Abstract
The blood-brain barrier (BBB), a selective barrier formed by brain microvascular endothelial cells (BMEC), represents a major challenge for the efficient accumulation of pharmaceutical drugs into the brain. The receptor-mediated transcytosis (RMT) has recently gained increasing interest for pharmaceutical industry as it shows a great potential to shuttle large-sized therapeutic cargos across the BBB. Confirming the presence of the RMT pathway by BMEC is therefore important for the screening of peptides or antibody libraries that bind RMT receptors. Herein, a comparative study was performed between a human cell line of BMEC (HBEC) and human induced pluripotent stem cells-derived BMEC-like cells (hiPS-BMEC). The significantly higher gene and protein expressions of transporters and tight junction proteins, excepting CD31 and VE-cadherin were exhibited by hiPS-BMEC than by HBEC, suggesting more biomimetic BBB features of hiPS-BMEC. The presence and functionality of transferrin receptor (TfR), known to use RMT pathway, were confirmed using hiPS-BMEC by competitive binding assays and confocal microscopy observations. Finally, cysteine-modified T7 and cysteine modified-Tfr-T12 peptides, previously reported to be ligands of TfR, were compared regarding their permeability using hiPS-BMEC. The hiPS-BMEC could be useful for the identification of therapeutics that can be transported across the BBB using RMT pathway.
Keywords: Blood-brain barrier, In vitro model, Tight junction, Transferrin receptor, Receptor-mediated transcytosis
Graphical abstract
1. Introduction
Pharmaceutical drug development is a lengthy and highly expensive process, whose costs have steadily increased over recent decades [1], due to the development of new modalities such as RNA therapeutics, antibody drug conjugates, or gene therapy [2]. Drug candidate libraries must be tested against potential targets, as well as drug side effects and toxicity towards human tissue to allow their commercialization [3]. Drug candidates for the treatment of neurological pathologies tend to have higher failure rates at the “bench-to-bedside” transition than any other drugs [4], most often because of insufficient evidences of the clinical efficacy of the drugs. It is also partly due to the lack of reliable human equivalent models to effectively screen potential therapeutic molecules, especially the brain. The permeation mechanism and the delivery efficiency of intravenously injected drugs to the brain are also poorly understood. Increasing efforts have been focused on the implementation of biologically relevant models of the brain vasculature to simulate the passage of anti-cancer drugs from the bloodstream to the brain.
Notably, the in vitro blood-brain barrier (BBB) models can provide a valuable tool for understanding intracellular trafficking and receptor binding related to the transport of drugs into the brain [5]. The BBB is a protective barrier located at the interface between the microvascular blood stream and the brain. It regulates the transport of nutrients and ions in the brain parenchyma, which is essential for brain homeostasis. This selective barrier also impedes the brain uptake of potentially harmful substances and neurotherapeutics [6]. The restrictive permeability mainly arises from the presence of highly specialized brain microvascular endothelial cells (BMEC), which are different from the endothelial cells (ECs) in other organs [7,8]. They are characterized by the absence of fenestration, a low level of pinocytic vesicles and high expression of tight junctions (TJs) between adjacent ECs. The presence of specific carriers and receptors that regulates the transport of ions, small and large molecules is also a hallmark of the BBB endothelium [9,10]. The source of BMEC is important to consider for the reproduction of all of these properties in vitro and is crucial for a better clinical translation of engineered BBB models [[11], [12], [13]]. Immortalized cell lines provide a readily available source of human brain microvascular endothelial cells (BMEC) but do not exhibit some of the characteristics of the in vivo BBB endothelium, being impaired by their inadequate barrier function and low expression of BBB-specific transporters and enzymes [14,15]. Human induced pluripotent stem cells derived BMEC-like cells (hiPS-BMEC) have recently generated great opportunities for the BBB modeling [[16], [17], [18], [19], [20], [21]]. Although the formation of strong TJs and the expression of some transporter proteins were demonstrated for hiPS-BMEC, the functions of the receptor-mediated transcytosis (RMT) receptors remains to be elucidated. The RMT pathway has recently attracted increasing interest due to the possibility of transporting certain large molecules across the BBB [6]. An ever-increasing list of protein, such as insulin, leptin, and lipoproteins, essential for brain functions has been proposed to be transported using the RMT [[22], [23], [24], [25]]. One of the most studied RMT receptors is the transferrin receptor (TfR) which mediates the delivery of iron to the brain [25]. TfR has gained particular attention due to its high expression level by both brain ECs and brain cancer cells [[26], [27], [28]], making it an attractive target for the specific delivery of drugs into the brain and to tumor site. Various drug delivery systems (DDS) have been designed for improving the delivery efficiency of drugs into the brain by adding moieties targeting RMT receptors for an enhanced transport across the BBB [[29], [30], [31], [32], [33]]. HiPS-BMEC-based models could be a valuable complementary tool prior to in vivo studies, as it could easily and rapidly screen the DDS functionalized with different ligands for RMT based on their in vitro permeation rate. Assessing the possibility of hiPS-BMEC to use RMT pathway is therefore highly desired, as it could create opportunities for finding novel treatments of central nervous system diseases.
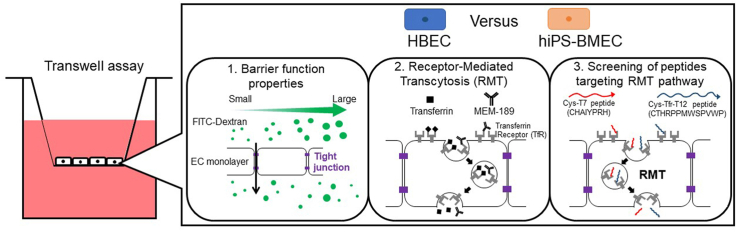
In this study, hiPS-BMEC [18] were compared to an immortalized human brain microvascular endothelial cell line (HBEC) [34] with respect to their gene and protein expression profile, as well as their barrier function properties. The functionality of the TfR-mediated transcytosis on hiPS-BMEC was then confirmed using the native ligand of the TfR, transferrin. Finally, as an applicative validation, two candidate peptides previously reported to be internalized using TfR [35], cysteine-modified T7 (Sequence CHAIYPRH) or cysteine-modified Tfr-T12 peptide (Sequence CTHRPPMWSPVWP), were screened with respect to their permeability ability across the hiPS-BMEC monolayer (Fig. 1).
Fig. 1.
Schematic overview of the comparison study between human BMEC/conditionally immortalized, clone ci18 (HBEC) and human iPS-derived BMEC-like (hiPS-BMEC).
2. Materials and methods
2.1. Reagents
Cysteine-modified T7 peptide (8 AA, CHAIYPRH, cys-T7) and cysteine-modified Tfr-T12 peptide (13 AA, CTHRPPMWSPVWP, cys-TfR-T12) were custom peptides synthesized by Scrum (Tokyo, Japan). (4-(4, 6-dimethoxy-1, 3, 5 -triazine-2-yl)-4-methyl-morpholinium chloride (DMT-MM, nH2O) was purchased from Watanabe Chemical (Hiroshima, Japan) (Ref. 3945-69-5). Unconjugated fluoresceinyl glycine amide (FGA) was obtained from Setareh Biotech (Eugene, OR, USA) (Ref. 136091-82-2). Dimethylsulfoxide (DMSO) was purchased from Nacalai Tesque (Kyoto, Japan) (Ref. 08489–45). Spectra/Por™ Biotech Cellulose Ester (CE) Dialysis Membrane Tubing (MWCO 500–1000Da) was purchased from Spectrum (New Brunswick, NJ, USA) (Ref. 131096). Lysotracker deep red (Ref. L12492) and FITC-dextran of 9.5 kDa (Ref. FD10S), 70 kDa (Ref. FD70S), 250 kDa (Ref. FD250S), 2000 kDa (Ref. FD2000S) were purchased from Sigma-Aldrich (St Louis, MO, USA). PitStop®2 (Ref. ab120687) was purchased from Abcam (Cambridge, UK).
2.2. DLS measurement
The zeta potential and size of FITC dextran molecular weight (MW) 9.5 kDa, 70 kDa, 250 kDa and 2000 kDa at 1 mg/mL were measured in PBS. The analysis was performed three times with an acquisition at 37 °C using dynamic light scattering (DLS) instruments (Malvern Instruments, Nano-ZS, UK). The data were analyzed by Malvern ZetaSizer Software (Version 7.13.1, UK) and values were reported as the means of triplicate measurements.
2.3. Cell culture
Human brain microvascular endothelial cells/conditionally immortalized clone ci18 (HBEC) used for this work were kindly provided by Prof. Furihata, generated as previously reported [34]. HBEC were cultured on collagen-coated dishes (100 mm, collagen type I, Iwaki, Tokyo, Japan). Cells were cultured in a humidified incubator (33 °C, 5% CO2) for the maintenance of growth activity. Culture medium was supplemented with 4 μg/mL of Blasticidin S HCl (Invitrogen, Waltham, MA, USA) to maintain selective pressure for routine culture. HBEC were grown in Vasculife (VEGF-Mv, LifeLine, Carlsbad, CA, USA) supplemented with 0.5 mL rh FGF-b, 0.5 mL ascorbic acid, 0.5 mL hydrocortisone hemisuccinate, 25 mL l-glutamine, 0.5 mL rh IGF-1, 0.5 mL rh EGF, 0.5 mL rh VEGF, 0.5 mL heparin sulfate, 25 mL fetal bovine serum (kit LifeFactor VEGF-Mv, LifeLine, Carlsbad, CA, USA), 25 mL supplementary fetal bovine serum (FBS, Gibco ThermoFisher, Waltham, MA, USA), and 1% penicillin-streptomycin (10,000 U.mL-1 - 10,000 μg/mL, Nacalai Tesque, Kyoto, Japan). For transport studies, 1.0 × 105 HBEC were seeded in a 24 well insert (Ref. 662641, Greiner Bio-One, Monroe, NC, USA) pre-coated with 100 μL of fibronectin at 0.04% (fibronectin from human plasma, F2006-5G, Sigma-Aldrich, St. Louis, MO, USA). Insert cultures of HBEC were then incubated at 37 °C for 2 days to induce their differentiation before their use in subsequent experiments, also performed at 37 °C.
