Summary
Background
Previous findings for the genetic and environmental contributions to DNA methylation variation were for limited age ranges only. We investigated the lifespan contributions and their implications for human health for the first time.
Methods
1,720 monozygotic twin (MZ) pairs and 1,107 dizygotic twin (DZ) pairs aged 0-92 years were included. Familial correlations (i.e., correlations between twins) for 353,681 methylation sites were estimated and modelled as a function of twin pair cohabitation history.
Findings
The methylome average familial correlation was around zero at birth (MZ pair: -0.01; DZ pair: -0.04), increased with the time of twins living together during childhood at rates of 0.16 (95%CI: 0.12-0.20) for MZ pairs and 0.13 (95%CI: 0.07-0.20) for DZ pairs per decade, and decreased with the time of living apart during adulthood at rates of 0.026 (95%CI: 0.019-0.033) for MZ pairs and 0.027 (95%CI: 0.011-0.043) for DZ pairs per decade. Neither the increasing nor decreasing rate differed by zygosity (both P>0.1), consistent with cohabitation environment shared by twins, rather than genetic factors, influencing the methylation familial correlation changes. Familial correlations for 6.6% (23,386/353,681) sites changed with twin pair cohabitation history. These sites were enriched for high heritability, proximal promoters, and epigenetic/genetic associations with various early-life factors and late-life health conditions.
Interpretation
Early life strongly influences DNA methylation variation across the lifespan, and the effects are stronger for heritable sites and sites biologically relevant to the regulation of gene expression. Early life could affect late-life health through influencing DNA methylation.
Funding
Victorian Cancer Agency, Cancer Australia, Cure Cancer Foundation.
Keywords: DNA methylation, Twin study, DOHaD hypothesis, Heritability
Research in context.
Evidence before this study
DNA methylation is proposed to play a critical role in the aetiology of complex human traits and diseases. Previous twin studies reported the average heritability of DNA methylation ranged 3%-20%, depending on the age of participants. However, these studies individually investigated specific and limited age ranges; therefore, they could only provide evidence for the investigated age range, but not for the whole lifespan.
Added value of this study
We investigated the genetic and environmental contributions to DNA methylation variation across the lifespan. Familial correlation in DNA methylation increased with the time of twins living together and decreased with the time of twins living apart, consistent with cohabitation-related early-life environmental factors influencing the familial correlation change. In addition, the effects are stronger for methylation sites affected by genetic factors, and sites associated with various early-life exposures and late-life health conditions.
Implications of all the available evidence
Early life strongly influences DNA methylation variation across the lifespan, and could affect late-life health through influencing DNA methylation.
Alt-text: Unlabelled box
Introduction
Epigenetic modifications regulate gene expression without changing the underlying DNA sequence, and have been proposed to play a critical role in the aetiology of complex human traits and diseases.1,2 DNA methylation, one of the most studied epigenetic modifications, has been found to be associated with several human traits and diseases, such as smoking3, 4, 5 and obesity.6, 7, 8
Understanding the causes of DNA methylation variation could inform the determinants and biological mechanisms that affect human health through influencing DNA methylation. Twin studies are classic designs for understanding the contributions of (unmeasured) genetic and environmental causes to the variation of human traits, including DNA methylation. Although twin studies might not necessarily measure any genetic or environmental factors, they infer the genetic and environmental contributions to variation by comparing the twin resemblance in the trait of interest between monozygotic (MZ) and dizygotic (DZ) twin pairs. Under the null hypothesis that genetic factors do not influence variation in the trait, MZ pairs will have the same resemblance as DZ pairs. If only additive genetic factors influence familial correlation in the trait (i.e., there is no effect of having a shared environment), the MZ pair correlation will be twice the DZ pair correlation. Under this latter assumption, the heritability of the trait (expressed as a percentage) can be estimated as 100 times the minimum of: (i) twice the difference between the MZ and DZ pair correlations and (ii) the MZ pair correlation.
Twin studies have considered a measure of the overall proportion of DNA methylation variation explained by genetic factors, which is defined as the average of the heritability estimates across the measured sites. The average heritability of the sites measured by the HumanMethylation27 and HumanMethylation450 BeadChip arrays has been found to range from 3% to 20% and vary with the age of twins, being highest in adolescence and young adulthood and lowest at birth and in middle age.9, 10, 11, 12, 13, 14, 15 There is, however, evidence that environmental factors shared between twins explain a substantial proportion of methylation variation at age of 18 years and middle age.14,15 These studies investigated specific and limited age ranges; therefore, they could only provide evidence for the investigated age range, but not for the whole lifespan.
We previously pooled DNA methylation data from studies in which the age of twins covered the whole lifespan to solve the issues that studies of limited age ranges found different estimates of the genetic and environmental contributions to the variation in methylation-based measures.16,17 We found evidence that the variation in both genome-wide average methylation and epigenetic age are consistent with being influenced by environmental factors shared by twins when they cohabit, and that these effects can persist across the whole lifespan. Such findings are not possible to be found by studies focusing on limited age ranges.
To address the issue that different genetic and environmental contributions to methylation variation have been found in different age ranges,9, 10, 11, 12, 13, 14, 15 here we investigated the determinants of methylation variation across the lifespan by combining and analysing data for 5,654 twins from nine twin studies. We also considered the implications of our findings for human health by leveraging published epigenetic and genetic associations with human diseases and traits.
Methods
Study sample
The sample was from nine studies (Table 1). 23 MZ pairs and 22 DZ pairs from the Peri/postnatal Epigenetic Twins Study (PETS).18 67 MZ pairs and 111 DZ pairs from the Brisbane Systems Genetics Study (BSGS).12,19 426 MZ pairs and 306 DZ pairs from the E-Risk Longitudinal Twin Study (E-Risk).15 150 MZ pairs from the Danish Twin Registry (DTR), which included two age groups, younger adults (mean age 33 years) and older adults (mean age 63 years).20 769 MZ pairs and 424 DZ pairs from the Netherlands Twin Register (NTR).13 66 MZ pairs and 66 DZ pairs from the Australian Mammographic Density Twins and Sisters Study (AMDTSS).21,22 93 MZ pairs and 153 DZ pairs from Multiple Tissue Human Expression Resource (MuTHER) Study.11 108 MZ pairs from the Older Australian Twins Study (OATS).23 18 MZ pairs and 25 DZ pairs from the Longitudinal Study of Aging Danish Twins (LSADT), which included longitudinal measurements that were conducted using blood samples collected in years 1997 and 2007, respectively.24 The samples of BSGS, E-Risk AMDTSS, MuTHER and OATS were of European descendent, while the samples of the other studies were from countries where the vast majority of population is of European descendent.
Table 1.
