Abstract
Plasmid pTKH11, originally obtained by electroporation of a Klebsiella oxytoca plasmid preparation into Escherichia coli XAC, expressed a high level of an AmpC-like β-lactamase. The enzyme, designated CMY-5, conferred resistance to extended-spectrum β-lactams in E. coli; nevertheless, the phenotype was cryptic in the K. oxytoca donor. Determination of the complete nucleotide sequence of pTKH11 revealed that the 8,193-bp plasmid encoded seven open reading frames, including that for the CMY-5 β-lactamase (blaCMY-5). The blaCMY-5 product was similar to the plasmidic CMY-2 β-lactamase of K. pneumoniae and the chromosomal AmpC of Citrobacter freundii, with 99.7 and 97.0% identities, respectively; there was a substitution of phenylalanine in CMY-5 for isoleucine 105 in CMY-2. blaCMY-5 was followed by the Blc and SugE genes of C. freundii, and this cluster exhibited a genetic organization identical to that of the ampC region on the chromosome of C. freundii; these results confirmed that C. freundii AmpC was the evolutionary origin of the plasmidic cephamycinases. In the K. oxytoca host, the copy number of pTKH11 was very low and the plasmid coexisted with plasmid pNBL63. Analysis of the replication regions of the two plasmids revealed 97% sequence similarity in the RNA I transcripts; this result implied that the two plasmids might be incompatible. Incompatibility of the two plasmids might explain the cryptic phenotype of blaCMY-5 in K. oxytoca through an exclusion effect on pTKH11 by resident plasmid pNBL63.
Extended-spectrum β-lactamases cause bacterial resistance to β-lactam antibiotics and contribute to therapeutic problems (16). Many of them are plasmid mediated and confer resistance to oxyimino-β-lactams, such as cefotaxime, ceftazidime, and the monobactam aztreonam (16). Plasmid-mediated β-lactamases are often expressed in large amounts and are encoded by transposons which can be easily transferred from one replicon to another (15). Chromosomally encoded β-lactamases are present in most gram-negative bacteria; these enzymes are often expressed at low levels and therefore may not contribute to clinical β-lactam resistance (22). Additionally, plasmid-mediated β-lactamases, such as MIR-1, CMY-1, and CMY-2, with high isoelectric points and cephalosporinase activity have been described (2, 4, 26). These findings have raised concern that chromosomal β-lactamases may cause serious therapeutic problems if their genes are translocated onto plasmids.
In previous studies (41, 42), a group of Klebsiella oxytoca isolates resistant to aztreonam (MIC, 32 μg/ml) and cefuroxime (MIC, 128 μg/ml) was characterized, and the resistance was found to be caused by an extended-spectrum β-lactamase with a pI of 5.25 and designated OXY-2a. The gene for this β-lactamase (blaOXY-2a) was located on the chromosome of the K. oxytoca isolates (40). However, in an effort to transfer the β-lactam resistance along with plasmid DNA from the K. oxytoca isolates by electroporation into ampicillin-susceptible Escherichia coli XAC, transformants resistant to extended-spectrum β-lactams were obtained (42). The transformants produced a β-lactamase with a pI of 8.4 and designated CMY-5 and showed increased resistance to aztreonam, cefuroxime, cefotaxime, and ceftazidime (MIC, >32 μg/ml). The substrate and inhibition profiles of the CMY-5 β-lactamase were highly similar to those of the chromosomal AmpC enzyme in E. coli (42). However, the enzyme activity in the transformants was much higher than that in the susceptible recipient (42). The transformants harbored a 7.8-kb plasmid which differed from the plasmids seen in the donors and could be transferred into E. coli XAC or C600 recipients by the conventional transformation technique (42). Nevertheless, the gene for the CMY-5 β-lactamase (blaCMY-5) was poorly expressed in K. oxytoca, as determined by the β-lactam resistance phenotype and enzymatic properties (42). Additionally, the extremely low frequency of transformation of pTKH11 by electroporation implied that pTKH11 existed in K. oxytoca at a low copy number.
In this study, the cryptic plasmid in K. oxytoca and the AmpC-like β-lactamase carried by this plasmid were further investigated.
MATERIALS AND METHODS
Bacteria, plasmids, and growth conditions.
