Abstract
Background
Colorectal cancer (CRC) develops through chromosomal instability (CIN) or microsatellite instability (MSI) due to deficient mismatch-repair (dMMR). We aimed to characterise novel cancer-associated genes that are downregulated upon malignant transformation in microsatellite stable (MSS) CRCs, which typically exhibit CIN with proficient mismatch-repair (pMMR).
Methods
Comprehensive screening was conducted on adenomas, MSI/MSS CRCs and cell lines, followed by copy number analysis, and their genetic and prognostic relevance was confirmed in microarray and RNA-seq cohorts (n = 3262, in total). Immunohistochemistry for SH2D4A was performed in 524 specimens of adenoma, carcinoma in situ and dMMR/pMMR CRC. The functional role of SH2D4A was investigated using CRC cell lines.
Results
A set of 11 genes, including SH2D4A, was downregulated during the adenoma-carcinoma sequence in MSS/CIN CRCs, mainly due to chromosome 8p deletions, and their negative prognostic impact was validated in independent cohorts. All adenomas were SH2D4A positive, but a subset of CRCs (5.3%) lacked SH2D4A immunohistochemical staining, correlating with poor prognosis and scarce T cell infiltration. SH2D4A depletion did not affect cell proliferation or IL-6-induced STAT3 phosphorylation.
Conclusions
Our findings suggest that downregulation of multiple genes on chromosome 8p, including SH2D4A, cooperatively contribute to tumorigenesis, resulting in the immune cold tumour microenvironment and poor prognosis.
Subject terms: Colorectal cancer, Cancer genomics, Tumour immunology, Tumour biomarkers
Background
Colorectal cancer (CRC) remains one of the most common causes of cancer-related death worldwide [1]. CRC is a highly heterogeneous disease with diverse clinical and biological behaviour; however, the genomic basis of this heterogeneity remains poorly understood. Colorectal carcinogenesis is driven by genomic instability accompanying progressive accumulation of genetic and epigenetic changes that activate oncogenes and inactivate tumour-suppressor genes [2]. As an early event in the tumorigenic process of CRC, genomic instability occurs in the form of either microsatellite instability (MSI) caused by deficiency in the DNA mismatch-repair (MMR) system or chromosomal instability (CIN), each associated with distinct molecular pathways [3–5]. Most CRCs arise from neoplastic precursor lesions (so-called adenoma), and 65–90% of CRCs develop through the adenoma-carcinoma sequence, commonly accompanied by CIN, in which CIN exists at the adenoma stage, becoming more prominent along with tumour progression [3, 4, 6–8]. The CIN pathway is characterised by structural and numerical chromosomal changes that result in various types of somatic copy number alterations (SCNAs), aneuploidy and loss of heterozygosity (LOH), whereas CIN tumours are considered non-hypermutated and microsatellite stable (MSS) with proficient MMR (pMMR) [4, 5]. CIN phenotypes are commonly coupled with mutational inactivation of the tumour-suppressor genes APC and TP53 and activating mutations in the KRAS oncogene [6, 8]. By contrast, ~15% of CRCs are deficient MMR (dMMR), exhibiting MSI and hypermutation but typically lacking CIN [8, 9]. The Cancer Genome Atlas (TCGA) Network showed that CRCs can be broadly categorised as hypermutated or non-hypermutated [5]. A new classification system of CRC, comprising 4 consensus molecular subtypes (CMS1-4), has also been proposed to encompass transcriptomic profiles that arise from cancer cells as well as from the tumour microenvironment (TME) [10]. Indeed, genomic instability crucially contributes to the creation of distinct TME characteristics. It is well understood that MSI CRCs exhibit the immune “hot” TME that are heavily infiltrated by tumour-infiltrating lymphocytes (TILs) due to the recognition of a high number of mutation-induced tumour neoantigens, along with the expression of immune checkpoint molecules, thereby patients with metastatic MSI CRC can benefit from immune checkpoint inhibitors (ICIs) [9]. By contrast, it has become increasingly clear that CIN also has multiple and diverse roles in the recognition of cancer cells by the immune system. The presence of arm-level and chromosome-level SCNAs, but not focal SCNAs, was correlated with immune evasion, in which the expression of cytotoxic infiltrating immune cell markers were reduced [11, 12]. Specific SCNAs were also associated with altered immune cell infiltration [13]. Besides, CIN appeared to be involved in the crosstalk between cancer cells and their immune microenvironment, for instance, as mediated through the cGAS-STING pathway [14, 15]. However, as SCNAs can impact gene dosage and gene expression of a larger number of neighboring genes, it remains unknown whether any genes targeted by SCNAs specifically promote tumour progression and generate the immunosuppressive TME in CIN CRC.
In the current study, we aimed to identify novel cancer-associated genes that are dysregulated through genomic deletions upon malignant transformation particularly in MSS/CIN CRC. Here we performed comprehensive multi-omics data analysis using an extensive number of precancerous and cancerous samples from multiple cohorts by combining with genomic and clinical data sources. We then focused on a candidate tumour suppressor gene SH2D4A on the short arm of chromosome 8 (8p) that were deleted and downregulated in a subgroup of MSS/CIN CRCs exhibiting poor survival outcomes and low levels of immune infiltration.
Methods
Multi-omics data analysis
All microarray data are publicly available from the Gene Expression Omnibus (GEO) repository (http://www.ncbi.nlm.nih.gov/geo), and GEO datasets used in this study were summarised in Supplementary Table 1. We utilised the preprocessed values obtained from each microarray dataset. If a gene is represented by multiple probe sets, only the probe with the highest mean expression was used. For TCGA data analysis, genomic, epigenomic, transcriptomic and clinical data, including somatic copy number analysis (SNP6 Array-based data), Illumina RNA-seq analysis and methylation array analysis (Infinium HumanMethylation450 BeadChip) for both colon and rectal adenocarcinoma (COADREAD), were downloaded through cBioPortal (http://www.cbioportal.org/) [5, 16]. MSI status, molecular subtypes (CIN, GS, HM-SNV and MSI), CMS subtypes (CMS1-4) and the tumour purity estimated by the ABSOLUTE algorithm were further obtained as described previously [10, 17, 18]. We also used proteomic data from the Clinical Proteomic Tumor Analysis Consortium (CPTAC), which conducted mass spectrometry-based proteomic analysis of TCGA specimens [19].
For genome-wide screening of genes, we initially used a dataset GSE41258 that contained a large number of colorectal adenoma and carcinoma samples with available clinical information on MSI status and relapse-free survival (RFS) [20]. This was followed by analysis on a microarray dataset of MSI and MSS CRC cell lines, GSE59857 [21], and then SCNAs for each genes were examined by TCGA DNA copy number analysis data for deep deletion. For validation purposes, additional datasets were obtained from GEO, including GSE77953, GSE20916, GSE37364, GSE4183, GSE71187, GSE39582, GSE26682, GSE33113, GSE13294, GSE4554, GSE18088, GSE39084, GSE24551 and GSE42284 as listed in Supplementary Table 1. They were discovered by carefully searching the GEO database according to the availability of at least 15 adenomas or 15 MSI CRCs in each dataset. To build multi-gene signatures, we computed signature scores by taking the mean expression values for each sample. We defined the 11-gene signature based on the expression of SH2D4A, CNOT7, PCM1, NUDT18, R3HCC1, XPO7, SLC25A37, KCTD9, PPP2R2A, ELP3 and CCDC25. Immune infiltration was evaluated using the 141-immune gene signature according to ESTIMATE [22]. For survival analysis in gene expression datasets, patients were dichotomised into high and low groups on the basis of the median expression values for SH2D4A or the 11-gene within each cohort. For prognostic validation of the expression of SH2D4A and the 11-gene, six independent GEO datasets of CRC, including GSE39582, GSE24551, GSE30378, GSE33113, GSE39084 and GSE17538 with RFS information, were aggregated as a microarray meta-cohort (n = 1111).