Human iPS cells (hiPS) were successfully differentiated to hiPS-BMEC as previously reported [18] (Fig. S1). Briefly, hiPS were detached with Accutase (SCR005, Merck, Branchburg, NJ, USA) and seeded on Matrigel-coated 6-well plate at a density of 1–1.5 × 105 cells/well and cultured in mTeSR1 medium (Ref. 85850, STEMCELL Technologies, Vancouver, BC, Canada) containing 10 μM ROCK inhibitor Y27632 (Ref. 259–00613, Fujifilm Wako, Osaka, Japan) (day −3). After 24h, the medium was replaced with fresh mTeSR1 medium without ROCK inhibitor Y27632 (day −2). On day 0, the medium was replaced with unconditioned medium (UM). On day 6, the medium was replaced by human endothelial serum-free medium (SFM, Ref. 11111044, Gibco ThermoFisher, Waltham, MA, USA) containing 1% human platelet-derived serum (hPDS, Ref. P2918 Sigma-Aldrich, St. Louis, MO, USA), 10 μM all-trans-retinoic acid (ATRA, Ref. 182–01116, Fujifilm Wako, Osaka, Japan) and 20 ng/mL human fibroblast growth factor 2 (FGF2, Ref. 161-0010-3, Katayama Chemical Industries Co., Osaka, Japan). On day 8, the cells were detached using Accutase and seeded at a density of 3.3 × 105 onto 24-well inserts with 0.4 μm pore size (Ref. 353095, Falcon, Flowery Branch, GA, USA) coated with fibronectin/collagen IV (FN/Col IV, Pharmaco-cell, Nagasaki, Japan) and cultured in SFM with hPDS, ATRA, and FGF2. On day 9, floating cells were removed by washing with SFM, and the medium was replaced with fresh SFM with hPDS. The hiPS-BMEC insert cultures at day 10 of the differentiation were used in subsequent experiments.
2.4. Assessment of transendothelial electrical resistance (TEER)
The TEER was monitored for 1 h at different time points (0, 10, 30, 45, 60 min) in SFM medium using the cultures of HBEC and hiPS-BMEC seeded in the 24-well insert. The TEER was measured using a Millicell® ERS-2 Volt-Ohm Meter (Millipore, Bedford, MA, USA) equipped with a STX01 chopstick electrode (Millipore, Bedford, MA, USA). The TEER value was calculated from the following equation (1) [36]:
| (1) |
Where Rmonolayer is the resistance of the cell monolayer along with the filter membrane; Rblank is the resistance of the cell-free insert membrane in SFM medium, and A is the surface area of the membrane (0.33 cm2).
2.5. Quantitative reverse Transcription-PCR (qRT-PCR)
The HBEC and hiPS-BMEC cultured in inserts were rinsed with PBS, and RNA was extracted using PureLink RNA Micro Kit (Invitrogen, Waltham, MA, USA). RNA content was assessed with a NanodropTM spectrometer (N1000, Thermo Fisher Scientific, Waltham, MA, USA). A reverse transcription of the RNA samples into cDNA using iSCRIPT cDNA synthesis kit (Bio-Rad, Hercules, USA) was performed. A real-time quantitative polymerase chain reaction (RT-qPCR) was then conducted on the cDNA samples using Taqman Fast Advanced Mix (Taqman Gene Expression Assays, Thermo Fisher Scientific, Waltham, MA, USA) (cf. Supplementary Data, Table S1 for probes reference) in the StepOnePlus Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA, USA). Peptidylprolyl Isomerase A (PPIA) was used as the housekeeping gene to normalize the gene expression.
2.6. Immunofluorescence
The HBEC and hiPS-BMEC insert cultures were rinsed three times in PBS then fixed in 4% paraformaldehyde (Fujifilm Wako, Osaka, Japan) at room temperature for 15 min. Permeabilization was carried out using 0.2% Triton X-100 for 15 min (Sigma-Aldrich, St. Louis, MO, USA) diluted in PBS. After PBS rinsing, blocking was performed for 1 h at room temperature with 1% bovine serum albumin (BSA, Sigma-Aldrich, St. Louis, MO, USA) in PBS, to minimize non-specific staining. The samples were incubated with primary antibodies overnight at 4 °C: anti-CD31 antibody (mouse anti-human, NCL-CD31-1A10, Leica, Wetzlar, Germany) was diluted in 1% BSA in PBS at 1/100, while antibodies against tight-junction proteins, adherens junction protein and transporters were diluted in 1% BSA in PBS at 1/50 (rabbit anti-human TfR, NB500-493AF647, NovusBio, Centennial, USA) (mouse anti-human ZO-1, ZO1-1A12, Invitrogen, Waltham, MA, USA), 1/100 (mouse anti-human Claudin-5, 35–2500, Thermo Fisher Scientific, Waltham, MA, USA) or at 1/500 (rabbit anti-human VE-cadherin, ab33168, Abcam, Cambridge, UK). Double staining was usually carried out at the same time, except for the anti-TfR antibody which was already conjugated with Alexa Fluor 647. After PBS rinsing, samples were incubated for 2 h at room temperature in the dark with secondary antibodies diluted at 1/100 in 1% BSA in PBS (goat anti-mouse, Alexa Fluor 647, A21235, Thermo Fisher Scientific, Waltham, MA, USA; or goat anti-rabbit, Alexa Fluor 488, ab150077, Abcam, Cambridge, UK). The nuclei were counterstained with Hoechst (Thermo Fisher Scientific, Waltham, MA, USA). After washing three times with PBS, the samples were observed using confocal laser scanning microscope FluoView FV3000 (Olympus, Tokyo, Japan) using ×10 or ×40 magnification. The images were taken by keeping the same exposure time and excitation power for each sample for fluorescence quantification assessment. Fluorescence intensity of ×10 magnification pictures was quantified using Fiji software [37] and normalized by cell number.
2.7. Fluorescent labeling of peptides
A stock solution at 1 mg/mL of each peptide was prepared by dissolving 5 mg of cys-T7 or cys-Tfr-12 peptide in MilliQ water. A 500 mM stock solution of DMT-MM was prepared in MilliQ water. 10.04 μL of DMT-MM stock solution (5.02 μmol, 1 eq.) and 6.275 μL of DMT-MM stock solution (3.137 μmol, 1 eq.) were respectively added dropwise to the 5 mL of the stock solution of cys-T7 peptide or cys-Tfr-T12 peptide stock solution (5.02 μmol, 1 eq.). These solutions were stirred at room temperature for 10 min. In parallel, a stock solution of 25 mM of FGA were prepared in DMSO. 7.53 μmol (1.5 eq.) or 4.70 μmol (1.5 eq.) of 25 mM of FGA stock solution are respectively added to the DMT-MM/cys-T7 peptide and DMT-MM/cys-Tfr-T12 reaction mixture and stirred in an ice bath for 1 h and then at room temperature for 24 h. The mixture was then dialyzed for 3 days with a cellulose ester dialysis tubing (MWCO 500–1000 Da) and freeze dried (Freeze Dryer FDU-2200, Eyela, Tokyo, Japan) for 3 days to afford the FGA-labeled peptides.
2.8. Permeability studies
Permeability studies were performed as similarly reported in previous studies [38,39]. For all the permeability studies, HBEC or hiPS-BMEC cultures were rinsed three times with PBS, Human Endothelial Serum Free Medium (SFM) (Ref. 11111044, ThermoFisher, Waltham, MA, USA) was then added to the upper side with the tested molecules (200 μL) and to the lower side (1000 μL) of the inserts and the system was incubated at 37 °C for the permeability assays for 1 h. Different molecular weights (MW) (9.5, 70, 250, 2000 kDa) of fluorescein isothiocyanate (FITC) labeled dextran at a final concentration of 1 mg/mL were added to the upper side before beginning the assay. For the competition binding assays, Alexa Fluor 488-transferrin (AF 488-Tf) (009-540-050, Jackson ImmunoResearch, West Grove, PA, USA) (final concentration 125 nM) was co-incubated with different concentrations of unlabeled transferrin (Tf) (009-000-050, Jackson ImmunoResearch, West Grove, PA, USA) (final concentration 0; 12.5; 125; 1250 nM). Alexa Fluor 647-MEM-189 (AF 647-MEM-189) (NB500-493AF647, NovusBio, Centennial, USA), Alexa Fluor 647-immunoglobulin G1 (IgG1) (NBP1-97005AF647, NovusBio, Centennial, USA) and Alexa Fluor 647-13E4 (NB100-73092, NovusBio, Centennial, USA) were incubated at a final concentration of 64 nM with HBEC or hiPS-BMEC. FGA-cys-T7 peptide or FGA-cys-Tfr-T12 peptide at a final concentration of 10 μg/mL were co-incubated with different amounts of unlabeled transferrin (Tf) (final concentration 0; 12.5; 125; 1250 nM) using hiPS-BMEC.
Following the incubation with the tested molecule, 10 μL of the medium was collected in the lower chambers and replaced with the same volume of fresh medium at t = 10, 15, 30, 45, 60 min. The fluorescence in the lower chamber was measured using a NanodropTM fluorospectrometer (N3300, Thermo Fisher Scientific, Waltham, MA, USA) from which was deduced the amount of transported compound. The cumulative amount transported across the membrane was plotted against time, and the slopes of the linear regions were used to calculate the permeability coefficients, as previously described [39].
The apparent permeability (Papp) was calculated using the following equation (2):
| (2) |
Where dQ/dt is the transport rate, defined as the slope obtained from linear regression of the transported amount, C0 is the initial concentration on the donor side and A is the surface area of the inserts (0.33 cm2).
The effective permeability (Pe) of the EC monolayer was calculated using the following equation (3):
| (3) |
Where PS, dQ/dt, and D0 are, respectively, the permeability surface area product, the slope of the linear region of a plot of the amount of permeant in the receiver chamber over time, and the initial concentration of the tested molecule on the donor side.
| (4) |
| (5) |
Where PStotal and PSm are respectively the permeability surface area product corresponding to the transwell membrane with and without the EC monolayer and PSe is the surface area product value for the endothelial monolayer. A is the surface area of the insert membrane.
2.9. Confirmation of TfR-mediated endocytosis by confocal microscopy imaging
HBEC or hiPS-BMEC were co-incubated with 125 nM of AF488-Tf and 64 nM of AF647 MEM-189 for 1h at 37 °C. Cells were then fixed with PFA 4%, washed three times with PBS prior to the observation. Images were then taken with a confocal laser scanning microscope AX (Nikon, Tokyo, Japan).
2.10. Co-localization studies
HiPS-BMEC insert cultures were washed with PBS three times and incubated for 45 min with a solution of 10 μM of each FGA-labeled peptide (cys-T7 or cys-Tfr-T12) or 125 nM of Alexa Fluor 488-transferrin (AF 488-Tf) diluted in SFM medium, followed by staining with 50 nM of lysotracker deep red diluted in SFM medium for 15 min prior to fixation with PFA 4%.