The strains and plasmids used are listed in Table 1. Bacteria were grown in Luria-Bertani medium (Difco Laboratories, Detroit, Mich.) or on Luria Bertani agar plates supplemented with appropriate antibiotics.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant properties and commentsa | Source or reference(s) |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | recA endA1 gyrA96 thi-1 hsdR17 supE44 relA1 φ80 lacZΔM15 | Bethesda Research Laboratories |
| SN01 | ampA1+ ampC+ pyrB thr leu recA rpsL | 25 |
| SN03 | ampA1 ampC8 pyrB thr leu recA rpsL; spontanous SN01 mutant (mutations in structural and regulatory regions of ampC) | 25 |
| K-12 XAC | Nar Rir | 41 |
| TKH11 | E. coli XAC transformant from plasmid preparation of KH11 | 42 |
| TNBL63 | E. coli XAC transformant from plasmid preparation of NBL63 | 42 |
| K. oxytoca | ||
| KH11 | Azr Xmr clinical isolate | 41, 42 |
| KH55 | Azr Xmr clinical isolate | 41, 42 |
| KH78 | Azr Xmr clinical isolate | 41, 42 |
| KH93 | Azr Xmr clinical isolate | 41, 42 |
| NBL63 | Azr Xmr clinical isolate | 41, 42 |
| Plasmids | ||
| pGEM-3Z | Ampr subcloning vector | Promega |
| pNU6 | Ampr harboring ampC | 10 |
| pKH11 | Plasmid from K. oxytoca KH11 (subgroup 2) | 41, 42 |
| pNBL63 | Plasmid from K. oxytoca NBL63 (subgroup 3) | 41, 42 |
| pTKH11 | Plasmid from transformant TKH11 | 42 |
| pTKH78 | Plasmid from transformant TKH78 | 42 |
| pTNBL63 | Plasmid from transformant TNBL63 | 42 |
| pSW-21 | pGEM-3Z with 0.85-kb PstI fragment of pTKH11 | This study |
| pSW-22 | pGEM-3Z with 2.74-kb PstI-BglII fragment of pTKH11 | This study |
| pSW-24 | pGEM-3Z with 2.24-kb BglII fragment of pTKH11 | This study |
| pSW-28 | pGEM-3Z with 5.96-kb BglII fragment of pTKH11 | This study |
| pSW-26 | pGEM-3Z with 2.45-kb BglII fragment of pNBL63 | This study |
Amp, ampicillin; Az, aztreonam; Na, nalidixic acid; Ri, rifampin; Xm, cefuroxime.
Recombinant DNA techniques.
Plasmid DNA was prepared by use of Mini and Midi columns (Qiagen GmbH, Dusseldorf, Germany) as recommended by the manufacturer. Transformation of plasmid DNA was performed as described by Kushner (18). The antibiotics were used at 4 or 100 μg/ml for ampicillin, 8 μg/ml for aztreonam, and 100 μg/ml for cefotaxime and ceftazidime. Restriction enzymes, calf intestine alkaline phosphatase, and T4 DNA ligase were purchased from New England Biolabs, Inc. (Beverly, Mass.), and the conditions for enzymatic reactions were those suggested by the manufacturer. Analysis of plasmid DNA fragments was performed by electrophoresis in 0.7 to 1.5% agarose gels with TBE buffer (100 mM Tris base, 89 mM boric acid, 50 mM EDTA [pH 8.0]). As a size standard for the linear restriction fragments, a 1-kb DNA molecular weight marker (GIBCO BRL, Grand Island, N.Y.) was used.
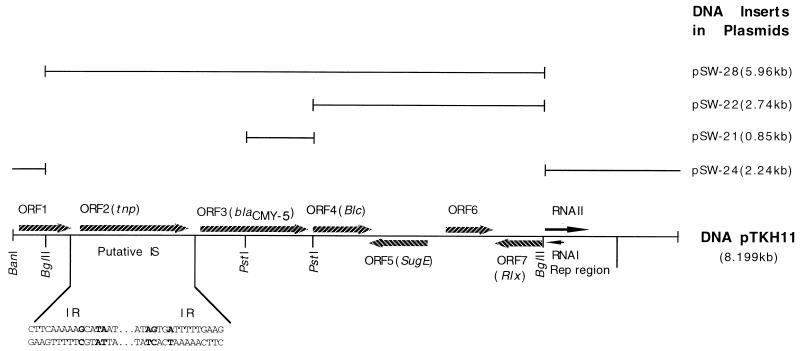
Cloning of pTKH11 and the replication region of pNBL63.
For the purpose of DNA sequencing the entire pTKH11, the plasmid was digested with BglII and with BglII plus PstI, and the 0.85-kb PstI, 2.74-kb PstI-BglII, 2.24-kb BglII, and 5.96-kb BglII DNA fragments were separately ligated into plasmid vector pGEM-3Z to form recombinant plasmids pSW-21, pSW-22, pSW-24, and pSW-28, respectively (Table 1; see also Fig. 3). The 2.45-kb BglII fragment of plasmid pNBL63, which may include the replication region for this plasmid, was also ligated into pGEM-3Z to generate plasmid pSW-26.
FIG. 3.
Genetic organization of the genes identified on plasmid pTKH11. The direction of gene transcription is shown by the arrows. The DNA inserts in various recombinant plasmids are represented by lines of different lengths, and the significant restriction sites are indicated. The inverted repeat (IR) in the putative insertion element (IS) is shown, and mismatched nucleotides are indicated by bold letters.
DNA sequencing.
The DNA sequence was determined by the dideoxy chain termination method (31) with an automated DNA sequencing system (model 377; PE/ABI, Foster City, Calif.). The template DNAs used for sequencing were plasmids pSW-21, pSW-22, pSW-24, pSW-26, and pSW-28; the primer walking was started with two vector-based primers specific for pGEM-3Z. Nucleotide and derived amino acid sequences were analyzed with the GCG program (Genetics Computer Group, Inc., Madison, Wis.) and DNA Star software (Lasergene, Madison, Wis.).
PCR.