Gene enrichment analysis was performed using The Database for Annotation, Visualization and Integrated Discovery (DAVID) Bioinformatics Resources6.8 (http://david.abcc.ncifcrf.gov/home.jsp), as described elsewhere [23].
Patient samples
We enrolled 20 patients with adenoma, 32 patients with carcinoma in situ (Tis), and 472 consecutive patients with stage I to IV primary CRC who underwent surgical resection at Fukushima Medical University hospital between 2002 and 2013 without preoperative chemotherapy or radiotherapy. Their available formalin-fixed paraffin-embedded (FFPE) whole tissue sections were used for immunohistochemistry. Tumours were classified according to the Japanese Classification of Colorectal, Appendiceal and Anal Carcinoma [24]. Clinical and pathological information was retrospectively obtained from medical records. For survival analysis, 256 patients with stage II and III CRC who underwent curative resection (R0) were used for RFS, which was defined as time from the date of surgery to the date of first relapse. The study was conducted in accordance with the Declaration of Helsinki and was approved by the Institutional Review Board of Fukushima Medical University.
Immunohistochemistry
Four-µm-thick sections were deparaffinised in xylene and rehydrated in a series of ethanol. Endogenous peroxidases were blocked with 0.3% hydrogen peroxide in methanol. Antigens were retrieved by autoclave, and slides were incubated with the following primary antibodies: SH2D4A (Rabbit polyclonal; HPA001871; Prestige Antibodies® Powered by Atlas Antibodies, Sigma–Aldrich; 1:200), CD4 (mouse; clone 4B12; M7310, Dako/Agilent Technologies; 1:100), CD8 (mouse; clone C8/144B; M7103; Dako/Agilent Technologies; 1:100), and Foxp3 (mouse; clone 236 A/E7; ab20034; abcam; 1:200). Sections were subsequently incubated with horseradish peroxidase (HRP)-coupled anti-mouse or anti-rabbit secondary antibodies (Envision+System, K4003 or K4001; Dako/Agilent Technologies). Peroxidase was visualised with DAB (Dojindo), and nuclei were counterstained with Mayer’s Hematoxylin Solution (131-09665; Wako/Fujifilm). Negative controls were done by replacing primary antibodies with PBS.
Assessment of staining
For SH2D4A staining, cytoplasmic SH2D4A immunoreactivity in tumour cells was evaluated, and tumours were classified into two categories, SH2D4A negative and SH2D4A positive. SH2D4A negative was defined as no staining of SH2D4A in tumour cells. Evaluation of CD4, CD8 and Foxp3 staining was described previously [18]. Briefly, the invasive front region of the tumour was reviewed in four independent areas, and evaluated by counting the number of stained lymphocytes at a magnification of ×400. For Foxp3 staining, four independent hotspot areas were selected at a magnification of ×40, and then counted at a magnification of ×400, as described previously. The immunostains were evaluated by two observers (K.S and T.M) who were blinded all of the clinical data.
Determination of MSI/MMR status
For gene expression datasets, we defined MSI-high as MSI, and combined MSI-low and microsatellite stable as MSS. For FFPE specimens, immunohistochemistry for MMR proteins (MLH1, MSH2, MSH6 and PMS2) was performed as described previously [18]. Loss of at least one MMR protein were defined as dMMR, and tumours with intact MMR protein expression as pMMR.
Cell culture, siRNA transfection, qRT-PCR and western blotting
Thirteen short tandem repeat (STR)-authenticated human colorectal cancer cell lines were used. Cell lines were purchased from ATCC (SW480, SW620 and RKO), RIKEN Cell bank (HCT116), JCRB Cell Bank (SW837, HT29 and LoVo) and KCLB (SNU503, SNU81 and SNU407). These cell lines were STR-authenticated and characterised by suppliers. LS180 and Colo205 were obtained as previously described and authenticated by STR analysis (Promega) [25]. Frozen aliquots were used in experiments within 6 months after resuscitation. Cells were maintained with RPMI-1640 (ThermoFisher Scientific) containing 10% fetal bovine serum (FBS) and penicillin/streptomycin (100 IU/ml) (ThermoFisher Scientific) at 37 °C in a humidified atmosphere of 5% CO2. Cells were stimulated with recombinant human IL-6 protein (206-IL, R&D systems). Cell proliferation was measured using the Cell Counting Kit-8 (CCK-8, DOJINDO) according to the manufacturer’s instructions.
For knockdown of SH2D4A, cells were plated and transfected with 10 nmol/L of siRNA oligonucleotides for SH2D4A or scramble control (Silencer Select; s34231, s34232 and negative control#1) using Lipofectamine RNAiMAX Reagent (ThermoFisher Scientific) according to the manufacture’s protocol.
Total RNA from cultured cells 72 h after transfection was isolated using TRIzol Reagent (ThermoFisher Scientific) and 200 ng of total RNA was reverse transcribed to cDNA using ReverTra Ace qPCR RT Master Mix with gDNA Remover (TOYOBO) according to the manufacturer’s instruction. qRT-PCR was conducted using Taqman assays (ThermoFisher Scientific), including SH2D4A (Hs00222955_m1) and ACTB (Hs99999903_m1) with PrimeTime Gene Expression Master Mix (Integrated DNA Technologies) on the QuantStudio 3 (ThermoFisher Scientific) in triplicate. Relative expression levels were determined by the 2-ΔΔCt method as described by manufacturer’s instruction.
Total protein was extracted using Pierce RIPA Buffer (ThermoFisher Scientific) supplemented with Halt Protease Inhibitor Cocktail (ThermoFisher Scientific). The concentration of the protein lysates was measured and the lysates were boiled in SDS-PAGE Sample Loading Buffer (GBiosciences). Equal amount of protein was separated by 8–16% Tris-Glycine Gel (ThermoFisher Scientific) and then transferred onto PVDF membrane using iBlot2 Dry Blotting System (ThermoFisher Scientific). The membrane was blocked with 5% nonfat dried skimmed milk (Cell Signaling Technology), and incubated with primary antibodies, including SH2D4A (Rabbit polyclonal; HPA001919; Prestige Antibodies® Powered by Atlas Antibodies, Sigma–Aldrich; 1:1000), STAT3 (Rabbit; clone D1B2J; #30835; Cell Signaling Technology; 1:1000), pSTAT3 (Rabbit; clone Tyr705/D3A7; #9145; Cell Signaling Technology; 1:2000), PCNA (Rabbit polyclonal; 10205-2-AP; Proteintech, Rosemont, IL, USA; 1:5000) and β-actin (Mouse; clone AC-15; sc-69879; Santa Cruz Biotechnology; 1:5000). Then the membrane was incubated with goat anti-mouse or anti-rabbit HRP secondary antibody (Cell Signaling Technology) and developed with ECL Prime Western Blotting Detection Reagents (GE Healthcare) using LAS4000 imager (GE Healthcare). For fractionation assay, cytoplasmic and nuclear protein fractions were extracted using ProteoExtract® Subcellular Proteome Extraction Kit (539790, Merck) according to the manufacturer’s protocol.
