Abstract
The rapid spread of the SARS-CoV-2 Omicron (B.1.1.529) variant, including in highly vaccinated populations, has raised important questions about the efficacy of current vaccines. In this study, we show that the mRNA-based BNT162b2 vaccine and the adenovirus-vector-based Ad26.COV2.S vaccine provide robust protection against high-dose challenge with the SARS-CoV-2 Omicron variant in cynomolgus macaques. We vaccinated 30 macaques with homologous and heterologous prime-boost regimens with BNT162b2 and Ad26.COV2.S. Following Omicron challenge, vaccinated macaques demonstrated rapid control of virus in bronchoalveolar lavage, and most vaccinated animals also controlled virus in nasal swabs. However, 4 vaccinated animals that had moderate Omicron-neutralizing antibody titers and undetectable Omicron CD8+ T cell responses failed to control virus in the upper respiratory tract. Moreover, virologic control correlated with both antibody and T cell responses. These data suggest that both humoral and cellular immune responses contribute to vaccine protection against a highly mutated SARS-CoV-2 variant.
Keywords: SARS-CoV-2, Omicron, macaque
Graphical abstract
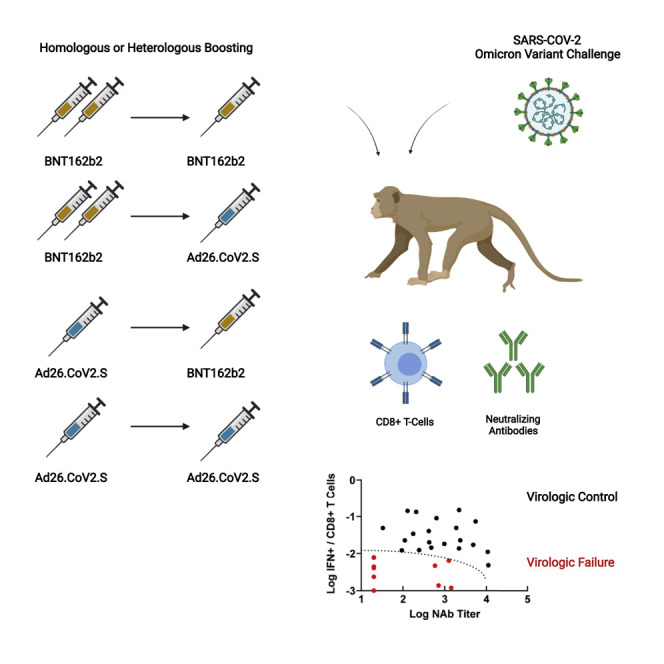
Heterologous as well as homologous prime-boosting with the mRNA vaccine BNT162b2 and adenovirus-vector-based Ad26.COV2.S vaccine provides robust protection against SARS-CoV-2 Omicron in cynomolgus macaques, with both humoral and cellular immune responses being critical for overall protection.
Introduction
The highly mutated SARS-CoV-2 Omicron variant has been shown to evade neutralizing antibody (NAb) responses induced by current vaccines, although a third immunization augments Omicron-specific NAb responses (Carreño et al., 2022; Cele et al., 2022; Liu et al., 2022; Nemet et al., 2022; Schmidt et al., 2022). In contrast, T cell responses induced by current vaccines are highly cross-reactive to SARS-CoV-2 variants including Omicron (Keeton et al., 2022; Liu et al., 2022; Tarke et al., 2022).
Recent clinical effectiveness studies have shown that the mRNA-based BNT162b2 vaccine (Polack et al., 2020) and the adenovirus-vector-based Ad26.COV2.S vaccine (Sadoff et al., 2021) provided 70% and 85% protection, respectively, against hospitalization with Omicron in South Africa (Collie et al., 2022; Gray et al., 2021). This robust protection by both vaccine platforms against severe disease with the SARS-CoV-2 Omicron variant in the absence of high titers of Omicron-specific NAbs suggest the possible relevance of other immune effector mechanisms. In this study, we evaluated the immunogenicity and protective efficacy of BNT162b2 and Ad26.COV2.S, including homologous and heterologous boost regimens, against SARS-CoV-2 Omicron challenge in nonhuman primates.
Results
Humoral immune responses
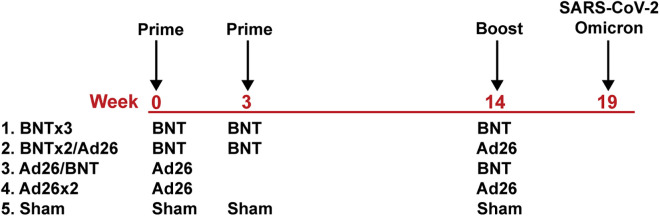
We immunized 30 adult cynomolgus macaques with homologous and heterologous regimens with BNT162b2 and Ad26.COV2.S or sham vaccine (N = 6/group; Figure 1 ). Groups of animals were primed with either two immunizations of 30 μg BNT162b2 at weeks 0 and 3 or a single immunization of 5 × 1010 vp Ad26.COV2.S at week 0. At week 14, animals received a homologous or heterologous boost with 30 μg BNT162b2 or 5 × 1010 vp Ad26.COV2.S.
Figure 1.
Study schema
Vaccine groups and timing of immunization and challenge are shown.
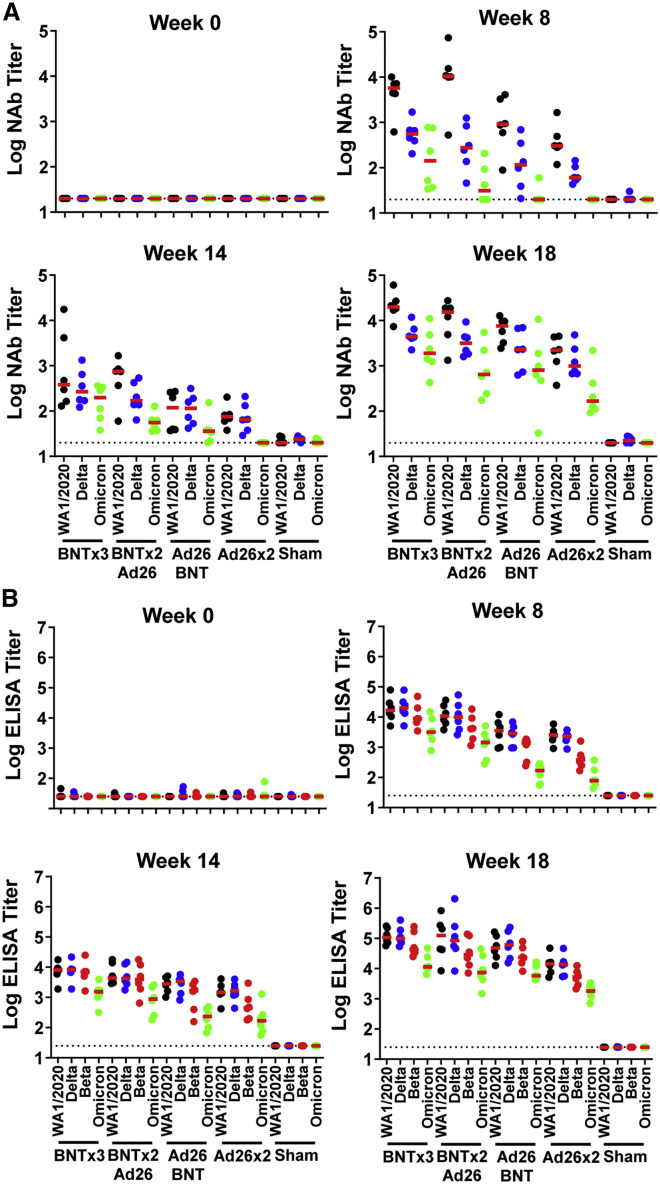
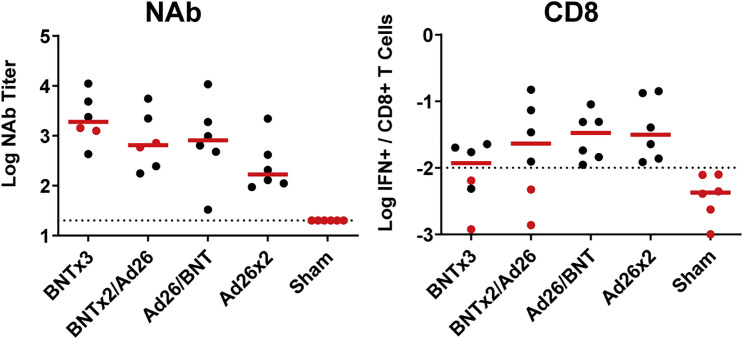
NAb responses were evaluated by luciferase-based pseudovirus-neutralizing antibody assays (Yu et al., 2021a). Vaccine-matched WA1/2020 NAbs were induced in all animals after the priming immunization at week 8 and were 13.3-fold higher in the BNT162b2-primed animals compared with the Ad26.COV2.S-primed animals. The WA1/2020 NAb titers in the BNT162b2 vaccinated groups declined more than 10-fold by week 14 (Figure 2 A), consistent with immune kinetics following BNT162b2 vaccination in humans, although mRNA vaccines are more potent in macaques than in humans (Collier et al., 2021; Falsey et al., 2021). Omicron NAbs were low in all groups prior to the boost. At week 18 after the homologous and heterologous boosts, median WA1/2020 NAb titers were 19,901, 15,451, 7,461, 2,215, and <20 in the BNTx3, BNTx2/Ad26, Ad26/BNT, Ad26x2, and sham groups, respectively. Median Omicron NAb titers at week 18 were 1,901, 650, 810, 168, and <20, respectively, reflecting a 9- to 23-fold reduction compared with WA1/2020 NAb titers (Figure 2A). All four vaccinated groups showed higher Omicron NAb titers than sham controls at week 18 (p = 0.0022 for all four vaccinated groups, two-tailed Mann-Whitney tests; Figure 2A).
Figure 2.
