Abstract
Objective:
The hypothesis of the present study is that overexpression of osteoprotegerin (OPG) promotes preosteoblast maturation.
Materials and Methods:
The preosteoblast cell line MC3T3-E1 was transfected with OPG overexpression. OPG expression was confirmed by enzyme-linked immunosorbent assay (ELISA) and Western blot. Changes in the transcription factors in OPG-expressing cells were assessed by real-time polymerase quantitative polymerase chain reaction (RT-qPCR). Alkaline phosphate (ALP) expression was measured by ELISA.
Results:
The success of stable transfection of MC3T3-E1 cells with OPG overexpression was confirmed by MoFlow sorting followed by G418 selection. RT-qPCR showed that expression of RunX2, the most important osteoblast differentiation controlling factor, was suppressed. Smad1 and Akt1, as well as ALP, were upregulated in the OPG overexpressing cells.
Conclusion:
Results from the present study provide evidence that overexpression of OPG in preosteoblasts promotes its differentiation into mature osteoblasts.
Keywords: Osteoprotegerin (OPG), Osteoblast, Preosteoblast, Alkaline phosphatase, Runx2, Orthodontics
INTRODUCTION
Teeth move to designed optimal position induced in orthodontic treatment by mechanical force, which leads to bone remodeling through bone resorption by osteoclasts in the compression side and new bone formation by osteoblasts in the tension side. In a whole biological cycle of bone remodeling, osteoclasts are recruited and activated to resorb the old bone, after which preosteoblast attaches and matures on the surface of the bone to secrete bone matrix subsequently mineralized in the resorption pits to modify bone shape.1 Bone formation is included in distinct stages of proliferation, matrix maturation, and finally mineralization.2 Preosteoblast originates from mesenchymal stem cells, and its proliferation and differentiation are notably divided into three phases: preosteoblast, mature osteoblast, and osteocyte, which are regulated by transcription factors and express specific phenotypic genes.2–4
The differentiation of preosteoblasts into mature osteoblasts goes through different distinct stages and is under the control of transcription factors such as Runx2, Smad1, and Akt1. Osteoprotegerin (OPG) inhibits osteoclast differentiation and activation by competing with receptor activator NF-κB (RANK) to combine with RANK ligand (RANKL), but its role in osteoblast differentiation is still unclear.
Several transcription factors are known to be essential in osteoblast differentiation. Runx2 is a normal transcriptional factor that contributes to regulatory mechanisms during osteoblast differentiation and is required to modulate target gene expression at key developmental transitions.5,6 Akt1 and Smad1 interact with Runx2 to regulate and control transcriptional levels of osteoblast differentiation.7–9 Alkaline phosphate (ALP) is a key early marker of matrix maturation that is expressed in preosteoblasts during osteoblast differentiation and upregulated expression in mature osteoblasts and is downregulated in osteocytes.3
Osteoblasts are able to modulate osteoclastogenesis by changing the ratio of OPG and the receptor activator of RANKL during bone remodeling.10 Osteoclasts are activated by RANKL/RANK pathways and are inhibited by OPG, a decoy factor competing with RANK for RANKL engagement.11 The essential role of OPG has been well described in osteoclast differentiation both in vitro and in vivo.11 Mice deficient in OPG expression exhibit severe osteoporosis, while overexpression of OPG results in osteopetrosis and a decreased number of osteoclasts.12,13 However the direct effects of OPG on osteoblast differentiation are still unknown. Previous studies showed that osteoblast maturation was suppressed when OPG expression was inhibited by treatment with granulocyte colony-stimulating factor,14 and mature osteoblasts have an inhibitory role in osteoclastogenesis15 by vitamin D acting through the vitamin D receptor followed by OPG upregulated expression. On the basis of previous studies, we hypothesize that overexpression of OPG promotes preosteoblast maturation.
