Abstract
Background
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron (B.1.1.529) variant has proven to be highly transmissible and has outcompeted the Delta variant in many regions of the world. Early reports have also suggested that Omicron may result in less severe clinical disease in humans. Here, we show that Omicron is less pathogenic than prior SARS-CoV-2 variants in Syrian golden hamsters.
Methods
Hamsters were inoculated with either SARS-CoV-2 Omicron or other SARS-CoV-2 variants. Animals were followed for weight loss, and upper and lower respiratory tract tissues were assessed for viral loads and histopathology.
Findings
Infection of hamsters with the SARS-CoV-2 WA1/2020, Alpha, Beta, or Delta strains led to 4%–10% weight loss by day 4 and 10%–17% weight loss by day 6. In contrast, infection of hamsters with two different Omicron challenge stocks did not result in any detectable weight loss, even at high challenge doses. Omicron infection led to substantial viral replication in both the upper and lower respiratory tracts but demonstrated lower viral loads in lung parenchyma and reduced pulmonary pathology compared with WA1/2020 infection.
Conclusions
These data suggest that the SARS-CoV-2 Omicron variant may result in robust upper respiratory tract infection, but less severe lower respiratory tract clinical disease, compared with prior SARS-CoV-2 variants.
Funding
Funding for this study was provided by NIH grant CA260476, the Massachusetts Consortium for Pathogen Readiness, the Ragon Institute, and the Musk Foundation.
Keywords: SARS-CoV-2, COVID-19, omicron, hamster, pathogenicity
Graphical abstract
Context and significance
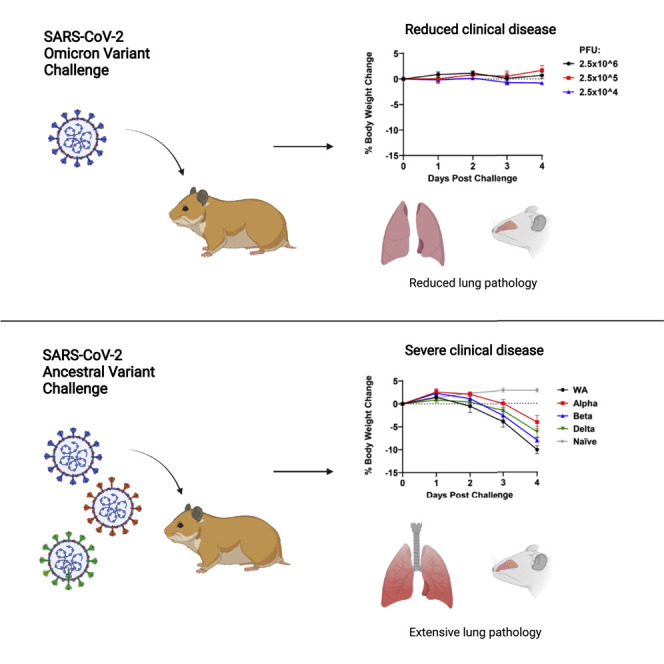
The SARS-CoV-2 Omicron variant has proven highly transmissible and has outcompeted the Delta variant in many regions of the world. In this study, investigators studied Omicron infection in Syrian golden hamsters. Although infection with prior SARS-CoV-2 variants led to substantial weight loss following infection in hamsters, infection with the Omicron variant did not result in any detectable weight loss, even at high challenge doses. Moreover, Omicron infection led to lower viral loads and reduced pathology in the lung compared with prior SARS-CoV-2 variants. These findings show that the SARS-CoV-2 Omicron variant led to less severe lower respiratory tract disease compared with prior SARS-CoV-2 variants in hamsters.
Syrian golden hamsters infected with the SARS-CoV-2 Omicron variant did not lose weight and showed less virus in the lung and reduced lung pathology compared with hamsters infected with prior SARS-CoV-2 variants. These observations are consistent with emerging human data suggesting that Omicron infection is less severe than infection with prior variants.
Introduction
The highly mutated severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron variant has led to rapid global spread, including in individuals who have been fully vaccinated.1 However, early reports from South Africa and the United Kingdom suggest that the severity of clinical disease with Omicron may be lower than for prior variants. It is unclear whether this lower clinical severity is due to the Omicron virus itself or if it reflects population immunity due to prior vaccination and/or infection.
Syrian golden hamsters provide a robust model to study SARS-CoV-2 disease, with reproducible weight loss and pneumonia following SARS-CoV-2 infection.2 , 3 We developed two SARS-CoV-2 Omicron challenge stocks and assessed clinical disease, viral loads, and histopathology in hamsters.
