Abstract
Objective
The minor allele (A) of the rs373863828 variant (p.Arg457Gln) in CREBRF is restricted to indigenous peoples of the Pacific islands (including New Zealand Māori and peoples of Polynesia), with a frequency of up to 25% in these populations. This allele associates with a large increase in body mass index (BMI) but with significantly lower risk of type-2 diabetes (T2D). It remains unclear whether the increased BMI is driven by increased adiposity or by increased lean mass.
Methods
We undertook body composition analysis using DXA in 189 young men of Māori and Pacific descent living in Aotearoa New Zealand. Further investigation was carried out in two orthologous Arg458Gln knockin mouse models on FVB/NJ and C57BL/6j backgrounds.
Results
The rs373863828 A allele was associated with lower fat mass when adjusted for BMI (p < 0.05) and was associated with significantly lower circulating levels of the muscle inhibitory hormone myostatin (p < 0.05). Supporting the human data, significant reductions in adipose tissue mass were observed in the knockin mice. This was more significant in older mice in both backgrounds and appeared to be the result of reduced age-associated increases in fat mass. The older male knockin mice on C57BL/6j background also had increased grip strength (p < 0.01) and lower levels of myostatin (p < 0.05).
Conclusion
Overall, these results prove that the rs373863828 A-allele is associated with a reduction of myostatin levels which likely contribute to an age-dependent lowering of fat mass, at least in males.
Keywords: Body mass index, Obesity, Body composition, Genetics, Type-2 diabetes, Myostatin, ER stress, Golgi stress, CREB3
Highlights
-
•
The CREBRF p.457Gln variant associates with decreased fat mass in males.
-
•
Consistent with this CREBRF p.457Gln associates with decreased myostatin levels.
-
•
These effects are more obvious with age.
1. Introduction
Many genetic variants have been identified that contribute to the risk of developing either type-2 diabetes (T2D) [[1], [2], [3]] or obesity [[3], [4], [5]]. However, most common gene variants only have a small effect size. One exception is the rs373863828 variant in the CREBRF gene. This gene codes for the highly conserved but poorly understood protein CREBRF (CREB3 Regulatory Factor, also known as Luman Recruitment Factor (LRF) of C5orf41) [6]. The rs373863828 minor (A) allele causes the p.Arg457Gln substitution, and the functional consequences of this are poorly understood. This variant is unique to isolated populations in the islands of the Pacific where it is present at minor allele frequencies (MAF) of up to 25% in Polynesia (including Samoa, Aotearoa NZ Māori) [[7], [8], [9]] and at lower MAF in some areas of Melanesia and Micronesia [9,10]. Importantly, of single gene variants common in a population, it has the biggest impact on BMI, being associated with an increase of up to 3 kg/m2 per rs373863828 A allele [[7], [8], [9], [10]].
Increases in BMI in adults are commonly associated with a relative increase in adiposity [11], and initial evidence suggested the impact of the rs373863828 A allele on BMI was due to increased adiposity [8]. This suggests that this variant was an evolutionary adaptation for facilitating energy storage and thus was acting as a thrifty gene [8]. However, this would be inconsistent with Māori and other peoples from Polynesia having higher relative lean mass for a given BMI compared to people of European or Asian ancestry [12,13]. Recent follow-up studies undertaken in Samoa have indicated that in males, the rs373863828 A allele is associated with increased lean mass, not adiposity, suggesting the effects on BMI may be driven by greater increases in lean mass rather than fat mass [14,15]. To address these inconsistencies, we examined the impact of the minor allele of rs373863828 p.Arg457Gln CREBRF on body composition in a cohort of young Māori and Pacific men and in two mouse knockin models.
2. Materials and methods
2.1. Human studies
Healthy men of NZ Māori and/or Pacific Island descent (n = 189) were recruited in Auckland and Wellington, Aotearoa New Zealand, and gave written informed consent to participate. Eligibility criteria included being free from chronic illnesses, including cardiovascular or metabolic disease, 18–45 years of age, with a BMI of 20–45 kg/m2. This study was approved by the Health and Disability Ethics Committee, New Zealand (17STH79).
Body composition was determined by dual energy x-ray absorptiometry (DXA, model iDXA, GE Healthcare, Madison, WI and Hologic Horizon A, Hologic, Marlborough, MA). Resting metabolic rate (RMR) was measured in overnight fasted participants by indirect calorimetry (Parvo Medics True One 2400, UT, USA and Promethion, Sable Systems, NV, USA) using a ventilated hood following 30 min resting in a supine position. An overnight fasted venous blood sample was collected between 8:00 am and 10 am in the morning, with plasma collected in a tube containing EDTA. Plasma and serum were recovered before being frozen at −80 °C. Circulating myostatin/GDF-8 in both human (plasma) and mouse (serum) samples was measured by commercial ELISA (R&D systems, Minneapolis, MN, USA, catalogue number RDSDGDF80). DNA was from whole blood isolated using a commercially available kit (DNA extraction minikit, Qiagen, Hilden Germany). rs373863828 genotype was determined using a custom designed Taqman probe set (Applied Biosystems, Foster City, CA, USA) as described previously [16].