To detect the presence of plasmid pTKH11 in the K. oxytoca isolates, the following primer pair was used: primer S28R2M (5′-CAA TGT GTG AGA AGC AGT CT-3′; nucleotides [nt] 2186 to 2205 in the pTKH11 sequence) and primer S21R1 (5′-GTA CAT ATC GCC AAT ACG-3′; complementary to nt 3114 to 3231). This primer pair generates a 1,045-bp amplicon containing 188 bp upstream of blaCMY-5 and a 770-bp sequence 5′ of blaCMY-5. A primer pair specific for the region and including a 298-bp sequence 3′ of blaCMY-5 and 231 nt downstream of blaCMY-5 was also used: primer S21F1 (5′-TTG GCG ATA TGT ACC AGG-3′; nt 3218 to 3235) and primer S22R1M (5′-TTC ATA CCA GGT TCC CAG-3′; complementary to nt 3730 to 3747). This primer pair produces a 529-bp PCR fragment.
Preparation of total DNA was previously described (39). The PCR mixture was prepared with a PCR reagent kit according to the standard method recommended by the manufacturer (Perkin-Elmer Cetus, Branchburg, N.J.). One to 4 μg of plasmid or total DNA from the K. oxytoca isolates and 40 pmol of each primer were included. PCR amplification was performed with a DNA Thermal Cycler 480 (Perkin-Elmer Cetus) at the following temperature profiles: 94°C for 5 min; 30 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 1 min; 72°C for 5 min; and 4°C for the remainder of the reaction.
Southern blot analysis.
Transfer of DNA to nylon filters was performed essentially as described by Southern (33). Labeling of DNA probes (as shown in Fig. 1) was performed with digoxigenin as described by the manufacturer (Boehringer Mannheim Biochemicals, Indianapolis, Ind.). Hybridization was performed at 70°C with buffers recommended in the instructions included in the digoxigenin kit from Boehringer.
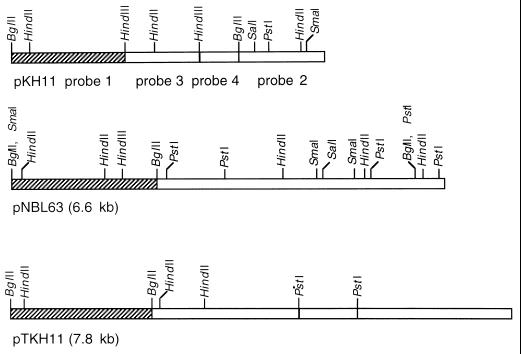
FIG. 1.
Restriction maps of plasmids pKH11, pNBL63, and pTKH11. The restriction enzymes used in the construction of the maps are shown above the bars. The four fragments of pKH11 used as probes are indicated under the bars. The bars represent the DNA sequences of the plasmids. DNA fragments showing sequence similarities are indicated by hatched bars.
Preparation of β-lactamase extracts and Western blotting.
The preparation of crude β-lactamase extracts was previously described (41, 42). The protein content of crude enzyme preparations was determined by use of an AB Kemila-preparat protein assay (Bio-Rad, Sollentuna, Sweden) with lyophilized bovine serum albumin as a standard. Electrophoresis was performed with a 12% polyacrylamide slab gel containing 0.1% sodium dodecyl sulfate (20). A low-molecular-weight protein standard from Pharmacia (Uppsala, Sweden) was run in parallel. Immunoblot transfer experiments were carried out essentially as described by Swanson et al. (34) with 1:1,000-diluted polyclonal antiserum raised against E. coli K-12 AmpC β-lactamase (5) as the primary antibody. Affinity-purified, alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G (Organon Teknika Corp., West Chester, Pa.) was used as the secondary antibody.
Nucleotide sequence accession numbers.
The nucleotide sequences described in this communication can be found in EMBL-GenBank under accession no. Y17716 for pTKH11 and Y17846 for the replication region of pNBL63.
RESULTS
Analysis of plasmid DNA with restriction endonucleases.
To reveal the relationships among the plasmids in K. oxytoca donor strains (KH11 and NBL63) and the E. coli XAC transformants (42), restriction maps of the K. oxytoca plasmids and the plasmids from the E. coli transformants were constructed by single and double digestions with a series of restriction enzymes. As shown in Fig. 1, the restriction pattern of pKH11 was different from that of the E. coli transformant plasmid pTKH11 and that of pNBL63. However, the plasmid from each of the E. coli transformants had the same restriction map as pTKH11.
DNA hybridization.
Southern blot analysis was performed to further examine the relationships among the plasmids. The 4.8-kb plasmid pKH11 was digested with BglII and HindIII, generating four DNA fragments: 1.75 kb by BglII-HindIII, 1.35 kb by BglII, 1.1 kb by HindIII, and 0.6 kb by BglII-HindIII (Fig. 1). Each of the fragments was labeled as a probe to hybridize with the DNA fragments produced by restriction digestion with BglII plus HindIII for pNBL63 and BglII plus PstI for pTKH11. Under stringent conditions, the 1.75-kb BglII-HindIII fragment of pKH11 hybridized with the 2.4-kb BglII fragment of pNBL63 and the 2.2-kb BglII fragment of pTKH11 (Fig. 1). These homologous fragments may be the areas where the replication origins for the plasmids are located. The other probes from pKH11 failed to hybridize with any DNA fragment from either pTKH11 or pNBL63.