Statistical analysis
Fisher’s exact test, χ2 test, unpaired t-test with Welch’s correction or Mann–Whitney U test were used to determine differences between two variables, where appropriate. For comparisons across multiple groups, one-way analysis of variance (ANOVA) was performed. Correlations were assessed using the Spearman’s coefficient. Cumulative survival was estimated by the Kaplan–Meier method, and differences between two groups were analysed by the log-rank test. Cox proportional hazards regression were used to compute Univariate and multivariate models. All statistical analyses were conducted using GraphPad Prism v6.04 (Graphpad Software Inc.) or SPSS Statistics version 26 (IBM Corporation). All P-values were two-sided, and P-values less than 0.05 were considered statistically significant.
Results
Identification of a set of genes that are downregulated in MSS CRC
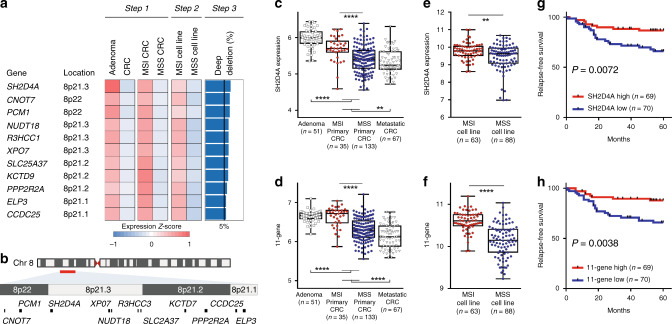
To identify cancer-associated genes that are downregulated during the adenoma-carcinoma sequence through the CIN pathway due to SCNAs in MSS CRCs, we first analysed comprehensive gene expression profiles of pre-malignant and malignant colorectal epithelial tissues using an Affymetrix microarray dataset (GSE41258) [20], in which colorectal adenomas and primary CRCs with MSI status were deposited (Step 1, Fig. 1a and Supplementary Fig. 1). This resulted in a total of 190 unique genes (225 probes) that were not only significantly downregulated in primary CRCs (n = 186) compared to adenomas (n = 49) but also significantly decreased in MSS CRCs (n = 133) compared to those of MSI CRCs (n = 35). We then utilised a microarray dataset of CRC cell lines based on Illumina platform (GSE59857) [21], showing that 68 of the 190 genes were significantly downregulated in MSS cell lines compared to those of MSI cell lines (Step 2). We next adopted an additional criterion to determine whether the downregulation of those genes was potentially attributed to SCNAs. To this end, deep deletions for those 68 genes were examined by means of TCGA DNA copy number analysis data (n = 640) (Step 3). This led us to identify a set of 11 genes, including SH2D4A, CNOT7, PCM1, NUDT18, R3HCC1, XPO7, SLC25A37, KCTD9, PPP2R2A, ELP3 and CCDC25, that exhibited recurrent deep deletions in more than 5% of primary CRCs, among which the highest percentage of deep deletions were SH2D4A (6.56%) (Fig. 1a). Intriguingly, all 11 genes were found to be located on chromosome 8p.21-22 (Fig. 1a, b), while the expression of 11 genes showed significant moderate to strong correlations between one another with Spearman’s r ranging from 0.206 to 0.746 (Supplementary Fig. 2). We then focused on SH2D4A and we also developed the 11-gene signature based on the expression of 11 genes (designated the 11-gene) for further analysis. Figure 1c–f demonstrated the levels of SH2D4A and the 11-gene in adenomas and primary CRCs with MSI status (GSE41258, Fig. 1c, d) and in CRC cell lines with MSI status (GSE59857, Fig. 1e, f). We also found that the levels of SH2D4A and the 11-gene in metastatic CRCs were significantly lower than those of adenomas and primary CRCs (Fig. 1c, d), further suggesting the potential contribution of downregulation of SH2D4A and the 11-gene to tumour progression. As demonstrated in Fig. 1g, h, it is also worth noting that decreased levels of SH2D4A expression and the 11-gene were each significantly associated with poor RFS in GSE41258 (n = 139). In addition, the expression of SH2D4A and the 11-gene remained prognostic, when the analysis was restricted to MSS CRCs (Supplementary Fig. 3a, b).
Fig. 1. Identification of 11 genes on chromosome 8p, including SH2D4A, that were downregulated through the adenoma-carcinoma sequence particularly in microsatellite stable (MSS) colorectal cancer (CRC) compared to microsatellite unstable (MSI) CRC.
a Heatmap depicting the expression z-scores for the 11 genes in the analyses of adenomas vs CRCs, MSI CRCs vs MSS CRCs (Step 1, GSE41258) and MSI cell lines vs MSS cell lines (Step 2, GSE59857), with the frequency (%) of deep deletions in the 11-gene loci (Step 3, TCGA). b Chromosomal locations of the 11 genes spanning 8p21-22. c, d The levels of SH2D4A or the 11-gene signature in adenomas, primary CRCs (MSI or MSS) and metastatic CRCs (Step 1, GSE41258). e, f The levels of SH2D4A or the 11-gene signature in MSI or MSS CRC cell lines (Step 2, GSE59857). ****P < 0.0001, ***P < 0.001, **P < 0.01. g, h Kaplan–Meier curves illustrating relapse-free survival according to the expression of SH2D4A or the 11-gene signature (n = 139, GSE41258). Patients were dichotomised as high or low based on the median expression of SH2D4A (g) and the 11-gene (h).
SH2D4A was downregulated in MSS CRC
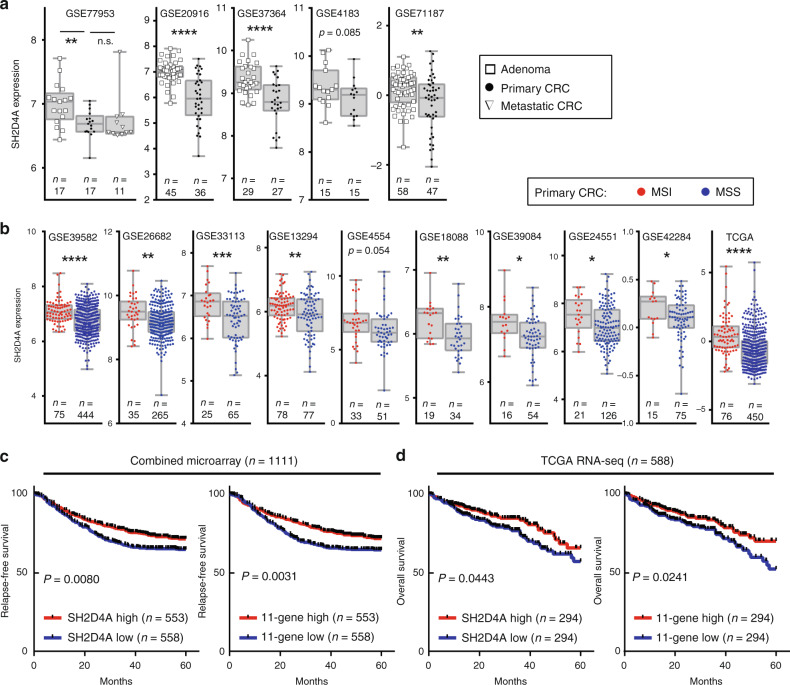
We next attempted to validate the downregulation of SH2D4A and the 11-gene using multiple datasets that contained adenomas and MSI/MSS CRCs. As shown in Fig. 2a, five microarray datasets were used and SH2D4A expression was found to be decreased significantly with the adenoma-to-carcinoma transition in most of the analysis. Furthermore, 10 independent datasets of CRC analysed on microarray or RNA-seq revealed that MSS CRCs had significantly lower levels of SH2D4A expression than those of MSI CRCs (Fig. 2b). Correspondingly, significantly decreased levels of the 11-gene in CRCs compared to adenomas, as well as in MSS CRCs compared MSI CRCs, were confirmed in all cohorts we analysed (Supplementary Fig. 4). Although MSI CRCs frequently carry BRAF mutations, SH2D4A expression was not specifically altered by BRAF mutation status in both MSI and MSS CRCs (Supplementary Fig. 5). These results further prompted us to validate the prognostic impact of SH2D4A and the 11-gene in CRC. For this purpose, we obtained two large cohorts of CRC, including a meta-cohort (n = 1111) by combining 6 microarray datasets with RFS data available (GSE39582, GSE24551, GSE30378, GSE33113, GSE39084 and GSE17538) and the TCGA cohort based on RNA-seq analysis (n = 588). Patients with lower expression of SH2D4A and the 11-gene had significantly shorter RFS in the microarray meta-cohort (Fig. 2c). The TCGA cohort further confirmed that decreased levels of SH2D4A and the 11-gene were each significantly associated with poor OS (Fig. 2d). Our finding consistently indicates that SH2D4A and the 11-gene were downregulated through the adenoma-carcinoma progression particularly in MSS CRC, and that the downregulation of SH2D4A and the 11-gene had negative prognostic impact in CRC.