HiPS-BMEC insert cultures were washed with PBS three times and incubated with a solution of 10 μM of FGA-cys-T7 or FGA-cys-Tfr-T12 and 125 nM Alexa Fluor 647-transferrin (AF 647-Tf) diluted in SFM medium for 1 h prior to fixation with PFA 4%.
Fixed cells were observed with a confocal laser scanning microscope FluoView FV3000 (Olympus, Tokyo, Japan). Images were taken by keeping the same exposure time and excitation power for each sample. The colocalization ratio between lysotracker deep red and FGA-peptides or AF 488-Tf, as well as the colocalization ratio of AF 647-Tf and FGA-cys-T7 or FGA-cys-Tfr-T12 peptide was quantified by IMARIS software (Oxford Instruments, Version 9.2.1, Bitplane, Belfast, UK).
2.11. Inhibition study
HiPS-BMEC insert cultures were treated with 25 μM of PitStop®2 dissolved in SFM medium for 30 min at 37 °C. Cells were washed three times with PBS prior to incubation with 10 μM of FGA-cys-T7 peptide or FGA-cys-TfR-T12 peptide dissolved in SFM medium for 45 min at 37 °C. After the incubation with FGA-peptides, cells were fixed with PFA 4%, washed three times with PBS, stained with DAPI and again washed three times with PBS prior to the observation. Images were then taken with a confocal laser scanning microscope FluoView FV3000 (Olympus, Tokyo, Japan) by keeping the same exposure time and excitation power for each sample.
2.12. Statistical analysis
All values are presented as means ± standard deviation (SD). Statistical analysis of the data was performed with Student's t-test or one-way ANOVA using EzAnova software when more than two samples were compared (Version 0.985, University of South Carolina, Columbia, SC, USA) with Tukey multiple comparison post-hoc tests. Differences were considered statistically significant at p ≤ 0.05.
3. Results and discussion
3.1. Evaluation of an endothelial marker, tight junctions and transporter expressions
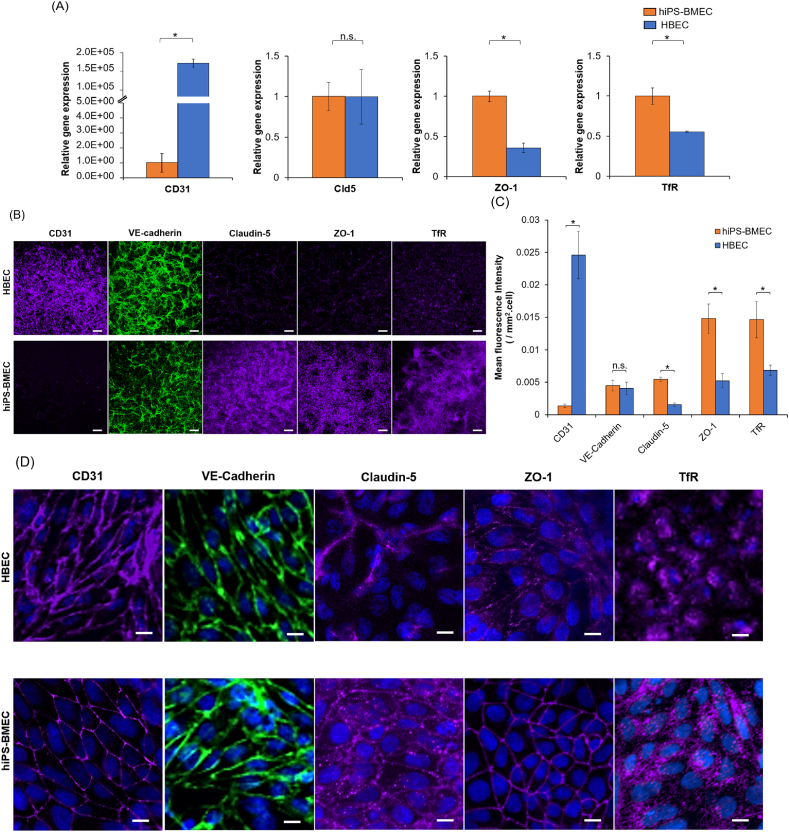
The gene expression and phenotypical features of BMEC/conditionally immortalized clone ci18 (HBEC) or human iPS-derived BMEC-like cells (hiPS-BMEC) were first investigated, focusing on the expression of endothelial marker CD31, tight junction proteins (TJ) and transporter. CD31 is an adherent molecule expressed at the inter-endothelial junctions and is involved in blood vessel formation, vascular integrity and remodeling [[40], [41], [42], [43]]. The expression of complex TJs between the BMEC is also very important for barrier function properties, as they control the paracellular transport and the maintenance of cell polarity [11]. Claudin 5 (Cld5) is the most abundant form of the claudin family in the brain endothelium and is needed for the macromolecular assembly of the TJ [44]. VE-cadherin is an endothelial-specific transmembrane protein which play a major role in the establishment and development of endothelial cell-cell junction [45,46]. Zonula occludens-1 (ZO-1) is involved in the maintenance of TJ stability and functionality by interacting with the TJ components and the actin cytoskeleton [[47], [48], [49]]. The delivery of nutrients, ions and proteins into the brain is ensured by the presence of carriers and transporters expressed on the EC membrane. Among them, transferrin receptor (TfR) is highly expressed by BMEC and is responsible for the transport of transferrin-bound iron into the brain tissue [50]. Iron is required for several fundamental biological processes that maintain normal neurological functions (oxygen transport, neurotransmitter metabolism, DNA synthesis) [25]. As seen in Fig. 2A, RT-qPCR showed that the expression of CD31 was significantly higher (15,000-fold) in HBEC than hiPS-BMEC. Both cell types expressed the gene for Cld5 in a similar manner. However, the mRNA for ZO-1 and TfR were significantly higher for hiPS-BMEC than HBEC. The protein expression of the same endothelial marker, TJ proteins, and transporter was also assessed by immunofluorescence staining (Fig. 2B). The comparison of the fluorescence intensity (Fig. 2C) revealed a similar tendency as the RT-qPCR results, with an increased expression of ZO-1 for hiPS-BMEC compared to HBEC. These observations are in line with the previously reported characteristics for hiPS-BMEC, particularly the high expression of TJ proteins, including ZO-1 and Claudin-5 [16,17,51]. Although differences of Cld5 gene expression could not be detected by RT-qPCR, it was more expressed by hiPS-BMEC than HBEC at the protein level. Moreover, hiPS-BMEC showed uniform and continuous staining for Claudin-5 and ZO-1, while punctate contacts with intercellular gaps was detected with HBEC (Fig. 2D), suggesting differences in barrier stability and integrity. TfR showed higher expression at both gene and protein levels by hiPS-BMEC, as compared to HBEC. Both hiPS-BMEC and HBEC were positive for CD31 and VE-cadherin (Fig. 2D) which is in accordance with previous reports [17,18,34]. Even though CD31 expression at gene level was detected with hiPS-BMEC (Fig. 2D), it remains relatively very low as compared to HBEC (Fig. 2C). This higher expression of CD31 by HBEC as compared to hiPS-BMEC is similarly found at the protein level, but to a lesser extent than the gene expression level. The observed differences between expression at gene and protein levels are expected to be due to transcriptional and post-transcriptional regulation processes [52].
Fig. 2.
Gene and protein expression of endothelial marker, tight junction proteins and transporters between HBEC (blue) and hiPS-BMEC (orange) (n = 3). (A) RT-qPCR results. Data are presented as means ± S.D. The biological replicates of HBEC and hiPS-BMEC come from three independent differentiations. (B) Immunostaining observed by confocal light scanning microscopy. Scale bar = 150 μm. The images were taken using same exposure time and excitation power for each protein with HBEC and hiPS-BMEC. (C) Immunostaining fluorescence intensity comparison using pictures from Fig. 2B. Data are presented as means ± S.D. Statistical analysis was performed using the Student's t-test (∗p ≤ 0.05, n. s. p > 0.05). The biological replicates of HBEC and hiPS-BMEC come from three independent differentiations. (D) Zoom in pictures of immunostaining observed by confocal light scanning microscopy. The images were taken using different exposure time and excitation power for each sample. Scale bar = 10 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Although the comparison of HBEC to primary human BMEC has been already initiated in a previous work [34], the degree of phenotypical similarities to the in vivo brain endothelium for hiPS-BMEC remains however to be elucidated.
3.2. Evaluation of the barrier function properties
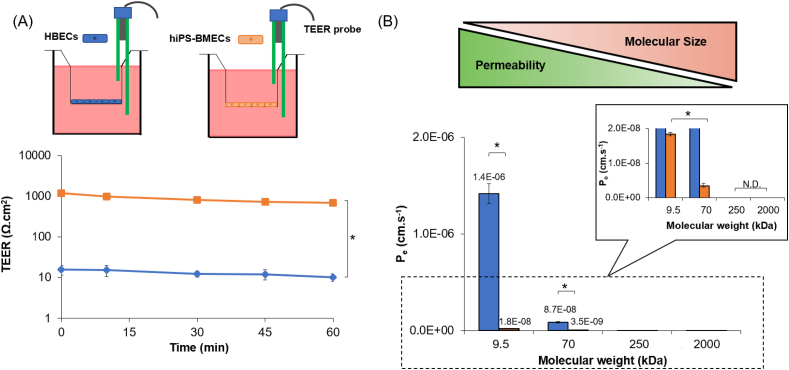
In addition to the expression of BBB markers and vascular phenotype, BMEC should exhibit tight barrier properties and express functional transport systems. One of the hallmarks of the BBB is its high trans-endothelial electrical resistance (TEER), mainly resulting from the presence of TJ protein interactions between adjacent brain microvascular ECs [16,53]. For both cell types, the TEER value remains stable for a period of 1 h (Fig. 3A), confirming the possibility of performing permeability assays without any variability in the barrier function properties in this lapse of time. The TEER value for hiPS-BMEC exceeded 1000 Ω cm2, around 100-fold higher than the value obtained for HBEC, independently of the time point measurement. The TEER value observed for hiPS-BMEC is in accordance with previously reported values [16]. Although the in vivo TEER values across the BBB in human brain cannot be easily measured, values from the brains of other species such as rat [54] and frog [55] have been estimated to exceed 1000 Ω cm2. Moreover, a study demonstrated that if the TEER can be maintained above 900 Ω cm2, the model has the potential to be used to predict in vivo human permeability of both small and large molecules [56]. The TEER value obtained for hiPS-BMEC is closer to physiological TEER than HBEC (Fig. 2B and C). Although the TEER is a versatile and non-invasive approach to confirm the barrier integrity of the EC monolayer, several factors such as temperature, medium composition, type of electrode probe or surface area can greatly affect the value of the measurement. The TEER value is thus subject to considerable variations between experiments and laboratories [53].