Omicron infection leads to less severe clinical disease
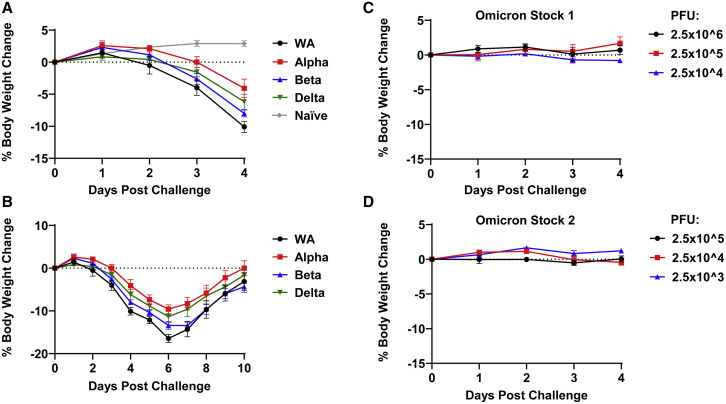
We generated two independent SARS-CoV-2 Omicron stocks in VeroE6-TMPRSS2 cells (see STAR Methods). Syrian golden hamsters (n = 6/group) were inoculated by the intranasal route with 100 μL virus containing 5 × 104 PFU WA1/2020, Alpha (B.1.1.7), Beta (B.1.351), and Delta (B.1.617.2) stocks, essentially as we previously reported.2 , 4 Infected hamsters showed a mean reduction of 4%–10% of body weight by day 4 (Figure 1 A) and 10%–17% of body weight by day 6 (Figure 1B), while uninfected hamsters showed a mean increase of 1% body weight.2 , 4 One animal infected with WA1/2020 reached the 20% weight loss criteria for humane euthanization, but the rest of the hamsters recovered their body weights by approximately day 10.
Figure 1.
Weight loss in hamsters infected with SARS-CoV-2 variants
(A and B) Mean body-weight change following infection of hamsters with SARS-CoV-2 WA1/2020, Alpha, Beta, and Delta variants, along with weight change in uninfected control hamsters.
(C and D) Mean body-weight change following infection of hamsters with SARS-CoV-2 Omicron stocks 1 and 2. Mean body-weight changes with standard errors are shown.
Additional groups of hamsters (n = 4/group) were inoculated by the intranasal route with 100 μL of 2.5 × 106, 2.5 × 105, or 2.5 × 104 PFU of Omicron stock 1 or 2.5 × 105, 2.5 × 104, or 2.5 × 103 PFU of stock 2. Hamsters inoculated with Omicron stock 1 showed a mean reduction of -1%, -2%, and 1% of body weight by day 4 for these challenge doses, respectively (Figure 1C). Hamsters inoculated with Omicron stock 2 showed a mean reduction of 0%, 0%, and 1% of body weight by day 4 for these challenge doses, respectively (Figure 1D). These data demonstrate that two SARS-CoV-2 Omicron stocks did not lead to clinical weight loss in hamsters, even when inoculated at high doses.
Reduced virus in lung following Omicron infection
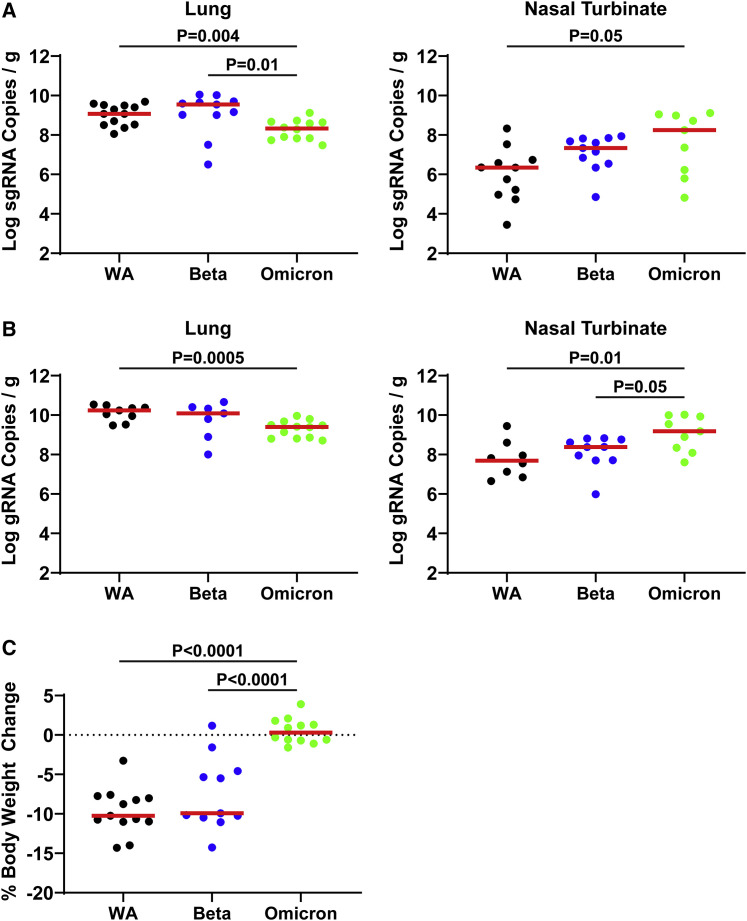