A multivariable linear regression model was used to test for associations with rs373863828 A allele (assuming an additive effect of the A-allele) with all parameters presented in Tables 2 and Supplementary Table 2 (RStudio v1.2.1335 statistical software, www.rstudio.com). The proportion of self-reported grandparents of Polynesian ancestry (ANC), age, weight, height and BMI were included as covariates where indicated [16].
Table 2.
Impact of rs373863828 A allele on body composition on men of Māori and Pacific ancestry. Adjustments were made for age, ancestry (ANC) and Body Mass Index (BMI) as indicated.
| Age, ANC adjusted |
BMI, Age, ANC adjusted |
|||||
|---|---|---|---|---|---|---|
| Beta | SE | p-value | Beta | SE | p-value | |
| Myostatin (pg/ml) | −384 | 193 | 0.048 | −396 | 191 | 0.040 |
| Fat mass (%) | −1.13 | 0.96 | 0.240 | |||
| Lean mass (%) | 1.10 | 0.94 | 0.241 | |||
| Fat mass (kg) | −1.39 | 1.50 | 0.358 | −1.84 | 0.85 | 0.031 |
| Lean mass (kg) | 1.68 | 1.18 | 0.159 | 1.43 | 0.99 | 0.149 |
| BMC (kg) | 0.09 | 0.07 | 0.228 | 0.08 | 0.07 | 0.252 |
| BMD (g/cm2) | 0.02 | 0.02 | 0.218 | 0.02 | 0.02 | 0.240 |
| RMR (kcal) | 38.0 | 43.1 | 0.379 | 32.5 | 38.6 | 0.401 |
2.2. Mouse model
Orthologous mouse CREBRF p.458Q gene variant knock-in mice were generated on both FVB/NJ and C57BL/6j backgrounds. The FVB/NJ mice were generated as previously reported [17]. The mice on the C57BL/6j background were generated by the Rodent Genetics for Research (ROGER) Facility (Faculty of Medical and Health Sciences, University of Auckland, New Zealand) using CRISPR/Cas9 technology based on the targeting strategy shown in Suppl Figure 1. Sequencing was used to verify the presence of the variant and lack of non-intended changes (sequencing primers AAGAACCCCATACCAGATAGC and TACAGTGACAAAGACAGGTATGG). Crispon was used to check the specificity of guide RNA sequence, and no non-specific exonic cut sites were found [18]. Mouse genotyping was performed either (i) in-house via High Resolution Meltcurve (HRM) analysis on the Quant Studio 6 (QS6, Thermo Scientific) with the primers GGATTCTGAGGCCTTCTGA and CCTCTTACCATGATGTAAGCCA, or (ii) commercially, genotyped via real-time PCR (Transnetyx, TN, USA). Further validation of genotypes was performed using an RFLP assay in which the knock-in removes a TaqI restriction site (primers TGGCTGAAAACCCAAGTACACT and AGCTTGACAATTGTGGGACCA).
Protocols for animal studies were approved by the University of Auckland Animal Ethics Committee and performed in accordance with the NZ Animal Welfare Act (1999) and the Guide for the Care and Use of Laboratory Animals [19]. We also complied with the ARRIVE guidelines [20]. Animals were maintained in a 12 h light/dark cycle with a 30-minute dawn/dusk phase. Temperature was set at 22 °C with a tolerance of +/−2 °C. Humidity was 55% with a tolerance of +/− 10%. Mice had food and water ad libitum, and the diet was the Teklad 2018, 18% protein rodent diet. Body composition in 18-month-old male FVB/NJ was measured with a magnetic resonance imaging technique (EchoMRI 100H, EchoMRI LLC Houston TX, USA). Measurements were taken following calibration with three primary data accumulations at the same time of day on non-fasted mice. Body composition for all other groups was measured by DXA using a Lunar PIXImus densitometer (DEXA-GE). The PIXImus was calibrated before each session with a phantom provided by the manufacturer. Mice were anesthetized by isoflurane inhalation during the measurement, and mouse length was measured in the supine position prior to the anaesthetic wearing off. Analysis of images was performed with the Lunar PIXImus v2.10 software. All mice were measured at the same time of day.
Grip strength was assessed using a Harvard Apparatus Grip Strength Meter with the grid for mice attachment (Harvard Apparatus, Holliston, MA, USA). Each mouse underwent a familiarisation, 2 days prior to the experiment, and all mice were tested in triplicate to arrive at an average value. Balance/coordination was assessed using the rotarod technique with the RotamexX-5 (Columbus Instruments, USA).