Fig. 3.
Characterization of the barrier function properties of HBEC (blue) and hiPS-BMEC (orange) (n = 3). (A) TEER measurement. The biological replicates of HBEC and hiPS-BMEC come from three different differentiations. (B) Effective permeability coefficients (Pe) of FITC-dextran with different molecular weights. Data are presented as means ± S.D. Statistical analysis was performed using one-way ANOVA (∗p ≤ 0.05, n. s. p > 0.05). N.D. means “not detected”. The biological replicates of HBEC and hiPS-BMEC are issued from the same differentiation. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3. Size-dependent molecular permeability of fluorescent dextran
Paracellular fluorescent tracer compounds, such as fluorescein isothiocyanate-dextran (FITC-dextran), have also been widely used to detect potential leakages in the endothelial barrier [57]. To explore the potential molecular size effect on the permeability of fluorescent tracers, a range of different molecular weights (MW) of FITC-dextran (9.5 kDa, 70 kDa, 250 kDa and 2000 kDa) were chosen to represent the small, intermediate- and large-sized agents but with a similar surface charge (Fig. S2). The apparent (Papp) and effective (Pe) permeability coefficient values were inversely correlated with the size of the tracer. As shown in Fig. 3B, both HBEC and hiPS-BMEC were totally impermeable to 250 and 2000 kDa FITC-dextran. However, a drastic increase in the permeability was observed with smaller sizes of FITC-dextran (9.5 and 70 kDa) for HBEC. Conversely, hiPS-BMEC showed a significantly lower permeability to fluorescent tracers than HBEC, even with the smallest ones such as 9.5 kDa FITC dextran. For example, the Papp value of 9.5 kDa FITC-dextran was 6.2 × 10−8 cm/s and 1.0 × 10−6 cm/s for hiPS-BMEC and HBEC respectively (Fig. S3). The Pe value of 9.5 kDa FITC-dextran showed a similar trend in the fluctuation changes, with 1.8 × 10−8 cm/s for hiPS-BMEC and 1.4 × 10−6 cm/s for HBEC (Fig. 3B). The junctions of the confluent HBEC cell monolayers appeared to be tight enough to block the transfer of large-sized dextran, 250 and 2000 kDa FITC dextran, in the lower compartment of the insert but remained permeable to the small- and intermediate-sized ones, 9.5 and 70 kDa FITC dextran. These results are consistent with previous reports, demonstrating the low barrier function properties of EC cell lines compared to hiPS-BMEC. It is generally well accepted that human brain EC lines such as hCMEC/D3 are relatively leaky and exhibit low paracellular resistance. For example, it has been reported that the Papp value of hCMEC/D3 for 4 kDa and 2000 kDa FITC dextran was about 6.2 × 10−6 cm/s and 3.2 × 10−6 cm/s respectively [58]. Linville et al. reported a similar low permeability of 10 kDa FITC dextran using hiPS-BMEC with a Papp about 5.0 × 10−8 cm/s [59]. Taken together, these results demonstrated that hiPS-BMEC displayed a restrictive permeability ability of compounds through the cell monolayer that resemble to the in vivo brain endothelium [60]. The high TEER value and enhanced size-dependent selectivity of FITC dextran observed with hiPS-BMEC could result from their ability to form stronger and more stable TJs between adjacent cells than HBEC, as seen by ZO-1, VE-cadherin and Claudin-5 junctional continuity and barrier integrity (Fig. 2D).
3.4. Confirmation of the transferrin receptor-mediated transcytosis
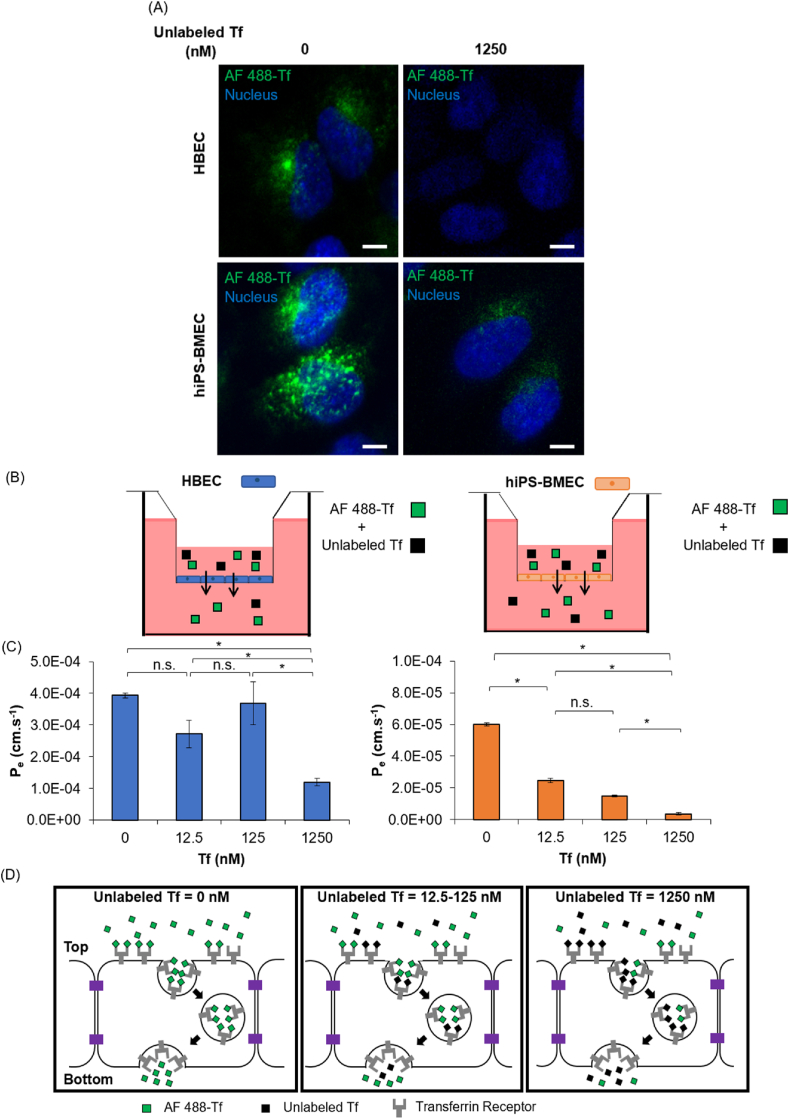
The ability of HBEC and hiPS-BMEC to use RMT pathway for the transport of molecules was investigated, with a particular focus on the transferrin receptor (TfR)-mediated transcytosis. In order to validate the interaction between Alexa Fluor 488-transferrin (AF 488-Tf) and TfR, a competition binding was performed using AF 488-Tf in the presence of an excess of unlabeled Tf, the native ligand of TfR [25]. The exact localization of AF 488-Tf was evaluated with HBEC and hiPS-BMEC using confocal laser scanning microscopy. Interestingly, higher amounts of AF 488-Tf can be observed inside hiPS-BMEC than with HBEC in the absence of unlabeled Tf (Fig. 4A, Fig. S4), which can be explained by the higher expression of TfR by hiPS-BMEC (Fig. 2B). Both HBEC and hiPS-BMEC showed higher fluorescent signals of AF 488-Tf in the cytoplasm in the absence of unlabeled Tf than when co-incubated with a 10-fold excess of unlabeled Tf (1250 nM) as compared to that of AF 488-Tf (125 nM). The reduced internalization of AF 488-Tf observed inside HBEC and hiPS-BMEC potentially resulted from the competition between AF 488-Tf and unlabeled Tf for the same or overlapping binding region on TfR. The co-incubation with an excess of unlabeled Tf impaired the possibility of AF 488-Tf to bind to TfR, leading to a decrease of the fluorescence signal inside the cells. A similar phenomenon was previously reported, where human serum Tf showed a dose-dependent inhibition of the binding of radiolabeled human serum Tf (125I-hTf) to TfR [61],[62]. By increasing the unlabeled Tf, a lower signal for the radiolabeled Tf was found in the cell lysates, confirming the lower cell internalization of 125I-hTf. Collectively, these results suggest that the fluorescent labeling of Tf does not affect the possibility to bind to TfR, thus AF 488-Tf can be used for monitoring the binding to TfR.
Fig. 4.
(A) Localization of Alexa Fluor 488 labeled Tf (AF 488-Tf) with the co-incubation without or with 1250 nM of unlabeled transferrin (Tf) using HBEC and hiPS-BMEC, images were taken after 1 h incubation at 37 °C. Scale bar = 10 μm. (B) Schematic illustration of the method. The medium in the bottom compartment was collected at predetermined time to measure fluorescence intensity. (C) Effective permeability of 125 nM of AF 488-Tf with co-incubation at various concentrations (0; 12.5; 125; 1250 nM) of unlabeled Tf with HBEC (blue) and hiPS-BMEC (orange). Data are presented as means ± S.D. Statistical analysis was performed using one-way ANOVA (∗p ≤ 0.05, n. s. p > 0.05). The biological replicates of HBEC and hiPS-BMEC come from three different differentiations. (D) Proposed mechanism of the permeation of 125 nM of AF 488-Tf when co-incubated with or without 1250 nM of unlabeled Tf by hiPS-BMEC. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Since the cellular uptake of AF 488-Tf could be clearly visualized by confocal imaging when co-incubated with unlabeled Tf, we sought to investigate the possibility to modulate the permeation rate of AF 488-Tf across the monolayer of HBEC or hiPS-BMEC by using a quantitative competition binding assay to TfR. For that, we quantified the permeability of AF 488-Tf during the co-incubation with an increasing concentration of unlabeled Tf in a transwell assay (Fig. 4B). The permeability of AF 488-Tf reached a maximum Pe of 3.9 × 10−4 cm/s and 6.0 × 10−5 cm/s in the absence of Tf with HBEC and hiPS-BMEC respectively. When co-incubated with a concentration of unlabeled Tf of 12.5 and 125 nM, a high fraction of AF 488-Tf was transported in the bottom compartment of the insert for HBEC but significantly decreased for hiPS-BMEC (Fig. 4C). The Pe value of AF 488-Tf significantly decreased only above the concentration of 125 nM of unlabeled Tf for HBEC, whereas a concentration-dependent decrease of the Pe value of AF 488-Tf was found for hiPS-BMEC, indicating AF 488-Tf transport was dependent on the binding to TfR. The PSm values of AF 488-Tf was in the same range, independently of the concentration of unlabeled Tf which was co-incubated during the competition assay to TfR (Fig. S5). It confirmed that the actual concentration of AF 488-Tf in the donor side of the transwell was not lowered due to the adsorption in the culture plate or membrane of the insert or potential aggregation between transferrin molecules, but mainly resulted from the transportation efficiency of AF 488-Tf by HBEC or hiPS-BMEC.