Fig. 2. Validation of clinical and prognostic association of SH2D4A and the 11-gene signature in multiple independent cohorts.
a SH2D4A expression in adenoma, primary colorectal cancer (CRC) and metastatic CRC in five cohorts. b SH2D4A expression in MSI and MSS CRC in 10 cohorts. ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05, n.s. P > 0.1. c Kaplan–Meier curves depicting relapse-free survival according to the expression of SH2D4A or the 11-gene signature in the combined microarray cohort (n = 1111). d Kaplan–Meier curves depicting overall survival according to the expression of SH2D4A or the 11-gene signature in TCGA RNA-seq cohort (n = 588). For survival analysis, patients were dichotomised as high or low based on the median in each dataset.
SH2D4A downregulation through chromosome 8p deletion in chromosomally unstable CRCs
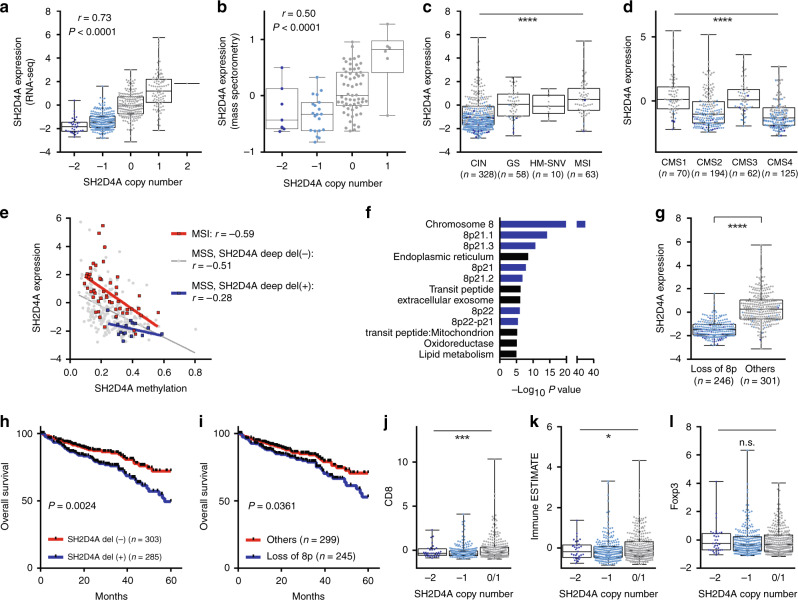
We examined somatic mutations, SCNAs, genomic and transcriptomic molecular subtypes and DNA hypermethylation, to more clarify the genetic mechanisms responsible for the downregulation of SH2D4A (Fig. 3a–i and Supplementary Fig. 6a–g). The TCGA cohort demonstrated a strong positive correlation between SH2D4A mRNA expression by RNA-seq and SH2D4A SCNAs, including deep deletion (copy number, −2), shallow deletion (−1), diploid (0), gain (1) and amplification (2) (Fig. 3a, Spearman’s r = 0.73). Likewise, in the CPTAC dataset, we confirmed the significant correlation of SH2D4A protein expression by mass spectrometry with SH2D4A SCNAs (Fig. 3b, r = 0.50). Moreover, among 4 molecular subtypes [17], including MSI, HM-SNV, GS and CIN, CRCs with CIN showed significantly decreased levels of SH2D4A expression and the 11-gene, which were particularly remarkable in tumours with genetic deletion of SH2D4A (Fig. 3c and Supplementary Fig. 6a). On the other hand, low levels of SH2D4A and the 11-gene were preferentially found in CMS2 and CMS4 tumours, both of which were characterised by high SCNA levels (Fig. 3d and Supplementary Fig. 6b) [10]. Those findings were confirmed in GSE39582 (Supplementary Fig. 6d–g). We also noticed that SH2D4A levels varied considerably even within MSI tumours, in which SH2D4A was not deleted, and that some MSI tumours showed decreased levels of SH2D4A expression comparable to those of MSS tumours with SH2D4A deletions (Figs. 1c, 2b, and 3c). As shown in Fig. 3e, both MSI tumours and MSS tumours without deep deletion of SH2D4A demonstrated significant inverse relationship between SH2D4A methylation and expression (r = −0.59 and −0.51, respectively), which appeared to be much stronger than that of MSS tumours exhibiting SH2D4A deep deletion (r = −0.28). Those results indicate that SH2D4A downregulation was mostly attributed to SCNAs in CIN CRCs, whereas DNA hypermethylation might also contribute to the silencing of SH2D4A in a subset of MSI and MSS CRCs.
Fig. 3. The association of SH2D4A expression with genetic, epigenetic and clinical features.
a Correlation between SH2D4A mRNA levels by RNA-seq and SH2D4A copy number in the TCGA cohort. b Correlation between SH2D4A protein levels by mass spectrometry and SH2D4A copy number in the CPTAC cohort. c SH2D4A expression in 4 molecular subtypes, including CIN, GS, HM-SNV and MSI. d SH2D4A expression in CMS1-4 subtypes. e Correlation between SH2D4A expression and methylation levels in MSI CRCs, MSS CRCs without SH2D4A deep deletion and MSS CRCs exhibiting SH2D4A deep deletion. f SH2D4A-co-expressed genes identified significantly enriched GO terms by DAVID analysis. g SH2D4A expression in tumours with and without loss of chromosome 8p. h Kaplan–Meier curves illustrating overall survival according to SH2D4A deletion status. i Kaplan–Meier curves illustrating overall survival according to loss of chromosome 8p status. J–l Association between SH2D4A copy number and immune signatures, including CD8A (j), Immune ESTIMATE (k), and FOXP3 (l). ****P < 0.0001, ***P < 0.001, *P < 0.05, n.s. P > 0.05.