We next assessed tissue viral loads on day 4 following infection of additional groups of hamsters with 5 × 104 PFU WA1/2020 (n = 13), Beta (n = 11), and Omicron stock 1 (n = 12). We selected Omicron stock 1 for this experiment because of its higher titer. Levels of E subgenomic RNA (sgRNA) and N genomic RNA (gRNA) were assessed by RT-PCR in lungs and nasal turbinates.5 , 6 In lung tissue, hamsters infected with WA1/2020, Beta, and Omicron had a median of 9.07, 9.55, and 8.33 log sgRNA copies/g tissue, respectively. Median levels of lung sgRNA were 0.74 log lower in Omicron-infected hamsters compared with WA1/2020-infected hamsters (p = 0.004, two-tailed Mann-Whitney test; Figure 2 A), and sgRNA levels correlated with mean tissue culture infectious dose (TCID50) titers (data not shown). In nasal turbinates, hamsters infected with WA1/2020, Beta, and Omicron had a median of 6.35, 7.34, and 8.25 log sgRNA copies/g tissue, respectively. Median levels of nasal turbinate sgRNA were 1.90 log higher in Omicron-infected hamsters compared with WA1/2020-infected hamsters (p = 0.05, two-tailed Mann-Whitney test; Figure 2A). Similarly, median levels of lung gRNA were 0.84 log lower in Omicron-infected hamsters compared with WA1/2020-infected hamsters (p = 0.005, two-tailed Mann-Whitney test; Figure 2B), but median levels of nasal turbinate sgRNA were 1.49 log higher in Omicron-infected hamsters (p = 0.01, two-tailed Mann-Whitney test; Figure 2B). Body weights were reduced in the WA1/2020- and Beta-infected animals, as expected, but not the Omicron-infected animals on day 4 (Figure 2C). These data suggest that Omicron infection in hamsters leads to lower levels of virus in the lower respiratory tract compared with WA1/2020 infection.
Figure 2.
Tissue viral loads in hamsters on day 4 following SARS-CoV-2 infection
(A) E subgenomic RNA (sgRNA) levels in lung and nasal turbinates following infection of hamsters with SARS-CoV-2 WA1/2020, Beta, and Omicron variants (limit of detection 100 viral copies/g tissue).
(B) N genomic RNA (gRNA) levels in lung and nasal turbinates following infection of hamsters with SARS-CoV-2 WA1/2020, Beta, and Omicron variants (limit of detection 100 viral copies/g tissue).
(C) Weight loss at time of necropsy of hamsters in (A) and (B).
Data for WA1/2020 reflect N = 13 hamsters pooled from four different challenge experiments, data for Beta reflect N = 11 hamsters pooled from three different challenge experiments, and data for Omicron reflect N = 12 hamsters pooled from one experiment. Log sgRNA copies per gram tissue are shown. Medians (red bars) are depicted. p values represent two-sided Mann-Whitney tests.