Tissues and serum were harvested at euthanasia at around 21.5 months of age. Serum samples were frozen at −80 °C until use. Gastrocnemius tissue was preserved in RNALater (Invitrogen) and stored at −20 °C prior to mRNA preparation using RNeasy Mini Kit (Qiagen) with pre-treatment with Proteinase K as recommended for fibrous tissue. Quantification of mRNA was done using nanodrop (Thermo Scientific). The presence of genomic-DNA contamination was assessed using power Syber Green RNA to CT one-step kit (Thermofisher, 4389986, USA) to compare no-RT enzyme controls to those with the RT enzyme present and primers detecting beta-actin (CACTGTCGAGTCGCGTCC and TCATCCATGGCGAACTGGTG). There was no amplification from no-RT control samples. One-step TB green primer script RT-PCR kit II (Takara, RR086A, Japan) was used for measuring myostatin by RT-qPCR. Primers used for myostatin were AGAAGATGGGCTGAATCCCT and CATCGCAGTCAAGCCCAAAG. Three reference genes were selected from a test panel of 7: Beta-actin (as above), SDHA (TCGACAGGGGAATGGTTTGG and CTTCCGAGCTTCTGCACCAT) and HPRT (AGTCCCAGCGTCGTGATTAG and TTTCCAAATCCTCGGCATAATGA). Efficiency of all primer pairs were determined using calibration curves, and final relative gene expression (RGE) was calculated using the Pfaffle method, with geometric averaging of relative quantities of reference genes [21,22].
3. Results
To investigate the effect of the A allele of rs373863828 on relative levels of lean and fat mass in humans, we performed DXA scans on a cohort of 189 young Māori and Pacific men. Characteristics of the participants by rs373863828 genotype are presented in Table 1. The presence of the rs373863828 A allele was not associated with bone mineral density, bone mineral content or resting metabolic rate (Table 2). Total fat mass was significantly lower when adjusted for BMI, age and ancestry (Table 2, p = 0.031) or when adjusted for weight, age and ancestry (p = 0.024 Suppl. Table 1). One possible candidate for such an effect on body composition is myostatin, as the hormone plays a role in regulating the development of muscle and fat tissue and thus also plays a key role in altering the balance between lean and fat mass [[23], [24], [25], [26], [27]]. We found that carriers of rs373863828-A allele had significantly lower plasma concentrations of myostatin when adjusted for BMI, age and ancestry (β −396 pg/ml p 0.04) (Table 2). The effect on fat mass held when adjusted for weight, age and ancestry but not when adjusted for height, age and ancestry (Suppl. Table 1). This indicated the mechanisms by which the rs373863828-A allele affects body composition may involve a relationship between height and fat mass.
Table 1.
Characteristics of Human Participants.
| GG | AG | AA | |
|---|---|---|---|
| n | 139 | 44 | 6 |
| Age (y) | 27 ± 7 | 28 ± 6 | 28 ± 7 |
| Weight (kg) | 103.7 ± 17.9 | 107.2 ± 14.7 | 105.3 ± 8.1 |
| Height (cm) | 181 ± 6 | 181 ± 5 | 182 ± 6 |
| BMI (kg/m2) | 31.52 ± 5.08 | 32.76 ± 4.02 | 31.68 ± 3.84 |
| Myostatin (pg/ml) | 3495 ± 1515 | 3322 ± 1147 | 2456 ± 763 |
| Fat mass (%) | 30.1 ± 7.1 | 30.2 ± 6.9 | 25.4 ± 6.7 |
| Lean mass (%) | 67.2 ± 7 | 66.9 ± 6.5 | 71.9 ± 6.4 |
| Fat mass (kg) | 31.3 ± 11.3 | 31.8 ± 10.3 | 26.1 ± 8.3 |
| Lean mass (kg) | 69.1 ± 9 | 71.2 ± 8 | 75.5 ± 5.7 |
| BMC (kg) | 3.52 ± 0.54 | 3.66 ± 0.56 | 3.77 ± 0.44 |
| BMD (g/cm2) | 1.34 ± 0.14 | 1.38 ± 0.11 | 1.39 ± 0.1 |
| RMR (kcal) | 2034 ± 307 | 2071 ± 313 | 2176 ± 178 |
Identifying the associations of specific gene variants with body composition in humans is confounded by the diversity of genetic backgrounds, that variants generally have weak effect sizes (reducing study power) and differences in environmental exposures among individuals. Therefore, to address the role of rs373863828 on an isogenic background, we created two knockin mouse models. CRISPR/Cas9 was used to convert the orthologous mouse CREBRF Arg at position 458 to Gln in both C57BL/6j and FVB/NJ mice using CRISPR/Cas9 (Suppl. Figure 1). Mating pairs where both were homozygous for the glutamine allele produced litter sizes that were not different from mating pairs carrying the arginine allele; this indicates there was no effect on in utero or postnatal survival (data not shown). All mice used in this study came from mating heterozygous mice.