Humoral immune responses following vaccination
Antibody responses at weeks 0 (baseline), 8 (post-prime), 14 (pre-boost), and 18 (post-boost) following vaccination with BNTx3, BNTx2/Ad26, Ad26/BNT, Ad26x2, or sham (N = 30; N = 6/group).
(A) Neutralizing antibody (NAb) titers by a luciferase-based pseudovirus neutralization assay.
(B) Receptor-binding domain (RBD)-specific binding antibody titers by ELISA. Responses were measured against the SARS-CoV-2 WA1/2020 (black), B.1.617.2 (Delta; blue), B.1.351 (Beta; red), and B.1.1.529 (Omicron; green) variants. Dotted lines represent limits of quantitation. Medians (red bars) are shown. Omicron-specific NAbs in the vaccinated groups were compared with the sham controls by two-sided Mann-Whitney tests. ∗p < 0.05.
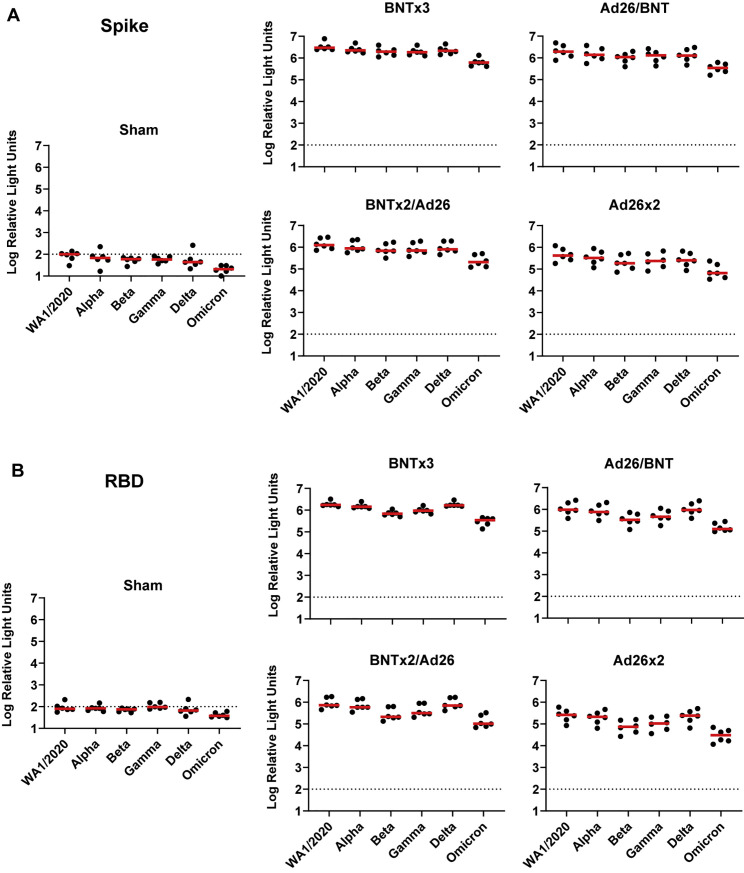
Receptor-binding domain (RBD)-specific binding antibodies were assessed by ELISA. At week 18, median WA1/2020 ELISA titers were 107,705, 125,694, 60,634, 14,193, and <25 in the BNTx3, BNTx2/Ad26, Ad26/BNT, Ad26x2, and sham groups, respectively. Median Omicron ELISA titers were 11,333, 7,452, 5,805, 1,783, and <25, respectively, reflecting a 8- to 17-fold reduction compared with WA1/2020 ELISA titers (Figure 2B). Similar trends were observed in multiplex spike- and RBD-specific binding assays using the Meso Scale Discovery electrochemiluminescence assay (ECLA) (Jacob-Dolan et al., 2021) (Figure S1 ). These data show that homologous and heterologous boosts substantially increased antibody responses in all groups, although Omicron-binding and -neutralizing antibody responses remained approximately 10-fold lower than WA1.2020 antibody responses.
Figure S1.
Spike- and RBD-specific binding antibody responses following vaccination, related to Figure 1
(A and B) Spike-specific (A) and RBD-specific (B) antibody responses against multiple variants are shown at week 18 (post-boost) following vaccination with BNTx3, BNTx2/Ad26, Ad26/BNT, Ad26x2, or sham (N = 30; N = 6/group) with the Meso-Scale Discovery electrochemiluminescence assay (ECLA). Dotted lines represent limits of quantitation. Medians (red bars) are shown.
Cellular immune responses
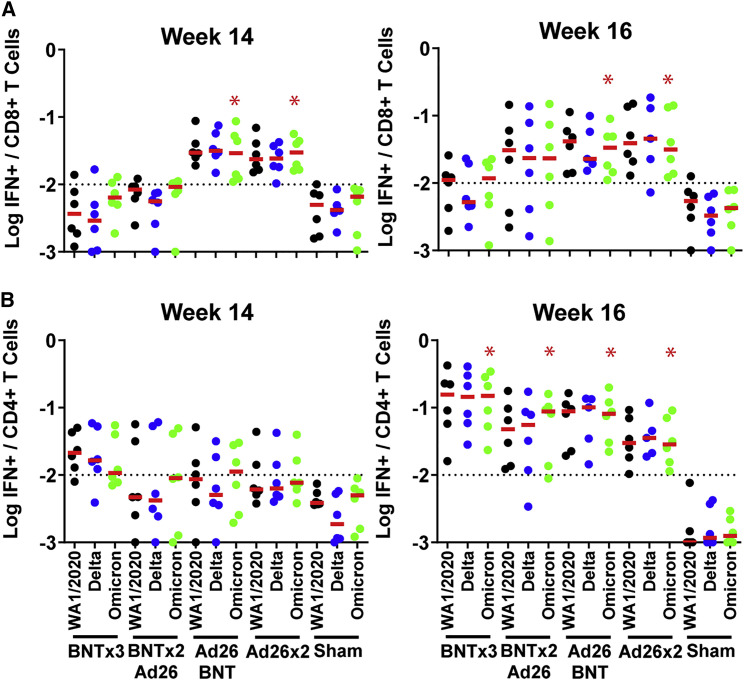
We next assessed spike-specific CD8+ and CD4+ T cell responses by multiparameter flow cytometry. At week 14 prior to the boost, WA1/2020 spike-specific IFN-γ CD8+ T cell responses were 13.1-fold higher in the Ad26.COV2.S-primed animals compared with the BNT162b2-primed animals (Figure 3 A), consistent with cellular immune data in humans (Atmar et al., 2022; Collier et al., 2021; Munro et al., 2021). The two groups that received Ad26.COV2.S, but not the two groups that received BNT162b2, showed higher Omicron CD8+ T cell responses than sham controls at week 14 (p = 0.0022 for the two groups that received Ad26.COV2.S, two-tailed Mann-Whitney tests; Figure 3A). In contrast, WA1/2020 spike-specific IFN-γ CD4+ T cell responses were comparable across groups (Figure 3B). Moreover, for both CD8+ and CD4+ T cell responses, Omicron responses were similar to WA1/2020 responses, indicative of substantial cross-reactivity of T cell responses (Alter et al., 2021; Keeton et al., 2022; Liu et al., 2022; Tarke et al., 2022). At week 16 after the homologous and heterologous boosts, median Omicron-spike-specific IFN-γ CD8+ T cell responses were 0.012%, 0.023%, 0.034%, 0.031%, and 0.004% in the BNTx3, BNTx2/Ad26, Ad26/BNT, Ad26x2, and sham groups, respectively (Figure 3A). Median Omicron-spike-specific IFN-γ CD4+ T cell responses were 0.150%, 0.088%, 0.081%, 0.028%, and 0.001% in the BNTx3, BNTx2/Ad26, Ad26/BNT, Ad26x2, and sham groups, respectively (Figure 3B).
Figure 3.
Cellular immune responses following vaccination
T cell responses at weeks 14 (pre-boost) and 18 (post-boost) following vaccination with BNTx3, BNTx2/Ad26, Ad26/BNT, Ad26x2, or sham (N = 30; N = 6/group).
(A and B) Pooled peptide spike-specific IFN-γ (A) CD8+ T cell responses and (B) CD4+ T cell responses by intracellular cytokine staining assays. Responses were measured against the SARS-CoV-2 WA1/2020 (black), B.1.617.2 (Delta; blue), and B.1.1.529 (Omicron; green) variants. Dotted lines represent limits of quantitation. Medians (red bars) are shown. Omicron-specific CD8+ and CD4+ T cell responses in the vaccinated groups were compared with the sham controls by two-sided Mann-Whitney tests. ∗p < 0.05.
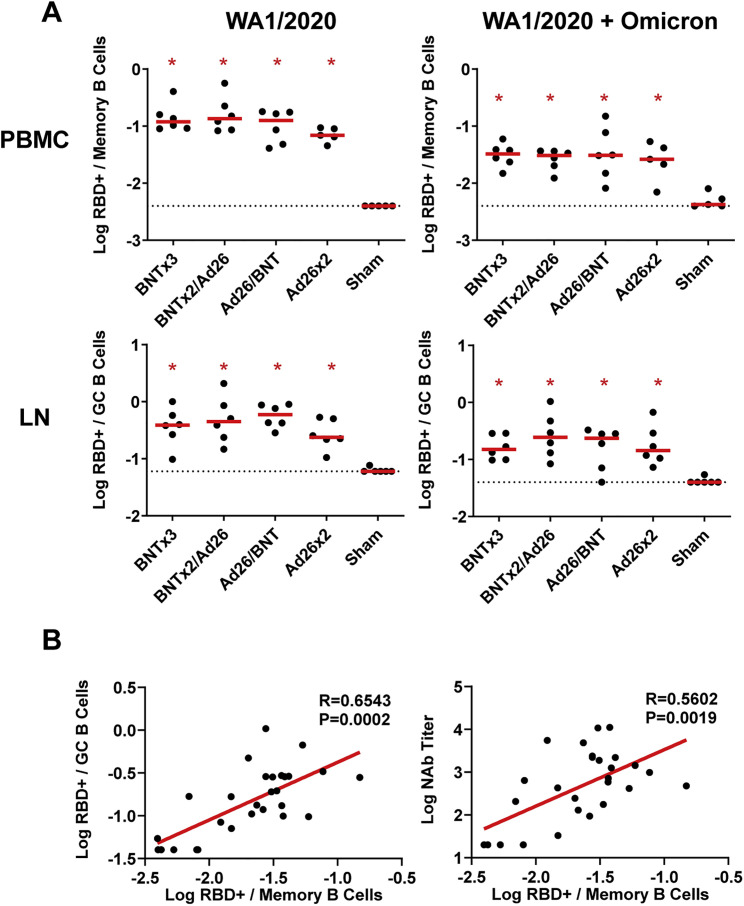
We also assessed memory IgG+ B cells in peripheral blood as well as germinal center CD20+IgD-IgG+Ki67+Bcl6+ B cells in lymph nodes at week 16 by multiparameter flow cytometry. WA1/2020 and cross-reactive WA1/2020 and Omicron-RBD-specific memory B cells and germinal center B cells were induced at comparable levels in all vaccinated groups (Figure S2 A). Peripheral Omicron-RBD-specific memory B cells correlated with lymph node Omicron-RBD-specific germinal center B cells (R = 0.6543, p = 0.0002, two-sided Spearman rank-correlation test) and serum Omicron NAb titers (R = 0.5602, p = 0.0019, two-sided Spearman rank-correlation test) at week 16 (Figure S2B).