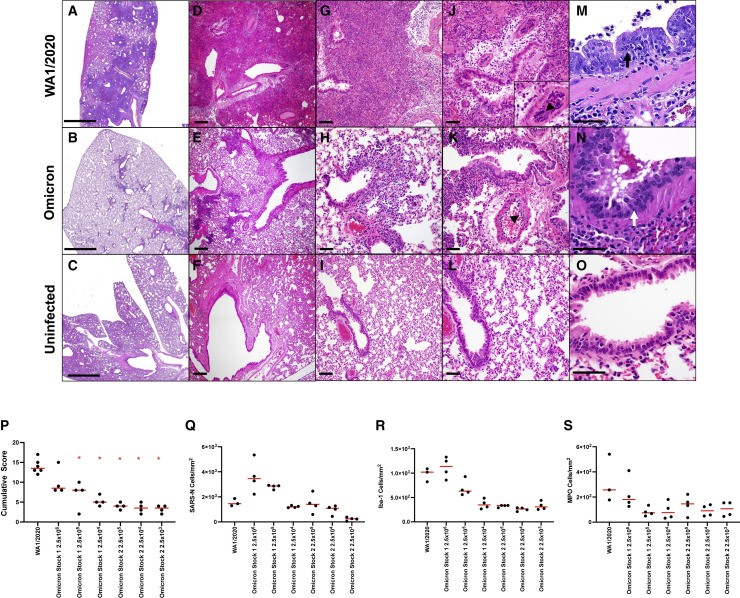
Reduced lung pathology following Omicron challenge
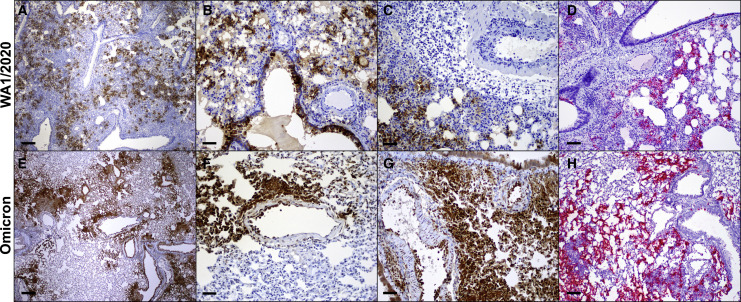
Omicron-infected hamsters demonstrated reduced bronchiolar epithelial changes, interstitial inflammation and consolidation, and endothelialitis as compared with WA1/2020 infected hamsters on day 4 following infection (Figures 3A–3O and S1), with lower lung pathology scores in animals infected with Omicron at doses of 2.5 × 105 PFU as compared with animals infected with WA1/2020 at a dose of 5 × 104 PFU (Figure 3P). Despite significantly reduced overall pathology, however, Omicron-infected hamsters had similar numbers of SARS nucleocapsid (SARS-N)-positive cells in lung as compared with WA1/2020-infected hamsters (Figure 3Q) but showed a trend toward fewer Iba-1-positive cells (macrophages) and myeloperoxidase-positive cells (neutrophils) per unit area of lung (Figures 3R, 3S, and S1). Both WA1/2020- and Omicron-infected animals had SARS-N-positive bronchiolar epithelium and pneumocytes (Figure 4 ). Nasal turbinate pathology was pronounced in Omicron-infected animals but did not differ in distribution, severity, or viral positivity compared with WA1/2020-infected hamsters (Figure S1).
Figure 3.
Histopathology in hamsters on day 4 following SARS-CoV-2 infection
Lung tissue from hamsters infected with 5 × 104 PFU SARS-CoV-2 WA1/2020 (top) and 2.5 × 105 PFU Omicron (middle) compared with uninfected hamsters (bottom) was stained with H&E.
(A–C) Low power-representative images of lung. There are multifocal to locally extensive areas of interstitial inflammation and consolidation associated with bronchioles in WA1/2020-infected animals that are reduced in Omicron-infected animals.
(D–L) Medium power-representative images of bronchioles and lumina showing presence of intraluminal necrotic epithelium in both WA1/2020- and Omicron-infected animals (J–L) and perivascular edema and marginating inflammatory cells along the endothelium of medium-sized arterioles (arrowheads).
(M–O) High magnification images of bronchiolar epithelium showing cellular atypia, hypertrophy, and loss of basal nuclear polarity in degenerative bronchiolar epithelium that is more pronounced in WA1/2020-infected hamsters compared with Omicron-infected hamsters.
(P) Cumulative pathology scoring of (1) airways (bronchi, bronchioles), (2) interstitium, (3) alveoli, (4) vessels, (5) edema, and (6) regeneration. Each feature received a score of 0–3 with a maximum possible score of 18 per animal.
(Q–S) Quantitative image analysis of immunohistochemistry using HALO (Indicalabs)-optimized algorithms to enumerate (Q) SARS-N-positive (R), Iba-1-positive (macrophages), and (S) myeloperixade-positive (neutrophils) cells per unit area. ∗p = 0.0048, two-tailed Mann-Whitney test. Scale bars: 2 mm (A–C), 200 μm (D–F), 100 μm (G–I), and 50 μM (J–O).
Figure 4.
SARS-CoV-2 Omicron variant distribution in lung
(A–H) Immunohistochemistry for nucleocapsid protein (brown) and RNAscope in situ hybridization for viral RNA (red) in hamsters infected with SARS-CoV2 WA1/2020 (A–D) and Omicron (E–H). Scale bars: 200 μm (A and E) and 50 μm (B–D and F–H).
