Abstract
Calorie restriction (CR) and the activation of autophagy extend healthspan by delaying the onset of age-associated diseases in most living organisms. Because protein kinase CK2 (CK2) downregulation induces cellular senescence and nematode aging, we investigated CK2’s role in CR and autophagy. This study indicated that CR upregulated CK2’s expression, thereby causing SIRT1 and AMP-activated protein kinase (AMPK) activation. CK2α overexpression, including antisense inhibitors of miR-186, miR-216b, miR-337-3p, and miR-760, stimulated autophagy initiation and nucleation markers (increase in ATG5, ATG7, LC3BII, beclin-1, and Ulk1, and decrease in SQSTM1/p62). The SIRT1 deacetylase, AKT, mammalian target of rapamycin (mTOR), AMPK, and forkhead homeobox type O (FoxO) 3a were involved in CK2-mediated autophagy. The treatment with the AKT inhibitor triciribine, the AMPK activator AICAR, or the SIRT1 activator resveratrol rescued a reduction in the expression of lgg-1 (the Caenorhabditis elegans ortholog of LC3B), bec-1 (the C. elegans ortholog of beclin-1), and unc-51 (the C. elegans ortholog of Ulk1), mediated by kin-10 (the C. elegans ortholog of CK2β) knockdown in nematodes. Thus, this study indicated that CK2 acted as a positive regulator in CR and autophagy, thereby suggesting that these four miRs’ antisense inhibitors can be used as CR mimetics or autophagy inducers.
Keywords: autophagy, calorie restriction, CK2, SIRT1
INTRODUCTION
Calorie restriction (CR), which consists of chronic reduction in total calorie intake without malnutrition, is the most successful strategy to delay cellular senescence. The CR extends lifespan, improving the healthspan by counteracting the onset of age-associated diseases in most, if not all, living organisms including human (López-Lluch and Navas, 2016; Madeo et al., 2019; Sinclair, 2005; Testa et al., 2014) CR’s hallmark feature is the decrease in cell components’ oxidative damage (Gouspillou and Hepple, 2013; Kowaltowski, 2011). Besides, CR induces macroautophagy (herein referred to as autophagy), which plays an essential role in the beneficial effects exerted by CR (Madeo et al., 2019; Testa et al., 2014). Autophagy’s lysosomal degradation pathway is a conserved cellular recycling process that eliminates damaged organelles and protein aggregates, thereby protecting cellular functionality and integrity. Thus, autophagy activation extends lifespan and/or healthspan in numerous model organisms. In contrast, dysfunctional autophagy can result in advanced aging and quickened the onset of several diseases, including neurodegeneration, cancer, and cardiovascular diseases (Choi et al., 2013; Ravanan et al., 2017). Autophagy is mediated by evolutionarily conserved autophagy-related genes (ATG) (Levine and Kroemer, 2019). LC3BI is converted to its lapidated form, LC3BII, during autophagy, and is incorporated in autophagosomes, which then function as docking sites for adapter proteins, such as SQSTM1/p62 (Hanna et al., 2012; Sahani et al., 2014). Also, various protein kinases and transcription factors are involved in controlling the autophagy apparatus. For example, the transcription factor forkhead homeobox type O (FoxO) 3a plays a vital role in regulating several ATG, including beclin-1 and ulk1 (Mammucari et al., 2007; Zhao et al., 2007). The AMP-activated protein kinase (AMPK) stimulates autophagy as well by activating Ulk1 through direct Ulk1’s phosphorylation (Salminen and Kaarniranta, 2012). In contrast, the high activity of the mammalian target of rapamycin (mTOR) inhibits Ulk1’s activation by phosphorylating Ulk1 (Heras-Sandoval et al., 2014).
Cellular senescence is proliferation’s terminal arrest triggered by various cellular stresses such as telomere shortening, oncogenic activation, and oxidative stress. Although senescence has been regarded as an effective cancer suppression mechanism, it is also involved in some pathological conditions, such as aging, age-associated diseases, and tumorigenesis (Childs et al., 2015; McHugh and Gil, 2018). Protein kinase CK2 (CK2) is a serine/threonine kinase that catalyzes the phosphorylation of several nuclear and cytoplasmic proteins. Furthermore, the CK2’s holoenzyme is a heterotetramer composed of two catalytic (α and/or α’) subunits and two regulatory β subunits, where the β subunit stimulates the α or α’ subunit’s catalytic activity, thereby mediating tetramer formation and substrate recognition (Litchfield, 2003). Our previous study identified CK2 as a senescence regulator. CK2 down-regulation induces several senescence markers, including senescence-associated β-galactosidase activity (Ryu et al., 2006), the p53-p21Cip1/WAF1 axis’ activation (Kang et al., 2009), senescence-associated heterochromatin foci (SAHF) formation (Park et al., 2018), and senescence-associated secretory phenotype (SASP) expression (Song and Bae, 2021). The miR-186, miR-216b, miR-337-3p, and miR-760 promote cellular senescence by inhibiting CK2α (Kim et al., 2012; Lee et al., 2014). Besides, knockdown of kin-10—the Caenorhabditis elegans ortholog of CK2β—led to a short lifespan phenotype and induced age-related biomarkers in nematodes (Park et al., 2017).
Therefore, because CR and autophagy are broadly believed to suppress cellular senescence, we examined CK2’s role in CR and autophagy. Our results indicated that CK2 was upregulated in CR conditions and that CK2’s downregulation inhibited autophagy through AMPK, AKT, mTOR, SIRT1, and FoxO3a. We also established that AKT, AMPK, and SIRT1 was associated with decreased autophagy markers, mediated by CK2 downregulation in nematodes. Moreover, findings established the antisense inhibitors of miR-186, miR-216b, miR-337-3p, and miR-760, which upregulated CK2, as autophagy inducers and putative CR mimetics.
MATERIALS AND METHODS
Chemicals, siRNA, and antibodies
We obtained triciribine, rapamycin, AICAR, resveratrol, spermidine, and antibody against SQSTM1/p62 from Sigma-Aldrich (USA). We also purchased antibodies against CK2α, CK2β, β-actin, SIRT1, and siRNA for SIRT1 (sc-40986) from Santa Cruz Biotechnology (USA). We obtained antibodies against LC3, Atg5, Atg7, Atg5 (D5F5U), AMPK, phospho-AMPK (T172), FoxO3a, LKB1, and phospho-LKB1 (S428), from Cell Signaling Technology (USA).