Characteristics and DNA methylation twin pair correlations of the sample
| Study | Country | Twins | Number of twins (number of females) | Mean age (SD), range | Twin pair correlations of the 353,681 sites |
|||
|---|---|---|---|---|---|---|---|---|
| 25th percentile | Median | Mean | 75th percentile | |||||
| PETS | Australia | MZ | 46 (24) | 0 (0), 0-0 | -0.21 | -0.04 | -0.01 | 0.16 |
| DZ | 44 (21) | 0 (0), 0-0 | -0.19 | -0.05 | -0.04 | 0.10 | ||
| BSGS | Australia | MZ | 134 (62) | 13.8 (1.9), 12-18 | -0.03 | 0.09 | 0.14 | 0.27 |
| DZ | 222 (107) | 13.2 (2.0), 10-24 | -0.01 | 0.06 | 0.08 | 0.16 | ||
| E-Risk | UK | MZ | 852 (414) | 18 (0), 18-18 | -0.12 | 0.21 | 0.26 | 0.37 |
| DZ | 612 (300) | 18 (0), 18-18 | 0.09 | 0.17 | 0.20 | 0.28 | ||
| DTR young adults | Denmark | MZ | 146 (66) | 33.1 (2.0), 30-37 | 0.10 | 0.24 | 0.26 | 0.39 |
| NTR | The Netherlands | MZ | 1,538 (1082) | 36.1 (12.4), 18-78 | 0.05 | 0.12 | 0.20 | 0.32 |
| DZ | 848 (511) | 33.9 (10.5), 17-79 | 0.01 | 0.06 | 0.09 | 0.15 | ||
| AMDTSS | Australia | MZ | 132 (132) | 55.6 (8.4), 43-72 | -0.06 | 0.05 | 0.08 | 0.19 |
| DZ | 132 (132) | 57.0 (7.2), 43-74 | -0.09 | -0.001 | 0.001 | 0.09 | ||
| MuTHER | UK | MZ | 186 (186) | 61.0 (9.3), 44-85 | 0.01 | 0.12 | 0.14 | 0.26 |
| DZ | 306 (306) | 57.4 (9.3), 39-83 | -0.01 | 0.06 | 0.07 | 0.14 | ||
| DTR old adults | Denmark | MZ | 154 (78) | 63.2 (4.1), 57-74 | 0.06 | 0.18 | 0.20 | 0.31 |
| OATS | Australia | MZ | 216 (136) | 71.2 (6.0), 65-90 | -0.004 | 0.09 | 0.12 | 0.22 |
| LSADT 1997 | Denmark | MZ | 36 (22) | 76.3 (2.0), 73-82 | -0.13 | 0.07 | 0.08 | 0.28 |
| DZ | 50 (40) | 76.2 (1.6), 74-80 | -0.10 | 0.05 | 0.05 | 0.20 | ||
| LSADT 2007 | Denmark | MZ | 36 (22) | 86.2 (2.0), 83-92 | -0.12 | 0.07 | 0.08 | 0.27 |
| DZ | 50 (40) | 86.1 (1.6), 84-90 | -0.09 | 0.05 | 0.05 | 0.20 | ||
PETS – Peri/postnatal Epigenetic Twins Study; BSGS – Brisbane System Genetics Study; E-Risk – Environmental Risk Longitudinal Twin Study; DTR – Danish Twin Registry, with participants of two age groups: younger and older adults; NTR – Netherlands Twin Register; AMDTSS – Australian Mammographic Density Twins and Sisters Study; MuTHER – Multiple Tissue Human Expression Resource Study; OATS – Older Australian Twins Study; LSADT – Longitudinal Study of Aging Danish Twins, with samples collected in years 1997 and 2007, respectively; MZ – monozygotic; DZ – dizygotic; SD – standard deviation.
Ethics
This study was approved by the Human Research Ethics Committee of The University of Melbourne. Informed consent was obtained from all participants by the original studies. PETS was approved by the Human Research Ethics Committees of the Royal Women's Hospital, Mercy Hospital for Women, and Monash Medical Centre, Melbourne. BSGS was approved by the Human Research Ethics Committee of the Queensland Institute for Medical Research. E-Risk was approved by the NRES Committee London — Camberwell St Giles Ethics Committee, and the Joint South London and Maudsley and the Institute of Psychiatry Research Ethics Committee approved each phase of the E-Risk study. DTR was approved by the Regional Scientific Ethical Committees for Southern Denmark. AMDTSS approved by the Australian Twin Registry and the Human Research Ethics Committee of The University of Melbourne. MuTHER was approved by the Research Ethics Committee of St. Thomas’ Hospital, London. OATS was approved by the Australian Twin Registry and the Ethics Committees of University of New South Wales, The University of Melbourne, Queensland Institute of Medical Research and the Southeastern Sydney and Illawarra Area Health Service. LSADT was approved by the Danish Scientific Ethics Committees.
DNA methylation data
For NTR, we used the methylation site-specific twin pair correlation estimates from van Dongen et al.;13 no individual-level methylation data were accessed. For the other studies, individual-level methylation data were used, most of which were accessed through public repositories (Table S1). The data of PETS and OATS are available from the relevant authors on request. In PETS, methylation was measured using DNA extracted from cord blood samples and the HumanMethylationEPIC array. Data were background corrected, normalised using quantile normalisation25 within the minfi package.26 In BSGS, DNA was extracted from peripheral blood and methylation was measured using the HumanMethylation450 (HM450) array. Individual probes were normalised across all samples using a generalized linear model with a logistic link function; see McRae et al. for more details.12 In E-Risk, DNA was extracted from blood samples and methylation was measured using the HM450 array. Data were normalised with the dasen function from the wateRmelon package;27 see Hannon et al. for more details.15 In DTR, DNA was extracted from blood samples and methylation was measured using the HM450 array. Data were normalised using the subset-quantile within-array normalisation (SWAN) method28 within the minfi package;26 see Tan et al. for more details.20 In AMDTSS, DNA was extracted from dried blood spots stored on Guthrie cards and methylation was measured using the HM450 array. Data was processed by Bioconductor minfi package,26 which included normalisation of data using Illumina's reference factor-based normalization methods and the SWAN method28 and an empirical Bayes batch-effects removal method ComBat29 to minimise the technical variation across batches; see Li et al. for more details.22 In MuTHER, DNA was extracted from adipose tissue samples and methylation was measured using the HM450 array. Data were quantile normalised;25 see Grundberg et al. for more details.11 In OATS, DNA was extracted from blood samples and methylation was measured using the HM450 array. Data were normalised using the SWAN method.28 In LSADT, methylation was measured using the HM450 array and data were normalised using the SWAN method;28 see Tan et al. for more details.24 There were 353,681 autosomal methylation sites common to the nine studies, and these sites were included in analysis.
Statistical methods
Site-specific twin pair correlation
Within each study, we estimated the correlation between twins (i.e., familial correlation) for each of the 353,681 methylation sites. Correlation was defined as the ratio of the twin pair covariance in methylation to the methylation variance of the total sample. For each site and within each study, the methylation Beta-values were adjusted for age, sex, factors related to study design, batch effects, and blood cell proportions (CD8+ T cells, CD4+ T cells, B cells, natural killer cells, monocytes and granulocytes, and nucleated red blood cells additionally for PETS) estimated from the methylation data30,31 using linear regression (Table S1). Residuals from the regression were inverse-normal transformed and used to estimate twin pair correlations for MZ pairs and DZ pairs, respectively.
We examined the twin pair correlations for sites suggested to be affected by genetic factors or being at loci that have been reported to be associated with smoking or body mass index (BMI), as the two traits have been studied for epigenetic association by various studies and several loci have been consistently reported. For sites affected by genetic factors, we used the methylation site heritability estimates found by van Dongen et al. by analysing NTR data,13 and the methylation quantitative trait locus (mQTL) results by Min et al.32 Loci associated with smoking or BMI included AHRR, F2RL3, 2q27.1, 6p21.33, HIF3A, SOCS3 and ABCG1.3, 4, 5, 6, 7, 8
Negative control
We used pairs of unrelated samples as negative control. Within each study, unrelated samples were randomly paired resulting in the same number of pairs as the number of twin pairs, and site-specific methylation correlation was estimated for the pairs using the same methods as above. We repeated this process for 10 times to avoid that the results are biased by a single randomisation.
Overall twin pair correlation and cohabitation
Suggested by the pattern of age-specific twin pair correlation and following previous theoretical and empirical studies,16,17,33,34 we modelled the average twin pair correlation of the 353,681 investigated sites, , as a function of the twin pair's postnatal cohabitation history using the pooled data across all studies. The model was fitted using piecewise linear splines with a knot point at t0:
where t is the time in years since the twin pair started living together, t0 is the time when the twin pair started living separately, ε is the error term of the regression, α is the intercept of the regression which reflects the average correlation when the pair started living together, and λ and ν are indicators for cohabitation history: λ=1 and ν=0 when t≤t0, and λ=0 and ν=1 when t>t0; therefore, when the twin pair lived together, when the twin pair lived apart, and β1 and β2 reflect the rates at which changes with the time of living together and the time of living apart, respectively. A twin pair was assumed to start living together since birth and become separated at age of 18 years (i.e., t = the average age of the twin pair, and t0 = 18). Age of 18 years was chosen based on previous theoretical and empirical evidence16,17,33,34 and comparing the modelling likelihoods under different values of t0; the likelihood was the greatest when t0 = 18 (Figure S1).
