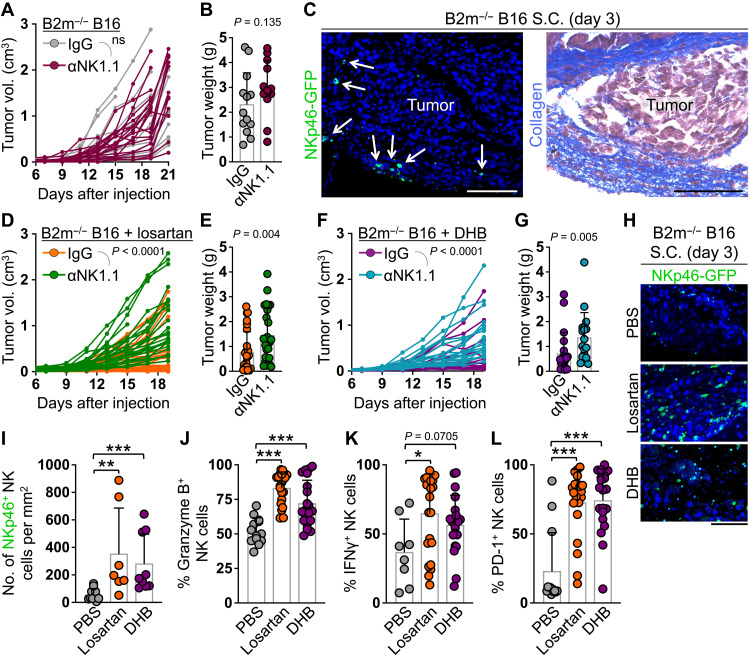
Considering the essential role of ECM proteins in skin homeostasis and wound healing, our attempts to remove or degrade ECM proteins in the skin transplantation system led to the failure of skin engraftment. Thus, we examined the role of ECM proteins in suppressing NK cell cytotoxicity against cancer cells in the skin. We subcutaneously injected B2m−/− B16-F10 melanoma cells (B16) into the flanks of WT mice and treated them with control IgG or anti-NK1.1 antibody to determine whether NK cells inhibited the growth of MHC-I–deficient tumors in the skin (fig. S9A). Similar to WT melanoma, NK cell depletion did not affect the growth or terminal weight of B2m−/− melanoma in the skin (Fig. 8, A and B, and fig. S9B). NK cells infiltrated B2m−/− melanoma as early as day 3 after tumor injection but were embedded in a collagen-rich ECM surrounding the tumor foci (Fig. 8C and fig. S9C). To investigate the role of tumor collagen deposition in suppressing NK cell immunity against melanoma in the skin, we treated mice that received B2m−/− B16 subcutaneously with losartan (60 mg/kg) to block collagen synthesis or collagen prolyl 4-hydroxylase inhibitor, 3,4-dihydroxybenzoic acid (DHB; 40 mg/kg), which blocks collagen assembly into a triple helix (fig. S9A) (42, 43). Losartan and DHB treatment protected the mice from B2m−/− melanoma growth in the skin in an NK cell–dependent manner, which led to a significant survival benefit (Fig. 8, D to G, and fig. S10, A to D). NK cells highly infiltrated the tumor parenchyma in losartan- and DHB-treated mice and showed markedly elevated expression of granzyme B, IFNγ, and PD-1 (Fig. 8, H to L). Unlike NK cells, losartan and DHB treatment did not induce T cell recruitment into the B2m−/− B16 tumors (fig. S10, E and F). Furthermore, the proportions of tumor-infiltrating regulatory T cells, macrophages, and myeloid-derived suppressor cells remained unaltered or increased following losartan and DHB treatments (fig. S10, G to J). To further confirm that the NK cell immunity unleashed upon losartan and DHB treatment was related to their effects on blocking collagen deposition in the tumor, we treated a group of mice that received B2m−/− B16 subcutaneously with collagenase and hyaluronidase mixture and found a similar dependency of tumor growth on NK cells (fig. S10K). NK cell depletion in phosphate-buffered saline (PBS) (carrier control)–, losartan-, and DHB-treated mice resulted in lymph node metastasis of B2m−/− melanoma, suggesting a role for NK cells in the clearance of metastasizing melanoma cells (fig. S10L). Consistent with this effect, and in contrast to the lack of NK cell’s ability to control melanoma in the skin, NK cells effectively killed intravenously injected WT and B2m−/− B16 cells and suppressed melanoma lung metastasis (fig. S11). Collectively, these data indicate strong NK cell killing of MHC-I–deficient tumor cells in the circulation, which is likely blocked by ECM proteins in peripheral tissues.
Fig. 8. Collagen deposition blocks NK cells from eliminating B2m-deficient melanoma cells in the skin.
(A and B) B2m−/− B16 melanoma subcutaneous growth over time (A) and terminal weight (B) in WT mice treated with control IgG [n = 17 for (A) and 13 for (B)] or anti-NK1.1 antibody [n = 19 for (A) and n = 13 for (B)]. (C) Representative images of NKp46-GFP+ NK cell (white arrows) IF and collagen stain in adjacent sections of a B2m−/− B16 melanoma at day 3 after subcutaneous (S.C.) injection in Ncr1iCre,ROSAmT-mG mice. (D to G) B2m−/− B16 melanoma subcutaneous growth over time (D and F) and terminal weight (E and G) in WT mice treated with losartan (D and E) or DHB (F and G) plus control IgG [n = 28 (D), 20 (F), 27 (E), and 20 (G)] or anti-NK1.1 antibody [n = 26 (D), 20 (F), 25 (E), and 18 (G)]. (H and I) Representative images (H) and quantification (I) of NKp46-GFP+ NK cells in B2m−/− B16 melanoma at day 3 after subcutaneous injection in PBS-treated (n = 10), losartan-treated (n = 7), and DHB-treated (n = 9) Ncr1iCre,ROSAmT-mG mice. (J to L) Quantification of granzyme B (J), IFNγ production (K), and PD-1 surface expression (L) on NK cells isolated from B2m−/− B16 melanoma at days 19 to 21 after subcutaneous injection in PBS-treated [n = 13 (J and L) and 8 (K)], losartan-treated [n = 21 (J) and 22 (K and L)], and DHB-treated [n = 20 (J, K, and L)] mice. (A, D, and F) Two-way analysis of variance (ANOVA). (B, E, G, and I to L) Graphs show means ± SD, Mann-Whitney U test. *P < 0.05, **P < 0.01, and ***P < 0.001. (C and H) Scale bars, 100 μm.
To translate our fundamental findings to human cancers, we analyzed The Cancer Genome Atlas (TCGA) and other publicly available datasets for the frequency of B2M gene alteration and mutations in multiple types of solid cancers and leukemias. While B2M gene mutations and deletion were found across solid cancer types including those with low mutational burden, leukemias did not carry any aberrations in the B2M gene (fig. S12, A and B). Furthermore, B2M gene expression in acute myeloid leukemia samples was consistently high and showed the least variance compared with solid cancer types (fig. S12C). These findings indicate a remarkable resistance to down-regulation of the B2M gene in blood cancers even when confronted with CD8+ T cell pressure. Thus, circulating cytotoxic NK cells likely eliminate leukemias that lose MHC-I, while solid cancers that down-regulate MHC-I in peripheral tissues are protected because of the impaired NK cell cytotoxicity caused by ECM proteins.