MATERIALS AND METHODS
Materials
The plasmid pIRES2-EGFP was a gift from Dr Horton (University Medical Center, Groningen, The Netherlands). Digestion enzyme, T4 DNA liganase, and pfu DNA polymerase from fermentas were used (St Leon-Rot, Germany); in addition, one-shot top 10 chemically competent Escherichia coli cells from Invitrogen (Breda, The Netherlands), plasmids and reverse transcriptase polymerase chain reaction (PCR) products, purification kits from Qiagen (Valencia, Calif); FuGENE 6 transfection reagents from Roche (Indianapolis, Ind), a real-time quantitative PCR one-step kit from Applied Biosystems (Foster City, Calif), an OPG enzyme-linked immunosorbent assay (ELISA) kit from R&D Systems (R&D Systems, Minneapolis, Minn), an ALP ELISA kit from ANASpec (San Jose, Calif), and OPG and β-actin primary and secondary antibodies from Santa Cruz (Heidelberg, Germany) were used. Preosteoblast cell line MC3TC-E1 was supplied by the European Collection of Cell Cultures (ECACC; Salisbury, United Kingdom).
Cell Culture
MC3T3-E1 subclone 14 cell line was cultured in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) at 37°C in a humid atmosphere with 5% carbon dioxide (CO2). Cells were passaged every 3 days with 1∶3 ratios. MC3T3-E1, transfected with empty plasmids or constructed OPG recombinant, was maintained in DMEM supplemented with 10% FBS and 600 ng/mL G418 after MoFlow (BD Biosciences, San Jose, Calif) sorting.
Cell Proliferation
Cells were seeded in 96-well plates at a density of 1000 cells/well with 100 µL DMEM supplemented with 10% FBS and 600 ng/mL G418. Relative absorbance optical density (OD) values were measured daily for 6 consecutive days. Briefly, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide–based assay (MTT) was added to the cells at a final concentration of 2 mg/mL and was incubated for 3 hours at 37°C; then medium was aspirated from the wells, and 100 µL dimethyl sulfoxide (DMSO) was added and incubated at 37°C for 5 minutes. Subsequently, the samples were mixed and were read in an ELISA reader at a wavelength of 540 nm. Relative absorbance OD values were plotted as a function of time.
Plasmids Construct and Cell Transfection
pIRES2-EGFP contains the internal ribosome entry site of the encephalomyocarditis virus (ECMV) between the multiple cloning site (MCS) and the enhanced green fluorescent protein (EGFP) coding region. This permits inclusion of both the gene of interest (cloned into the MCS) and the EGFP gene to be translated from a single bicistronic mRNA. pIRES2-EGFP is designed for efficient selection by flow cytometry.
OPG primers, including restriction enzyme sites (NhEI and BamH1), were designed by Clone Managers (Cary, NC) and were used in PCR to amplify the OPG gene fragment (Table 1). Subsequently, the OPG fragment was inserted into the plasmids by T4 DNA ligases. The sequence of OPG construct was confirmed by automated sequencing in the laboratory. Plasmids were transfected into MC3T3-E1 cells with FuGENE 6 transfection reagent according to the manufacturer's instructions. A stable cell line was established by sorting with MoFlow and was selected by 600 µg/mL G418 for 3 weeks. Single cell–derived cell colonies were expanded and tested for green fluorescent protein (GFP) expression by fluorescence activated cell sorting (FACS) and for secreted OPG protein by ELISA kit and Western blot according to the manufacturer's instructions.
Table 1.