Figure S2.
RBD-specific B cell responses following vaccination, related to Figure 1
(A) Total WA1/2020 and cross-reactive WA1/2020 and Omicron RBD-specific memory B cell responses in peripheral blood mononuclear cells (PBMC) and germinal center B cell responses in lymph nodes (LN) are shown at week 16 (post-boost) following vaccination with BNTx3, BNTx2/Ad26, Ad26/BNT, Ad26x2, or sham (N = 30; N = 6/group). Dotted lines represent limits of quantitation. Medians (red bars) are shown. Vaccinated groups were compared with the sham controls by two-sided Mann-Whitney tests. ∗p < 0.05.
(B) Correlations of Omicron-RBD-specific memory B cell responses in peripheral blood mononuclear cells (PBMC) with Omicron germinal center B cell responses in lymph nodes (LN) (left) and Omicron serum NAb titers are shown at week 16 (post-boost) (right). Correlations were assessed by two-sided Spearman rank-correlation tests. R and p values and a regression line of best fit are shown.
Protective efficacy following SARS-CoV-2 Omicron challenge
At week 19, all animals were challenged with 106 PFU SARS-CoV-2 Omicron by the intranasal and intratracheal routes. This challenge stock was generated in VeroE6-TMPRSS2 cells and had a titer of 2.3 × 109 TCID50/mL and 2.5 × 107 PFU/mL in VeroE6-TMPRSS2 cells, and the sequence of the challenge stock was fully verified (EPI_ISL_7171744; Mehul Suthar, Emory University). Following challenge, viral loads were assessed in bronchoalveolar lavage (BAL) and nasal swab (NS) samples by RT-PCR for E subgenomic RNA (sgRNA) (Dagotto et al., 2021; Wolfel et al., 2020), and infectious virus titers were quantitated by TCID50 assays.
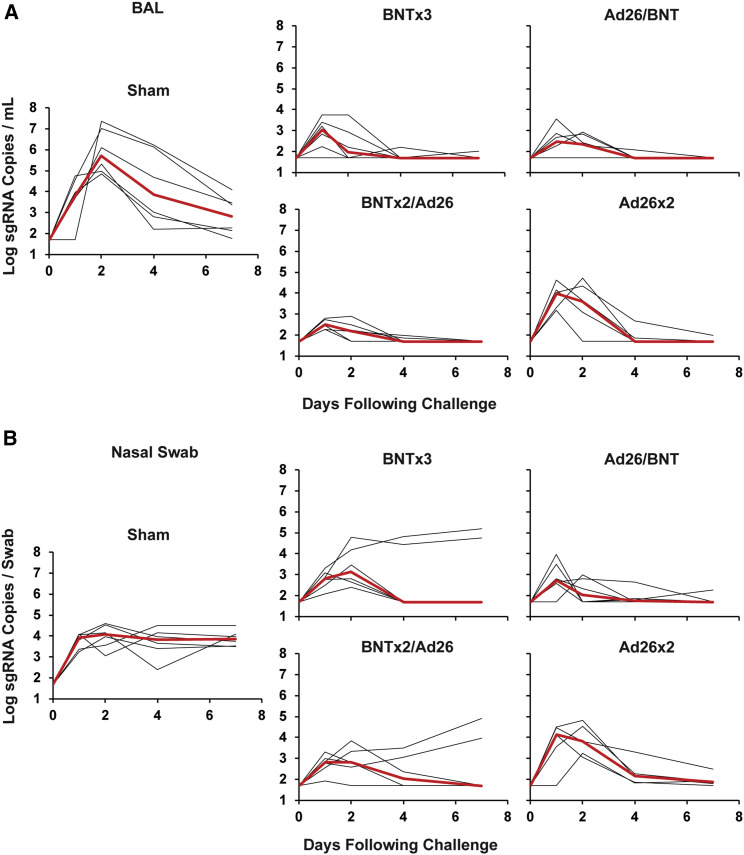
Sham controls showed high median viral loads of 5.70 (range 4.84–7.36) log sgRNA copies/mL in BAL on day 2, and these levels declined substantially by day 7 to median levels of 2.82 (range 1.78–4.10) log sgRNA copies/mL (Figure 4 A). Nearly all vaccinated animals demonstrated breakthrough infection in BAL, but viral loads were substantially lower in vaccinated animals compared with sham controls on day 2 and mostly resolved by day 4 (Figure 4A). In NS, sham controls showed lower median virus levels of 4.06 (range 3.05–4.59) log sgRNA copies/mL on day 2, but these levels only declined minimally by day 7 to median levels of 3.85 (range 3.50–4.49) log sgRNA copies/mL (Figure 4B). All vaccinated animals showed breakthrough infection in NS, but viral loads resolved in most vaccinated animals by day 4, with the exception of 2 animals in the BNTx3 group and 2 animals in the BNTx2/Ad26 group that showed persistent high levels of virus in NS through day 7, which was comparable with sham controls (Figure 4B).
Figure 4.
Viral loads following SARS-CoV-2 Omicron challenge
(A) Log subgenomic RNA (sgRNA) copies/mL in bronchoalveolar lavage (BAL) following SARS-CoV-2 Omicron challenge.
(B) Log subgenomic RNA (sgRNA) copies/swab in nasal swabs (NS) following SARS-CoV-2 Omicron challenge. Medians (red lines) are shown.
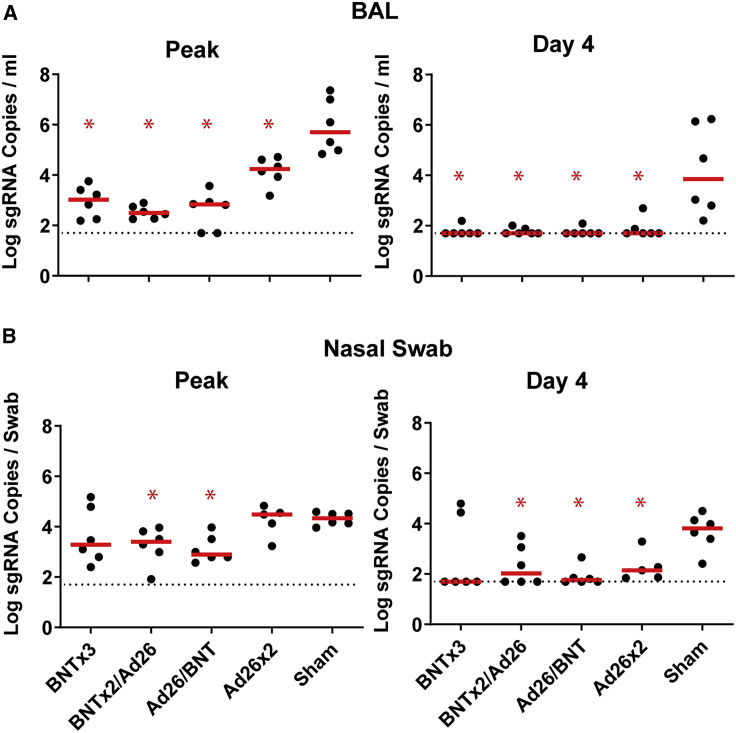
Median log peak viral loads in BAL were reduced by 2.68-, 3.21-, 2.87-, and 1.46-fold in the BNTx3, BNTx2/Ad26, Ad26/BNT, and Ad26x2 groups, respectively, compared with sham controls (p = 0.0022, p = 0.0022, p = 0.0022, and p = 0.0022, respectively, two-tailed Mann-Whitney tests; Figure 5 A). Median log day 4 viral loads in BAL were also significantly reduced to undetectable levels in all groups compared with sham controls (p = 0.0022, p = 0.0022, p = 0.0022, and p = 0.0043, respectively, two-tailed Mann-Whitney tests; Figure 5A). Median log peak viral loads in NS were only reduced in the heterologous BNTx2/Ad26 and Ad26/BNT groups compared with sham controls (p = 0.0043 and p = 0.0043, respectively, two-tailed Mann-Whitney tests; Figure 5B). Median log day 4 viral loads in NS were reduced in the BNTx2/Ad26, Ad26/BNT, and Ad26x2 groups compared with sham controls (p = 0.0152, p = 0.0043, and p = 0.0087, respectively, two-tailed Mann-Whitney tests; Figure 5B).
Figure 5.
Comparison of peak and day 4 viral loads
(A) Log subgenomic RNA (sgRNA) copies/mL in bronchoalveolar lavage (BAL) at peak and on day 4 following SARS-CoV-2 Omicron challenge.
