Summary
How different sensory stimuli are collected, processed, and further transformed into a coordinated motor response is a fundamental question in neuroscience. In particular, the internal and external conditions that drive animals to switch to backward walking and the mechanisms by which the nervous system supports such behavior are still unknown. In fruit flies, moonwalker descending neurons (MDNs) are considered command-type neurons for backward locomotion as they receive visual and mechanosensory inputs and transmit motor-related signals to downstream neurons to elicit backward locomotion. Whether other modalities converge onto MDNs, which central brain neurons activate MDNs, and whether other retreat-driving pathways exist is currently unknown. Here, we show that olfactory stimulation can elicit MDN-mediated backward locomotion. Moreover, we identify the moonwalker subesophageal zone neurons (MooSEZs), a pair of bilateral neurons, which can trigger straight and rotational backward locomotion. MooSEZs act via postsynaptic MDNs and via other descending neurons. Although they respond to olfactory input, they are not required for odor-induced backward walking. Thus, this work reveals an important modality input to MDNs, a novel set of neurons presynaptic to MDNs driving backward locomotion and an MDN-independent backward locomotion pathway.
Keywords: Drosophila, sensorimotor integration, motor control, neural pathways, backward locomotion, olfaction, SEZ, descending neurons, MDNs, MooSEZs
Graphical abstract
Highlights
-
•
MooSEZs elicit backward locomotion via MDN-dependent and MDN-independent pathways
-
•
MooSEZs are connected to MDNs and other descending neurons
-
•
MooSEZs and MDNs both respond to olfactory input
-
•
MooSEZs can trigger rotational backward locomotion
Israel et al. describe olfactory-driven backward locomotion that is mediated by MDNs. They also identify a pair of bilateral central brain neurons, MooSEZs, which respond to odor input and contribute to rotational backward locomotion via postsynaptic MDNs and additional descending neurons.
Introduction
Walking is a fundamental behavioral feature of many terrestrial organisms, allowing them to respond adaptably to their environment. In order to support motor-based behaviors, and specifically rhythmic activities such as walking, the nervous system needs to perform a sequential multi-step process: collect relevant sensory information, integrate incoming information with internal state, and efficiently execute the selected motor program. Although the nervous system of invertebrates is compact in comparison with that of vertebrates, their walking repertoire is highly diverse and complex.1 Thus, insect nervous systems can serve as experimental models to study the neuronal mechanisms underlying coordinated motor activity. In particular, the fruit fly seems to be an ideal choice, as individual cells within its brain can be precisely targeted and easily manipulated using genetic techniques.2
Several decades of research have led to substantial knowledge about basic motor pattern generation in insects, such as reflex-activated leg movements,3,4 jumping,5 wing movement in flying,6,7 and leg stepping in walking.8,9 In addition, more involved behaviors were examined, such as coordination of antennae and leg movement during obstacle negotiation,10,11 gap crossing,12 grooming,13 chemotaxis orientation,14 and courtship behavior.15,16 Although land animals have the ability to walk backward upon sensing an obstacle or danger in their path, the neural mechanisms underlying the initiation and execution of backward walking are still poorly understood.1
By exploiting the powerful genetic toolkit available for fruit flies,17 a specific cluster of neurons, moonwalker descending neurons (MDNs), which triggers backward walking, was identified.18 In support of MDNs role in driving backward locomotion, it was demonstrated that MDN activity correlates with episodes of backward walking.19 In addition, it was shown that a population of neurons in the fly’s visual system induce backward walking via MDNs through indirect synaptic connections20 and that ascending mechanosensory neurons in the ventral nerve cord (VNC) activate MDNs to mediate touch-evoked backward walking.21 However, other sensory pathways such as gustation, thermoreception, and in particular, olfaction, a major sensory input on which insects rely heavily,22 have not been associated with MDN-mediated backward walking. Furthermore, while significant progress has been recently made in characterizing the downstream targets of MDNs in the VNC of adult flies,23,24 little is known about the upstream neural circuits in the central brain which directly control and modulate MDN activity and thus backward walking.
In the case of flies, odors are sensed by first-order olfactory sensory neurons (OSNs) expressing either odorant receptors (ORs) or ionotropic receptors (IRs).25, 26, 27, 28, 29 There are 51 types of OSNs defined by the type of OR or IR they express.30 OSNs project to the antennal lobe (AL), where the axons of each type of OSN target a single glomerulus.31, 32, 33 Second-order projection neurons (PNs) send their dendrites to a single glomerulus and project to two higher brain regions: the mushroom body (MB), where associative olfactory memories occur and the lateral horn (LH).34
In this study, we show that olfactory input can trigger backward locomotion mediated by MDNs. In addition, we identify a pair of bilateral neurons originating in the subesophageal zone (SEZ), which trigger straight backward walking upon short symmetric activation and backward turning following asymmetric activation. We name these neurons Moonwalker SEZ neurons (MooSEZs). MooSEZs are monosynaptically connected to MDNs and can trigger backward walking in an MDN-dependent and -independent manner. While both MDNs and MooSEZs respond to odors, odor-driven backward walking is only eliminated by blockade of MDN activity. Thus, this work reveals an important modality input to MDNs, a novel set of MDN-presynaptic neurons driving backward locomotion and an MDN-independent pathway.
Results
Olfactory input triggers MDN-dependent backward walking
Recently, it was demonstrated that visual input to MDNs generates backward walking.20 It was suggested that this backward locomotion is part of a repertoire of escape responses to a visual threat. Similar to visual inputs, olfactory cues can also signal threat.35, 36, 37 For instance, the odorant geosmin serves flies as an alarm signal for the presence of toxic microbes in their environment.38 We first examined whether 2-butanone, a strongly aversive odor,39 can trigger backward walking. To this end we placed flies in an open-field arena and examined their responses when exposed to 2-butanone. While exposure to 2-butanone elicited turning and forward walking in most flies, in ∼40% of the flies turning was combined with a pronounced backward walking component (Figures 1A–1C). To verify that the observed backward walking response is indeed an odor-specific effect, we used the more appetitive apple cider vinegar (ACV) and geranyl acetate (GA).39 Following exposure to ACV and GA, we observed negligible retreat responses across tested flies (Figures 1A–1C). Yet, backward walking responses evoked by 2-butanone in the open-field arena were rather transient, consisting of only a number of retreat steps coupled to odor presentation. We therefore sought to enhance the backward walking phenotype and repeated the experiment in a linear chamber assay system that was previously shown to effectively restrict flies’ lateral and rotational movements (Figure 1D).18 Using this apparatus, all three tested odors induced backward walking responses among flies. However, strong and robust backward walking was observed only for 2-butanone (Figures 1E and 1F; Video S1). To fully characterize odor-evoked motor responses in the linear chambers, we exposed flies to an additional set of nine odorants, with different levels of odor valence, as previously reported.39 Interestingly, it seems that in this setting, all examined odors elicit backward locomotion to some extent (Figures S1A and S1B). However, it appears that the magnitude of the motor effect depends on odor valence since the extent to which flies walked backward in the linear chambers was tightly correlated with odor valence (Figure S1C). Thus, it seems that the linear chamber assay drives a hard-wired innate response of odor-driven backward walking which is modulated by odor valence.
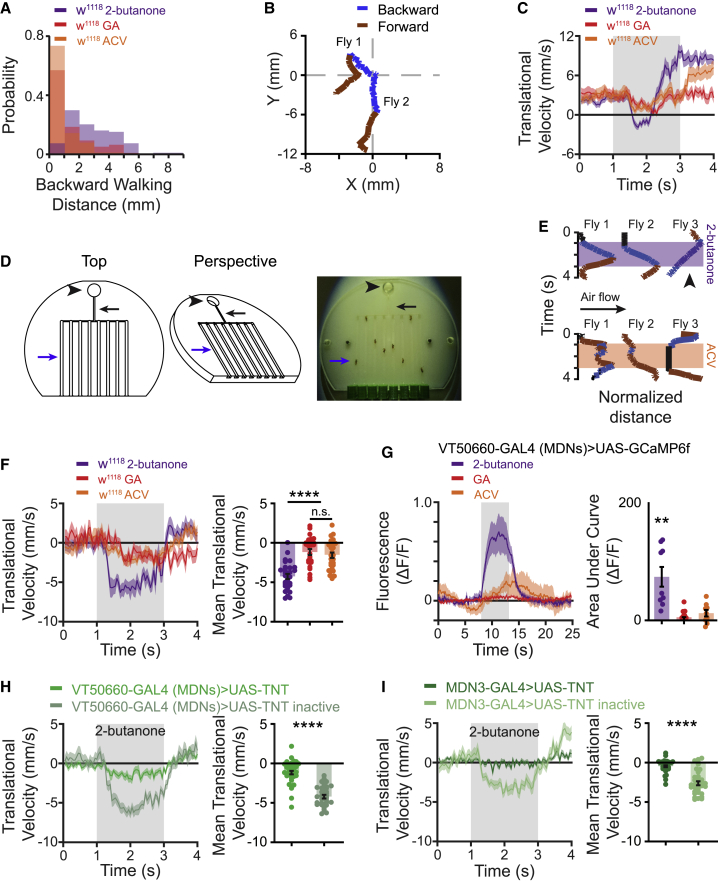
Figure 1.
Odor-induced backward walking is mediated by MDNs
(A) The probability of observing backward locomotion in response to a 2-s odor pulse in an open arena. Backward walking distance is defined as the integrated area under the negative values of the translational velocity versus time curve during the odor pulse. Translational velocity is defined as the projection of the fly’s velocity vector on its orientation direction. For ACV and GA the probability of a fly to perform odor-evoked backward retreat is lower compared with the aversive odor 2-butanone (61 ≤ n ≤ 103).
(B) Examples of two fly trajectories during application of 2-butanone for 2 s in the open arena. Blue and brown lines designate backward and forward locomotion respectively. The (0, 0) coordinate represents fly’s trajectory onset at t = 0. Flies responded in a transient backward locomotion.
(C) Translational velocity ± SEM (shading) following application of 2-butanone (dark purple), ACV (orange) and GA (red) in the open arena. The 2-s odor pulse is labeled in light gray (61 ≤ n ≤ 103).
(D) Left, schematic of the linear chamber behavior apparatus. Top and perspective views are presented. Right, a picture of the linear chamber behavior apparatus. Black arrowheads indicate odor inlet, black arrows indicate narrow airstream path to the linear grooves, and blue arrows indicate a single linear groove.
(E) Examples of flies’ trajectory in the linear chambers. Blue denotes backward walking, brown forward motion, and black stalling. 2-butanone is labeled in purple, ACV is labeled in orange. Note that evenly paced backward walking is robustly induced by 2-butanone but not ACV, irrespective of airflow direction. This can be observed in the 2-butanone trace of Fly 3 (labeled with a black arrowhead) where backward locomotion toward the odor source is observed.
(F) Left, translational velocity ± SEM (shading) following application of 2-butanone (dark purple), ACV (orange) and GA (red) in the linear chambers. The 2-s odor pulse is labeled in light gray. A clear backward motion is observed following 2-butanone but not following ACV or GA. Right, mean translational velocity during the 2-s odor pulse obtained from traces on the left. A significant difference is observed between the backward walking velocity generated by 2-butanone and that elicited by ACV or GA (25 ≤ n ≤ 30, ∗∗∗∗ p < 0.0001, Kruskal-Wallis test followed by Dunn’s post-hoc test; see Table S1).
(G) Left, averaged traces ± SEM (shading) of Ca2+ responses following an odor pulse as designated. The 5-s odor pulse is labeled in light gray. The broad MDN driver line VT50660-GAL4 was used to drive GCaMP6f. Right, area under the curve of ΔF/F during the 5-s odor pulse for the traces presented in the left panel. A significant Ca2+ response in the MDNs is elicited by 2-butanone but not by ACV or GA (9 ≤ n ≤ 11 flies, ∗∗ p < 0.01, one-sample t test; see Table S1).
(H and I) Left, translational velocity ± SEM (shading) following application of 2-butanone for flies expressing TNT under the control of VT50660-GAL4 (MDNs) (H, green), or under the control of the narrow split GAL4 driver line MDN3-GAL4 (I, dark green) in the linear chambers. The 2-s odor pulse is labeled in light gray. The inactive TNT is labeled in light green. In both cases the expression of TNT dramatically decreases backward walking. Right, mean translational velocity during the 2-s odor pulse obtained from traces on the left. A significant difference is observed between the backward walking velocities generated by 2-butanone when TNT is expressed and when the inactive TNT is expressed in MDNs (MDN3-GAL4: 35 ≤ n ≤ 37, VT50660-GAL4 (MDNs): 32 ≤ n ≤ 40, ∗∗∗∗p < 0.0001, Mann-Whitney test; see Table S1).
Example of w1118 responses to ACV (left), GA (middle) and 2-butanone (right).
As MDNs were demonstrated to be causally linked to backward walking,18 we examined whether an odor pulse can trigger neuronal responses in MDNs. We expressed GCaMP6f40 in MDNs using the broad MDN driver line VT50660-GAL418 and performed two-photon in vivo Ca2+ imaging, focusing on the dendritic arbors of the MDNs. As expected from the results above, MDNs showed strong responses to application of 2-butanone and relatively weak or no responses to application of ACV and GA (Figure 1G). To examine whether MDNs are necessary for olfactory-driven backward locomotion, we expressed in MDNs the tetanus toxin light chain (TNT), which is an inhibitor of synaptic transmission,41 using either the broad MDN driver, VT050660-GAL4, or the more specific split-GAL4, MDN3-GAL4.18 Expression of TNT in MDNs abolished odor-driven backward locomotion. Expression of the inactive tetanus toxin (TNT-inactive) had no effect on the odor-elicited backward walking (Figures 1H and 1I). Taken together, we found a backward walking response elicited by odors that requires MDNs.
Optogenetic activation of GH146II-, NP225-, and NP5288-GAL4 triggers backward walking
The results above suggest that an olfactory input can trigger backward walking via MDNs. In an attempt to identify the underlying neurons responsible for this effect, we screened 43 driver lines covering different populations of olfactory neurons by acute optogenetic activation with ChR2-XXM.42 Since optogenetic stimulations can induce complex motor responses20,21 that can be masked in one-dimensional linear chambers, this set of experiments was performed in the open arena assay in which flies can walk freely. We first verified that we can optogenetically trigger and identify backward walking by activating MDNs with a blue light pulse. Activation of MDNs using both the broad and narrow MDN driver lines indeed elicited sustained backward walking, as previously observed18 (Figure S2A). Out of the 43 drivers examined, we observed robust and prolonged backward walking in three broad driver lines which cover most of the PNs: GH146II-GAL4 (GH146-GAL4 on the second chromosome), NP5288-GAL4, and NP225-GAL4 (Figures 2A and 2B; Video S2). Importantly, MDNs are not labeled by these lines (Figures S2B and S3). Surprisingly, optogenetic activation of GH146-GAL4 on the X chromosome (GH146X-GAL4)43 did not elicit backward locomotion (Figures 2A and 2B), suggesting that GH146II-GAL4-induced backward locomotion is not mediated by PNs covered by both GH146 driver lines. In some driver lines, optogenetic activation resulted in a seizure-like behavior and thus these driver lines could not be further examined (Figure 2A). To verify that the observed backward walking phenotype persists independent of optogenetic actuator and light wavelength, we also tested red light activated CsChrimson44 using the driver lines GH146II-GAL4 and NP5288-GAL4. Indeed, activation of CsChrimson elicited robust backward walking, similarly to blue light activation of ChR2-XXM (Figures 2A, 2B, and 2D). We also characterized the sensitivity of the elicited backward walking to the light stimulus intensity. Whereas activation using blue light (470 nm) was intensity sensitive and ranged from eliciting forward motion at low intensities to eliciting backward walking at high intensities, red light (617 nm) excitation evoked strong and efficient activation of backward walking at all tested intensities (Figure S2C). It was demonstrated that red light penetrance through adult fly cuticle is significantly higher than that of blue light.45 Thus, flies’ lack of backward walking at low blue light intensities presumably results from weak optogenetic activation that failed to induce noticeable motor responses.
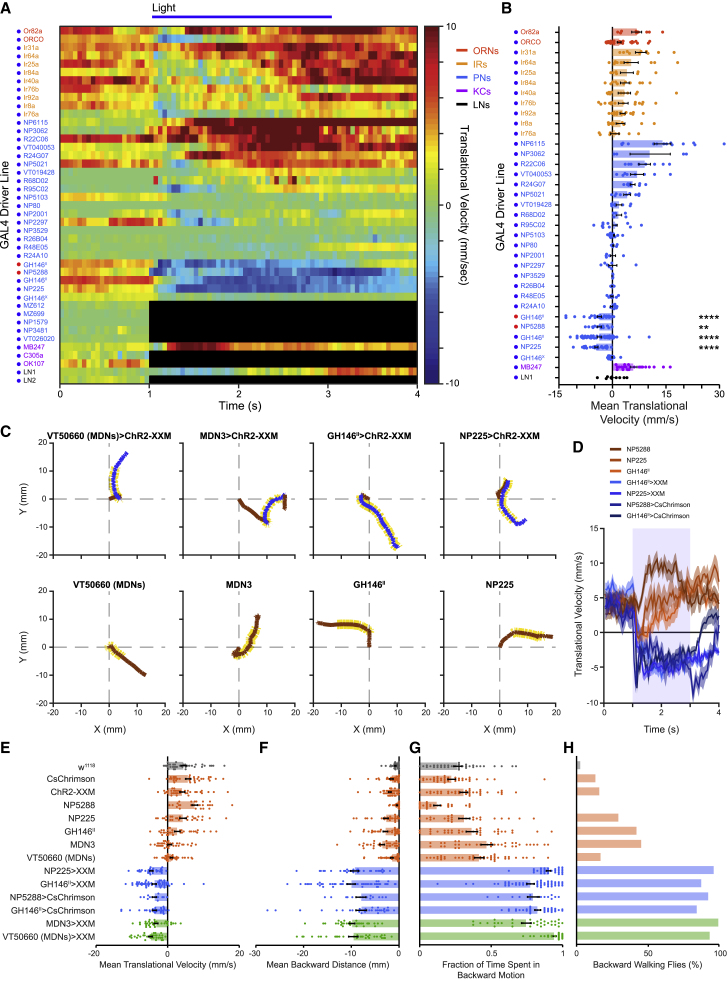
Figure 2.