DISCUSSION
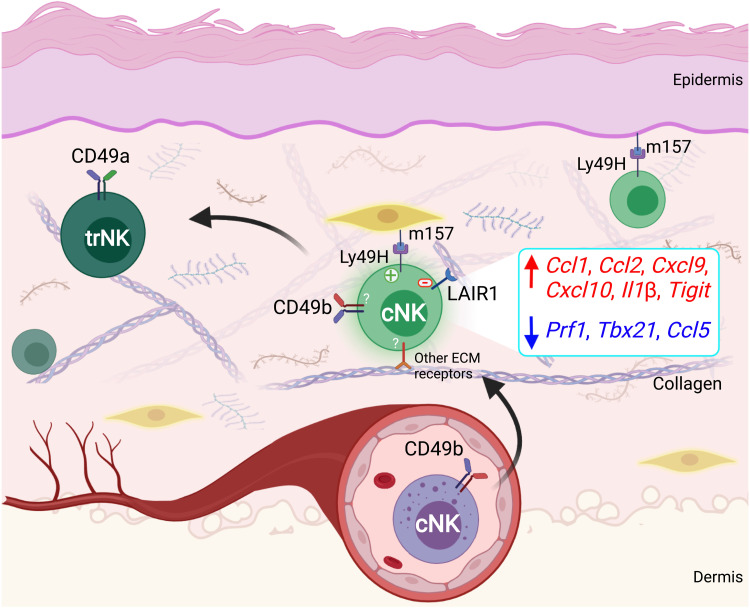
NK cell killing of virally infected or malignant cells is a vital aspect of innate immunity. Yet, the mechanisms behind the inability of NK cells to directly eliminate NK cell–sensitive targets in peripheral tissues have remained undetermined. We show here, using skin transplantation and cancer models, that, once NK cells exit the circulation and enter the target tissue, they experience a profound change in their function (Fig. 9). Our data strongly suggest that the interplay between several ECM proteins and NK cell receptors plays a cooperative role in this functional shift immediately upon entry of cNK cells into the skin. NK cell–ECM protein interactions suppress NK cell’s direct cytotoxicity and instead promote its helper function to recruit and activate adaptive immunity. Although this regulation of NK cell cytotoxicity may limit nonspecific tissue damage in the context of localized viral infection in peripheral tissues, it allows the outgrowth of cancer cells that have managed to evade cytotoxic T cell immunity.
Fig. 9. Schematic diagram of ECM protein–mediated switch in cNK cell effector function in the skin.
Created with BioRender.com
Large numbers of NK cells are recruited into m157-expressing donor skin; however, NK cells are unable to directly reject the skin grafts even with the loss of MHC-I and the addition of NK cell–stimulating poly(I:C) and cytokines in a syngeneic model. This demonstrates that the MHC-I–independent inhibition of NK cell’s cytotoxicity dominated over multiple strong activation signals tested. Skin graft rejection is accomplished using F1 recipients, which provides missing self for NK cell activation while simultaneously preserving MHC-I expression on the donor cells. In this system, NK cells licensed by BALB/c MHC-I are fully activated against B6 MHC-I–expressing m157+ donor skin cells (i.e., missing self) (25), while CD8+ T cells are able to contribute to the rejection as the MHC-I signaling axis remains operational. Graft rejection begins after day 12 after transplant, which is approximately 7 days later than the initial accumulation of NK cells in the graft. Most of the rejection is completed by day 20 after transplant, which is in agreement with the timing of an adaptive immune response. The timing of NK and T cell infiltration into the skin graft, the kinetics of graft rejection, and the requirement for both NK and T cells in this process suggest that NK cells activate the adaptive immune response and T cells function as effectors to reject m157tg donor skin grafts in F1 recipients.
NK cell activation has been associated with the production of high levels of IFNγ and TNFα in addition to CCL4, CCL5, XCL1, and granulocyte-macrophage colony-stimulating factor (GM-CSF) (44–46). These cytokines and chemokines promote macrophage activation together with recruitment and activation of dendritic cells and T cells at sites of inflammation (47–51). We find that activated cNK cells entering the skin from circulation immediately up-regulate the CXCR3 ligands, CXCL9 and CXCL10, in addition to CCL2 and CXCL5, chemokines that profoundly induce T cell recruitment. In contrast, CCL5 is down-regulated and CCL4, XCL1, and GM-CSF expression is not altered in cNK cells as they exit the circulation. A transient increase in CCR7 is also observed, which may drive newly recruited cNK cells deeper into the skin and, subsequently, the draining lymph nodes, where CCL21+ lymphatic endothelial cells reside. A concomitant reduction in the expression of T-bet and eomesodermin (EOMES) and an increase in inducible T-cell costimulator (ICOS) are noted, which may drive a helper cNK cell functional program. Furthermore, the cytotoxic effectors Gzma, Gzmb, and Prf1 are either reduced or not changed in activated cNK cells as they exit the circulation, supporting our observations of limited NK cell direct cytotoxicity in the skin. Although NK cells may directly activate CD8+ T cells in the skin graft, an indirect path through dendritic cells and/or CD4+ T cell activation likely plays an important role in the observed NK cell helper function, which will be further investigated in future studies (46, 51, 52). While direct killing of some target cells by NK cells may occur, the rejection of m157tg skin graft is driven by CD8+ and CD4+ T cells, which likely recognize m157 as foreign antigen expressed by the donor skin cells (53–55).
Previous studies have shown a role for TGFβ in impairing NK cell function in cancer models by promoting the conversion of cNK cells to trNK cells (12, 13, 37). We show that the impairment in NK cell cytotoxicity is an immediate consequence of cNK cell exit from the circulation into a peripheral tissue microenvironment. In response to skin transplantation, CD49a+CD49b− trNK cells develop from cNK cells that have exited the circulation and populated the skin graft over time. We find that cNK cells up-regulate TGFβ receptor as soon as they exit circulation, which may promote their conversion into trNK cells in the TGFβ-rich environment of the skin graft. Although they may provide cytokine-mediated help to T cells later in the response, trNK cells are not seen in notable numbers until after day 14 after transplant. In addition, blocking trNK cell development by deleting Tgfbr2 (12, 13) or Hobit transcription factor (38) does not result in NK cell cytotoxicity or graft rejection. These findings demonstrate that the switch in NK cell function from killer to helper is an immediate consequence of its exit from circulation and entry into the peripheral tissue microenvironment.
Cancers in peripheral tissues that are expected to be strong targets for NK cells have often failed to show objective responses (56–58). Our results suggest that NK cells experience inhibition of their cytotoxicity likely mediated by collagens and elastin once they exit the circulation and enter the stroma. This may explain the distinct selection for loss of MHC-I by solid cancers but not leukemias in humans. We speculate that NK cell’s rapid cytotoxic response in the circulation and delayed helper response in tissues can be explained by selection pressure to prolong host survival. The suppression of a cytotoxic innate immune response in the peripheral tissues may prevent overreaction to localized pathological insults, which could predispose to excessive tissue damage and the development of chronic inflammation. Meanwhile, a “helper” immune response for the development of an overall more targeted, strength-appropriate adaptive immunity may be best suited to combat viral infections in peripheral tissues. In contrast, infection of the blood requires immediate control to ensure host survival. These evolutionary choices may explain the switch in NK cell function as they exit circulation and enter peripheral tissues. Future studies are required to examine this concept and determine the role of each ECM protein and its receptor in regulating NK cell function in peripheral tissues.
MATERIALS AND METHODS
Study approval
Animal studies were approved by the Massachusetts General Hospital Institutional Animal Care and Use Committee (IACUC).
Mice
C57BL/6 albino WT, B2m−/−, m157tg, m157tg,B2m−/−, C57BL/6 Ncr1iCre,ROSAmT-mG, Ncr1iCre,Tgfbr2fl/fl, Act-mOVAtg, C57BL/6 × BALB/c F1 Ncr1iCre,ROSAmT-mG, C57BL/6 Ncr1iCre,ROSADTR, C57BL/6 × BALB/c F1 Ncr1iCre,ROSADTR, and C57BL/6 × BALB/c F1 CD4Cre,ROSADTR were bred in house. C57BL/6 mice (Charles River Laboratories, Wilmington, MA, USA; strain code: 207) were used in the parabiosis, LEGENDplex, and tumor experiments. C57BL/6-Ly5.1 mice (Charles River Laboratories) were used for in vivo BM rejection experiments. C57BL/6 Hobit−/− mice and Lair1−/− mice were gifts from K. P. van Gisbergen (Sanquin Research and Landsteiner Laboratory, Amsterdam UMC, Amsterdam, The Netherlands) and S. Komarova (Faculty of Dentistry, McGill University, Quebec, Canada), respectively. Mutant mice were genotyped using the primers listed in table S1A. All mice were housed under specific pathogen–free conditions and given water and food ad libitum, in the animal facility at the Massachusetts General Hospital in accordance with animal care regulations. All mice were closely monitored by the authors, facility technicians, and an independent veterinarian when necessary. All procedures were performed according to the protocols approved by the IACUC at the Massachusetts General Hospital.