Since the 11 genes, including SH2D4A, on chromosome 8p were significantly correlated with one another (Supplementary Fig. 2), it is speculated that the expression of those 11 genes were simultaneously regulated mainly through large-scale genomic deletions but not resulted from focal SCNAs. Co-expression analysis in the TCGA cohort identified 313 genes to be significantly positively correlated with SH2D4A with Spearman’s r > 0.3 and P < 1.00E-8 (Supplementary Table 2). DAVID bioinformatics analysis revealed that the 313 SH2D4A-correlated genes were strongly enriched for chromosome 8 as well as several cytobands of chromosome 8p (Fig. 3f, terms with P < 1.00E-5 are shown). Also, downregulation of SH2D4A and the 11-gene were closely associated with loss of chromosome 8p (Fig. 3g and Supplementary Fig. 6c), indicating that loss of 8p was the major contributor to the concomitant downregulation of those 11 genes on 8p, including SH2D4A, particularly in CIN tumours. Besides chromosomal locations, DAVID also identified significantly enriched keywords and terms that are particularly involved in cellular metabolism, such as lipid metabolism, oxidoreductase, endoplasmic reticulum and mitochondrion (Fig. 3f and Supplementary Table 3). We also found that SH2D4A expression and the 11-gene were not affected by the levels of estimated tumour purity (Supplementary Fig. 7a, b).
SH2D4A deletion was associated with poor prognosis and immune suppression
We then asked whether deletions of SH2D4A and loss of 8p could have prognostic roles in CRC. Strikingly, not only SH2D4A deletions (deep and shallow deletions were combined) but also loss of chromosome 8p were significantly associated with poor OS (Fig. 3h, i), which was highly consistent with the negative prognostic impact of the downregulation of SH2D4A and the 11-gene in multiple cohorts of CRC (Figs. 1g, h and 2c, d). Analogous to our findings in CRC, previous studies reported that a cluster of six genes on chromosome 8p, including SH2D4A, were deleted and downregulated frequently in hepatocellular carcinoma (HCC) and that lower expression of those genes were associated with poorer prognosis in patients with HCC [26, 27]. Intriguingly, in HCC, SH2D4A was correlated with regulatory and cytotoxic T cell infiltration [28]. We therefore sought to determine whether there was any association between immune infiltration and SH2D4A deletions in CRC. As shown in Fig. 3j, k, the levels of CD8A expression (as a marker of CD8 + T cells) and an immune infiltration signature based on immune ESTIMATE were significantly lower in tumours with SH2D4A deletions, whereas the expression of FOXP3 (regulatory T cells; Tregs) did not significantly differ among tumours with and without SH2D4A deletions (Fig. 3l).
Loss of SH2D4A was associated with immune suppression and poor prognosis
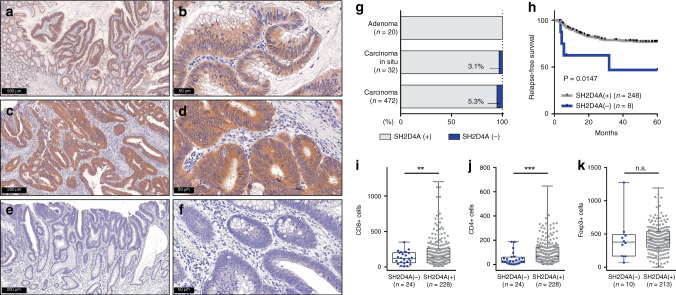
We conducted immunohistochemistry for SH2D4A using 524 surgically resected specimens of adenoma (n = 20), carcinoma in situ (Tis) (n = 32) and stage I–IV primary CRC (n = 472). Immunoreactivity for SH2D4A protein was typically seen in the epithelial cell cytoplasm in both non-neoplastic colonic mucosa and tumour tissues (Fig. 4a and Supplementary Fig. 8a). All adenomas (100.0%) as well as the vast majority of Tis tumours (96.9%) and CRCs (94.7%) were determined to be positive for SH2D4A staining (Fig. 4a–g). By contrast, we found that only 25 CRCs (5.3%) lacked SH2D4A staining (Fig. 4e–g). Supplementary Table 4 shows clinicopathological characteristics of 472 CRCs according to SH2D4A staining. We found no significant association of SH2D4A expression with common features, including age, sex, tumour location, differentiation, tumour invasion, lymphatic/venous invasion, lymph node metastasis, distant metastasis or disease stage. Of 25 CRCs with negative SH2D4A expression, 23 (92.0%) were pMMR, while only 2 (8.0%) were dMMR, however, there was no significant association between SH2D4A expression and MMR status. Strikingly, loss of SH2D4A immunostaining was significantly associated with poorer RFS compared to those of SH2D4A positive, in patients with stage II and III CRC (Fig. 4h). As shown in Table 1, univariate and multivariate Cox analysis identified SH2D4A negative as a significant prognostic factor for poor RFS when adjusted for clinical covariates and MMR status (HR 3.65; 95% CI 1.28–10.44; P = 0.016). We also performed immunohistochemistry for CD8, CD4 and Foxp3, to evaluate the association between SH2D4A expression and T cell infiltration in the TME. SH2D4A-negative tumours showed significantly lesser number of CD8 + and CD4 + T cell infiltration compared to those of SH2D4A positive (Fig. 4i, j and Supplementary Fig. 8b). By contrast, infiltration of Foxp3+ cells was not correlated with SH2D4A expression (Fig. 4k). As dMMR tumours are known to be immunologically hot and to exhibit dense T cell infiltration, tumours were further stratified by SH2D4A expression and MMR status. We found that significantly different levels of CD8 + and CD4 + infiltrating cells between groups, displaying particularly lesser number of CD8 and CD4 lymphocytes in SH2D4A-negative tumours, irrespective of MMR status (Supplementary Fig. 9a–c).
Fig. 4. Immunohistochemistry for SH2D4A expression in adenoma and carcinoma specimens.
a–f Representative images of resected specimens of adenoma (a, b), pMMR CRC with positive SH2D4A expression (c, d) and pMMR CRC lacking SH2D4A staining (e, f). g Relative proportion of SH2D4A positive and SH2D4A negative, in adenoma, carcinoma in situ and carcinoma specimens. h Kaplan–Meier curve depicting relapse-free survival according to the expression of SH2D4A. i–k Immune infiltration of CD8 + (i), CD4 + (j) and Foxp3 + (k) cells according to the expression of SH2D4A. ***P < 0.001, **P < 0.01, n.s. P > 0.05.
Table 1.
Univariate and multivariate Cox analysis for relapse-free survival in patients with stage II and III colorectal cancer (n = 256).
