Abstract
The coenzyme A (CoA) transferases are a superfamily of proteins central to the metabolism of acetyl‐CoA and other CoA thioesters. They are diverse group, catalyzing over a 100 biochemical reactions and spanning all three domains of life. A deeply rooted idea, proposed two decades ago, is these enzymes fall into three families (I, II, and III). Here we find they fall into different families, which we achieve by analyzing all CoA transferases characterized to date. We manually annotated 94 CoA transferases with functional information (including rates of catalysis for 208 reactions) from 97 publications. This represents all enzymes we could find in the primary literature, and it is double the number annotated in four protein databases (BRENDA, KEGG, MetaCyc, UniProt). We found family I transferases are not closely related to each other in terms of sequence, structure, and reactions catalyzed. This family is not even monophyletic. These problems are solved by regrouping the three families into six, including one family with many non‐CoA transferases. The problem (and solution) became apparent only by analyzing our large set of manually annotated proteins. It would have been missed if we had used the small number of proteins annotated in UniProt and other databases. Our work is important to understanding the biology of CoA transferases. It also warns investigators doing phylogenetic analyses of proteins to go beyond information in databases.
Keywords: coenzyme A transferases, databases, enzymology, evolution, phylogenetics
1. INTRODUCTION
Acetyl coenzyme A (CoA) and other acyl‐CoA compounds are central in metabolism. Reactions that form or consume these compounds are thus of great interest in biochemistry. The CoA transferases (EC 2.8.3._) are a superfamily that catalyze one such set of reactions. Specifically, they transfer of a CoA group from an acyl‐CoA donor to a carboxylate acceptor. 1 In total, they catalyze over 100 such reactions, each involving a different donor/acceptor pair.
The diversity of these enzymes and the reactions they catalyze has led to several ways of classifying them. 1 , 2 , 3 , 4 , 5 , 6 , 7 , 8 , 9 One of the first and most enduring ways is now two decades old. In 2001, Heider 1 named three families of transferases (I, II, and III). Members of family I are classical CoA transferases and use an acyl‐CoA as a substrate. Members of family II are distinct by being able to use acyl‐[acyl‐carrier protein], also. Members of family III have amino acid sequences that differ from other families. Prior to Heider, 1 it was thought different families may exist, 3 , 10 but Heider 1 was crucial to naming the differences. Heider 1 used only 16 named enzymes, which reflected the amount of evidence then available.
Since that time, investigators have proposed changes to the three families, but changes have been minor. For example, investigators have examined family I transferases and proposed splitting them into different subfamilies. 4 , 8 , 9 One problem is that rather than analyze all CoA transferases, investigators have focused on only a few enzymes, usually those closely related to an enzyme just discovered. 4 , 6 , 7 , 8 , 9 There has been no attempt to go back, find all CoA transferases, and determine if the original families are still appropriate.
The situation with CoA transferases represents a common problem in analyses of protein families. It is laborious to go back to the primary literature and find all members of a family or superfamily, and so analyses often rely on a few well‐known proteins and sequence homologs. Another approach is to find proteins through UniProt 11 or other databases. 12 , 13 , 14 Databases are popular because they have large numbers of proteins, and some proteins are already annotated with functional information from the literature. Indeed, several analyses of protein families have used databases as a major or sole source of functional information. 15 , 16 , 17 , 18 , 19 However, it is not clear how complete is information in databases and if it can substitute for a search of the primary literature.
Here we find, annotate, and analyze nearly 100 experimentally characterized CoA transferases from the primary literature. This analysis shows the three traditional families of CoA transferases are not appropriate, and they need to be regrouped into six to reflect evolutionary relationships. Importantly, the six families were readily apparent when only analyzing our large number of manually annotated enzymes. If we used the small numbers of proteins annotated in UniProt and three other databases, the families were not as apparent. Our study of CoA transferases serves as a warning and shows phylogenetic analyses need to go beyond the small numbers of proteins in databases.
2. RESULTS
2.1. Our analysis involves nearly 100 experimentally characterized enzymes
The CoA transferases have been divided into three families, but this classification is based on analyzing few enzymes. 1 We aimed to do an analysis with all experimentally characterized enzymes reported in the literature. Accordingly, we looked for all enzymes with (a) experimental evidence for catalyzing at least one reaction and (b) an amino acid sequence. Enzymes with catalytic activity inferred by homology, but with no experimental evidence, were not considered (except where noted).
In total, we found 94 enzymes described in 97 publications (Table 1 and Table S1). These publications measured rates of 208 CoA transferase reactions (Table S2). Of these, 105 reactions occurred at rates we considered biochemically significant and were included subsequently in our analysis (see Table S2 and Materials and Methods). In addition to bona fide CoA transferases, we included n = 5 enzymes that do not catalyze CoA transferase reactions but are closely related (indistinguishable from sequence alone).
TABLE 1.