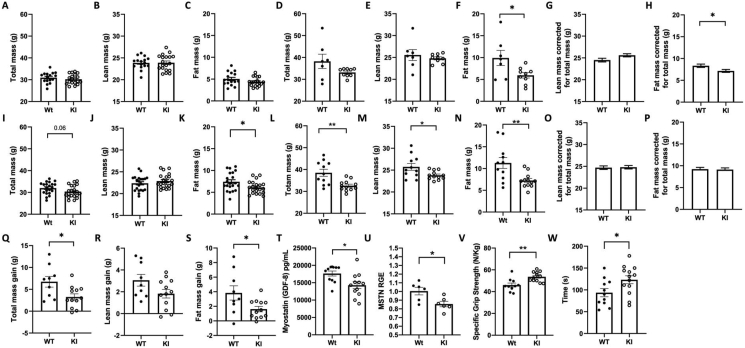
We studied body composition in male mice homozygous for either the arginine or the glutamine allele at 25-weeks (young) and 20-months (old) of age. While the knockin had different effects on overall body weight between the strains, a consistent pattern of body composition was observed (Suppl. Table 2). In the younger mice homozygous for the variant, there was no difference in lean or fat mass in FVB/NJ mice (Figure 1A–C), but in C57BL/6j background, CREBRF KI mice had lower fat mass (Figure 1I–K). There was also a statistically significant reduction in body fat percentage in the older mice in both lines (Suppl. Table 2). This is largely due to a reduced level of fat mass in the older male knockin mice on both C57BL/6j and FVB/NJ backgrounds (Figure 1 D-F and L-N). Linear modelling using total mass as a covariate (Figure 1 G,H, O, P) provided further evidence there was a reduction in fat mass for a given total mass (body weight) in the older FVB/NJ mice. While this was not significant in the older C57BL/6j6 knockin mice, which were smaller overall (with lower lean, fat and total mass), it was notable that as these mice aged (25 weeks vs 93 weeks; Figure 1 Q-S and Suppl. Figure 2), there was a significantly lower age-associated gain in fat tissue.
Figure 1.
Effects of R458Q knock-in in the CREBRF gene on body composition in mouse models. Body size and composition in 25-week-old FVB males (A–C) and 18-month-old FVB males (D–H). Body size and composition in 25-week-old C57 males (I–K) and 20-month-old C57 males (L–P). Comparison of gain in total mass (Q), lean mass (R) or fat mass (S) from 25 to 93 weeks of age in the same group of C57 male (WT n = 9, KI n = 12). Circulating myostatin (T) gastrocnemius myostatin mRNA relative gene expression (RGE) (U) in 20-month-old C57 male mice. Specific grip strength (V), and Rotarod, latency to fall (W) in 20-month-old C57 male mice. All error bars are SEM. All statistics are two-way unpaired t-test other than G, H, O and P; where linear modelling was performed to correct for total mass as a covariate, adjusted means with SEM are shown. ∗p < 0.05 and ∗∗p < 0.01 vs Wt of same age and background strain.
We examined levels of myostatin and found that consistent with human data, circulating concentrations were significantly lower in 20-month-old C57BL/6j knockin mice homozygous for the variant (p < 0.05) (Figure 1T), and myostatin mRNA levels in gastrocnemius muscle were also significantly reduced (p < 0.05) (Figure 1U). Twenty-month-old male C57BL/6j knockin mice had greater grip strength (Figure 1 V) and increased latency time on the rotarod (Figure 1W). Both responses are known to be improved by reductions in myostatin signalling in mice [28,29].
4. Discussion
The minor A allele of rs373863828 in the CREBRF gene can be considered a favourable BMI allele because of it's association with such a large increase in BMI whilst simultaneously being associated with a significant decrease in risk of T2D [7,8]. To provide further insights into the contribution of the CREBRF rs373863828 minor allele to adiposity and lean mass, we studied the impact of this allele on body composition in a cohort of New Zealand-based young, healthy men of various Māori and Pacific ancestries. We find significantly lower levels of fat tissue in carriers of the rs373863828 allele. This pattern would be consistent with previous body composition studies showing that a lower level of fat mass for a given BMI is observed in Māori and Pacific peoples compared with people of European or Asian ancestry in New Zealand [12,13]. However, we did not observe statistically significant differences in relative lean or fat as was found in recent studies in Samoan men [15]. One plausible reason for this lack of statistical significance in our studies is inadequate power due to numbers in the study. It is also possible that age may influence the effect rs373863828 has on body composition since the more significant effects, reported previously in Samoan subjects, were from an older cohort (48–53 years) [15] compared with the cohort reported here (with an average age of 28 years). This is supported by our findings in mice where the orthologous version of the human CREBRF rs373863828 variant resulted in age-dependent differences in fat levels.
One strength of the current study is that we can analyse body composition in knockin mouse models and at two different ages, as well as study the impact of the variant in two different isogenic backgrounds. This allowed us to study the impact in the context of being homozygous for the variant and without the influences of environment. Notably, the effects seen here are more obvious in mice whose age was equivalent to late middle age in humans [30], which is consistent with the observations in humans [15]. These mouse studies provide independent evidence to support the case that the p.Arg457Gln coding variant of CREBRF promotes change in fat mass, and this is consistent with the observation that the rs373863828 minor allele is associated with a reduced risk of T2D [31]. Given that CREBRF and its target CREB3 are widely expressed, other mechanisms are likely to also contribute to the reduced risk of T2D. For example, we recently demonstrated in human studies in males that the rs373863828 A allele associates with increased plasma insulin in response to a glucose load in males, independent of differences in insulin sensitivity [16].