Although the presence of RMT transport was confirmed with HBEC, these cells are probably not the most suitable endothelial cell source for the measurement of the modulation of the permeation of RMT ligands, such as AF 488-Tf, due to the contribution of a potential additional paracellular transportation of Tf. Fig. 4D showed the expected mechanism to explain the competition binding assay to TfR by hiPS-BMEC. The presence of an increased concentration of unlabeled Tf diminished the probability of AF 488-Tf binding to TfR, leading to the decrease of endocytosis of AF 488-Tf. It is of note that the addition of 125 nM of AF 488-Tf was strongly inhibited by only 12.5 nM of unlabeled Tf, as observed in Fig. 4A, which may be attributed to a higher affinity of unlabeled Tf as compared to AF 488-Tf. This potential change of affinity of AF 488-Tf for TfR could be explained by the presence of the fluorescent labeling. Some precautions may need to be taken when using fluorescent labeling as a detection method for the measurement of the permeability of Tf, as it could potentially affect the affinity of Tf to TfR, thus the permeation rate. The radiolabeling of Tf has been indeed reported to decrease the affinity to TfR [63], such phenomenon may also occur with the fluorescence labeling of Tf. A comparison of the binding affinity between unlabeled Tf or AF 488-Tf to the TfR by the measurement of their equilibrium dissociation constant could be helpful to validate such hypothesis.
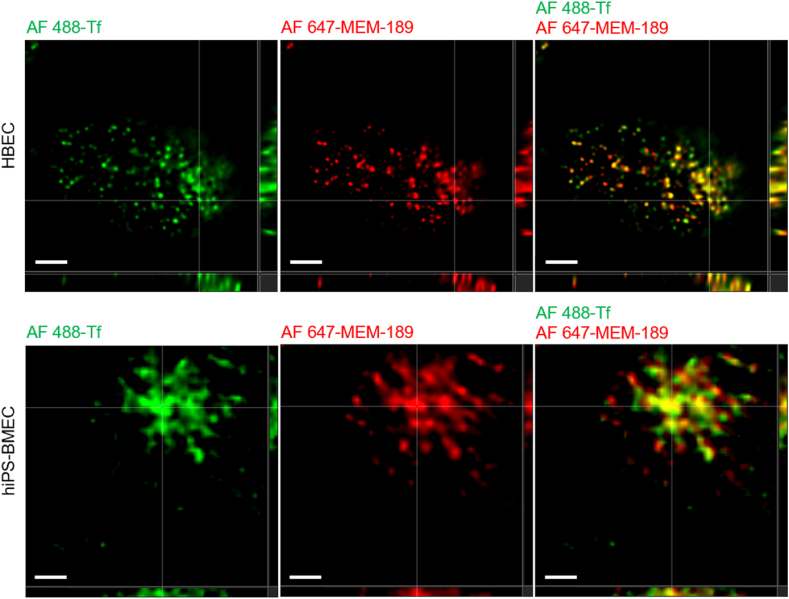
To confirm that AF 488-Tf is able to interact with the TfR despite the fluorescence labeling, we performed a colocalization study with Alexa Fluor 647-MEM-189 (AF 647-MEM-189), a transportable antibody targeting TfR. As observed in Fig. 5, Alexa Fluor 488-transferrin (AF 488-Tf) could be transported into their cytoplasm (Fig. S6). The colocalization of AF 488-Tf and AF 647-MEM-189 confirm that AF 488-Tf was endocytosed by HBEC and hiPS-BMEC by binding to the TfR. The internalization of the Tf-TfR complex leading to release of iron from the ligand is associated with an endosomal acidification [64],[65]. The colocalization ratio between lysotracker deep red, a specific stain of acidic vesicles, and AF 488-Tf further validate that both cell types used this specific transportation route for the intracellular accumulation of Tf in their cytoplasm (Fig. S7). Additionally, AF 488-Tf was shown to be transported faster from apical to basolateral side than basolateral to apical side of hiPS-BMEC (Fig. S8). It confirms the polarized expression of the TfR on cell membrane, with a predominance on the apical side, similarly reported in previous study for brain endothelial cells [66].
Fig. 5.
Confocal images of HBEC (top) and hiPS-BMEC (bottom) after the co-incubation of 125 nM of Alexa Fluor 488-transferrin (AF 488-Tf) and 64 nM of Alexa Fluor 647-MEM-189 (Anti-Human Transferrin Receptor antibody) for 1 h at 37 °C. Scale bar = 2 μm.
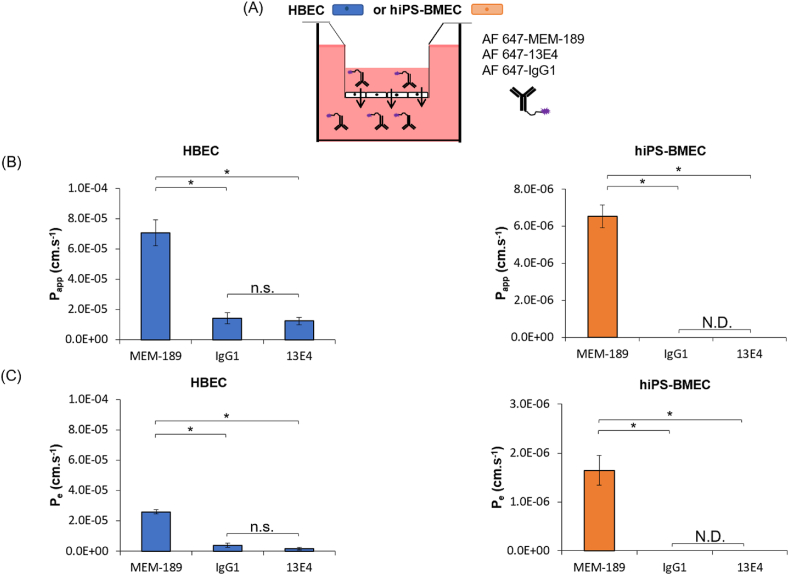
To gain further insights of the permeability of large-sized TfR ligands with HBEC and hiPS-BMEC, we next compared the permeability of AF 647-MEM-189, with the high-affinity human anti-TfR antibody (13E4) and the mouse immunoglobulin G1 isotype control (IgG1), molecules reported to have a poor transportability [67],[68]. As seen in Fig. 6, hiPS-BMEC showed the possibility to efficiently transport AF 647-MEM-189 with a Papp value of 6.5 × 10−6 cm/s and Pe value of 1.7 × 10−6 cm/s, further validating the presence of TfR-mediated transport. Some differences in the permeability of large-sized TfR ligands were observed between HBEC and hiPS-BMEC. AF 647-MEM-189 showed indeed a higher permeability with HBEC as compared to hiPS-BMEC with a Papp value around 7.1 × 10−5 cm/s (Fig. 6B). It could be explained by the “leaky” properties of HBEC, which showed an increased permeability for FITC-dextran with a MW < 70 kDa as compared to hiPS-BMEC (Fig. 3B). It suggests that AF 647-MEM-189, which has a MW∼156.2 kDa, may be transported by both transcellular and paracellular transport with HBEC, but to a lesser extent with hiPS-BMEC. The AF 647-13E4 antibody and AF 647-IgG1 also showed a high permeability for HBEC, with a Papp value of 1.23 × 10−6 cm/s and 1.41 × 10−6 cm/s and with a Pe value of and 1.8 × 10−6 cm/s and 3.9 × 10−6 cm/s respectively. Conversely, hiPS-BMEC were totally impermeable for AF 647-13E4 and AF 647-IgG1, during the permeability assay, as the fluorescence signal was below the detection limit for these two fluorescent molecules (<100 ng/mL). These results confirm the hypothesis about the additional paracellular transport of large-sized molecules by HBEC, such as AF 647-MEM-189, AF 647-IgG1 or AF 647-13E4, as compared to hiPS-BMEC. The fact that HBEC also have an unstable RMT-mediated permeability was already observed from Fig. 4 when Tf was labeled with AF 488, the fluorescent labeling potentially changing its transport efficiency towards an increased paracellular transport. Taken together, these results confirmed that hiPS-BMEC should be preferred for the accurate measurement of the permeation rates of TfR ligands due to the reduced contribution of the paracellular route as compared to HBEC. Especially, hiPS-BMEC seem to be more reliable for the measurement of the permeation of large-sized molecules such as antibodies.
Fig. 6.
Permeation of antibodies binding to TfR using HBEC and hiPS-BMEC (n = 3). (A) Schematic illustration of the method. The medium in the bottom compartment was collected at determined time to measure fluorescence intensity. (B) Apparent permeability coefficient of 64 nM of AF 647-MEM-189, AF 647-13E4 or AF 647-IgG1. Data are presented as means ± S.D. N.D. means “not detected”. (C) Effective permeability coefficient of 64 nM of AF 647-MEM-189, AF 647-13E4 or AF 647-IgG1 after 1 h incubation at 37 °C. Data are presented as means ± S.D. N.D. means “not detected”. The biological replicates of HBEC and hiPS-BMEC come from three and two different differentiations respectively.
3.5. Screening of cysteine modified T7 or TfR-T12 peptides targeting TfR by competition binding assay with unlabeled Tf
We next investigated whether HiPS-BMEC could be also useful for investigating the permeation of molecules with smaller size, such as peptides binding to TfR. The HAIYPRH (T7) peptide and the THRPPMWSPVWP (Tfr-T12) peptide, discovered by phage display, have been reported as potential ligands of TfR [35]. These peptides have been reported to exhibit a high binding affinity to TfR in the nM range, with a dissociation constant of Kd = 10 and 15 nM for the T7 peptide [69] and Tfr-T12 peptide [35], respectively. These values are comparable to the native ligand, Tf (Kd = 2.8 nM) [35]. The T7 and Tfr-T12 peptides have thus been widely used as ligands for constructing tumor-targeted nanodrug delivery systems [[69], [70], [71], [72], [73], [74]].