Coenzyme A transferases that have been experimentally characterized in the primary literature a
| Enzyme | Organism | References | Enzyme | Organism | References |
|---|---|---|---|---|---|
| AarC (Aac) | Acetobacter aceti | [20, 21] | FldA (Csp) | Clostridium sporogenes | [22] |
| AbfT (Cam) | Clostridium aminobutyricum | [23, 24, 25] | Frc (Lac) | Lactobacillus acidophilus | [26] |
| ACH1 (Sce) | Saccharomyces cerevisiae | [27] | Frc (Ofo) | Oxalobacter formigenes | [29, 30, 31] |
| Act (Fsp) | Firmicutes sp. | [32] | GctAB (Afe) | Acidaminococcus fermentans | [33, 34, 35, 36] |
| Act (Vpa) | Variovorax paradoxus | [37] | HadA (Cdi) | Clostridium difficile | [38] |
| ActA (Cgl) | Corynebacterium glutamicum | [39] | IaaL (Aar) | Aromatoleum aromaticum | [7] |
| AibAB (Mxa) | Myxococcus xanthus | [40] | IctA (Ate) | Aspergillus terreus | [41] |
| Asct (Fhe) | Fasciola hepatica | [42] | IpdAB (Mtu) | Mycobacterium tuberculosis | [43] |
| AtoDA (Eco) | Escherichia coli | [44, 45] | IpdAB (Rjo) | Rhodococcus jostii | [43] |
| BaiF (Csc) | Clostridium scindens | [46] | MadA (Mru) | Malonomonas rubra | [3, 28] |
| BaiK (Csc) | Clostridium scindens | [46] | Mcr (Mtu) | Mycobacterium tuberculosis | [47] |
| BbsEF (Tar) | Thauera aromatica | [48, 49] | Mct (Cau) | Chloroflexus aurantiacus | [50] |
| Bct (Gme) | Geobacter metallireducens | [51] | Mct (Hhi) | Haloarcula hispanica | [52] |
| CaiB (Atu) | Agrobacterium tumefaciens | [53] | MdcA (Aca) | Acinetobacter calcoaceticus | [54] |
| CaiB (Eco) | Escherichia coli | [55, 56, 57] | MdcA (Kpn) | Klebsiella pneumoniae | [58, 59] |
| CaiB (Psp) | Proteus sp. | [60] | MdcA (Ppu) | Pseudomonas putida | [61] |
| CarA (Awo) | Acetobacterium woodii | [62] | OXCT1 (Hsa) | Homo sapiens | [63, 64] |
| Cat1 (Aba) | Acinetobacter baumannii | [65] | Oxct1 (Mmu) | Mus musculus | [66] |
| Cat1 (Aca) | Anaerostipes caccae | [67] | Oxct1 (Rno) | Rattus norvegicus | [68] |
| Cat1 (Ace) | Acetobacter cerevisiae | [65] | OXCT1 (Ssc) | Sus scrofa | [69, 70, 71, 72] |
| Cat1 (Asp) | Anaerostipes sp. | [30] | p49 (Afr) | Artemia franciscana | [73] |
| Cat1 (Bfr) | Bacteroides fragilis | [65] | PcaIJ (Aba) | Acinetobacter baylyi | [74, 75] |
| Cat1 (Bsp) | Butyricicoccus sp. | [76] | PcaIJ (Atu) | Agrobacterium tumefaciens | [77] |
| Cat1 (Cdi) | Corynebacterium diphtheriae | [65] | PcaIJ (Ppu) | Pseudomonas putida | [78] |
| Cat1 (Ckl) | Clostridium kluyveri | [79] | PcaIJ (Sme) | Sinorhizobium meliloti | [80] |
| Cat1 (Ibu) | Intesimonas butyriciproducens | [76] | Pct (Cpr) | Clostridium propionicum | [8, 81] |
| Cat1 (Mca) | Moraxella catarrhalis | [65] | Pct (Mel) | Megasphaera elsdenii | [32] |
| Cat1 (Mel) | Megasphaera elsdenii | [76] | Pct (Reu) | Ralstonia eutropha | [4] |
| Cat1 (Pgi) | Porphyromonas gingivalis | [82] | RipA (Ype) | Yersinia pestis | [83, 84, 85] |
| Cat1 (Psp) | Peptoniphilus sp. | [76] | SCACT (Cgr) | Cutibacterium granulosum | [86] |
| Cat1 (Rho) | Roseburia hominis | [67] | ScoAB (Bsu) | Bacillus subtilis | [87] |
| Cat1 (Rsp) | Roseburia sp. | [76] | ScoAB (Hpy) | Helicobacter pylori | [88, 89] |
| Cat1 (Sal) | Snodgrassella alvi | [65] | Scot (Ala) | Anaerotignum lactatifermentans | [32] |
| Cat2 (Aca) | Anaerostipes caccae | [67] | SCOT (Dme) | Drosophila melanogaster | [90] |
| Cat2 (Asp) | Anaerostipes sp. | [76] | Scot (Msp) | Megasphaera sp. | [32] |
| Cat2 (Pgi) | Porphyromonas gingivalis | [82] | ScpC (Eco) | Escherichia coli | [91] |
| Cat2 (Rsp) | Roseburia sp. |
[76] |
Sct (Cau) | Chloroflexus aurantiacus | [92] |
| Cat3 (Pgi) | Porphyromonas gingivalis | [82] | SmtAB (Cau) | Chloroflexus aurantiacus | [93] |
| CatIJ (Pkn) | Pseudomonas knackmussii | [94] | SptAB (Aar) | Aromatoleum aromaticum | [95] |
| CitF (eco) | Escherichia coli | [96, 97] | SUGCT (Hsa) | Homo sapiens | [98] |
| CitF (Kpn) | Klebsiella pneumoniae | [10, 99] | TbASCT (Tbr) | Trypanosoma brucei | [100] |
| CoAT (Aac) | Acidipropioni. acidipropionici | [101] | TvASCT (Tva) | Trichomonas vaginalis | [102] |
| CoaT (Ani) | Aspergillus nidulans | [103] | UctB (Aac) | Acetobacter aceti | [104] |
| CoaT (Cty) | Clostridium tyrobutyricum | [105] | UctC (Aac) | Acetobacter aceti | [6] |
| CoAT (Pfr) | Propionibacterium freudenreichii | [106] | YdiF (Eco) | Escherichia coli | [107] |
| CoAT (Rsp) | Ruminococcaceae sp. | [105] | YfdE (Eco) | Escherichia coli | [6] |
| CtfAB (Cac) | Clostridium acetobutylicum | [108, 109] | YfdW (Eco) | Escherichia coli | [30, 110] |
See Table S1 for more details.
The number of enzymes and reactions we found is double that in UniProt and three other protein databases (see below). Our search of the literature was thus exhaustive.
2.2. Phylogeny of CoA transferases reveals six, not three, families
With the large number of experimentally characterized enzymes in hand, we built a phylogenetic of their protein sequences. We constructed these trees using W‐IQ‐TREE 111 after aligning sequences with Clustal Omega. 112 We also constructed a sequence similarity network, which is another way to explore how sequences are related. 113 We constructed this network using Cytoscape 114 after aligning sequences using the Needleman–Wunsch algorithm. 115
The phylogenetic tree revealed problems with dividing the CoA transferases into the three traditional families (I, II, and III) (Figure 1a). The sequences of family I were not closely related, as was apparent in the phylogenetic tree. Indeed, some members of family I were more closely related to members of other families than each other. Worst of all, family I (or family II) is not even monophyletic. These problems were most apparent after including all family II enzymes. Previous analyses either omitted family II enzymes entirely, 4 , 8 , 9 or they did not report a phylogenetic tree of their sequences. 1 , 2 , 5
FIGURE 1.