Individual twin pairs across all studies were included in the modelling with each twin pair as a data point. Within each study, each twin pair was assumed to have a correlation the same as the study's zygosity-specific ; because we did not have access to NTR individual-level data, we did not use the observed twin pair ages, but assumed that, within each study, the average age of a twin pair was equal to the study's zygosity-specific average age. For example, in BSGS, all 67 MZ pairs were assumed to have a correlation of 0.14 and an age of 13.8 years, and all 111 DZ pairs were assumed to have a correlation of 0.08 and an age of 13.2 years. Robust standard errors, calculated using the R sandwich package, were used in the statistical tests to account for clustering by study. The difference in a regression parameter by zygosity was investigated by modelling both zygosities together and testing an interaction term with the zygosity.
We conducted sensitivity analyses: 1) Using the observed twin pair ages, in which we simulated the NTR ages using a beta distribution and the summary statistics (twin pair sample size, and mean, minimum, maximum and standard deviation in age) reported by van Dongen et al.13; 2) Using the average twin pair age of NTR and the observed twin pair ages for the other studies; 3) Excluding NTR and using the observed twin pair ages for the other studies; 4) Modelling the median correlation of the investigated sites the same as the main analysis.
Site-specific twin pair correlation and cohabitation
We investigated for which individual methylation sites the twin pair correlations also changed with cohabitation, by using the same modelling methods as those for , with being replaced as the site-specific twin pair correlation. Statistical tests for β1 and β2 were one-sided. False Discovery Rate (FDR) was used to adjust for testing across the 353,681 investigated sites, and results with a FDR<0.05 were treated as statistically significant.
Genomic location enrichment analysis
For the cohabitation-dependent methylation sites identified above, we investigated the enrichment of them for genomic features in terms of chromosomes, CpG density regions and gene-centric regions. The annotations by Slieker et al.35 were used: four CpG density regions – CpG island (CGI; CG content >50%, length >200 bp and observed/expected ratio of CpGs >0.6), CGI shore (2-kb region flanking CGI), CGI shelf (2-kb region flanking CGI shore) and non-CGI region; five gene-centric regions – intergenic region (>10 kb from the nearest transcription start site [TSS]), distal promoter (−10 kb to 1.5 kb from the nearest TSS), proximal promoter (−1.5 kb to +500 bp from the nearest TSS), gene body (+500 bp to 3’ end of the gene) and downstream region (3’ end to +5 kb from 3’ end). A two-sided Fisher's exact test was performed to investigate whether the observed number of the cohabitation-dependent methylation sites in each genomic location is different from the expected number by chance, and results with a FDR<0.05 were considered statistically significant.
Pathway analysis
We conducted pathway analysis for the cohabitation-dependent sites via a hypergeometric test taking into account the number of sites per gene, using the gometh function of Bioconductor missMethyl package36 with the 353,681 investigated methylation sites as the background set of sites. Both Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathway and gene ontology enrichments were analysed.
Epigenetic association enrichment analysis
We intersected the cohabitation-dependent sites with sites reported by epigenome-wide association studies to investigate epigenetic association enrichment. Studies published before 2021 were searched on PubMed using term “epigenome-wide association study” and included if they are original research about epigenome-wide association studies of human traits/exposures and if there were at least 10 genome-wide significant (FDR<0.05, or Bonferroni-adjusted P<0.05 if FDR is not available) sites reported. There were no criteria on the sample size of the original studies. The threshold of 10 was chosen to avoid the analysis being biased by the results of studies reporting only few significant sites. A total of 40 traits/exposures reported by 57 studies were analysed (Table S2). A one-sided Fisher's exact test was performed for each trait/exposure to assess the enrichment.
Genetic association enrichment analysis
We intersected the genes annotated by the cohabitation-dependent sites with the genes found by genome-wide association studies (GWAS) of human traits/exposures to investigate genetic association enrichment. The cohabitation-dependent sites were annotated to genes using Illumina's annotation file. Genes found by GWAS were retrieved from the GWAS Catalog (https://www.ebi.ac.uk/gwas/; accessed on 13 January 2021), which contained 87,777 genetic variants mapped to 27,621 genes associated with 3,292 traits/exposures at P<5 × 10−8 (Table S3). A one-sided Fisher's exact test was performed for each trait/exposure to assess the enrichment.
Role of funding source
The funders had no role in study design, data collection, data analyses, interpretation, or writing of report.
Results
The sample included 5,654 twins (3,619 females, 2,035 males), including 1,720 MZ pairs (2,202 females, 1,238 males) and 1,107 DZ pairs (1,417 females, 797 males; Table 1). The sample is representative of the general population in terms of age range: the twins aged 0-92 years (mean: 34.4 years; standard deviation: 19.4 years), covering the whole human lifespan. The twins were from four countries: Australia, Denmark, The Netherlands and UK.
Across the lifespan, the average methylation value of the 353,681 investigated sites slightly increased from 0.47 at birth to 0.51 at age >80 years (Figure S2a). The overall methylation variation, assessed by the average standard deviation in methylation value across sites, increased with age – from 0.020 at birth to 0.024 at age 18 years, then to 0.027 at age >80 years – the increase was more significant before adulthood (Figure S2b), suggesting cumulative non-genetic effects on methylation variation during the lifespan, especially before adulthood. For studies with both MZ and DZ twins available, there was no apparent difference in the average methylation value or methylation variation by zygosity.
Methylation familial correlation across the lifespan
Consistently across all studies, there was no correlation for pairs of unrelated samples: for both MZ and DZ pairs, the average correlation of the 353,681 sites, , was close to zero within each study (Table S4). Such results also suggest that there were unlikely cohort effects between the included studies either due to different biological samples, different data pre-processing methods or other unknown factors.
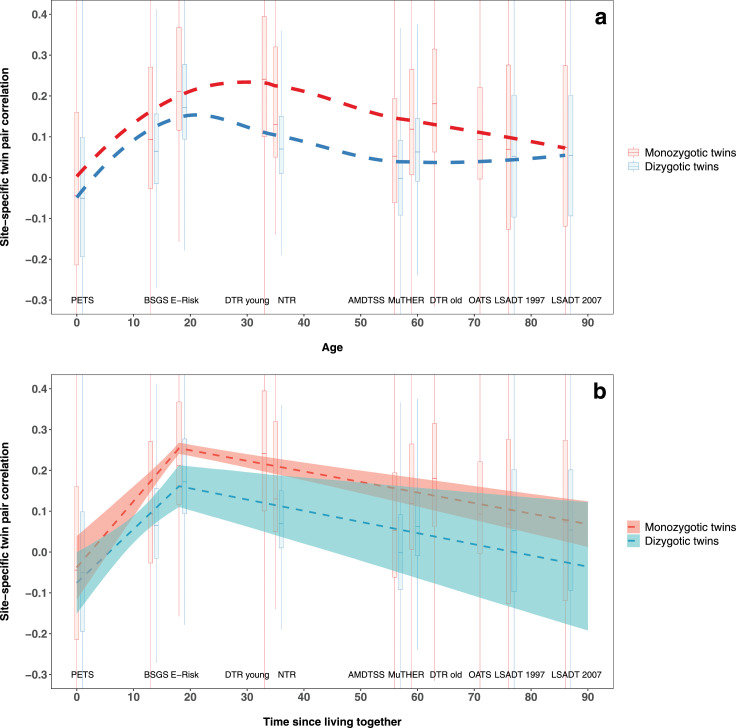
As to twin pairs, across all studies, there was a clear pattern: for both MZ and DZ pairs, was close to zero at birth (-0.01 and -0.04 for MZ and DZ pairs, respectively) and similar to the for unrelated samples, increased with age to 18 years (0.26 and 0.20 for MZ and DZ pairs, respectively), and then decreased with age in adulthood to 0.08 and 0.05 for MZ and DZ pairs at ages >80 years, respectively (Table 1; Figures 1a, S3 and S4).