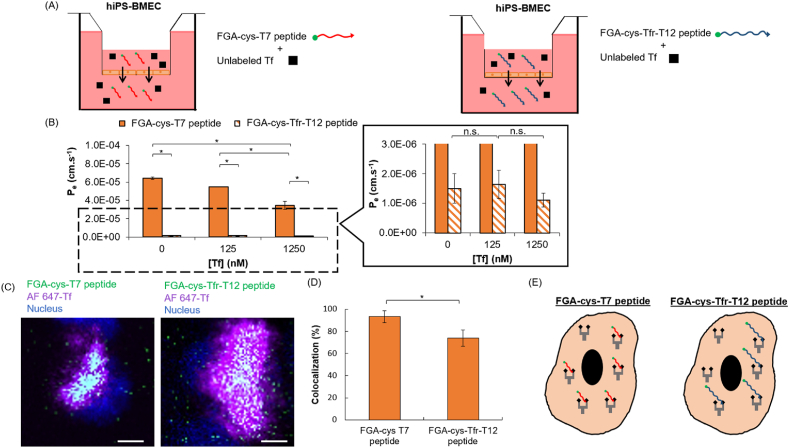
Although it has already been reported in the literature that the binding site of the T7 and the Tfr-T12 peptide on TfR was identified to be different from that of the Tf to TfR [35], we wanted to confirm that the site of the fluorescein glycine amide (FGA) labeled cysteine-modified peptides (FGA-cys-T7 and FGA-cys-Tfr-T12 peptide) also does not overlap with that of the Tf on TfR. To verify this, a competition binding assay for TfR was performed by the co-incubation of an increasing concentration of unlabeled Tf and by maintaining the same concentration of the FGA-cys-T7 or FGA-cys-Tfr-T12 peptide (Fig. 7A). As previously stated, both HBEC and hiPS-BMEC are both available to visualize TfR-mediated transcytosis (Fig. 5), but for this permeability assessment, hiPS-BMEC were chosen instead of HBEC since the RMT transport of HBEC was previously found unstable when using labeled proteins such as the AF 488-Tf in the transwell permeability assay (Fig. 4). For hiPS-BMEC, the permeability of both peptides was drastically reduced when co-incubated with a high concentration of unlabeled Tf (1250 nM) (Fig. 7B, Fig. S9). The dose-dependent inhibition of the FGA-cys-T7 peptide permeability showed a similar trend to the one found in the competition binding assay between AF 488-Tf and unlabeled Tf with hiPS-BMEC (Fig. 4). Conversely, the FGA-cys-Tfr-T12 peptide did not show any drastic changes in its permeability. This suggests that the cell transport of the FGA-cys-T7 peptide by hiPS-BMEC may follow the same mechanism as Tf, namely the RMT pathway. To further validate this possibility, the exact localization of FGA-cys-T7 and FGA-cys-Tfr-T12 peptide was then investigated using hiPS-BMEC. The colocalization percentage with AF 647-Tf was higher for the FGA-cys-T7 peptide than for FGA-cys-Tfr-T12 using hiPS-BMEC (Fig. 7C and D). Additionally, the colocalization ratio between lysotracker deep red, indicator of the endocytic process, and the FGA-cys-T7 peptide was higher than with the FGA-cys-Tfr-T12 peptide (Figs. S10–S11). Interestingly, the FGA-cys-T7 peptide and AF 488-Tf showed a similar colocalization ratio with lysotracker deep red (Fig. S11), demonstrating that the FGA-cys-T7 peptide and AF 488-Tf shared a similar ability to be endocytosed. Conversely, the FGA-cys-Tfr-T12 peptide showed a lower colocalization ratio with lysotracker deep red. The FGA-cys-T7 peptide seems slightly more likely able to bind the TfR in presence of AF 488-Tf on TfR, as compared to FGA-cys-Tfr-T12 peptide (Fig. 7E). To shed light on the cell internalization mechanism of these peptides, hiPS-BMEC were incubated with either the FGA-cys-T7 peptide or the FGA-cys-Tfr-T12 peptide after inhibiting the clathrin-mediated pathway. It is well established that clathrin-coated pits of the plasma membrane are involved in the RMT pathway, including TfR-mediated endocytosis. In this experiment, hiPS-BMEC were next used to demonstrate the potential involvement of clathrin-mediated pathway (Fig. 5). For this purpose, hiPS-BMEC insert cultures were treated with the potent clathrin inhibitor PitStop®2, which is known to interfere with receptor-mediated endocytosis, including the cell uptake of Tf using TfR [75]. As seen in Fig. S12, no shape of vesicles containing aggregates of FGA-peptides could be found in the cell cytoplasm, which is different as than the results found without inhibition treatment (Fig. S10), suggesting a successful inhibition of the endocytosis of both peptides. These results confirm that these peptides are preferentially internalized by the RMT pathway by the binding to TfR. Interestingly, a higher fluorescence signal could still be found with FGA-cys-Tfr-T12 than FGA-cys-T7 peptide after clathrin inhibition, suggesting FGA-cys-Tfr-T12 peptide could also potentially use the clathrin-independent endocytosis for its internalization by hiPS-BMEC.
Fig. 7.
Competitive binding assay between 10 μg/mL of FGA-cys-T7 and FGA-cys-Tfr-T12 peptide and various concentrations (0; 125; 1250 nM) of unlabeled Tf with hiPS-BMEC (n = 3). (A) Schematic illustration of the method. The medium in the bottom compartment was collected at determined time to measure fluorescence intensity. (B) Effective permeability coefficient of 10 μg/mL FGA-cys-T7 peptide (full bars) and FGA-cys-Tfr-T12 peptide (hatched bars) when co-incubated with various concentrations of unlabeled Tf by hiPS-BMEC after 1 h incubation at 37 °C. Data are presented as means ± S.D. Statistical analysis was performed using one-way ANOVA (∗p ≤ 0.05, n. s. p > 0.05). The biological replicates of hiPS-BMEC are issued from the same differentiation. (C) Colocalization of FGA-cys-T7 or FGA-cys-Tfr-T12 peptide and Alexa Fluor 647- transferrin (AF 647-Tf) with hiPS-BMEC. Scale bar = 10 μm. The biological replicates of hiPS-BMEC are issued from the same differentiation. (D) Quantification of the colocalization percentage between FGA-peptides and AF 647-Tf using hiPS-BMEC calculated with IMARIS software. Data are presented as means ± S.D. Statistical analysis was performed using the Student's t-test (∗p ≤ 0.05). (E) Schematic illustration of the interaction between FGA-cys-T7 or FGA-cys-Tfr-T12 peptide and Tf for the binding to TfR with hiPS-BMEC.
4. Conclusion
Overall, this work showed that hiPS-BMEC could represent a better alternative than the immortalized cell line HBEC to mimic the functional properties of the BBB endothelium. The gene and protein expression of tight junction proteins and transporters was globally upregulated with hiPS-BMEC compared to HBEC, excepting for the endothelial markers CD31 and VE-cadherin. Several works have actually identified that the current differentiation procedures to obtain hiPS-BMEC actually led to a mixed endothelial-epithelial profile of hiPS-BMEC [17,[76], [77], [78]]. While the hiPS-BMEC in vitro model superiority was confirmed in this study, compared to the immortalized cell line HBEC, further validations concerning the degree of accuracy of the data might be needed regarding their genes and proteins expression profile versus complementary system or experimental techniques, such as in vivo models and human tissue, as recommended by Lippmann et al. [79]. Although precautions should be taken when using hiPS-BMEC for mimicking the physiological phenomenon of human BMEC, the ability to recapitulate the functional properties of human BBB must be regarded as an important criterion for judging their suitability for the development of BBB models.
Even though both cell types globally showed the same trend in the permeability of the tested molecules, HBEC exhibited an additional paracellular transport of large-sized molecules compared to hiPS-BMEC, due to their weak expression of tight junctions. The leakiness properties of HBEC are therefore not suitable for the accurate measurement of the permeability of tested molecules, which is fundamental for improving in vitro drug testing accuracy and the “bench-to-bedside’’ transition of brain cancer drugs. Additionally, the permeability of two peptides transported by the RMT pathway was also confirmed, with a higher permeability of the cys-T7 peptide than the cys-Tfr-T12 peptide by hiPS-BMEC. The cys-T7 peptide could therefore serve as an efficient targeting moiety to enhance the delivery of therapeutics through the BBB endothelium.
Finally, although cell monolayer is a handful model to directly study inherent properties of HBEC and hiPS-BMEC, the incorporation of astrocytes and pericytes could be useful to represent more closely native-like environment. The BBB functions of cocultured BMEC with astrocytes and/or pericytes were found to be greater than those of monocultured ones [39,80], including the upregulation of the expression of TJ and transporters, such as the TfR. The incorporation of human astrocytes and pericytes could enhance even more the functional properties of the monoculture model of hiPS-BMEC used in this study. The increased complexity of this in vitro model would greatly benefit the study of the passage of molecules mediated by the TfR, such as the TfR-targeted antibodies and peptides used in this study, as it would more closely recapitulate the native structure of the brain endothelium.
Author contributions
MP carried out the experiments. MP and FL wrote and edited the manuscript. TY, KKa provided hiPS-BMEC. MP, FL, YSM, KKi, KS, KKa, SY, SI, HH, MM contributed to the design and implementation of the research and to the analysis of the results. All authors have seen and approved the final manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
The authors wish to thank Prof. Furihata from the School of Pharmacy, Tokyo University of Pharmacy and Life Sciences (Hachioji, Tokyo, Japan) for providing human brain microvascular endothelial cells/conditionally immortalized clone ci18 (HBEC) which were used in this work.
This work was supported by a Grant-in-Aid for Scientific Research (A) (20H00665), and Bilateral Joint Research Projects of the JSPS (20199946), as well as AMED-MPS (21be0304207h0005) and COCKPI-T funding of Takeda Pharmaceutical Company Limited.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mtbio.2022.100232.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
DLS measurements, apparent and effective permeability coefficients of the tested molecules, immunofluorescence staining are displayed on Supporting Information.