Skin transplantation
C57BL/6 albino ear skin from WT, B2m−/−, m157tg, m157tg,B2m−/−, and Act-mOVAtg mice was harvested after euthanasia, cartilage was removed, and skin was placed in a petri dish with 1× PBS (Life Technologies, Thermo Fisher Scientific, Grand Island, NY, USA; catalog no. 14190144). Recipient mice were anesthetized for the procedure with an intraperitoneal injection of 4.5 μl/g body weight of ketamine (10 mg/ml) (KetaVed, Boehringer, Ingelheim Vetmedica, Fort Dodge, IA, USA; catalog no. 045-290) combined with xylazine (0.5 mg/ml) (AnaSed, Akorn, Lake Fortes, IL, USA; catalog no. 139-236) sterile solution in addition to isofluorane (Baxter Healthcare Corporation, Deerfield, IL, USA; catalog no. 1001936040) as required. Mice were placed on a heating pad during and after the surgery. Backs of recipient mice were shaved and cleansed with 70% ethanol. A circular piece of recipient skin approximately 1.5 times the area of ear skin to be transplanted was excised from the back of recipient mice, and subcutaneous fascia was removed. Donor ear skin was placed split side down and sutured using no. 6-0 silk surgical sutures (Ethicon, Puerto Rico; catalog no. K889H). Approximately one dozen of equidistant sutures were made around the donor skin to attach the ear to the recipient tissue. Next, the skin transplant was coated with antibiotic ointment (Medline Industries, Northfield, IL, USA; catalog no. CUR001231) and protected with a 2 cm by 2 cm patch of Vaseline gauze (Convidien, Mansfield, MA, USA; catalog no. 884413605). A bandage (Tensoplast, BSN Medical, Charlotte, NC, USA; catalog no. 02115-00) was wrapped around the torso of the mouse and sutured in place using no. 4-0 silk surgical sutures (Ethicon, Puerto Rico; catalog no. K871) on the dorsal region suturing into the back skin adjacent to the shoulders and hips. Seven days after surgery, the bandage was removed, and donor skin was monitored daily for any sign of inflammation and/or rejection. Pictures were taken every other day, and ImageJ (version 1.52a) was used to measure graft size. Recipient and donor mice were sex- and aged-matched.
Tissue harvesting
Mice were anesthetized using ketamine/xylazine, and 1 μg of monoclonal anti–CD45-Brilliant Violet (BV) 605 (clone 30-F11, BioLegend, San Diego, CA; catalog no. 103155) was injected by retro-orbital injection to label circulating CD45+ cells. After 3 min had elapsed, peripheral blood was collected by retro-orbital bleeding, and, following euthanasia, the liver, spleen, lymph nodes, and donor and recipient skin were collected for analysis. Red blood cells (RBCs) in peripheral blood, spleen, and liver were lysed using RBC lysis buffer (10×; BioLegend, catalog no. 420301). After washing with 1× PBS/2% fetal calf serum (FCS)/5 mM EDTA, 5 × 106 cells were prepared for flow cytometry staining.
Skin harvesting
The back of the mouse was shaved, and a large rectangle of shaved skin encompassing the donor graft and recipient skin was excised. Large regions of fat on the underside of the skin were carefully scraped away, and then the donor and recipient skin was separated with a razor blade. These were placed in separate dishes and chopped with scissors into ~1-mm pieces. Each sample was then placed into a 15-ml tube with 10-ml digestion buffer [RPMI 1640 (Life Technologies, catalog no. 21870076) and collagenase IV (200 U/ml; Worthington Biochemical, Lakewood, NJ, USA)]. The skin was incubated for 2 hours at 37°C with shaking. Following incubation, digested skin was poured onto a 70 μM strainer placed in a 50-ml tube. The 15-ml tube was rinsed with ~3 ml of 1× PBS/2% FCS/5 mM EDTA and added to the digested sample. Skin was mashed with a plunger through the 70 μM strainer that was then rinsed approximately three times with a small volume of 1× PBS/2% FCS/5 mM EDTA. The cells were centrifuged at 300g for 5 min at 4°C and resuspended in 2.4G2 blocking solution before flow cytometry staining. NK cells were stained for activating, inhibitory receptors and CD107a surface expression for baseline evaluation. For the ex vivo NK cell stimulation assay, donor skin graft–derived cells were resuspended in RPMI/10% fetal bovine serum (FBS)/1% penicillin and streptomycin (P/S; Thermo Fisher Scientific, Waltham, MA; catalog no. 14140122)/20 mM glutamine and were plated on control IgG (clone MOPC-173, BioLegend, catalog no. 400203) or anti-NK1.1 (clone PK136, BioLegend, catalog no. 108703) precoated plate (Nunc MaxiSorp plate, BioLegend, catalog no. 423501) or treated with 1× PMA/Ion solution (BioLegend, catalog no. 423301). The cells were incubated at 37°C/5% CO2 for a total of 4 hours with the last 3 hours in the presence of 1 μl of anti–CD107a-BV421 (1D4B, BioLegend, catalog no. 121618), 1× brefeldin A (BioLegend, catalog no. 420601), and 1× monensin (BioLegend, catalog no. 420701). Following the coculture, cells were washed with 1× PBS/2% FCS/5 mM EDTA, and the pellet was resuspended in 2.4G2 blocking solution before flow cytometry staining.
Liver harvesting
The peritoneal cavity was opened to expose the liver, and 5 to 10 ml of 1× PBS were injected into the hepatic portal vein using a 27G needle to perfuse the liver. The gall bladder was then removed, and all lobes of the liver were collected and mashed through a 70-μm filter sitting in a 50-ml tube using a syringe plunger. The filter was rinsed with 1× PBS/2% FCS/5 mM EDTA three to four times during the mashing procedure. The cell suspension was centrifuged at 300g for 5 min at 4°C and then supernatant-aspirated, and the cell pellet washed with 10 ml of 1× PBS/2% FCS/5 mM EDTA. Following centrifugation and aspiration of the supernatant, the pellet was resuspended in 25 ml of isotonic Percoll [8.44 ml of Percoll (Healthcare Biosciences, Uppsala, Sweden; catalog no. 17-0891-01)/0.47 ml of 20× PBS/16.09 ml of 1× PBS] at room temperature (RT). Liver cell suspensions were centrifuged at 693g for 12 min at RT with no brake; the leukocyte pellet at the bottom of the tube was then washed with 10 ml of 1× PBS/2% FCS/5 mM EDTA and RBC-lysed (RBC lysis buffer 10×, BioLegend, catalog no. 420301). Following washing with 1× PBS/2% FCS/5 mM EDTA, the pellet was resuspended in 2.4G2 blocking solution before flow cytometry staining.