Cell culture and CR
We cultured HCT116 human colon cancer cells and MCF-7 human breast cancer cells (ATCC, USA) in Dulbecco’s modified Eagle medium (Thermo Fisher Scientific, USA), with 25 mM glucose and all amino acids, containing 10% (v/v) fetal bovine serum under a humidified atmosphere of 5% (v/v) CO2 at 37°C. For cell’s starvation, we washed the cells with phosphate-buffered saline twice and then cultured them in an Earle’s balanced salt solution (Thermo Fisher Scientific), with 5.5 mM glucose without amino acid, according to the indicated time.
RNA and DNA transfection
We transfected the pcDNA-HA-CK2α, pECE-Flag-SIRT1, pECE-Flag-SIRT1 (H363Y), and pECE-HA-FoxO3a into cells using Polyfect (Qiagen, Germany) as per the manufacturer’s instructions. We purchased the mimics for miR-186, miR-216b, miR-337-3p, miR-760, and the control miRNA from Genolution (Korea). We obtained the antisense inhibitors for miR-186, miR-216b, miR-337-3p, and miR-760 from Panagene (Korea). The CK2α siRNA’s sequence was 5′-UCAAGAUGACUACCAGCUGdTdT-3′, whereas, that of the negative control was 5′-GCUCAGAUCAAUACGGAGAdTdT-3′. We transfected the RNAs into cells using Lipofectamine (Invitrogen, USA) for 48 h.
Immunoblotting
We separated proteins on 12% or 15% polyacrylamide gels in the presence of sodium dodecyl sulfate (SDS), and then transferred them onto nitrocellulose membranes. We subsequently blocked the membranes with 5% (w/v) non-fat, dried skim milk in TBST (20 mM Tris-HCl [pH 7.4], 150 mM NaCl, and 0.05% Tween 20) for 2 h and then incubated them with specific antibodies in 1% (w/v) non-fat, dried skim milk for 1 h. Next, we washed the membranes three times with TBST, and then treated them with the ECL system (GE Healthcare, UK) to develop the signals. When deemed necessary, we stripped the membranes with a stripping buffer (2% SDS, 100 mM β-mercaptoethanol, and 50 mM Tris-HCl [pH 7.0]) at 50°C for 1 h with gentle shaking, after which we re-probed them with an anti-β-actin antibody as an internal loading control.
Reverse transcription polymerase chain reaction
We extracted total RNA from human cancer cells or nematodes. We reverse transcribed the RNA using gene-specific primers and reverse transcriptase (Takara Bio, Japan), after which we polymerase chain reaction (PCR)-amplified the resulting cDNA. We have listed the primers we used in our assays in Supplementary Table S1. We subsequently resolved the PCR products on a 1.5% agarose gel. Next, we performed reverse transcription PCR (RT-PCR) bands’ quantification using densitometry. Primers for β-actin (human cells) and act-2 (nematodes) RNA were used to standardize the amounts of RNA in each sample.
Culture of C. elegans and RNAi experiment
We acquired the C. elegans DA2123 [lgg-1p::gfp::lgg-1+rol-6(su1006)] strain and N2 (wild-type) strain from the Caenorhabditis Genetics Center (CGC, USA). We grew the nematodes at 21°C on nematode growth medium (NGM) agar plates with Escherichia coli strain OP50 as a food source. E. coli HT115 cells expressing double-stranded kin-10 RNA were obtained from the C. elegans ORFeome RNAi library. We used the E. coli HT115 strain carrying the empty L4440 vector as control RNAi. We fed the nematodes with kin-10 RNAi at the L4 larval stage for one day to deactivate the kin-10 function. For some experiments, we treated the nematodes with AICAR, rapamycin, resveratrol, or spermidine, respectively.
Reporter gene assay
We transferred the synchronized (L4 larva) reporter nematodes [lgg-1p::gfp::lgg-1+rol-6(su1006)] to the HT115-seeded NGM plates. After one day, we measured the fluorescence of green fluorescent protein (GFP) using a fluorescence microscope (Zeiss AxioCam MRc; Zeiss, Germany) at excitation and emission wavelengths of 490 nm, respectively. After that, we quantified the relative fluorescence intensity of the setup using ImageJ software to determine the GFP levels. We conducted three independent experiments.
Statistical analysis
We analyzed our data with one-way ANOVA using the IBM SPSS Statistics (ver. 24.0; IBM, USA). We considered the results significant when the P value was <0.05. We also used Duncan’s multiple-range test to identify the differences between the groups as α = 0.05.
RESULTS
CK2 was upregulated, and induced SIRT1 and AMPK activation during CR in human cancer cells
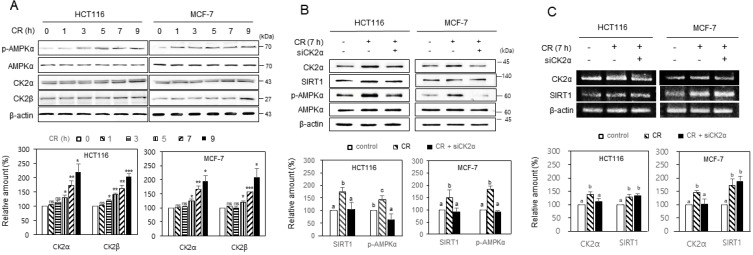
From previous studies because CK2 downregulation induced cellular senescence and nematode aging (Park et al., 2017; Ryu et al., 2006), and the CR provides an anti-aging effect (López-Lluch and Navas, 2016; Madeo et al., 2019; Sinclair, 2005; Testa et al., 2014), we supposed that CK2 would be upregulated under CR conditions. Therefore, we incubated HCT116 and MCF-7 cells in CR conditions to test this hypothesis. Western blot analysis showed that CK2α’s and CK2β’s protein levels increased under the CR condition than the normal condition (Fig. 1A). Furthermore, consistent with previous reports (Cohen et al., 2004; Salminen and Kaarniranta, 2012), the CR condition elevated p-AMPK (T172) and SIRT1 protein levels (Fig. 1B). Thus, to determine the relationship between CK2, SIRT1, and AMPK under CR conditions, we treated cells with CK2α siRNA and incubated them under CR conditions. We observed that treatment with CK2α siRNA completely abolished the CR-induced increase of SIRT1 and p-AMPK, thereby indicating CK2 as an upstream regulator of SIRT1 and AMPK in CR conditions (Fig. 1B). We then examined whether the CR-mediated increase in the protein levels of CK2α and SIRT1 correlated with an increase at the transcriptional level. RT-PCR indicated that CR upregulated the mRNA levels of CK2α and SIRT1 (Fig. 1C, Supplementary Fig. S1A). The knockdown of CK2α did not suppress the CR-mediated increase in the mRNA levels of SIRT1 (Fig. 1C). Furthermore, CK2α overexpression did not change the mRNA level of SIRT1 as well (Supplementary Fig. S1B). Taken together, the data suggested that CR upregulated CK2α and SIRT1 at the transcription level, whereas, CK2 upregulated SIRT1 at the post-transcription level under CR conditions.