References
- 1.DiMasi J.A., Feldman L., Seckler A., Wilson A. Trends in risks associated with new drug development: success rates for investigational drugs. Clin. Pharmacol. Therapeut. 2010;87(3):272–277. doi: 10.1038/clpt.2009.295. [DOI] [PubMed] [Google Scholar]
- 2.Blanco M.-J., Gardinier K.M. New chemical modalities and strategic thinking in early drug discovery. ACS Med. Chem. Lett. Mar. 2020;11(3):228–231. doi: 10.1021/acsmedchemlett.9b00582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Culot M., et al. An in vitro blood-brain barrier model for high throughput (HTS) toxicological screening. Toxicol. Vitro. Apr. 2008;22(3):799–811. doi: 10.1016/j.tiv.2007.12.016. [DOI] [PubMed] [Google Scholar]
- 4.Gribkoff V.K., Kaczmarek L.K. The need for new approaches in CNS drug discovery: why drugs have failed, and what can Be done to improve outcomes. Neuropharmacology. Jul. 2017;120:11–19. doi: 10.1016/j.neuropharm.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cecchelli R., et al. In vitro model for evaluating drug transport across the blood–brain barrier. Adv. Drug Deliv. Rev. Apr. 1999;36(2):165–178. doi: 10.1016/S0169-409X(98)00083-0. [DOI] [PubMed] [Google Scholar]
- 6.Pardridge W.M. The blood-brain barrier: bottleneck in brain drug development. NeuroRx. Jan. 2005;2(1):3–14. doi: 10.1602/neurorx.2.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joó F. Endothelial cells of the brain and other organ systems: some similarities and differences. Prog. Neurobiol. Feb. 1996;48(3):255–273. doi: 10.1016/0301-0082(95)00046-1. [DOI] [PubMed] [Google Scholar]
- 8.Uwamori H., Ono Y., Yamashita T., Arai K., Sudo R. Comparison of organ-specific endothelial cells in terms of microvascular formation and endothelial barrier functions. Microvasc. Res. Mar. 2019;122:60–70. doi: 10.1016/j.mvr.2018.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abbott N.J., Rönnbäck L., Hansson E. Astrocyte–endothelial interactions at the blood–brain barrier. Nat. Rev. Neurosci. Jan. 2006;7(1) doi: 10.1038/nrn1824. [DOI] [PubMed] [Google Scholar]
- 10.Wong A.D., Ye M., Levy A.F., Rothstein J.D., Bergles D.E., Searson P.C. The blood-brain barrier: an engineering perspective. Front. Neuroeng. Aug. 2013;6 doi: 10.3389/fneng.2013.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reichel A., Begley D.J., Abbott N.J. In: The Blood-Brain Barrier: Biology and Research Protocols. Nag S., editor. Humana Press; Totowa, NJ: 2003. An overview of in vitro techniques for blood-brain barrier studies; pp. 307–324. [DOI] [PubMed] [Google Scholar]
- 12.Neuhaus W. Handb Exp Pharmacol; Jun. 2020. In Vitro Models of the Blood-Brain Barrier. [DOI] [PubMed] [Google Scholar]
- 13.Piantino M., Figarol A., Matsusaki M. Three-dimensional in vitro models of healthy and tumor brain microvasculature for drug and toxicity screening. Front. Toxicol. 2021;3 doi: 10.3389/ftox.2021.656254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang S., et al. Identification of two immortalized cell lines, ECV304 and bEnd3, for in vitro permeability studies of blood-brain barrier. PLoS One. Oct. 2017;12(10):e0187017. doi: 10.1371/journal.pone.0187017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weksler B., Romero I.A., Couraud P.-O. The hCMEC/D3 cell line as a model of the human blood brain barrier. Fluids Barriers CNS. Mar. 2013;10(1):16. doi: 10.1186/2045-8118-10-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lippmann E.S., et al. Human blood-brain barrier endothelial cells derived from pluripotent stem cells. Nat. Biotechnol. Aug. 2012;30(8):783–791. doi: 10.1038/nbt.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lippmann E.S., Al-Ahmad A., Azarin S.M., Palecek S.P., Shusta E.V. A retinoic acid-enhanced, multicellular human blood-brain barrier model derived from stem cell sources. Sci. Rep. Feb. 2014;4(1) doi: 10.1038/srep04160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurosawa T., et al. Expression and functional characterization of drug transporters in brain microvascular endothelial cells derived from human induced pluripotent stem cells. Mol. Pharm. Dec. 2018;15(12):5546–5555. doi: 10.1021/acs.molpharmaceut.8b00697. [DOI] [PubMed] [Google Scholar]
- 19.Campisi M., Shin Y., Osaki T., Hajal C., Chiono V., Kamm R.D. 3D self-organized microvascular model of the human blood-brain barrier with endothelial cells, pericytes and astrocytes. Biomaterials. Oct. 2018;180:117–129. doi: 10.1016/j.biomaterials.2018.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cecchelli R., et al. A stable and reproducible human blood-brain barrier model derived from hematopoietic stem cells. PLoS One. juin 2014;9(6):e99733. doi: 10.1371/journal.pone.0099733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ribecco-Lutkiewicz M., et al. A novel human induced pluripotent stem cell blood-brain barrier model: applicability to study antibody-triggered receptor-mediated transcytosis. Sci. Rep. Jan. 2018;8(1):1873. doi: 10.1038/s41598-018-19522-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dehouck B., Fenart L., Dehouck M.-P., Pierce A., Torpier G., Cecchelli R. A new function for the LDL receptor: transcytosis of LDL across the blood–brain barrier. JCB (J. Cell Biol.) Aug. 1997;138(4):877–889. doi: 10.1083/jcb.138.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Golden P.L., Maccagnan T.J., Pardridge W.M. Human blood-brain barrier leptin receptor. Binding and endocytosis in isolated human brain microvessels. J. Clin. Invest. Jan. 1997;99(1):14–18. doi: 10.1172/JCI119125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duffy K.R., Pardridge W.M. Blood-brain barrier transcytosis of insulin in developing rabbits. Brain Res. Sep. 1987;420(1):32–38. doi: 10.1016/0006-8993(87)90236-8. [DOI] [PubMed] [Google Scholar]
- 25.Jones A.R., Shusta E.V. Blood-brain barrier transport of therapeutics via receptor-mediation. Pharm. Res. (N. Y.) Sep. 2007;24(9):1759–1771. doi: 10.1007/s11095-007-9379-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qian Z.M., Li H., Sun H., Ho K. Targeted drug delivery via the transferrin receptor-mediated endocytosis pathway. Pharmacol. Rev. Dec. 2002;54(4):561–587. doi: 10.1124/pr.54.4.561. [DOI] [PubMed] [Google Scholar]
- 27.Calzolari A., et al. Transferrin receptor 2 is frequently and highly expressed in glioblastomas. Transl. Oncol. Apr. 2010;3(2):123–134. doi: 10.1593/tlo.09274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnsen K.B., Burkhart A., Thomsen L.B., Andresen T.L., Moos T. Targeting the transferrin receptor for brain drug delivery. Prog. Neurobiol. Oct. 2019;181:101665. doi: 10.1016/j.pneurobio.2019.101665. [DOI] [PubMed] [Google Scholar]
- 29.Pang Z., et al. Enhanced intracellular delivery and chemotherapy for glioma rats by transferrin-conjugated biodegradable polymersomes loaded with doxorubicin. Bioconjugate Chem. Jun. 2011;22(6):1171–1180. doi: 10.1021/bc200062q. [DOI] [PubMed] [Google Scholar]
- 30.Prades R., et al. Delivery of gold nanoparticles to the brain by conjugation with a peptide that recognizes the transferrin receptor. Biomaterials. Oct. 2012;33(29):7194–7205. doi: 10.1016/j.biomaterials.2012.06.063. [DOI] [PubMed] [Google Scholar]
- 31.Kang T., et al. Enhancing glioblastoma-specific penetration by functionalization of nanoparticles with an iron-mimic peptide targeting transferrin/transferrin receptor complex. Mol. Pharm. Aug. 2015;12(8):2947–2961. doi: 10.1021/acs.molpharmaceut.5b00222. [DOI] [PubMed] [Google Scholar]
- 32.Fan K., et al. Ferritin nanocarrier traverses the blood brain barrier and kills glioma. ACS Nano. May 2018;12(5):4105–4115. doi: 10.1021/acsnano.7b06969. [DOI] [PubMed] [Google Scholar]
- 33.Ruan S., et al. Acid-responsive transferrin dissociation and GLUT mediated exocytosis for increased blood–brain barrier transcytosis and programmed glioma targeting delivery. Adv. Funct. Mater. 2018;28(30) doi: 10.1002/adfm.201802227. [DOI] [Google Scholar]
- 34.Ito R., et al. A human immortalized cell-based blood–brain barrier triculture model: development and characterization as a promising tool for Drug−Brain permeability studies. Mol. Pharm. Nov. 2019;16(11):4461–4471. doi: 10.1021/acs.molpharmaceut.9b00519. [DOI] [PubMed] [Google Scholar]
- 35.Lee J.H., Engler J.A., Collawn J.F., Moore B.A. Receptor mediated uptake of peptides that bind the human transferrin receptor. Eur. J. Biochem. 2001;268(7):2004–2012. doi: 10.1046/j.1432-1327.2001.02073.x. [DOI] [PubMed] [Google Scholar]
- 36.Srinivasan B., Kolli A.R., Esch M.B., Abaci H.E., Shuler M.L., Hickman J.J. TEER measurement techniques for in vitro barrier model systems. J. Lab. Autom. Apr. 2015;20(2) doi: 10.1177/2211068214561025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schindelin J., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. Jul. 2012;9(7) doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sade H., Baumgartner C., Hugenmatter A., Moessner E., Freskgård P.-O., Niewoehner J. A human blood-brain barrier transcytosis assay reveals antibody transcytosis influenced by pH-dependent receptor binding. PLoS One. Apr. 2014;9(4) doi: 10.1371/journal.pone.0096340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakagawa S., et al. A new blood–brain barrier model using primary rat brain endothelial cells, pericytes and astrocytes. Neurochem. Int. Mar. 2009;54(3):253–263. doi: 10.1016/j.neuint.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 40.DeLisser H.M., et al. Involvement of endothelial PECAM-1/CD31 in angiogenesis. Am. J. Pathol. Sep. 1997;151(3):671–677. [PMC free article] [PubMed] [Google Scholar]