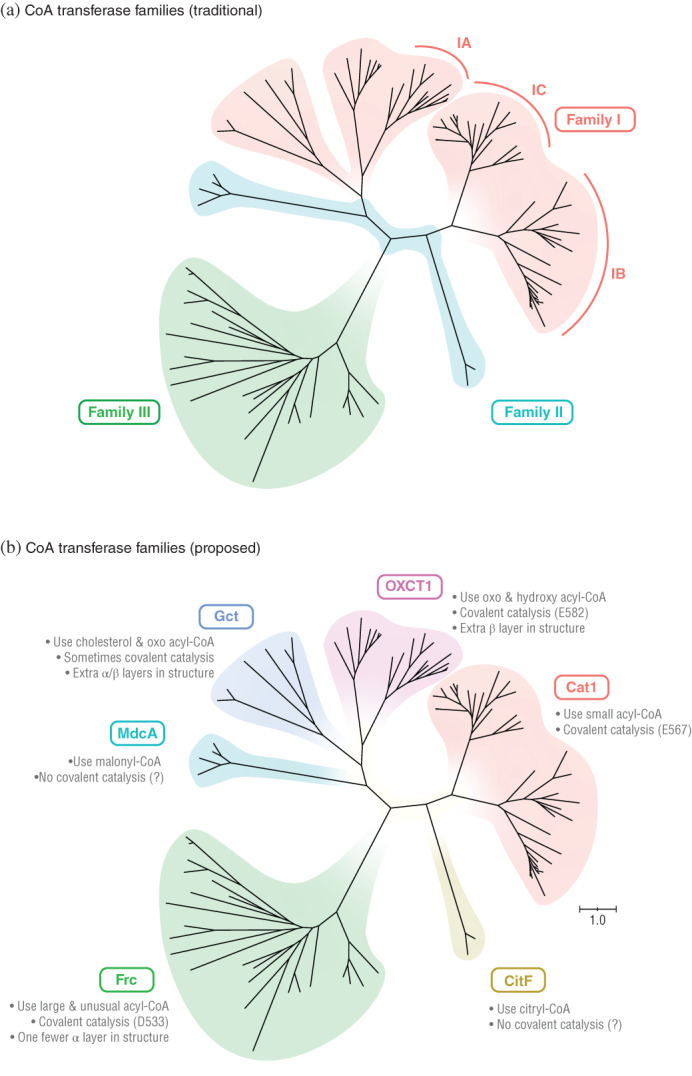
Phylogenetic tree of CoA transferase sequences shows the superfamily should be divided into six, not three families. (a) The three traditional CoA transferase families. Subfamilies of family I also shown. (b) Our six proposed families. The proposed families are consistent with other properties noted (see Figure 3 for names of enzymes and branch support values). Sequences in panel (a) were assigned to families based on descriptions in Reference [1] and to subfamilies based on Figure S1
Some analyses have proposed splitting family I into different subfamilies. 4 , 8 , 9 The phylogenetic tree (Figure 1A) shows one proposed split (according to Reference [9]), but it does not solve all problems. First, not all sequences from family I fall into a defined subfamily (IA, IB, and IC) (see also Figure S1). Second, no matter how it is split into subfamilies, family I is not monophyletic.
The tree suggests dividing the CoA transferases into six, not three, families (Figure 1b). These six families are monophyletic (Figure 1b), and members within the same family are more closely related to each other than to members of other families. This division is also consistent with other properties, such as reactions catalyzed, type of catalysis, and crystal structure (see below).
The sequence similarity network also showed that CoA transferases fall into six or more families (Figure 2). We constructed the network to show clusters of sequences with ≥25% identity (the minimum value observed for most homologous sequences). 116 , 117 Sequences fell into six clusters, with one additional sequence not falling into any cluster. This supports the idea that CoA transferases form more than three and at least six families.
FIGURE 2.
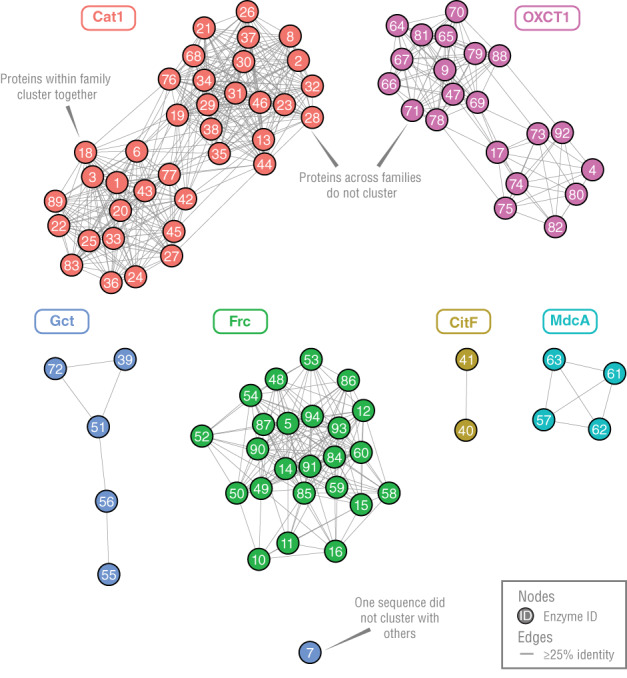
Sequence similarity network of CoA transferases show they cluster into six groups, which correspond to the six families we propose (see Table S1 for enzyme IDs)
2.3. The families of CoA transferases differ in reactions catalyzed
We explored if our six proposed families of enzymes would differ in the reactions they catalyzed. We created a heat map of the n = 94 enzymes and n = 105 reactions we identified earlier, and it revealed members within a given family generally catalyze similar reactions (Figure 3).
FIGURE 3.