Primer Sequences
Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
Cells, transfected with control plasmids (pEGFP) or OPG recombinant (pOPG), were harvested and total mRNA was isolated using a Stratagene Kit (Stratagene Products, La Jolla, Calif). One-step real-time qPCR reactions were performed using Power SYBR Green RNA-to-CT One-Step Kit (Applied Biosystems, Foster City, Calif) in a final volume of 10 µL. Reactions, including 20 ng total RNA, were performed by Applied Biosystems 7500 Fast PCR Systems. Data were analyzed by SDS Software, version 2.3 (Applied Biosystems). Total RNA was reverse-transcripted at 48°C for 30 minutes. RT-qPCR was performed with DNA polymerase, including an activation step at 95°C for 10 minutes, followed by 40 cycles of denaturation and annealing/extension (95°C for 10 seconds and 60°C for 1 minute). A melt curve (denaturation 95°C for 15 seconds, annealing 65°C for 15 seconds, and denaturation 95°C for 15 seconds) was performed for specific amplification analysis. Data were read by SDS 2.3 software from Applied Biosystems. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as the reference gene.
OPG and Alkaline Phosphate ELISA
Secreted OPG protein was measured by an OPG ELISA kit. Briefly, the cells were seeded at a density of 5 × 10 E5 cells per well in a 12-well plate and were cultured for 24 hours. Cells were starved in DMEM without FBS for another 24 hours. Then 200 µL DMEM with 10% FBS was added to the culture for 48 hours. The supernatant was collected for ELISA testing, and cell numbers were counted. Secreted OPG protein concentration per 10 E5 cells was calculated from the standard curve.
Alkaline phosphate (ALP) protein was measured by an ELISA kit. Briefly, 96-well plates were coated with coating protein overnight; then 200 µL blocking buffer was added, and the plates were incubated for 30 minutes; 100 µL samples were added and incubated for 1 hour with a plate shaker after washing with the washing buffer. Enzyme-conjugated secondary antibody was added to the wells and was incubated for 1 hour before substrate was added, and reading was done with an absorbance ELISA reader with 405 nm wavelength. All procedures were performed at room temperature.
Western Blot
Cells were lysed by RIPA lysis buffer (Thermo Fisher Scientific, Rockford, Ill). A total of 30 micrograms lysate was subjected to denatured 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and was transferred to a nitrocellulose membrane. The membrane was incubated with OPG or β-actin antibodies.16
Statistical Analysis
Expression of ALP and the gene of interest in OPG-expressing cells and control cells were calculated by paired Student's t-test. P ≤ .05 was considered statistically significant.
RESULTS
Recombinant OPG DNA Construction and Cell Transfection
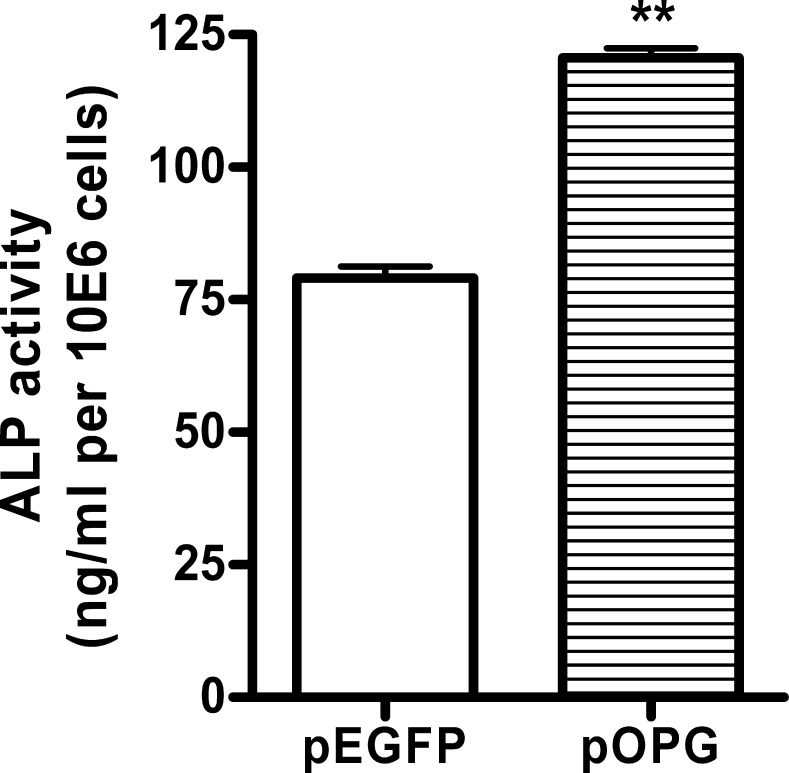
pIRES2-EGFP permits both the gene of interest and the EGFP gene to be translated from a single bicistronic mRNA. FACS analysis showed that the cells shifted to the right with EGFP expression compared with the control group, indicating that the plasmids had been successfully introduced into MC3T3-E1 cells (Figure 1).