Flow cytometry
Single-cell suspensions from all samples were prepared by straining through a 70-μm filter. 2.4G2 supernatant–treated cells were stained in 1× PBS/2% FCS/5 mM EDTA with the appropriate surface antibodies (table S1B) for 30 min at 4°C, washed, and analyzed by flow cytometry. For intracellular staining, cells were fixed and permeabilized using the True-Nuclear Transcription Buffer Set (BioLegend, catalog no. 424401) according to the manufacturer’s protocol, then incubated with appropriate antibodies (table S1B) for 60 min at RT, washed, and analyzed by flow cytometry. For in vitro signaling experiments, following the specified time points, cells were fixed with an equal volume of prewarmed 1× aldehyde-based fixation buffer (BioLegend, catalog no. 420801) for 20 min at RT. Fixed cells were permeabilized by dropwise addition of chilled methanol-based solution True-Phos Perm Buffer (BioLegend, catalog no. 425401) while mixed on a vortex. Cells were stored in −20°C and in the dark for 1 to 3 days before washing them two times with ~3 ml of 1× PBS/2% FCS/5 mM EDTA. Cells were subsequently stained with appropriate antibodies against surface markers and specific phosphorylated signaling proteins (table S1B) for 60 min at RT, washed, and analyzed by flow cytometry. Cells were assayed on a BD LSRFortessa X-20 flow cytometer (BD Bioscience, Billerica, MA, USA), and data were analyzed using FlowJo software version 10 (Tree Star, Ashlad, OR, USA). NK cells in WT nonreporting mice were identified as CD3−NK1.1+NKp46+.
BM transplantation
BM cells from B6-Ly5.1 (CD45.1), B6 WT (CD45.2), and B6 B2m−/− (CD45.2) were collected from the tibia and femur by flushing bones with RPMI 1640 into a petri dish under sterile conditions. Cells were then counted and resuspended in RPMI 1640. Ly5.1:WT (control) and Ly5.1:B2m−/− (test) BM cells were mixed 1:1 and intravenously injected into sublethally irradiated (450 cGy) Ncr1icre,ROSAmT-mG recipients. Three days later, spleens of recipient mice were harvested, and the ratio of WT:Ly5.1 and WT:B2m−/− cells was determined by flow cytometry. The percentage of rejection was calculated as follows
Adoptive NK cell transfer
m157tg,B2m−/− donor skin was transplanted onto the back of C57BL/6 WT mice following the abovementioned skin transplant procedure. On day 1 after transplant, six spleens from Ncr1iCre,ROSAmTmG mice were harvested and mashed through a 70-μm filter. Spleens were resuspended in RBC lysis for 2 min and washed with 1× PBS/2% FCS/5 mM EDTA. NKp46-GFP+ROSA− cells were sorted using a Sony FX500 cell sorter (Sony Biotechnology, San Jose, CA, USA). Sorted cells were centrifuged, and 8.5 × 105 NKp46-GFP+ cells resuspended in 200 μl of sterile RPMI 1640 were filtered through a 40-μm strainer followed by intravenous injection into recipient mice using a sterile 28G syringe (0.5-ml BD insulin syringe, Franklin Lakes, NJ, USA; catalog no. 329461). Negative controls consisted of mice injected with 200 μl of RPMI 1640 alone. Twenty days after transplant, mice were euthanized, and peripheral blood, spleen, liver, and donor and recipient skin were collected for analysis.
Parabiosis
Parabiont partners were cohoused in the same cage for a week before the surgery. Eight- to 12-week-old female C57BL/6 WT mice were surgically connected to Ncr1iCre,ROSAmTmG weight- and age-matched partner female mice. Surgery was performed on a heating pad, and animals were anesthetized using isoflurane. Longitudinal skin incisions were made on the shaved sides of each animal. Knee and elbow joints from each animal were first sutured using a no. 4-0 surgical suture. Following attachment of joints, the skin of the animals was connected using no. 6-0 sutures ending with a double surgical knot. To minimize pain, 0.1 mg of carprofen (Rimadyl, Zoetis, Brazil; catalog no. 141-199) was injected intraperitoneally each day for 2 days after surgery, in addition to close monitoring every day for signs of pain or stress. On day 21 after parabiosis, ear skin from m157tg,B2m−/− mice was transplanted onto the back of the C57BL/6 WT mouse of each pair. After 20 days, mice were euthanized, and peripheral blood, spleen, liver, and donor and recipient skin from the C57BL/6 WT parabiont were collected for analysis.
NK cell sorting
At day 20 after m157tg,B2m−/− transplantation onto Ncr1iCre,ROSAmTmG B6 mice and day 10 after m157tg transplantation onto Ncr1iCre,ROSAmTmG F1 mice, NK cells from blood, spleen and donor skin (cNK and trNK), and recipient skin (trNK cells) were sorted for RNA-seq. First, mice were anesthetized using ketamine/xylazine, and 1 μg of monoclonal anti–CD45-BV605 (clone 30-F11, BioLegend) was injected via the retro-orbital route 3 min before tissue harvest to label circulating CD45+ cells. Tissues were harvested as described above, and, following staining, NKp46+ROSA−CD45+CD49a−CD49b+ (cNK) cells from peripheral blood and spleen; CD3−NKp46+ROSA−CD45−CD49a−CD49b+ (cNK), CD3−NKp46+ROSA−CD45−CD49a+CD49b+ (dpNK), and CD3−NKp46+ROSA−CD45−CD49a+CD49b− (trNK) cells from donor skin; and CD3−NKp46+ROSA−CD45−CD49a+CD49b− (trNK) cells from recipient skin were sorted using a BD FACSAria II (BD Bioscience, Billerica, MA, USA). Sorted NK cells were collected in 15-ml tubes containing 3 ml of RPMI 1640 supplemented with 10% FBS (Corning, Manassas, VA, USA) and 5% P/S.
RNA-seq and analysis
Sorted cells were centrifuged, medium-aspirated and resuspended in 5 μl of TCL lysis buffer with 1% β-mercaptoethanol (Thermo Fisher Scientific, catalog no. 21-985-023), and added to a 96-well Eppendorf twin-tec barcoded plate (Eppendorf, NY, USA). Plates were stored at −80°C until sequencing as described (59). Modified SmartSeq2 complementary DNA and Illumina Nextera XT library construction and sequencing were conducted at the Broad Institute of MIT and Harvard using an Illumina NextSeq 500 system (Boston, MA, USA). All samples were quasi-mapped to GRCm38 (mm10) using Salmon with the “gcBias” and “seqBias” options and collapsed down to gene-level abundance estimates using the “EnsDb.Mmusculus.v79” annotation package. All downstream differential expression analysis was carried out using DESeq2. The results table was restricted to genes with a minimum of 10 total counts across the dataset, and mitochondrial, pseudo-, and ribosomal genes were removed. Original data are available at the National Center for Biotechnology Information Gene Expression Omnibus, accession number: GSE148600. Gene set enrichment analysis (GSEA) (www.broad.mit.edu/gsea/) was used to identify significantly up- or down-regulated pathways in CD49b+ cNK cells in donor skin in comparison with CD49+ cNK cells in circulation (blood and spleen). GSEA was performed using molecular signatures database (MSigDB) 50 hallmark gene sets (h.all), and the curated 6226 gene sets (c2.all) and pathways containing enriched genes between 15 and 1000 genes were considered.
Cell lines
B16-F10 WT and B2m−/− cell lines were maintained in RPMI 1640 supplemented with 10% FBS and 1% P/S and cultured at 37°C/5% CO2. WT-MEFs and m157-MEFs were a gift from W. Yokoyama, Washington University, St. Louis, MO, USA. MEFs were cultured in RPMI 1640 containing 10% FBS and 1% P/S/20 mM glutamine (Life Technologies, catalog no. 25030-081).
Melanoma experiments
All tumor experiments were performed in 8- to 12-week-old C57BL/6 female WT or Ncr1iCre,ROSAmT-mG mice. For the subcutaneous tumor model, 2.5 × 105 B16-F10 WT or B2m−/− cells were subcutaneously injected into the shaved flanks of mice. Tumor growth was measured every other day using a digital caliper starting from day 7. Tumor volume was calculated as follows: . To inhibit collagen deposition in and around the tumor, mice were administered with losartan potassium (60 mg/kg) (Thermo Fisher Scientific, catalog no. L02325G) or DHB (40 mg/kg) (Sigma-Aldrich, catalog no. 37580) in 100 μl of sterile 1× PBS, while the control mice were administered with the equivalent volume of sterile 1× PBS. These drugs were injected intraperitoneally every day for the duration of the study starting the day of tumor inoculation.