Short activation of GH146II-, NP225-, and NP5288-GAL4 induces similar backward walking as MDNs
(A) Translational velocity matrix of different GAL4 driver lines (as designated) expressing ChR2-XXM (blue dots) or CsChrimson (red dots) in an open arena. A light pulse (horizontal blue line) was given between 1 and 3 s. In some cases, light activation of ChR2-XXM resulted in a seizure-like behavior which did not allow assessing walking velocity. Such cases are labeled in black.
(B) Mean translational velocity during the 2 s light pulse obtained from traces used to compose the matrix in (A). A significant backward locomotion is observed for GH146II-, NP225- and NP5288-GAL4 (4 ≤ n ≤ 50, ∗∗p < 0.01, ∗∗∗∗p < 0.0001, Wilcoxon signed rank test (one-sided) with Bonferroni correction; see Table S1).
(C) Examples of single fly walking trajectories in the open arena for MDNs, GH146II-, NP225-GAL4 driving UAS-ChR2-XXM and parental controls before, during, and after optogenetic stimulation. 2-s blue light pulse is designated by yellow, blue denotes backward walking and brown forward motion. The (0, 0) coordinate represents flies’ trajectory onset at t = 0. A short 2-s optogenetic activation of flies expressing ChR2-XXM using GH146II- or NP225-GAL4 driver lines elicits motor response of persistent backward retreat, which is similar to the motor pattern observed for flies expressing ChR2-XXM in MDNs using VT50660-GAL4 (MDNs) or the narrow split GAL4 driver line MDN3-GAL4. In contrast, light stimulation of the respective parental control flies fails to elicit backward walking.
(D) Translational velocity ± SEM (shading) for GH146II-, NP225-, or NP5288-GAL4 driving either UAS-ChR2-XXM or UAS-CsChrimson as designated in the open arena. Sustained backward walking is observed throughout the 2-s light pulse (labeled by light blue).
(E–H) Analysis of the following parameters during the 2-s light pulse delivered in the open arena: (E) mean translational velocity, (F) mean backward distance covered by the flies, (G) fraction of time spent in backward motion, and (H) percentage of flies that covered a minimal 3-mm backward walking distance for MDN driver lines (green), GH146II-, NP225-, or NP5288-GAL4 driver lines (blue), parental controls (brown), and w1118 flies (gray). For all computed parameters, GH146II-, NP225-, and NP5288-GAL4 driver lines resemble MDN driver lines for the short light stimulation that was delivered (14 ≤ n ≤ 50, p < 0.0001 for all comparisons to respective parental controls and w1118 except for mean translational velocity, MDN3-GAL4 p < 0.05, mean backward distance, GH146II-GAL4 p < 0.01, fraction of time spent in backward motion, MDN3-GAL4 p < 0.01. Kruskal-Wallis test followed by Dunn’s post-hoc test. Percentage of backward walking flies: p < 0.001 for all comparisons to respective parental controls and w1118, chi-squared test with Bonferroni correction; see Table S1).
Example of backward locomotion following a 2-s blue light pulse (470 nm) in flies carrying GH146II-GAL4 and UAS-ChR2-XXM.
Activation of MDNs elicits robust and sustained backward walking with a weak angular change (Figures 2C and S2A). In contrast, activation of the visual and mechanosensory pathways that activate MDNs elicits pronounced but transient backward walking with a strong turning component.20,21 We replicated these response dynamics by optogenetically activating LC16-1-GAL4, which drives expression in the lobula columnar 16 (LC16) cells, a distinct class of visual projection neurons (VPNs),46 and TLA-GAL4, which drives expression in TwoLumps ascending (TLA) neurons21 (Figures S2D and S2E). Interestingly, the backward locomotion following a short 2-s optogenetic activation of GH146II-, NP225-, and NP5288-GAL4 was distinctively similar to the persistent, straight motor response evoked by a short optogenetic activation of MDNs (Figures 2A, 2C, and 2D), as opposed to the transient and curved responses generated for LC16 VPNs and TLA short optogenetic activation (Figures S2D and S2E). We also compared a number of parameters describing the efficiency of a short light stimulation on backward walking and found no significant differences between MDN activation using MDN3 and VT50660 and the three GAL4 driver lines, GH146II, NP225, and NP5288 (Figures 2E–2H).
Backward locomotion induced by GH146II has MDN-dependent and -independent components
We have found that odors can elicit MDN-dependent backward locomotion and that neurons labeled by the broad PN driver line GH146II-GAL4 (as well as NP225- and NP5228-GAL4) can drive backward locomotion upon optogenetic stimulation. However, optogenetic activation using the GH146X-GAL4 driver line, which targets the PNs labeled by GH146II-GAL4, did not induce backward locomotion. This suggests that GH146II-GAL4 induced backward locomotion is not mediated by olfactory-related neurons and thus raises the question whether the observed backward locomotion is MDN-dependent. To examine the involvement of MDNs in GH146II-GAL4 evoked backward locomotion, we asked whether GH146II-GAL4 neurons could trigger neuronal responses in MDNs. We optogenetically activated ex vivo the GH146II-GAL4 neurons with CsChrimson and examined calcium transients in the dendritic arbors of MDNs using the genetically encoded Ca2+ indicator GCaMP6m40 (Figure S1D). Overall, we observed Ca2+ responses in MDNs following optogenetic activation of GH146II-GAL4 neurons (Figure 3A). However, while calcium transients were easily detected in MDN dendritic arbors of some of the flies, only weak responses were observed in others upon GH146II-GAL4 optogenetic stimulation. Stochastic variation in CsChrimson expression levels in GH146II-GAL4-targeted neurons across flies may underlie MDN response variability following GH146II-GAL4 optogenetic activation.
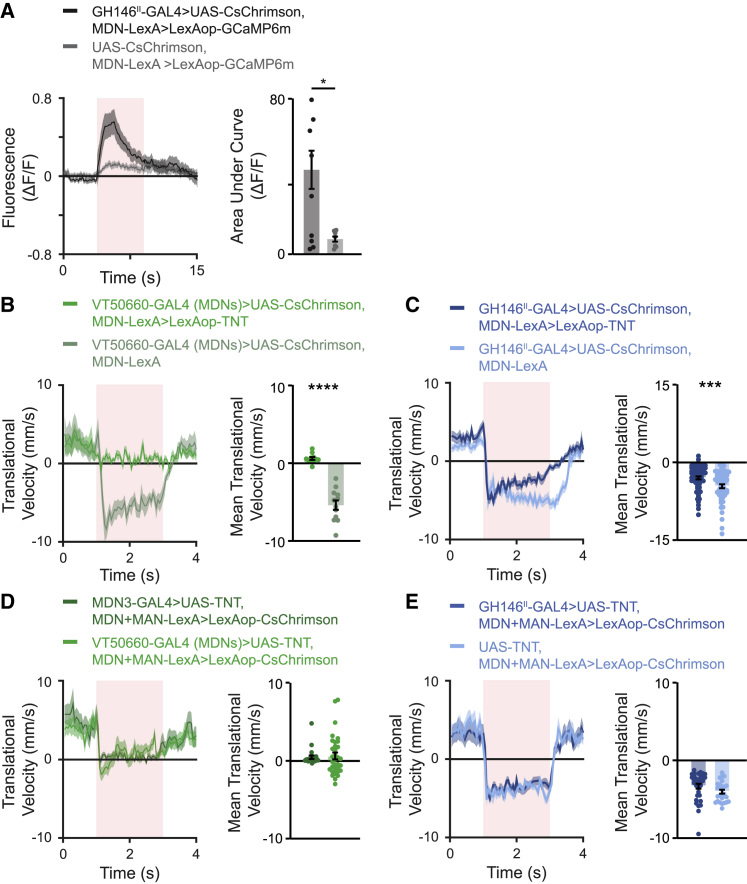
Figure 3.
GH146II-GAL4 neurons mediating backward locomotion are both MDN dependent and independent
(A) Left, averaged traces ± SEM (shading) of Ca2+ responses measured in MDNs following optogenetic activation of GH146II-GAL4 neurons. For the experimental group, labeled in black, the MDN-LexA driver line was used to drive LexAop-GCaMP6m and GH146II-GAL4 to drive UAS-CsChrimson. As a control, labeled in gray, the same genotype was used except that the GH146II-GAL4 was omitted. The 5-s light pulse is labeled in light red. Right, area under the curve of ΔF/F during the 5-s light pulse for the traces presented in the left panel. Optogenetic activation of the GH146II-GAL4 neurons leads to a significant Ca2+ response in MDNs (7 ≤ n ≤ 9, ∗p < 0.05, two-sample t test; see Table S1).
(B) Left, translational velocity ± SEM (shading) following optogenetic activation of MDNs in the presence (dark green) or absence (light green) of TNT in the open arena. The VT50660-GAL4 (MDNs) driver was used to drive UAS-CsChrimson, and the MDN-LexA (VT44845-LexA) driver was used to drive LexAop-TNT (when required). The 2-s light pulse is labeled in light red. Right, mean translational velocity during the 2-s light pulse obtained from traces on the left. TNT in MDNs completely blocks backward walking and a significant difference is observed between the mean translational velocities when TNT is expressed and when TNT is not expressed in the MDNs (11 ≤ n ≤ 12, ∗∗∗∗ p < 0.0001, Mann-Whitney test; see Table S1).
(C) Left, translational velocity ± SEM (shading) following optogenetic activation of GH146II-GAL4 neurons in the presence (dark blue) or absence (light blue) of TNT in MDNs in the open arena. The GH146II-GAL4 driver was used to drive UAS-CsChrimson and the MDN-LexA (VT44845-LexA) driver was used to drive LexAop-TNT (when required). The 2-s light pulse is labeled in light red. Right, mean translational velocity during the 2-s light pulse obtained from traces on the left. Backward locomotion is not abolished in flies expressing TNT in the MDNs upon light stimulation of GH16II-GAL4 neurons (58 ≤ n ≤ 64, ∗∗∗ p < 0.001, Mann-Whitney test; see Table S1).
(D) Left, translational velocity ± SEM (shading) following optogenetic activation of MDNs and MANs while blocking MDNs activity with UAS-TNT using MDN3 (dark green) or VT50660-GAL4 (MDNs) (light green) driver lines in the open arena. The split MDN+MAN-LexA (VT49484-ZpLexADBD; VT50660-p65ADZp) driver was used to drive LexAop-CsChrimson. The 2-s light pulse is labeled in light red. Right, mean translational velocity during the 2-s light pulse obtained from traces on the left. TNT in MDNs suppresses backward walking and a significant difference is observed between the mean translational velocities when TNT is not expressed (E, light blue) and when TNT is expressed (22 ≤ n ≤ 34, ∗∗∗∗ p < 0.0001, Kruskal-Wallis test followed by Dunn’s post-hoc test; see Table S1).
(E) Left, translational velocity ± SEM (shading) following optogenetic activation of MDNs and MANs in the presence (dark blue) or absence (light blue) of TNT in GH146II-GAL4 neurons in the open arena. The split MDN+MAN-LexA (VT49484-ZpLexADBD; VT50660-p65ADZp) driver was used to drive LexAop-CsChrimson. The 2-s light pulse is labeled in light red. Right, mean translational velocity during the 2-s light pulse obtained from traces on the left. Backward locomotion is not reduced in flies expressing TNT in GH146II-GAL4 neurons upon light stimulation of MDNs and MANs (22 ≤ n ≤ 34, Kruskal-Wallis test followed by Dunn’s post-hoc test; see Table S1).
We then examined whether the established MDN neural pathway is necessary for GH146II-GAL4-evoked backward locomotion. To do this, we inactivated MDNs by expressing TNT using MDN-LexA and lexAoP-TNT18 while artificially activating GH146II-GAL4. As a positive control, the efficiency of the LexAop-TNT was confirmed by direct optogenetic activation of MDNs. As expected, TNT efficiently silenced MDNs and eliminated backward walking (Figure 3B; Video S3). However, silencing MDNs while activating GH146II-GAL4 neurons did not abolish backward locomotion. Rather, excitation of GH146II-GAL4 neurons in flies with blocked MDNs elicited backward retreat, although to a weaker extent (Figure 3C; Video S3). Thus, these results suggest that GH146II-GAL4-labeled neurons are upstream of MDNs but also control motor circuits that mediate backward walking independent of MDNs. These neurons are either downstream of MDNs or participate in a parallel pathway to MDNs to support backward walking.
Left, example of the efficiency of TNT in silencing MDNs. The VT50660-GAL4 (MDNs) driver was used to drive UAS-CsChrimson, and the VT44845-LexA (MDN-LexA) was used to drive LexAop-TNT. Right, example of backward locomotion generated by activation of GH146II-GAL4 with UAS-CsChrimson while VT44845-LexA (MDN-LexA) driving LexAop-TNT was used to silence MDNs.
To examine whether the neuronal pathways underlying GH146II-GAL4-elicited backward walking are downstream of MDNs and moonwalker ascending neurons (MANs), which were previously shown to facilitate sustained backward locomotion,18 we optogenetically co-activated MDNs and MANs and silenced the neurons labeled by GH146II-GAL4 using TNT. As positive controls, we expressed TNT only in MDNs or in both MDNs and MANs. While backward walking was strongly reduced in positive control flies (Figure 3D), it was not impaired in flies expressing TNT in GH146II-GAL4 neurons (Figure 3E). Taken together, our results demonstrate that GH146II-GAL4 neurons are neither reciprocally connected to MDNs, nor downstream of MANs. Rather, they provide feedforward inputs to MDNs and participate in an additional backward locomotion pathway in parallel to MDNs.
Anatomical analysis of GH146II-, NP225-, and NP5288-GAL4 lines
To identify candidate neurons covered by GH146II-, NP225-, and NP5288-GAL4 driver lines (other than the olfactory pathway), we conducted a comprehensive anatomical analysis of these lines. All lines exhibit very similar expression patterns in both brain and VNC; notable exceptions are the anterior paired lateral (APL) neuron in the MB that is consistently labeled by GH146II-GAL4 (Figure S3), as summarized previously,47 and additional somata in the central brain and VNC labeled by NP5288-GAL4. The additional expression shared between all three lines includes three paired lateral somata and one pair of medioventral somata in the SEZ, as well as several paired neurons across VNC segments, including a paired neuron in the posterior VNC that ascends into the contralateral protocerebrum (Figure S3).
GH146II-, NP225-, and NP5288-GAL4 VNC neurons do not contribute to backward walking
The anatomical analysis revealed labeling in the VNC. Thus, it is possible that VNC neuron activity underlies the observed backward locomotion. We addressed this option using two approaches. In order to isolate VNC neurons from central brain neurons, flies were decapitated. Such chirurgical manipulation removes any central brain neurons but maintains ascending and descending tracts, such as axons of MDNs. Decapitated flies were shown to maintain coordinated walking following application of octopamine to the cervical connective.48 We first verified that decapitated flies have the ability to perform backward walking. Indeed, optogenetically activating MDNs using ChR2-XXM still resulted in robust backward walking (Figures 4A and 4B; Video S4). We then optogenetically activated neurons covered by GH146II-, NP225-, and NP5288-GAL4 in decapitated flies and found that backward walking was eliminated (Figures 4C and 4D; Video S4). However, we did observe behavioral responses to the optogenetic activation of GH146II-, NP225-, NP5288-GAL4 VNC neurons. Flies changed their posture by shifting their body weights to meso- and metathoracic legs, lifting their forelegs and moving them in an oscillatory fashion (Video S4). Investigation of this behavior was out of the scope of the current study and therefore we did not further analyze it. Thus, optogenetic activation of GH146II-, NP225-, and NP5288-GAL4 VNC neurons affects the flies’ behavior but does not generate any backward walking. As expected, no behavioral response whatsoever was observed in decapitated w1118 flies or any of the parental controls (Figures 4B and 4D). The above results suggest that optogenetic activation of GH146II-, NP225-, and NP5288-GAL4 neurons triggers two behavioral responses: the first is the backward walking that probably depends on labeled neurons in the central brain, and the second is the shift in body posture and foreleg movement, which involves labeled neurons in the VNC. We therefore revisited the experiments used for Figure 2A and indeed observed a combination of the two behavioral responses (Video S2). We then used tsh-GAL80 to block expression of CsChrimson in VNC neurons15,49 driven by GH146II-, NP225-, and NP5288-GAL4, which abolished expression in most VNC neurons (Figure 4E). As expected from the above conclusion, such manipulation eliminated the shift in body posture and movement of the foreleg in decapitated flies (Video S5) indicating the efficiency of tsh-GAL80 in blocking the expression of CsChrimson. In intact flies, backward walking was maintained for CsChrimson activation with GH146II-, NP225-, and NP5288-GAL4 when tsh-GAL80 was expressed (Figure 4F; Video S5). Taken together, the combined results of the above experiments suggest that GH146II, NP225-, and NP5288-GAL4 neurons in the central brain contribute to the backward walking we observed. These neurons function upstream and in parallel to MDNs.