Figure 1.
DNA methylation familial correlation across the lifespan for the 1,720 monozygotic twin pairs and 1,107 dizygotic twin pairs
a) Pattern of the overall twin pair correlation changed with age.
Study-, zygosity- and site-specific twin pair correlation across the 353,681 investigated sites are showed as boxplots. The dashed curves are the zygosity-specific Locally Weighted Scatterplot Smoothing curves fitted across all studies. The plot is cropped along Y axis to better present the pattern. The full plot is in Figure S3.
PETS – Peri/postnatal Epigenetic Twins Study; BSGS – Brisbane System Genetics Study; E-Risk – Environmental Risk Longitudinal Twin Study; DTR – Danish Twin Registry, with participants of two age groups: younger and older adults; NTR – Netherlands Twin Register; AMDTSS – Australian Mammographic Density Twins and Sisters Study; MuTHER – Multiple Tissue Human Expression Resource Study; OATS – Older Australian Twins Study; LSADT – Longitudinal Study of Aging Danish Twins, with samples collected in years 1997 and 2007, respectively.
b) The overall twin pair correlation as a function of cohabitation history.
The dashed lines are the zygosity-specific fitted lines from modelling the overall twin pair correlation of the 353,681 investigated sites as a function twin pair cohabitation history, with confidence intervals showed as ribbons.
Methylation familial correlation for sites affected by genetic factors, or associated with smoking or BMI
The pattern of methylation familial correlation with age for both MZ and DZ pairs above was also evident for heritable methylation sites or sites associated with mQTL: Figures S5a-S5c show the correlations of sites having a heritability estimate <20% (224,027 sites), 20-50% (95,383 sites) and >50% (31,180 sites), respectively, based on the heritability estimates from van Dongen et al.;13 Figure S5d shows the correlations of 113,013 sites associated with mQTL discovered and replicated by Min et al.32 Similar pattens of the methylation familial correlation with age were also evident for the loci previously established to be associated with smoking or BMI, including AHRR, F2RL3, 2q27.1, 6p21.33, HIF3A, SOCS3 and ABCG1 (Figure S6).
Methylation familial correlation changed with cohabitation history
Suggested by the pattern in methylation familial correlation above and following previous theoretical and empirical studies,16,17,33,34 we modelled the average correlation of the 353,681 investigated sites, , as a function of the twin pair's postnatal cohabitation history using the pooled data across studies (Figure 1b, Table 2). We found that increased with cohabitation time up to age 18 years (when the vast majority would have been living together): the increasing rates per 10 years were 0.16 (95% confidence interval [CI]: 0.12, 0.20, P<2.8 × 10−14, t-test) for MZ pairs and 0.13 (95% CI: 0.07, 0.20, P=2.0 × 10−5, t-test) for DZ pairs. The rate for MZ pairs was not different from the rate for DZ pairs (P=0.13, t-test; null hypothesis: the rate is due to common environmental effects only), but less than twice the rate for DZ pairs (P=0.03, t-test; null hypothesis: the rate is due to additive genetic effects only). For ages >18 years (when the vast majority of twin pairs would have been living apart), decreased with time: the decreasing rates per 10 years were -0.026 (95% CI: -0.033, -0.019, P=1.5 × 10−12, t-test) for MZ pairs and -0.027 (95% CI: -0.043, -0.011, P=8.1 × 10−4, t-test) for DZ pairs. The two rates were not significantly different (P=0.84, t-test; null hypothesis: the rate is due to common environmental effects only), but the MZ pair rate was less than twice the DZ pair rate (P=0.048, t-test; null hypothesis: the rate is due to additive genetic effects only). The confidence intervals of the average correlation for MZ and DZ pairs overlapped at all ages across the lifespan except at ages of 10-35 years, when MZ pairs had a higher average correlation than DZ pairs (Figure 1b). Sensitivity analyses of using alternative samples, twin pair ages or correlation measures gave similar results (Table 2).
Table 2.
Modelling results for the overall DNA methylation correlation as a function of twin pair cohabitation history
| Twin pair | Intercept α |
Increasing rate β1 |
Decreasing rate β2 |
|||
|---|---|---|---|---|---|---|
| Estimate (95% CI) | P | Estimate (95% CI) | P | Estimate (95% CI) | P | |
| Main analysis | ||||||
| MZ pairs | -0.038 (-0.114, 0.038) | 0.33 | 0.162 (0.121, 0.204) | 2.8 × 10−14 | -0.026 (-0.033, -0.019) | 1.5 × 10−12 |
| DZ pairs | -0.075 (-0.150, -0.0003) | 0.05 | 0.132 (0.071, 0.192) | 2.0 × 10−5 | -0.027 (-0.043, -0.011) | 8.1 × 10−4 |
| MZ & DZ pairs combined | -0.071 (-0.174, 0.033) | 0.18 | 0.159 (0.099, 0.219) | 2.0 × 10−7 | -0.025 (-0.036, -0.014) | 8.0 × 10−6 |
| Sensitivity analysis: Observed twin pair age used in modelling (NTR age simulated) | ||||||
| MZ pairs | -0.025 (-0.070, 0.019) | 0.27 | 0.149 (0.118, 0.181) | <1 × 10−15 | -0.020 (-0.031, -0.010) | 2.2 × 10−4 |
| DZ pairs | -0.062 (-0.114, -0.009) | 0.02 | 0.119 (0.061, 0.177) | 6.7 × 10−5 | -0.023 (-0.039, -0.007) | 6.2 × 10−3 |
| MZ & DZ pairs combined | -0.057 (-0.127, 0.014) | 0.12 | 0.146 (0.097, 0.195) | 5.1 × 10−9 | -0.020 (-0.033, -0.007) | 3.1 × 10−3 |
| Sensitivity analysis: Observed twin pair age used in modelling (NTR age assumed to be the average) | ||||||
| MZ pairs | -0.029 (-0.081, 0.023) | 0.28 | 0.156 (0.127, 0.185) | <1 × 10−15 | -0.025 (-0.032, -0.018) | 2.4 × 10−11 |
| DZ pairs | -0.065 (-0.119, -0.011) | 0.02 | 0.124 (0.071, 0.177) | 5.2 × 10−6 | -0.026 (-0.042, -0.010) | 1.7 × 10−3 |
| MZ & DZ pairs combined | -0.060 (-0.138, 0.017) | 0.13 | 0.152 (0.105, 0.199) | 2.3 × 10−10 | -0.024 (-0.035, -0.013) | 1.9 × 10−5 |
| Sensitivity analysis: Observed twin pair age used in modelling (NTR data excluded) | ||||||
| MZ pairs | -0.031 (-0.086, 0.025) | 0.28 | 0.159 (0.129, 0.189) | <1 × 10−15 | -0.025 (-0.032, -0.019) | 9.0 × 10−14 |
| DZ pairs | -0.078 (-0.148, -0.008) | 0.03 | 0.142 (0.096, 0.188) | 1.9 × 10−9 | -0.027 (-0.038, -0.016) | 1.2 × 10−6 |
| MZ & DZ pairs combined | -0.066 (-0.153, 0.022) | 0.14 | 0.160 (0.112, 0.209) | 1.5 × 10−10 | -0.025 (-0.034, -0.016) | 5.9 × 10−8 |
| Sensitivity analysis: Median correlation of the investigated sites used in modelling | ||||||
| MZ pairs | -0.069 (-0.131, -0.007) | 0.03 | 0.143 (0.099, 0.187) | 2.6 × 10−10 | -0.020 (-0.031, -0.009) | 5.3 × 10−4 |
| DZ pairs | -0.072 (-0.133, -0.012) | 0.02 | 0.116 (0.060, 0.173) | 5.8 × 10−5 | -0.024 (-0.039, -0.008) | 3.7 × 10−3 |
| MZ & DZ pairs combined | -0.078 (-0.151, -0.004) | 0.04 | 0.136 (0.083, 0.189) | 5.0 × 10−7 | -0.020 (-0.033, -0.008) | 1.7 × 10−3 |
MZ – monozygotic; DZ – dizygotic; CI – confidence interval; NTR – Netherlands Twin Register.