An important finding of the current study is the association of the rs373863828 A allele with reduction in myostatin levels in both the mice and the human participants. This is likely to contribute to differences in body composition as lower myostatin is associated with increased muscle mass [23,32] and reductions in fat mass [26]. It is known that lower levels of myostatin result in changes in body composition with ageing, and this is consistent with our findings [33]. In addition, reduced myostatin levels are associated with a lower risk of T2D [23,24,34], which is consistent with the effects of the rs373863828 A allele on risk of T2D.
To date, two main roles have been identified for CREBRF; the first is to bind to the transcription factor CREB3 and regulate CREB3 levels and activity [6]. The transcription factor component CREB3 is cleaved and thus activated by ER/Golgi stress [35]; in turn, it plays a key role in regulating endoplasmic and Golgi stress responses in cells [36,37]. CREBRF has also been shown to regulate the cellular location and activity of the glucocorticoid receptor [38]. The rs373863828 A allele could affect myostatin, either through glucocorticoid- or through ER/Golgi stress-mediated pathways. Both ER stress [39] and glucocorticoid signalling [40] are known to play important roles in regulating muscle mass. Further studies will be required to resolve this issue.
One limitation of our study is that our human cohort did not include females. Given that it is known there exists sexual dimorphism in the regulation of myostatin [25,32,41], it is possible that different effects will be observed in women. Other sexually dimorphic effects of rs373863828 have been observed, for example, the male-specific effects observed on height [17,42] and differences in the pattern of body composition between males and females [15,43]. Further studies will be required to resolve these issues.
Our results also provide further insights into the thrifty gene hypothesis [44,45]. One interpretation of this hypothesis is that there is positive selection for fat accumulation as an energy store for long periods of starvation [46]. In the case of rs373863828 evidence for natural selection occurring around this locus in Polynesian people has been suggested as evidence that this is a thrifty gene [8]. Our results do not support this, as there is no evidence it is driving an increase in adiposity. Similarly, a variant of PPARGC1 has been proposed as a thrifty gene in Polynesian people [47], but this has been rejected following further analysis of the locus in the genomes of Polynesian people for a signature of selection [48]. This hypothesis has also been questioned based on logic and lack of evidence [49]. Reasons for natural selection at the CREBRF locus will require further study and other hypotheses can be proposed. For example, it has been suggested that recent genetic selection in indigenous populations may have been caused by the arrival of novel Western diseases [50]. Given the known roles of CREB3 and CREBRF in regulating viral infectivity [51,52], it is plausible that recent natural selection at the CREBRF locus relates to such effects.
5. Conclusions
Using mouse models and human phenotyping, we provide evidence that in males the CREBRF p.457Gln allele does not drive increased adiposity and is hence not likely to be a thrifty gene as previously proposed [8]. Our results support a model where in males this gene variant drives a reduction in age-dependent accumulation of fat, and this is supported by the observation of lower myostatin levels in human participants with a p.457Gln allele and in knockin mice that model this variant. Such a phenotype is consistent with improvements in glucose metabolism and consequent reduction in risk of T2D in carriers of the CREBRF p.457Gln allele, despite their increased BMI [24,34,53].
Funding sources
This study was funded by the Auckland Medical Research Foundation (AMRF), the Health Research Council (HRC) Programme Grant 18-681 and the Maurice Wilkins Centre (MWC). TLM is supported by a Rutherford Discovery Fellowship.
Credit authorship contribution statement
Kate Lee Conceptualization, supervision, formal analysis, writing—original draft, review and editing, Investigation; Sanaz Vakili Methods, investigation; Hannah Burden Methods, investigation; Shannon Adams Methods, investigation; Greg C. Smith Methods; Braydon Kulatea Investigation; Robert D. Atiola Methods, investigation; Conor Watene-O’Sullivan Investigation, conceptualization; Morag Wright-McNaughton Investigation; Danielle Sword Investigation; Ryan Paul Writing—review and editing; Frances King Investigation, conceptualization; Phillip Wilcox Writing —review and editing; Prasanna Kallingappa Methods; Jeremy D. Krebs Conceptualization, Writing—review and editing; Rosemary M. Hall Conceptualization, writing—review and editing; Lindsay D. Plank Conceptualization, Writing—review and editing; Tony R. Merriman Conceptualization, writing—review and editing; Rinki Murphy Conceptualization, writing—review and editing; Troy L. Merry Conceptualization, supervision, formal analysis, writing—original draft, review and editing, investigation; Peter R. Shepherd Supervision, formal analysis, conceptualization, writing—original draft, review and editing.
Acknowledgements
We thank Dr Nigel Turner for facilitating production and access to knockin animals. He mihi nui tēnei ki ngā kai tuku taonga (We would like to thank the study participants for their time and tissue samples).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2022.101464.