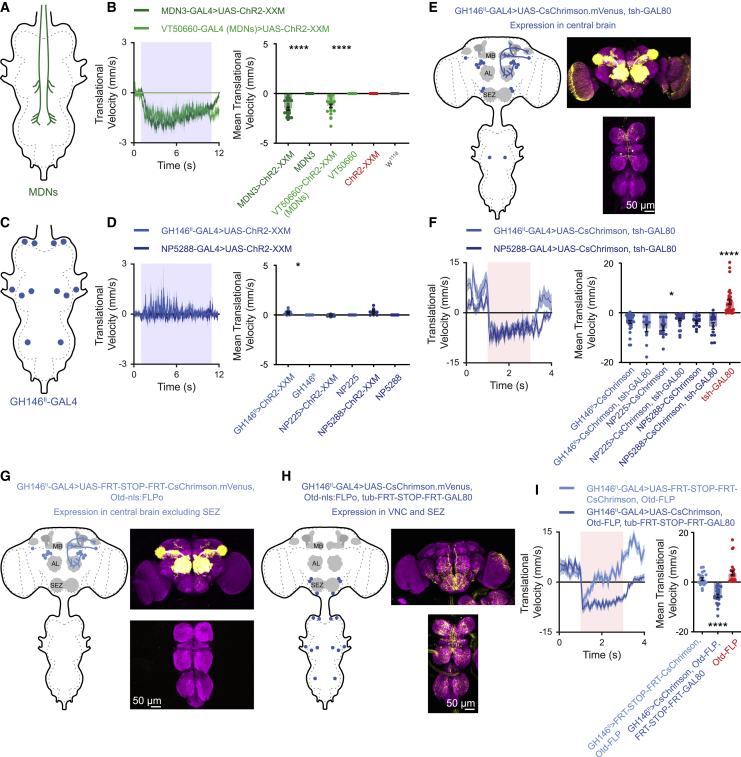
Figure 4.
SEZ neurons underlie GH146II-GAL4 optogenetically induced backward locomotion
(A) Schematic drawing of the expression pattern for the flies used in (B).
(B) Decapitated flies in the open arena show backward locomotion in the case of MDN driver lines as designated. ChR2-XXM was used to activate the neurons. Left, translational velocity ± SEM (shading). The 10-s light pulse is labeled in light blue. Right, mean translational velocity during the 10-s light pulse obtained from flies such as presented on the left for MDN driver lines driving UAS-ChR2-XXM, parental controls, and w1118 flies (12 ≤ n ≤ 28, ∗∗∗∗ p < 0.0001 for all comparisons to respective parental controls and w1118, Kruskal-Wallis test followed by Dunn’s post-hoc test; see Table S1).
(C) Schematic drawing of the expression pattern for the flies used in (D).
(D) Decapitated flies in the open arena show no backward locomotion in the case of GH146II-, NP225- and NP5288-GAL4 driver lines. UAS-ChR2-XXM was used to activate the neurons. Left, translational velocity ± SEM (shading). The 10-s light pulse is labeled in light blue. Right, mean translational velocity during the 10-s light pulse obtained from flies such as presented on the left for GH146II-, NP225-, and NP5288-GAL4 driver lines driving UAS-ChR2-XXM and parental controls (9 ≤ n ≤ 29, ∗p < 0.05, Kruskal-Wallis test followed by Dunn’s post-hoc test; see Table S1).
(E) Left, schematic drawing of the expression pattern for the flies used in (F). Right, expression pattern of GH146II-GAL4 driving UAS-CsChrimson.mVenus in the presence of tsh-GAL80. Maximum intensity projection of 106 confocal sections (1 μm) through the central brain and ventral nerve cord is presented.
(F) Blocking expression in the VNC does not abolish backward locomotion in the case of GH146II-, NP225-, and NP5288-GAL4 driver lines in the open arena. CsChrimson was used to activate the neurons. Left, translational velocity ± SEM (shading). The 2-s light pulse is labeled in light red. Right, mean translational velocity during the 2-s light pulse obtained from flies such as presented on the left for GH146II-, NP225-, and NP5288-GAL4 driving either UAS-CsChrimson, or UAS-CsChrimson along with tsh-GAL80 which blocks expression in the VNC, and tsh-GAL80 control. (12 ≤ n ≤ 33, ∗p < 0.05, ∗∗∗∗p < 0.0001 for all comparisons to respective parental controls, Kruskal-Wallis test followed by Dunn’s post-hoc test; see Table S1).
(G) Left, schematic drawing of the expression pattern for the flies used to compose the light blue trace in (I). Right, expression pattern of GH146II-GAL4 driving UAS-FRT-STOP-FRT-CsChrimson.mVenus in the presence of Otd-nls:FLPo. Maximum intensity projection of 117 confocal sections (1 μm) through the central brain and ventral nerve cord is presented.
(H) Left, schematic drawing of the expression pattern for the flies used to compose the dark blue trace in (I). Right, expression pattern of GH146II-GAL4 driving UAS-CsChrimson.mVenus in the presence of Otd-nls:FLPo and tub-FRT-STOP-FRT-GAL80. Maximum intensity projection of 145 confocal sections (1 μm) through the central brain and ventral nerve cord is presented.
(I) Blocking expression in GH146II-GAL4 SEZ neurons affects backward locomotion in the open arena. CsChrimson was used to activate the neurons. Left, translational velocity ± SEM (shading). The 2-s light pulse is labeled in light red. Right, mean translational velocity during the 2-s light pulse obtained from flies as presented on the left for GH146II-GAL4 driving UAS-FRT-STOP-FRT-CsChrimson.mVenus in the presence of Otd-nls:FLPo (G, light blue), GH146II-GAL4 driving UAS-CsChrimson in the presence of both Otd-nls:FLPo and tub-FRT-STOP-FRT-GAL80 (H, dark blue), and Otd-nls:FLPo control. Activation of GH146II-GAL4 SEZ neurons led to significant backward locomotion (21 ≤ n ≤ 28, ∗∗∗∗ p < 0.0001, Kruskal-Wallis test followed by Dunn’s post-hoc test; see Table S1).
Left, example of backward locomotion in MDN3-GAL4 driving UAS-ChR2-XXM in decapitated flies. Right, example of lack of backward locomotion in GH146II-GAL4 driving UAS-ChR2-XXM in decapitated flies. Note the oscillatory movement of the forelegs.
Left, example of the efficiency of tsh-GAL80 to block VNC expression in decapitated flies carrying GH146II-GAL4, UAS-CsChrimson and tsh-GAL80. No foreleg movement is observed following optogenetic activation. Right, example of backward locomotion in NP5288-GAL4 driving UAS-CsChrimson in intact flies when expression in VNC was blocked using tsh-GAL80.
GH146II-GAL4 SEZ neurons induce backward walking
The anatomical analysis of the GH146II-, NP225-, and NP5288-GAL4 driver lines (Figure S3) revealed two brain regions with shared expression pattern, the AL and the SEZ. In order to isolate the brain region responsible for the robust backward walking, we took advantage of the Otd-nls:FLPo transgene that expresses flippase (FLP) specifically in the brain50 with the exception of the gnathal ganglia in the SEZ.51 Due to the strong similarity between GH146II-, NP225-, and NP5288-GAL4 expression patterns (Figure S3), for this set of experiments, we only used the GH146II-GAL4 driver line. Otd-nls:FLPo was used along with GH146II-GAL4 and UAS-FRT-STOP-FRT-CsChrimson to drive activity mostly in PNs (Figure 4G). Flies expressing CsChrimson in this restricted manner showed no persistent backward walking, although they did transiently walk backward at the onset of the light pulse (Figure 4I). This response can arise from either weak expression of CsChrimson in the SEZ due to residual stochastic expression of the Otd-nls:FLPo transgene, or from the activity of neurons labeled by the GH146II-GAL4 driver line in brain regions other than the SEZ. Studying this transient backward component was out of the scope of the current study, and therefore, we did not further analyze it. In a complementary experiment, we used Otd-nls:FLPo, GH146II-GAL4, UAS-CsChrimson and tub-FRT-STOP-FRT-GAL80 in order to drive expression selectively in the SEZ and the VNC, by expressing the GAL4 suppressor GAL8052 in the entire central brain except for the gnathal ganglia (Figure 4H). Expression of CsChrimson in this subset resulted in robust and sustained backward walking (Figure 4I). Recalling that the VNC neurons are not required for backward walking (Figures 4C–4F), the combined results point to the SEZ as the brain region underlying the sustained backward walking observed in GH146II-, NP225-, and NP5288-GAL4 flies.
A single SEZ neuron in each brain hemisphere drives backward locomotion
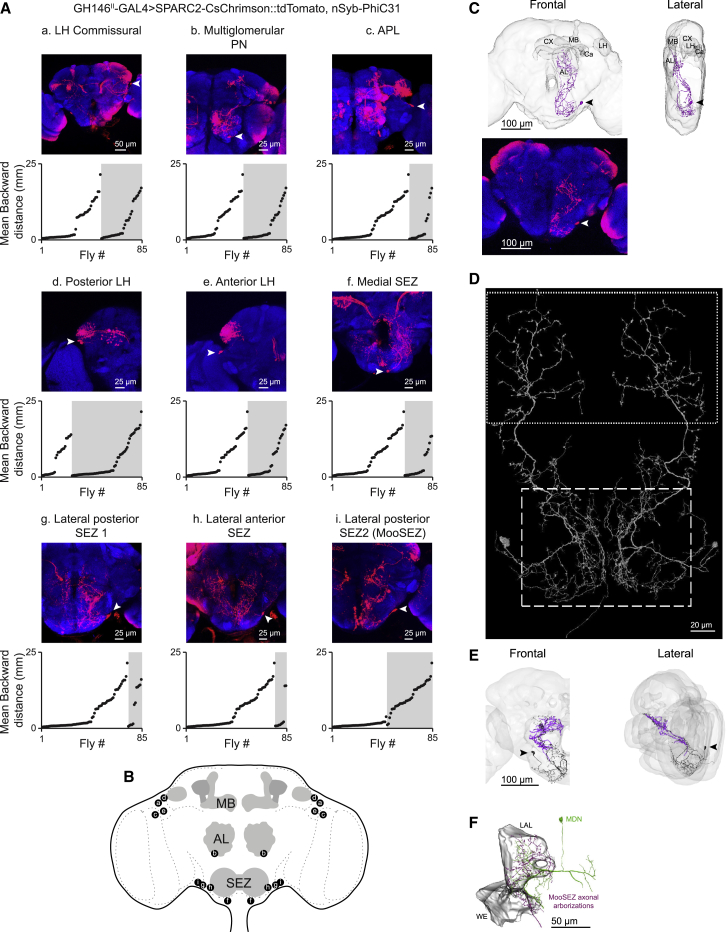
To determine which of the SEZ neurons triggers backward walking, we used the recently developed sparse predictive activity through recombinase competition (SPARC) method.53 By expressing nSyb-PhiC31 recombinase pan-neuronally together with each of the three available SPARC2-CsChrimson::tdTomato variants (s, sparse; i, intermediate; d, dense) and GH146II-GAL4, we generated flies that expressed CsChrimson::tdTomato in different fractions of GH146II-GAL4 labeled neurons in a stochastically distributed manner. Individual flies were then subjected to behavior experiments followed by dissection and staining to determine which neurons were labeled. We defined nine expression clusters, which also included bilateral SEZ neurons (Figures 5A and 5B; Table S2). We performed a multiple linear regression analysis (n = 85) in which the flies’ average backward walking distance was regressed against the nine defined independent variables.21 Only a single cluster, containing a single lateral SEZ neuron in each brain hemisphere, correlated significantly with the observed backward locomotion (Figures 5A and 5B; Table S2). We name these two neurons MooSEZs, for Moonwalker SEZ neurons.
Figure 5.
A pair of bilateral SEZ neurons, MooSEZs, underlies GH146II-GAL4 optogenetically induced backward locomotion
(A) GH146II-GAL4 expression pattern is composed of nine different neuronal clusters (a–i) in the central brain (as designated, arrowheads indicate somata). Maximum intensity projections of 24–111 confocal sections (1 μm) through the central brain are presented. The SPARC method was used to generate mosaic flies expressing CsChrimson::tdTomato in stochastically distributed subsets of neurons within the GH146II-GAL4 driver line. SPARC generated flies were individually exposed to a 2-s red light pulse in the open arena. For each of the identified neuronal cluster, the mean backward translations of individual flies during the light pulse averaged over three trials as a function of expression are presented. The gray shading indicates expression in the cluster of interest. Only for the MooSEZ cluster (lateral posterior SEZ 2) a clear lack of backward walking is observed when there is no expression and a clear backward locomotion when there is expression. Multiple linear regression analysis in which each cell cluster is set as a predictor of the mean backward translation indicates that MooSEZs exclusively correlate with backward locomotion (Table S2).
(B) Schematic drawing of the cell clusters identified in (A) using the SPARC genetic method.
(C) Top, three-dimensional light microscope reconstruction of a single MooSEZ. Frontal (left) and lateral (right) views are presented. The MooSEZ soma is labeled with a black arrowhead. MB, mushroom body; CX, central complex; AL, antennal lobe; Ca, calyx; LH, lateral horn. Bottom, expression pattern of a stochastically targeted MooSEZ expressing CsChrimson::tdTomato labeled by GH146II-GAL4 using the SPARC “Sparse” effector. MooSEZ morphology is faithfully recapitulated by the LM reconstructed MooSEZ presented on top. The MooSEZ soma is labeled with a white arrowhead. Maximum intensity projection of 69 confocal sections (1 μm) through the central brain is presented. For visualization purposes, the projection image is vertically flipped.
(D) Three-dimensional EM reconstruction of MooSEZ in both hemispheres identified in the FAFB using FlyWire. MooSEZ SEZ branches are thinner and show a higher degree of arborization typical of dendritic regions (dashed box), whereas protocerebral branches exhibit a higher probability of varicose and globular swellings, typical of presynaptic areas (stippled box).
(E) MooSEZ three-dimensional EM reconstructions in FlyWire (black), and in FlyEM hemibrain (purple). The partially reconstructed hemibrain MooSEZ highly resembles the fully reconstructed FlyWire MooSEZ within the hemibrain volume. Both EM reconstructions are similar to the fully reconstructed LM MooSEZ in (C). FlyWire MooSEZ soma is labeled with a black arrowhead. Frontal (left) and lateral (right) views are presented.
(F) Three-dimensional EM reconstructions of MooSEZ neurites (fragment #5813046185, purple) and a representative post synaptic MDN (fragment #5813021075, light green) identified in the FlyEM hemibrain using NeuPrint+. MooSEZ is synaptically coupled to MDNs. Frontal view of the lateral accessory lobe (LAL) and the wedge (WE) brain regions in the right brain hemisphere is presented.
MooSEZs are presynaptic partners of MDNs
Having unequivocally identified MooSEZs as underlying GH146II-GAL4-mediated backward walking, we used light microscopy (LM) images of SPARC generated flies in which a single MooSEZ was stochastically and sparsely targeted to reconstruct its structure (Figure 5C; Video S6). Since LM imaging suggested spatial proximity between MooSEZ neurites and MDN dendritic arbors and thus possible direct connectivity, we sought to characterize MooSEZ and its synaptic contacts in a higher structural resolution provided by electron microscopy (EM). To do so, we used NeuronBridge54, 55, 56 to match a SPARC LM image of MooSEZ to specific neuronal fragment in the FlyEM hemibrain dataset, an EM connectome of the dorsal-right part of an adult female Drosophila brain.57 We identified fragment #5813046185 as an excellent match to MooSEZ neurites located within the FlyEM hemibrain volume. Furthermore, using FlyWire,58 we identified the complete anatomical structure of MooSEZ with its characteristic morphology in both hemispheres of the female adult fly brain (FAFB)59 (Figure 5D). MooSEZ morphology implies dendritic regions mostly in the SEZ, and putative presynaptic projections in the lower lateral accessory lobe (LAL)60 located posterior to the AL and extending lateroventrally—a major input region of MDNs.18 The putative presynaptic arborizations are organized in a characteristic two-dimensional vertical sheet that slopes from anterolateral to posteriomedial (Figures 5C and 5D; Video S6). Using natverse,61 we cross-matched the FlyEM hemibrain MooSEZ fragment with the fully reconstructed FlyWire MooSEZ in the FAFB (Figure 5E). Importantly, the FlyWire MooSEZ shows great morphological similarity to the FlyEM hemibrain MooSEZ fragment, and these two independent EM reconstructions highly resemble the MooSEZ LM reconstruction (Figures 5C and 5E). Next, using neuPrint+,62 we visualized the identified MooSEZ fragment and searched for MDNs among all MooSEZ mapped postsynaptic partners in the FlyEM hemibrain. In line with the LM imaging results, we found that MooSEZ provides direct inputs onto MDNs by forming 13 to 21 synapses with each of the four MDNs in the hemibrain (Figure 5F). Taken together, we identify a single SEZ neuron in each brain hemisphere, MooSEZ, which is presynaptic to MDNs and drives backward locomotion (Figure 5). Combined with the results that show clear activation of MDNs by GH146II-GAL4 neurons, and the existence of an MDN-independent pathway (Figure 3), we conclude that MooSEZs act directly upstream of MDNs and via other descending neurons (DNs) to evoke backward walking.
Translational movie running through the entire central brain from the posterior to the anterior aspect (left) or from the anterior to the posterior aspect (right) of a fly in which the left MooSEZ was sparsely targeted to express CsChrimson::tdTomato using the SPARC method. To generate the movies, each confocal section (1 μm) of the original stacks (144 slices) was converted into a video frame.