Figure 1.
Fluorescence activated cell sorter (FACS) analysis of enhanced green fluorescent protein (EGFP) expression. MC3T3-E1 (A) was transfected with empty plasmids (pEGFP) (B) or constructed osteoprotegerin (OPG) recombinant (pOPG) (C) and was selected by G418. FACS analysis showed that most of the stable transfection cells shifted to the right with EGFP expression compared with the nontransfected cells.
OPG Expression
Both cytoplasmic OPG and secreted OPG were upregulated in pOPG groups compared with the control group (P < .05). Supernatants were collected at 24 hours and were tested by ELISA. OPG protein concentration secreted by transfected cells (pOPG) was almost two-fold higher than in the control (pEGFP, Figure 2A). Western blot showed that OPG protein in cytoplasm increased compared with control cells (Figure 2B).
Figure 2.
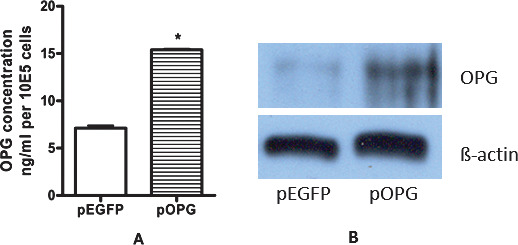
Osteoprotegerin (OPG) expression: (A) Secreted OPG protein was measured by enzyme-linked immunosorbent assay (ELISA). Data were the means of triplicates indicated by standard deviation (t-test; *P < .05). (B) Western blot for OPG protein in cytoplasm. OPG protein was confirmed to overexpress in MC3T3-E1.
Expression of Transcriptional Factors
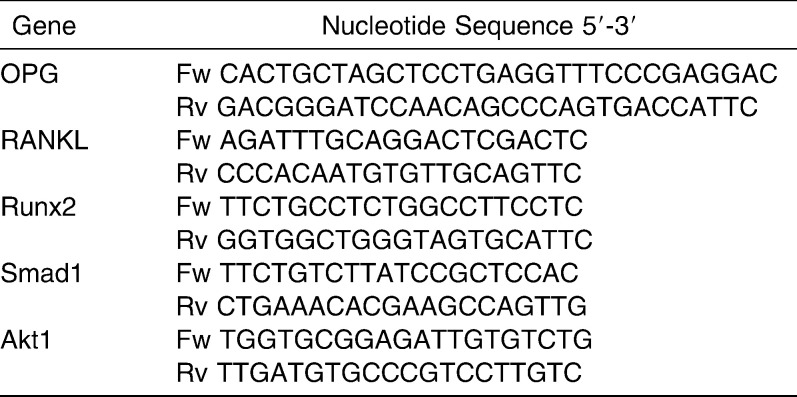
Transcriptional factors were differently expressed in pOPG and pEGFP cells. Runx2, a transcription factor blocking osteoblast differentiation, was suppressed to half that of the control group (pEGFP) (P < .01). Expressions of Smad1 and Akt1 were almost two times higher in the pOPG group than in the control group (Figure 3). RANKL mRNA expression was not significantly different between groups.
Figure 3.