- 41.Privratsky J.R., Newman P.J. PECAM-1: regulator of endothelial junctional integrity. Cell Tissue Res. Mar. 2014;355(3):607–619. doi: 10.1007/s00441-013-1779-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chistiakov D.A., Orekhov A.N., Bobryshev Y.V. Endothelial PECAM-1 and its function in vascular physiology and atherogenic pathology. Exp. Mol. Pathol. Jun. 2016;100(3):409–415. doi: 10.1016/j.yexmp.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 43.Wimmer I., et al. PECAM-1 stabilizes blood-brain barrier integrity and favors paracellular T-cell diapedesis across the blood-brain barrier during neuroinflammation. Front. Immunol. 2019;10 doi: 10.3389/fimmu.2019.00711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hewitt K.J., Agarwal R., Morin P.J. The claudin gene family: expression in normal and neoplastic tissues. BMC Cancer. Jul. 2006;6(1):186. doi: 10.1186/1471-2407-6-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luissint A.-C., Artus C., Glacial F., Ganeshamoorthy K., Couraud P.-O. Tight junctions at the blood brain barrier: physiological architecture and disease-associated dysregulation. Fluids Barriers CNS. Nov. 2012;9(1):23. doi: 10.1186/2045-8118-9-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li W., Chen Z., Chin I., Chen Z., Dai H. The role of VE-cadherin in blood-brain barrier integrity under central nervous system pathological conditions. Curr. Neuropharmacol. Nov. 2018;16(9):1375–1384. doi: 10.2174/1570159X16666180222164809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fanning A.S., Jameson B.J., Jesaitis L.A., Anderson J.M. The tight junction protein ZO-1 establishes a link between the transmembrane protein occludin and the actin cytoskeleton. J. Biol. Chem. Nov. 1998;273(45):29745–29753. doi: 10.1074/jbc.273.45.29745. [DOI] [PubMed] [Google Scholar]
- 48.Umeda K., et al. ZO-1 and ZO-2 independently determine where claudins are polymerized in tight-junction strand formation. Cell. Aug. 2006;126(4):741–754. doi: 10.1016/j.cell.2006.06.043. [DOI] [PubMed] [Google Scholar]
- 49.Van Itallie C.M., Fanning A.S., Holmes J., Anderson J.M. Occludin is required for cytokine-induced regulation of tight junction barriers. J. Cell Sci. Aug. 2010;123(16):2844–2852. doi: 10.1242/jcs.065581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jefferies W.A., Brandon M.R., Hunt S.V., Williams A.F., Gatter K.C., Mason D.Y. Transferrin receptor on endothelium of brain capillaries. Nature. Nov. 1984;312(5990) doi: 10.1038/312162a0. [DOI] [PubMed] [Google Scholar]
- 51.Katt M.E., Xu Z.S., Gerecht S., Searson P.C. Human brain microvascular endothelial cells derived from the BC1 iPS cell line exhibit a blood-brain barrier phenotype. PLoS One. avr 2016;11(4):e0152105. doi: 10.1371/journal.pone.0152105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu P., Vogel C., Wang R., Yao X., Marcotte E.M. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat. Biotechnol. Jan. 2007;25(1) doi: 10.1038/nbt1270. [DOI] [PubMed] [Google Scholar]
- 53.Srinivasan B., Kolli A.R., Esch M.B., Abaci H.E., Shuler M.L., Hickman J.J. TEER measurement techniques for in vitro barrier model systems. J. Lab. Autom. Apr. 2015;20(2):107–126. doi: 10.1177/2211068214561025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Butt A.M., Jones H.C., Abbott N.J. Electrical resistance across the blood-brain barrier in anaesthetized rats: a developmental study. J. Physiol. 1990;429(1):47–62. doi: 10.1113/jphysiol.1990.sp018243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Crone C., Olesen S.P. Electrical resistance of brain microvascular endothelium. Brain Res. Jun. 1982;241(1):49–55. doi: 10.1016/0006-8993(82)91227-6. [DOI] [PubMed] [Google Scholar]
- 56.Mantle J.L., Min L., Lee K.H. Minimum transendothelial electrical resistance thresholds for the study of small and large molecule drug transport in a human in vitro blood–brain barrier model. Mol. Pharm. Dec. 2016;13(12):4191–4198. doi: 10.1021/acs.molpharmaceut.6b00818. [DOI] [PubMed] [Google Scholar]
- 57.Yu F., Kumar N.D.S., Foo L.C., Ng S.H., Hunziker W., Choudhury D. A pump-free tricellular blood–brain barrier on-a-chip model to understand barrier property and evaluate drug response. Biotechnol. Bioeng. 2020;117(4):1127–1136. doi: 10.1002/bit.27260. [DOI] [PubMed] [Google Scholar]
- 58.De Jong E., Williams D.S., Abdelmohsen L.K.E.A., Van Hest J.C.M., Zuhorn I.S. A filter-free blood-brain barrier model to quantitatively study transendothelial delivery of nanoparticles by fluorescence spectroscopy. J. Contr. Release. Nov. 2018;289:14–22. doi: 10.1016/j.jconrel.2018.09.015. [DOI] [PubMed] [Google Scholar]
- 59.Linville R.M., et al. Long-term cryopreservation preserves blood–brain barrier phenotype of iPSC-derived brain microvascular endothelial cells and three-dimensional microvessels. Mol. Pharm. Sep. 2020;17(9):3425–3434. doi: 10.1021/acs.molpharmaceut.0c00484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Easton A.S., Sarker M.H., Fraser P.A. Two components of blood-brain barrier disruption in the rat. J. Physiol. 1997;503(3):613–623. doi: 10.1111/j.1469-7793.1997.613bg.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang D., Lee H.-F., Pettit S.C., Zaro J.L., Huang N., Shen W.-C. Characterization of transferrin receptor-mediated endocytosis and cellular iron delivery of recombinant human serum transferrin from rice (Oryza sativaL.) BMC Biotechnol. Nov. 2012;12(1):92. doi: 10.1186/1472-6750-12-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pardridge W.M., Eisenberg J., Yang J. Human blood-brain barrier transferrin receptor. Metabolism. Sep. 1987;36(9):892–895. doi: 10.1016/0026-0495(87)90099-0. [DOI] [PubMed] [Google Scholar]
- 63.Fuchs H., Gessner R. Iodination significantly influences the binding of human transferrin to the transferrin receptor. Biochim. Biophys. Acta. Feb. 2002;1570(1) doi: 10.1016/s0304-4165(02)00146-0. [DOI] [PubMed] [Google Scholar]
- 64.Steere A.N., Byrne S.L., Chasteen N.D., Mason A.B. Kinetics of iron release from transferrin bound to the transferrin receptor at endosomal pH. Biochim. Biophys. Acta Gen. Subj. Mar. 2012;1820(3):326–333. doi: 10.1016/j.bbagen.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kleven M.D., Jue S., Enns C.A. The transferrin receptors, TfR1 and TfR2 bind transferrin through differing mechanisms. Biochemistry. Mar. 2018;57(9):1552–1559. doi: 10.1021/acs.biochem.8b00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Roberts R.L., Fine R.E., Sandra A. Receptor-mediated endocytosis of transferrin at the blood-brain barrier. J. Cell Sci. Feb. 1993;104(2):521–532. doi: 10.1242/jcs.104.2.521. [DOI] [PubMed] [Google Scholar]
- 67.Sade H., Baumgartner C., Hugenmatter A., Moessner E., Freskgård P.-O., Niewoehner J. A human blood-brain barrier transcytosis assay reveals antibody transcytosis influenced by pH-dependent receptor binding. PLoS One. avr 2014;9(4):e96340. doi: 10.1371/journal.pone.0096340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Park T.-E., et al. Hypoxia-enhanced Blood-Brain Barrier Chip recapitulates human barrier function and shuttling of drugs and antibodies. Nat. Commun. Jun. 2019;10(1) doi: 10.1038/s41467-019-10588-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Oh S., Kim B.J., Singh N.P., Lai H., Sasaki T. Synthesis and anti-cancer activity of covalent conjugates of artemisinin and a transferrin-receptor targeting peptide. Cancer Lett. Feb. 2009;274(1):33–39. doi: 10.1016/j.canlet.2008.08.031. [DOI] [PubMed] [Google Scholar]
- 70.Han L., Huang R., Liu S., Huang S., Jiang C. Peptide-conjugated PAMAM for targeted doxorubicin delivery to transferrin receptor overexpressed tumors. Mol. Pharm. Dec. 2010;7(6):2156–2165. doi: 10.1021/mp100185f. [DOI] [PubMed] [Google Scholar]
- 71.Tang J., et al. A stabilized retro-inverso peptide ligand of transferrin receptor for enhanced liposome-based hepatocellular carcinoma-targeted drug delivery. Acta Biomater. Jan. 2019;83:379–389. doi: 10.1016/j.actbio.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 72.Wang S., Sun H. Transferrin receptors targeting peptide (T7 peptide) surface-modified sorafenib nanoliposomes enhance the anti-tumor effect in colorectal cancer. Pharmaceut. Dev. Technol. Oct. 2020;25(9):1063–1070. doi: 10.1080/10837450.2020.1786116. [DOI] [PubMed] [Google Scholar]
- 73.Mu L.-M., et al. Lipid vesicles containing transferrin receptor binding peptide TfR-T 12 and octa-arginine conjugate stearyl-R 8 efficiently treat brain glioma along with glioma stem cells. Sci. Rep. Jun. 2017;7(1) doi: 10.1038/s41598-017-03805-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wängler C., et al. In vitro and initial in vivo evaluation of 68Ga-labeled transferrin receptor (TfR) binding peptides as potential carriers for enhanced drug transport into TfR expressing cells. Mol. Imag. Biol. Apr. 2011;13(2):332–341. doi: 10.1007/s11307-010-0329-6. [DOI] [PubMed] [Google Scholar]
- 75.von Kleist L., et al. Role of the clathrin terminal domain in regulating coated pit dynamics revealed by small molecule inhibition. Cell. Aug. 2011;146(3):471–484. doi: 10.1016/j.cell.2011.06.025. [DOI] [PubMed] [Google Scholar]
- 76.Vatine G.D., et al. Human iPSC-derived blood-brain barrier chips enable disease modeling and personalized medicine applications. Cell Stem Cell. Jun. 2019;24(6):995–1005. doi: 10.1016/j.stem.2019.05.011. e6. [DOI] [PubMed] [Google Scholar]
- 77.Delsing L., Herland A., Falk A., Hicks R., Synnergren J., Zetterberg H. Models of the blood-brain barrier using iPSC-derived cells. Mol. Cell. Neurosci. Sep. 2020;107:103533. doi: 10.1016/j.mcn.2020.103533. [DOI] [PubMed] [Google Scholar]
- 78.Lu T.M., et al. Pluripotent stem cell-derived epithelium misidentified as brain microvascular endothelium requires ETS factors to acquire vascular fate. Proc. Natl. Acad. Sci. U.S.A. Feb. 2021;118(8) doi: 10.1073/pnas.2016950118. p. e2016950118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lippmann E.S., Azarin S.M., Palecek S.P., Shusta E.V. Commentary on human pluripotent stem cell-based blood–brain barrier models. Fluids Barriers CNS. Oct. 2020;17(1):64. doi: 10.1186/s12987-020-00222-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Canfield S.G., et al. An isogenic blood–brain barrier model comprising brain endothelial cells, astrocytes, and neurons derived from human induced pluripotent stem cells. J. Neurochem. 2017;140(6) doi: 10.1111/jnc.13923. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.