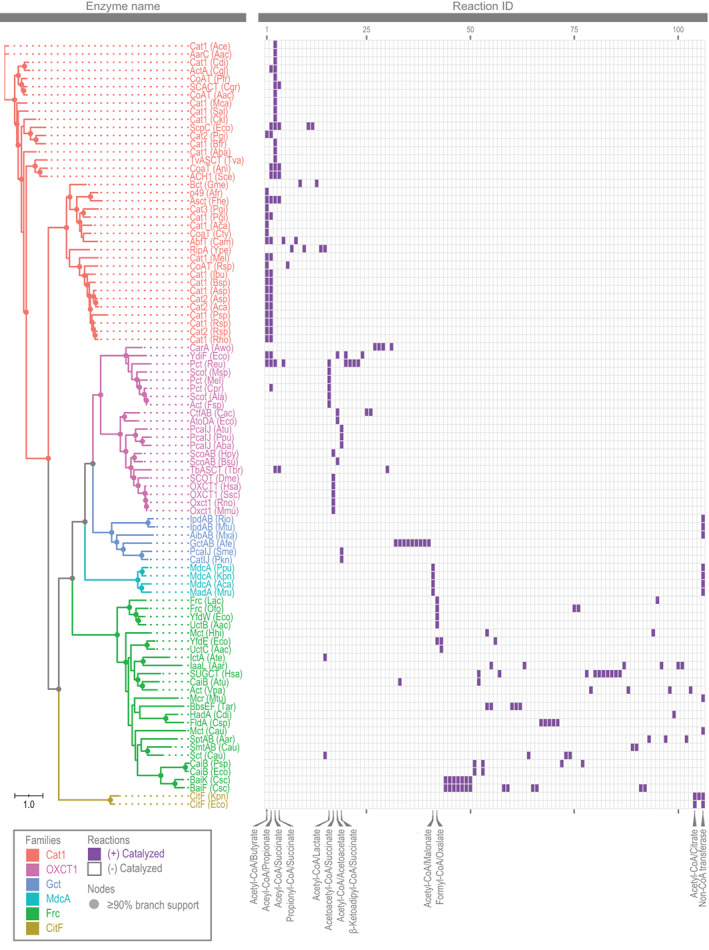
Heat map of CoA transferases and reactions catalyzed, affirming division into six families (see Table S1 for information on these enzymes and Table S2 for their reactions)
The Cat1 family catalyzes reactions involving small acyl‐CoA (Figure 3). Acetyl‐CoA/butyrate was the most common pair of substrates and used by 51% of enzymes. Other common substrates were acetyl‐CoA/propionate (49% enzymes), acetyl‐CoA/succinate (46% enzymes), and propionyl‐CoA/succinate (14% enzymes).
The OXCT1 family is different in using oxo and hydroxy acyl‐CoA (Figure 3). The most common substrates were acetyl‐CoA/lactate (29% of enzymes), acetoacetyl‐CoA/succinate (29% of enzymes), acetyl‐CoA/acetoacetate (19% of enzymes), and β‐ketoadipyl‐CoA/succinate (14% of enzymes).
Half of all members of the Gct family catalyze only nontransferase reactions (Figure 3). For example, two members cleave the ring of a cholesterol‐CoA derivative. 43 All reactions still involve acyl‐CoA as a substrate.
All members of CitF and MdcA families can catalyze CoA transferase reactions (Figure 3). Specifically, CitF uses acetyl‐CoA/citrate, and MdcA uses acetyl‐CoA/malonate. However, they also catalyze transferase reactions involving acyl‐ACP. 10 , 118 , 119 Acyl‐ACP is the likely substrate in cells, as CitF and MdcA are part of larger enzymes systems containing ACP. 54 , 97 , 118 , 119 , 120
The reactions of the Frc family differ from those of other families, but they otherwise have little in common (Figure 3). Formyl‐CoA/oxalate is used by 21% of enzymes, but all other pairs of substrates are used by two or fewer enzymes.
In sum, there are several differences in the reactions catalyzed by the CoA transferases. These differences provide further support for dividing the transferases into six families.
2.4. The families of CoA transferases have different amino acid residues in the active site
In a number of CoA transferases, the residues of the active site have been identified by mutation or crystallography (see Table S1). We wanted to see if one key residue—that involved in covalent catalysis—and the surrounding region differed across our proposed families. We aligned sequences of the enzymes and highlighted the residue involved in covalent catalysis.
Our analysis showed some similarities across families, but these are punctuated by clear differences (Figure 4). In the Cat1 family, the residue involved in covalent catalysis was a glutamate (E567). Glutamate served this role in families OXCT1 and Gct, also, but the glutamate occupies a completely different position in the alignment (E582). This resulted from a number of indels between the glutamate and a conserved glycine (G584). In the Frc family, the residue involved in covalent catalysis was aspartate (D533). In CitF and MdcA families, no residues for covalent catalysis were identified, as covalent catalysis is not thought to occur. 10 , 121 In the Gct family, some members only catalyze nontransferase reactions, and for these, no residues for covalent catalysis were identified. Indeed, these members are not thought to use covalent catalysis. 40 , 43
FIGURE 4.
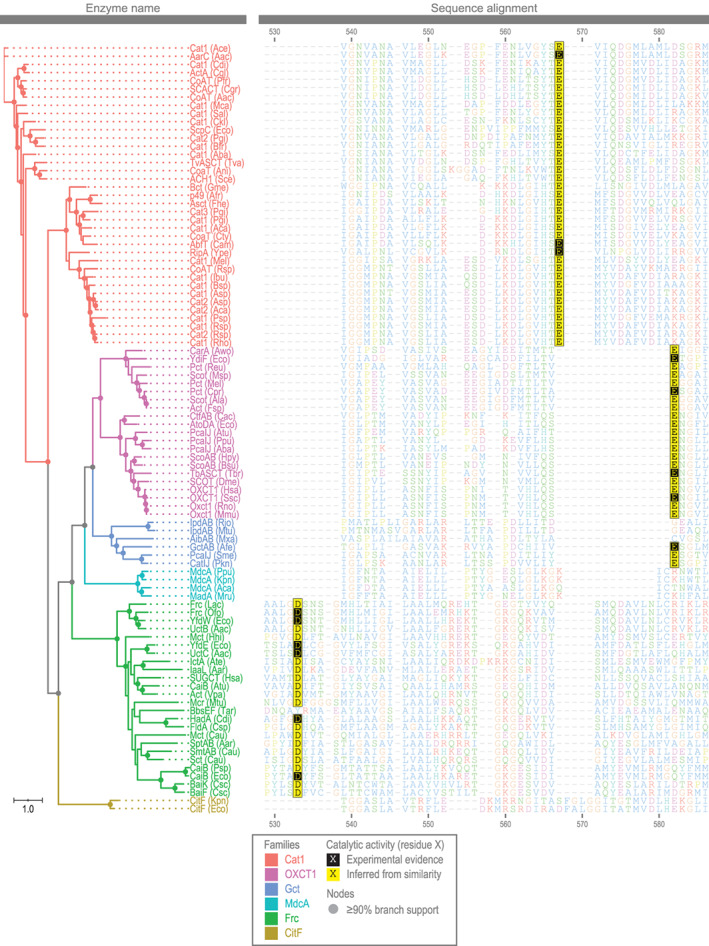
Sequence alignment of the active site of CoA transferases reveals differences among families (see Table S1 for information on these enzymes, their sequences, and their active site residues)
This analysis shows our proposed families differ in residues of the active site. These differences, such as in the region surrounding the catalytic glutamate, become clear by analyzing a large number of proteins where the catalytic residue was annotated.