Gene expression in empty plasmids (pEGFP) and osteoprotegerin (OPG) recombinant (pOPG) cells. For gene expression analysis, ΔΔCt was used; gene fold changes were calculated as the difference in gene expression between control group (empty plasmid transfected cells) and experiment group (OPG recombinant transfected cells). Gene fold changes were considered significant when P < .05 (**P < .01). In the control group, the gene expression level was normalized to 1, and the experiment group results indicated relative fold changes to the control group. Data were the means of triplicates with standard deviation indicated. The data showed that Runx2 was significantly suppressed, and Smad1 and Akt1 were significantly upregulated in the experiment group, but the RANKL expression was not significantly changed (t-test; **P < .001).
Alkaline Phosphate Expression
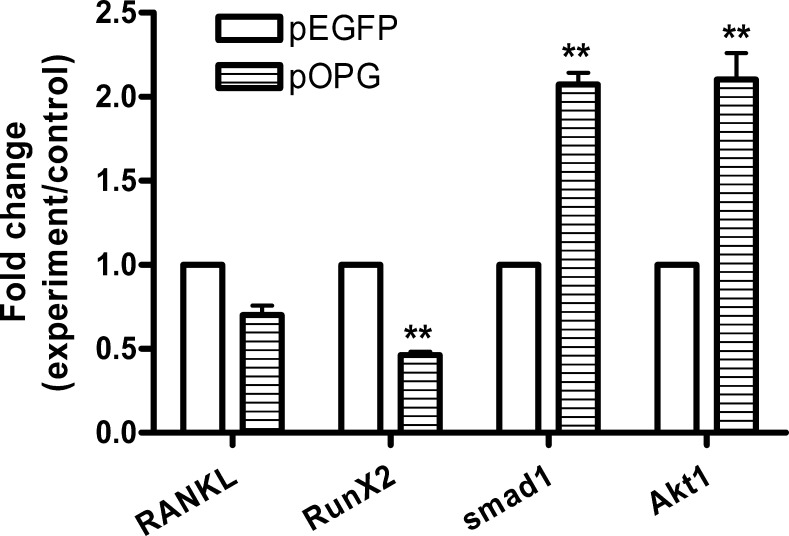
ALP was intensively expressed in pOPG cells compared with pEGFP cells. ELISA results showed that ALP activity was significantly higher in OPG-expressing cells (pOPG) (P < .001) (Figure 4). When the preosteoblasts differentiated to mature osteoblasts, ALP expression was upregulated. These data support that OPG promotes ALP expression.
Figure 4.
Alkaline phosphate (ALP) expression in osteoprotegerin (OPG)-expressing cells (pOPG) and the control group (pEGFP). Cells were collected and lysed to subject enzyme-linked immunosorbent assay (ELISA) measurement. Data were the means of triplicates with the standard deviation bar. ALP activity in the OPG-expressing cells (pOPG) was significantly higher than in the control group (pEGFP) (t-test; **P < .01).
Cell Proliferation
The proliferation of OPG-expressing (pOPG) and control cells (pEGFP) was assessed by MTT assay. The growth rate of OPG-expressing cells was slower than in the control group (Figure 5). Cell proliferation was significantly inhibited in OPG-expressing cells after 3 days of culturing.
Figure 5.
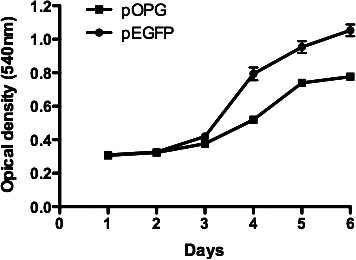
MC3T3-E1 cell proliferation in the presence of empty plasmids (pEGFP) or osteoprotegerin (OPG) recombinant (pOPG) transfection. Data were the means of triplicates at each time point with the standard deviation indicated. A 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide–based assay (MTT) on cell proliferation showed that OPG-expressing cells (pOPG) grew slower than the control group (pEGFP) after 3 days of culturing.