| Univariate | Multivariate | |||
|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P-value | |
| Age | ||||
| Continuous | 1.021 (0.996–1.047) | 0.101 | ||
| Gender | ||||
| Male (n = 160) | Reference | |||
| Female (n = 96) | 1.175 (0.697–1.981) | 0.546 | ||
| Tumour location | ||||
| Colon (n = 169) | Reference | |||
| Rectum (n = 87) | 1.239 (0.732–2.097) | 0.424 | ||
| Tumour differentiation | ||||
| Well (n = 109) | Reference | Reference | ||
| Moderately (n = 132) | 1.749 (0.993–3.080) | 0.053 | 1.956 (1.088–3.515) | 0.025 |
| Poorly (n = 15) | 2.221 (0.751–6.565) | 0.149 | 2.274 (0.747–6.922) | 0.148 |
| Tumour invasion | ||||
| T1–3 (n = 168) | Reference | Reference | ||
| T4 (n = 88) | 2.480 (1.480–4.157) | 0.001 | 2.326 (1.366–3.960) | 0.002 |
| Lymphatic invasion | ||||
| Absent (n = 42) | Reference | |||
| Present (n = 214) | 1.887 (0.810–4.394) | 0.141 | ||
| Venous invasion | ||||
| Absent (n = 27) | Reference | Reference | ||
| Present (n = 229) | 3.675 (0.897–15.063) | 0.071 | 2.238 (0.533–9.396) | 0.271 |
| Lymph node metastasis | ||||
| Absent (n = 143) | Reference | Reference | ||
| Present (n = 113) | 2.635 (1.542–4.503) | <0.001 | 2.129 (1.230–3.683) | 0.007 |
| Adjuvant chemotherapy | ||||
| Absent (n = 149) | Reference | |||
| Present (n = 107) | 1.348 (0.803–2.247) | 0.262 | ||
| MMR status | ||||
| pMMR (n = 231) | Reference | Reference | ||
| dMMR (n = 25) | 0.304 (0.074–1.246) | 0.098 | 0.236 (0.057–0.974) | 0.046 |
| SH2D4A by immunohistochemistry | ||||
| Positive (n = 248) | Reference | Reference | ||
| Negative (n = 8) | 3.299 (1.193–9.118) | 0.021 | 3.653 (1.278–10.442) | 0.016 |
Bold values indicate statistical significance p < 0.05.
In the multivariate model, only variables with P < 0.1 in univariate analysis were included.
HR hazard ratio, 95% CI 95% confidence interval, MMR DNA mismatch-repair, dMMR deficient MMR, pMMR proficient MMR
The effect of SH2D4A depletion in CRC cell lines
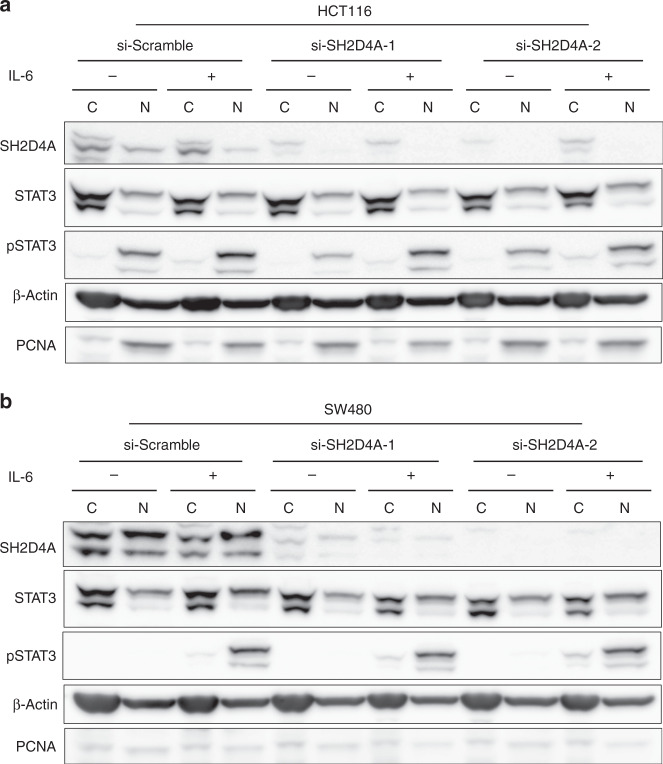
In HCC, SH2D4A appeared to be a candidate tumour suppressor that can inhibit IL-6-responsive STAT3 transcriptional activity [28]. Hence, the downregulation of SH2D4A could enhance IL-6/STAT3 signaling and thus likely lead to promoting tumour cell proliferation and repressing immune surveillance [28]. We therefore hypothesised that SH2D4A may have a tumour-suppressive function in CRC via inhibition of endogenous STAT3 signaling. To address this hypothesis, we utilised two CRC cell lines, including HCT116 and SW480, harboring no deletion or mutation at the SH2D4A locus, with relatively higher SH2D4A protein expression among 13 cell lines examined by western blotting (Supplementary Fig. 10). HCT116 and SW480 cells were transfected with siRNAs targeting SH2D4A or scramble negative control and cells were treated with IL-6 (Supplementary Fig. 11a–e). We found that siRNA-mediated SH2D4A depletion had no clear effect on cell proliferation (Supplementary Fig. 11c, d). In addition, IL-6 stimulated STAT3 phosphorylation in whole-cell lysates was not altered by SH2D4A depletion (Supplementary Fig. 11e). In HCC, it was shown that SH2D4A can directly interact with STAT3 to retain it in the cytoplasm, leading to inhibition of nuclear translocation of STAT3. We therefore extracted cytoplasmic and nuclear protein fractions from siRNA-transfected cells, and STAT3 and phosphorylated STAT3 (pSTAT3) were analysed by western blotting. As shown in Fig. 5a, b, although pSTAT3 was predominantly detected in the nuclear extracts in response to IL-6 stimulation, SH2D4A depletion did not significantly change the levels of pSTAT3 in both cell lines. Also, total STAT3 levels in cytoplasmic and nuclear fractions appeared to be unaffected by knockdown of SH2D4A.
Fig. 5. SH2D4A depletion and IL-6-induced nuclear and cytoplasmic STAT3 phosphorylation in CRC cell lines.
a, b HCT116 and SW480 cells transfected with siRNAs targeting SH2D4A or scramble negative control in the presence or absence of 100 ng/mL IL-6 for 30 min. Cell fractions of cytosol (C) and nucleus (N) were analysed with indicated antibodies by western blot.
Discussion
In the present study, we demonstrated for the first time that SH2D4A was downregulated during the adenoma-carcinoma transition in MSS CRCs, which typically exhibited CIN, most likely due to chromosome 8p loss or large deletions involving 8p.21-22. Notably, decreased levels of SH2D4A mRNA expression were significantly associated with poor clinical outcomes, not only in the discovery dataset (n = 139), but also in two large validation cohorts based on microarray (n = 1111) and RNA-seq (n = 588). Correspondingly, deletions of SH2D4A were also associated with poor prognosis. This prognostic impact was further recapitulated by an immunohistochemistry cohort (n = 256), showing that a subset of patients with CRCs lacking SH2D4A staining had significant poorer RFS than those with positive SH2D4A staining. Furthermore, low levels of the 11-gene signature based on chromosome 8p genes, as well as loss of chromosome 8p itself had significant poor prognostic impact. Those results showed a phenotypic overlap between CRCs exhibiting chromosome 8p loss, SH2D4A deletion, decreased SH2D4A mRNA expression and loss of SH2D4A protein expression. It is worth noting that loss of SH2D4A protein expression provided strong prognostic stratification, independent of conventional clinical factors. Therefore, although further validation studies are needed, immunohistochemistry for SH2D4A with resected FFPE specimens might be used as a practical assay for prognostic stratification of CRC. Since the lack of SH2D4A immunostaining correlated with poor RFS in stage II and III patients after surgery, those “high-risk” patients may benefit from intensive postoperative management, including adjuvant chemotherapy, to reduce the likelihood of disease recurrence.