For the collagenase/hyaluronidase experiment, 2.5 × 105 B16-F10 B2m−/− cells were resuspended in 1× collagenase and hyaluronidase mix (STEMCELL Technologies, catalog no. 07912) and were subcutaneously injected into the shaved flanks of mice. To deplete collagen within the tumor, mice were administered with 1× collagenase and hyaluronidase mix (STEMCELL Technologies, catalog no. 07912) in 100 μl of sterile 1× PBS, while control mice were administered with the equivalent volume of sterile 1× sPBS. These enzymes were injected via intratumoral route starting on the day of tumor inoculation (day 0), days 1 to 4, and then every other day (days 6, 8, 10, etc.) until the end of the study. Except for day 3 tumor harvest, mice were euthanized when a tumor reached 2 cm in diameter in accordance with IACUC protocols. For early time point tumor analysis, specific Ncr1iCre,ROSAmT-mG mice from the PBS, losartan, and DHB group were euthanized on day 3 after tumor inoculation; the implanted tumors were harvested and fixed in prechilled 95% ethanol overnight at 4°C before subjecting them to subsequent processing.
For metastatic melanoma studies, mice were intravenously injected via tail vein with 2 × 105 B16-F10 WT or B2m−/− cells. Mice were euthanized 14 days after injection; lungs were then harvested, fixed in 4% paraformaldehyde (PFA; Sigma-Aldrich, catalog no. P6148), and collected for histological analysis. Blinded quantification of the metastatic lung foci was performed on macroscopic images, and the size of metastatic foci was measured using ImageJ software.
Tumor harvesting
Terminal subcutaneous tumors were excised with the skin and weighed before cutting in half. One-half was placed in an appropriate fixative, while the other half was separated from the skin and chopped with scissors into ~1-mm pieces. These were transferred to a 15-ml tube with 10 ml of digest buffer [RPMI 1640 (Life Technologies, catalog no. 21870076) and collagenase IV (200 U/ml) (Worthington Biochemical, Lakewood, NJ, USA)] and incubated for 2 hours at 37°C with shaking. Following incubation, the digested tumor was poured onto a 70 μM strainer placed in a 50-ml tube. The 15-ml tube was rinsed with ~3 ml of 1× PBS/2% FCS/5 mM EDTA and added to the digested sample. The tumor was mashed with a plunger through the 70 μM strainer, which was then rinsed approximately three times with a small volume of 1× PBS/2% FCS/5 mM EDTA. The tumor-infiltrating CD45+ immune cells were isolated by magnetic-based positive selection using CD45 microbeads (Miltenyi Biotec, catalog no. 130-052-301) according to the manufacturer’s protocol. The stained cells were passed through the LS columns (Miltenyi Biotec, catalog no. 130-042-401) placed between magnets for isolation of immune cells. The isolated CD45+ cells were centrifuged at 300g for 5 min at 4°C and resuspended in 2.4G2 blocking solution before flow cytometry staining.
Cytokine, poly(I:C), and DT treatment
For cytokine treatment, mice were injected with 100 ng per mouse of mouse IL-12 (mIL-12) (BioLegend, catalog no. 577002), 2 μg per mouse of mIL-15 (Shenandoah Biotechnology, Warwick, PA; catalog no. 200-07), and 100 ng per mouse of mIL-18 (Shenandoah Biotechnology, catalog no. 200-83) in 200 μl of sterile 1× PBS. For the combination poly(I:C) and cytokines, mice were injected with 200 μg per mouse of poly(I:C) (Sigma-Aldrich, catalog no. P9582), 2 μg per mouse of mIL-15 (Shenandoah Biotechnology, catalog no. 200-07), and 100 ng per mouse of mIL-18 (Shenandoah Biotechnology, catalog no. 200-83) in 200 μl of sterile 1× PBS. Treatments were injected intraperitoneally starting the day of the skin transplant (day 0), day 2, and day 4 and then injected subcutaneously at days 7, 10, 13, 16, and 19 after transplant. DT (Sigma-Aldrich, catalog no. D0564) was injected intraperitoneally at 500 ng per mouse the day before the transplant and 200 ng per mouse at days indicated in fig. S5E combined with NK or T cell depletion.
NK cell depletion, T cell depletion, and checkpoint blockade
Mice were intraperitoneally injected with 500 μg per mouse of IgG isotype control (Southern Biotech, Birmingham, AL, USA; catalog no. 0107-01) or depleting anti-NK1.1 antibody (clone PK136, Bio X Cell, West Lebanon, NH, USA; catalog no. BE0036) 2 days before tumor cell injection and 250 μg per mouse every other day starting the day of the tumor cell injection. For depletion of NK or T cells (anti-CD4 antibody, clone GK1.5, Bio X Cell; anti-CD8α antibody, clone YTS 169.4, Bio X Cell) in a skin transplant setting, 500 μg per mouse of each depleting antibody were injected the day of the transplant and 250 μg at days indicated in fig. S5E. Anti-mouse PD-1 (clone 29F.1A12, Bio X Cell, catalog no. BE2073), anti-TIGIT (clone 1G9, Bio X Cell, catalog no. BE0274), and anti-CTLA4 (clone 9D9, Bio X Cell, catalog no. BE0164) antibodies were intraperitoneally injected at 200 μg each per mouse at day −2 and then every 3 days beginning at day 0 of skin transplantation.
In vitro experiments
Before the experiment, blood from C57BL/6 WT and m157tg mice was collected, incubated overnight at 4°C, and centrifuged, and serum was isolated from the pellet. Spleens from C57BL/6 mice were harvested and mashed through a 70-μm filter, and cells were resuspended in RBC lysis for 2 min. After washing with 1× PBS/2% FCS/5 mM EDTA, cells were counted, and 5 × 106 splenocytes were resuspended in 500 μl of RPMI 10% FBS/1% P/S/20 mM glutamine and 10% serum of either WT or m157tg mice, then plated in a 24-well plate, and incubated for 3 hours at 37°C/5% CO2. Cells were then washed and replated, and 200 μl of a mixture containing 1 × 105 WT-MEFs or m157-MEFs, mIL-12 (10 ng/ml; BioLegend, catalog no. 577002), or mIL-12/mIL-18 (12.5 ng/ml; Shenandoah Biotechnology, catalog no. 200-83) was added to the well. After incubation for another hour at 37°C/5% CO2, 1 μl of anti–CD107a-eF660 (clone 1D4B, eBioscience, catalog no. 50-1071-82), 1× brefeldin A (BioLegend, catalog no. 420601), and 1× monensin (BioLegend, catalog no. 420701) were added, and cells were incubated for 6 hours. Then, the supernatant was removed, and cells were washed and resuspended in 2.4G2 blocking solution before flow cytometry staining. Cells were assayed on a BD FACSCanto (BD Biosciences).
For in vitro experiments in the presence of ECM components, collagen I (rat tail; Sigma-Aldrich, catalog no. 08-115), collagen III (human placenta; Sigma-Aldrich, catalog no. C4407), collagen IV (mouse; Sigma-Aldrich, catalog no. C0543), elastin (mouse lung; Sigma-Aldrich, catalog no. E6402), laminin (mouse; Thermo Fisher Scientific, catalog no. 23017015), and fibronectin (rat plasma; Sigma-Aldrich, catalog no. F0635) were diluted in 1× PBS and coated at 20 μg/cm2 in a 24-well tissue culture plate for 30 min at RT with gentle shaking followed by overnight at 4°C. Spleens from Ncr1iCre,ROSAmT-mG, WT, or Lair1−/− mice were harvested as described above. Then, 5 × 106 splenocytes resuspended in RPMI 10% FBS/1% P/S/20 mM glutamine were mixed with 20 μg of respective ECM protein per well and were incubated at RT for 10 min. Splenocytes and ECM mix were plated onto the ECM-coated well and were incubated at 37°C/5% CO2 for 2 hours (short ECM exposure) or 17 hours (long ECM exposure). At the end of the incubation, 1 × 105 MEFs (WT or m157) were added to each well to obtain a 50:1 splenocyte:MEF ratio. For cell signaling experiments, hydrogen peroxide (H2O2) at a final concentration of 2.5 mM or 1 × 105 m157-MEFs were added to each well except in “0 min” wells, which were unstimulated and fixed immediately. Splenocytes were stimulated with 2.5 mM H2O2 for 2 and 5 min or were cocultured with m157-MEFs for 30 and 60 min (signaling) or 7 hours (CD107a and IFNγ) at 37°C/5% CO2. Splenocytes and MEFs in coculture were supplemented with mIL-12 (10 ng/ml; BioLegend, catalog no. 577002), mIL-15 (1 ng/ml; Shenandoah Biotechnology, catalog no. 200-07), and human IL-2 (BioLegend, catalog no. 589104; only long ECM exposure) as well as for CD107a/IFNγ experiments. One microliter of anti–CD107a-BV421 (1D4B, BioLegend, catalog no. 121618), 1× brefeldin A (BioLegend, catalog no. 420601), and 1× monensin (BioLegend, catalog no. 420701) were added. Following the coculture, cells were washed with 1× PBS/2% FCS/5 mM EDTA, and the pellet was resuspended in 2.4G2 blocking solution before flow cytometry staining.