(B) Log subgenomic RNA (sgRNA) copies/swab in nasal swabs (NS) at peak and on day 4 following SARS-CoV-2 Omicron challenge. Dotted lines represent limits of quantitation. Medians (red bars) are shown. Vaccinated groups were compared with the sham controls by two-sided Mann-Whitney tests. ∗p < 0.05.
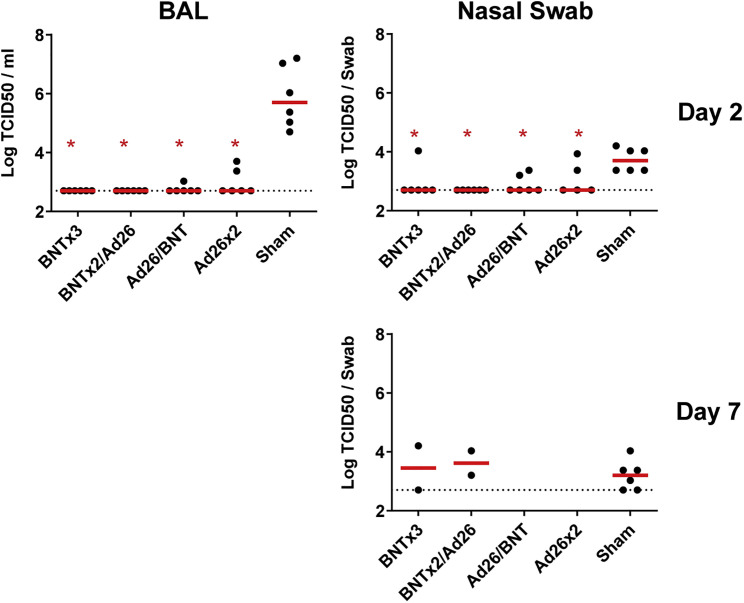
Consistent with the sgRNA viral load data, vaccinated animals also showed substantial reductions of infectious virus titers compared with sham controls in BAL and NS by TCID50 assays on day 2 (Figure S3 ). The 4 vaccinated animals and the 6 sham controls that showed persistent high levels of sgRNA in NS on day 7 also mostly showed persistent infectious virus titers by TCID50 assays (Figure S3).
Figure S3.
TCID50 titers, related to Figure 3
Log TCID50/mL in bronchoalveolar lavage (BAL) and nasal swabs on day 2 following SARS-CoV-2 Omicron challenge (top). Log TCID50/mL is also shown in nasal swabs on day 7 following SARS-CoV-2 Omicron challenge in the 4 vaccinated animals in the BNTx3 and BNTx2/Ad26 groups and in the 6 sham controls with persistently positive sgRNA levels on day 7 (bottom). Dotted lines represent limits of quantitation. Medians (red bars) are shown. Vaccinated groups were compared with the sham controls by two-sided Mann-Whitney tests. ∗p < 0.05.
Correlates of protection
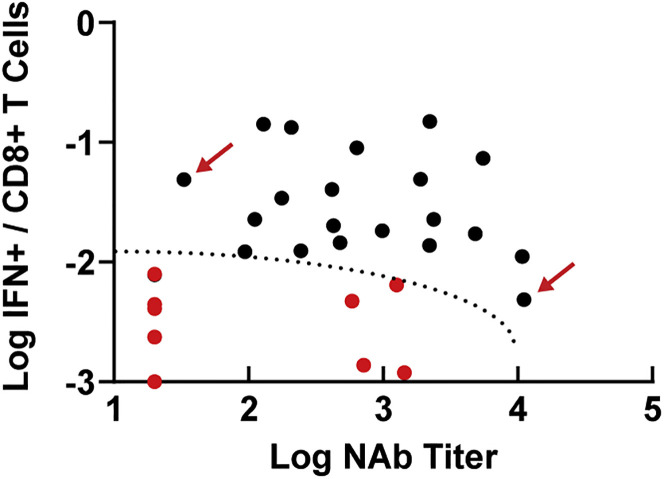
We evaluated the immunologic profiles of the 4 vaccinated animals that failed to control viral replication in NS following challenge. These animals had moderate Omicron-specific NAb titers (586–1,434) but negligible Omicron-specific CD8+ T cell responses (0.001%–0.006%) prior to challenge (red dots, Figure S4 ). These 4 vaccinated animals and the 6 sham controls fell into a defined region of “immunologic space” defined by low to moderate Omicron NAbs and low Omicron CD8+ T cell responses (Figure 6 ), suggesting that virologic failure following Omicron challenge was associated with simultaneously low humoral and cellular immunity to the challenge virus. In contrast, animals with a low NAb titer but a high CD8+ T cell response, or a particularly high NAb titer but a low CD8+ T cell response, demonstrated virologic control following challenge (red arrows; Figure 6).
Figure S4.
Omicron-specific NAb and CD8+ T cell responses following the boost immunization, related to Figures 1 and 2
The 4 animals that failed to show virologic control in NS are highlighted in red (2 in the BNTx3 group, 2 in the BNTx2/Ad26 group).
Figure 6.
Immunologic space defined by Omicron NAb titer and Omicron CD8+ T cell responses
Plot of all 30 animals by their post-boost Omicron NAb titer and Omicron CD8+ T cell responses. Red dots represent the 10 animals that failed to control virus by day 7 in NS (6 controls, 4 vaccinated animals). The dotted line represents the region of immunologic space, defined post-hoc, which was associated with failure of virologic control. Red arrows show representative animals with low NAb titers but high CD8+ T cell responses, or high NAb titers but low CD8+ cell responses, which showed rapid virologic control.
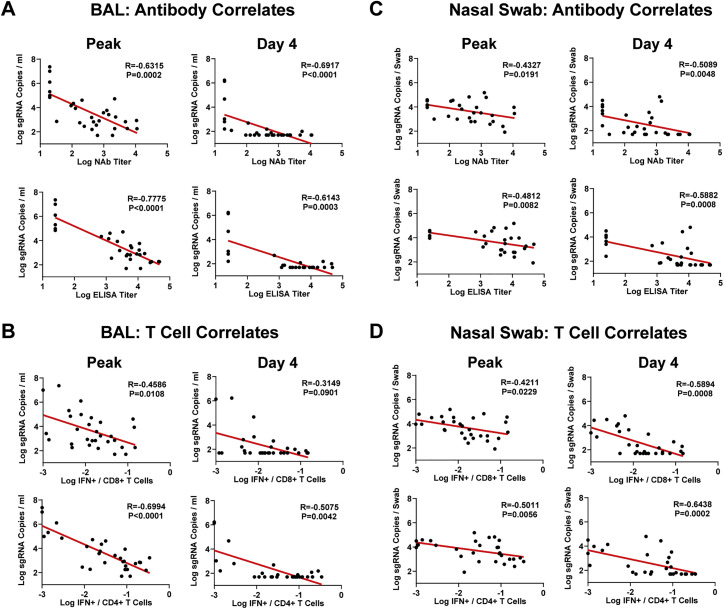
The variability of immune responses prior to challenge and viral loads following challenge allowed for a detailed immune correlates analysis. NAb titers, ELISA titers, CD8+ T cell responses, and CD4+ T cell responses all inversely correlated with sgRNA copies/mL in both BAL and NS (Figure S5 ). Since NAb titers and CD8+ T cell responses were not correlated (Figure 6), these data suggest that both humoral and cellular immunity separately contributed to virologic control following Omicron challenge.
Figure S5.
Correlates of protection, related to Figure 5
(A and B) Correlations of week 18 NAb and ELISA titers (A) and week 16 CD8+ and CD4+ T cell responses (B) with peak and day 4 sgRNA copies/mL in BAL are shown.
(C and D) Correlations of week 18 NAb and ELISA titers (C) and week 16 CD8+ and CD4+ T cell responses (D) with peak and day 4 sgRNA copies/swab in NS are also shown. Correlations were assessed by two-sided Spearman rank-correlation tests. R and p values and a regression line of best fit are shown.
Histopathology and immunohistochemistry
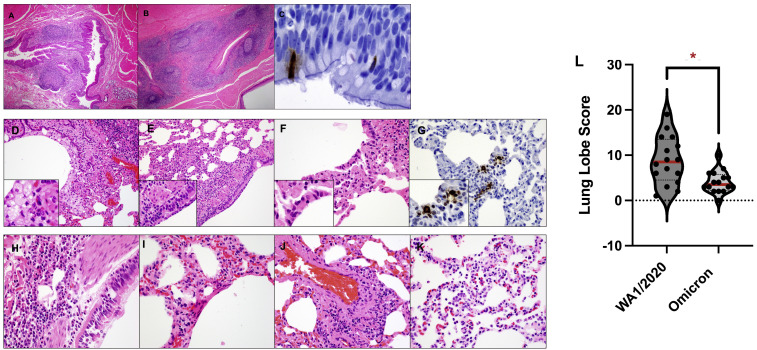
In a separate pilot study, we evaluated histopathology and immunohistochemistry on day 2 following infection of unvaccinated macaques with the SARS-CoV-2 Omicron variant. On day 2 following Omicron infection, we observed lymphoid hyperplasia in the submucosa and rare SARS-CoV-2-positive ciliated epithelial cells in the nasopharynx (Figures S6 A–S6C). Interstitial inflammation, expansion of septae, syncytial formation, and endothelialitis were observed in the lung in Omicron infected animals (Figures S6D–S6K). Lung histopathology scores were lower in macaques infected with Omicron compared with macaques infected with WA1/2020 (Chandrashekar et al., 2020) (p = 0.0054, two-tailed Mann-Whitney test; Figure S6L). In the main study, histopathology was minimal in both vaccinated animals and sham controls at necropsy on day 10 following Omicron challenge (data not shown).
Figure S6.