Fig. 1. CK2 upregulated and induced SIRT1 and AMPK activation during calorie restriction in human cancer cells.
(A) HCT116 and MCF-7 cells were incubated under CR conditions for the indicated time. (B and C) Cells were transfected with CK2α siRNA for 1.5 days, then incubated under CR conditions for 7 h. The levels of each protein (A and B) and mRNA (C) were then determined by immunoblotting using specific antibodies, and by RT-PCR analysis using specific primers, respectively (top). Representative data from three independent experiments are shown. β-Actin was used as a control. The graphs represent the quantitation of CK2α, CK2β, and SIRT1 relative to β-actin, and p-AMPK (T172) relative to the unphosphorylated AMPK (bottom). Data were reported as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001. Bars that do not have the same letter (a, b, c) were significantly different among groups at P < 0.05.
CK2 positively regulated autophagy in human cancer cells
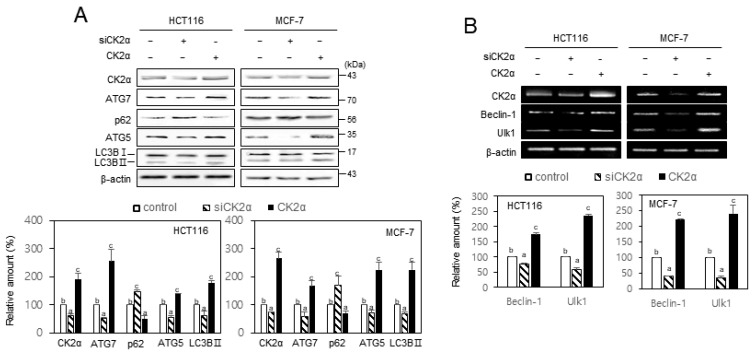
Given that CR is a key inducer of autophagy (López-Lluch and Navas, 2016; Madeo et al., 2019; Sinclair, 2005; Testa et al., 2014), we investigated CK2’s role in autophagy in HCT116 and MCF-7 cells. Western blotting and RT-PCR indicated that treatment with CK2α siRNA reduced several autophagy markers’ expression, including the reduction of ATG5, ATG7, LC3BII, beclin-1, and Ulk1, and the induction of SQSTM1/p62. In contrast, CK2α’s overexpression upregulated the expression of ATG5, ATG7, LC3BII, beclin-1, and Ulk1, but decreased the level of SQSTM1/p62 (Fig. 2). Moreover, additional treatment with CK2α siRNA after CR abrogated both the induction of ATG5, ATG7, LC3BII, beclin-1, and Ulk1 and the reduction of SQSTM1/p62, caused by CR (Supplementary Fig. S2). These results suggested that CK2 stimulated autophagy in HCT116 and MCF-7 cells. We have previously shown that miR-186, miR-216b, miR-337-3p, and miR-760 promoted cellular senescence by targeting CK2α, whereas, these four miRNAs’ antisense inhibitors increased CK2α expression and suppressed CK2 inhibition-mediated senescence (Kim et al., 2012; Lee et al., 2014). Thus, we investigated the effects of these four miRs and antisense inhibitors (4 miRi) on autophagy. Western blotting and RT-PCR indicated that treatment with the four miRs decreased autophagy, whereas treatment with the four miR inhibitors (4 miRi) resulted in increased autophagy (Supplementary Fig. S3). Taken together, these results indicated that CK2 positively regulated autophagy in human cancer cells, and suggested that these antisense inhibitors can be used as autophagy inducers and putative CR mimetics.
Fig. 2. CK2 positively regulated autophagy in human cancer cells.
HCT116 and MCF-7 cells were transfected with CK2α siRNA or pcDNA3.1-HA-CK2α for 2 days. (A) Immunoblotting was then used to determine the level of each protein using specific antibodies (top). (B) The level of each mRNA was determined by RT-PCR using specific primers (top). Representative data from three independent experiments are shown. β-Actin was used as a control. The graphs represent quantitation of each protein and mRNA relative to β-actin (bottom). Data were reported as mean ± SEM. Bars that do not have the same letter (a, b, c) were significantly different among groups at P < 0.05.