Phenotype analysis of plasmids in an E. coli AmpC− background.
To distinguish enzyme activity encoded by the plasmid from that encoded by the chromosome, plasmid pTKH11 was transformed into both E. coli SN01, with a wild-type ampC gene, and its ampA1 ampC8 mutant derivative, SN03 (25). Transformants were obtained from both strains selected on agar plates containing either aztreonam (8 μg/ml) or ceftazidime (100 μg/ml). Preparations of pKH11 and pNBL63 failed to transform E. coli SN01 and SN03 to β-lactam resistance.
Analysis of ampC β-lactamase expression.
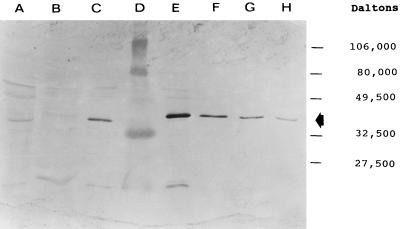
The expression of AmpC-like proteins was tested by Western blot analysis (Fig. 2). AmpC-like proteins were observed in crude β-lactamase preparations from K. oxytoca KH11, E. coli SN03/pTKH11, and E. coli SN03/pNU6. Depending on the dilution factor of the samples, the amount of the AmpC-like protein in E. coli SN03/pTKH11 was approximately 400-fold larger than that in K. oxytoca KH11 and 10-fold larger than that in E. coli SN03/pNU6 (10). No expression of the AmpC-like protein was seen in the E. coli SN03 recipient strain.
FIG. 2.
Western blot of crude β-lactamase from an E. coli recipient, a K. oxytoca donor, and their transformants with AmpC β-lactamase-specific antisera. The AmpC β-lactamase-specific band is indicated by an arrow. Lane A, 50 μg of crude β-lactamase from K. oxytoca KH11; lane B, 50 μg of crude β-lactamase from E. coli SN03; lane C, 5 μg of crude β-lactamase from E. coli SN03/pNU6; lane D, molecular weight standards; lane E, 5 μg of crude β-lactamase from E. coli SN03/pTKH11; lane F, 0.5 μg of crude β-lactamase from E. coli SN03/pTKH11; lane G, 0.25 μg of crude β-lactamase from E. coli SN03/pTKH11; lane H, 0.125 μg of crude β-lactamase from E. coli SN03/pTKH11.
DNA sequence of pTKH11.
The size of plasmid pTKH11 was determined to be 8,193 bp, and a BanI restriction site was designated as the first nucleotide to linearize the circle sequence in order to facilitate description. The whole DNA sequence was analyzed for open reading frames (ORFs), and the amino acid sequences deduced from the ORFs were compared with the sequences of known polypeptides in both the Tblastn and the Blastp data banks. The regions containing no significant ORFs (more than 50 amino acid residues) were sent to the data banks for DNA sequence comparison. As shown in Fig. 3, the plasmid contained seven ORFs, five of which were transcribed in the same direction as the DNA sequence of pTKH11 and the other two of which were located on the opposite DNA strand. Each of the ORFs was preceded by putative Shine-Dalgarno and promoter consensus sequences, except for ORF1.
The 188-amino-acid product of ORF1 showed no similarities with any protein in the data banks. Located 97 bp downstream of ORF1, ORF2, which encoded a polypeptide of 420 amino acid residues, showed 12% identity and 38% similarity to the transposase of IS1247 (accession no. X84038) from Xanthobacter autotrophicus (37). An inverted repeat of 16 bp was identified at 151 nt upstream (nt 628 to 644) and 163 nt downstream (complementary to nt 2220 to 2235) of ORF2 (Fig. 3). The inverted repeat was imperfect, with three nucleotide mismatches. No direct repeat which might serve as the target site for an insertion element could be found at the junction regions of the inverted repeat.
The 381-amino-acid product of ORF3, which followed ORF2 at an interval of 317 nt, showed 99.7 and 97% identities to the CMY-2 β-lactamase (1) and AmpC of Citrobacter freundii (13), respectively. Thus, ORF3 was identified as blaCMY-5. The only difference noted in the two plasmidic enzymes was the substitution of Leu-105 in CMY-2 with Phe-105 in CMY-5 (Fig. 4). Analysis of the blaCMY-5 flanking region, including 466 upstream and 124 downstream nt, revealed that the DNA sequence of the blaCMY-5 region was similar to that of CMY-2 by over 99%, and the same consensus sequences were shared by both β-lactamases.
FIG. 4.
Multiple amino acid sequence alignments of the CMY-5 β-lactamase with K. pneumoniae CMY-2 and C. freundii AmpC (BLACF) β-lactamases. The amino acid stretches in bold letters are the active site (SVSK), the conserved triad (KTG), and the class C-typical motif (YXN). Amino acid substitutions are indicated with asterisks, and Phe-105 in CMY-5 and Leu-105 in CMY-2 are shown in bold letters.