Similar modelling results were found for the average correlation of heritable methylation sites or sites associated with mQTL (Table S5): the average correlation significantly changed with age, and nor the increasing rate or decreasing rate differed by zygosity. Notably, the higher the heritability of the sites, the greater the increasing and decreasing rates, i.e., the increasing and decreasing trends were more significant for high heritable sites.
Site-specific methylation familial correlation changed with cohabitation history
Suggested by the pattern of changing with twin pair cohabitation described above, we investigated for which individual methylation sites twin pair correlation also changed with cohabitation. At the 5% level of FDR, for MZ pairs the correlations for 222,950 (63.0% of the 353,681 investigated sites; the same definition for the proportions below) increased with the time living together and the correlations for 124,348 sites (35.2%) decreased with the time living apart. There were 89,555 sites in common; that is, MZ pair correlations for 89,555 sites (25.3%) changed with cohabitation in the same way as the average. For DZ pairs, the corresponding numbers and percentages were 153,276 (43.3%), 103,568 (29.3%) and 62,274 (17.6%), respectively. Common to MZ and DZ pairs, twin pair correlations for 23,386 sites (6.6%) changed with cohabitation (Table S6). Ignoring statistical test, for MZ pairs, 317,602 (89.8%) sites numerically increased with the time living together, 302,811 (85.6%) sites numerically decreased with the time living apart, and 276,647 (78.2%) sites numerically changed with cohabitation; for DZ pairs, the corresponding numbers and percentages were 304,757 (86.2%), 304,434 (86.1%) and 270,441 (76.5%), respectively; common to MZ and DZ pairs, there were 223,585 sites (63.2%) numerically changed with cohabitation. The significantly cohabitation-dependent sites accounted for 10.5% (23,386/223,585) of the numerically cohabitation-dependent sites.
The average correlation of the 23,386 significant cohabitation-dependent sites also increased with the time living together and decreased with the time living apart, and the increasing and decreasing trends were stronger than those of all the investigated sites, or those of the investigated sites excluding the cohabitation-dependent sites (Figure S7). These were also supported by the modelling results, which showed that the increasing and decreasing rates in the familial correlation of the cohabitation-dependent sites were larger (Table S5).
Methylation value, variation and heritability of the cohabitation-dependent sites
Consistently across all studies, the 23,386 cohabitation-dependent sites were enriched for intermediate methylated sites and depleted for hypermethylated sites (Figure S8). They were also enriched for sites with higher variation and depleted for sites with lower variation (Figure S9).
Similar to the results for all the investigated sites, the average methylation value of the cohabitation-dependent sites remained relatively constant with age across the lifespan (Figure S10a). The overall variation of the cohabitation-dependent sites also increased with age, and the increasing trend was more significant than those of all the investigated sites (Figure S10b).
The cohabitation-dependent sites were enriched for heritable sites (Figure S11): the sites had an average heritability estimate of 31%, higher than the average heritability estimate of 19% for the rest of the sites (P<10−15, Mann-Whitney U test).
Genomic feature enrichment of the cohabitation-dependent sites
We investigated the enrichment of the 23,386 cohabitation-dependent sites for genomic features in terms of chromosomes, CpG density regions and gene-centric regions (Table S6). These sites were located on all 22 autosomes; enriched for being located on chromosome 19, and depleted for chromosomes 2, 12 and 13 (all FDR<0.04 after adjusting for 22 Fisher's exact tests). With respect to CpG density, these sites were enriched for being located in CpG islands (CGIs) and CpG shores, and depleted for being located in non-CGI regions and CpG shelves (all FDR<2.1 × 10−18 after adjusting for four Fisher's exact tests). With respect to gene-centric regions, these sites were enriched in proximal promoters, and depleted in distal promoters and gene bodies (all FDR<2.9 × 10−3after adjusting for five Fisher's exact tests).
Pathway analysis for the cohabitation-dependent sites
The 23,386 cohabitation-dependent sites were enriched for 18 gene ontology terms (all FDR<0.05, hypergeometric test), most of which were related to nervous system development (Table S7). No significant KEGG pathway was found for these cohabitation-dependent sites (Table S8). The top pathway was axon guidance (FDR=0.05, hypergeometric test), which is also related to nervous system development.
Epigenetic association enrichment of the cohabitation-dependent sites
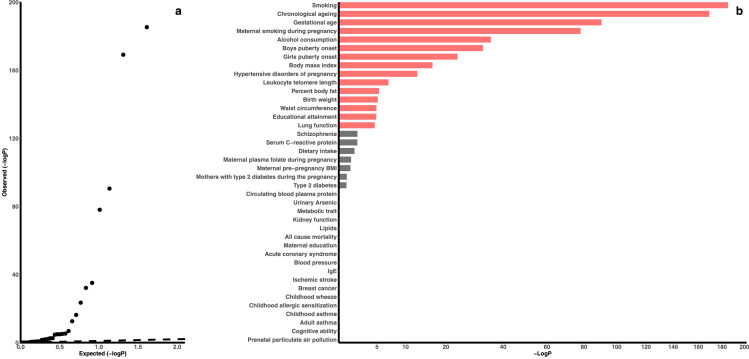
The 23,386 cohabitation-dependent sites were enriched for the epigenetic associations of 15 traits/exposures (all FDR<0.05, Fisher's exact test; Figure 2, Table S2), including health-related lifestyle factors (smoking and alcohol consumption), ageing measures (chronological ageing and leukocyte telomere length), prenatal exposures (gestational age, maternal smoking during pregnancy, hypertensive disorders of pregnancy and birth weight), puberty onset, adiposity measures (BMI, body fat percentage and waist circumference), educational attainment and lung function. Seven traits/exposures had a nominal P<0.05 but a FDR>0.05, including schizophrenia (P=0.0003, Fisher's exact test), serum C-reactive protein level (P=0.003, Fisher's exact test), dietary intake (P=0.005, Fisher's exact test), prenatal exposures (maternal pre-pregnancy BMI, maternal plasma folate level and type 2 diabetes during pregnancy; all P<0.02, Fisher's exact test) and type 2 diabetes (P=0.03, Fisher's exact test).
Figure 2.
Epigenetic association enrichment of the 23,386 cohabitation-dependent sites
a) QQ plot for the enrichment analysis of the 40 investigated human traits/exposures. b) P-values for the 40 investigated human traits/exposures from the enrichment analysis. The P-values (Fisher's exact test) are shown as -log10(P). Red bars are the significant human traits/exposures with a false discovery rate <5%.
Genetic association enrichment of the cohabitation-dependent sites
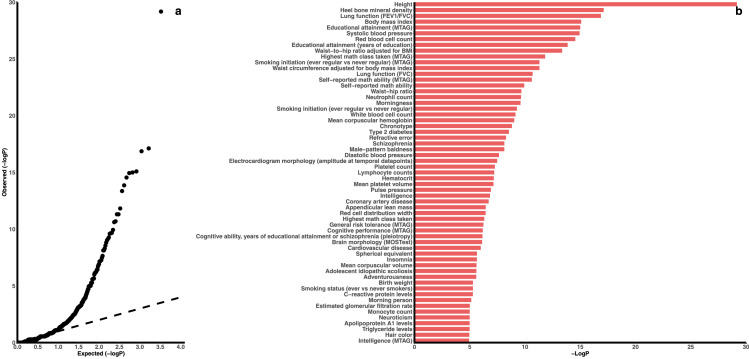
The 23,386 cohabitation-dependent sites were enriched for genetic associations with 59 traits/exposures (all FDR<0.05, Fisher's exact test; Figure 3, Table S3), such as anthropometric or adiposity traits (e.g., height, BMI, waist-to-hip ratio and waist-hip ratio), lung function, cognitive traits (e.g., educational attainment, math ability and intelligence), blood cell counts, smoking, blood pressure, lipids levels, and diseases such as type 2 diabetes and schizophrenia. 247 traits/exposures had a nominal P<0.05 but a FDR>0.05, such as puberty onset (e.g., age at menarche, P=3.0 × 10−5, Fisher's exact test ), sex hormone levels (e.g., sex hormone-binding globulin levels, P=2.1 × 10−5; total testosterone levels, P=2.6 × 10−5; both from Fisher's exact test), cardiovascular diseases (e.g., atrial fibrillation, P=5.9 × 10−5; coronary artery disease, P=9.0 × 10−5; both from Fisher's exact test) and cancers (e.g., colorectal, P=1.5 × 10−5; prostate, P=9.0 × 10−4; breast, P=4.2 × 10−3; all from Fisher's exact test).