Discussion
Our data demonstrate that the SARS-CoV-2 Omicron variant infected Syrian golden hamsters but did not result in detectable weight loss, even at doses that were 50 times higher than the doses of WA1/2020, Alpha, Beta, and Delta that led to substantial weight loss. The consistency of these findings with two independent Omicron challenge stocks suggests the generalizability of these conclusions.7 , 8
Omicron infection led to robust viral replication in both the upper and lower respiratory tracts, with higher viral loads in nasal turbinates and lower viral loads in the lung compared with WA1/2020. Moreover, Omicron led to reduced lung histopathology compared with WA1/2020, suggesting that Omicron infection leads to reduced lower respiratory tract disease in hamsters. Future studies could compare tissue viral loads of Omicron with Delta and additional variants.
Our findings in hamsters are consistent with emerging reports suggesting that Omicron is highly transmissible but induces less severe clinical pneumonia in humans compared with prior SARS-CoV-2 variants. Future studies could further define the pathogenesis of infection with the SARS-CoV-2 Omicron variant and the mechanisms associated with its reduced pathogenicity.
Limitations of study
It is not known whether the SARS-CoV-2 Omicron model in hamsters is predictive for humans. It is also not known whether weight loss or viral loads in the lower respiratory tract is more predictive of severe clinical disease.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Human SARS Coronavirus Nucleoprotein/NP Monoclonal Antibody, Clone R040 1:1500 | Sino Biological | REF 40143-R040 Lot MA14AP2104; RRID:AB_2827976 |
| Anti-Iba1 Rabbit Polyclonal 1:4000 | Wako Pure Chemical Corporation | REF 019-19741 Lot CAE1308; RRID:AB_839504 |
| Anti-Human Myeloperoxidase, Polyclonal 1:1500 | Dako | REF A0398 Lot 41321498; RRID:AB_2335676 |
| Bacterial and virus strains | ||
| SARS-CoV-2 | BEI Repository | Isolate: USA-WA1/2020 |
| SARS-CoV-2 | BEI Repository | Isolate: B.1.351 |
| SARS-CoV-2 | BEI Repository | Isolate: B.1.1.529 |
| SARS-CoV-2 | BEI Repository | Isolate: B.1.617.2 |
| SARS-CoV-2 | BEI Repository | B.1.1.7 |
| Biological samples | ||
| Hamster SARS-CoV2 infected lung tissue, fixed, embedded | Bioqual, Inc. | N/A |
| Hamster SARS-CoV2 infected nasal turbinate, fixed, embedded | Bioqual, Inc. | N/A |
| Oral swabs from Hamster | Bioqual, Inc. | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Citrate buffer antigen retrieval | Thermo | AP-9003-500 |
| Protein block | Biocare | BE965H |
| DaVinci Green Antibody diluent | Biocare | PD900M |
| Rabbit Mach-2 HRP-Polymer | Biocare | RHRP520L |
| Critical commercial assays | ||
| RNAscope® 2.5 HD Detection Reagents-RED | Advanced Cell Diagnostics | Cat. No.322360 |
| SARS-CoV-2, S gene probe | Advanced Cell Diagnostics | 848561; V-CoV2019-S |
| DapB Negative control probe | Advanced Cell Diagnostics | 310043 |
| Experimental models: Organisms/strains | ||
| Syrian golden hamsters | Envigo | HsdHan:AURA https://www.envigo.com/model/hsdhan-aura |
| Oligonucleotides | ||
| Primer:sgLeadSARSCoV2-F Forward: CGAT CTCTTGTAGATCTGTTCTC |
Wolfel et al., 2020 | ThermoFisher Scientific:4448510 |
| Primer: E_Sarbeco_R Reverse: ATATTGC AGCAGTACGCACACA |
Wolfel et al., 2020 | ThermoFisher Scientific:4448510 |
| Probe:E_Sarbeco_P1 : VIC-ACACTAG CCATCCTTACTGCGCTTCG-MGBNFQ |
Wolfel et al., 2020 | ThermoFisher Scientific:4448510 |