ORF4 was located 94 bp downstream of blaCMY-5 and its 177-amino-acid peptide product exhibited 98% identity with the outer membrane lipoprotein encoded by gene Blc from C. freundii (7). ORF5, which encoded a product of 105 amino acids, was located on the complementary strand of the pTKH11 sequence, and the stop codon of this ORF overlapped that of ORF4 by 2 nt. The deduced amino acid sequence of the product of ORF5 was identical to that of the SugE protein of C. freundii (unpublished data; accession no. U21727). ORF6, which encodes a putative protein with unknown function, was transcribed in the same direction and was located between ORF5 and ORF7. At 56 bp downstream of ORF6, ORF7 was located in the same orientation as ORF5 and encoded a putative protein with 70% identity and 77% similarity to the relaxation protein (accession no. X01654) of Salmonella typhimurium (6).
Within the 1.8-kb DNA sequence at the 3′ region (nt 6254 to 8199) of pTKH11, no significant ORFs could be identified. By DNA sequence comparison, it was shown that the 1- to 1.5-kb region 5′ of ORF7, coordinating nt 6371 to 7370 or nt 7870 (Fig. 3), was significantly homologous to replication regions of various plasmids (9, 23, 24, 28). The highest similarity was shown by the replicon of pJHCMW1, which was first identified in a pathogenic multiresistant K. pneumoniae strain isolated from the cerebrospinal fluid of a neonate with meningitis (38). These sequence similarities indicated that the 733-bp sequence from nt 6371 to nt 7105 might be the replicon of pTKH11. The RNA transcripts RNA I and RNA II, which serve as functional elements for plasmid replication (27), were identified (Fig. 3). RNA I and RNA II of pTKH11 were 70% and 81% similar, respectively, to the corresponding transcripts from pJHCMW1 (9).
DNA sequence of the replication region of plasmid pNBL63.
The 1,423-bp 3′ part of the 2,444-bp BglII fragment of pNBL63 was highly similar to the replication regions of several RNA priming plasmids (8, 9, 24). By DNA comparison with these well-characterized plasmids, the replication region of pNBL63 was identified (Fig. 5). The region can be divided into three functionally different parts. The first part, from nt 1037 to nt 1731, was the DNA sequence containing genes for RNA I and RNA II, which showed 84% similarity with the corresponding region in pJHCMW1. The 103-bp RNA I and the 521-bp RNA II transcripts of pNBL63 exhibited 71 and 87% similarities to those of pJHCMW1, respectively. The promoters for RNA I and RNA II were completely conserved in the two plasmids. A consensus sequence for the replication origin (oriV) of ColE1 (36) was also found at an appropriate location (nt 1599 to 1601). In the second part, the DNA sequence encoding a peptide of 63 amino acids was observed 178 bp downstream of oriV and showed 65% identity and 76% similarity to the genetic product of Rom (17). In the third part, the DNA sequence encoding a putative protein of 133 amino acid residues was identified downstream of that encoding the Rom homolog and was 29% identical and 44% similar in the amino terminus to entry exclusion protein 1 (Exc1) produced by the ColE1 plasmid (8).
FIG. 5.
Nucleotide sequence of the 1,424-bp replication region of pNBL63. The sequence includes the 1,424-bp 3′ part of the 2,444-bp BglII fragment of pNBL63. The consensus sequences are underlined and labeled. The putative transcription start sites are indicated by bold letters. The putative start codons are underlined, the stop codons are designated with asterisks, and the gene designations are given below the deduced amino acids, which are specified by standard one-letter abbreviations.
Similarity between the replication regions of pNBL63 and pTKH11.
The regions containing genes for RNA I and RNA II, which were 745 bp for pNBL63 (nt 1035 to 1780) and 784 bp for pTKH11 (nt 6370 to 7154), were 83% similar between the two plasmids. The RNA I transcript of pNBL63 was highly similar to that of pTKH11, with an identity of 97% (Fig. 6a). The RNA II transcripts of the two plasmids were estimated to be 521 nt for pNBL63 and 560 nt for pTKH11, with an identity of 90% (Fig. 6b). Nevertheless, no Rom- and Exc1-like genes were identified in the region downstream of the RNA I-RNA II region of pTKH11.
FIG. 6.
Bestfit comparison of the pNBL63 and pTKH11 RNA Is and RNA IIs. Identical nucleotides are indicated by vertical bars, and gaps are shown by periods.
Detection of plasmid pTKH11 in DNA preparations from K. oxytoca isolates by PCR amplification.
To confirm the presence of pTKH11 in the K. oxytoca isolates, a plasmid preparation from strain NBL63 and total DNA samples from strains KH11, KH55, KH78, and KH93 were used as templates for the detection of pTKH11. PCR products of 1.05 and 0.5 kb were obtained with primer pairs S28R2M-S21R1 and S21F1-S22R1M, respectively. Nevertheless, template DNAs had to be used in relatively large amounts (2 to 4 μg/100-μl reaction). The PCR products were confirmed to be the DNA fragments of the blaCMY-5 region in pTKH11 by DNA sequencing.