2.5. The families of CoA transferases differ in structure
Our analysis showed families differ by sequence, and we wanted to see if crystal structures would also differ. Accordingly, we aligned structures of n = 24 enzymes, and we made a tree of the alignment.
We found our proposed families have different structures (Figure 5). In the tree, structures clustered by family. Differences were also clear by comparing structures visually. All transferases had two domains, each with several layers of α helices and β sheets. However, they differed in the number and arrangement of layers. Domain 1 of Gct, for example, had a large number of layers, with three layers of α helices and two β sheets (Figure 5 and Figure S2).
FIGURE 5.
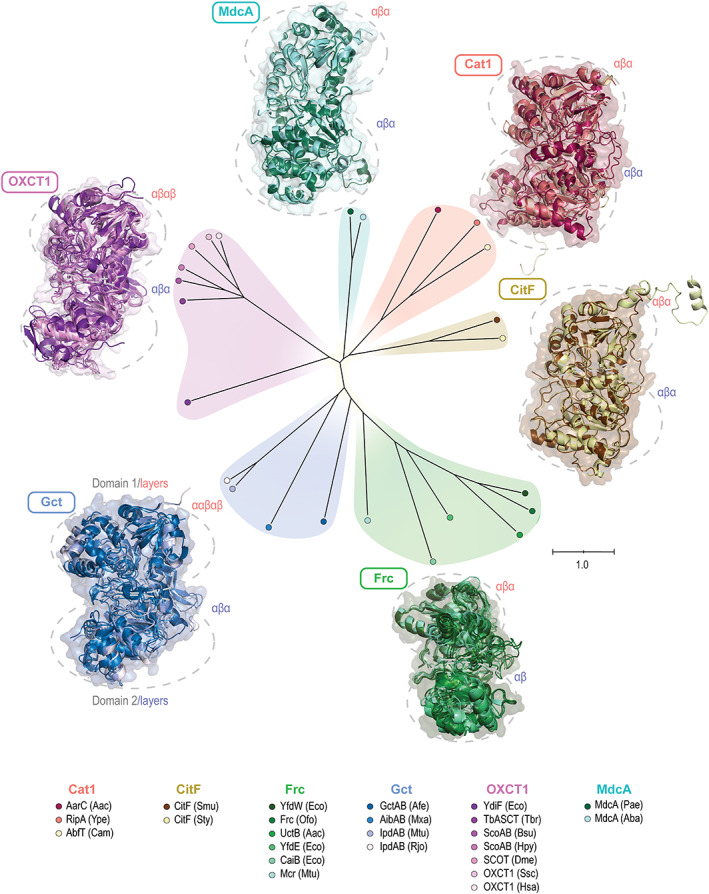
Phylogenetic tree of CoA transferase structures shows that differences among families extend to the structural level. Structures were aligned together, colored individually, then overlaid within family (see Table S1 for information on these enzymes and PDB accession numbers for structures)
Compared to the phylogenetic tree built with protein sequences, the tree built with structures differed somewhat. For example, families CitF and MdcA were distantly related according to their sequences, but appeared more closely related from their structures. The families were still distinct, showing our proposed families are valid at both sequence and structural levels.
One caveat is there were few structures available for families CitF and MdcA, and those available are for putative enzymes (Table S3). As putative enzymes, they have no experimental evidence of activity, and instead their catalytic activity is inferred by homology. If we exclude them, the position of the remaining families in the tree is not affected (Figure S3). We did not use putative enzymes elsewhere in our analysis. In sum, despite some caveats, this analysis agrees with the sequence analysis and suggests dividing the CoA transferases into six families.
2.6. The structure of CoA transferases differs around the active site
Having found the CoA transferases differ in overall structure, we wanted to see if differences extended to the active site. We were most interested in comparing Cat1, Gct, and OXCT1 families, given glutamate serves as a catalytic residue in all three. The crystal structures showed differences around the catalytic residue (Figure S4). In the Cat1 family, the residue was followed by an alpha helix. In Gct, and OXCT1 families, the helix was replaced by part of a β sheet. The helix and sheet correspond to a region of indels in the sequence alignment (from E567 to G584; see Figure 4). These results point to structural elements gained or lost during the evolution of the CoA transferases. Despite clear similarities, there are also differences around the active site for Cat1, Gct, and OXCT1 families.
2.7. Enzyme databases are missing half of all enzymes and reactions
Our analysis used the primary literature as a source for information on enzymes. Searching for information in this way is laborious, and many investigators search protein databases instead. We wanted to see if these databases had comparable information and would have led us to the same conclusions.
We searched four widely used databases (BRENDA, KEGG, MetaCyc, UniProt) for CoA transferases. 11 , 12 , 13 , 14 We searched for enzymes according to their enzyme commission (EC) numbers and three other identifiers (Rhea ID, KEGG REACTION ID, MetaCyc Reaction ID/BioCyc ID) 12 , 14 , 122 , 123 (Table S4). We counted enzymes annotated with a reaction, amino acid sequence, and literature reference.
We found information in databases was far from complete. The four databases we searched had information annotated for only n = 48 enzymes (Figure 6a and Table S5). Half (49%) of the enzymes we found in the primary literature were thus missing information. The databases reported the enzymes catalyzed only n = 57 CoA transferase reactions (Figure 6a and Table S6). About half (46%) of the reactions we found were missing.
FIGURE 6.