Conflict of interest
All authors declare no competing interests.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Cole J.B., Florez J.C. Genetics of diabetes mellitus and diabetes complications. Nature Reviews Nephrology. 2020;16(7):377–390. doi: 10.1038/s41581-020-0278-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krentz N.A.J., Gloyn A.L. Insights into pancreatic islet cell dysfunction from type 2 diabetes mellitus genetics. Nature Reviews Endocrinology. 2020;16(4):202–212. doi: 10.1038/s41574-020-0325-0. [DOI] [PubMed] [Google Scholar]
- 3.Yaghootkar H., Whitcher B., Bell J.D., Thomas E.L. Ethnic differences in adiposity and diabetes risk - insights from genetic studies. Journal of Internal Medicine. 2020;288(3):271–283. doi: 10.1111/joim.13082. [DOI] [PubMed] [Google Scholar]
- 4.Goodarzi M.O. Genetics of obesity: what genetic association studies have taught us about the biology of obesity and its complications. Lancet Diabetes & Endocrinology. 2018;6(3):223–236. doi: 10.1016/S2213-8587(17)30200-0. [DOI] [PubMed] [Google Scholar]
- 5.Speakman J.R., Loos R.J.F., O’Rahilly S., Hirschhorn J.N. GWAS for BMI: a treasure trove of fundamental insights into the genetic basis of obesity. International Journal of Obesity. 2018;42(8):1524–1531. doi: 10.1038/s41366-018-0147-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Audas T.E., Li Y., Liang G., Lu R. A novel protein, Luman/CREB3 recruitment factor, inhibits Luman activation of the unfolded protein response. Molecular and Cellular Biology. 2008;28(12):3952–3966. doi: 10.1128/MCB.01439-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krishnan M., Major T.J., Topless R.K., Dewes O., Yu L., Thompson J.M.D., et al. Discordant association of the CREBRF rs373863828 A allele with increased BMI and protection from type 2 diabetes in Maori and Pacific (Polynesian) people living in Aotearoa/New Zealand. Diabetologia. 2018;61(7):1603–1613. doi: 10.1007/s00125-018-4623-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Minster R.L., Hawley N.L., Su C.-T., Sun G., Kershaw E.E., Cheng H., et al. A thrifty variant in CREBRF strongly influences body mass index in Samoans. Nature Genetics. 2016;48(9):1049–1054. doi: 10.1038/ng.3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naka I., Furusawa T., Kimura R., Natsuhara K., Yamauchi T., Nakazawa M., et al. A missense variant, rs373863828-A (p.Arg457Gln), of CREBRF and body mass index in Oceanic populations. Journal of Human Genetics. 2017;62:847–849. doi: 10.1038/jhg.2017.44. [DOI] [PubMed] [Google Scholar]
- 10.Hanson R.L., Safabakhsh S., Curtis J.M., Hsueh W.-C., Jones L.I., Aflague T.F., et al. Association of CREBRF variants with obesity and diabetes in pacific islanders from Guam and Saipan. Diabetologia. 2019;62(9):1647–1652. doi: 10.1007/s00125-019-4932-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hocking S., Samocha-Bonet D., Milner K.-L., Greenfield J.R., Chisholm D.J. Adiposity and insulin resistance in humans: the role of the different tissue and cellular lipid depots. Endocrine Reviews. 2013;34(4):463–500. doi: 10.1210/er.2012-1041. [DOI] [PubMed] [Google Scholar]
- 12.Lim U., Monroe K.R., Buchthal S., Fan B., Cheng I., Kristal B.S., et al. Propensity for intra-abdominal and hepatic adiposity varies among ethnic groups. Gastroenterology. 2019;156(4):966–975 e10. doi: 10.1053/j.gastro.2018.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rush E.C., Freitas I., Plank L.D. Body size, body composition and fat distribution: comparative analysis of European, Maori, Pacific Island and Asian Indian adults. British Journal of Nutrition. 2009;102(4):632–641. doi: 10.1017/S0007114508207221. [DOI] [PubMed] [Google Scholar]
- 14.Arslanian K.J., Fidow U.T., Atanoa T., Unasa-Apelu F., Naseri T., Wetzel A.I., et al. A missense variant in CREBRF, rs373863828, is associated with fat-free mass, not fat mass in Samoan infants. International Journal of Obesity. 2021;45(1):45–55. doi: 10.1038/s41366-020-00659-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hawley N.L., Duckham R.L., Carlson J.C., Naseri T., Reupena M.S., Lameko V., et al. medRxiv; 2021. The association of CREBRF variant rs373863828 with body composition in adult Samoans medRxiv. https://doi.org/10.1101/2021.02.11.21251582. [Google Scholar]
- 16.Burden H.J., Adams S., Kulatea B., Wright-McNaughton M., Sword D., Ormsbee J.J., et al. The CREBRF diabetes-protective rs373863828-A allele is associated with enhanced early insulin release in men of Maori and Pacific ancestry. Diabetologia. 2021;64(12):2779–2789. doi: 10.1007/s00125-021-05552-x. [DOI] [PubMed] [Google Scholar]