An intersection approach to narrow down GH146II-, NP225-, and NP5288-GAL4 expression pattern
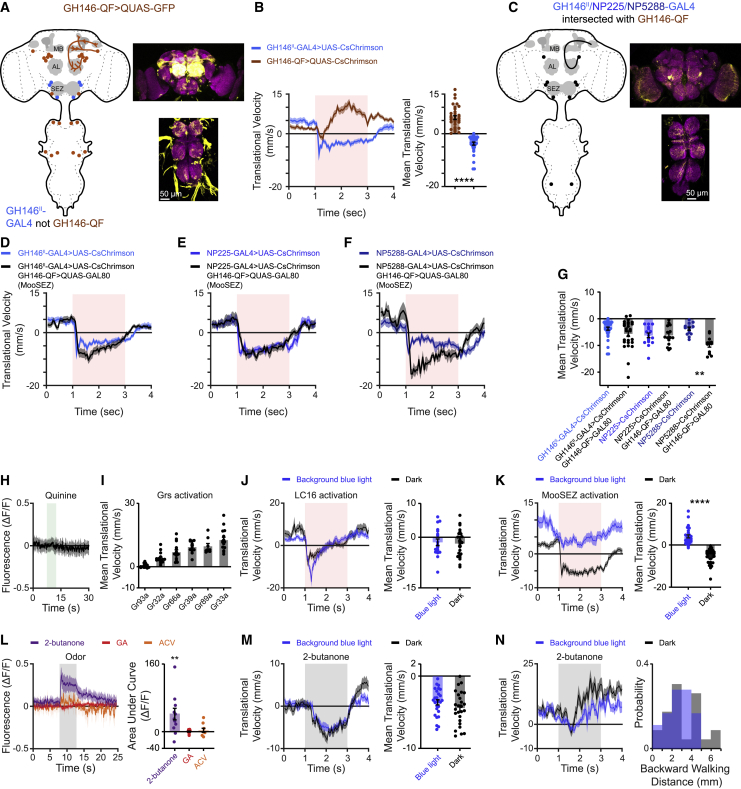
The results thus far demonstrate that odors can trigger backward locomotion mediated by MDNs and that MooSEZs, when activated, can trigger backward movement whether via direct connections with MDNs or through a parallel pathway. However, the physiological stimulus which activates MooSEZs is not clear. GH146II-, NP225-, and NP5288-GAL are all broad labeling driver lines, which are unsuitable for behavioral and SEZ functional imaging experiments. To try and limit the number of neurons covered by these driver lines, we used another driver line which should have a strong overlap with these three driver lines. It is well known that different transcription systems (i.e., GAL4,63 LexA,64 and QF65) or genomic insertion sites can affect the expression pattern.66,67 We therefore examined to what extent GH146-QF recapitulates the labeled cells in GH146II-GAL4, which revealed notable exceptions in the SEZ (Figure 6A).
Figure 6.
A genetic intersection approach to narrow GH146II-, NP225-, and NP5288-GAL4 expression pattern to examine MooSEZ physiological input
(A) Left, schematic drawing of the expression pattern for GH146-QF. Right, expression pattern of GH146-QF driving QUAS-GFP. Maximum intensity projection of 150 confocal sections (1 μm) through the central brain and VNC is presented.
(B) Left, translational velocity ± SEM (shading) for GH146II-GAL4 and GH146-QF driving CsChrimson as designated in the open arena. Backward walking is observed only for GH146II-GAL4. The 2-s light pulse is labeled by light red. Right, mean translational velocity during the 2-s light pulse obtained from flies as presented on the left (n = 33, ∗∗∗∗p < 0.0001, Mann-Whitney test; see Table S1).
(C) Left, schematic drawing of the expression pattern for GH146II-, NP225-, and NP5288-GAL4 when intersected with GH146-QF. Right, expression pattern of an intersection between GH146II-GAL4 driving CsChrimson.mVenus and GH146-QF driving the GAL4 inhibitor QUAS-GAL80. Maximum intensity projection of 128 confocal sections (1 μm) through the central brain and VNC is presented.
(D–F) Translational velocity ± SEM (shading) for GH146II- (D), NP225- (E), and NP5288-GAL4 (F) driving UAS-CsChrimson (blue) or when intersected with GH146-QF driving QUAS-GAL80 (black) in the open arena. Backward walking is maintained in intersection flies and even enhanced (NP5288-GAL4). The 2-s light pulse is labeled by light red.
(G) Mean translational velocity during the 2-s light pulse obtained from traces in (D) to (F). No significant difference is observed between the mean backward walking velocities of intersection flies compared with non-intersection flies, except for NP5288-GAL4 intersection flies which exhibit a significant increase in backward locomotion (11 ≤ n ≤ 33, ∗∗ p < 0.01, Kruskal-Wallis test followed by Dunn’s post-hoc test; see Table S1).
(H) Averaged traces ± SEM (shading) of Ca2+ responses following application of quinine for 5 s (labeled in light green). GH146II-GAL4 was used to drive UAS-GCaMP6f and GH146-QF was used to drive QUAS-GAL80 to limit the expression pattern as presented in (C). No Ca2+ response to quinine was observed (n = 8; see Table S1).
(I) Mean translational velocity during a 2-s red light pulse activating bitter-sensing taste cells in the open arena. Six different Gr-GAL4 driver lines (as designated) targeting bitter-sensing neurons were used to drive UAS-CsChrimson. Activation of bitter neurons for 2 s does not elicit backward locomotion (8 ≤ n ≤ 15, Wilcoxon signed rank test (one-sided) with Bonferroni correction; see Table S1).
(J) Left, translational velocity ± SEM (shading) following optogenetic activation of LC16 visual projection neurons with CsChrimson (black) or when the inhibitory GtACR2 channel was co-activated in GH146II-GAL4 neurons (blue) in the open arena. The red light activating CsChrimson and the blue light activating GtACR2 were provided between the first and the third seconds. The 2-s red light pulse is labeled in light red. GH146II-GAL4 was used to drive UAS-GtACR2 and LC16-LexA was used to drive LexAop-CsChrimson. Silencing MooSEZs using GtACR2 does not affect LC16-triggered backward locomotion. Right, mean translational velocity during the 2-s red light pulse obtained from traces on the left. No significant difference is observed between the mean translational velocities when GtACR2 is co-activated and when GtACR2 is not co-activated in MooSEZs (18 ≤ n ≤ 25, Mann-Whitney test; see Table S1).
(K) Left, translational velocity ± SEM (shading) following optogenetic activation of MooSEZs with CsChrimson (black) or when the inhibitory GtACR2 channel was co-activated (blue) in the open arena. The red light activating CsChrimson was provided between the first and third seconds (light red shading). When GtACR2 was co-activated the blue light was on throughout the experiment. GtACR2 efficiently blocks MooSEZ activation by CsChrimson. Right, mean translational velocity during the 2-s red light pulse obtained from traces on the left. A significant difference is observed between the backward walking velocities generated with and without the co-activation of GtACR2 in MooSEZs (29 ≤ n ≤ 45, ∗∗∗∗ p < 0.0001, Mann-Whitney test; see Table S1).
(L) Left, averaged traces ± SEM (shading) of Ca2+ responses in intersection flies following an odor pulse as designated. The 5-s odor pulse is labeled in light gray. GH146II-GAL4 was used to drive UAS-GCaMP6f and GH146-QF was used to drive QUAS-GAL80 to limit the expression pattern as presented in (C). Right, area under the curve of ΔF/F during the 5-s odor pulse for the traces presented in the left panel. A significant odor-evoked Ca2+ response is observed only for 2-butanone (7 ≤ n ≤ 11, ∗∗p < 0.01, one-sample t test; see Table S1).
(M) Left, translational velocity ± SEM (shading) of intersection flies following application of 2-butanone when MooSEZs were blocked using blue light activation of GtACR2 or in control experiment performed in the dark in the linear chambers. The 2-s odor pulse is labeled in gray. When GtACR2 was activated the blue light was on throughout the experiment. GH146II-GAL4 was used to drive UAS-GtACR2 and GH146-QF was used to drive QUAS-GAL80 to limit the expression pattern as presented in (C). Right, mean translational velocity during the 2-s odor pulse obtained from traces on the left. No significant difference is observed between the mean translational velocities when GtACR2 is activated and when GtACR2 is not activated in MooSEZs (26 ≤ n ≤ 27, Mann-Whitney test).
(N) Left, translational velocity ± SEM (shading) of intersection flies following application of 2-butanone when MooSEZs were blocked using blue light activation of GtACR2 or in control experiment performed in the dark in the open arena. The 2-s odor pulse is labeled in gray. When GtACR2 was activated the blue light was on throughout the experiment. GH146II-GAL4 was used to drive UAS-GtACR2 and GH146-QF was used to drive QUAS-GAL80 to limit the expression pattern as presented in (C). Right, the probability of observing backward locomotion among intersection flies in response to a 2-s 2-butanone pulse in the open arena. The probability of a fly to perform backward retreat in response to application of 2-butanone is similar whether GtACR2 is activated in MooSEZs or not (18 ≤ n ≤ 23).
The anatomical analysis suggested that GH146-QF does not overlap with GH146II-, NP225-, and NP5288-GAL4 in the SEZ neuron relevant for backward walking. Indeed, optogenetic activation with CsChrimson of GH146-QF did not elicit backward walking (Figure 6B). We then performed an intersection between GH146II-, NP225-, and NP5288-GAL4 with GH146-QF. GH146II-, NP225-, and NP5288-GAL4 were used to drive CsChrimson and GH146-QF was used to drive QUAS-GAL80. Subtracting GH146-QF from GH146II-GAL4 strongly decreased the expression in the VNC (Figure 6C). In addition, AL labeling was mostly abolished. In all brains examined, we observed that only a single PN, which labels glomerulus VL2a, was reliably expressed (Figure S4). This PN receives input from neurons expressing the ionotropic receptor 84a (IR84a), which couples food presence to the activation of the courtship circuitry and increases male courtship.68 In most cases, we observed stochastic labeling of few other PNs; however, these were not consistently labeled across brains (Figure S4). As expected, the expression pattern of SEZ neurons was unaffected (Figure 6C). Optogenetic activation using CsChrimson of all three GAL4 drivers with subtracted GH146-QF resulted in clear and sustained backward walking (Figures 6D–6G). Importantly, contrary to the stochastic nature of AL glomeruli labeling, all flies reliably performed backward walking (Figure 6G). Thus, these results further support our conclusion that MooSEZs underlie this sustained backward walking. In some cases (GH146II- and NP5288-GAL4), we actually observed a stronger backward walking phenotype (Figures 6D, 6F, and 6G). This may arise from reduced appetitive olfactory input due to the reduced expression in AL, as activating all ORNs using ORCO-GAL469 resulted in a forward movement (Figures 2A and 2B), or, more likely, from smoother backward locomotion as the oscillatory foreleg activity we observed (Video S2) was totally abolished in these flies. Indeed, NP5288-GAL4 flies showed the strongest foreleg movement and showed the largest change in backward walking following the intersection (Figures 6F and 6G; Video S7). Taken together, our intersection approach dramatically reduces the expression pattern by GH146II-, NP225-, and NP5288-GAL4 and maintains expression in MooSEZs.
Left, example of backward locomotion in NP5288-GAL4 driving UAS-CsChrimson. Note the strong oscillatory foreleg movement. Right, example of backward locomotion in NP5288-GAL4 with subtracted GH146-QF driving UAS-CsChrimson. Note the absence of oscillatory foreleg movement and increase in backward locomotion.
Left, example of backward turning in GH146II-GAL4 SPARC mosaic fly expressing CsChrimson in the right MooSEZ. Right, example of backward turning in GH146II-GAL4 SPARC mosaic fly expressing CsChrimson in the left MooSEZ.
MooSEZs respond to odor input
We now turn to examine the physiological input that activates MooSEZs. MooSEZs reside in the SEZ, a brain region receiving mostly gustatory and mechanosensory input.70 Hence, we examined whether MooSEZs respond to quinine that is an aversive gustatory input. We generated flies carrying GCaMP6f40 using the intersection approach and performed two-photon in vivo Ca2+ imaging. Application of quinine did not elicit any Ca2+ response in MooSEZs (Figure 6H). In line with this result, optogenetic activation of bitter-sensing taste cells expressing gustatory receptors (Grs), which were shown to be required for responses to various bitter tastants,71 did not elicit backward walking (Figure 6I).
The lack of backward walking following activation of bitter-sensing neurons, and of a gustatory-related signal in MooSEZs led us to also examine other modalities. Recently, it was demonstrated that the MDNs mediate visually driven backward locomotion triggered by activation of the LC16 neuronal population.20 Thus, we tested whether LC16-evoked backward locomotion is MooSEZ dependent. We optogenetically stimulated the visual neurons with CsChrimson while simultaneously blocking MooSEZs. Since expression of TNT resulted in movement impairment (data not shown), for this set of experiments we used the anion channelrhodopsin GtACR272 for the induction of a transient and temporally controlled neuronal silencing. Inhibiting MooSEZs did not have any apparent effect on LC16-evoked backward locomotion (Figure 6J). To verify that GtACR2 efficiently blocked MooSEZs, we co-expressed it with CsChrimson under the control of GH146II-GAL4. Such manipulation blocked upon blue light stimulation the backward walking of the flies normally elicited by the activation of CsChrimson using red light pulse (Figure 6K). Thus, MooSEZs do not seem to receive visual input driving backward locomotion from the LC16 VPNs.
The apparent lack of visual and gustatory signal in MooSEZs has led us to also examine olfactory input. As shown earlier, olfactory cues can trigger backward locomotion under certain conditions (Figures 1, S1A, and S1B; Video S1). Interestingly, we observed a small but significant Ca2+ signal following application of the aversive odor 2-butanone, which was not detected following applications of ACV and GA (Figure 6L).
The fact that odors activate MooSEZs and that optogenetic activation of MooSEZs results in backward locomotion suggests that MooSEZs may mediate odor-evoked backward locomotion. To examine this, we used the intersection approach to express GtACR2 in MooSEZs and exposed flies to 2-butanone. Silencing MooSEZs had little or no effect on odor-driven backward locomotion in both the linear chambers (Figure 6M) and the open arena (Figure 6N). Thus, MooSEZs seem to receive olfactory information but are dispensable for odor-evoked backward locomotion in the open arena or the linear chamber behavioral assays. However, this result does not rule out the possibility that MooSEZs do participate in odor-driven backward locomotion under different behavioral circumstances.
MooSEZs contribute to rotational backward locomotion
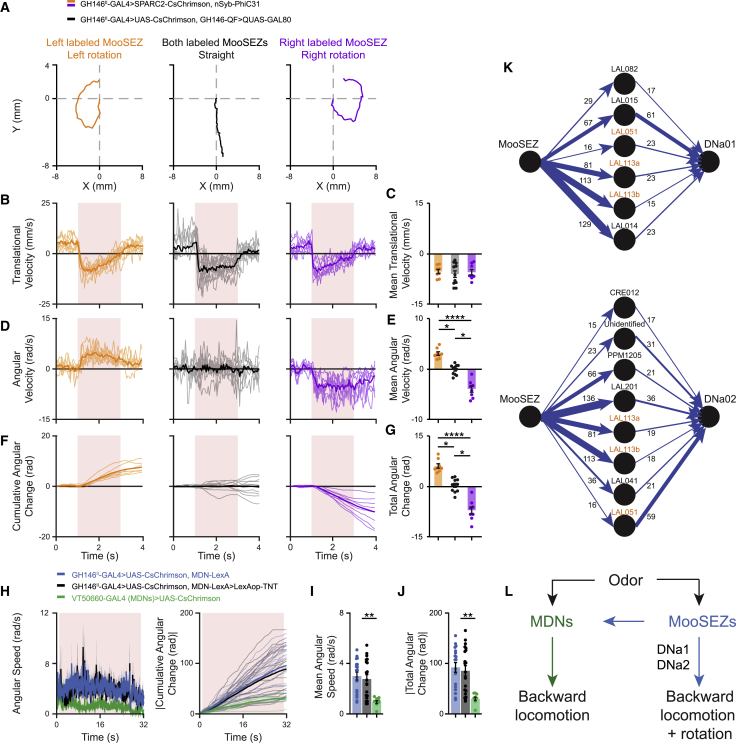
The above result, which demonstrates that MooSEZs receive olfactory input but do not mediate odor-driven backward locomotion, raises the question about the ethological relevance of the MooSEZs. It was previously shown that asymmetric activation of the MDNs leads to backward turning.20 Since the backward walking phenotype following a short MooSEZ activation had similar features as the backward locomotion induced by MDNs (Figures 2E–2H), we sought to examine whether asymmetric activation of MooSEZs could also drive backward turning. The stochastic labeling of GH146II-GAL4 neurons by the SPARC method used above (Figure 5A) often labeled a single MooSEZ in each brain, thus enabling us to test the motor responses evoked by unilateral activation of a single MooSEZ. Interestingly, contrary to the straight backward walking observed upon short bilateral activation of MooSEZs, asymmetric activation of MooSEZs for 2 s led to robust ipsilateral backward rotation (with respect to the location of MooSEZ soma in the brain) without affecting the overall backward translation (Figures 7A–7G; Video S8). In contrast to the relatively weak and transient backward turning generated upon unilateral activation of MDNs,20 backward rotation following MooSEZ unilateral activation seemed to be stronger and continuous. Furthermore, we also found significant differences in backward rotation for long periods of bilateral activation. While MDN activation using a 30-s light stimulation typically resulted in straight backward walking, the same activation protocol generated a pronounced rotational component when MooSEZs were stimulated. Similar rotational movements were also observed when MooSEZs were activated and MDNs were silenced using TNT (Figures 7H–7J). Thus, these results indicate that MooSEZs induce MDN-independent backward turning.
Figure 7.
MooSEZs drive MDN-independent rotational backward locomotion
(A) Example trajectories of flies in which MooSEZs were optogenetically activated unilaterally (right, purple; left, orange) or bilaterally (black) for 2 s in the open arena. CsChrimson was expressed in left, right or both MooSEZs.
(B, D, and F) Single fly traces (light) of translational velocity (B), angular velocity (D), cumulative angular change (F), and their respective means (dark), for flies in which MooSEZs were optogenetically activated unilaterally (right, purple; left, orange) or bilaterally (black) for 2 s in the open arena. Angular velocity is defined as a change in the fly’s orientation angle in relation to a global orientation coordinate system. Left rotations are defined as positive values and right rotations as negative values. Cumulative angular change is defined as the cumulative integrated area under the angular velocity versus time curve. Total angular change is defined as the integrated area under the angular velocity versus time curve during the light pulse. The light pulse is labeled in light red. CsChrimson was expressed in left, right or both MooSEZs.
(C, E, and G) Mean translational velocity (C), mean angular velocity (E), and total angular change (G), during the 2-s light pulse for flies presented in (B, D, and F, respectively). Whereas translational velocity is similar across conditions, angular movement is tightly correlated with MooSEZ side of activation (8 ≤ n ≤ 11, ∗p < 0.05, ∗∗∗∗p < 0.0001, Kruskal-Wallis tests followed by Dunn’s post-hoc tests; see Table S1).