DISCUSSION
In a previous study, a plasmid having a low copy number and a cryptic phenotype with respect to β-lactam resistance in clinical isolates of K. oxytoca was found to express an AmpC-like β-lactamase in E. coli XAC electrotransformants (42). In this investigation, the plasmid, designated pTKH11, was identified as being harbored by each of the electrotransformants and as existing in most of the K. oxytoca donors. Each of the K. oxytoca isolates also contained an additional plasmid, such as pNBL63, which could be readily extracted by a routine procedure. pTKH11 was obtained only through electroporation of the plasmid into E. coli (42), and the detection of pTKH11 in K. oxytoca by PCR required a large amount of template DNA. These results suggested that pNBL63 was predominantly replicated, while pTKH11 obviously existed in very few copies in the K. oxytoca isolates. A similar finding was recently reported by other investigators (21).
Plasmid pTKH11 was introduced into the ampC β-lactamase null mutant SN03 (25), and the resultant transformant showed resistance to aztreonam and ceftazidime and hyperproduction of an AmpC-like protein, designated CMY-5 (Fig. 2). The level of production of the CMY-5 β-lactamase was low in K. oxytoca KH11. These data strongly support the notions that CMY-5 was encoded by pTKH11 and that the plasmid indeed existed in the K. oxytoca isolates. Moreover, the low level of expression of CMY-5 was consistent with the extremely low copy number of plasmid pTKH11 in K. oxytoca.
A comparison of amino acid sequences showed that CMY-5 differed from CMY-2 by only one residue and showed 97% identity with C. freundii AmpC (Fig. 4). The high degree of amino acid identity conferred the same catalytic activities on the enzymes (1, 42) and implied a close relationship among the three β-lactamases examined here.
The β-lactamases CMY-2, LAT-1, and BIL-1 are members of the plasmidic cephamycinases (CMYs) of amber class C β-lactamases (1, 11) and are believed to have a very close evolutionary relationship with C. freundii AmpC because of their high homologies (>94%) (1). Furthermore, a plasmid carrying a CMY-2 β-lactamase gene was characterized in C. freundii, and the plasmid was transferable to K. pneumoniae and E. coli (3). In this investigation, it was shown that blaCMY-5 was followed by Blc and SugE (Fig. 3); blaCMY-5 was 98% similar to C. freundii Blc and 100% identical to C. freundii SugE. Assembly of the DNA sequences for the ampC region of C. freundii GC3 (13) and the Blc and SugE regions of C. freundii (accession no. U21727) revealed that the genetic organization of these genes was identical to that found in pTKH11. The high degree of similarity in both the gene products and the genetic organization provided evidence that the chromosomally encoded AmpC of C. freundii is the origin of the plasmid-mediated CMYs. Thus, C. freundii AmpC should be regarded as the evolutionary precursor of CMYs rather than a homologous enzyme in the same classification group.
The Blc product may provide a starvation response function (7). The product of SugE is involved in nitrogen fixation in bacterial cells (12). Therefore, neither Blc nor SugE is related to the expression of the ampC gene. The insertion sequence-like element observed upstream of blaCMY-5 might be responsible for the translocation of the β-lactamase. ORF7 of pTKH11 specified a protein similar to an S. typhimurium relaxation protein which is involved in the formation of the relaxation complex during the mobilization of plasmid pSC101 (6). Accordingly, the product of ORF7 may also play a role in the spread of pTKH11.
blaCMY-5 was preceded by an intact promoter sequence; hence, the CMY-5 β-lactamase could be highly expressed in E. coli XAC and E. coli SN03. Thus, it was surprising that the blaCMY-5 gene product failed to confer resistance in the K. oxytoca isolates. Analysis of the replication regions for pNBL63 and pTKH11 showed that they both belonged to the RNA priming category of plasmids (29, 30, 32). In these plasmids, the replication process is regulated by the frequency of RNA I priming (14). The sequence homology between RNA Is also determines plasmid incompatibility through the cross suppression of replication by RNA I (19, 35). The extremely low copy number of pTKH11 in K. oxytoca might be due to incompatibility between pNBL63 and pTKH11, since a high degree of sequence similarity in the RNA I transcripts (97%) was noted. pNBL63 may have originated from K. oxytoca based on the fact that its RNA II transcript showed a high degree of similarity to that of pJHCMW1 of K. pneumoniae; pTKH11, on the other hand, may have been transferred from C. freundii into K. oxytoca. Thus, pTKH11 could not be stably maintained in the K. oxytoca host.
In conclusion, K. oxytoca plasmid pTKH11 was characterized as harboring a β-lactamase from the CMY family. CMY-5 was cryptic in the K. oxytoca clinical isolates but was highly expressed in E. coli. The cryptic phenotype was probably caused by the presence of an incompatible plasmid in the K. oxytoca host cell. The CMY β-lactamases were demonstrated to have originated from the chromosomal ampC gene of C. freundii based on the identical genetic organizations seen in the blaCMY-5 and C. freundii ampC gene regions. These results present direct evidence for the translocation of a β-lactamase-associated gene region from the chromosome to a plasmid.
ACKNOWLEDGMENTS
M. Norgren was supported by the Medical Research Council (grant 08675) and the medical faculty at Umeå University. S. W. Wu was partially supported by the Kempe Foundation.