DISCUSSION
In the present study, we showed that OPG plays an important role in both osteoblastogenesis and osteoclastogenesis by controlling the differentiation and maturation of osteoclast and osteoblast. In osteoclastogenesis, OPG is a decoy factor competing with RANK for RANKL, which results in osteoclastogenesis inhibition.1 The ratio of RANKL to OPG is critical for osteoclastogenesis. Previous studies showed that bone mass increased significantly along with downregulation of Runx2 and upregulation of OPG in BMP receptor-1α conditioned knockout mice.17 LaminA/C knockout mice also showed decreased OPG expression and impaired osteoblast differentiation.18 However, no studies have been performed up to now on the direct effect of OPG expression on osteoblastogenesis. To our knowledge, this is the first study that addressed the effects of OPG expression on preosteoblast maturation. Our results show that OPG expression promotes osteoblast differentiation accompanied by increased ALP expression.
Our results also indicate that OPG can promote preosteoblast to differentiate into mature osteoblast, which was associated with the regulation of Runx2, Smad1, and Akt1 gene expression. When OPG was upregulated, Runx2 was inhibited while ALP expression was upregulated, indicating that OPG promotes the terminal differentiation of osteoblast progenitor to mature osteoblast.
Runx2 is an essential transcription factor in osteoblasts and plays an important role in governing physiologically responsive control of skeletal genes.5 Runx2 plays different roles in different stages of osteoblast differentiation and proliferation.19 Runx2 accumulates in the immature osteoblast in the early stage of osteoblast differentiation while blocking the maturation of osteoblast in the later stage.20 Runx2 interacts with different proteins to control osteoblast differentiation and proliferation.5 The effect of Runx2 function on osteoblast differentiation and OPG expression is controversial.21–23 Some studies show that Runx2 induces RANKL expression and suppresses OPG expression,24 and osteoblast differentiation was arrested in Runx2 overexpression cells.22 Another study shows that Runx2 contributes to OPG activities.21 The reason is probably that Runx2 regulates its target genes together with other factors in different osteoblast maturation stages and different cell lineages. Akt1 is an important transcription gene for controlling posttranscriptional levels of osteoblast differentiation7; it interacts with Runx2 to regulate osteocalcin transcription.8 Smad1 is another gene that interacts with Runx2. Runx2/Smad1 interaction is one of the physiologic pathways to transmit bone morphogenetic protein receptor signals to target genes for osteoblast differentiation.9 Smads may modify the transcriptional prosperities of cell growth of Runx2 by interacting directly with COOH terminals of Runx2.23 In our study, when OPG was overexpressed in preosteoblasts, both Smad1 and Akt1 genes were upregulated, but the Runx2 gene was downregulated, suggesting that upregulated smad1 and Akt1 interacted with Runx2 to promote osteoblast phenotypes by activating genes specific for osteoblast maturation. Suppressed Runx2 expression probably promoted the preosteoblast to move out from the cell cycle for proliferation.20,22,23 This was supported by our results that OPG-overexpressing osteoblasts showed a slower growth rate. It is disputable about the Runx2 expression level in osteoblast maturation stage.20,23 In our study, Runx2 mRNA suppression probably occurred because Runx2 reduces its biological stimulation of OPG activities to compromise the introduction of OPG overexpression. We suggest that OPG may play a key role in preosteoblast maturation.
Osteoblasts are originally derived from mesenchymal stem cells.25,26 Different osteoblast populations have some common features but exhibit unique phenotype genes.11 ALP is the marker of matrix maturation. During differentiation from mesenchymal cells to mature osteoblasts, ALP starts to be expressed in osteoprogenitors and is highly expressed in mature osteoblasts.2,3 It has been shown that ALP expression is significantly reduced in Runx2 overexpression osteoblasts, resulting in its differentiation inhibition.22 In the present study, ALP expression was significantly higher in OPG-expressing cells, indicating that OPG promotes matrix maturation in preosteoblast.