CK2 regulated autophagy via mTOR and SIRT1 in human cancer cells
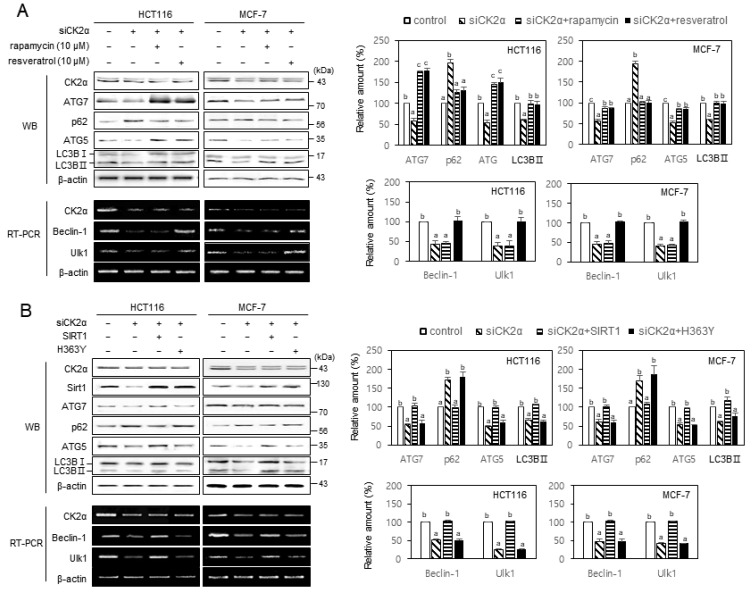
We previously showed that mTOR and SIRT1 are involved in CK2 downregulation-mediated senescence (Ham et al., 2019; Jang et al., 2011; Park et al., 2013). Therefore, in this study, we treated the cells with CK2α siRNA in the presence of the mTOR inhibitor rapamycin (10 μM) or the SIRT1 activator resveratrol (10 μM) to examine the role of mTOR and SIRT1 in autophagy, mediated by CK2. Rapamycin treatment suppressed both the reduction of ATG5, ATG7, and LC3BII and induction of SQSTM1/p62 caused by CK2 downregulation, maintaining the beclin-1’s and Ulk1’s levels unchanged. However, treatment with resveratrol successfully suppressed both the reduction of ATG5, ATG7, LC3BII, beclin-1, and Ulk1 and the induction of SQSTM1/p62 caused by CK2 downregulation (Fig. 3A). Furthermore, we transfected cells with pECE-Flag-SIRT1 or pECE-Flag-SIRT1 (H363Y) in the presence of CK2α siRNA to further confirm SIRT1’s impact on reduction of autophagy caused by CK2 downregulation. The overexpression of SIRT1’s wild-type, but not that of the catalytically inactive mutant of SIRT1 (H363Y), successfully abrogated both the reduction of ATG5, ATG7, LC3BII, beclin-1, and Ulk1 and the induction of SQSTM1/p62, all caused by CK2 downregulation (Fig. 3B). Taken together, these data suggested that SIRT1 is an upstream regulator of ATG5, ATG7, LC3BII, SQSTM1/p62, beclin-1, and ULK1 in CK2-mediated autophagy. Moreover, mTOR is an upstream regulator of ATG5, ATG7, LC3BII, and SQSTM1/p62, but not the transcription of beclin-1 and Ulk1 genes.
Fig. 3. CK2 positively regulated autophagy via mTOR and SIRT1 in human cancer cells.
(A) HCT116 and MCF-7 cells were transfected with CK2α siRNA for 2 days in the absence or presence of 10 μM rapamycin or 10 μM resveratrol. The levels of each protein (left-top) and mRNA (left-bottom) were determined by immunoblotting using specific antibodies and by RT-PCR using specific primers, respectively. Representative data from three independent experiments are shown. β-Actin was used as a control. The graphs represent the quantitation of each protein (right-top) and mRNA (right-bottom) relative to β-actin. WB, Western blotting. (B) Cells were transfected with CK2α siRNA for 2 days in the absence or presence of pECE-flag-SIRT1 or pECE-flag-SIRT1 mutant (H363Y). The levels of each protein (left-top) and mRNA (left-bottom) were determined by immunoblotting using specific antibodies and by RT-PCR using specific primers, respectively. Representative data from three independent experiments are shown. β-Actin was used as a control. The graphs represent the quantitation of each protein (right-top) and mRNA (right-bottom) relative to β-actin. Data are reported as mean ± SEM. Bars that do not have the same letter (a, b, c) were significantly different among groups at P < 0.05.
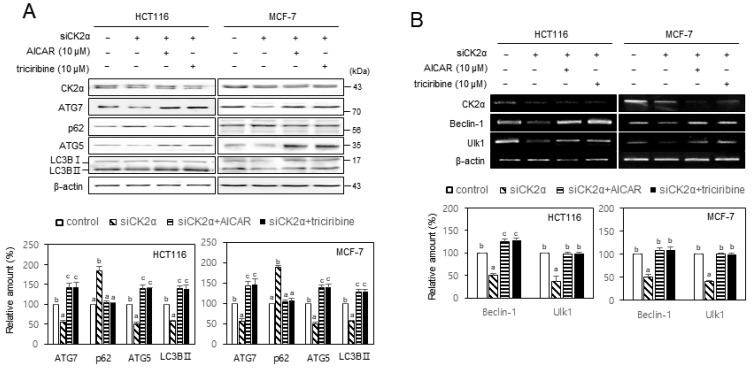
CK2 regulated autophagy via AMPK and AKT in human cancer cells
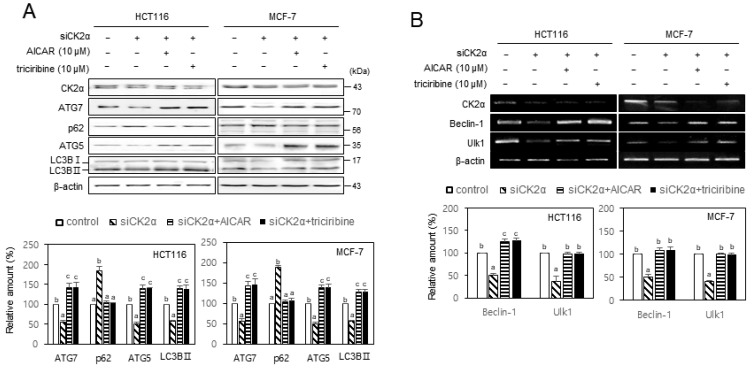
Given that AKT and AMPK are involved in CK2 downregulation-mediated senescence (Jang et al., 2020; Park et al., 2013), we investigated the role of AKT and AMPK in CK2-induced autophagy. We treated HCT116 and MCF-7 cells with CK2α siRNA in the presence of the AMPK activator AICAR (10 μM) or the AKT inhibitor triciribine (10 μM) to examine AKT’s and AMPK’s role in autophagy decline as mediated by CK2 downregulation. As shown in Fig. 4, treatment with AICAR or triciribine successfully impeded both the reduction of ATG5, ATG7, LC3BII, beclin-1, and Ulk1 and the induction of SQSTM1/p62, caused by CK2 downregulation, indicating that AMPK and AKT were upstream regulators of ATG5, ATG7, LC3BII, SQSTM1/p62, beclin-1, and Ulk1 in CK2-mediated autophagy. Furthermore, because LKB1 is a well-known AMPK upstream regulator (Mirouse and Billaud, 2011), we transfected cells with CK2α cDNA or CK2α siRNA to establish LKB1’s role in CK2-induced AMPK activation. CK2 overexpression elevated the levels of p-LKB1 (S428) and p-AMPK (T172) compared with control cells, whereas CK2 downregulation reduced the levels of p-LKB1 and p-AMPK (Supplementary Fig. S4A). It has been shown that SIRT1 activated LKB1 via deacetylation of Lys 48 within LKB1 (Lan et al., 2008). Therefore, additional treatment with SIRT1 siRNA after CK2 overexpression attenuated the increase in the levels of p-LKB1 and p-AMPK (T172), mediated by CK2 overexpression (Supplementary Fig. S4B). Taken together, these results suggested that CK2 activated autophagy through AKT and SIRT1–LKB1–AMPK pathways in human cancer cells.