NK cell enrichment
NK cells purified from spleens under sterile conditions were used for LEGENDplex experiments. Spleens from 8- to 9-week-old C57BL/6 female WT mice were harvested and mashed through a 70-μm filter. Cells were resuspended in RBC lysis for 2 min and washed with 1× PBS/2% FCS/5 mM EDTA. NK cells were enriched by negative selection using an NK cell isolation kit (Milteny Biotec, catalog no. 130-115-818) and magnetic LS columns (Milteny Biotec, catalog no. 130-042-401) according to the manufacturer’s instructions. Enrichment of isolated NK cells was confirmed by flow cytometry and was 88 to 90% CD3−NK1.1+NKp46+ cells. Enriched NK cells were resuspended in RPMI 1640 supplemented with 10% FBS and 1% P/S.
LEGENDplex
ECM protein coating on wells was performed as previously described. A total of 1 × 105 enriched splenic NK cells were mixed with 20 μg of respective ECM protein per well in RPMI 1640/10% FBS/1% P/S/20 mM glutamine and incubated at RT for 10 min. NK cells and ECM mix were plated onto the ECM-coated well and were incubated at 37°C/5% CO2 for 2 hours. At the end of the incubation, 1 × 105 MEF (WT or m157) cells were added to the respective wells to obtain a 1:1 NK:MEF ratio. NK cells and MEFs were cocultured at 37°C 5% CO2 for 24 hours supplemented with mIL-12 (10 ng/ml; BioLegend, catalog no. 577002) and mIL-15 (1 ng/ml; Shenandoah Biotechnology, catalog no. 200-07-100). The supernatant was collected at 12- and 24-hour intervals, centrifuged to remove cells/cell debris, and subsequently frozen at −80°C until further use. Quantification of murine cytokines and chemokines (CCL2, CCL5, CXCL9, and CXCL10) in supernatants was performed with LEGENDplex kits (BioLegend, catalog nos. 740451 and 740622) according to the manufacturer’s instructions. Briefly, a mix of capture beads with distinct size and fluorescence was incubated with undiluted supernatant or standard dilutions for 2 hours at RT with constant shaking. Beads were washed, a biotinylated detection antibody was added, and the plate was incubated for 1 hour at RT with constant shaking. The streptavidin-phycoerythrin conjugate was added and further incubated for 30 min at RT on a shaker. Beads were washed and subsequently acquired on a BD LSRFortessa X-20 flow cytometer. Analysis and quantification of the results were done using LEGENDplex data analysis software (BioLegend). Quantification was reported as picograms per milliliter for CCL2, CCL5, and CXCL10 and as mean fluorescence intensity for CXCL9.
Histology, collagen, elastin, and IF staining
Skin grafts were collected and incubated in 4% PFA/25% sucrose (Thermo Fisher Scientific, Hampton, NH, USA; catalog no. S5-3) solution at 4°C for 16 hours. The next day, slices were equilibrated for 4 hours in 50% sucrose solution, embedded in optimal temperature cutting compound (Thermo Fisher Scientific, catalog no. 23-730-571), snap-frozen, and stored at −80°C. For paraffin embedding, lungs and tumor were collected and fixed in 4% PFA overnight at 4°C. Subsequently, tissues were dehydrated in ethanol, processed, and embedded in paraffin according to standard histology processes. For paraffin embedding of Ncr1icre,RosamT-mG, a different fixation protocol was used to maintain the NKp46-GFP and Tdt-Tomato-ROSA fluorescence. Tumors from Ncr1icre,RosamT-mG mice were collected and fixed in prechilled 95% ethanol overnight at 4°C. Tissues were subsequently dehydrated in 100% ethanol, cleared in 100% xylene, and embedded in paraffin according to standard histology processes. For IF staining of frozen tissue, 7-μm sections of skin were cut on a cryostat, rehydrated in three washes of 1× tris-buffered saline (TBS) for 2 min each, followed by blocking of nonspecific protein with 5% bovine serum albumin (Thermo Fisher Scientific, catalog no. BP1600) and 5% goat serum (Sigma-Aldrich, catalog no. G9023) in 1× TBS. Sections were stained overnight at 4°C with primary antibodies (table S1B). The following day, slides were washed as above and incubated for 2 hours at RT with secondary antibodies conjugated to fluorochromes (table S1B). After washing as above, sections were incubated with a 1:4000 dilution of 4′,6-diamidino-2-phenylindole (Invitrogen, Carlsbad, CA; catalog no. D3571) in 1× TBS for 5 min at RT, washed, and air-dried, and coverslips were mounted with the ProLong Gold Antifade Reagent (Invitrogen, catalog no. P36930). Five or six randomly selected fields of view at ×200 total magnification were obtained for each section using a Zeiss Axioscan (Zeiss, Oberkochen, Germany). Blinded manual counting of NKp46+, CD3+, CD4+, and CD8+ T cells was performed using the ZEN Blue Software (Zeiss, Oberkochen, Germany). Automated counting was performed for CD11b, F4/80, and Arginase1 stains in the whole tumor region of the scanned slides by Halo3.0 software (Indica Labs, Albuquerque, USA). For IF of paraffin-embedded tissues, 5-μm sections were rehydrated and permeabilized with 1× PBS supplemented with 0.2% Triton X-100 (Thermo Fisher Scientific, catalog no. BP151) for 5 min. Antigen retrieval was then performed using a Cuisinart pressure cooker for 20 min at high pressure in antigen unmasking solution (Vector Laboratories, Burligame, CA; catalog no. H-3300). Slides were then washed three times for 3 min each in 1× PBS supplemented with 0.1% Tween 20 (Sigma-Aldrich, catalog no. P1379). Sections were blocked, stained (table S1B), and mounted as described above. Elastin fiber staining was performed using the Verhoeff Van Gienson Elastin Stain Kit (ab150667, Abcam) following the manufacturer’s protocol. For collagen staining, Masson’s trichrome staining (Polysciences, Warrington, PA; catalog no. 25088) was performed following the instructions of the manufacturer. Whole slide imaging was performed using a NanoZoomer S60 digital slide scanner (Hamamatsu, Japan) or Axioscan.Z1 (Zeiss, Germany) and analyzed with the NDP.view2 software (Hamamatsu) or ZEN Blue Software (Zeiss, Oberkochen, Germany), respectively.
B2M analysis in human cancers
B2M gene alterations and expression in solid cancer and leukemia samples in TCGA and other public databases were obtained and analyzed through cBioPortal for Cancer Genomics at www.cbioportal.org/.