- 17.Metcalfe L.K., Krishnan M., Turner N., Yaghootkar H., Merry T.L., Dewes O., et al. The Maori and Pacific specific CREBRF variant and adult height. International Journal of Obesity. 2020;44(3):748–752. doi: 10.1038/s41366-019-0437-6. [DOI] [PubMed] [Google Scholar]
- 18.Concordet J.P., Haeussler M. CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Research. 2018;46(W1):W242–W245. doi: 10.1093/nar/gky354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Press T.N.A. 2011. Guide for the Care and use of laboratory animals.https://www.nap.edu/catalog/12910/guide-for-the-care-and-use-of-laboratory-animals-eighth Eighth Edition [Internet]. Washington, DC: Available from: [Google Scholar]
- 20.Percie du Sert N., Hurst V., Ahluwalia A., Alam S., Avey M.T., Baker M., et al. The ARRIVE guidelines 2.0: updated guidelines for reporting animal research. Journal of Physiology. 2020;598(18):3793–3801. doi: 10.1113/JP280389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology. 2002;3(7) doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allen D.L., Hittel D.S., McPherron A.C. Expression and function of myostatin in obesity, diabetes, and exercise adaptation. Medicine & Science in Sports & Exercise. 2011;43(10):1828–1835. doi: 10.1249/MSS.0b013e3182178bb4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brandt C., Nielsen A.R., Fischer C.P., Hansen J., Pedersen B.K., Plomgaard P. Plasma and muscle myostatin in relation to type 2 diabetes. PLoS One. 2012;7(5):e37236. doi: 10.1371/journal.pone.0037236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McMahon C.D., Popovic L., Jeanplong F., Oldham J.M., Kirk S.P., Osepchook C.C., et al. Sexual dimorphism is associated with decreased expression of processed myostatin in males. American Journal of Physiology. Endocrinology and Metabolism. 2003;284(2):E377–E381. doi: 10.1152/ajpendo.00282.2002. [DOI] [PubMed] [Google Scholar]
- 26.McPherron A.C., Lee S.J. Suppression of body fat accumulation in myostatin-deficient mice. Journal of Clinical Investigation. 2002;109(5):595–601. doi: 10.1172/JCI13562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paul R.G., McMahon C., Elston M., Conaglen J. GH replacement titrated to serum IGF-1 does not reduce concentrations of myostatin in blood or skeletal muscle. Growth Hormone & IGF Research. 2019;44:11–16. doi: 10.1016/j.ghir.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 28.Whittemore L.A., Song K., Li X., Aghajanian J., Davies M., Girgenrath S., et al. Inhibition of myostatin in adult mice increases skeletal muscle mass and strength. Biochemical and Biophysical Research Communications. 2003;300(4):965–971. doi: 10.1016/s0006-291x(02)02953-4. [DOI] [PubMed] [Google Scholar]
- 29.Jin Q., Qiao C., Li J., Xiao B., Li J., Xiao X. A GDF11/myostatin inhibitor, GDF11 propeptide-Fc, increases skeletal muscle mass and improves muscle strength in dystrophic mdx mice. Skeletal Muscle. 2019;9(1):16. doi: 10.1186/s13395-019-0197-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flurkey K., Currer J.M., H D. 2nd ed. 2007. The mouse in aging research. The mouse in biomedical Research; pp. 637–672. [Google Scholar]
- 31.Lee D.H., Keum N., Hu F.B., Orav E.J., Rimm E.B., Willett W.C., et al. Comparison of the association of predicted fat mass, body mass index, and other obesity indicators with type 2 diabetes risk: two large prospective studies in US men and women. European Journal of Epidemiology. 2018;33(11):1113–1123. doi: 10.1007/s10654-018-0433-5. [DOI] [PubMed] [Google Scholar]
- 32.Paul R., Whiteman K., Falconer S.J., Oldham J.M., Jeanplong F., Matthews K.G., et al. IGF1 does not overcome sexual dimorphism of body and muscle size in Mstn−/− mice. Journal of Endocrinology. 2021;248:207–220. doi: 10.1530/JOE-20-0485. [DOI] [PubMed] [Google Scholar]
- 33.Jackson M.F., Luong D., Vang D.D., Garikipati D.K., Stanton J.B., Nelson O.L., et al. The aging myostatin null phenotype: reduced adiposity, cardiac hypertrophy, enhanced cardiac stress response, and sexual dimorphism. Journal of Endocrinology. 2012;213(3):263–275. doi: 10.1530/JOE-11-0455. [DOI] [PubMed] [Google Scholar]
- 34.Wang F., Liao Y., Li X., Ren C., Cheng C., Ren Y. Increased circulating myostatin in patients with type 2 diabetes mellitus. J Huazhong Univ Sci Technolog Med Sci. 2012;32(4):534–539. doi: 10.1007/s11596-012-0092-9. [DOI] [PubMed] [Google Scholar]
- 35.Liang G., Audas T.E., Li Y., Cockram G.P., Dean J.D., Martyn A.C., et al. Luman/CREB3 induces transcription of the endoplasmic reticulum (ER) stress response protein Herp through an ER stress response element. Molecular and Cellular Biology. 2006;26(21):7999–8010. doi: 10.1128/MCB.01046-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Penney J., Mendell A., Zeng M., Tran K., Lymer J., Turner P.V., et al. LUMAN/CREB3 is a key regulator of glucocorticoid-mediated stress responses. Molecular and Cellular Endocrinology. 2017;439:95–104. doi: 10.1016/j.mce.2016.10.022. [DOI] [PubMed] [Google Scholar]