Fig. 4. CK2 positively regulated autophagy via AMPK and AKT pathways in human cancer cells.
HCT116 and MCF-7 cells were transfected with CK2α siRNA for 2 days in the absence or presence of 10 μM AICAR or 10 μM triciribine. (A) The level of each protein was determined by immunoblotting using specific antibodies (top). (B) The level of each mRNA was determined by RT-PCR using specific primers (top). Representative data from three independent experiments are shown. β-Actin was used as a control. The graphs represent the quantitation of each protein and mRNA relative to β-actin (bottom). Data are reported as mean ± SEM. Bars that do not have the same letter (a, b, c) were significantly different among groups at P < 0.05.
CK2 regulated autophagy via FoxO3a in human cancer cells
Transcriptional activity of FOXO3a is also involved in CK2 downregulation-mediated senescence (Ham et al., 2019; Park and Bae, 2016). Therefore, we treated cells with CK2α siRNA in the presence of FoxO3a cDNA to examine FoxO3a’s role in autophagy, mediated by CK2. FoxO3a’s overexpression suppressed both the reduction of ATG5, ATG7, LC3BII, beclin-1, and Ulk1, and the induction of SQSTM1/p62, caused by CK2 downregulation (Fig. 5). These data suggested that FoxO3a is an upstream regulator of ATG5, ATG7, LC3BII, p62, beclin-1, and ULK1 during CK2-mediated autophagy.
Fig. 5. Overexpression of FoxO3a rescued CK2 downregulation-mediated autophagy inhibition in human cancer cells.
HCT116 and MCF-7 cells were transfected with CK2α siRNA for 2 days in the absence or presence of pECE-HA-FoxO3a. (A) The level of each protein was determined by immunoblotting using specific antibodies (top). (B) The level of each mRNA was determined by RT-PCR using specific primers (top). Representative data from three independent experiments are shown. β-Actin was used as a control. The graphs represent the quantitation of each protein and mRNA relative to β-actin (bottom). Data are reported as mean ± SEM. Bars that do not have the same letter (a, b, c) were significantly different among groups at P < 0.05.
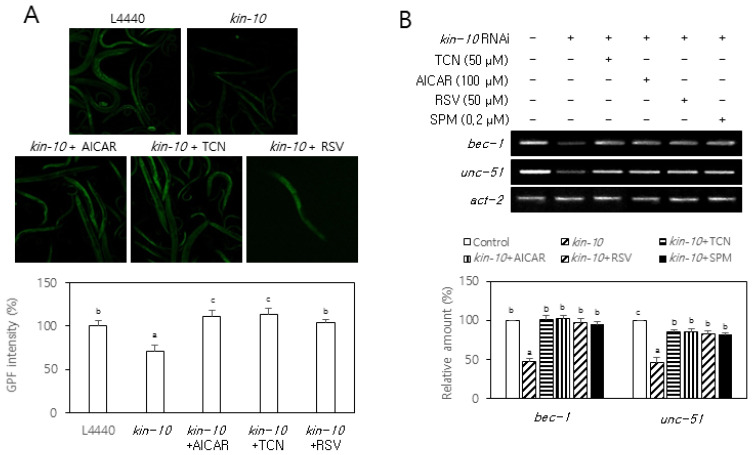
Kin-10 RNAi decreased autophagy markers in nematodes
We have previously shown that kin-10 (the C. elegans ortholog of CK2β) knockdown results in reduced longevity and onset of aging-related biomarkers via the AGE-1/PI3K–AKT-1/AKT–DAF-16/FoxO pathway in nematodes (Park et al., 2017). Therefore, to examine whether CK2 regulates LGG-1 (the C. elegans ortholog of LC3B) in C. elegans, we measured the fluorescence of a reporter worms that were specific for LGG-1. The kin-10 RNAi decreased the fluorescence of lgg-1::gfp from the reporter worms compared with the control RNAi (L4440). The treatment with triciribine (50 μM), AICAR (100 μM), and resveratrol (50 μM), also rescued the fluorescence reduction of the lgg-1::gfp reporter mediated by kin-10 knockdown (Fig. 6A). Moreover, kin-10 knockdown reduced the mRNA levels of bec-1 (the C. elegans ortholog of beclin-1) and unc-51 (the C. elegans ortholog of Ulk1) in nematodes compared with the control RNAi. Additionally, treatment with triciribine (50 μM), AICAR (100 μM), and resveratrol (50 μM), and the putative CK2 activator spermidine (0.2 μM) significantly abrogated the reduction of these mRNA levels mediated by kin-10 knockdown (Fig. 6B). These results collectively suggest that CK2 can positively regulate autophagy via AKT, AMPK, and SIRT1 in C. elegans.
Fig. 6. Kin-10 RNAi decreased autophagy markers in nematodes.