(H) Angular speed (left) ± SEM (shading) and single fly traces (light) of absolute cumulative angular change (right) and their respective means (dark) following 30 s long bilateral optogenetic stimulation of MDNs (green) and of GH146II-GAL4 neurons in the presence (black) or absence (blue) of TNT in MDNs in the open arena. Angular speed is defined as the absolute value of angular velocity. Absolute cumulative angular change is defined as the cumulative integrated area under the angular speed versus time curve. VT50660-GAL4 (MDNs) or GH146II-GAL4 were used to drive UAS-CsChrimson and MDN-LexA (VT44845-LexA) was used to drive LexAop-TNT (when required). The light pulse is labeled in light red. Continuous bilateral activation of MooSEZs generates MDN-independent sustained backward rotation.
(I and J) Mean angular speed (I) and absolute total angular change (J) during the 30-s light pulse for flies presented in (H). Absolute total angular change is defined as the integrated area under the angular speed versus time curve during the light pulse. Significantly higher angular speed and absolute total angular change are observed following MooSEZ long bilateral activation in the presence or absence of TNT in MDNs relative to MDN long bilateral activation (9 ≤ n ≤ 20, ∗∗ p < 0.01, Kruskal-Wallis test followed by Dunn’s post-hoc test; see Table S1).
(K) Connectivity diagram between MooSEZ and DNa01 (fragment #1170939344, top) or DNa02 (fragment #1140245595, bottom) in the FlyEM hemibrain. MooSEZ is strongly connected to DNa01 and DNa02 via various neurons. Each circle represents a cell type, and each arrow represents a directional synaptic connection. Arrow width represents synaptic connection strength derived from number of synapses forming the connection (indicated by the number above the arrow). Cell types connecting MooSEZ to both DNa01 and DNa02 are labeled in orange. For simplification purposes, connection with fewer than 15 synapses were omitted.
(L) Schematic of proposed MDN and MooSEZ pathways. Odors activate MDNs that are required for odor-induced backward locomotion. Odors also activate MooSEZs that activate MDNs and can also add a significant rotational component to the motor response presumably via DNa01 and DNa02.
MooSEZs are upstream of DNa01 and DNa02
Recently, it was demonstrated that two types of DNs, DNa01 and DNa02,73 contribute to rotational locomotion.73 Having identified MooSEZ in EM, we investigated whether DNa01 and DNa02 are downstream targets of MooSEZ. We therefore mapped the neuronal connectivity between MooSEZ and DNa01 as well as between MooSEZ and DNa02 in the FlyEM hemibrain using neuPrint. The connectivity analysis identified multiple disynaptic neural pathways connecting MooSEZ onto both DNa01 and DNa02, among them some major inputs mediated by strong synaptic connections (Figure 7K). For instance, LAL201 and LAL014 neurons, which connect MooSEZ onto DNa02 and DNa01, respectively, are MooSEZ first- and second-ranked postsynaptic partners in the FlyEM hemibrain in terms of number of synapses. Overall, about 16% and 18% of all MooSEZ synaptic outputs in the FlyEM hemibrain are connected onto DNa01 and DNa02 presynaptic neurons respectively. Inversely, about 16% of all DNa01, and 12% of all DNa02 synaptic inputs in the FlyEM hemibrain are formed by MooSEZ postsynaptic neurons. Taken together, our connectomics data analysis demonstrates that MooSEZs contribute an additional angular component to the motor output, presumably via DNa01 and DNa02, to perform complex curved backward responses.
Discussion
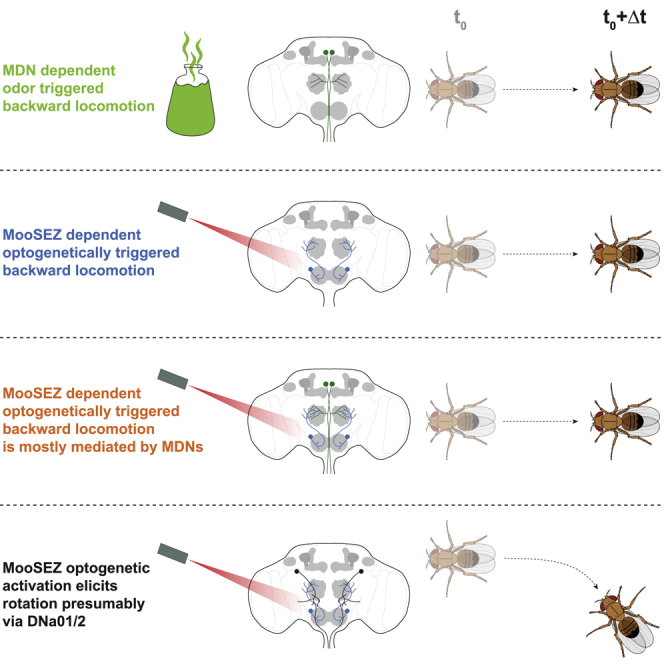
In this study, we showed that olfactory stimulus can trigger backward locomotion, which is mediated by MDNs (Figure 1). We discovered the MooSEZs, which trigger backward locomotion and unequivocally identified them in the fly brain (Figure 5). We showed that MooSEZs synapse onto MDNs (Figure 5) and act both upstream and in parallel to them (Figure 3). Although located in the SEZ, MooSEZs do not seem to respond to gustatory stimuli. Rather, MooSEZs respond to olfactory input. We further showed that MooSEZs are not necessary for olfactory-driven backward locomotion (Figure 6). However, we demonstrated that MooSEZs contribute to strong rotational backward locomotion, presumably via DNa01 and DNa02, which are downstream of MooSEZs (Figure 7).
The anatomy of the MooSEZs suggests mild polarity, with dendritic regions mostly in the ventral SEZ, and presynaptic projections predominantly in the inferior protocerebrum. The protocerebral innervation is found in the inferior posterior slope (IPS) and the crepine, but also reaching into wedge, vest, and the lower LAL, posterior to the AL. In the SEZ, the MooSEZs stay in the gnathal ganglia, sparing the prow. The SEZ predominantly receives mechanosensory and gustatory sensory input70 and SEZ output controls movements involved in feeding behavior.74,75 Although the main dendritic region of MooSEZs is in the ventral SEZ, they do not seem to respond to bitter taste. Rather, MooSEZs respond to olfactory input (Figure 6).
Interestingly, a significant portion of MooSEZ axonal projections terminate within the LAL, a major premotor region. The LAL receives inputs from the central complex (CX), the navigation hub of the insect brain, as well as from a variety of sensory processing regions, and projects output signals mainly through descending tracts to the VNC.76 The LAL was shown to be involved in a wide range of orientation-related behaviors across different insect species such as pheromone orientation in moths,77 flight in locusts,78 turning in cockroaches,79 and phonotaxis in crickets.80 Given the exceptional functional similarities of the LAL across insect species, it will be interesting to find whether MooSEZs are part of a canonical locomotor circuit and thus conserved across insect species.
MDNs were demonstrated to receive sensory information from the fly’s visual system20 and mechanosensory input from the VNC.21 Whether other sensory inputs converge onto MDNs was not resolved yet. Our results add olfaction to the multi-modality input of MDNs (Figure 1). In addition, we identify the SEZ as participating in MDN input (Figure 3). A recent study in larvae reported the existence of two SEZ neurons, AMB neurons, which mediate backward crawling in response to aversive blue light via activation of MDNs.81 Our results combined with the results from larvae,81,82 suggest that SEZ neurons may also act as modality integrators. The participation of the SEZ as an input source to MDNs also raises the option that SEZ gustatory or tactile input can activate MDNs.
Our results indicate that MooSEZs add an additional MDN-independent component to the backward walking motor response (Figures 3 and 7). Indeed, some experimental evidence from previously published work in both adult flies and larvae are in line with this conclusion. For instance, in adult flies, optogenetic activation of TLAs while blocking MDNs21 resulted in residual backward walking. In addition, backward movement was correlated with a large membrane voltage difference in DNa01 and DNa02, two non-MDN descending neurons,83,73 which are downstream of MooSEZs. In larvae, it was recently demonstrated that MDNs were not necessary for dead end evoked backward crawling.81 Taken together, it seems that rather than a single command circuit originating in MDNs, additional neural circuits are recruited to allow for an adaptive backward locomotion response (Figure 7L).
In this study, we show that unilateral activation of MooSEZs elicits ipsilateral backward turning (Figure 7). This result is analogous to recent findings demonstrating that unilateral activation of specific motor-related neurons can induce walking with a significant ipsilateral turning component.46,73,84 In addition, it was previously demonstrated that unilateral activation of MDNs also led to a steering bias.20 Thus, as MDNs are postsynaptic to MooSEZs, it is possible that the rotational response elicited by MooSEZ unilateral activation might be mediated to some extent by them. However, our data show that blocking MDNs had no effect on MooSEZ-induced backward turning (Figures 7H–7J) suggesting that the dominant pathway driving the observed rotational backward walking is MDN independent. This conclusion is further supported by the anatomical data demonstrating that both DNa01 and DNa02, which participate in steering maneuvers and descend to the VNC,83,73 are downstream targets of MooSEZs (Figure 7K). Flies usually encounter in their natural habitat complex odor plumes,85 which are constantly changing as a function of the flies’ spatial locations relative to the positions of the varied odor sources around them, and the wind structure in their surroundings. Thus, flies are frequently exposed to asymmetrical olfactory cues in their natural environment. The ability of MooSEZs to recruit downstream motor circuits to perform a directional retreat response upon asymmetrical activation may enable flies to effectively guide their behavior under natural conditions.
Our study demonstrates that an olfactory cue can trigger backward locomotion via MDNs. We also show that olfactory input activates MooSEZs that can trigger backward locomotion by themselves independent of MDNs (Figure 7L). However, silencing MooSEZs had no effect on odor-evoked backward locomotion (Figure 6). These observations suggest that MooSEZs do not function as command-type neurons for odor-evoked backward locomotion, but rather seem to be part of a distributed neural circuit architecture controlling backward locomotion. Thus, odors may activate multiple parallel neural pathways driving backward locomotion. Indeed, functional redundant signaling also exists in other neuronal circuits in the fly brain. For example, optogenetic activation of LC16 VPNs, which were shown to respond to a visual looming stimulus,46 elicited backward locomotion via MDNs.20 However, silencing LC16 neurons had no effect on backward locomotion in response to a visual looming stimulus.46 Thus, although optogenetic activation of LC16 neurons drives backward locomotion, silencing these neurons has no effect on the behavioral output to the visual stimulus that activates them. Another example are neurons expressing SIFamide (SIFa). These neurons have elevated activity in starved flies and are involved in hunger-mediated behavior. Acute activation of SIFa neurons was sufficient to increase the response of fed flies to a food-related odor and to enhance their food consumption. However, silencing SIFa neurons did not decrease food intake in starved flies.86,87 Taken together, contrary to optogenetic activation of MooSEZs, which generates a robust and constant activation that further activates MDN-independent motor circuits, it seems that ecological odor stimulation elicits a physiologically adequate response in MooSEZs, which is presumably used to support odor-evoked backward locomotion but cannot sustain odor-evoked backward locomotion by itself.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-GFP Polyclonal antibody |
Thermo Fisher Scientific | Cat#A-11122 RRID;AB_221569 |
| Mouse anti-GFP [9F9.F9] Monoclonal antibody |
Abcam | Cat#ab1218; RRID:AB_298911 |
| Rabbit anti-RFP Polyclonal antibody |
Abcam | Cat#ab62341; RRID:AB_945213 |
| Mouse anti-Bruchpilot Monoclonal antibody |
DSHB | Cat# nc82; RRID:AB_2314866 |
| Alexa Fluor 488 goat anti-mouse Polyclonal antibody |
Abcam | Cat#ab150113; RRID:AB_2576208 |
| Alexa Fluor 488 goat anti-rabbit Polyclonal antibody |
Thermo Fisher Scientific | Cat#A-11034; RRID:AB_2576217 |
| Alexa Fluor 568 goat anti-rabbit Polyclonal antibody |
Abcam | Cat#ab175471; RRID:AB_2576207 |
| Alexa Fluor 647 goat anti-mouse Polyclonal antibody |
Abcam | Cat#ab150115; RRID:AB_2687948 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| All-trans retinal | Sigma-Aldrich | Cat#R2500; CAS: 116-31-4 |
| Quinine hemisulfate monohydrate | Alfa Aesar | Cat#A17036; CAS: 6119-70-6 |
| 1-Hexanol | Sigma-Aldrich | Cat#H13303; CAS: 111-27-3 |
| Ethyl benzoate | Sigma-Aldrich | Cat#E12907; CAS: 93-89-0 |
| Ethyl acetate | Sigma-Aldrich | Cat#270989; CAS: 141-78-6 |
| 2-heptanone | Sigma-Aldrich | Cat#W254401; CAS: 110-43-0 |
| Methyl acetate | Sigma-Aldrich | Cat#W267600; CAS: 79-20-9 |
| 2-butanone | Sigma-Aldrich | Cat#34861; CAS: 78-93-3 |
| 3-octanol | Sigma-Aldrich | Cat#W358126; CAS: 589-98-0 |
| MCH (4-Methylcyclohexanol) | Sigma-Aldrich | Cat#153095; CAS: 589-91-3 |
| Geranyl acetate | Sigma-Aldrich | Cat#W250902; CAS: 105-87-3 |
| Ethyl lactate | Sigma-Aldrich | Cat#W244015; CAS: 97-64-3 |
| Hexyl acetate | Sigma-Aldrich | Cat#108154; CAS: 142-92-7 |
| Apple cider vinegar | Rauch Fruchtsäfte GmbH & Co OG | N/A |
| Mineral oil | Sigma-Aldrich | Cat#330779; CAS: 8042-47-5 |
| Experimental Models: Organisms/Strains | ||
| D. melanogaster: GH146II-GAL4 | BDSC | Cat#30026; RRID:BDSC_30026 |
| D. melanogaster: GH146X-GAL4 | Gift from Dr. Christopher J. Potter43 | Cat#91812; RRID:BDSC_91812 |
| D. melanogaster: GH146-QF, QUAS-mtdTomato | BDSC | Cat#30037; RRID:BDSC_30037 |
| D. melanogaster: GH146-QF, QUAS-mCD8-GFP | BDSC | Cat#30038; RRID:BDSC_30038 |
| D. melanogaster: GH146-QF | BDSC | Cat#30015; RRID:BDSC_30015 |
| D. melanogaster: MDN3-GAL4: VT50660-p65ADZp (attP40); VT44845-ZpGAL4DBD (attP2) | Gift from Dr. Barry J. Dickson18,88 | N/A |
| D. melanogaster: MDN+MAN-LexA: UAS-TNT-E, VT49484-ZpLexADBD (JK22C); VT50660-p65ADZp (attP2), 13XLexAop-CsChrimson-tdTomato (VK00005) | Gift from Dr. Barry J. Dickson21 | N/A |
| D. melanogaster: w1118 | BDSC | Cat#5905; RRID:BDSC_5905 |
| D. melanogaster: Or82a-GAL4 | BDSC | Cat#23125; RRID:BDSC_23125 |
| D. melanogaster: Ir31a-GAL4 | BDSC | Cat#41726; RRID:BDSC_41726 |
| D. melanogaster: Ir40a-GAL4 | BDSC | Cat#41727; RRID:BDSC_41727 |
| D. melanogaster: Ir25a-GAL4 | BDSC | Cat#41728; RRID:BDSC_41728 |
| D. melanogaster: Ir76b-GAL4 | BDSC | Cat#41730; RRID:BDSC_41730 |
| D. melanogaster: Ir8a-GAL4 | BDSC | Cat#41731; RRID:BDSC_41731 |
| D. melanogaster: Ir64a-GAL4 | BDSC | Cat#41732; RRID:BDSC_41732 |
| D. melanogaster: Ir92a-GAL4 | BDSC | Cat#41733; RRID:BDSC_41733 |
| D. melanogaster: Ir84a-GAL4 | BDSC | Cat#41734; RRID:BDSC_41734 |
| D. melanogaster: Ir76a-GAL4 | BDSC | Cat#41735; RRID:BDSC_41735 |
| D. melanogaster: GMR22C06-GAL4 (attP2) | BDSC | Cat#48974; RRID:BDSC_48974 |
| D. melanogaster: GMR24G07-GAL4 (attP2) | BDSC | Cat#49095; RRID:BDSC_49095 |
| D. melanogaster: GMR68D02-GAL4 (attP2) | BDSC | Cat#39471; RRID:BDSC_39471 |
| D. melanogaster: GMR26B04-GAL4 (attP2) | BDSC | Cat#49158; RRID:BDSC_49158 |
| D. melanogaster: GMR48E05-GAL4 (attP2) | BDSC | Cat#50370; RRID:BDSC_50370 |
| D. melanogaster: GMR24A10-GAL4 (attP2) | BDSC | Cat#49059; RRID:BDSC_49059 |
| D. melanogaster: GMR95C02-GAL4 (attP2) | BDSC | Cat#48431; RRID:BDSC_48431 |
| D. melanogaster: NP6115-GAL4 | DGGR | Cat#113851; RRID:DGGR_113851 |
| D. melanogaster: NP3062-GAL4 | DGGR | Cat#113083; RRID:DGGR_113083 |
| D. melanogaster: NP5021-GAL4 | DGGR | Cat#113545; RRID:DGGR_113545 |
| D. melanogaster: NP5103-GAL4 | DGGR | Cat#113584; RRID:DGGR_113584 |
| D. melanogaster: NP80-GAL4 | DGGR | Cat#103518; RRID:DGGR_103518 |