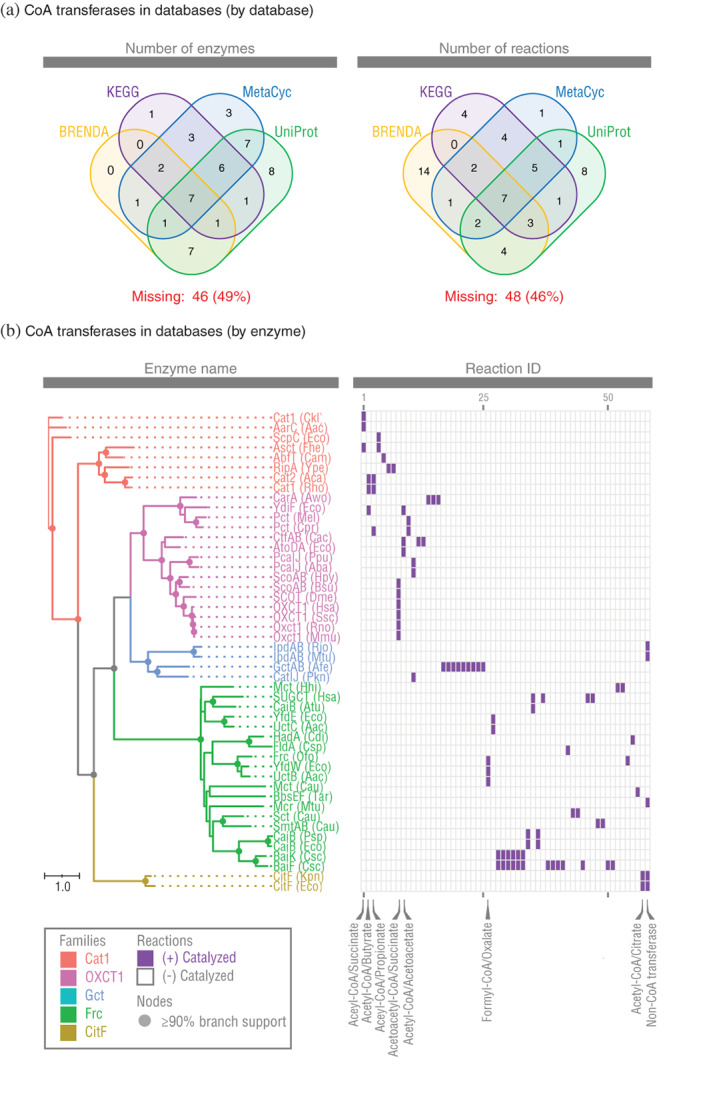
Protein databases are missing large numbers of CoA transferases in the primary literature. (a) Venn diagram of CoA transferases annotated by databases. (b) Heat map of CoA transferases and reactions annotated by databases (see Table S5 for information on these enzymes and Table S6 for their reactions)
We built a reaction heat map of all enzymes and reactions available in databases (Figure 6b). The result looked different from the heat map built using information from the primary literature (see Figure 2). When using information in databases, all families had fewer members, and the MdcA family was completely missing. Furthermore, reactions catalyzed by families Cat1 and OXCT1 were no longer clearly different. Consequently, it is not obvious that family I proposed by Heider 1 should be divided into separate families, as it is when using information from the primary literature. Without digging into the primary literature, we would have reached incorrect conclusions about the CoA transferase superfamily.
2.8. Additional phylogenetic trees shed more insight into the CoA transferases
Crystal structures show CoA transferases have two protein domains. We examined how these domains are organized within each enzyme, and then we built a phylogenetic tree to see if the organization differed by family (Figure S5). We found domains were organized in different ways (Figure S5a). In some enzymes, domains were separated (on two polypeptide chains), but in others, they were fused (on one chain). In rare cases, domains were fused and duplicated (two polypetide chains with two domains each). A phylogenetic tree (Figure S5b) showed members of Gct had only separated domains, whereas Cat1, MdcA, and CitF had only fused domains. OXCT1 had both separated and fused subunits. Frc had both fused domains and fused domains that had been duplicated. These differences in organization are small, but help further define the families of CoA transferases.
To this point, we have built phylogenetic trees using one amino acid sequence per enzyme. Alternatively, we could build trees using two separate sequences per enzyme, with one sequence per protein domain. Such a tree would reveal if domains evolved separately from a common ancestor, as proposed for some CoA transferases. 2 , 34 We thus built such a tree, and it showed sequences did not always cluster by domain (Figure S6). For example, domain 1 of CitF clustered with domain 2 of Gct. If the domains evolved separately, all sequences for domain 1 should cluster together, and sequences for domain 2 should cluster apart. This suggests that domains did not evolve separately, at least for some families.
We built phylogenetic trees using CoA transferases and very few (n = 5) additional enzymes. We in fact considered many more proteins, but they turned out not to be closely related to CoA transferases. In one phylogenetic tree, we included proteins considered by Pfam, 124 SCOP, 125 and other authorities 2 to be related to CoA transferases (see Table S7). The tree showed these proteins were not as closely related to the CoA transferases as the CoA transferases were to themselves (Figure S7). Furthermore, none are known to use acyl‐CoA as a substrate or perform transferase reactions. It makes sense to set the boundaries of the CoA superfamily around the proteins of our original tree (Figure 1).
3. DISCUSSION
Our analysis is important to the biology of CoA transferases and to phylogenetic analyses of proteins. It is important to the biology of the CoA transferases because it uproots the idea that these enzymes fall into three families. 1 This idea has stood for two decades, and others have proposed only minor changes as scores of enzymes have been discovered. 4 , 5 , 6 , 7 , 8 , 9 The strength of our approach is it uses all experimentally characterized enzymes and examines several properties (e.g., sequences, reactions, and structures). It is by using this comprehensive approach that we can conclude CoA transferases fall into six families.
Our work is important to the biology of CoA transferases, also, by shedding insight into their evolutionary history. Specifically, it suggests that the superfamily evolved (or lost) the ability to catalyze their reactions multiple times. Three families (MdcA, CitF, and Gct) have members that catalyze non‐CoA transferase reactions (either alone or in addition to CoA transferase reactions). The other three families (Frc, Cat1, and OXCT1) catalyze CoA transferase reactions, and few others. Given their complex phylogeny, the CoA transferases must have gained (or lost) the ability to catalyze CoA reactions at least twice. Reflecting this, the three families that catalyze mostly CoA transferase reactions (Frc, Cat1, and OXCT1) show key differences around the active site. This complex history is apparent only from analyzing our large set of manually annotated enzymes and reactions.
Besides its importance to the CoA transferases, our analysis is important to phylogenetic analyses. Namely, it shows the importance of conducting phylogenetic analyses with all proteins reported in the primary literature. It is convenient to focus on only a few proteins, or to use proteins with information reported in UniProt or other databases. Unfortunately, using only databases would lead to an incomplete picture. It is already known that databases rely on few publications for most of their information. 126 For gene ontology annotations in UniProt, the top 0.14% publications are used as a source of information for 25% of the proteins. 126 Our study shows that databases are missing information outright; half of all enzymes were missing the reaction they catalyze, their amino acid sequence, or a literature reference. Databases can be useful for starting a literature search, but they are not a replacement for one.
In sum, our work is important to the CoA transferase superfamily and to phylogenetic analyses in general. It warns investigators doing these analyses to go beyond proteins annotated in databases. Though our work on CoA transferases is comprehensive, a phylogenetic analysis is never done—it simply awaits discovery of the next protein. Digging deep in the literature for proteins ensures analyses are as accurate as possible until the next discovery.