Figure 3.
Genetic association enrichment of the 23,386 cohabitation-dependent sites
a) QQ plot for the enrichment analysis of the 3,292 investigated human traits/exposures. b) P-values for the 59 significant human traits/exposures from the enrichment analysis. The P-values (Fisher's exact test) are showed as -log10(P). Red bars are the significant human traits/exposures with a false discovery rate <5%. Only significant traits/exposures are showed in the plot. The results for all the traits/exposures are in Table S3.
Discussion
By conducting a twin study investigating DNA methylation familial correlation across the human lifespan, we have provided insights into the sources of methylation variation: early-life environmental factors shared by twins when they are cohabitating critically determine DNA methylation variation across the lifespan.
The overall methylation familial correlation, assessed as the average twin pair correlation of the investigated sites, for both MZ and DZ pairs are around zero at birth, similar to those for pairs of unrelated samples; that is, not correlated when twins first start postnatal cohabiting. The longer twins live together, the more similar they become, and the overall familial correlation reaches a peak at the age of 18 years. The MZ pair increasing rate was not different from the DZ pair increasing rate, but significantly less than twice the DZ pair increasing rate, suggesting that the increase in methylation familial correlation with age is inconsistent with genetic effects. It is, however, consistent with twins sharing a cohabitation environment in early life, the effects of which become stronger with the length of cohabitation. When twins live apart in adulthood, they no longer share a cohabitation environment, so the cohabitation-related environmental effects dissipate with time and result in methylation familial correlation decreasing at similar rates for MZ and DZ pairs across adulthood. That early-life environmental factors play a major role in determining methylation variation across the lifespan is also supported by our observation that the overall methylation variation increases dramatically with age before adulthood and is relatively stable across adulthood (Figure S2b).
We have found that, not only for all the investigated sites but also for sites with a high heritability estimate or sites associated with mQTL, the methylation familial correlation changed with twin pair cohabitation. These observations are consistent with cohabitation-related environmental factors consistently affecting the variation at a variety of methylation sites, even the sites affected by genetic factors (i.e., the so-called heritable sites). We further found that the 23,386 cohabitation-dependent sites had a higher average heritability estimate than the other sites, and the average twin pair correlation of the cohabitation-dependent sites changed with cohabitation more significantly than those of the other sites. These observations suggest that methylation sites affected by genetic factors are not immune from environmental effects, but rather the opposite – they are more likely to be affected by the cohabitation-related environmental factors. It would not be possible to make this insight by analysing individual twin studies with limited age ranges (i.e., fragments of the lifespan). Individuals twin studies, therefore, are likely to have underestimated the impact of cohabitation-related environment on methylation variation across the lifespan.
Although we did not find a statistically significant difference by zygosity in the increasing rate of methylation familial correlation, the point estimate for the MZ pairs was greater than for the DZ pairs. Consequently, the average MZ pair correlation was greater than the DZ pair correlation at the ages of 10-35 years, but not at any other ages (Figure 1b). This is consistent with previous studies reporting the highest average methylation heritability estimate at the ages of 10-35 years9, 10, 11, 12, 13, 14, 15 and suggests that previous heritability results at this age range might be confounded by cohabitation-related environmental effects. Notably, Hannon et al. found that the average proportion of methylation variation explained by environmental factors shared between twins of 18 years old was approximately 17%,15 consistent with our observation of cohabitation-related environmental effects at that age.
Fraga et al. assessed the similarity in epigenetic profile between twins using the Euclidean squared distance (different from the familial correlation we used), and found that older MZ twins had less similar epigenetic profiles than younger MZ twins and called this divergence in epigenome with age as epigenetic drift.37 Epigenetic drift has been attributed to changes in lifestyle and environment as well as to internal stochastic factors. Fraga et al. mainly compared twins aged <28 years with those aged >28 years, rather than across the whole lifespan as we did. Two hypotheses could be generated from comparing our finding of twins (both MZ and DZ) being more similar with age in childhood and less similar with age in adulthood with those of Fraga et al.: (i) epigenetic drift may only exist in adulthood, as twins become more similar, rather than less similar, with age before adulthood, and epigenetic drift may be due to not sharing the cohabitation-related environmental effects anymore; or (ii) epigenetic drift may exist throughout the lifespan, but the cohabitation-related environmental effects are more stronger than the effects of epigenetic drift so that twins are more similar with age before adulthood. More research is needed to test these hypotheses.
Cohabitation-related environmental factors appear to affect methylome unevenly, as the cohabitation-dependent sites are not distributed evenly but are more likely to be located at CpG enriched regions (CpG islands and shores) and proximal promoters. More importantly, given DNA methylation at proximal promoters is biologically relevant to the regulation of gene expression, enrichment in these regions has an important biological implication: cohabitation-related environmental factors could potentially affect gene expression regulation and downstream protein production via influencing methylation.
The cohabitation-dependent sites are enriched for biological pathways related to nervous system development, and for genetic and/or epigenetic associations with a variety of human traits/exposures. Notably, smoking, adiposity, education attainment and lung function were found in both epigenetic and genetic association enrichment analyses; smoking and adiposity are also supported by the observation that the methylation familial correlation at their associated loci changed with twin pair cohabitation (Figure S6). Enrichments for associations with a variety of human conditions, such as adiposity, metabolic traits/disorders, cardiovascular diseases, cancers and neurodevelopmental pathways/traits/disorders, imply that early-life environment could affect late-life human health via influencing methylation at disease-associated genes, consistent with the developmental origins of health and disease hypothesis38, 39, 40 and a gene-environment interaction view of disease aetiology. Identifying the environmental factors affecting methylation prior to adulthood might provide novel insights into which, and how, early-life factors affect late-life health outcomes, and provide obvious implications for disease intervention and its timing, especially given that the period before adulthood is thought as the window of developmental plasticity.41 These early-life environmental factors could be maternal smoking during pregnancy, gestational age, birth weight, puberty onset and educational attainment, the epigenetic associations of which have also been found to be enriched by the cohabitation-dependent sites. Associations between these early-life factors and the risks of aforementioned late-life health conditions have well been suggested,42, 43, 44, 45, 46, 47 and our study has provided mechanistic and biological insights into the relationships from the perspective of epigenetics. These insights are in line with that epigenetic mechanisms are proposed to underlie the developmental origins of health and disease hypothesis.39,48,49
Although our results are consistent with cohabitation-related environmental factors influencing methylation familial correlation change across the lifespan, other possible explanations could not be ruled out. Early life, especially adolescence, is the time of developmental plasticity41 when human developmental programs, which are controlled by genetic factors and affected by environment factors, reach the peak. The observed methylation familial correlation change is also consistent with genetic control interacting with environment underlying the developmental programs in early life.
Our study has several strengths. One strength is that it is the first twin study to our knowledge investigating DNA methylation familial correlation across the human lifespan, so it could provide evidence into the genetic and environmental causes of DNA methylation variation which are unable to be provided by studies focusing on limited ages only. The other strength is that it innovatively models the familial correlation as a function of twin pair cohabitation history and leverages the published epigenetic and genetic associations, so it could provide insights how early-life environmental factors influence late-life health potentially via DNA methylation.