| D. melanogaster: NP2001-GAL4 | DGGR | Cat#104059; RRID:DGGR_104059 |
| D. melanogaster: NP2297-GAL4 | DGGR | Cat#112868; RRID:DGGR_112868 |
| D. melanogaster: NP3529-GAL4 | DGGR | Cat#113327; RRID:DGGR_113327 |
| D. melanogaster: NP5288-GAL4 | DGGR | Cat#104937; RRID:DGGR_104937 |
| D. melanogaster: NP225-GAL4 | DGGR | Cat#112095; RRID:DGGR_112095 |
| D. melanogaster: NP1579-GAL4 | DGGR | Cat#104036; RRID:DGGR_104036 |
| D. melanogaster: Orco-GAL4 | BDSC | Cat#26818; RRID:BDSC_26818 |
| D. melanogaster: VT050660-GAL4 (attP2) | VDRC | Cat#200107 |
| D. melanogaster: VT040053-GAL4 (attP2) | VDRC | Cat#201352 |
| D. melanogaster: VT019428-GAL4 (attP2) | VDRC | Cat#200084 |
| D. melanogaster: VT026020-GAL4 (attP2) | VDRC | Cat#203479 |
| D. melanogaster: MZ612-GAL4 | Gift from Dr. Liria Masuda-Nakagawa89,90 | N/A |
| D. melanogaster: MB247-GAL4 | BDSC | Cat#50742; RRID:BDSC_50742 |
| D. melanogaster: c305a-GAL4 | BDSC | Cat#30829; RRID:BDSC_30829 |
| D. melanogaster: OK107-GAL4 | BDSC | Cat#854; RRID:BDSC_854 |
| D. melanogaster: LN1-GAL4 | DGGR | Cat#103945; RRID:DGGR_103945 |
| D. melanogaster: LN2-GAL4 | DGGR | Cat#104198; RRID:DGGR_104198 |
| D. melanogaster: MZ699-GAL4 | Gift from Dr. Liqun Luo91 | N/A |
| D. melanogaster: LC16-1-GAL4: R26A03-p65ADZp (attP40); R54A05-ZpGAL4DBD (attP2) | BDSC | Cat#68331; RRID:BDSC_68331 |
| D. melanogaster: TLA-GAL4: VT012330-p65ADZp (attP40); VT020742-ZpGAL4DBD (attP2) | BDSC21,88 | Cat#74205; RRID:BDSC_74205 Cat#72912; RRID:BDSC_72912 |
| D. melanogaster: LC16-LexA: R28F07-LexAp65 (JK22C) | Gift from Dr. Barry J. Dickson20 | N/A |
| D. melanogaster: MDN-LexA: UAS-CsChrimson-mCherry (attP5), VT044845-LexAp65 (attP40) | Gift from Dr. Barry J. Dickson20 | N/A |
| Drosophila melanogaster: nSyb-IVS-PhiC31 (attP18) | BDSC | Cat#84151; RRID:BDSC_84151 |
| D. melanogaster: SPARC2-D-CsChrimson::tdTomato | BDSC | Cat#84143; RRID:BDSC_84143 |
| D. melanogaster: SPARC2-I-CsChrimson::tdTomato | BDSC | Cat#84144; RRID:BDSC_84144 |
| D. melanogaster: SPARC2-S-CsChrimson::tdTomato | BDSC | Cat#84145; RRID:BDSC_84145 |
| D. melanogaster: UAS-CsChrimson.mVenus (attP40) | BDSC | Cat#55135; RRID:BDSC_55135 |
| D. melanogaster: UAS-CsChrimson.mVenus (attP2) | BDSC | Cat#55136; RRID:BDSC_55136 |
| D. melanogaster: UAS-ChR2-XXM | Gift from Dr. Robert J. Kittel42 | N/A |
| D. melanogaster: UAS-GtACR2.EYFP (attP2) | Gift from Dr. Adam Claridge-Chang72 | Cat#92987; RRID:BDSC_92987 |
| D. melanogaster: UAS-TNT | BDSC | Cat#28838; RRID:BDSC_28838 |
| D. melanogaster: UAS-TNT inactive | BDSC | Cat#28839; RRID:BDSC_28839 |
| D. melanogaster: UAS-FRT-STOP-FRT-CsChrimson.mVenus (attP2) | Gift from Dr. Galit Shohat-Ophir44 | N/A |
| D. melanogaster: tub-FRT-STOP-FRT-GAL80 | BDSC | Cat#39213; RRID:BDSC_39213 |
| D. melanogaster: LexAop-CsChrimson.mVenus (attP40) | BDSC | Cat#55138; RRID:BDSC_55138 |
| D. melanogaster: UAS-GCaMP6f (attP40) | BDSC | Cat#42747; RRID:BDSC_42747 |
| D. melanogaster: UAS-GCaMP6f (VK00005) | BDSC | Cat#52869; RRID:BDSC_52869 |
| D. melanogaster: LexAop-mCD8::GFP (attP2) | BDSC | Cat#32203; RRID:BDSC_32203 |
| D. melanogaster: LexAop-GCaMP6m (attP1) | BDSC | Cat#44275; RRID:BDSC_44275 |
| D. melanogaster: LexAop-GCaMP6m (VK00005) | BDSC | Cat#44276; RRID:BDSC_44276 |
| D. melanogaster: Otd-nls:FLPo | Gift from Dr. Stephen F. Goodwin50 | N/A |
| D. melanogaster: QUAS-GAL80 | BDSC92 | N/A |
| D. melanogaster: tsh-GAL80 | Gift from Dr. Gero Miesenböck, and Dr. Christian Wegener15 | N/A |
| D. melanogaster: LexAop-TNT::HA (VK00033) | Gift from Dr. Andrew Lin93 | N/A |
| D. melanogaster: QUAS-CsChrimson.mVenus#32c | Gift from Dr. Christopher J. Potter | Cat#91994; RRID:BDSC_91994 |
| D. melanogaster: Gr93a-GAL4 | BDSC | Cat#57679; RRID:BDSC_57679 |
| D. melanogaster: Gr89a-GAL4 | BDSC | Cat#57676; RRID:BDSC_57676 |
| D. melanogaster: Gr66a-GAL4 | BDSC | Cat#57670; RRID:BDSC_57670 |
| D. melanogaster: Gr39a.a-GAL4 | BDSC | Cat#57630; RRID:BDSC_57630 |
| D. melanogaster: Gr33a-GAL4 | BDSC | Cat#57623; RRID:BDSC_57623 |
| D. melanogaster: Gr32a-GAL4 | BDSC | Cat#57622; RRID:BDSC_57622 |
| Software and Algorithms | ||
| MATLAB | MathWorks | https://www.mathworks.com/products/matlab.html; RRID:SCR_001622 |
| GraphPad Prism | GraphPad Software | https://www.graphpad.com/scientific-software/prism/; RRID:SCR_002798 |
| Fiji | Schindelin et al.94 |
https://fiji.sc/; RRID:SCR_002285 |
| neuPrint+ | HHMI's Janelia Research Campus62 | https://neuprint.janelia.org/?dataset=hemibrain:v1.2.1&qt=findneurons |
| FlyWire | Branson et al.95 | https://flywire.ai/; RRID:SCR_019205 |
| Natverse | Bates et al.61 | https://natverse.org/ |
| NeuronBridge | HHMI's Janelia Research Campus56 | https://neuronbridge.janelia.org/ |
| Amira | Thermo Fisher Scientific | https://www.thermofisher.com/il/en/home/electron-microscopy/products/software-em-3d-vis/amira-software.html; RRID:SCR_007353 |
| Ctrax | Branson et al.95 | http://ctrax.sourceforge.net/ |
| MScan | Sutter Instrument | https://www.sutter.com/MICROSCOPES/mcs.html |
| LabVIEW | National Instruments |
https://www.ni.com/en-il/shop/labview.html; RRID:SCR_014325 |
| Pixelink Capture OEM | Pixelink | https://pixelink.com/products/software/pixelink-capture-software/pixelink-capture-software-download/ |
| LAS AF | Leica Microsystems | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Moshe Parnas (mparnas@tauex.tau.ac.il).
Materials availability
This study did not generate new unique reagents. Requests of fly stocks used in the study should be directed to and will be fulfilled by the lead contact, Moshe Parnas (mparnas@tauex.tau.ac.il).
Experimental model and subject details
Fly strains
Fly strains were raised on cornmeal agar under a 12 h light/12 h dark cycle at 25°C.
Fly strains are detailed in the Key Resources Table.
Method details
Odors used
Odors and mineral oil were purchased from Sigma-Aldrich (Rehovot, Israel) except for ACV which was purchased locally (Rauch, Austria). Odors were diluted in mineral oil except for ACV which was diluted in double distilled water (DDW). Diluted odors were prepared on a daily basis.
Adult flies
For activation of CsChrimson or GtACR2, flies were collected 2-5 days post eclosion and grown for another 3-5 days on 1 mM all-trans retinal (R2500; Sigma-Aldrich) supplemented food in complete darkness before experimental testing was performed.
Behavioral assays
Experiments were conducted in two behavioral chambers: an open arena96 of 60 mm diameter and linear chambers,18 40∗1.5∗1.5 mm Length\width\height (8 linear grooves in a plate). Both chambers were composed of polyoxymethylene and covered with a transparent acrylic plastic.
Flies were allowed to acclimatize to the behavioral setups and walk freely for ∼1-3 minutes before start of an experimental trial. For optogenetic experiments, flies were illuminated in the open arena with either 617 nm LED (M617L3; THORLABS) for CsChrimson activation, or with 470 nm LED (M470L3; THORLABS) for ChR2-XXM or GtACR2 activation. For the majority of the experiments in which CsChrimson was activated, flies were illuminated with a red light intensity of ∼9.5 mW/cm2. In experiments in which ChR2-XXM or GtACR2 were activated, flies were illuminated with a blue light intensity of ∼35 mW/cm2. For olfactory experiments, a two second odor pulse was delivered either through a 5 mm diameter hole in the plastic lid located at the center of the open arena, or through a 5 mm hole that was further connected by a narrow airstream path to one end of the linear chambers. Odors were streamed by passing air flow through vials containing liquid odorants. Odors at 5∗10-2 final dilution were delivered by switching mass-flow controlled air carrier and stimulus streams at a final flow of 0.8 l/min (CMOSens Performance Line, Sensirion) via software controlled solenoid valves (The Lee Company) through a 1/16 inch ultra-chemical-resistant Versilon PVC tubing (Saint-Gobain, NJ, USA). Throughout performed experiments, the open arena and linear chambers were illuminated from the bottom by a high intensity 810 nm IR LED (SFH 4786S; OSRAM), while flies' behavioral responses were recorded from above by a camera (PL-D795MU-T; PIXELINK), equipped with 16 mm focal length lens (NMV-16M11; NAVITAR) coupled to 800 nm long-pass filter (LP800; MIDOPT), at 20 frames per second and 832x832 pixel resolution. Videos were captured using Pixelink Capture OEM software. A custom written software (LabVIEW 7.1, National Instruments) controlled the delivery of light and odor pulses and synchronized them with the recording camera via TTL inputs.
For decapitation experiments, flies were briefly anesthetized by placing them on ice and decapitated with a sharp bladed scalpel. Then, they were allowed to recover for at least ∼10 minutes before experimental testing.
In open arena experiments, groups of 3-15 female and male flies were loaded, and presented with either a two, ten or thirty second light stimulus, or a two second odor pulse, except for SPARC53 stochastic activation experiments, in which female flies were tested individually and illuminated with a two second light pulse for 3 consecutive trials. In linear chamber experiments, 8 flies were loaded in 8 separate grooves in a plate and were exposed to a two second odor stimulus.
Functional imaging
Functional imaging was performed on 5-10 days post-eclosion females using a two-photon laser-scanning microscopy (DF-Scope installed on an Olympus BX51WI microscope) as previously described.39,97,98 The brain was superfused with carbonated solution (95% O2, 5% CO2) containing 103 mM NaCl, 3 mM KCl, 5 mM trehalose, 10 mM glucose, 26 mM NaHCO3, 1 mM NaH2PO4, 3 mM CaCl2, 4 mM MgCl2, 5 mM N-Tris (TES), pH 7.3. Fluorescence was excited by a Ti-Sapphire laser (Mai Tai HP DS, 100 fs pulses) centered at 910 nm, attenuated by a Pockels cell (Conoptics) and coupled to a galvo-resonant scanner. Excitation light was focused by a 20X, 1.0 NA objective (Olympus XLUMPLFLN20XW), and emitted photons were detected by GaAsP photomultiplier tubes (Hamamatsu Photonics, H10770PA-40SEL), whose currents were amplified (Hamamatsu HC-130-INV) and transferred to the imaging computer (MScan 2.3.01). Images were acquired at 30 frames per second. When odor-evoked Ca2+ responses were examined, odors at 5∗10-2 final dilution were delivered by switching mass-flow controlled carrier and stimulus streams at a final flow of 0.8 l/min (Sensirion) via software controlled solenoid valves (The Lee Company) for five seconds. Air-streamed odor was delivered through a 1/16 inch ultra-chemical-resistant Versilon PVC tubing (Saint-Gobain, NJ, USA) that was placed ~5 mm from the fly’s antenna. For quinine application, an electrode with a picospritzer at low pressure was used to deliver the liquid (6 mM) to the proboscis of the fly. When necessary, movies were motion-corrected using the TurboReg99 ImageJ plugin. For Ca2+ imaging analysis, regions of interests (ROIs) were hand-drawn in Fiji/ImageJ.94 ΔF/F was calculated as previously described.39,97,98
Structural imaging
Brain dissections, fixation, and immunostaining were performed as described.39,98 To visualize native GFP, tdTomato, and mVenus, we followed our previous adult brain protocol.100 In short, dissected brains were fixed in 4 % [w/v] paraformaldehyde in PBS (1.86 mM NaH2PO4, 8.41 mM Na2HPO4, 175 mM NaCl) for 20 minutes at room temperature. Samples were washed for 3×20 minutes in PBS containing 0.3% (v/v) Triton-X-100 (PBT) and blocked with 5% normal goat serum for at least 30 minutes. Samples were then incubated for 2 days with primary antibodies. After being washed three times with PBT, samples were incubated for 2 days with secondary antibodies in PBT followed by additional washing in PBT and embedding in Vectashield. Primary antibodies used: rabbit anti-GFP (Thermo Fisher Scientific A-11122, 1:1000), mouse anti-GFP (Abcam ab1218, 1:250), rabbit anti-RFP (Abcam ab62341, 1:250), mouse anti-Bruchpilot (DSHB, 1:50). Secondary antibodies used: Alexa Fluor 488 goat anti-mouse (Abcam ab150113, 1:500), Alexa Fluor 488 goat anti-rabbit (Thermo Fisher Scientific A-11034, 1:500), Alexa Fluor 568 goat anti-rabbit (Abcam ab175471, 1:500), Alexa Fluor 647 goat anti-mouse (Abcam ab150115, 1:500). Images were collected on a Leica TCS SP5 (LAS AF software v2.7.3.9723) with HCX PL APO lambda blue 20.0x0.70 IMM UV objective and processed in Fiji/ImageJ.94
LM 3D reconstruction
The LM 3D reconstruction of MooSEZ morphology from SPARC experiments was performed in Amira 5.2 as described before,47 including a custom-written skeletonization add-on.101
EM 3D reconstruction and analysis
Fragment #5813046185 in the FlyEM hemibrain volume57 was matched to MooSEZ neurites by performing anatomical similarity search between an LM image of MooSEZ neurites (labeled by the SPARC method) and candidate neuronal fragments in the FlyEM hemibrain dataset using NeuronBridge54, 55, 56 3D search tool. Right (segment ID: 720575940622871283; nucleus coordinates: 110481, 71647, 5418) and left (segment ID: 720575940633628664; nucleus coordinates: 153740, 76085, 5035) MooSEZs were identified in the FAFB volume using FlyWire.58 The natverse package was used to access, compare and visualize FlyEM hemibrain57 and FlyWire58 EM datasets and brain surface meshes for reference.61 To identify and visualize MooSEZ and its postsynaptic cells in the FlyEM hemibrain dataset, the neuPrint+ web application (hemibrain:v1.2.1) “Simple connections” and "Find neurons" plugins62 were used. The percentage of MooSEZ synaptic outputs onto DNa01 (or DNa02) presynaptic neurons in the FlyEM hemibrain was calculated by dividing the number of MooSEZ synaptic outputs onto DNa01 (or DNa02) presynaptic neurons by the total number of MooSEZ synaptic outputs in the hemibrain and multiplying by 100. The number of MooSEZ synaptic outputs onto DNa01 (or DNa02) presynaptic neurons in the FlyEM hemibrain was computed by executing a custom Cypher query in neuPrint+ mapping all disynaptic neural pathways connecting MooSEZ onto DNa01 (or DNa02) in the hemibrain. The total number of MooSEZ synaptic outputs in the FlyEM hemibrain was obtained using the neuPrint+ plugin "Simple connections". Similarly, the percentage of DNa01 (or DNa02) synaptic inputs formed by MooSEZ postsynaptic neurons in the FlyEM hemibrain was calculated by dividing the number of DNa01 (or DNa02) synaptic inputs formed by MooSEZ postsynaptic neurons by the total number of DNa01 (or DNa02) synaptic inputs in the hemibrain and multiplying by 100. The number of DNa01 (or DNa02) synaptic inputs formed by MooSEZ postsynaptic neurons in the FlyEM hemibrain was computed by executing a custom Cypher query in neuPrint+ mapping all disynaptic neural pathways connecting MooSEZ onto DNa01 (or DNa02) in the hemibrain. The total number of DNa01 (or DNa02) synaptic inputs in the FlyEM hemibrain was obtained using neuPrint+ plugin "Simple connections". Finally, to generate connectivity diagram between MooSEZ and DNa01, as well as between MooSEZ and DNa02 in the FlyEM hemibrain, neuPrint+ “Shortest paths” plugin was used. The connectivity diagrams fulfill an inclusion criterion of at least 15 synapses for each directed neural path.