(A) Synchronized (L4 larva) reporter nematodes lgg-1::gfp were fed on control RNAi or kin-10 RNAi plates containing AICAR (100 μM), triciribin (TCN, 0.2 μM), or resveratrol (RSV, 50 μM) under standard conditions. Representative fluorescence images at day 1 of adulthood were obtained at 10× magnification (top). The fluorescence intensity of GFP was quantified using ImageJ software by determining the average pixel intensity (n = 30 per condition). (B) Synchronized (L4 larva) nematodes were fed on control RNAi or kin-10 RNAi plates containing AICAR (100 μM), triciribine (TCN, 0.2 μM), resveratrol (RSV, 50 μM), or spermine (SPM, 0.2 μM) under standard conditions. Subsequently, lysates from nematodes were used in RT-PCR using specific primers. The level of each mRNA was determined by RT-PCR (top). Representative data from the three independent experiments are shown. Act-2 was used as a control. The graphs represent the quantitation of each protein and mRNA relative to act-2 (bottom). Data are reported as mean ± SEM. Bars that do not have the same letter (a, b, c) were significantly different among groups at P < 0.05.
DISCUSSION
Aging is associated with an increase in reactive oxygen species (ROS) generation and a decreased antioxidant defense system (Luo et al., 2020). CR extends longevity in several organisms from yeasts to mammals (López-Lluch and Navas, 2016; Madeo et al., 2019; Sinclair, 2005; Testa et al., 2014), and a CR’s hallmark feature is a decrease in oxidative damage of cell components (Gouspillou and Hepple, 2013; Kowaltowski, 2011). We previously showed as well that CK2 downregulation stimulated ROS accumulation through both superoxide anion generation in the cytoplasm by NADPH oxidase activation (Jeon et al., 2010) and antioxidant reduction by inhibiting transcription factors, such as FoxO3a and Nrf2 via AKT, SIRT1, and AMPK (Ham et al., 2019; Jang et al., 2020; Park and Bae, 2016; Park et al., 2013). The present study clearly shows that CR upregulates CK2 (Fig. 1, Supplementary Fig. S1). Thus, our previous and present studies collectively suggested that CR reduced intracellular ROS, at least in part, through CK2 activation. Furthermore, CR promoted AMPK activation by increasing the cellular AMP/ATP ratio (Salminen and Kaarniranta, 2012). However, the present study suggested a novel mechanism for CR-mediated AMPK activation, because CK2 induced AMPK activation under CR conditions by activating the SIRT1–LKB1 pathway (Fig. 1, Supplementary Figs. S1 and S4). This study also importantly indicated that CR upregulated CK2 and SIRT1 at the transcription level, whereas CK2 upregulated SIRT1 at the post-transcription level (Fig. 1, Supplementary Fig. S1). This study is consistent with a previous report demonstrating that CR increased the mRNA level of SIRT1 (Yu et al., 2014). Despite our findings, we were curious how CR upregulated CK2α’s mRNA levels. Although at present, we were unable to precisely explain CK2α’s regulatory mechanism under CR conditions, long noncoding RNAs were proposed to be involved in CR-mediated CK2α upregulation (Y.S. Lee and Y.S. Bae, unpublished data). Also how does CK2 upregulate SIRT1 at the post-transcription level? Our previous studies have shown that CK2 increased both the deacetylase activity of SIRT1 by CK2 phosphorylation (Jang et al., 2011) and the protein levels of SIRT1 through inhibiting proteasome-dependent degradation (Ham et al., 2019).
Another CR’s hallmark feature is autophagy induction (López-Lluch and Navas, 2016; Madeo et al., 2019; Sinclair, 2005; Testa et al., 2014). Autophagy suppresses senescence through homeostatic action, such as eliminating damaged cellular components. Additionally, autophagy occurs in sequential steps, including initiation (membrane isolation), nucleation, autophagosome formation, cargo selection, autophagolysosome formation, and degradation (Choi et al., 2013; Ravanan et al., 2017). Furthermore, the ULK1, beclin-1, ATG5, ATG7, LC3BII, and SQSTM1/p62 play a significant role in autophagy initiation and nucleation (Hanna et al., 2012; Levine and Kroemer, 2019; Sahani et al., 2014). The present study indicated that CK2 downregulation by CK2α siRNA and four miRs (miR-186, miR-216b, miR-337-3p, and miR-760) inhibited autophagy in human cancer cells. Consistently, CK2α overexpression and the antisense inhibitors of the four miRs stimulated autophagy (Fig. 2, Supplementary Figs. S2 and S3). SIRT1, AKT, mTOR, AMPK and FoxO3a were established as upstream regulators of ATG5, ATG7, LC3BII, and SQSTM1/p62 during CK2-mediated autophagy (Figs. 3-5). Moreover, we demonstrated here that SIRT1, AKT, AMPK, and FoxO3a affects (directly or indirectly) the transcription of beclin-1 and ulk1 genes during CK2-mediated autophagy (Figs. 3-5). In contrast, mTOR was uninvolved in the transcription of beclin-1 and ulk1 genes during CK2-mediated autophagy (Fig. 3). Consistent with these data, it has been reported that mTORC1 acts as an autophagy’s negative regulator by phosphorylating Ulk1 (Heras-Sandoval et al., 2014). Finally, treatment with triciribine, AICAR, and resveratrol rescued the reduction of both the fluorescence of the lgg-1::gfp reporter and mRNA levels of bec-1 and unc-51, mediated by kin-10 knockdown in C. elegans. These results proposed that CK2 also positively regulated autophagy via AKT, AMPK, and SIRT1 in nematodes (Fig. 6). Thus, judging by the mechanism by which ULK1, becli-1, ATG5, ATG7, LC3BII, and SQSTM1/p62 function, this study suggested that CK2 promoted autophagy initiation (membrane isolation) and nucleation stages before the autophagosome formation. We have also previously shown that CK2 downregulation activated the AKT–mTOR pathway through PI3K (Park et al., 2013) or SIRT1 (Song and Bae, 2021) and inhibited the SIRT1–FoxO3a pathway (Ham et al., 2019) during senescence. Therefore, our previous and present studies propose that CK2, induced by CR, stimulates autophagy initiation and nucleation by inhibiting the AKT–mTOR pathway, and activating the SIRT1–FoxO3a and SIRT1–LKB1–AMPK pathways.