In good agreement with the prognostic roles of chromosome 8p genes in CRC demonstrated in our study, Roessler S et al. reported the association of poor outcomes in HCC with 6 genes on chromosome 8p that were deleted and downregulated, in which 3 genes, including SH2D4A, ELP3 and CCDC25, overlapped with the 11 genes that we identified in CRC in this study [26]. Roessler S et al. further demonstrated that SH2D4A can work as a tumour suppressor that suppresses cell proliferation and inhibits IL-6-induced nuclear STAT3 transcriptional activity in HCC cells by directly interacting with STAT3 to retain it in the cytoplasm [28]. Hence, downregulation of SH2D4A may enhance IL-6 responsive STAT3 activation that can result in tumour cell proliferation. This prompted us to determine the effect of SH2D4A inactivation in CRC, using two CRC cell lines without SCNAs or mutations at the SH2D4A locus. However, we found no clear impact of SH2D4A depletion in cell proliferation, while total and phosphorylated STAT3 levels, either in the nucleus or in the cytoplasm, were not clearly affected by SH2D4A depletion. Therefore, functional roles of SH2D4A in CRC remains inconclusive. Although we were not able to explain this discrepancy of in vitro SH2D4A roles between CRC and HCC, this study provided several important findings that are consistent with previous studies. We found that CRCs with SH2D4A deletions exhibited low levels of immune infiltration signatures, and that tumours lacking SH2D4A protein expression displayed poor prognosis and the immune cold TME with scarce CD4 + and CD8 + T cell infiltration. Likewise, in HCC, CD4 + and CD8 + T cell infiltration was significantly reduced in tumours with low SH2D4A protein expression [28]. Moreover, across cancer types, arm-level and chromosome-level SCNAs, but not focal SCNAs, were significantly correlated with reduced immune signatures, especially CD8 + T cells, and poor survival [11]. It is also worth noting that chromosome 8p is one of the most commonly deleted genomic regions in solid cancer [12, 29], and that deletions of chromosome 8p were associated with poor prognosis in HCC, breast cancer and prostate cancer [26, 30–33]. Taken together, these data suggest that highly aneuploid (i.e. CIN) tumours exhibiting loss of chromosome 8p are characterised by SH2D4A downregulation, reduced immune infiltration and poor prognosis. Meanwhile, unlike the strong correlation between SH2D4A expression and Foxp3+ cells in HCC tissues [28], we found that SH2D4A expression was not associated with infiltrating Foxp3+ cells in CRC. This discrepancy between HCC and CRC may in part result from the different clinical and prognostic impact of infiltrating Foxp3+ cells among cancer types [34–36].
Although 25% of the genome is known to be affected by arm-level SCNAs [37], their functional consequences are still not fully determined, as large SCNAs can exert widespread effects on gene expression and function. Recurrent deletions of chromosome 8p and its association with poor survival in human cancer suggest that cancer-associated genes with tumour-suppressive properties reside in this region. Despite several gene candidates located on chromosome 8p that have been proposed, none of them fulfilled the two-hit criteria for bona fide tumour suppressor genes [2, 30]. By contrast, Xue W et al. revealed that multiple genes on chromosome 8p can cooperatively inhibit tumorigenesis and their co-suppression can synergistically promote tumour growth using a murine model with in vivo RNAi targeting [38]. Cai Y et al. generated a human mammary epithelial cell model carrying chromosome 8p hemizygous deletion, showing that large-scale 8p deletions create phenotypes associated with lipid metabolism and metastasis that fundamentally differ from those arising from loss of a single gene [30]. Indeed, the present study revealed that a large number of chromosome 8p genes were co-suppressed with SH2D4A, which were involved in metabolic pathways, such as lipid metabolism, oxidoreductase, endoplasmic reticulum and mitochondrion. Alternatively, arm-level SCNAs may concomitantly require other specific SCNAs, as 8p deletion frequently co-occurs with gains of chromosome 8q involving the MYC oncogene and losses of the TP53 tumour suppressor gene on chromosome 17p [31, 33, 38, 39]. It is therefore plausible that although alterations of single genes, such as SH2D4A, on chromosome 8p are often insufficient to explain the phenotypic impact caused by large deletions within 8p, multiple genes on chromosome 8p are cooperatively responsible for tumorigenesis in CRC in a haploinsufficient manner, where the consequences of large deletions in chromosome 8p are context-dependent.
This study has several limitations. First, the exact functions of SH2D4A in CRC was not elucidated, and it remains unclear whether SH2D4A downregulation alone can functionally affect tumorigenesis. Second, although we used a large number of samples from multiple public datasets, they were based on exploratory and retrospective analysis, in which some cohorts lacked sufficient genomic or clinical information. Also, the prognostic value of SH2D4A staining by immunohistochemistry remains to be validated in independent FFPE cohorts. Thirdly, only about 5% of CRCs exhibiting SH2D4A-negative protein by immunohistochemistry appeared to be discrepant from the incidence of SH2D4A deletions or loss of chromosome 8p, which was found in up to 45% of CRCs. In addition, the proportion of SH2D4A protein loss was not clearly different between pMMR and dMMR tumours in our FFPE cohort, although statistical power was limited. Those findings suggest that additional epigenetic and post-translational mechanisms regulate the expression of SH2D4A protein in CRC. Future studies would need a large number of FFPE tissue cohorts, by combining immunohistochemistry with genetic and epigenetic analysis.
In conclusion, we identified a subgroup of CRCs exhibiting SH2D4A downregulation and arm-level deletion of chromosome 8p. Our findings suggest that downregulation of multiple genes on chromosome 8p cooperatively contribute to tumorigenesis, characterised by the immune cold tumour microenvironment and poor prognosis.
Supplementary information
Author contributions
HO and KK provided the study concept and design. HO, YK, HO, SH, SF, WS, MS, ZS and TM collected the data. TM, SN, KS, MI and AK performed the experiments. TM, HO, SN and KM interpret the data. TM, HO and KK wrote the paper.
Funding information
This work was supported by grants from Japan Society for the Promotion of Science (JSPS) KAKENHI Grant Numbers 20K09061 and 20K08963.
Data availability
The public datasets used in this study are available from the GEO database (http://www.ncbi.nlm.nih.gov/geo).