Histopathology and immunohistochemistry of Omicron infection, related to Figure 1
(A–K) (A–C) Pharynx and (D–K) lungs from macaques on day 2 following Omicron infection demonstrated lymphoid hyperplasia of the pharynx (A and B), SARS-N positive ciliated epithelial cells in the pharynx (C), foamy macrophages and degenerating neutrophils in bronchiole lumen (D), cellular necrotic debris adhering to bronchiolar ciliated epithelium (E), alveolar syncytia (F), SARS-N-positive ciliated epithelial cells in the pulmonary interstitium (G), neutrophilic bronchitis (H), hyaline membranes (I), endothelialitis (J), and type II pneumocyte hyperplasia (K). Scoring involved assessment of the following lesions: interstitial inflammation and septal thickening, interstitial infiltrate (eosinophils), interstitial infiltrate (neutrophils), hyaline membranes, interstitial fibrosis, alveolar infiltrate (macrophages), bronchoalveolar infiltrate (neutrophils), epithelial syncytia, type II pneumocyte hyperplasia, bronchi infiltrate (macrophages), bronchi infiltrate (neutrophils), bronchi (hyperplasia of bronchus-associated lymphoid tissue), bronchiolar or peribronchiolar infiltrate (mononuclear cells), perivascular infiltrate (mononuclear cells), and endothelialitis. Each feature assessed was assigned a score of: 0, no substantial findings; 1, minimal; 2, mild; 3, moderate; 4, moderate to severe; 5, marked or severe. Scores were added for all lesions across all lung lobes for each macaque, for a maximum possible score of 600 for each macaque.
(L) Summary of lung pathology scores from SARS-CoV-2 WA1/2020- and Omicron-infected macaques. Medians (red bars) are shown. Dotted line represents no pathology. Lung pathology scores were compared in macaques infected with Omicron versus WA1/2020 by two-sided Mann-Whitney tests. ∗p < 0.05.
Discussion
In this study, we demonstrate that the BNT162b2 and Ad26.COV2.S vaccines led to rapid virologic control in the upper and lower respiratory tracts following high-dose, heterologous challenge with the SARS-CoV-2 Omicron variant in the majority of macaques. However, 4 vaccinated animals with moderate Omicron-specific NAb titers but negligible Omicron-specific CD8+ T cell responses failed to control viral replication in NS by day 7. These data suggest the importance of vaccine-elicited CD8+ T cell responses and indicate that both humoral and cellular immune responses likely contribute to protection against the highly mutated SARS-CoV-2 Omicron variant in macaques. NAb responses in the absence of adequate CD8+ T cell responses may be insufficient for virologic control following Omicron challenge.
Correlates of vaccine protection against SARS-CoV-2 infection have to date largely focused on neutralizing antibody titers (Feng et al., 2021; Gilbert et al., 2022). Correlates of protection against severe disease, however, may be different than correlates of protection against infection, and the potential importance of vaccine-elicited T cell responses may be greater for SARS-CoV-2 variants such as Omicron that largely escape NAb responses. In this study, Omicron-specific NAbs were markedly lower than WA1/2020 NAbs, whereas Omicron-specific T cell responses were comparable WA1/2020 T cell responses, indicating substantial cross-reactivity of cellular immune responses against SARS-CoV-2 variants. Moreover, while BNT162b2 induced higher NAb responses than Ad26.COV2.S, Ad26.COV2.S induced higher CD8+ T cell responses than BNT162b2, which is consistent with human data (Atmar et al., 2022; Collier et al., 2021; Munro et al., 2021). The different immune profiles induced by mRNA and Ad26 vaccine platforms suggest possible advantages of heterologous prime-boost (“mix-and-match”) vaccine regimens for diversifying immune responses.
We observed that virus persisted longer in NS compared with BAL in sham controls following Omicron challenge, which differs from prior SARS-CoV-2 variants in macaques (Chandrashekar et al., 2020, 2021; He et al., 2021; Mercado et al., 2020; Yu et al., 2020, 2021b). Although the implications of this observation remain to be determined, prolonged duration of virus shedding in the upper respiratory tract, together with substantial escape from NAbs, may contribute to the high degree of transmissibility of the SARS-CoV-2 Omicron variant.
Recent studies have shown that BNT162b2 and Ad26.COV2.S provided robust 70% and 85% protection, respectively, against hospitalization with Omicron in South Africa (Collie et al., 2022; Gray et al., 2021), largely in the absence of Omicron-specific NAbs. These data suggest that immune parameters other than NAb responses likely contribute to protection against severe disease. We previously reported that CD8+ T cells contributed to protection against re-challenge with SARS-CoV-2 in convalescent macaques, particularly when antibody responses were suboptimal (McMahan et al., 2021). Taken together, our data suggest that protection against a highly mutated SARS-CoV-2 variant involves the combination of humoral and cellular immunity, and not NAbs alone unless antibody titers are exceptionally high. Specifically, moderate NAb titers without CD8+ T cell responses may be insufficient for virologic control. Future studies could also compare the relative importance of neutralizing versus functional non-neutralizing antibodies for protection. Taken together, these data have important implications for understanding immune correlates of protection against highly mutated SARS-CoV-2 variants.
Limitations of the study
This study utilizes viral loads and infectious virus titers following challenge to assess protective efficacy, and thus, immunologic correlates may not apply to protection against clinical disease, which is mild in macaques. This study also does not evaluate vaccination protection against transmission. Finally, the macaque model of SARS-CoV-2 infection may not be fully representative of human infection.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| mouse anti-human IFN-γ monoclonal antibody | BD Pharmigen | Cat # 554699 |
| Rabbit polyclonal anti-human IFN-γ Biotin | U-Cytech | Cat # CT243 |
| Streptavidin-alkaline phosphatase antibody | Southern Biotech | Cat # 7100-04 |
| Anti-macaque IgG HRP | Nonhuman Primate Reagent Resource | Cat# 1b3-HRP: 0320K235 / 070920SC |
| Mouse anti-human CD45 BUV805 | BD Pharmingen | Cat # 742055 RRID:AB_2871344 |
| Mouse anti-human CD11c Alexa700 | Affymetrix | Cat # 56-0116-42 RRID:AB_10547281 |
| Mouse anti-human CD123 Alexa700 | FISHER/NOVUS | Cat # NB6001185AF700 |
| Mouse anti-human CD7 Alexa700 | BD Pharmingen | Cat # 561603 RRID:AB_10898348 |
| Mouse anti-human CD3 APC-Cy7 | BD Pharmingen | Cat # 557757 RRID:AB_396863 |
| Mouse anti-human CD14 BV570 | BIOLEGEND | Cat # 301832 RRID:AB_2563629 |
| Mouse anti-human CD21 BV605 | BD Pharmingen | Cat # 740395 RRID:AB_2740125 |
| Mouse anti-human CD95 BV711 | BIOLEGEND | Cat # 305644 RRID:AB_2632623 |
| Mouse anti-human CD80 BV786 | BD Pharmingen | Cat # 564159 RRID:AB_2738631 |
| Mouse anti-human IgM BUV395 | BD Pharmingen | Cat # 563903 RRID:AB_2721269 |
| Mouse anti-human CD27 BUV563 | BD Pharmingen | Cat # 741366 RRID:AB_2870866 |
| Mouse anti-human IgG BUV737 | BD Pharmingen | Cat # 612819 RRID:AB_2870143 |
| Goat anti-human IgD PE | Southern Biotec | Cat # 2030-09 RRID:AB_2795630 |
| Mouse anti-human Ki-67 PerCP-cy5.5 | BD Pharmingen | Cat # 561284 RRID:AB_10611574 |
| Mouse Anti-Bcl-6 PE-CF594 | BD Pharmingen | Cat # 562401 |
| SARS CoV Nucleoprotein / NP mouse monoclonal antibody (Clone MM05) | Sino Biological | Cat # 40143-MM05 RRID:AB_2827977 |
| Biological samples | ||
| Non-human primate SARS-CoV2 infected lung tissue, paraformaldehyde fixed, paraffin embedded | Bioqual, Inc.; Tufts Cummings School of Veterinary Medicine | N/A |
| Bronchoalveolar lavage from Non-Human Primates | Bioqual, Inc. | N/A |
| Nasal swabs from Non-Human Primates | Bioqual, Inc. | N/A |
| EDTA, SST, Paxgene collection tubes with whole blood, from Non-Human Primates | Bioqual, Inc. | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Peptides for WA1/2020, Omicron, Delta, Beta | 21st Century Biochemicals | Custom |
| Nitroblue Tetrazolium Chloride/5-bromo-4-chloro 3 ‘indolyl phosphate p-toludine salt | Thermo Scientific | Cat #34042 |
| SARS-CoV-2 Receptor Binding Domain protein (WA1/2020, Omicron, Delta, Beta) | Aaron Schmidt Laboratory; Harvard Medical School | https://doi.org/10.1126/sciimmunol.abe0367 |
| Biotinylated SARS-CoV-2 (WA1/2020) RBD | Sino Biological | Cat # 40592-V08B-B |
| SARS-CoV-2 (WA1/2020) RBD | Sino Biological | Cat # 40592-V08B |
| SARS-CoV-2 (Omicron) RBD | Sino Biological | Cat # 40592-V08H121 |
| BV650 streptavidin | BD Pharmingen | Cat # 563855 RRID:AB_2869528 |
| Fixation Medium (Medium A) | ThermoFisher Scientific | Cat # GAS001S100 |
| Permeabilization Medium (Medium B) | ThermoFisher Scientific | Cat # GAS002S100 |
| Citrate buffer antigen retrieval | ThermoFischer Scientific | Cat # AP-9003-500 |
| Protein block | Biocare | Cat # BP974M |
| DaVinci Green Antibody diluent | Biocare | Cat # PD900M |
| Mach-2 Mouse HRP-Polymer | Biocare | Cat # MHRP520L |
| Critical commercial assays | ||
| V-PLEX SARS-CoV-2 Panel 22 Kit – Human IgG | Meso Scale Discovery | K15559U |
| V-PLEX SARS-CoV-2 Panel 23 Kit – Human IgG | Meso Scale Discovery | K15567U |
| Experimental models: Cell lines | ||
| HEK293T-hACE2 | This paper | N/A |
| Experimental models: Organisms/strains | ||
| Macaca mulatta | https://www.unmc.edu/rhesusgenechip/index.htm | https://www.unmc.edu/rhesusgenechip/index.htm |
| Oligonucleotides | ||
| Primer:sgLeadSARSCoV2-F Forward: CGATCTCTTGTAGATCTGTTCTC | Wolfel et al., 2020 | ThermoFisher Scientific:4448510 |
| Primer: E_Sarbeco_R Reverse: ATATTGCAGCAGTACGCACACA | Wolfel et al., 2020 | ThermoFisher Scientific:4448510 |
| Probe:E_Sarbeco_P1 : VIC-ACACT AGCCATCCTTACTGCGCTTCG-MGBNFQ |