4. MATERIALS AND METHODS
4.1. Search for CoA transferases in primary literature
We searched for CoA transferases in the primary literature. As explained in Results, we considered only enzymes with (a) experimental evidence for catalyzing at least one reaction and (b) an amino acid sequence. An important resource was Zhang et al. 86 which had manually annotated information for n = 51 enzyme from Cat1, OXCT1, and Gct families. We also used databases 11 , 12 , 13 , 14 as starting points.
We identified reactions as biochemically significant if they occurred at fast rates. We arbitrarily defined these as reactions that occur at ≥10% of the fastest CoA transferase reaction for a given enzyme. If an enzyme catalyzed only one CoA transferase reaction, the reaction was significant by default. This definition is arbitrary, and we wanted to identify reactions that are statistically different from 0 instead (p < .05). However, many literature studies do not report statistical significance.
Some publications did not give report gene or protein names. We assigned a name in cases where it was missing. Information is otherwise as reported in the publication.
4.2. Construction of phylogenetic tree of sequences
To construct phylogenetic trees of amino acid sequences, we first performed multiple sequence alignment with Clustal Omega. 112 Following References [127, 128], we ran Clustal Omega using the package msa 129 of R 130 and the default parameter values.
With the aligned sequences, we calculated the tree using maximum‐likelihood with W‐IQ‐TREE 111 and default parameter values. The substitution model used was LG + F + G4, which gave the lowest AICc value. Branch support values were calculated using the ultrafast bootstrap analysis 131 with 1,000 maximum iterations.
We visualized the final tree with the package ggtree 132 of R and the Interactive Tree of Life (iTOL). 133 We used ggtree for rectangular trees (Figures 3, 4, and 6 and Figures S1, S5, S6, and S7) and the iTOL for equal‐daylight trees (Figures 1 and 5 and S3).
As mentioned, CoA transferases show different organization of domains (Figure S5), and we had to take this into account before alignment. For enzymes with fused domains, we used the amino acid sequence as is. For enzymes with separated domains, we concatenated the sequences of the two subunits before alignment. For enzymes with fused domains that had been duplicated, we arbitrarily chose the first subunit for alignment and discarded the second. If we chose the second subunit instead, the tree was not affected. This procedure ensured that each domain was represented once and only once in the alignment.
Protein sequences are available in FASTA format on Figshare (https://figshare.com/articles/dataset/Sequences_and_phylogenetic_trees_of_coenzyme_A_transferases/16617892). Phylogenetic trees are available in Newick format at the same resource.
4.3. Construction of sequence similarity networks
To construct sequence similarity networks, we first performed pairwise sequence alignment using the Needleman–Wunsch algorithm. 115 We ran this algorithm using the package Biostrings 134 in R. We used BLOSUM62 substitution matrix, gap opening penalty of 10, and gap extension penalty of 0.5. We then calculated pairwise identity of the sequences.
With the pairwise identity of the aligned sequences, we constructed the sequence similarity network in Cytoscape. 114 We filtered out (removed) edges with <25% identity. We visualized the network using the yFiles organic layout.
4.4. Construction of heat map of reactions
We constructed a heat map of CoA transferase reactions using data from the primary literature. We took reactions we found in the literature and ordered them according to their frequency within family. Following this order, we assigned reactions IDs, created a heat map using ggplot2, 135 and laid it next to the phylogenetic tree constructed with ggtree.
4.5. Construction of sequence alignment of active site
We constructed a sequence alignment of the active site using the same alignment for the phylogenetic trees. The sequence alignment was visualized using ggplot2.
4.6. Construction of phylogenetic tree of crystal structures
To construct the tree of crystal structures, we first aligned the structures with SALIGN. 136 We chose this alignment tool because it can accommodate proteins with multiple subunits. For proteins with two subunits (A and B), we used the command “FIRST:A:LAST:B” to ensure all residues were included. The tool outputted (a) the aligned structures in pdb format and (b) pairwise distances between structures.
With the pairwise distances, we calculated the tree using the minimum evolution method. We used the package ape 137 of R and default parameter values. The pairwise distances were from the log file of SALIGN, and they were for the guide tree during the last alignment iteration.
We visualized the aligned structures using PyMOL (v. 2.0, Schrödinger, LLC). We loaded aligned structures (pdb format) into PyMOL, colored them using palettes generated by package colorspace 138 of R, and then captured images as ray traces. For best comparison, we captured all ray traces from the same view for a given figure.
4.7. Searching for CoA transferases in databases
To find CoA transferase reactions in databases, we searched for EC numbers for CoA transferase reactions (Table S4). For databases that allowed them, we searched for additional reaction IDs (Table S4). For UniProt, we also searched for Rhea IDs. For MetaCyc, we also searched for Rhea IDs and MetaCyc Reaction IDs/BioCyc IDs. For KEGG, we also searched for KEGG REACTION IDs.
Our analysis also included n = 5 enzymes that catalyze non‐CoA transferase reactions only. For fair comparison, we searched databases for these enzymes, too.
We counted enzymes annotated with a reaction, amino acid sequence, and literature reference. We did not count enzymes with partial or incorrect information (see Table S5). For UniProt, we counted only enzymes with “Experimental evidence at protein level” (see Table S5).
The Venn diagram of reactions by database was generated using package venn 139 in R. The phylogenetic tree and heat map were generated as described previously.
4.8. Prediction of sequence regions of protein domains
To build a phylogenetic tree of the two different domains of CoA transferases (Figure S6), we had to predict sequences regions for each domain. First, we identified sequence regions in enzymes where they were known (or obvious). These enzymes included n = 24 with crystal structures and n = 12 where domains were part of separate subunits. Second, we built profile hidden Markov models (pHMMs) of these sequence regions. We used the hmmbuild command of HMMER, 140 using the known sequence regions as the input. Third, we predicted sequence regions of domains in all remaining CoA transferases. We used the hmmsearch command of HMMER, using the pHMMs and sequences of CoA transferases as an input.
Supporting information
Figure S1. Phylogenetic tree of CoA transferases that shows subfamilies IA, IB, and IC proposed in Tielens et al. (2010). Sequences included are from Fig. 3 of Tielens et al. (2010), plus all sequences from our own analysis. Some sequences from Tielens et al. (2010) belong to putative enzymes (ones with no biochemical evidence of activity), and for this reason were not included in our own analysis.
Figure S2. Crystal structure of a Gct family protein, showing the large number of layers in domain 1 that distinguishes this family. Structure of a Cat1 family protein, with typical three layers, shown for comparison. Proteins shown are GctAB (Afe) and AarC (Aac). Structures are presented in stereo and can be seen in 3D.
