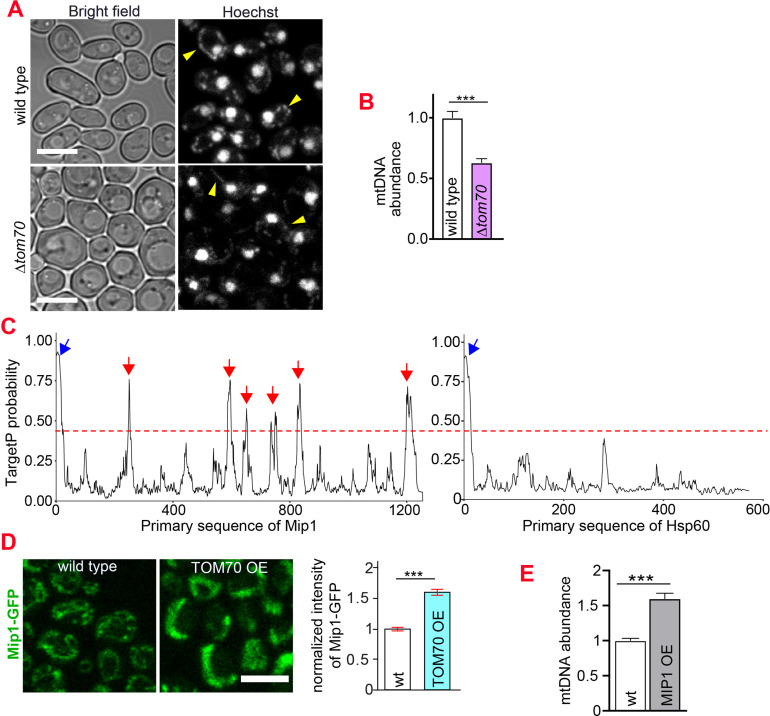
Figure 3. Tom70 regulates the abundance of mtDNA.
(A, B) Representative images and quantification of mtDNA in wild type and ∆tom70 cells were stained by Hoechst dye. Yellow arrowheads point to the mtDNA. Bar graph are the mean and s.e.m. from 87 and 109 cells. (C) iMTS probability profiles of Mip1 and Hsp60 predicted by TargetP algorithm. The consecutively N-terminally truncated sequences of the proteins were used as input to calculate the TargetP scores for each residue, which shows internal regions with presequence-like properties, or iMTS. Hsp60, another mitochondrial protein does not depend on Tom70, was also plotted in the same way for comparison (Backes et al., 2018). Red line (0.4) indicates the cutoff defined in previous study (Backes et al., 2018). Blue and red arrows indicate the N-terminal signal peptide and iMTS, respectively. (D) Representative images and quantification of Mip1-GFP in control and TOM70 OE cells. Bar graphs are the mean and s.e.m. from 300 and 438 cells. (E) Quantification of mtDNA in control and MIP1 OE cells from Hoechst staining. Bar graphs are the mean and s.e.m. from 253 and 209 cells. Scale bar for all images: 5 μm. Images are representative of at least two independent experiments.Bar graphs are the normalized mean and s.e.m. Data were analyzed with unpaired two-tailed t test: ***, p < 0.001.