REFERENCES
- 1.Bauernfeind A, Stemplinger I, Jungwirth R, Giamarellou H. Characterization of the plasmidic beta-lactamase CMY-2, which is responsible for cephamycin resistance. Antimicrob Agents Chemother. 1996;40:221–224. doi: 10.1128/aac.40.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauernfeind A, Horl G. Novel R-factor born beta-lactamase of Escherichia coli conferring resistance to cephalosporins. Infection. 1987;15:257–259. doi: 10.1007/BF01644127. [DOI] [PubMed] [Google Scholar]
- 3.Bauernfeind A, Ang O, Bal C, Jungwirth R. Presented at the Scientific Meeting of the European Society of Chemotherapy, abstr. OC2. Coimbra, Portugal. 1994. Plasmidic cephamycinase gene in Citrobacter freundii transferable to Klebsiella pneumoniae and Escherichia coli. [Google Scholar]
- 4.Bauernfeind A, Schweighart S, Dornbusch K, Giamarellou H. Program and abstracts of the 30th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1990. A transferable cephamycinase (CMY-ase) in Klebsiella pneumoniae (K. pn.), abstr. 190; p. 118. [Google Scholar]
- 5.Bergstrom S, Normark S. Beta-lactam resistance in clinical isolates of Escherichia coli caused by elevated production of the ampC-mediated chromosomal beta-lactamase. Antimicrob Agents Chemother. 1979;16:427–433. doi: 10.1128/aac.16.4.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernardi A, Bernardi F. Complete sequence of pSC101. Nucleic Acids Res. 1984;12:9415–9426. doi: 10.1093/nar/12.24.9415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bishop R E, Penfold S S, Frost L S, Holtje J V, Weiner J H. Stationary phase expression of a novel Escherichia coli outer membrane lipoprotein and its relationship with mammalian apolipoprotein D. Implications for the origin of lipocalins. J Biol Chem. 1995;270:23097–23103. doi: 10.1074/jbc.270.39.23097. [DOI] [PubMed] [Google Scholar]
- 8.Chan P T, Ohmori H, Tomizawa J, Lebowitz J. Nucleotide sequence and gene organization of ColE1 DNA. J Biol Chem. 1985;260:8925–8935. [PubMed] [Google Scholar]
- 9.Dery K J, Chavideh R, Waters V, Chamorro R, Tolmasky L S, Tolmasky M E. Characterization of the replication and mobilization regions of the multiresistance Klebsiella pneumoniae plasmid pJHCMW1. Plasmid. 1997;38:97–105. doi: 10.1006/plas.1997.1303. [DOI] [PubMed] [Google Scholar]
- 10.Edlund T, Grundstrom T, Normark S. Isolation and characterization of DNA repetitions carrying the chromosomal beta-lactamase of Escherichia coli K-12. Mol Gen Genet. 1979;173:115–125. doi: 10.1007/BF00330301. [DOI] [PubMed] [Google Scholar]
- 11.Fosberry A P, Payne D J, Lawlor E J, Hodgson J E. Cloning and sequencing analysis of blaBIL-1, a plasmid-mediated class C β-lactamase gene in Escherichia coli BS. Antimicrob Agents Chemother. 1994;38:1182–1185. doi: 10.1128/aac.38.5.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greener T, Govezensky D, Zamir A. A novel multicopy suppressor of a groEL mutation includes two nested open reading frames transcribed from different promoters. EMBO J. 1993;12:889–896. doi: 10.1002/j.1460-2075.1993.tb05729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haruta, S., K. Taniguchi, M. Nukaga, and T. Sawai. 1996. Unpublished data.
- 14.Itoh T, Tomizawa J. Formation of an RNA primer for initiation of replication of ColE1 DNA by ribonuclease H. Proc Natl Acad Sci USA. 1980;77:2450–2454. doi: 10.1073/pnas.77.5.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacoby G A. Extrachromosomal resistance in Gram-negative organisms: the evolution of β-lactamase. Trends Microbiol. 1994;2:357–360. doi: 10.1016/0966-842x(94)90611-4. [DOI] [PubMed] [Google Scholar]
- 16.Jacoby G A, Medeiros A A. More extended-spectrum beta-lactamases. Antimicrob Agents Chemother. 1991;35:1697–1704. doi: 10.1128/aac.35.9.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keenleyside W J, Whitfield C. Lateral transfer of rfb genes: a mobilizable ColE1-type plasmid carries the rfbO:54 (O:54 antigen biosynthesis) gene cluster from Salmonella enterica serovar Borreze. J Bacteriol. 1995;177:5247–5253. doi: 10.1128/jb.177.18.5247-5253.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kushner S R. An improved method for transformation of Escherichia coli with ColE1-derived plasmids. In: Boyer H B, Nicosia S, editors. Genetic engineering. Amsterdam, The Netherlands: Elsevier/North-Holland Publishing Co.; 1987. p. 17. [Google Scholar]
- 19.Lacatena R M, Cesareni G. Base pairing of RNAI with its complementary sequence in the primer precursor inhibits ColE1 replication. Nature. 1981;294:623–626. doi: 10.1038/294623a0. [DOI] [PubMed] [Google Scholar]