A reduced proteolytic activity has been considered responsible for accumulating damaged macromolecules and organelles in most aged organisms’ tissues (Cuervo, 2008). Thus, autophagy was initially thought to inhibit cellular senescence by removing damaged cellular components (an anti-senescence mechanism). However, recent studies suggested that autophagy also stimulates cellular senescence by facilitating senescence-associated secretory proteins (a pro-senescence mechanism) (Kwon et al., 2017). Similarly, our previous and present studies indicate that CK2 suppresses cellular senescence and promoted autophagy. Therefore, we suggested that CK2 was involved in autophagy’s anti-senescence function, but not in autophagy’s pro-senescence function. It was also shown that CR mimetics mimicked the biochemical effects of nutrient deprivation, thereby triggering autophagy (Madeo et al., 2019; Testa et al., 2014). Therefore, because many human diseases are associated with cellular senescence, CK2 activators, including the antisense inhibitors of miR-186, miR-216b, miR-337-3p, and miR-760, are proposed to provide a strategy for anti-aging interventions, such as CR mimetics and autophagy inducers.
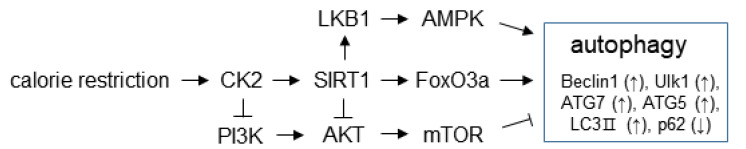
Our experimental data can be summarized as shown in Fig. 7. CR activates CK2, which activates SIRT1. Subsequently, SIRT1 stimulates both the LKB1–AMPK pathway and FoxO3a, which are positive regulators of the autophagy initiation and nucleation stages. Additionally, CK2 inhibits the AKT–mTOR pathway, which is a negative regulator of autophagy, through PI3K or SIRT1. Thus, CR induces autophagy, at least in part, via CK2 activation.
Fig. 7. Schematic of the mechanism proposed for the calorie restriction-mediated autophagy induction.
Supplemental Materials
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
ACKNOWLEDGMENTS
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and Future Planning (NRF-2019R1A2C1005219).
Footnotes
AUTHOR CONTRIBUTIONS
Y.S.B. conceived and designed the experiments. J.W.P. and J.J. performed the experiments. Y.S.B. wrote the paper. All authors have read and agreed to the published version of the manuscript.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Childs B.G., Durik M., Baker D.J., van Deursen J.M. Cellular senescence in aging and age-related disease: from mechanisms to therapy. Nat. Med. 2015;21:1424–1435. doi: 10.1038/nm.4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi A.M., Ryter S.W., Levine B. Autophagy in human health and disease. N. Engl. J. Med. 2013;368:651–662. doi: 10.1056/NEJMra1205406. [DOI] [PubMed] [Google Scholar]
- Cohen H.Y., Miller C., Bitterman K.J., Wall N.R., Hekking B., Kessler B., Howitz K.T., Gorospe M., de Cabo R., Sinclair D.A. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- Cuervo A.M. Autophagy and aging: keeping that old broom working. Trends Genet. 2008;24:604–612. doi: 10.1016/j.tig.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouspillou G., Hepple R.T. Facts and controversies in our understanding of how caloric restriction impacts the mitochondrion. Exp. Gerontol. 2013;48:1075–1084. doi: 10.1016/j.exger.2013.03.004. [DOI] [PubMed] [Google Scholar]
- Ham H.J., Park J.W., Bae Y.S. Defect of SIRT1-FoxO3a axis is associated with the production of reactive oxygen species during protein kinase CK2 downregulation-mediated cellular senescence and nematode aging. BMB Rep. 2019;52:265–270. doi: 10.5483/BMBRep.2019.52.4.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna R.A., Quinsay M.N., Orogo A.M., Giang K., Rikka S., Gustafsson Å.B. Microtubule-associated protein 1 light chain 3 (LC3) interacts with Bnip3 protein to selectively remove endoplasmic reticulum and mitochondria via autophagy. J. Biol. Chem. 2012;287:19094–19104. doi: 10.1074/jbc.M111.322933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heras-Sandoval D., Pérez-Rojas J.M., Hernández-Damián J., Pedraza-Chaverri J. The role of PI3K/AKT/mTOR pathway in the modulation of autophagy and the clearance of protein aggregates in neurodegeneration. Cell. Signal. 2014;26:2694–2701. doi: 10.1016/j.cellsig.2014.08.019. [DOI] [PubMed] [Google Scholar]
- Jang D.E., Song J., Park J.W., Yoon S.H., Bae Y.S. Protein kinase CK2 activates Nrf2 via autophagic degradation of Keap1 and activation of AMPK in human cancer cells. BMB Rep. 2020;53:272–277. doi: 10.5483/BMBRep.2020.53.5.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S.Y., Kim S.Y., Bae Y.S. p53 deacetylation by SIRT1 decreases during protein kinase CK2 downregulation-mediated cellular senescence. FEBS Lett. 2011;585:3360–3366. doi: 10.1016/j.febslet.2011.09.027. [DOI] [PubMed] [Google Scholar]
- Jeon S.M., Lee S.J., Kwon T.K., Kim K.J., Bae Y.S. NADPH oxidase is involved in protein kinase CKII down-regulation-mediated senescence through elevation of the level of reactive oxygen species in human colon cancer cells. FEBS Lett. 2010;584:3137–3142. doi: 10.1016/j.febslet.2010.05.054. [DOI] [PubMed] [Google Scholar]
- Kang J.Y., Kim J.J., Jang S.Y., Bae Y.S. The p53-p21Cip1/WAF1 pathway is necessary for cellular senescence induced by the inhibition of protein kinase CKII in human colon cancer cells. Mol. Cells. 2009;28:489–494. doi: 10.1007/s10059-009-0141-9. [DOI] [PubMed] [Google Scholar]
- Kim S.Y., Lee Y.H., Bae Y.S. miR-186, miR-216b, miR-337-3p, and miR-760 cooperatively induce cellular senescence by targeting α subunit of protein kinase CKII in human colorectal cancer cells. Biochem. Biophys. Res. Commun. 2012;429:173–179. doi: 10.1016/j.bbrc.2012.10.117. [DOI] [PubMed] [Google Scholar]