Quantification and statistical analysis
Quantification of behavioral experiments
Acquired videos were tracked with Ctrax106 which allows to determine the locations and orientations of flies along a recorded video. Following initial tracking with Ctrax, the "FixErrors Matlab GUI" was used to compensate for tracking errors. Only flies that could walk in an unconstrained manner and were accurately tracked were considered in the analysis. The script "compute_perframe_stats" from Ctrax "BehavioralMicroarray" MATLAB toolbox was used to compute per-frame parameters of the flies' locomotor activity. Custom MATLAB scripts were used to perform final analysis based on the parameters outputted by the "compute_perframe_stats" script. Translational velocity was defined as the projection of the fly’s velocity vector on its forward-backward orientation axis. Mean translational velocity was computed as the mean of per-fly translational velocity mean values during the odor or light pulse. Backward walking distance was computed by integrating the area under translational velocity versus time curve for negative translational velocity values during the odor or light pulse. Mean backward distance was computed as the mean of backward distances covered by flies during the light pulse. For SPARC stochastic activation experiments, mean backward distance of each fly was computed as the mean of the backward distances covered by each fly during the light pulse over three trials. Angular velocity was defined as the change in the fly’s orientation angle in relation to a global orientation coordinate system. Left rotations were defined as positive values whereas right rotations as negative values. Mean angular velocity was computed as the mean of per-fly angular velocity mean values during the light pulse. Cumulative angular change was computed by calculating the cumulative integral of the angular velocity versus time curve. Total angular change was computed by integrating the area under the angular velocity versus time curve during the light pulse. Angular speed was defined as the absolute value of angular velocity. Mean angular speed was computed as the mean of per-fly angular speed mean values during the light pulse. Absolute cumulative angular change was computed by calculating the cumulative integral of the angular speed versus time curve. Absolute total angular change was computed by integrating the area under the angular speed versus time curve during the light pulse. Fraction of time spent in backward motion was computed by dividing the number of time frames in which flies were measured with negative translational velocity during the light pulse by the number of time frames in which light was delivered. Percentage of backward walking flies was calculated by defining a minimal backward walking distance of 3 mm as a threshold for walking activity that had a pronounced backward component during the light stimulation. Accordingly, backward walking percentage was calculated as the ratio between the number of flies that walked backwards 3 mm and above to the total number of tested flies multiplied by 100. For Regression analysis of SPARC53 stochastic flies, each cluster was set to 1 if at least one cell within the cluster was targeted, and to 0 in case in which none of the cells within the cluster were labeled.
Statistics and data analysis
Statistical testing and parameter extraction were done using GraphPad Prism 9.2.0 and custom MATLAB codes (The MathWorks, Inc.). All statistical tests details can be found in Table S1. Significance was defined as a p-value smaller than 0.05 and all statistical tests were two-sided, except for (left) one-sided Wilcoxon signed rank tests performed in Figures 2B and 6I.
For presentation of imaging results, bar plots with dots were generated using the UnivarScatter MATLAB ToolBox (https://www.mathworks.com/matlabcentral/fileexchange/54243-univarscatter) and the shadedErrorBar function (https://github.com/raacampbell/shadedErrorBar) for shaded errors on imaging traces. For presentation of translational velocity and angular speed traces with shaded error bars, the boundedline MATLAB function was used (https://github.com/kakearney/boundedline-pkg).
Acknowledgments
We thank Dr. Robert J. Kittel, Dr. Christopher J. Potter, Dr. Barry J. Dickson, Dr. Adam Claridge-Chang, Dr. Andrew Lin, Dr. Gero Miesenböck, Dr. Christian Wegener, Dr. Stephen F. Goodwin, Dr. Liria Masuda-Nakagawa, Dr. Liqun Luo, Dr. Galit Shohat-Ophir, the Bloomington Drosophila Stock Center (BDSC), the Vienna Drosophila Resource Center (VDRC), and the Department of Drosophila Genomics and Genetic Resources (DGGR) at Kyoto Institute of Technology for fly strains. We also thank Dr. Anton Sheinin and Eran Rosen for providing technical guidance and assistance, Dr. Hadas Lerner Nussbaum for providing feedback on the manuscript and technical assistance, and Bar Gitter for providing technical assistance. Members Alex Javier, Imaan Tamimi, and Dr. Katharina Eichler of the Cambridge “Drosophila Connectomics Group” and Alexis E. Santana Cruz contributed to proofreading MooSEZs in FlyWire. Dr. Katharina Eichler also provided expert assistance with natverse usage and EM data interpretation. This work was supported by the European Research Council (676844, M.P.) and the Deutsche Forschungsgemeinschaft (HU 2474/1-1, W.H.).
Author contributions
Project initiation, S.I, M.P.; conceptualization, S.I., M.P.; methodology, S.I., M.P.; investigation, S.I., E.R., D.W., W.H., M.P.; formal analysis, S.I., E.R., M.P.; software, S.I.; writing—original draft, M.P.; writing—reviewing & editing, S.I., E.R., W.H., M.P.; visualization, S.I., E.R., D.W., W.H., M.P.; funding acquisition, W.H., M.P.; supervision, M.P.
Declaration of interests
The authors declare no competing interests.
Published: February 8, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cub.2022.01.035.
Supplemental information
Data and code availability
The data and code used to generate Figures 1, 2, 3, 4, 5, 6, 7, and S1–S4, Tables S1 and S2, and Video S1. Differential odor-induced backward locomotion, related to Figure 1, Video S2. Backward locomotion following optogenetic activation of GH146II-GAL4, related to Figure 2, Video S3. GH146II-GAL4-induced, MDN-independent, backward locomotion, related to Figure 3, Video S4. GH146II-GAL4 VNC neurons cannot induce backward locomotion, related to Figure 4, Video S5. Blocking expression in VNC neurons does not impair optogenetically induced backward locomotion, related to Figure 4, Video S6. Confocal stack of the GH146II-GAL4 SPARC mosaic fly used for MooSEZ three-dimensional light microscope reconstruction, related to Figure 5, Video S7. Blocking expression in VNC neurons enhances backward locomotion in NP5288-GAL4 flies, related to Figure 6, Video S8. Unilateral activation of MooSEZs triggers ipsilateral backward rotation, related to Figure 7 are available from the corresponding author upon request. The study did not generate any new code or dataset. Any additional information required to reanalyze the data reported in the paper is available from the lead contact upon request.
References
- 1.Bidaye S.S., Bockemühl T., Büschges A. Six-legged walking in insects: how CPGs, peripheral feedback, and descending signals generate coordinated and adaptive motor rhythms. J. Neurophysiol. 2018;119:459–475. doi: 10.1152/jn.00658.2017. [DOI] [PubMed] [Google Scholar]
- 2.Wilson R.I. Early olfactory processing in Drosophila: mechanisms and principles. Annu. Rev. Neurosci. 2013;36:217–241. doi: 10.1146/annurev-neuro-062111-150533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hedwig B., Burrows M. Presynaptic inhibition of sensory neurons during kicking movements in the locust. J. Neurophysiol. 1996;75:1221–1232. doi: 10.1152/jn.1996.75.3.1221. [DOI] [PubMed] [Google Scholar]
- 4.Bässler U. The walking-(and searching-) pattern generator of stick insects, a modular system composed of reflex chains and endogenous oscillators. Biol. Cybern. 1993;69:305–317. [Google Scholar]
- 5.Burrows M. Jumping performance of froghopper insects. J. Exp. Biol. 2006;209:4607–4621. doi: 10.1242/jeb.02539. [DOI] [PubMed] [Google Scholar]
- 6.Robertson R.M., Pearson K.G. Neural circuits in the flight system of the locust. J. Neurophysiol. 1985;53:110–128. doi: 10.1152/jn.1985.53.1.110. [DOI] [PubMed] [Google Scholar]
- 7.Ausborn J., Stein W., Wolf H. Frequency control of motor patterning by negative sensory feedback. J. Neurosci. 2007;27:9319–9328. doi: 10.1523/JNEUROSCI.0907-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Büschges A., Akay T., Gabriel J.P., Schmidt J. Organizing network action for locomotion: insights from studying insect walking. Brain Res. Rev. 2008;57:162–171. doi: 10.1016/j.brainresrev.2007.06.028. [DOI] [PubMed] [Google Scholar]
- 9.Ariel G., Ayali A. Locust collective motion and its modeling. PLoS Comput. Biol. 2015;11 doi: 10.1371/journal.pcbi.1004522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krause A.F., Winkler A., Dürr V. Central drive and proprioceptive control of antennal movements in the walking stick insect. J. Physiol. Paris. 2013;107:116–129. doi: 10.1016/j.jphysparis.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 11.Harley C.M., English B.A., Ritzmann R.E. Characterization of obstacle negotiation behaviors in the cockroach, Blaberus discoidalis. J. Exp. Biol. 2009;212:1463–1476. doi: 10.1242/jeb.028381. [DOI] [PubMed] [Google Scholar]
- 12.Pick S., Strauss R. Goal-driven behavioral adaptations in gap-climbing Drosophila. Curr. Biol. 2005;15:1473–1478. doi: 10.1016/j.cub.2005.07.022. [DOI] [PubMed] [Google Scholar]
- 13.Seeds A.M., Ravbar P., Chung P., Hampel S., Midgley F.M., Mensh B.D., Simpson J.H. A suppression hierarchy among competing motor programs drives sequential grooming in Drosophila. eLife. 2014;3 doi: 10.7554/eLife.02951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao X.J., Potter C.J., Gohl D.M., Silies M., Katsov A.Y., Clandinin T.R., Luo L. Specific kinematics and motor-related neurons for aversive chemotaxis in Drosophila. Curr. Biol. 2013;23:1163–1172. doi: 10.1016/j.cub.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clyne J.D., Miesenböck G. Sex-specific control and tuning of the pattern generator for courtship song in Drosophila. Cell. 2008;133:354–363. doi: 10.1016/j.cell.2008.01.050. [DOI] [PubMed] [Google Scholar]
- 16.von Philipsborn A.C., Liu T., Yu J.Y., Masser C., Bidaye S.S., Dickson B.J. Neuronal control of Drosophila courtship song. Neuron. 2011;69:509–522. doi: 10.1016/j.neuron.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 17.Pfeiffer B.D., Jenett A., Hammonds A.S., Ngo T.T.B., Misra S., Murphy C., Scully A., Carlson J.W., Wan K.H., Laverty T.R., et al. Tools for neuroanatomy and neurogenetics in Drosophila. Proc. Natl. Acad. Sci. USA. 2008;105:9715–9720. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bidaye S.S., Machacek C., Wu Y., Dickson B.J. Neuronal control of Drosophila walking direction. Science. 2014;344:97–101. doi: 10.1126/science.1249964. [DOI] [PubMed] [Google Scholar]
- 19.Chen C.L., Hermans L., Viswanathan M.C., Fortun D., Aymanns F., Unser M., Cammarato A., Dickinson M.H., Ramdya P. Imaging neural activity in the ventral nerve cord of behaving adult Drosophila. Nat. Commun. 2018;9:4390. doi: 10.1038/s41467-018-06857-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sen R., Wu M., Branson K., Robie A., Rubin G.M., Dickson B.J. Moonwalker descending neurons mediate visually evoked retreat in Drosophila. Curr. Biol. 2017;27:766–771. doi: 10.1016/j.cub.2017.02.008. [DOI] [PubMed] [Google Scholar]
- 21.Sen R., Wang K., Dickson B.J. TwoLumps ascending neurons mediate touch-evoked reversal of walking direction in Drosophila. Curr. Biol. 2019;29:4337. doi: 10.1016/j.cub.2019.11.004. e5–4344.e5. [DOI] [PubMed] [Google Scholar]
- 22.Zhao Z., McBride C.S. Evolution of olfactory circuits in insects. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 2020;206:353–367. doi: 10.1007/s00359-020-01399-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng K., Sen R., Minegishi R., Dübbert M., Bockemühl T., Büschges A., Dickson B.J. Distributed control of motor circuits for backward walking in Drosophila. Nat. Commun. 2020;11:1–17. doi: 10.1038/s41467-020-19936-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee K.M., Doe C.Q. A locomotor neural circuit persists and functions similarly in larvae and adult Drosophila. eLife. 2021;10 doi: 10.7554/eLife.69767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gomez-Diaz C., Martin F., Garcia-Fernandez J.M., Alcorta E. The two main olfactory receptor families in Drosophila, ORs and IRs: a comparative approach. Front. Cell. Neurosci. 2018;12:253. doi: 10.3389/fncel.2018.00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hallem E.A., Carlson J.R. The odor coding system of Drosophila. Trends Genet. 2004;20:453–459. doi: 10.1016/j.tig.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 27.Hallem E.A., Carlson J.R. Coding of odors by a receptor repertoire. Cell. 2006;125:143–160. doi: 10.1016/j.cell.2006.01.050. [DOI] [PubMed] [Google Scholar]
- 28.Benton R., Vannice K.S., Gomez-Diaz C., Vosshall L.B. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell. 2009;136:149–162. doi: 10.1016/j.cell.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abuin L., Bargeton B., Ulbrich M.H., Isacoff E.Y., Kellenberger S., Benton R. Functional architecture of olfactory ionotropic glutamate receptors. Neuron. 2011;69:44–60. doi: 10.1016/j.neuron.2010.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bates A.S., Schlegel P., Roberts R.J.V., Drummond N., Tamimi I.F.M., Turnbull R., Zhao X., Marin E.C., Popovici P.D., Dhawan S., et al. Complete connectomic reconstruction of olfactory projection neurons in the fly brain. Curr. Biol. 2020;30:3183. doi: 10.1016/j.cub.2020.06.042. e6–3199.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao Q., Yuan B., Chess A. Convergent projections of Drosophila olfactory neurons to specific glomeruli in the antennal lobe. Nat. Neurosci. 2000;3:780–785. doi: 10.1038/77680. [DOI] [PubMed] [Google Scholar]
- 32.Mombaerts P., Wang F., Dulac C., Chao S.K., Nemes A., Mendelsohn M., Edmondson J., Axel R. Visualizing an olfactory sensory map. Cell. 1996;87:675–686. doi: 10.1016/s0092-8674(00)81387-2. [DOI] [PubMed] [Google Scholar]
- 33.Vosshall L.B., Wong A.M., Axel R. An olfactory sensory map in the fly brain. Cell. 2000;102:147–159. doi: 10.1016/s0092-8674(00)00021-0. [DOI] [PubMed] [Google Scholar]
- 34.Tanaka N.K., Endo K., Ito K. Organization of antennal lobe-associated neurons in adult Drosophila melanogaster brain. J. Comp. Neurol. 2012;520:4067–4130. doi: 10.1002/cne.23142. [DOI] [PubMed] [Google Scholar]
- 35.Sayin S., Boehm A.C., Kobler J.M., de Backer J.F., Grunwald Kadow I.C. Internal state dependent odor processing and perception—the role of neuromodulation in the fly olfactory system. Front. Cell. Neurosci. 2018;12:11. doi: 10.3389/fncel.2018.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haverkamp A., Hansson B.S., Knaden M. Combinatorial codes and labeled lines: how insects use olfactory cues to find and judge food, mates, and oviposition sites in complex environments. Front. Physiol. 2018;9:49. doi: 10.3389/fphys.2018.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huoviala P., Dolan M.-J., Love F.M., Myers P., Frechter S., Namiki S., Pettersson L., Roberts R.J.V., Turnbull R., Mitrevica Z., et al. Neural circuit basis of aversive odour processing in Drosophila from sensory input to descending output. eLife. 2020;9 [Google Scholar]
- 38.Stensmyr M.C., Dweck H.K.M., Farhan A., Ibba I., Strutz A., Mukunda L., Linz J., Grabe V., Steck K., Lavista-Llanos S., et al. A conserved dedicated olfactory circuit for detecting harmful microbes in Drosophila. Cell. 2012;151:1345–1357. doi: 10.1016/j.cell.2012.09.046. [DOI] [PubMed] [Google Scholar]
- 39.Lerner H., Rozenfeld E., Rozenman B., Huetteroth W., Parnas M. Differential role for a defined lateral horn neuron subset in naïve odor valence in Drosophila. Sci. Rep. 2020;10:6147. doi: 10.1038/s41598-020-63169-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen T.W., Wardill T.J., Sun Y., Pulver S.R., Renninger S.L., Baohan A., Schreiter E.R., Kerr R.A., Orger M.B., Jayaraman V., et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sweeney S.T., Broadie K., Keane J., Niemann H., O’Kane C.J. Targeted expression of tetanus toxin light chain in Drosophila specifically eliminates synaptic transmission and causes behavioral defects. Neuron. 1995;14:341–351. doi: 10.1016/0896-6273(95)90290-2. [DOI] [PubMed] [Google Scholar]