Wolfel et al., 2020 | ThermoFisher Scientific:4448510 |
| Primer:pcDNA.T7.NdeI.Fwd Forward: TGATCTCATATGGAACCCACTGCTTACTG | This paper | Integrated DNA Technologies |
| Primer: pcDNA.T7.Rev Reverse: CACTGTGCTGGATATCTGC | This paper | Integrated DNA Technologies |
| Recombinant DNA | ||
| psPAX2 | AIDS Resource and Reagent Program | Cat# 11348 |
| pLenti-CMV Puro-Luc | Addgene | Cat# 17477 |
| pcDNA3.1-SARS CoV-2 SΔCT | This paper | N/A |
| Software and algorithms | ||
| GraphPad Prism 8.4.2 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| FlowJo | BD Bioscience | https://www.flowjo.com/ |
| Diva | ||
| SoftMax Pro 6.5.1 | SoftMax Pro Software | https://www.moleculardevices.com/products/microplate-readers/acquisition-and-analysis-software/softmax-pro-software |
| BioRender | BioRender | https://biorender.com/ |
| Other | ||
| Ad26.COV2.S | Janssen | JNJ-78436735 |
| Comirnaty | Pfizer | BNT162b2 |
| RNA Standard: SARS-CoV-2 E gene subgenomic RNA (sgRNA) | This paper | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Dan Barouch (dbarouch@bidmc.harvard.edu).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Animals and study design
30 outbred adult male and female cynomolgus macaques ages 4-12 years old were randomly allocated to 5 experimental groups (N=6/group; Figure S1). Animals were 3-10 years old, and 3-10kg. All animals were singly housed at Bioqual, Inc. (Rockville, MD). Groups of animals were primed with either two immunizations of 30 μg BNT162b2 at weeks 0 and 3 or a single immunization of 5x1010 vp Ad26.COV2.S at week 0. At week 14, animals were boosted with either 30 μg BNT162b2 or 5x1010 vp Ad26.COV2.S. Clinical vaccines were obtained from pharmacies by the NIH SAVE Consortium. At week 19, all animals were challenged with 106 PFU SARS-CoV-2 Omicron by the intranasal and intratracheal routes in a total volume of 2 mls. This challenge stock was generated in VeroE6-TMPRSS2 cells and had a titer of 2.3x109 TCID50/ml and 2.5x107 PFU/ml in VeroE6-TMPRSS2 cells and was fully sequenced (EPI_ISL_7171744; Mehul Suthar, Emory University). Following challenge, viral loads were assessed in bronchoalveolar lavage (BAL) and nasal swab (NS) samples by RT-PCR for E subgenomic RNA (sgRNA), and infectious virus titers were quantitated by TCID50 assays. Animals were sacrificed on day 9 or 10 following challenge. Immunologic and virologic assays were performed blinded. All animal studies were conducted in compliance with all relevant local, state, and federal regulations and were approved by the Bioqual Institutional Animal Care and Use Committee (IACUC).
Method details
Pseudovirus neutralizing antibody assay
The SARS-CoV-2 pseudoviruses expressing a luciferase reporter gene were used to measure pseudovirus neutralizing antibodies (Yu et al., 2021a). In brief, the packaging construct psPAX2 (AIDS Resource and Reagent Program), luciferase reporter plasmid pLenti-CMV Puro-Luc (Addgene) and spike protein expressing pcDNA3.1-SARS-CoV-2 SΔCT were co-transfected into HEK293T cells (ATCC CRL_3216) with lipofectamine 2000 (ThermoFisher Scientific). Pseudoviruses of SARS-CoV-2 variants were generated by using WA1/2020 strain (Wuhan/WIV04/2019, GISAID accession ID: EPI_ISL_402124), B.1.617.2 (Delta, GISAID accession ID: EPI_ISL_2020950), or B.1.1.529 (Omicron, GISAID ID: EPI_ISL_7358094.2). The supernatants containing the pseudotype viruses were collected 48h after transfection; pseudotype viruses were purified by filtration with 0.45-μm filter. To determine the neutralization activity of human serum, HEK293T-hACE2 cells were seeded in 96-well tissue culture plates at a density of 2.0 × 104 cells per well overnight. Three-fold serial dilutions of heat-inactivated serum samples were prepared and mixed with 50 μl of pseudovirus. The mixture was incubated at 37 °C for 1 h before adding to HEK293T-hACE2 cells. After 48 h, cells were lysed in Steady-Glo Luciferase Assay (Promega) according to the manufacturer’s instructions. SARS-CoV-2 neutralization titers were defined as the sample dilution at which a 50% reduction (NT50) in relative light units was observed relative to the average of the virus control wells.
Enzyme-linked immunosorbent assay (ELISA)
SARS-CoV-2 spike receptor-binding domain (RBD)-specific binding antibodies in serum were assessed by ELISA. 96-well plates were coated with 1 μg/mL of similarly produced SARS-CoV-2 WA1/2020, B.1.617.2 (Delta), B.1.351 (Beta), or B.1.1.529 (Omicron) RBD protein in 1× Dulbecco phosphate-buffered saline (DPBS) and incubated at 4 °C overnight. Assay performance was similar for these four RBD proteins. After incubation, plates were washed once with wash buffer (0.05% Tween 20 in 1× DPBS) and blocked with 350 μL of casein block solution per well for 2 to 3 hours at room temperature. Following incubation, block solution was discarded and plates were blotted dry. Serial dilutions of heat-inactivated serum diluted in Casein block were added to wells, and plates were incubated for 1 hour at room temperature, prior to 3 more washes and a 1-hour incubation with a 1μg/mL dilution of anti–macaque IgG horseradish peroxidase (HRP) (Nonhuman Primate Reagent Resource) at room temperature in the dark. Plates were washed 3 times, and 100 μL of SeraCare KPL TMB SureBlue Start solution was added to each well; plate development was halted by adding 100 μL of SeraCare KPL TMB Stop solution per well. The absorbance at 450 nm was recorded with a VersaMax microplate reader (Molecular Devices). For each sample, the ELISA end point titer was calculated using a 4-parameter logistic curve fit to calculate the reciprocal serum dilution that yields an absorbance value of 0.2. Interpolated end point titers were reported.
Electrochemiluminescence assay (ECLA)
ECLA plates (MesoScale Discovery SARS-CoV-2 IgG, Panels 22, 23) were designed and produced for multiplex binding assays with up to 10 antigen spots in each well, including either Spike or RBD proteins from multiple SARS-CoV-2 variants (Jacob-Dolan et al., 2021). The plates were blocked with 50 uL of Blocker A (1% BSA in distilled water) solution for at least 30 minutes at room temperature shaking at 700 rpm with a digital microplate shaker. During blocking the serum was diluted to 1:5,000 or 1:50,000 in Diluent 100. The calibrator curve was prepared by diluting the calibrator mixture from MSD 1:10 in Diluent 100 and then preparing a 7-step 4-fold dilution series plus a blank containing only Diluent 100. The plates were then washed 3 times with 150 μL of Wash Buffer (0.5% Tween in 1x PBS), blotted dry, and 50 μL of the diluted samples and calibration curve were added in duplicate to the plates and set to shake at 700 rpm at room temperature for at least 2 h. The plates were again washed 3 times and 50 μL of SULFO-Tagged anti-Human IgG detection antibody diluted to 1x in Diluent 100 was added to each well and incubated shaking at 700 rpm at room temperature for at least 1 h. Plates were then washed 3 times and 150 μL of MSD GOLD Read Buffer B was added to each well and the plates were read immediately after on a MESO QuickPlex SQ 120 machine. MSD titers for each sample was reported as Relative Light Units (RLU) which were calculated as Sample RLU minus Blank RLU and then fit using a logarithmic fit to the standard curve. The upper limit of detection was defined as 2x10∧6 RLU for each assay and the signal for samples which exceeded this value at 1:5,000 serum dilution was run again at 1:50,000 and the fitted RLU was multiplied by 10 before reporting. The lower limit of detection was defined as 1 RLU and an RLU value of 100 was defined to be positive for each assay.
Intracellular cytokine staining (ICS) assay
CD4+ and CD8+ T cell responses were quantitated by pooled peptide-stimulated intracellular cytokine staining (ICS) assays. Peptide pools were 16 amino acid peptides overlapping by 11 amino acids spanning the SARS-CoV-2 WA1/2020, B.1.617.2 (Delta), or B.1.1.529 (Omicron; GISAID ID: EPI_ISL_7358094.2) Spike proteins (21st Century Biochemicals). 106 peripheral blood mononuclear cells well were re-suspended in 100 μL of R10 media supplemented with CD49d monoclonal antibody (1 μg/mL) and CD28 monoclonal antibody (1 μg/mL). Each sample was assessed with mock (100 μL of R10 plus 0.5% DMSO; background control), peptides (2 μg/mL), and/or 10 pg/mL phorbol myristate acetate (PMA) and 1 μg/mL ionomycin (Sigma-Aldrich) (100μL; positive control) and incubated at 37°C for 1 h. After incubation, 0.25 μL of GolgiStop and 0.25 μL of GolgiPlug in 50 μL of R10 was added to each well and incubated at 37°C for 8 h and then held at 4°C overnight. The next day, the cells were washed twice with DPBS, stained with aqua live/dead dye for 10 mins and then stained with predetermined titers of monoclonal antibodies against CD279 (clone EH12.1, BB700), CD38 (clone OKT10, PE), CD28 (clone 28.2, PE CY5), CD4 (clone L200, BV510), CD95 (clone DX2, BUV737), CD8 (clone SK1, BUV805) for 30 min. Cells were then washed twice with 2% FBS/DPBS buffer and incubated for 15 min with 200 μL of BD CytoFix/CytoPerm Fixation/Permeabilization solution. Cells were washed twice with 1X Perm Wash buffer (BD Perm/WashTM Buffer 10X in the CytoFix/CytoPerm Fixation/ Permeabilization kit diluted with MilliQ water and passed through 0.22μm filter) and stained with intracellularly with monoclonal antibodies against Ki67 (clone B56, FITC), CD69 (clone TP1.55.3, ECD), IL10 (clone JES3-9D7, PE CY7), IL13 (clone JES10-5A2, BV421), TNF-α (clone Mab11, BV650), IL4 (clone MP4-25D2, BV711), IFN-γ (clone B27; BUV395), CD45 (clone D058-1283, BUV615), IL2 (clone MQ1-17H12, APC), CD3 (clone SP34.2, Alexa 700)for 30 min. Cells were washed twice with 1X Perm Wash buffer and fixed with 250μL of freshly prepared 1.5% formaldehyde. Fixed cells were transferred to 96-well round bottom plate and analyzed by BD FACSymphony™ system. Data were analyzed using FlowJo v9.9.