Our study has several limitations. First, although our data covered the whole lifespan, they are a combination of several cross-sectional datasets rather than longitudinal; longitudinal data for twins are needed to validate our findings. Second, the sample size of some included studies might not be substantial, though we primarily analysed the point estimate of methylation site-specific familial correlation. Third, we did not have cohabitation history data of the twins and assumed they live together from birth to age of 18 years based on previous theoretical and empirical evidence16,17,33,34 and the fit of the data (Figure S1). We could not rule out the possibility that some twins were reared apart before adulthood, though such type of twins is likely to be rare. Fourth, we did not have data on factors such as socioeconomic status and education, so we were not able to investigate the impact of these factors. Our sample was of European descendent or from countries where the vast majority of population is of European descent; therefore, whether our findings are applicable to populations of non-European descendent needs investigations. Fifth, we did not have data on health-related exposures and health outcomes, so we were not able to directly investigate their relationships with the cohabitation-dependent sites, but used the published genetic and epigenetic associations; therefore, we could not provide direct evidence for the role of methylation linking exposures and outcomes. Future studies with relevant exposure and outcome data are needed to investigate the relationships, especially for the mediating role of cohabitation-dependent sites between exposures and outcomes using longitudinal data, to further understand the role of DNA methylation in health.
In conclusion, our findings are consistent with early life critically determining DNA methylation variation across the lifespan. The effects persist during the whole lifespan, and are stronger for methylation sites affected by genetic factors and sites biologically relevant to gene expression regulation. The variability of a substantial number of DNA methylation sites change with cohabitation and these sites are enriched for genetic and epigenetic associations with a variety of early-life factors and late-life health conditions, implying that early life could affect late-life health through influencing DNA methylation.
Declaration of interests
GSD is employed by Genetic Technologies Ltd. PSS receives payments for Advisory Board meetings for Biogen Australia and Roche Australia that are not related to this study. The other authors have no conflicts of interest to declare.
Acknowledgments
Contributors
SL & JLH were involved in study conception and design. SL and YZ analysed the data. SL and JLH drafted the first version of the manuscript. Data collection: PETS—SL, EMW, JMC, RS, MCS and JLH, AMDTSS—SL, EMW, TLN, GSD, GGG, MCS and JLH, OATS—NJA, KAM, PSS and AT, DTR and LSADT—QT. All authors participated manuscript revision and approved the final manuscript. SL has verified the data used in the analysis.
Acknowledgements
This work was supported by grant ECRF19020 from the Victorian Cancer Agency, and by grant 1187896 awarded through the 2019 Priority-driven Collaborative Cancer Research Scheme and funded by Cure Cancer with the support of Cancer Australia. SL is a Victorian Cancer Agency Early Career Research Fellow (ECRF19020). SL and TLN were supported by the Cancer Council Victoria Postdoctoral Research Fellowship and the Picchi Award from the Victorian Comprehensive Cancer Centre. TLN was supported by the Cure Cancer Australia (APP1159399), and received a Grants-in-Aid grant from the Cancer Council Victoria (AF7305). MCS is a National Health and Medical Research Council (NHMRC) Senior Research Fellow (APP1155163). JLH is a NHMRC Senior Principal Research Fellow. The PETS was supported by grants from the NHMRC (Grant No. 1146333 to JC). The AMDTSS was facilitated through access to Twins Research Australia, a national resource supported by a Centre of Research Excellence Grant (Grant No. 1079102) from the NHMRC. The AMDTSS was supported by NHMRC (Grant Nos. 1050561 and 1079102), Cancer Australia and National Breast Cancer Foundation (Grant No. 509307). The OATS was funded by a NHMRC and Australian Research Council (ARC) Strategic Award Grant of the Ageing Well, Ageing Productively Program (Grant No. 401162) and NHMRC Project Grants (Grant Nos. 1045325 and 1085606). The OATS was facilitated through Twins Research Australia, a national resource in part supported by a Centre for Research Excellence Grant (Grant No. 1079102) from the NHMRC.
We thank the participants of all studies. The PETS thanks all of the supportive families who participated in the PETS study throughout the years, and research staff, volunteers, and the phlebotomists from Royal Children's Hospital pathology department. The OATS thanks the participants for their time and generosity in contributing to this research and acknowledges the contribution of the OATS research team (https://cheba.unsw.edu.au/project/older-australian-twins-study) to this study.
Data Sharing
The data analysed in this study are available in Gene Expression Omnibus (GSE56105, GSE105018, GSE61496, GSE100227, GSE73115) and ArrayExpress (E-MTAB-1866). The NTR correlation data is available in van Dongen et al (Ref. 13). The data of PETS and OATS are available from the relevant authors on request.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.103927.
Appendix. Supplementary materials
References
- 1.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358(11):1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 2.Petronis A. Epigenetics as a unifying principle in the aetiology of complex traits and diseases. Nature. 2010;465(7299):721–727. doi: 10.1038/nature09230. [DOI] [PubMed] [Google Scholar]
- 3.Breitling LP, Yang R, Korn B, Burwinkel B, Brenner H. Tobacco-smoking-related differential DNA methylation: 27K discovery and replication. Am J Hum Genet. 2011;88(4):450–457. doi: 10.1016/j.ajhg.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Joehanes R, Just AC, Marioni RE, et al. Epigenetic Signatures of Cigarette Smoking. Circ Cardiovasc Genet. 2016;9(5):436–447. doi: 10.1161/CIRCGENETICS.116.001506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li S, Wong EM, Bui M, et al. Causal effect of smoking on DNA methylation in peripheral blood: a twin and family study. Clin Epigenetics. 2018;10(1):18. doi: 10.1186/s13148-018-0452-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dick KJ, Nelson CP, Tsaprouni L, et al. DNA methylation and body-mass index: a genome-wide analysis. Lancet. 2014;383(9933):1990–1998. doi: 10.1016/S0140-6736(13)62674-4. [DOI] [PubMed] [Google Scholar]
- 7.Wahl S, Drong A, Lehne B, et al. Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity. Nature. 2017;541(7635):81–86. doi: 10.1038/nature20784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li S, Wong EM, Bui M, et al. Inference about causation between body mass index and DNA methylation in blood from a twin family study. Int J Obes (Lond) 2019;43(2):243–252. doi: 10.1038/s41366-018-0103-4. [DOI] [PubMed] [Google Scholar]