| Primer:2019-nCoV_N1 Forward: GACCC CAAAATCAGCGAAAT |
CDC Research Use Only 2019-Novel Coronavirus (2019-nCoV) Real-time RT-PCR Primers and Probes | ThermoFisher Scientific:4448510 |
| Primer:2019-nCoV_N1 Reverse: TCTGGT TACTGCCAGTTGAATCTG |
CDC Research Use Only 2019-Novel Coronavirus (2019-nCoV) Real-time RT-PCR Primers and Probes | ThermoFisher Scientific:4448510 |
| Probe:2019-nCoV_N1 FAM- ACCCCGCAT TACGTTTGGTGGACC-BHQ1 |
CDC Research Use Only 2019-Novel Coronavirus (2019-nCoV) Real-time RT-PCR Primers and Probes | ThermoFisher Scientific:4448510 |
| Recombinant DNA | ||
| Plasmid: pcDNA3.1+. SARS-CoV-2 E gene subgenomic RNA (sgRNA) | This paper | N/A |
| Software and algorithms | ||
| QuantStudio Real-Time PCR Software v1.7.1 | Life Technologies | https://www.thermofisher.com/us/en/home/global/forms/life-science/quantstudio-6-7-flex-software.html |
| GraphPad Prism 8.4.2 | GraphPad software | https://www.graphpad.com/scientific-software/prism/ |
| BioRender | BioRender | https://biorender.com/ |
| Other | ||
| RNA Standard: SARS-CoV-2 E gene subgenomic RNA (sgRNA) | This paper | N/A |
| RNA Standard: SARS-CoV-2 N gene genomic RNA (gRNA) | This paper | N/A |
| AmpliCap-Max T7 High Yield Message Maker Kit | Cellscript | C-ACM04037 |
| RNeasy 96 QIAcube HT Kit | Qiagen | 74182 |
| SuperScript VILO Master Mix | Invitrogen | Life Technologies: 11755500 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact Dan Barouch (dbarouch@bidmc.harvard.edu).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Animals and study design
Eight week old inbred female and male golden Syrian hamsters (Mesocricetus auratus) (Envigo) were randomly allocated to groups and infected with SARS-CoV-2 variants in a volume of 100 μL (50 μL/nostril) by the intranasal route. Hamsters were healthy and drug-naïve with no history of previous procedures. All hamsters were housed at Bioqual, Inc. (Rockville, MD). Following challenge, body weights were assessed daily. Body weight loss that exceeded 20% of the weight on the day of challenge was established as a humane endpoint euthanasia criteria. A subset of animals were necropsied on day 4 for tissue viral loads and histopathology. All animal studies were conducted in compliance with all relevant local, state, and federal regulations and were approved by the BIOQUAL Institutional Animal Care and Use Committee (IACUC).
Method details
SARS-CoV-2 viral stocks. Seed stock information is as follows: WA1/2020 stock (USA-WA1/2020; BEI Resources NR-5228; 2.34 × 109 TCID50/mL), Alpha (B.1.1.7; USA/CA_CDC_5574/2020; BEI Resources NR-54011; 1.58 × 107 TCID50/mL), Beta (B.1.351; South Africa/KRISP-K005325/2020; BEI Resources NR-54974; 1.99 × 108 TCID50/mL), and Delta (B.1.617.2; USA/PHC658/2021; BEI Resources NR-55612; 5.00 × 108 TCID50/mL) challenge stocks have been previously described.4 We generated two independent SARS-CoV-2 Omicron stocks in VeroE6-TMPRSS2 cells inoculated with nasal swabs from Omicron-infected individuals. Omicron Stock one had a titer of 2.3 × 109 TCID50/mL and 2.5 × 107 PFU/mL in VeroE6-TMPRSS2 cells (EPI_ISL_7171744; Mehul Suthar, Emory University). Omicron Stock 2 had a titer of 2.3 × 108 TCID50/mL and 3.8 × 106 PFU/mL in VeroE6-TMPRSS2 cells (EPI_ISL_7263803; Yoshihiro Kawaoka, University of Wisconsin). All SARS-CoV-2 stocks were fully sequenced. All TCID50 and PFU assays were in VeroE6-TMPRSS2 cells. Note that TCID50 titers in VeroE6-TMPRSS2 cells are approximately 100-fold higher than TCID50 titers in VeroE6 cells.