- Kowaltowski A.J. Caloric restriction and redox state: does this diet increase or decrease oxidant production? Redox Rep. 2011;16:237–241. doi: 10.1179/1351000211Y.0000000014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon Y., Kim J.W., Jeoung J.A., Kim M.S., Kang C. Autophagy is pro-senescence when seen in close-up, but anti-senescence in long-shot. Mol. Cells. 2017;40:607–612. doi: 10.14348/molcells.2017.0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan F., Cacicedo J.M., Ruderman N., Ido Y. SIRT1 modulation of the acetylation status, cytosolic localization, and activity of LKB1. Possible role in AMP-activated protein kinase activation. J. Biol. Chem. 2008;283:27628–27635. doi: 10.1074/jbc.M805711200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.H., Kim S.Y., Bae Y.S. Upregulation of miR-760 and miR-186 is associated with replicative senescence in human lung fibroblast cells. Mol. Cells. 2014;37:620–627. doi: 10.14348/molcells.2014.0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Kroemer G. Biological functions of autophagy genes: a disease perspective. Cell. 2019;176:11–42. doi: 10.1016/j.cell.2018.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litchfield D.W. Protein kinase CK2: structure, regulation and role in cellular decisions of life and death. Biochem. J. 2003;369:1–15. doi: 10.1042/bj20021469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Lluch G., Navas P. Calorie restriction as an intervention in ageing. J. Physiol. 2016;594:2043–2060. doi: 10.1113/JP270543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J., Mills K., le Cessie S., Noordam R., van Heemst D. Ageing, age-related diseases and oxidative stress: what to do next? Ageing Res. Rev. 2020;57:100982. doi: 10.1016/j.arr.2019.100982. [DOI] [PubMed] [Google Scholar]
- Madeo F., Carmona-Gutierrez D., Hofer S.J., Kroemer G. Caloric restriction mimetics against age-associated disease: targets, mechanisms, and therapeutic potential. Cell Metab. 2019;29:592–610. doi: 10.1016/j.cmet.2019.01.018. [DOI] [PubMed] [Google Scholar]
- Mammucari C., Milan G., Romanello V., Masiero E., Rudolf R., Del Piccolo P., Burden S.J., Di Lisi R., Sandri C., Zhao J., et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- McHugh D., Gil J. Senescence and aging: causes, consequences, and therapeutic avenues. J. Cell Biol. 2018;217:65–77. doi: 10.1083/jcb.201708092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirouse V., Billaud M. The LKB1/AMPK polarity pathway. FEBS Lett. 2011;585:981–985. doi: 10.1016/j.febslet.2010.12.025. [DOI] [PubMed] [Google Scholar]
- Park J.H., Kim J.J., Bae Y.S. Involvement of PI3K-AKT-mTOR pathway in protein kinase CKII inhibition-mediated senescence in human colon cancer cells. Biochem. Biophys. Res. Commun. 2013;433:420–425. doi: 10.1016/j.bbrc.2013.02.108. [DOI] [PubMed] [Google Scholar]
- Park J.H., Lee J.H., Park J.W., Kim D.Y., Hahm J.H., Nam H.G., Bae Y.S. Downregulation of protein kinase CK2 activity induces age-related biomarkers in C. elegans. Oncotarget. 2017;8:36950–36963. doi: 10.18632/oncotarget.16939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.W., Kim J.J., Bae Y.S. CK2 downregulation induces senescence-associated heterochromatic foci formation through activating SUV39h1 and inactivating G9a. Biochem. Biophys. Res. Commun. 2018;505:67–73. doi: 10.1016/j.bbrc.2018.09.099. [DOI] [PubMed] [Google Scholar]
- Park S.Y., Bae Y.S. Inactivation of the FoxO3a transcription factor is associated with the production of reactive oxygen species during protein kinase CK2 downregulation-mediated senescence in human colon cancer and breast cancer cells. Biochem. Biophys. Res. Commun. 2016;478:18–24. doi: 10.1016/j.bbrc.2016.07.106. [DOI] [PubMed] [Google Scholar]
- Ravanan P., Srikumar I.F., Talwar P. Autophagy: the spotlight for cellular stress responses. Life Sci. 2017;188:53–67. doi: 10.1016/j.lfs.2017.08.029. [DOI] [PubMed] [Google Scholar]
- Ryu S.W., Woo J.H., Kim Y.H., Lee Y.S., Park J.W., Bae Y.S. Downregulation of protein kinase CKII is associated with cellular senescence. FEBS Lett. 2006;580:988–994. doi: 10.1016/j.febslet.2006.01.028. [DOI] [PubMed] [Google Scholar]
- Sahani M.H., Itakura E., Mizushima N. Expression of the autophagy substrate SQSTM1/p62 is restored during prolonged starvation depending on transcriptional upregulation and autophagy-derived amino acids. Autophagy. 2014;10:431–441. doi: 10.4161/auto.27344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen A., Kaarniranta K. AMP-activated protein kinase (AMPK) controls the aging process via an integrated signaling network. Ageing Res. Rev. 2012;11:230–241. doi: 10.1016/j.arr.2011.12.005. [DOI] [PubMed] [Google Scholar]
- Sinclair D.A. Toward a unified theory of caloric restriction and longevity regulation. Mech. Ageing Dev. 2005;126:987–1002. doi: 10.1016/j.mad.2005.03.019. [DOI] [PubMed] [Google Scholar]
- Song J., Bae Y.S. CK2 down-regulation increases the expression of senescence-associated secretory phenotype factors through NF-κB activation. Int. J. Mol. Sci. 2021;22:406. doi: 10.3390/ijms22010406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa G., Biasi F., Poli G., Chiarpotto E. Calorie restriction and dietary restriction mimetics: a strategy for improving healthy aging and longevity. Curr. Pharm. Des. 2014;20:2950–2977. doi: 10.2174/13816128113196660699. [DOI] [PubMed] [Google Scholar]
- Yu W., Zhou H.F., Lin R.B., Fu Y.C., Wang W. Short-term calorie restriction activates SIRT1-4 and -7 in cardiomyocytes in vivo and in vitro. Mol. Med. Rep. 2014;9:1218–1224. doi: 10.3892/mmr.2014.1944. [DOI] [PubMed] [Google Scholar]
- Zhao J., Brault J.J., Schild A., Cao P., Sandri M., Schiaffino S., Lecker S.H., Goldberg A.L. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.