- 20.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Liu Y, Mee B L, Mulgrave L. Identification of clinical isolates of indole-positive Klebsiella spp., including Klebsiella planticola, and a genetic and molecular analysis of their beta-lactamases. J Clin Microbiol. 1997;35:2365–2369. doi: 10.1128/jcm.35.9.2365-2369.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matthew M, Harris A M. Identification of beta-lactamases by analytical isoelectric focusing: correlation with bacterial taxonomy. J Gen Microbiol. 1976;94:55–67. doi: 10.1099/00221287-94-1-55. [DOI] [PubMed] [Google Scholar]
- 23.Mikiewicz D, Wrobel B, Wegrzyn G, Plucienniczak A. Isolation and characterization of a ColE1-like plasmid from Enterobacter agglomerans with a novel variant of rom gene. Plasmid. 1997;38:210–219. doi: 10.1006/plas.1997.1312. [DOI] [PubMed] [Google Scholar]
- 24.Nomura N, Murooka Y. Characterization and sequencing of the region required for replication of a non-selftransmissible plasmid pEC3 isolated from Erwinia carotovora subsp. carotovora. J Ferment Bioeng. 1994;78:250–254. [Google Scholar]
- 25.Normark S, Burman L G. Resistance of Escherichia coli to penicillins: fine mapping and dominance of chromosomal β-lactamase mutations. J Bacteriol. 1977;132:1–7. doi: 10.1128/jb.132.1.1-7.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Papanicolaou G A, Medeiros A A, Jacoby G A. Novel plasmid-mediated β-lactamase (MIR-1) conferring resistance to oxyimino- and α-methoxy β-lactams in clinical isolates of Klebsiella pneumoniae. Antimicrob Agents Chemother. 1990;34:2200–2209. doi: 10.1128/aac.34.11.2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Polisky B. ColE1 replication control circuitry: sense from antisense. Cell. 1988;55:929–932. doi: 10.1016/0092-8674(88)90235-8. [DOI] [PubMed] [Google Scholar]
- 28.Rose R E. The nucleotide sequence of pACYC177. Nucleic Acids Res. 1988;16:356. doi: 10.1093/nar/16.1.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakakibara Y, Tomizawa J I. Replication of colicin E1 plasmid DNA in cell extracts. Proc Natl Acad Sci USA. 1974;71:802–806. doi: 10.1073/pnas.71.3.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakakibara Y, Tomizawa J I. Replication of colicin E1 plasmid DNA in cell extracts. II. Selective synthesis of early replicative intermediates. Proc Natl Acad Sci USA. 1974;71:1403–1407. doi: 10.1073/pnas.71.4.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Selzer G, Som T, Itoh T, Tomizawa J. The origin of replication of plasmid p15A and comparative studies on the nucleotide sequences around the origin of related plasmids. Cell. 1983;32:119–129. doi: 10.1016/0092-8674(83)90502-0. [DOI] [PubMed] [Google Scholar]
- 33.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 34.Swanson J, Mayer L W, Tam M R. Antigenicity of Neisseria gonorrhoeae outer membrane protein(s) III detected by immunoprecipitation and Western blot transfer with a monoclonal antibody. Infect Immun. 1982;38:668–672. doi: 10.1128/iai.38.2.668-672.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tomizawa J, Itoh T. Plasmid ColE1 incompatibility determined by interaction of RNAI with primer transcripts. Proc Natl Acad Sci USA. 1981;78:6096–6100. doi: 10.1073/pnas.78.10.6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tomizawa J-I, Ohmori H, Bird R E. Origin of replication of colicin E1 plasmid DNA. Proc Natl Acad Sci USA. 1977;74:1865–1869. doi: 10.1073/pnas.74.5.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Der Ploeg J, Willemsen M, Van Hall G, Janssen D B. Adaption of Xanthobacter autotrophicus GJ10 to bromoacetate due to activation and mobilization of the haloacetate dehalogenase gene by insertion element IS1247. J Bacteriol. 1995;177:1348–1356. doi: 10.1128/jb.177.5.1348-1356.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Woloj M, Tolmasky M E, Roberts M, Crosa J H. Plasmid-encoded amikacin resistance in multiresistant strains of Klebsiella pneumoniae isolated from neonates with meningitis. Antimicrob Agents Chemother. 1986;29:315–319. doi: 10.1128/aac.29.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu S, de Lencastre H, Tomasz A. Genetic organization of the mecA region in methicillin-susceptible and methicillin-resistant strains of Staphylococcus sciuri. J Bacteriol. 1998;180:236–242. doi: 10.1128/jb.180.2.236-242.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu, S. W., K. Dornbusch, and G. Kronvall. Genetic characterization of resistance to extended-spectrum beta-lactams in Klebsiella oxytoca septicemia isolates recovered from hospitals in the Stockholm area. Antimicrob. Agents Chemother., in press. [DOI] [PMC free article] [PubMed]
- 41.Wu S W, Dornbusch K, Goransson E, Ransjo U, Kronvall G. Characterization of Klebsiella oxytoca septicaemia isolates resistant to aztreonam and cefuroxime. J Antimicrob Chemother. 1991;28:389–397. doi: 10.1093/jac/28.3.389. [DOI] [PubMed] [Google Scholar]
- 42.Wu S W, Dornbusch K, Norgren M, Kronvall G. Extended spectrum beta-lactamase from Klebsiella oxytoca, not belonging to the TEM or SHV family. J Antimicrob Chemother. 1992;30:3–16. doi: 10.1093/jac/30.1.3. [DOI] [PubMed] [Google Scholar]