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
This study was conducted in compliance with the declaration of Helsinki. The study was approved by the Institutional Review Board of Fukushima Medical University (No. 2289 and No. 2847), and samples were obtained with the patients’ informed consent.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-021-01660-y.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021 doi: 10.3322/caac.21660.. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–58. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sansregret L, Vanhaesebroeck B, Swanton C. Determinants and clinical implications of chromosomal instability in cancer. Nat Rev Clin Oncol. 2018;15:139–50. doi: 10.1038/nrclinonc.2017.198. [DOI] [PubMed] [Google Scholar]
- 4.Nguyen LH, Goel A, Chung DC. Pathways of Colorectal Carcinogenesis. Gastroenterology. 2020;158:291–302. doi: 10.1053/j.gastro.2019.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Network, T. C. G. A. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–7. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pino MS, Chung DC. The chromosomal instability pathway in colon cancer. Gastroenterology. 2010;138:2059–72. doi: 10.1053/j.gastro.2009.12.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shih IM, Zhou W, Goodman SN, Lengauer C, Kinzler KW, Vogelstein B. Evidence that genetic instability occurs at an early stage of colorectal tumorigenesis. Cancer Res. 2001;61:818–22. [PubMed] [Google Scholar]
- 8.Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet. 2019;394:1467–80. doi: 10.1016/S0140-6736(19)32319-0. [DOI] [PubMed] [Google Scholar]
- 9.Ganesh K, Stadler ZK, Cercek A, Mendelsohn RB, Shia J, Segal NH, et al. Immunotherapy in colorectal cancer: rationale, challenges and potential. Nat Rev Gastroenterol Hepatol. 2019;16:361–75. doi: 10.1038/s41575-019-0126-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guinney J, Dienstmann R, Wang X, de Reyniès A, Schlicker A, Soneson C, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21:1350–6. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davoli T, Uno H, Wooten EC, Elledge SJ. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science. 2017;355:eaaf8399. doi: 10.1126/science.aaf8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taylor AM, Shih J, Ha G, Gao GF, Zhang X, Berger AC, et al. Genomic and functional approaches to understanding cancer aneuploidy. Cancer Cell. 2018;33:676–89 e673. doi: 10.1016/j.ccell.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH, et al. The immune landscape of cancer. Immunity. 2018;48:812–30 e814. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bakhoum SF, Cantley LC. The multifaceted role of chromosomal instability in cancer and its microenvironment. Cell. 2018;174:1347–60. doi: 10.1016/j.cell.2018.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwon J, Bakhoum SF. The cytosolic DNA-sensing cGAS-STING pathway in cancer. Cancer Discov. 2020;10:26–39. doi: 10.1158/2159-8290.CD-19-0761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y, Sethi NS, Hinoue T, Schneider BG, Cherniack AD, Sanchez-Vega F, et al. Comparative molecular analysis of gastrointestinal adenocarcinomas. Cancer Cell. 2018;33:721–35 e728. doi: 10.1016/j.ccell.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Endo E, Okayama H, Saito K, Nakajima S, Yamada L, Ujiie D, et al. A TGFbeta-dependent stromal subset underlies immune checkpoint inhibitor efficacy in DNA mismatch repair-deficient/microsatellite instability-high colorectal cancer. Mol Cancer Res. 2020;18:1402–13. doi: 10.1158/1541-7786.MCR-20-0308. [DOI] [PubMed] [Google Scholar]
- 19.Vasaikar S, Huang C, Wang X, Petyuk VA, Savage SR, Wen B, et al. Proteogenomic analysis of human colon cancer reveals new therapeutic opportunities. Cell. 2019;177:1035–49 e1019. doi: 10.1016/j.cell.2019.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sheffer M, Bacolod MD, Zuk O, Giardina SF, Pincas H, Barany F, et al. Association of survival and disease progression with chromosomal instability: a genomic exploration of colorectal cancer. Proc Natl Acad Sci USA. 2009;106:7131–6. doi: 10.1073/pnas.0902232106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Medico E, Russo M, Picco G, Cancelliere C, Valtorta E, Corti G, et al. The molecular landscape of colorectal cancer cell lines unveils clinically actionable kinase targets. Nat Commun. 2015;6:7002. doi: 10.1038/ncomms8002. [DOI] [PubMed] [Google Scholar]
- 22.Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, Torres-Garcia W, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 24.Japanese Society for Cancer of the Colon and Rectum. Japanese Classification of Colorectal, Appendiceal, and Anal Carcinoma: the 3d English Edition [Secondary Publication]. J Anus Rectum Colon. 2019;3:175–95. [DOI] [PMC free article] [PubMed]
- 25.Noda M, Okayama H, Tachibana K, Sakamoto W, Saito K, Thar Min AK, et al. Glycosyltransferase gene expression identifies a poor prognostic colorectal cancer subtype associated with mismatch repair deficiency and incomplete glycan synthesis. Clin Cancer Res. 2018;24:4468–81. doi: 10.1158/1078-0432.CCR-17-3533. [DOI] [PubMed] [Google Scholar]
- 26.Roessler S, Long EL, Budhu A, Chen Y, Zhao X, Ji J, et al. Integrative genomic identification of genes on 8p associated with hepatocellular carcinoma progression and patient survival. Gastroenterology. 2012;142:957–66 e912. doi: 10.1053/j.gastro.2011.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quagliata L, Andreozzi M, Kovac M, Tornillo L, Makowska Z, Moretti F, et al. SH2D4A is frequently downregulated in hepatocellular carcinoma and cirrhotic nodules. Eur J Cancer. 2014;50:731–8. doi: 10.1016/j.ejca.2013.11.018. [DOI] [PubMed] [Google Scholar]
- 28.Ploeger C, Waldburger N, Fraas A, Goeppert B, Pusch S, Breuhahn K, et al. Chromosome 8p tumor suppressor genes SH2D4A and SORBS3 cooperate to inhibit interleukin-6 signaling in hepatocellular carcinoma. Hepatology. 2016;64:828–42. doi: 10.1002/hep.28684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Birnbaum D, Adelaide J, Popovici C, Charafe-Jauffret E, Mozziconacci MJ, Chaffanet M. Chromosome arm 8p and cancer: a fragile hypothesis. Lancet Oncol. 2003;4:639–42. doi: 10.1016/S1470-2045(03)01225-7. [DOI] [PubMed] [Google Scholar]
- 30.Cai Y, Crowther J, Pastor T, Abbasi Asbagh L, Baietti MF, De Troyer M, et al. Loss of chromosome 8p governs tumor progression and drug response by altering lipid metabolism. Cancer Cell. 2016;29:751–66. doi: 10.1016/j.ccell.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 31.Lebok P, Mittenzwei A, Kluth M, Ozden C, Taskin B, Hussein K, et al. 8p deletion is strongly linked to poor prognosis in breast cancer. Cancer Biol Ther. 2015;16:1080–7. doi: 10.1080/15384047.2015.1046025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kluth M, Amschler NN, Galal R, Moller-Koop C, Barrow P, Tsourlakis MC, et al. Deletion of 8p is an independent prognostic parameter in prostate cancer. Oncotarget. 2017;8:379–92. doi: 10.18632/oncotarget.13425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.El Gammal AT, Bruchmann M, Zustin J, Isbarn H, Hellwinkel OJ, Kollermann J, et al. Chromosome 8p deletions and 8q gains are associated with tumor progression and poor prognosis in prostate cancer. Clin Cancer Res. 2010;16:56–64. doi: 10.1158/1078-0432.CCR-09-1423. [DOI] [PubMed] [Google Scholar]
- 34.Shang B, Liu Y, Jiang SJ, Liu Y. Prognostic value of tumor-infiltrating FoxP3+ regulatory T cells in cancers: a systematic review and meta-analysis. Sci Rep. 2015;5:15179. doi: 10.1038/srep15179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saito T, Nishikawa H, Wada H, Nagano Y, Sugiyama D, Atarashi K, et al. Two FOXP3(+)CD4(+) T cell subpopulations distinctly control the prognosis of colorectal cancers. Nat Med. 2016;22:679–84. doi: 10.1038/nm.4086. [DOI] [PubMed] [Google Scholar]
- 36.Saleh R, Elkord E. FoxP3(+) T regulatory cells in cancer: prognostic biomarkers and therapeutic targets. Cancer Lett. 2020;490:174–85. doi: 10.1016/j.canlet.2020.07.022. [DOI] [PubMed] [Google Scholar]
- 37.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xue W, Kitzing T, Roessler S, Zuber J, Krasnitz A, Schultz N, et al. A cluster of cooperating tumor-suppressor gene candidates in chromosomal deletions. Proc Natl Acad Sci USA. 2012;109:8212–7. doi: 10.1073/pnas.1206062109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muleris M, Chalastanis A, Meyer N, Lae M, Dutrillaux B, Sastre-Garau X, et al. Chromosomal instability in near-diploid colorectal cancer: a link between numbers and structure. PLoS ONE. 2008;3:e1632. doi: 10.1371/journal.pone.0001632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The public datasets used in this study are available from the GEO database (http://www.ncbi.nlm.nih.gov/geo).