CONCLUSIONS
The present study provides for the first time evidence that OPG expression is associated with preosteoblast maturation and promotes matrix maturation.
Along with Akt1 and Smad1, Runx2 may be a crucial factor that controls OPG expression in both osteoblastogenesis and osteoclastogenesis.
Acknowledgments
This work was financially supported by the Dutch Organization of Scientific Research (NWO ZonMW 016.056.171).
REFERENCES
- 1.Hadjidakis D. J, Androulakis I. I. Bone remodeling. Ann N Y Acad Sci. 2006;1092:385–396. doi: 10.1196/annals.1365.035. [DOI] [PubMed] [Google Scholar]
- 2.Aubin J. E. Regulation of osteoblast formation and function. Rev Endocr Metab Disord. 2001;2:81–94. doi: 10.1023/a:1010011209064. [DOI] [PubMed] [Google Scholar]
- 3.Zhang X, Aubin J. E, Inman R. D. Molecular and cellular biology of new bone formation: insights into the ankylosis of ankylosing spondylitis. Curr Opin Rheumatol. 2003;15:387–393. doi: 10.1097/00002281-200307000-00004. [DOI] [PubMed] [Google Scholar]
- 4.Kassem M, Abdallah B. M, Saeed H. Osteoblastic cells: differentiation and trans-differentiation. Arch Biochem Biophys. 2008;473:183–187. doi: 10.1016/j.abb.2008.03.028. [DOI] [PubMed] [Google Scholar]
- 5.Stein G. S, Lian J. B, van Wijnen A. J, et al. Runx2 control of organization, assembly and activity of the regulatory machinery for skeletal gene expression. Oncogene. 2004;23:4315–4329. doi: 10.1038/sj.onc.1207676. [DOI] [PubMed] [Google Scholar]
- 6.Ducy P, Zhang R, Geoffroy V, Ridall A. L, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 7.Kawamura N, Kugimiya F, Oshima Y, et al. Akt1 in osteoblasts and osteoclasts controls bone remodeling. PLoS ONE. 2007;2:e1058. doi: 10.1371/journal.pone.0001058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao G, Jiang D, Ge C, et al. Cooperative interactions between activating transcription factor 4 and Runx2/Cbfa1 stimulate osteoblast-specific osteocalcin gene expression. J Biol Chem. 2005;280:30689–30696. doi: 10.1074/jbc.M500750200. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y. W, Yasui N, Ito K, et al. A RUNX2/PEBP2alpha A/CBFA1 mutation displaying impaired transactivation and Smad interaction in cleidocranial dysplasia. Proc Natl Acad Sci U S A. 2000;97:10549–10554. doi: 10.1073/pnas.180309597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khosla S. Minireview: the OPG/RANKL/RANK system. Endocrinology. 2001;142:5050–5055. doi: 10.1210/endo.142.12.8536. [DOI] [PubMed] [Google Scholar]
- 11.Boyle W. J, Simonet W. S, Lacey D. L. Osteoclast differentiation and activation. Nature. 2003;423:337–342. doi: 10.1038/nature01658. [DOI] [PubMed] [Google Scholar]
- 12.Bucay N, Sarosi I, Dunstan C. R, et al. Osteoprotegerin-deficient mice develop early onset osteoporosis and arterial calcification. Genes Dev. 1998;12:1260–1268. doi: 10.1101/gad.12.9.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ominsky M. S, Stolina M, Li X, et al. One year of transgenic overexpression of osteoprotegerin in rats suppressed bone resorption and increased vertebral bone volume, density, and strength. J Bone Miner Res. 2009;24:1234–1246. doi: 10.1359/jbmr.090215. [DOI] [PubMed] [Google Scholar]