- 42.Scholz N., Guan C., Nieberler M., Grotemeyer A., Maiellaro I., Gao S., Beck S., Pawlak M., Sauer M., Asan E., et al. Mechano-dependent signaling by latrophilin/CIRL quenches cAMP in proprioceptive neurons. eLife. 2017;6 doi: 10.7554/eLife.28360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berdnik D., Fan A.P., Potter C.J., Luo L. MicroRNA processing pathway regulates olfactory neuron morphogenesis. Curr. Biol. 2008;18:1754–1759. doi: 10.1016/j.cub.2008.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klapoetke N.C., Murata Y., Kim S.S., Pulver S.R., Birdsey-Benson A., Cho Y.K., Morimoto T.K., Chuong A.S., Carpenter E.J., Tian Z., et al. Independent optical excitation of distinct neural populations. Nat. Methods. 2014;11:338–346. doi: 10.1038/nmeth.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Inagaki H.K., Jung Y., Hoopfer E.D., Wong A.M., Mishra N., Lin J.Y., Tsien R.Y., Anderson D.J. Optogenetic control of Drosophila using a red-shifted channelrhodopsin reveals experience-dependent influences on courtship. Nat. Methods. 2014;11:325–332. doi: 10.1038/nmeth.2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu M., Nern A., Williamson W.R., Morimoto M.M., Reiser M.B., Card G.M., Rubin G.M. Visual projection neurons in the Drosophila lobula link feature detection to distinct behavioral programs. eLife. 2016;5 doi: 10.7554/eLife.21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pitman J.L., Huetteroth W., Burke C.J., Krashes M.J., Lai S.L., Lee T., Waddell S. A pair of inhibitory neurons are required to sustain labile memory in the Drosophila mushroom body. Curr. Biol. 2011;21:855–861. doi: 10.1016/j.cub.2011.03.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yellman C., Tao H., He B., Hirsh J. Conserved and sexually dimorphic behavioral responses to biogenic amines in decapitated Drosophila. Proc. Natl. Acad. Sci. USA. 1997;94:4131–4136. doi: 10.1073/pnas.94.8.4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu J.Y., Kanai M.I., Demir E., Jefferis G.S.X.E., Dickson B.J. Cellular organization of the neural circuit that drives Drosophila courtship behavior. Curr. Biol. 2010;20:1602–1614. doi: 10.1016/j.cub.2010.08.025. [DOI] [PubMed] [Google Scholar]
- 50.Asahina K., Watanabe K., Duistermars B.J., Hoopfer E., González C.R., Eyjólfsdóttir E.A., Perona P., Anderson D.J. Tachykinin-expressing neurons control male-specific aggressive arousal in Drosophila. Cell. 2014;156:221–235. doi: 10.1016/j.cell.2013.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aranha M.M., Herrmann D., Cachitas H., Neto-Silva R.M., Dias S., Vasconcelos M.L. apterous brain neurons control receptivity to male courtship in Drosophila Melanogaster females. Sci. Rep. 2017;7:46242. doi: 10.1038/srep46242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suster M.L., Seugnet L., Bate M., Sokolowski M.B. Refining GAL4-driven transgene expression in Drosophila with a GAL80 enhancer-trap. Genesis. 2004;39:240–245. doi: 10.1002/gene.20051. [DOI] [PubMed] [Google Scholar]
- 53.Isaacman-Beck J., Paik K.C., Wienecke C.F.R., Yang H.H., Fisher Y.E., Wang I.E., Ishida I.G., Maimon G., Wilson R.I., Clandinin T.R. SPARC enables genetic manipulation of precise proportions of cells. Nat. Neurosci. 2020;23:1168–1175. doi: 10.1038/s41593-020-0668-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meissner G.W., Dorman Z., Nern A., Forster K., Gibney T., Jeter J., Johnson L., He Y., Lee K., Melton B., et al. An image resource of subdivided Drosophila GAL4-driver expression patterns for neuron-level searches. bioRxiv. 2020 doi: 10.1101/2020.05.29.080473. [DOI] [Google Scholar]
- 55.Otsuna H., Ito M., Kawase T. Color depth MIP mask search: a new tool to expedite Split-GAL4 creation. bioRxiv. 2018 doi: 10.1101/318006. [DOI] [Google Scholar]
- 56.Clements J., Goina C., Kazimiers A., Otsuna H., Svirskas R.R., Rokicki K. 2020. NeuronBridge Codebase (Janelia Research Campus) [Google Scholar]
- 57.Scheffer L.K., Xu C.S., Januszewski M., Lu Z., Takemura S.Y., Hayworth K.J., Huang G.B., Shinomiya K., Maitlin-Shepard J., Berg S., et al. A connectome and analysis of the adult Drosophila central brain. eLife. 2020;9:1–74. doi: 10.7554/eLife.57443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dorkenwald S., McKellar C.E., Macrina T., Kemnitz N., Lee K., Lu R., Wu J., Popovych S., Mitchell E., Nehoran B., et al. FlyWire: online community for whole-brain connectomics. Nat. Methods. 2022;19:119–128. doi: 10.1038/s41592-021-01330-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zheng Z., Lauritzen J.S., Perlman E., Robinson C.G., Nichols M., Milkie D., Torrens O., Price J., Fisher C.B., Sharifi N., et al. A complete electron microscopy volume of the brain of adult Drosophila melanogaster. Cell. 2018;174:730. doi: 10.1016/j.cell.2018.06.019. e22–743.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ito K., Shinomiya K., Ito M., Armstrong J.D., Boyan G., Hartenstein V., Harzsch S., Heisenberg M., Homberg U., Jenett A., et al. A systematic nomenclature for the insect brain. Neuron. 2014;81:755–765. doi: 10.1016/j.neuron.2013.12.017. [DOI] [PubMed] [Google Scholar]
- 61.Bates A.S., Manton J.D., Jagannathan S.R., Costa M., Schlegel P., Rohlfing T., Jefferis G.S.X.E. The natverse, a versatile toolbox for combining and analysing neuroanatomical data. eLife. 2020;9 doi: 10.7554/eLife.53350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Clements J., Dolafi T., Umayam L., Neubarth N.L., Berg S., Scheffer L.K., Plaza S.M. neuPrint: analysis tools for EM connectomics. bioRxiv. 2020 doi: 10.1101/2020.01.16.909465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fischer J.A., Giniger E., Maniatis T., Ptashne M. GAL4 activates transcription in Drosophila. Nature. 1988;332:853–856. doi: 10.1038/332853a0. [DOI] [PubMed] [Google Scholar]
- 64.Lai S.L., Lee T. Genetic mosaic with dual binary transcriptional systems in Drosophila. Nat. Neurosci. 2006;9:703–709. doi: 10.1038/nn1681. [DOI] [PubMed] [Google Scholar]
- 65.Potter C.J., Tasic B., Russler E.V., Liang L., Luo L. The Q system: a repressible binary system for transgene expression, lineage tracing, and mosaic analysis. Cell. 2010;141:536–548. doi: 10.1016/j.cell.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen Y.J., Chang H.H., Lin S.H., Lin T.Y., Wu T.H., Lin H.J., Liou N.F., Yang C.J., Chen Y.T., Chang K.H., et al. Differential efficacy of genetically swapping GAL4. J. Neurogenet. 2019;33:52–63. doi: 10.1080/01677063.2018.1564289. [DOI] [PubMed] [Google Scholar]
- 67.Jenett A., Rubin G.M., Ngo T.T.B., Shepherd D., Murphy C., Dionne H., Pfeiffer B.D., Cavallaro A., Hall D., Jeter J., et al. A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2012;2:991–1001. doi: 10.1016/j.celrep.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grosjean Y., Rytz R., Farine J.P., Abuin L., Cortot J., Jefferis G.S.X.E., Benton R. An olfactory receptor for food-derived odours promotes male courtship in Drosophila. Nature. 2011;478:236–240. doi: 10.1038/nature10428. [DOI] [PubMed] [Google Scholar]
- 69.Wang J.W., Wong A.M., Flores J., Vosshall L.B., Axel R. Two-photon calcium imaging reveals an odor-evoked map of activity in the fly brain. Cell. 2003;112:271–282. doi: 10.1016/s0092-8674(03)00004-7. [DOI] [PubMed] [Google Scholar]
- 70.Kendroud S., Bohra A.A., Kuert P.A., Nguyen B., Guillermin O., Sprecher S.G., Reichert H., Vijayraghavan K., Hartenstein V. Structure and development of the subesophageal zone of the Drosophila brain. II. Sensory compartments. J. Comp. Neurol. 2018;526:33–58. doi: 10.1002/cne.24316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dweck H.K.M., Carlson J.R. Molecular logic and evolution of bitter taste in Drosophila. Curr. Biol. 2020;30:17. doi: 10.1016/j.cub.2019.11.005. e3–30.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mohammad F., Stewart J.C., Ott S., Chlebikova K., Chua J.Y., Koh T.W., Ho J., Claridge-Chang A. Optogenetic inhibition of behavior with anion channelrhodopsins. Nat. Methods. 2017;14:271–274. doi: 10.1038/nmeth.4148. [DOI] [PubMed] [Google Scholar]
- 73.Rayshubskiy A., Holtz S.L., D’Alessandro I., Li A.A., Vanderbeck Q.X., Haber I.S., Gibb P.W., Wilson R.I. Neural circuit mechanisms for steering control in walking Drosophila. bioRxiv. 2020 doi: 10.1101/2020.04.04.024703. [DOI] [Google Scholar]
- 74.Freeman E.G., Dahanukar A. Molecular neurobiology of Drosophila taste. Curr. Opin. Neurobiol. 2015;34:140–148. doi: 10.1016/j.conb.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wright G.A. To feed or not to feed: circuits involved in the control of feeding in insects. Curr. Opin. Neurobiol. 2016;41:87–91. doi: 10.1016/j.conb.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 76.Steinbeck F., Adden A., Graham P. Connecting brain to behaviour: a role for general purpose steering circuits in insect orientation? J. Exp. Biol. 2020;223:jeb212332. doi: 10.1242/jeb.212332. [DOI] [PubMed] [Google Scholar]
- 77.Olberg R.M. Pheromone-triggered flip-flopping interneurons in the ventral nerve cord of the silkworm moth, Bombyx mori. J. Comp. Physiol. 1983;152:297–307. [Google Scholar]
- 78.Homberg U. Flight-correlated activity changes in neurons of the lateral accessory lobes in the brain of the locust Schistocerca gregaria. J. Comp. Physiol. A. 1994;175:597–610. [Google Scholar]
- 79.Harley C.M., Ritzmann R.E. Electrolytic lesions within central complex neuropils of the cockroach brain affect negotiation of barriers. J. Exp. Biol. 2010;213:2851–2864. doi: 10.1242/jeb.042499. [DOI] [PubMed] [Google Scholar]
- 80.Zorović M., Hedwig B. Descending brain neurons in the cricket Gryllus bimaculatus (de Geer): auditory responses and impact on walking. J. Comp. Physiol. A. 2013;199:25–34. doi: 10.1007/s00359-012-0765-7. [DOI] [PubMed] [Google Scholar]
- 81.Omamiuda-Ishikawa N., Sakai M., Emoto K. A pair of ascending neurons in the subesophageal zone mediates aversive sensory inputs-evoked backward locomotion in Drosophila larvae. PLoS Genet. 2020;16 doi: 10.1371/journal.pgen.1009120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tastekin I., Riedl J., Schilling-Kurz V., Gomez-Marin A., Truman J.W., Louis M. Role of the subesophageal zone in sensorimotor control of orientation in Drosophila larva. Curr. Biol. 2015;25:1448–1460. doi: 10.1016/j.cub.2015.04.016. [DOI] [PubMed] [Google Scholar]
- 83.Namiki S., Dickinson M.H., Wong A.M., Korff W., Card G.M. The functional organization of descending sensory-motor pathways in Drosophila. eLife. 2018;7 doi: 10.7554/eLife.34272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bidaye S.S., Laturney M., Chang A.K., Liu Y., Bockemühl T., Büschges A., Scott K. Two brain pathways initiate distinct forward walking programs in Drosophila. Neuron. 2020;108:469–485.e8. doi: 10.1016/j.neuron.2020.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Murlis J., Elkinton J.S., Cardé R.T. Odor plumes and how insects use them. Annu. Rev. Entomol. 1992;37:505–532. [Google Scholar]
- 86.Martelli C., Pech U., Kobbenbring S., Pauls D., Bahl B., Sommer M.V., Pooryasin A., Barth J., Arias C.W.P., Vassiliou C., et al. SIFamide translates hunger signals into appetitive and feeding behavior in Drosophila. Cell Rep. 2017;20:464–478. doi: 10.1016/j.celrep.2017.06.043. [DOI] [PubMed] [Google Scholar]
- 87.Lin S., Senapati B., Tsao C.H. Neural basis of hunger-driven behaviour in Drosophila. Open Biol. 2019;9:180259. doi: 10.1098/rsob.180259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tirian L., Dickson B.J. The VT GAL4, LexA, and split-GAL4 driver line collections for targeted expression in the Drosophila nervous system. bioRxiv. 2017 doi: 10.1101/198648. [DOI] [Google Scholar]
- 89.Marin E.C., Watts R.J., Tanaka N.K., Ito K., Luo L. Developmentally programmed remodeling of the Drosophila olfactory circuit. Development. 2005;132:725–737. doi: 10.1242/dev.01614. [DOI] [PubMed] [Google Scholar]
- 90.Masuda-Nakagawa L.M., Tanaka N.K., O’Kane C.J. Stereotypic and random patterns of connectivity in the larval mushroom body calyx of Drosophila. Proc. Natl. Acad. Sci. USA. 2005;102:19027–19032. doi: 10.1073/pnas.0509643102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ito K., Sass H., Urban J., Hofbauer A., Schneuwly S. GAL4-responsive UAS-tau as a tool for studying the anatomy and development of the Drosophila central nervous system. Cell Tissue Res. 1997;290:1–10. doi: 10.1007/s004410050901. [DOI] [PubMed] [Google Scholar]
- 92.Potter C.J., Luo L. Using the Q system in Drosophila melanogaster. Nat. Protoc. 2011;6:1105–1120. doi: 10.1038/nprot.2011.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Karuppudurai T., Lin T.Y., Ting C.Y., Pursley R., Melnattur K.V., Diao F., White B.H., Macpherson L.J., Gallio M., Pohida T., Lee C.-H. A hard-wired glutamatergic circuit pools and relays UV signals to mediate spectral preference in Drosophila. Neuron. 2014;81:603–615. doi: 10.1016/j.neuron.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Branson K., Robie A.A., Bender J., Perona P., Dickinson M.H. High-throughput ethomics in large groups of Drosophila. Nat. Methods. 2009;6:451–457. doi: 10.1038/nmeth.1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Simon J.C., Dickinson M.H. A new chamber for studying the behavior of Drosophila. PLoS One. 2010;5 doi: 10.1371/journal.pone.0008793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bielopolski N., Amin H., Apostolopoulou A.A., Rozenfeld E., Lerner H., Huetteroth W., Lin A.C., Parnas M. Inhibitory muscarinic acetylcholine receptors enhance aversive olfactory learning in adult Drosophila. eLife. 2019;8 doi: 10.7554/eLife.48264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rozenfeld E., Lerner H., Parnas M. Muscarinic modulation of antennal lobe GABAergic local neurons shapes odor coding and behavior. Cell Rep. 2019;29:3253. doi: 10.1016/j.celrep.2019.10.125. e4–3265.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Thévenaz P., Ruttimann U.E., Unser M. A pyramid approach to subpixel registration based on intensity. IEEE Trans. Image Process. 1998;7:27–41. doi: 10.1109/83.650848. [DOI] [PubMed] [Google Scholar]
- 100.Huetteroth W., Perisse E., Lin S., Klappenbach M., Burke C., Waddell S. Sweet taste and nutrient value subdivide rewarding dopaminergic neurons in Drosophila. Curr. Biol. 2015;25:751–758. doi: 10.1016/j.cub.2015.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Schmitt S., Evers J.F., Duch C., Scholz M., Obermayer K. New methods for the computer-assisted 3-D reconstruction of neurons from confocal image stacks. Neuroimage. 2004;23:1283–1298. doi: 10.1016/j.neuroimage.2004.06.047. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Example of w1118 responses to ACV (left), GA (middle) and 2-butanone (right).
Example of backward locomotion following a 2-s blue light pulse (470 nm) in flies carrying GH146II-GAL4 and UAS-ChR2-XXM.
Left, example of the efficiency of TNT in silencing MDNs. The VT50660-GAL4 (MDNs) driver was used to drive UAS-CsChrimson, and the VT44845-LexA (MDN-LexA) was used to drive LexAop-TNT. Right, example of backward locomotion generated by activation of GH146II-GAL4 with UAS-CsChrimson while VT44845-LexA (MDN-LexA) driving LexAop-TNT was used to silence MDNs.
Left, example of backward locomotion in MDN3-GAL4 driving UAS-ChR2-XXM in decapitated flies. Right, example of lack of backward locomotion in GH146II-GAL4 driving UAS-ChR2-XXM in decapitated flies. Note the oscillatory movement of the forelegs.
Left, example of the efficiency of tsh-GAL80 to block VNC expression in decapitated flies carrying GH146II-GAL4, UAS-CsChrimson and tsh-GAL80. No foreleg movement is observed following optogenetic activation. Right, example of backward locomotion in NP5288-GAL4 driving UAS-CsChrimson in intact flies when expression in VNC was blocked using tsh-GAL80.
Translational movie running through the entire central brain from the posterior to the anterior aspect (left) or from the anterior to the posterior aspect (right) of a fly in which the left MooSEZ was sparsely targeted to express CsChrimson::tdTomato using the SPARC method. To generate the movies, each confocal section (1 μm) of the original stacks (144 slices) was converted into a video frame.
Left, example of backward locomotion in NP5288-GAL4 driving UAS-CsChrimson. Note the strong oscillatory foreleg movement. Right, example of backward locomotion in NP5288-GAL4 with subtracted GH146-QF driving UAS-CsChrimson. Note the absence of oscillatory foreleg movement and increase in backward locomotion.
Left, example of backward turning in GH146II-GAL4 SPARC mosaic fly expressing CsChrimson in the right MooSEZ. Right, example of backward turning in GH146II-GAL4 SPARC mosaic fly expressing CsChrimson in the left MooSEZ.
Data Availability Statement
The data and code used to generate Figures 1, 2, 3, 4, 5, 6, 7, and S1–S4, Tables S1 and S2, and Video S1. Differential odor-induced backward locomotion, related to Figure 1, Video S2. Backward locomotion following optogenetic activation of GH146II-GAL4, related to Figure 2, Video S3. GH146II-GAL4-induced, MDN-independent, backward locomotion, related to Figure 3, Video S4. GH146II-GAL4 VNC neurons cannot induce backward locomotion, related to Figure 4, Video S5. Blocking expression in VNC neurons does not impair optogenetically induced backward locomotion, related to Figure 4, Video S6. Confocal stack of the GH146II-GAL4 SPARC mosaic fly used for MooSEZ three-dimensional light microscope reconstruction, related to Figure 5, Video S7. Blocking expression in VNC neurons enhances backward locomotion in NP5288-GAL4 flies, related to Figure 6, Video S8. Unilateral activation of MooSEZs triggers ipsilateral backward rotation, related to Figure 7 are available from the corresponding author upon request. The study did not generate any new code or dataset. Any additional information required to reanalyze the data reported in the paper is available from the lead contact upon request.