B cell immunophenotyping
PBMCs or inguinal LN cells were stained with Aqua live/dead dye for 20 minutes, washed with 2% FBS/DPBS buffer, and cells were suspended in 2% FBS/DPBS buffer with Fc Block (BD Biosciences) for 10 minutes (He et al., 2021). After blocking, samples were stained with monoclonal antibodies against CD45 (clone D058-1283, brilliant ultraviolet (BUV) 805), CD3 (clone SP34.2, allophycocyanin (APC)-Cy7), CD7 (clone M-T701, Alexa Fluor700), CD123 (clone 6H6, Alexa Fluor 700), CD11c (clone 3.9, Alexa Fluor 700), CD19 (clone J3-119, phycoerythrin (PE)), CD20 (clone 2H7, PE-Cy5), IgD (IA6-2, PE), IgG (clone G18-145, BUV737), IgM (clone G20-127, BUV395), CD80 (clone L307.4, brilliant violet (BV) 786), CD95 (clone DX2, BV711), CD27 (clone M-T271, BUV563), CD21 (clone B-ly4, BV605), CD14 (clone M5E2, BV570). Samples were also stained with SARS-CoV-2 antigens, including biotinylated SARS-CoV-2 (WA1/2020) RBD proteins (Sino Biological), SARS-CoV-2 (WA1/2020) RBD proteins (Sino Biological) labeled with fluorescein isothiocyanate (FITC), SARS-CoV-2 (B.1.1.529) RBD proteins (Sino Biological) labeled with APC and DyLight 405. Staining was done at 4 °C for 30 minutes. After staining, cells were washed twice with 2% FBS/DPBS buffer, followed by incubation with BV650 streptavidin (BD Pharmingen) for 10 minutes, then washed twice with 2% FBS/DPBS buffer. For intracellular staining, cells were permeabilized using Caltag Fix & Perm (Thermo Fisher Scientific), then stained with monoclonal antibodies against Ki67 (clone B56, peridinin chlorophyll protein (PerCP)-Cy5.5) and Bcl6 (clone K112-91, PE-CF594). After staining, cells were washed and fixed by 2% paraformaldehyde. All data were acquired on a BD FACSymphony flow cytometer. Subsequent analyses were performed using FlowJo software (BD Bioscience, v.9.9.6). For analyses, in singlet gate, dead cells were excluded by Aqua dye and CD45 was used as a positive inclusion gate for all leukocytes. Within class-switched memory B cell populations, gated as CD20+IgG+CD27+IgM-CD3-CD14-CD11c-CD123-CD7-, SARS-CoV-2 WA1/2020 RBD-specific B cells were identified as double positive for SARS-CoV-2 (WA1/2020) RBD labeled with different fluorescent probes, and SARS-CoV-2 (B.1.1.529) RBD-specific B cells were identified as double positive for SARS-CoV-2 (B.1.1.529) RBD proteins labeled with different fluorescent probes. Within GC B cells gated as CD20+ IgD- IgG+ Ki67+ Bcl6+, SARS-CoV-2 RBD-specific GC B cells were identified as double positive for SARS-CoV-2 RBD with different fluorescent probes.
Subgenomic RT-PCR assay
SARS-CoV-2 E gene subgenomic RNA (sgRNA) was assessed by RT-PCR using primers and probes as previously described (Yu et al., 2021a). A standard was generated by first synthesizing a gene fragment of the subgenomic E gene. The gene fragment was subsequently cloned into a pcDNA3.1+ expression plasmid using restriction site cloning (Integrated DNA Technologies). The insert was in vitro transcribed to RNA using the AmpliCap-Max T7 High Yield Message Maker Kit (CellScript). Log dilutions of the standard were prepared for RT-PCR assays ranging from 1x1010 copies to 1x10-1 copies. Viral loads were quantified from bronchoalveolar lavage (BAL) fluid and nasal swabs (NS). RNA extraction was performed on a QIAcube HT using the IndiSpin QIAcube HT Pathogen Kit according to manufacturer’s specifications (Qiagen). The standard dilutions and extracted RNA samples were reverse transcribed using SuperScript VILO Master Mix (Invitrogen) following the cycling conditions described by the manufacturer. A Taqman custom gene expression assay (Thermo Fisher Scientific) was designed using the sequences targeting the E gene sgRNA. The sequences for the custom assay were as follows, forward primer, sgLeadCoV2.Fwd: CGATCTCTTGTAGATCTGTTCTC, E_Sarbeco_R: ATATTGCAGCAGTACGCACACA, E_Sarbeco_P1 (probe): VIC-ACACTAGCCATCCTTACTGCGCTTCG-MGBNFQ. Reactions were carried out in duplicate for samples and standards on the QuantStudio 6 and 7 Flex Real-Time PCR Systems (Applied Biosystems) with the thermal cycling conditions: initial denaturation at 95°C for 20 seconds, then 45 cycles of 95°C for 1 second and 60°C for 20 seconds. Standard curves were used to calculate subgenomic RNA copies per ml or per swab. The quantitative assay sensitivity was determined as 50 copies per ml or per swab.
TCID50 assay
Vero-TMPRSS2 cells (obtained from A. Creanga) were plated at 25,000 cells per well in DMEM with 10% FBS and gentamicin, and the cultures were incubated at 37 °C, 5.0% CO2. Medium was aspirated and replaced with 180 μl of DMEM with 2% FBS and gentamicin. Serial dilution of samples as well as positive (virus stock of known infectious titre) and negative (medium only) controls were included in each assay. The plates are incubated at 37 °C, 5.0% CO2 for 4 days. Cell monolayers were visually inspected for cytopathic effect. The TCID50 was calculated using the Read–Muench formula.
Histopathology and immunohistochemistry
Lungs from SARS CoV-2 WA1/2020 and Omicron infected macaques were evaluated on day 2 following challenge by histopathology (Chandrashekar et al., 2020). At the time of fixation, lungs were suffused with 10% formalin to expand the alveoli. All tissues were fixed in 10% formalin and blocks sectioned at 5 μm. Slides were incubated for 30–60 min at 65°C then deparaffinized in xylene and rehydrated through a series of graded ethanol to distilled water. Sections were stained with hematoxylin and eosin. For SARS-N immunohistochemistry, heat-induced epitope retrieval was performed using a pressure cooker on steam setting for 25 min in citrate buffer (Thermo Fisher Scientific, AP-9003–500), followed by treatment with 3% hydrogen peroxide. Slides were then rinsed in distilled water and protein blocked (Biocare, BP974M) for 15 min followed by rinses in 1× PBS. Primary mouse anti-SARS-CoV-nucleoprotein antibody (Sinobiological; 40143-MM05) at 1:1000, was applied for 60 min, followed by mouse Mach-2 HRP-Polymer (Biocare) for 30 min and then counterstained with hematoxylin followed by bluing using 0.25% ammonia water. Staining was performed using a Biocare intelliPATH autostainer. Blinded evaluation and histopathologic scoring of eight representative lung lobes from cranial, middle and caudal, left and right lungs from each monkey was performed by a board-certified veterinary pathologist (AJM).
Quantification and statistical analysis
Descriptive statistics and logistic regression were performed using GraphPad Prism 8.4.3, (GraphPad Software, San Diego, California). Immunologic data were generated in duplicate and were compared by two-sided Mann-Whitney tests. Correlations were assessed by two-sided Spearman rank-correlation tests. P values less than 0.05 were considered significant.
Acknowledgments
The authors thank S. Ducat, T. Hayes, A. Goode, G. Romero, J. Yalley-Ogunro, M. Cabus, B. Narvaez, E. Bouffard, S. Gardner, M. Gebre, V. Giffin, and Y. Tian for generous advice, assistance, and reagents. We thank MesoScale Discovery for providing the ECLA kits. Funding: we acknowledge NIH contract 75N93021C00014, NIH grant CA260476, the Massachusetts Consortium for Pathogen Readiness, the Ragon Institute, and the Musk Foundation (D.H.B.) and NIH contract 75N93021C00016 (A.C.M.B.).
Author contributions
This study was designed by D.H.B. Immunologic and virologic assays were led by A.C., J.L., K.M., C.J.-D., J.L., X.H., D.H., T.A., J.B., B.C., N.P.H., M.L., J.M., M. Sciacca, D.S., M. Siamatu, O.P., H.V., H.W., and C.W. Humoral immune responses were assessed by C.J.-D., K.M., and J.Y. Vaccines were provided by A.C.M.B. Omicron challenge stock was provided by M.S.S. Animal work was led by L.P., D.V., A.V.R., J.M., M.B., A.C., J.V., E.T., M.G.L., and H.A. Pathology was led by N.J. and A.J.M. The paper was written by D.H.B. with the involvement of all co-authors.