- 14.Christopher M. J, Link D. C. Granulocyte colony-stimulating factor induces osteoblast apoptosis and inhibits osteoblast differentiation. J Bone Miner Res. 2008;23:1765–1774. doi: 10.1359/JBMR.080612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baldock P. A, Thomas G. P, Hodge J. M, et al. Vitamin D action and regulation of bone remodeling: suppression of osteoclastogenesis by the mature osteoblast. J Bone Miner Res. 2006;21:1618–1626. doi: 10.1359/jbmr.060714. [DOI] [PubMed] [Google Scholar]
- 16.Chakravarti A, Marceau A. A, Flamand L, Poubelle P. E. Normal human primary CD4+ T lymphocytes synthesize and release functional osteoprotegerin in vitro. Lab Invest. 2008;88:171–184. doi: 10.1038/labinvest.3700701. [DOI] [PubMed] [Google Scholar]
- 17.Kamiya N, Ye L, Kobayashi T, et al. Disruption of BMP signaling in osteoblasts through type IA receptor (BMPRIA) increases bone mass. J Bone Miner Res. 2008;23:2007–2017. doi: 10.1359/JBMR.080809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rauner M, Sipos W, Goettsch C, Wutzl A, Foisner R, Pietschmann P, Hofbauer L. C. Inhibition of lamin A/C attenuates osteoblast differentiation and enhances RANKL-dependent osteoclastogenesis. J Bone Miner Res. 2009;24:78–86. doi: 10.1359/jbmr.080902. [DOI] [PubMed] [Google Scholar]
- 19.Cameron E. R, Blyth K, Hanlon L, et al. The Runx genes as dominant oncogenes. Blood Cells Mol Dis. 2003;30:194–200. doi: 10.1016/s1079-9796(03)00031-7. [DOI] [PubMed] [Google Scholar]
- 20.Liu W, Toyosawa S, Furuichi T, et al. Overexpression of Cbfa1 in osteoblasts inhibits osteoblast maturation and causes osteopenia with multiple fractures. J Cell Biol. 2001;155:157–166. doi: 10.1083/jcb.200105052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thirunavukkarasu K, Halladay D. L, Miles R. R, Yang X, Galvin R. J, Chandrasekhar S, Martin T. J, Onyia J. E. The osteoblast-specific transcription factor Cbfa1 contributes to the expression of osteoprotegerin, a potent inhibitor of osteoclast differentiation and function. J Biol Chem. 2000;275:25163–25172. doi: 10.1074/jbc.M000322200. [DOI] [PubMed] [Google Scholar]
- 22.Merciris D, Marty C, Collet C, de Vernejoul M. C, Geoffroy V. Overexpression of the transcriptional factor Runx2 in osteoblasts abolishes the anabolic effect of parathyroid hormone in vivo. Am J Pathol. 2007;170:1676–1685. doi: 10.2353/ajpath.2007.061069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomas D. M, Carty S. A, Piscopo D. M, Lee J. S, Wang W. F, Forrester W. C, Hinds P. W. The retinoblastoma protein acts as a transcriptional coactivator required for osteogenic differentiation. Mol Cell. 2001;8:303–316. doi: 10.1016/s1097-2765(01)00327-6. [DOI] [PubMed] [Google Scholar]
- 24.Enomoto H, Shiojiri S, Hoshi K, et al. Induction of osteoclast differentiation by Runx2 through receptor activator of nuclear factor-kappa B ligand (RANKL) and osteoprotegerin regulation and partial rescue of osteoclastogenesis in Runx2-/- mice by RANKL transgene. J Biol Chem. 2003;278:23971–23977. doi: 10.1074/jbc.M302457200. [DOI] [PubMed] [Google Scholar]
- 25.Karsenty G, Wagner E. F. Reaching a genetic and molecular understanding of skeletal development. Dev Cell. 2002;2:389–406. doi: 10.1016/s1534-5807(02)00157-0. [DOI] [PubMed] [Google Scholar]
- 26.Karsenty G. The complexities of skeletal biology. Nature. 2003;423:316–318. doi: 10.1038/nature01654. [DOI] [PubMed] [Google Scholar]