Statistical analysis
Graphs show means ± SD. The numbers of mice per group used in each experiment are annotated in the corresponding figure legend as n. Graphs and statistical analysis were performed using GraphPad Prism 8 (La Jolla, CA, USA) and RStudio. All tumor quantifications were performed blindly. Two-tailed Fisher’s exact test was used to compare skin graft rejection grades and lymph node metastatic load among groups. Two-way analysis of variance (ANOVA) with Sidak’s multiple comparison test was used to compare tumor growth over time between different groups. Comparisons of survival were performed with the log-rank test. Two-tailed Mann-Whitney U test was used for all the other comparisons. A P value of less than 0.05 was considered significant.
Acknowledgments
We thank W. Yokoyama for m157tg mice and WT/m157-MEF, E. Vivier for Ncr1iCre mice, K. van Gisbergen for Hobit−/− mice, and S. Komarova for Lair1−/− mice. We thank T. Cunningham and K. Ngo for technical assistance.
Funding: S.D. holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund and was supported by Dermatology Foundation Physician Scientist Career Development Award. M.D.B., M.V., M.R., E.B.S., and S.D. were supported by grants from the Burroughs Wellcome Fund, the Sidney Kimmel Foundation, and NIH (K08AR068619 and DP5OD021353).
Author contributions: S.D. conceived the study. M.D.B., M.V., M.R., and S.D. designed the experiments. M.D.B., M.V., M.R., A.L., and E.B.S. performed the experiments and analyzed the data. M.D.B., M.V., M.R., M.S.L., and S.D. interpreted the data. M.D.B., M.V., and S.D. wrote the manuscript. R.T.M. contributed melanoma cell lines.
Competing interests: R.T.M. consults for Bristol Myers Squibb. The other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S12
Table S1
REFERENCES AND NOTES
- 1.Garrido F., Aptsiauri N., Cancer immune escape: MHC expression in primary tumours versus metastases. Immunology 158, 255–266 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bessoles S., Grandclement C., Alari-Pahissa E., Gehrig J., Jeevan-Raj B., Held W., Adaptations of natural killer cells to self-MHC class I. Front. Immunol. 5, 349 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Long E. O., Kim H. S., Liu D., Peterson M. E., Rajagopalan S., Controlling natural killer cell responses: Integration of signals for activation and inhibition. Annu. Rev. Immunol. 31, 227–258 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karlhofer F. M., Ribaudo R. K., Yokoyama W. M., MHC class I alloantigen specificity of Ly-49+ IL-2-activated natural killer cells. Nature 358, 66–70 (1992). [DOI] [PubMed] [Google Scholar]
- 5.Bennett M., Yu Y. Y., Stoneman E., Rembecki R. M., Mathew P. A., Lindahl K. F., Kumar V., Hybrid resistance: ‘Negative’ and ‘positive’ signaling of murine natural killer cells. Semin. Immunol. 7, 121–127 (1995). [DOI] [PubMed] [Google Scholar]
- 6.Kumar V., George T., Yu Y. Y., Liu J., Bennett M., Role of murine NK cells and their receptors in hybrid resistance. Curr. Opin. Immunol. 9, 52–56 (1997). [DOI] [PubMed] [Google Scholar]
- 7.Manilay J. O., Sykes M., Natural killer cells and their role in graft rejection. Curr. Opin. Immunol. 10, 532–538 (1998). [DOI] [PubMed] [Google Scholar]
- 8.Zijlstra M., Auchincloss H. Jr., Loring J. M., Chase C. M., Russell P. S., Jaenisch R., Skin graft rejection by beta 2-microglobulin-deficient mice. J. Exp. Med. 175, 885–893 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riggan L., Freud A. G., O’Sullivan T. E., True detective: Unraveling group 1 innate lymphocyte heterogeneity. Trends Immunol. 40, 909–921 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sojka D. K., Tian Z., Yokoyama W. M., Tissue-resident natural killer cells and their potential diversity. Semin. Immunol. 26, 127–131 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seillet C., Belz G. T., Differentiation and diversity of subsets in group 1 innate lymphoid cells. Int. Immunol. 28, 3–11 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Cortez V. S., Ulland T. K., Cervantes-Barragan L., Bando J. K., Robinette M. L., Wang Q., White A. J., Gilfillan S., Cella M., Colonna M., SMAD4 impedes the conversion of NK cells into ILC1-like cells by curtailing non-canonical TGF-β signaling. Nat. Immunol. 18, 995–1003 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao Y., Souza-Fonseca-Guimaraes F., Bald T., Ng S. S., Young A., Ngiow S. F., Rautela J., Straube J., Waddell N., Blake S. J., Yan J., Bartholin L., Lee J. S., Vivier E., Takeda K., Messaoudene M., Zitvogel L., Teng M. W. L., Belz G. T., Engwerda C. R., Huntington N. D., Nakamura K., Holzel M., Smyth M. J., Tumor immunoevasion by the conversion of effector NK cells into type 1 innate lymphoid cells. Nat. Immunol. 18, 1004–1015 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Lanier L. L., NK cell recognition. Annu. Rev. Immunol. 23, 225–274 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Bern M. D., Parikh B. A., Yang L., Beckman D. L., Poursine-Laurent J., Yokoyama W. M., Inducible down-regulation of MHC class I results in natural killer cell tolerance. J. Exp. Med. 216, 99–116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yokoyama W. M., Kim S., How do natural killer cells find self to achieve tolerance? Immunity 24, 249–257 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Brodin P., Karre K., Hoglund P., NK cell education: Not an on-off switch but a tunable rheostat. Trends Immunol. 30, 143–149 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Elliott J. M., Yokoyama W. M., Unifying concepts of MHC-dependent natural killer cell education. Trends Immunol. 32, 364–372 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orange J. S., Formation and function of the lytic NK-cell immunological synapse. Nat. Rev. Immunol. 8, 713–725 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wallin R. P., Screpanti V., Michaelsson J., Grandien A., Ljunggren H.-G., Regulation of perforin-independent NK cell-mediated cytotoxicity. Eur. J. Immunol. 33, 2727–2735 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Lebbink R. J., Meyaard L., Non-MHC ligands for inhibitory immune receptors: Novel insights and implications for immune regulation. Mol. Immunol. 44, 2153–2164 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Bauer S., Groh V., Wu J., Steinle A., Phillips J. H., Lanier L. L., Spies T., Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science 285, 727–729 (1999). [DOI] [PubMed] [Google Scholar]
- 23.Smith H. R., Heusel J. W., Mehta I. K., Kim S., Dorner B. G., Naidenko O. V., Iizuka K., Furukawa H., Beckman D. L., Pingel J. T., Scalzo A. A., Fremont D. H., Yokoyama W. M., Recognition of a virus-encoded ligand by a natural killer cell activation receptor. Proc. Natl. Acad. Sci. U.S.A. 99, 8826–8831 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hammer Q., Ruckert T., Romagnani C., Natural killer cell specificity for viral infections. Nat. Immunol. 19, 800–808 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Karre K., Ljunggren H. G., Piontek G., Kiessling R., Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defence strategy. Nature 319, 675–678 (1986). [DOI] [PubMed] [Google Scholar]