- 9.Gordon L, Joo JE, Powell JE, et al. Neonatal DNA methylation profile in human twins is specified by a complex interplay between intrauterine environmental and genetic factors, subject to tissue-specific influence. Genome Res. 2012;22(8):1395–1406. doi: 10.1101/gr.136598.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bell JT, Tsai PC, Yang TP, et al. Epigenome-wide scans identify differentially methylated regions for age and age-related phenotypes in a healthy ageing population. PLoS Genet. 2012;8(4) doi: 10.1371/journal.pgen.1002629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grundberg E, Meduri E, Sandling JK, et al. Global analysis of DNA methylation variation in adipose tissue from twins reveals links to disease-associated variants in distal regulatory elements. Am J Hum Genet. 2013;93(5):876–890. doi: 10.1016/j.ajhg.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McRae AF, Powell JE, Henders AK, et al. Contribution of genetic variation to transgenerational inheritance of DNA methylation. Genome Biol. 2014;15(5):R73. doi: 10.1186/gb-2014-15-5-r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Dongen J, Nivard MG, Willemsen G, et al. Genetic and environmental influences interact with age and sex in shaping the human methylome. Nat Commun. 2016;7:11115. doi: 10.1038/ncomms11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li S, Wong EM, Nguyen TL, et al. Causes of blood methylomic variation for middle-aged women measured by the HumanMethylation450 array. Epigenetics. 2017;12(11):973–981. doi: 10.1080/15592294.2017.1384891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hannon E, Knox O, Sugden K, et al. Characterizing genetic and environmental influences on variable DNA methylation using monozygotic and dizygotic twins. PLoS Genet. 2018;14(8) doi: 10.1371/journal.pgen.1007544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li S, Nguyen TL, Wong EM, et al. Genetic and environmental causes of variation in epigenetic aging across the lifespan. Clin Epigenetics. 2020;12(1):158. doi: 10.1186/s13148-020-00950-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li S, Wong EM, Dugue PA, et al. Genome-wide average DNA methylation is determined in utero. Int J Epidemiol. 2018;47(3):908–916. doi: 10.1093/ije/dyy028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saffery R, Morley R, Carlin JB, et al. Cohort profile: The peri/post-natal epigenetic twins study. Int J Epidemiol. 2012;41(1):55–61. doi: 10.1093/ije/dyr140. [DOI] [PubMed] [Google Scholar]
- 19.Powell JE, Henders AK, McRae AF, et al. The Brisbane Systems Genetics Study: genetical genomics meets complex trait genetics. PLoS One. 2012;7(4):e35430. doi: 10.1371/journal.pone.0035430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan Q, Frost M, Heijmans BT, et al. Epigenetic signature of birth weight discordance in adult twins. BMC Genomics. 2014;15:1062. doi: 10.1186/1471-2164-15-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Odefrey F, Stone J, Gurrin LC, et al. Common genetic variants associated with breast cancer and mammographic density measures that predict disease. Cancer Res. 2010;70(4):1449–1458. doi: 10.1158/0008-5472.CAN-09-3495. [DOI] [PubMed] [Google Scholar]
- 22.Li S, Wong EM, Joo JE, et al. Genetic and Environmental Causes of Variation in the Difference Between Biological Age Based on DNA Methylation and Chronological Age for Middle-Aged Women. Twin Res Hum Genet. 2015;18(6):720–726. doi: 10.1017/thg.2015.75. [DOI] [PubMed] [Google Scholar]
- 23.Sachdev PS, Lammel A, Trollor JN, et al. A comprehensive neuropsychiatric study of elderly twins: the Older Australian Twins Study. Twin Res Hum Genet. 2009;12(6):573–582. doi: 10.1375/twin.12.6.573. [DOI] [PubMed] [Google Scholar]
- 24.Tan Q, Heijmans BT, Hjelmborg JV, Soerensen M, Christensen K, Christiansen L. Epigenetic drift in the aging genome: a ten-year follow-up in an elderly twin cohort. Int J Epidemiol. 2016;45(4):1146–1158. doi: 10.1093/ije/dyw132. [DOI] [PubMed] [Google Scholar]
- 25.Touleimat N, Tost J. Complete pipeline for Infinium((R)) Human Methylation 450K BeadChip data processing using subset quantile normalization for accurate DNA methylation estimation. Epigenomics. 2012;4(3):325–341. doi: 10.2217/epi.12.21. [DOI] [PubMed] [Google Scholar]
- 26.Aryee MJ, Jaffe AE, Corrada-Bravo H, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30(10):1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pidsley R, YW CC, Volta M, Lunnon K, Mill J, Schalkwyk LC. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genomics. 2013;14:293. doi: 10.1186/1471-2164-14-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maksimovic J, Gordon L, Oshlack A. SWAN: Subset-quantile within array normalization for illumina infinium HumanMethylation450 BeadChips. Genome Biol. 2012;13(6):R44. doi: 10.1186/gb-2012-13-6-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 30.Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bakulski KM, Feinberg JI, Andrews SV, et al. DNA methylation of cord blood cell types: Applications for mixed cell birth studies. Epigenetics. 2016;11(5):354–362. doi: 10.1080/15592294.2016.1161875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Min JL, Hemani G, Hannon E, et al. Genomic and phenotypic insights from an atlas of genetic effects on DNA methylation. Nat Genet. 2021;53(9):1311–1321. doi: 10.1038/s41588-021-00923-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lange K. Cohabitation, convergence, and environmental covariances. Am J Med Genet. 1986;24(3):483–491. doi: 10.1002/ajmg.1320240311. [DOI] [PubMed] [Google Scholar]
- 34.Hopper JL, Mathews JD. Extensions to multivariate normal models for pedigree analysis. II. Modeling the effect of shared environment in the analysis of variation in blood lead levels. Am J Epidemiol. 1983;117(3):344–355. doi: 10.1093/oxfordjournals.aje.a113547. [DOI] [PubMed] [Google Scholar]
- 35.Slieker RC, Bos SD, Goeman JJ, et al. Identification and systematic annotation of tissue-specific differentially methylated regions using the Illumina 450k array. Epigenetics Chromatin. 2013;6(1):26. doi: 10.1186/1756-8935-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phipson B, Maksimovic J, Oshlack A. missMethyl: an R package for analyzing data from Illumina's HumanMethylation450 platform. Bioinformatics. 2016;32(2):286–288. doi: 10.1093/bioinformatics/btv560. [DOI] [PubMed] [Google Scholar]
- 37.Fraga MF, Ballestar E, Paz MF, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A. 2005;102(30):10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barker DJ. The origins of the developmental origins theory. J Intern Med. 2007;261(5):412–417. doi: 10.1111/j.1365-2796.2007.01809.x. [DOI] [PubMed] [Google Scholar]
- 39.Wadhwa PD, Buss C, Entringer S, Swanson JM. Developmental origins of health and disease: brief history of the approach and current focus on epigenetic mechanisms. Semin Reprod Med. 2009;27(5):358–368. doi: 10.1055/s-0029-1237424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gillman MW. Developmental origins of health and disease. N Engl J Med. 2005;353(17):1848–1850. doi: 10.1056/NEJMe058187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bateson P, Barker D, Clutton-Brock T, et al. Developmental plasticity and human health. Nature. 2004;430(6998):419–421. doi: 10.1038/nature02725. [DOI] [PubMed] [Google Scholar]
- 42.Belbasis L, Savvidou MD, Kanu C, Evangelou E, Tzoulaki I. Birth weight in relation to health and disease in later life: an umbrella review of systematic reviews and meta-analyses. BMC Med. 2016;14(1):147. doi: 10.1186/s12916-016-0692-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cupul-Uicab LA, Skjaerven R, Haug K, Melve KK, Engel SM, Longnecker MP. In utero exposure to maternal tobacco smoke and subsequent obesity, hypertension, and gestational diabetes among women in the MoBa cohort. Environ Health Perspect. 2012;120(3):355–360. doi: 10.1289/ehp.1103789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Crump C, Howell EA, Stroustrup A, McLaughlin MA, Sundquist J, Sundquist K. Association of Preterm Birth With Risk of Ischemic Heart Disease in Adulthood. JAMA Pediatr. 2019;173(8):736–743. doi: 10.1001/jamapediatrics.2019.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Biro FM, Deardorff J. Identifying opportunities for cancer prevention during preadolescence and adolescence: puberty as a window of susceptibility. J Adolesc Health. 2013;52(5 Suppl):S15–S20. doi: 10.1016/j.jadohealth.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Day FR, Elks CE, Murray A, Ong KK, Perry JR. Puberty timing associated with diabetes, cardiovascular disease and also diverse health outcomes in men and women: the UK Biobank study. Sci Rep. 2015;5:11208. doi: 10.1038/srep11208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barker DJ, Osmond C, Forsen TJ, Kajantie E, Eriksson JG. Trajectories of growth among children who have coronary events as adults. N Engl J Med. 2005;353(17):1802–1809. doi: 10.1056/NEJMoa044160. [DOI] [PubMed] [Google Scholar]
- 48.Hoffman DJ, Reynolds RM, Hardy DB. Developmental origins of health and disease: current knowledge and potential mechanisms. Nutr Rev. 2017;75(12):951–970. doi: 10.1093/nutrit/nux053. [DOI] [PubMed] [Google Scholar]
- 49.Waterland RA, Michels KB. Epigenetic epidemiology of the developmental origins hypothesis. Annu Rev Nutr. 2007;27:363–388. doi: 10.1146/annurev.nutr.27.061406.093705. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.