Genomic and subgenomic viral load assays
SARS-CoV-2 N gene genomic RNA (gRNA) and E gene subgenomic RNA (sgRNA) was assessed by reverse transcription polymerase chain reactions (RT-PCR) using primers and probes as previously described.5 , 6 , 9 Standards were generated by first synthesizing a gene fragment of the genomic N gene or subgenomic E gene.6 The gene fragments were subsequently cloned into a pcDNA3.1+ expression plasmid using restriction site cloning (Integrated DNA Technologies). The inserts were in vitro transcribed to RNA using the AmpliCap-Max T7 High Yield Message Maker Kit (CellScript). Log dilutions of the standard were prepared for RT-PCR assays ranging from 1 × 1010 copies to 1 × 10−1 copies. Viral loads were quantified from lung tissue as follows; total RNA was extracted on a QIAcube HT instrument using the RNeasy 96 QIAcube HT Kit according to manufacturer’s specifications (Qiagen). Standard dilutions and extracted total RNA from samples were reverse transcribed using SuperScript VILO Master Mix (Invitrogen) according to manufacturer’s specifications. A Taqman custom gene expression assay (Thermo Fisher Scientific) was designed using the sequences targeting the E gene sgRNA.6 The sequences for the custom assay were as follows, forward primer, sgLeadCoV2.Fwd: CGATCTCTTGTAGATCTGTTCTC, E_Sarbeco_R: ATATTGCAGCAGTACGCACACA, E_Sarbeco_P1 (probe): VIC-ACACTAGCCATCCTTACTGCGCTTCG-MGB. For the genomic N assays, the sequences for the forward (F) and reverse (R) primes and probe (P) were: 2019-nCoV_N1-F :5′-GACCCCAAAATCAGCGAAAT-3′; 2019-nCoV_N1-R: 5′-TCTGGTTACTGCCAGTTGAATCTG-3′; 2019-nCoV_N1-P: 5′-FAM-ACCCCGCATTACGTTTGGTGGACC-BHQ1-3’. Reactions were carried out in duplicate for samples and standards on the QuantStudio 6 and 7 Flex Real-Time PCR Systems (Applied Biosystems). The following thermal cycling conditions were used; initial denaturation at 95°C for 20 seconds, then 45 cycles of 95°C for 1 second and 60°C for 20 seconds. Standard curves were used to calculate subgenomic RNA copies and copy number was normalized to the input weight of lung tissue (copies/g); the quantitative assay sensitivity was 100 copies per gram.
Histopathology
Tissues were fixed in freshly prepared 4% paraformaldehyde for 24 hours, transferred to 70% ethanol, paraffin embedded within 7 to 10 days, and blocks sectioned at 5 μm. Slides were baked for 30 to 60 min at 65 °C and then deparaffinized in xylene and rehydrated through a series of graded ethanol to distilled water. Slides were stained with hematoxylin and eosin. Blinded assessment of tissue pathology was performed by two boarded veterinary pathologist (AJM, CPM). The following six features were evaluated as previously detailed:1 1)bronchi and bronchioles 2) interstitium 3) endothelial changes/endothelialitis 4) alveolar spaces/syncytia 5) edema 6) regeneration, with scores 0–3 (0 = none, 1 = mild, 2 = moderate, 3 = severe).
Immunohistochemistry
Paraffin blocks were sectioned at 5 μm. Slides were baked for 30–60 min at 65° then deparaffinized in xylene and rehydrated through a series of graded ethanol to distilled water. Heat induced epitope retrieval (HIER) was performed using a pressure cooker on steam setting for 25 minutes in citrate buffer (Thermo: AP-9003-500) followed by treatment with 3% hydrogen peroxide. Slides were then rinsed in distilled water and protein blocked (BioCare, BE965H) for 15 min followed by rinses in 1× phosphate buffer saline. The following primary antibodies were used in DaVinci Green Diluent (BioCare, PD900 M): Rabbit anti-Iba-1 (Wako:019-19741) diluted 1:4000; rabbit anti-Myeloperoxidase antibody (DAKO, A0398) diluted 1:1500; rabbit anti-SARS-Nucleocapsid protein antibody (Sino Biological clone R040, 40143-R040) diluted 1:1500. All antibodies were incubated for 60 min at room temperature, followed by rabbit Mach-2 HRP-Polymer (BioCare; RHRP520L) for 30 minutes, then counterstained with hematoxylin followed by bluing using 0.25% ammonia water. Labeling was performed on a Biocare InterlliPATH autostainer.
RNAscope in situ hybridization
RNAscope in situ hybridization was performed as directed with the following modifications using a probe for SARS-CoV-2, S gene encoding the spike protein (ACD Cat. No. 848561; V-CoV2019-S) and DapB (ACD Cat.No 310043) as a negative control. In brief, after slides were deparaffinized in xylene and rehydrated through a series of graded ethanol to distilled water, retrieval was performed for 30 min in ACD P2 retrieval buffer (ACD Cat. No. 322000) at 95–98°C, followed by treatment with protease plus (ACD Cat. No. 322331) diluted 1:10 in PBS for 20 min at 40°C. All washes were performed in 0.5× kit provided SSC. Slides were developed using the RNAscope® 2.5 HD Detection Reagents-RED (ACD Cat. No.322360).