Declaration of interests
D.H.B. is a co-inventor on provisional vaccine patents licensed to Janssen (63/121,482; 63/133,969; 63/135,182) and serves as a consultant to Pfizer. The authors report no other conflict of interest. A.C.M.B. has received funding from Abbvie for the commercial development of SARS-CoV-2 mAbs.
Inclusion and diversity
We worked to ensure sex balance in the selection of non-human subjects. One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science.
Published: April 28, 2022
Data and code availability
There is no dataset/code associated with the paper.
References
- Alter G., Yu J., Liu J., Chandrashekar A., Borducchi E.N., Tostanoski L.H., McMahan K., Jacob-Dolan C., Martinez D.R., Chang A., et al. Immunogenicity of Ad26.COV2.S vaccine against SARS-CoV-2 variants in humans. Nature. 2021;596:268–272. doi: 10.1038/s41586-021-03681-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atmar R.L., Lyke K.E., Deming M.E., Jackson L.A., Branche A.R., El Sahly H.M., Rostad C.A., Martin J.M., Johnston C., Rupp R.E., et al. Homologous and heterologous Covid-19 booster vaccinations. N. Engl. J. Med. 2022;386:1046–1057. doi: 10.1056/NEJMoa2116414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreño J.M., Alshammary H., Tcheou J., Singh G., Raskin A.J., Kawabata H., Sominsky L.A., Clark J.J., Adelsberg D.C., Bielak D.A., et al. Activity of convalescent and vaccine serum against SARS-CoV-2 Omicron. Nature. 2022;602:682–688. doi: 10.1038/s41586-022-04399-5. [DOI] [PubMed] [Google Scholar]
- Cele S., Jackson L., Khoury D.S., Khan K., Moyo-Gwete T., Tegally H., San J.E., Cromer D., Scheepers C., Amoako D.G., et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 2022;602:654–656. doi: 10.1038/s41586-021-04387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrashekar A., Liu J., Martinot A.J., McMahan K., Mercado N.B., Peter L., Tostanoski L.H., Yu J., Maliga Z., Nekorchuk M., et al. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science. 2020;369:812–817. doi: 10.1126/science.abc4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrashekar A., Liu J., Yu J., McMahan K., Tostanoski L.H., Jacob-Dolan C., Mercado N.B., Anioke T., Chang A., Gardner S., et al. Prior infection with SARS-CoV-2 WA1/2020 partially protects rhesus macaques against reinfection with B.1.1.7 and B.1.351 variants. Sci. Transl. Med. 2021;13:eabj2641. doi: 10.1126/scitranslmed.abj2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collie S., Champion J., Moultrie H., Bekker L.G., Gray G. Effectiveness of BNT162b2 vaccine against Omicron variant in South Africa. N. Engl. J. Med. 2022;386:494–496. doi: 10.1056/NEJMc2119270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier A.Y., Yu J., McMahan K., Liu J., Chandrashekar A., Maron J.S., Atyeo C., Martinez D.R., Ansel J.L., Aguayo R., et al. Differential kinetics of immune responses elicited by Covid-19 vaccines. N. Engl. J. Med. 2021;385:2010–2012. doi: 10.1056/NEJMc2115596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagotto G., Mercado N.B., Martinez D.R., Hou Y.J., Nkolola J.P., Carnahan R.H., Crowe J.E., Jr., Baric R.S., Barouch D.H. Comparison of subgenomic and total RNA in SARS-CoV-2 challenged rhesus macaques. J. Virol. 2021 doi: 10.1128/JVI.02370-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falsey A.R., Frenck R.W., Jr., Walsh E.E., Kitchin N., Absalon J., Gurtman A., Lockhart S., Bailey R., Swanson K.A., Xu X., et al. SARS-CoV-2 neutralization with BNT162b2 vaccine dose 3. N. Engl. J. Med. 2021;385:1627–1629. doi: 10.1056/NEJMc2113468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Phillips D.J., White T., Sayal H., Aley P.K., Bibi S., Dold C., Fuskova M., Gilbert S.C., Hirsch I., et al. Correlates of protection against symptomatic and asymptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:2032–2040. doi: 10.1038/s41591-021-01540-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert P.B., Montefiori D.C., McDermott A.B., Fong Y., Benkeser D., Deng W., Zhou H., Houchens C.R., Martins K., Jayashankar L., et al. Immune correlates analysis of the mRNA-1273 COVID-19 vaccine efficacy clinical trial. Science. 2022;375:43–50. doi: 10.1126/science.abm3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray G.E., Collie S., Garrett N., Goga A., Champion J., Zylstra M., Reddy T., Yende N., Seocharan I., Takalani A., et al. Vaccine effectiveness against hospital admission in South African health care workers who received a homologous booster of Ad26.COV2 during an Omicron COVID19 wave: preliminary results of the Sisonke 2 study. medRxiv. 2021 doi: 10.1101/2021.12.28.21268436. [DOI] [Google Scholar]
- He X., Chandrashekar A., Zahn R., Wegmann F., Yu J., Mercado N.B., McMahan K., Martinot A.J., Piedra-Mora C., Beecy S., et al. Low-dose Ad26.COV2.S protection against SARS-CoV-2 challenge in rhesus macaques. Cell. 2021;184:3467–3473.e11. doi: 10.1016/j.cell.2021.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob-Dolan C., Feldman J., McMahan K., Yu J., Zahn R., Wegmann F., Schuitemaker H., Schmidt A.G., Barouch D.H. Coronavirus-specific antibody cross reactivity in rhesus macaques following SARS-CoV-2 vaccination and infection. J. Virol. 2021;95:e00117–e00121. doi: 10.1128/JVI.00117-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeton R., Tincho M.B., Ngomti A., Baguma R., Benede N., Suzuki A., Khan K., Cele S., Bernstein M., Karim F., et al. T cell responses to SARS-CoV-2 spike cross-recognize Omicron. Nature. 2022;603:488–492. doi: 10.1038/s41586-022-04460-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Chandrashekar A., Sellers D., Barrett J., Jacob-Dolan C., Lifton M., McMahan K., Sciacca M., VanWyk H., Wu C., et al. Vaccines elicit highly conserved cellular immunity to SARS-CoV-2 Omicron. Nature. 2022;603:493–496. doi: 10.1038/s41586-022-04465-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Iketani S., Guo Y., Chan J.F., Wang M., Liu L., Luo Y., Chu H., Huang Y., Nair M.S., et al. Striking antibody evasion manifested by the omicron variant of SARS-CoV-2. Nature. 2022;602:676–681. doi: 10.1038/s41586-021-04388-0. [DOI] [PubMed] [Google Scholar]
- McMahan K., Yu J., Mercado N.B., Loos C., Tostanoski L.H., Chandrashekar A., Liu J., Peter L., Atyeo C., Zhu A., et al. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature. 2021;590:630–634. doi: 10.1038/s41586-020-03041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercado N.B., Zahn R., Wegmann F., Loos C., Chandrashekar A., Yu J., Liu J., Peter L., McMahan K., Tostanoski L.H., et al. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature. 2020;586:583–588. doi: 10.1038/s41586-020-2607-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro A.P.S., Janani L., Cornelius V., Aley P.K., Babbage G., Baxter D., Bula M., Cathie K., Chatterjee K., Dodd K., et al. Safety and immunogenicity of seven COVID-19 vaccines as a third dose (booster) following two doses of ChAdOx1 nCov-19 or BNT162b2 in the UK (COV-BOOST): a blinded, multicentre, randomised, controlled, phase 2 trial. Lancet. 2021;398:2258–2276. doi: 10.1016/S0140-6736(21)02717-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemet I., Kliker L., Lustig Y., Zuckerman N., Erster O., Cohen C., Kreiss Y., Alroy-Preis S., Regev-Yochay G., Mendelson E., Mandelboim M. Third BNT162b2 vaccination neutralization of SARS-CoV-2 Omicron infection. N. Engl. J. Med. 2022;386:492–494. doi: 10.1056/NEJMc2119358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadoff J., Gray G., Vandebosch A., Cárdenas V., Shukarev G., Grinsztejn B., Goepfert P.A., Truyers C., Fennema H., Spiessens B., et al. Safety and efficacy of single-dose Ad26.COV2.S vaccine against Covid-19. N. Engl. J. Med. 2021;384:2187–2201. doi: 10.1056/NEJMoa2101544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt F., Muecksch F., Weisblum Y., Da Silva J., Bednarski E., Cho A., Wang Z., Gaebler C., Caskey M., Nussenzweig M.C., et al. Plasma neutralization of the SARS-CoV-2 Omicron variant. N. Engl. J. Med. 2022;386:599–601. doi: 10.1056/NEJMc2119641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarke A., Coelho C.H., Zhang Z., Dan J.M., Yu E.D., Methot N., Bloom N.I., Goodwin B., Phillips E., Mallal S., et al. SARS-CoV-2 vaccination induces immunological T cell memory able to cross-recognize variants from Alpha to Omicron. Cell. 2022;185:847–859.e11. doi: 10.1016/j.cell.2022.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Muller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Yu J., Li Z., He X., Gebre M.S., Bondzie E.A., Wan H., Jacob-Dolan C., Martinez D.R., Nkolola J.P., Baric R.S., Barouch D.H. Deletion of the SARS-CoV-2 spike cytoplasmic tail increases infectivity in pseudovirus neutralization assays. J. Virol. 2021;95 doi: 10.1128/JVI.00044-21. e00044–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Tostanoski L.H., Mercado N.B., McMahan K., Liu J., Jacob-Dolan C., Chandrashekar A., Atyeo C., Martinez D.R., Anioke T., et al. Protective efficacy of Ad26.COV2.S against SARS-CoV-2 B.1.351 in macaques. Nature. 2021;596:423–427. doi: 10.1038/s41586-021-03732-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Tostanoski L.H., Peter L., Mercado N.B., McMahan K., Mahrokhian S.H., Nkolola J.P., Liu J., Li Z., Chandrashekar A., et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. 2020;369:806–811. doi: 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
There is no dataset/code associated with the paper.