- 37.Penney J., Taylor T., MacLusky N., Lu R. LUMAN/CREB3 plays a dual role in stress responses as a cofactor of the glucocorticoid receptor and a regulator of secretion. Frontiers in Molecular Neuroscience. 2018;11:352. doi: 10.3389/fnmol.2018.00352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martyn A.C., Choleris E., Gillis D.J., Armstrong J.N., Amor T.R., McCluggage A.R.R., et al. Luman/CREB3 recruitment factor regulates glucocorticoid receptor activity and is essential for prolactin-mediated maternal instinct. Molecular and Cellular Biology. 2012;32(24):5140–5150. doi: 10.1128/MCB.01142-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bohnert K.R., McMillan J.D., Kumar A. Emerging roles of ER stress and unfolded protein response pathways in skeletal muscle health and disease. Journal of Cellular Physiology. 2018;233(1):67–78. doi: 10.1002/jcp.25852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma K., Mallidis C., Bhasin S., Mahabadi V., Artaza J., Gonzalez-Cadavid N., et al. Glucocorticoid-induced skeletal muscle atrophy is associated with upregulation of myostatin gene expression. American Journal of Physiology. Endocrinology and Metabolism. 2003;285(2):E363–E371. doi: 10.1152/ajpendo.00487.2002. [DOI] [PubMed] [Google Scholar]
- 41.Paul R.G., Hennebry A.S., Elston M.S., Conaglen J.V., McMahon C.D. Regulation of murine skeletal muscle growth by STAT5B is age- and sex-specific. Skeletal Muscle. 2019;9(1):19. doi: 10.1186/s13395-019-0204-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carlson J.C., Rosenthal S.L., Russell E.M., Hawley N.L., Sun G., Cheng H., et al. A missense variant in CREBRF is associated with taller stature in Samoans. American Journal of Human Biology. 2020;32(6):e23414. doi: 10.1002/ajhb.23414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bond S.T., Calkin A.C., Drew B.G. Sex differences in white adipose tissue expansion: emerging molecular mechanisms. Clinical Science. 2021;135(24):2691–2708. doi: 10.1042/CS20210086. [DOI] [PubMed] [Google Scholar]
- 44.Neel J.V. Diabetes mellitus: a "thrifty" genotype rendered detrimental by "progress"? The American Journal of Human Genetics. 1962;14:353–362. [PMC free article] [PubMed] [Google Scholar]
- 45.Neel J.V. The "thrifty genotype" in 1998. Nutrition Reviews. 1999;57(5 Pt 2):S2–S9. doi: 10.1111/j.1753-4887.1999.tb01782.x. [DOI] [PubMed] [Google Scholar]
- 46.Lev-Ran A. Human obesity: an evolutionary approach to understanding our bulging waistline. Diabetes Metab Res Rev. 2001;17(5):347–362. doi: 10.1002/dmrr.230. [DOI] [PubMed] [Google Scholar]
- 47.Myles S., Hradetzky E., Engelken J., Lao O., Nürnberg P., Trent R.J., et al. Identification of a candidate genetic variant for the high prevalence of type II diabetes in Polynesians. European Journal of Human Genetics. 2007;15(5):584–589. doi: 10.1038/sj.ejhg.5201793. [DOI] [PubMed] [Google Scholar]
- 48.Cadzow M., Merriman T.R., Boocock J., Dalbeth N., Stamp L.K., Black M.A., et al. Lack of direct evidence for natural selection at the candidate thrifty gene locus, PPARGC1A. BMC Medical Genetics. 2016;17(1):80. doi: 10.1186/s12881-016-0341-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang G., Speakman J.R. Analysis of positive selection at single nucleotide polymorphisms associated with body mass index does not support the "thrifty gene" hypothesis. Cell Metabolism. 2016;24(4):531–541. doi: 10.1016/j.cmet.2016.08.014. [DOI] [PubMed] [Google Scholar]
- 50.Lindo J., Huerta-Sánchez E., Nakagome S., Rasmussen M., Petzelt B., Mitchell J., et al. A time transect of exomes from a Native American population before and after European contact. Nature Communications. 2016;7:13175. doi: 10.1038/ncomms13175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Audas T.E., Hardy-Smith P.W., Penney J., Taylor T., Lu R. Characterization of nuclear foci-targeting of Luman/CREB3 recruitment factor (LRF/CREBRF) and its potential role in inhibition of herpes simplex virus-1 replication. European Journal of Cell Biology. 2016;95:611–622. doi: 10.1016/j.ejcb.2016.10.006. [DOI] [PubMed] [Google Scholar]
- 52.Lu R., Yang P., O’Hare P., Misra V. Luman, a new member of the CREB/ATF family, binds to herpes simplex virus VP16-associated host cellular factor. Molecular and Cellular Biology. 1997;17(9):5117–5126. doi: 10.1128/mcb.17.9.5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mesinovic J., Zengin A., De Courten B., Ebeling P.R., Scott D. Sarcopenia and type 2 diabetes mellitus: a bidirectional relationship. Diabetes Metab Syndr Obes. 2019;12:1057–1072. doi: 10.2147/DMSO.S186600. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.