Quantitative image analysis
Quantitative image analysis was performed using HALO software (v3.0.311.405; Indica Labs) on at least one lung lobe cross-section from each animal. In cases where more than one cross-section was available, all lung lobes were quantified as an individual data point. For SARS-N, Iba-1, MPO, and CD3 IHC positivity, the Indica Labs - Multiplex IHC algorithm (v3.1.4) was used for quantitation. In all instances, manual inspection of all images was performed on each sample to ensure that the annotations were accurate.
Quantification and statistical analysis
Analysis of virologic and body weight data was performed using GraphPad Prism 9.1.2 (GraphPad Software). Comparison of data between groups was performed using two-sided Mann–Whitney tests. p values of less than 0.05 were considered significant. Graphical Abstract was generated using BioRender.
Acknowledgments
The authors acknowledge NIH grant CA260476, the Massachusetts Consortium for Pathogen Readiness, the Ragon Institute, and the Musk Foundation (D.H.B.).
Author contributions
This study was designed by D.H.B. Omicron challenge stocks were prepared by M.S. and Y.K. Animal work was performed by H.A. and M.G.L. Virologic assays were performed and analyzed by K.M., V.G., L.H.T., B.C., and M.S. Histopathology was performed by C.P.-M., N.J., S.D., and A.J.M. D.H.B., K.M., and L.H.T. had unrestricted access to all data. D.H.B., K.M., L.H.T., and A.J.M. prepared the first draft of the manuscript, performed the statistical analyses, and reviewed and edited the manuscript. All authors agreed to submit the manuscript, read and approved the final draft, and take full responsibility of its content and accuracy of the data.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science.
Published: March 17, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.medj.2022.03.004.
Supplemental information
Data and code availability
-
•
No datasets or codes are associated with the paper.
-
•
Data from this study is available from the Lead Contact upon request.
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.
References
- 1.Cele S., Jackson L., Khoury D.S., Khan K., Moyo-Gwete T., Tegally H., San J.E., Cromer D., Scheepers C., Amoako D.G., et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 2021;602:654–656. doi: 10.1038/s41586-021-04387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tostanoski L.H., Wegmann F., Martinot A.J., Loos C., McMahan K., Mercado N.B., Yu J., Chan C.N., Bondoc S., Starke C.E., et al. Ad26 vaccine protects against SARS-CoV-2 severe clinical disease in hamsters. Nat. Med. 2020;26:1694–1700. doi: 10.1038/s41591-020-1070-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan J.F., Zhang A.J., Yuan S., Poon V.K., Chan C.C., Lee A.C., Chan W.M., Fan Z., Tsoi H.W., Wen L., et al. Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID-19) in golden syrian hamster model: implications for disease pathogenesis and transmissibility. Clin. Infect. Dis. 2020;71:2428–2446. doi: 10.1093/cid/ciaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tostanoski L.H., Yu J., Mercado N.B., McMahan K., Jacob-Dolan C., Martinot A.J., Piedra-Mora C., Anioke T., Chang A., Giffin V.M., et al. Immunity elicited by natural infection or Ad26.COV2.S vaccination protects hamsters against SARS-CoV-2 variants of concern. Sci. Transl. Med. 2021;13:eabj3789. doi: 10.1126/scitranslmed.abj3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dagotto G., Mercado N.B., Martinez D.R., Hou Y.J., Nkolola J.P., Carnahan R.H., Crowe J.E., Jr., Baric R.S., Barouch D.H. Comparison of subgenomic and total RNA in SARS-CoV-2 challenged rhesus macaques. J. Virol. 2021;95:e02370–e02420. doi: 10.1128/JVI.02370-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Muller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 7.Halfmann P.J., Iida S., Iwatsuki-Horimoto K., Maemura T., Kiso M., Scheaffer S.M., Darling T.L., Joshi A., Loeber S., Singh G., et al. SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters. Nature. 2022 doi: 10.1038/s41586-022-04441-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shuai H., Chan J.F., Hu B., Chai Y., Yuen T.T., Yin F., Huang X., Yoon C., Hu J.C., Liu H., et al. Attenuated replication and pathogenicity of SARS-CoV-2 B.1.1.529 Omicron. Nature. 2022 doi: 10.1038/s41586-022-04442-5. [DOI] [PubMed] [Google Scholar]
- 9.Yu J., Tostanoski L.H., Peter L., Mercado N.B., McMahan K., Mahrokhian S.H., Nkolola J.P., Liu J., Li Z., Chandrashekar A., et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. 2020;369:806–811. doi: 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
No datasets or codes are associated with the paper.
-
•
Data from this study is available from the Lead Contact upon request.
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.