- 26.Narni-Mancinelli E., Chaix J., Fenis A., Kerdiles Y. M., Yessaad N., Reynders A., Gregoire C., Luche H., Ugolini S., Tomasello E., Walzer T., Vivier E., Fate mapping analysis of lymphoid cells expressing the NKp46 cell surface receptor. Proc. Natl. Acad. Sci. U.S.A. 108, 18324–18329 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tripathy S. K., Keyel P. A., Yang L., Pingel J. T., Cheng T. P., Schneeberger A., Yokoyama W. M., Continuous engagement of a self-specific activation receptor induces NK cell tolerance. J. Exp. Med. 205, 1829–1841 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elliott J. M., Wahle J. A., Yokoyama W. M., MHC class I-deficient natural killer cells acquire a licensed phenotype after transfer into an MHC class I-sufficient environment. J. Exp. Med. 207, 2073–2079 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joncker N. T., Shifrin N., Delebecque F., Raulet D. H., Mature natural killer cells reset their responsiveness when exposed to an altered MHC environment. J. Exp. Med. 207, 2065–2072 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huntington N. D., The unconventional expression of IL-15 and its role in NK cell homeostasis. Immunol. Cell Biol. 92, 210–213 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Marcais A., Viel S., Grau M., Henry T., Marvel J., Walzer T., Regulation of mouse NK cell development and function by cytokines. Front. Immunol. 4, 450 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hydes T., Noll A., Salinas-Riester G., Abuhilal M., Armstrong T., Hamady Z., Primrose J., Takhar A., Walter L., Khakoo S. I., IL-12 and IL-15 induce the expression of CXCR6 and CD49a on peripheral natural killer cells. Immun. Inflamm. Dis. 6, 34–46 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ni X., Fu B., Zhang J., Sun R., Tian Z., Wei H., Cytokine-based generation of CD49a+Eomes−/+ natural killer cell subsets. Front. Immunol. 9, 2126 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cortez V. S., Cervantes-Barragan L., Robinette M. L., Bando J. K., Wang Y., Geiger T. L., Gilfillan S., Fuchs A., Vivier E., Sun J. C., Cella M., Colonna M., Transforming growth factor-β signaling guides the differentiation of innate lymphoid cells in salivary glands. Immunity 44, 1127–1139 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang L., Peng H., Zhou J., Chen Y., Wei H., Sun R., Yokoyama W. M., Tian Z., Differential phenotypic and functional properties of liver-resident NK cells and mucosal ILC1s. J. Autoimmun. 67, 29–35 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Sojka D. K., Plougastel-Douglas B., Yang L., Pak-Wittel M. A., Artyomov M. N., Ivanova Y., Zhong C., Chase J. M., Rothman P. B., Yu J., Riley J. K., Zhu J., Tian Z., Yokoyama W. M., Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. eLife 3, e01659 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Viel S., Marcais A., Guimaraes F. S., Loftus R., Rabilloud J., Grau M., Degouve S., Djebali S., Sanlaville A., Charrier E., Bienvenu J., Marie J. C., Caux C., Marvel J., Town L., Huntington N. D., Bartholin L., Finlay D., Smyth M. J., Walzer T., TGF-β inhibits the activation and functions of NK cells by repressing the mTOR pathway. Sci. Signal. 9, ra19 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Mackay L. K., Minnich M., Kragten N. A., Liao Y., Nota B., Seillet C., Zaid A., Man K., Preston S., Freestone D., Braun A., Wynne-Jones E., Behr F. M., Stark R., Pellicci D. G., Godfrey D. I., Belz G. T., Pellegrini M., Gebhardt T., Busslinger M., Shi W., Carbone F. R., van Lier R. A. W., Kallies A., van Gisbergen K. P. J. M., Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science 352, 459–463 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Lebbink R. J., de Ruiter T., Kaptijn G. J., Bihan D. G., Jansen C. A., Lenting P. J., Meyaard L., Mouse leukocyte-associated Ig-like receptor-1 (mLAIR-1) functions as an inhibitory collagen-binding receptor on immune cells. Int. Immunol. 19, 1011–1019 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Meyaard L., The inhibitory collagen receptor LAIR-1 (CD305). J. Leukoc. Biol. 83, 799–803 (2008). [DOI] [PubMed] [Google Scholar]
- 41.Corcoran A., Cotter T. G., Redox regulation of protein kinases. FEBS J. 280, 1944–1965 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Diop-Frimpong B., Chauhan V. P., Krane S., Boucher Y., Jain R. K., Losartan inhibits collagen I synthesis and improves the distribution and efficacy of nanotherapeutics in tumors. Proc. Natl. Acad. Sci. U.S.A. 108, 2909–2914 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiong G., Stewart R. L., Chen J., Gao T., Scott T. L., Samayoa L. M., O’Connor K., Lane A. N., Xu R., Collagen prolyl 4-hydroxylase 1 is essential for HIF-1α stabilization and TNBC chemoresistance. Nat. Commun. 9, 4456 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Narni-Mancinelli E., Ugolini S., Vivier E., Tuning the threshold of natural killer cell responses. Curr. Opin. Immunol. 25, 53–58 (2013). [DOI] [PubMed] [Google Scholar]
- 45.Shi F. D., Ljunggren H. G., La Cava A., Van Kaer L., Organ-specific features of natural killer cells. Nat. Rev. Immunol. 11, 658–671 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bottcher J. P., Bonavita E., Chakravarty P., Blees H., Cabeza-Cabrerizo M., Sammicheli S., Rogers N. C., Sahai E., Zelenay S., Reis e Sousa C., NK cells stimulate recruitment of cDC1 into the tumor microenvironment promoting cancer immune control. Cell 172, 1022–1037.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Degli-Esposti M. A., Smyth M. J., Close encounters of different kinds: Dendritic cells and NK cells take centre stage. Nat. Rev. Immunol. 5, 112–124 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Gasteiger G., Rudensky A. Y., Interactions between innate and adaptive lymphocytes. Nat. Rev. Immunol. 14, 631–639 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kalinski P., Giermasz A., Nakamura Y., Basse P., Storkus W. J., Kirkwood J. M., Mailliard R. B., Helper role of NK cells during the induction of anticancer responses by dendritic cells. Mol. Immunol. 42, 535–539 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Mailliard R. B., Son Y. I., Redlinger R., Coates P. T., Giermasz A., Morel P. A., Storkus W. J., Kalinski P., Dendritic cells mediate NK cell help for Th1 and CTL responses: Two-signal requirement for the induction of NK cell helper function. J. Immunol. 171, 2366–2373 (2003). [DOI] [PubMed] [Google Scholar]
- 51.Martin-Fontecha A., Thomsen L. L., Brett S., Gerard C., Lipp M., Lanzavecchia A., Sallusto F., Induced recruitment of NK cells to lymph nodes provides IFN-γ for TH1 priming. Nat. Immunol. 5, 1260–1265 (2004). [DOI] [PubMed] [Google Scholar]
- 52.Mocikat R., Braumuller H., Gumy A., Egeter O., Ziegler H., Reusch U., Bubeck A., Louis J., Mailhammer R., Riethmuller G., Koszinowski U., Rocken M., Natural killer cells activated by MHC class I(low) targets prime dendritic cells to induce protective CD8 T cell responses. Immunity 19, 561–569 (2003). [DOI] [PubMed] [Google Scholar]
- 53.Le Moine A., Goldman M., Abramowicz D., Multiple pathways to allograft rejection. Transplantation 73, 1373–1381 (2002). [DOI] [PubMed] [Google Scholar]
- 54.Wise M., Zelenika D., Bemelman F., Latinne D., Bazin H., Cobbold S., Waldmann H., CD4 T cells can reject major histocompatibility complex class I-incompatible skin grafts. Eur. J. Immunol. 29, 156–167 (1999). [DOI] [PubMed] [Google Scholar]
- 55.Marino J., Paster J., Benichou G., Allorecognition by T lymphocytes and allograft rejection. Front. Immunol. 7, 582 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Habif G., Crinier A., Andre P., Vivier E., Narni-Mancinelli E., Targeting natural killer cells in solid tumors. Cell. Mol. Immunol. 16, 415–422 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nayyar G., Chu Y., Cairo M. S., Overcoming resistance to natural killer cell based immunotherapies for solid tumors. Front. Oncol. 9, 51 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sandel M. H., Speetjens F. M., Menon A. G., Albertsson P. A., Basse P. H., Hokland M., Nagelkerke J. F., Tollenaar R. A. E. M., van de Velde C. J. H., Kuppen P. J. K., Natural killer cells infiltrating colorectal cancer and MHC class I expression. Mol. Immunol. 42, 541–546 (2005). [DOI] [PubMed] [Google Scholar]
- 59.Trombetta J. J., Gennert D., Lu D., Satija R., Shalek A. K., Regev A., Preparation of single-cell RNA-Seq libraries for next generation sequencing. Curr. Protoc. Mol. Biol. 107, 4.22.1–4.22.17 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S12
Table S1