Figure S3. Phylogenetic tree of CoA transferase structures after removing putative enzymes, showing positions of other families in tree are unchanged.
Figure S4. Crystal structures show the region around active site of Cat1 family differs from that of Gct and OXCT1 families. Structures were aligned together and then overlaid within family. The indel region refers to E567 to G584 in Figure 4. The catalytic residue refers to that shown in Figure 4. Only structures with an identifiable catalytic residue are shown. See Table S1 for information on these enzymes and PDB accession numbers for structures.
Figure S5. CoA transferases show different organizations of their domains. (A) Types of organization. The N‐terminus and C‐terminus of each polypeptide (subunit) is shown. (B) Enzyme by type of organization. See Table S1 for information on these enzymes.
Figure S6. Phylogenetic tree of CoA transferases with sequences split by domains, showing that sequences do not always cluster by domain. See Table S1 for information on these enzymes.
Figure S7. Phylogenetic tree of CoA transferase superfamily plus proteins from additional families, showing the additional families are not closely related. See Table S1 for information on enzymes of the CoA transferase superfamily (Cat1, OXCT1, Gct, MdcA, Frc, CitF), and see Table S7 for information on proteins from additional/other families.
Table S1. Coenzyme A transferases that have been experimentally characterized in the primary literature
Table S2. Reactions of coenzyme A transferases in the primary literature
Table S3. Coenzyme A transferases that areputative enzymes and included in this analysis only for their structures
Table S4. Accession numbers of reactions in databases
Table S5. Coenzyme A transferases that have been annotated in databases
Table S6. Reactions of coenzyme A transferases in databases
Table S7. Proteins that some authorities consider part of a CoA transferase superfamily (1 representative protein per family)
Table S8. Coenzyme A transferase sequences and proposed subfamilies from Tielens et al. (2010)
ACKNOWLEDGEMENTS
This work was supported by an Agriculture and Food Research Initiative Competitive Grant [grant no. 2018‐67015‐27495] and Hatch Project [accession no. 1019985] from the United States Department of Agriculture National Institute of Food and Agriculture.
Hackmann TJ. Redefining the coenzyme A transferase superfamily with a large set of manually annotated proteins. Protein Science. 2022;31:864–881. 10.1002/pro.4277
Our work is important to one superfamily of proteins, the coenzyme A transferases. By analyzing nearly 100 experimentally characterized proteins, it overturns the idea these proteins fall into three families. Our work is also important to investigators who rely on databases, such as UniProt, for information on proteins. Half of all proteins in our analysis had no functional information in databases. Our study shows that the need to dig into the literature before analyzing protein families.
Funding information National Institute of Food and Agriculture, Grant/Award Numbers: 1019985, 2018‐67015‐27495; United States Department of Agriculture
REFERENCES
- 1. Heider J. A new family of CoA‐transferases. FEBS Lett. 2001;509:345–349. [DOI] [PubMed] [Google Scholar]
- 2. Anantharaman V, Aravind L. Diversification of catalytic activities and ligand interactions in the protein fold shared by the sugar isomerases, eIF2B, DeoR transcription factors, acyl‐CoA transferases and methenyltetrahydrofolate synthetase. J Mol Biol. 2006;356:823–842. [DOI] [PubMed] [Google Scholar]
- 3. Hilbi H, Dimroth P. Purification and characterization of a cytoplasmic enzyme component of the Na+−activated malonate decarboxylase system of Malonomonas rubra: Acetyl‐S‐acyl carrier protein: Malonate acyl carrier protein‐SH transferase. Arch Microbiol. 1994;162:48–56. [PubMed] [Google Scholar]
- 4. Lindenkamp N, Schürmann M, Steinbüchel A. A propionate CoA‐transferase of Ralstonia eutropha H16 with broad substrate specificity catalyzing the CoA thioester formation of various carboxylic acids. Appl Microbiol Biotechnol. 2013;97:7699–7709. [DOI] [PubMed] [Google Scholar]
- 5. Macieira S, Zhang J, Velarde M, Buckel W, Messerschmidt A. Crystal structure of 4‐hydroxybutyrate CoA‐transferase from Clostridium aminobutyricum . Biol Chem. 2009;390:1251–1263. [DOI] [PubMed] [Google Scholar]
- 6. Mullins EA, Sullivan KL, Kappock TJ. Function and X‐ray crystal structure of Escherichia coli YfdE. PLoS One. 2013;8:e67901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schuhle K, Nies J, Heider J. An indoleacetate‐CoA ligase and a phenylsuccinyl‐CoA transferase involved in anaerobic metabolism of auxin. Environ Microbiol. 2016;18:3120–3132. [DOI] [PubMed] [Google Scholar]
- 8. Selmer T, Willanzheimer A, Hetzel M. Propionate CoA‐transferase from Clostridium propionicum. Cloning of gene and identification of glutamate 324 at the active site. Eur J Biochem. 2002;269:372–380. [DOI] [PubMed] [Google Scholar]
- 9. Tielens AG, van Grinsven KW, Henze K, van Hellemond JJ, Martin W. Acetate formation in the energy metabolism of parasitic helminths and protists. Int J Parasitol. 2010;40:387–397. [DOI] [PubMed] [Google Scholar]
- 10. Dimroth P, Loyal R, Eggerer H. Characterization of the isolated transferase subunit of citrate lyase as a CoA‐transferase. Evidence against a covalent enzyme‐substrate intermediate. Eur J Biochem. 1977;80:479–488. [DOI] [PubMed] [Google Scholar]
- 11. UniProt C. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49:D480–D489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Caspi R, Billington R, Keseler IM, et al. The MetaCyc database of metabolic pathways and enzymes—a 2019 update. Nucleic Acids Res. 2020;48:D445–D453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chang A, Jeske L, Ulbrich S, et al. BRENDA, the ELIXIR core data resource in 2021: New developments and updates. Nucleic Acids Res. 2021;49:D498–D508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kanehisa M, Furumichi M, Sato Y, Ishiguro‐Watanabe M, Tanabe M. KEGG: Integrating viruses and cellular organisms. Nucleic Acids Res. 2021;49:D545–D551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Baier F, Tokuriki N. Connectivity between catalytic landscapes of the metallo‐beta‐lactamase superfamily. J Mol Biol. 2014;426:2442–2456. [DOI] [PubMed] [Google Scholar]
- 16. Copp JN, Akiva E, Babbitt PC, Tokuriki N. Revealing unexplored sequence‐function space using sequence similarity networks. Biochemistry. 2018;57:4651–4662. [DOI] [PubMed] [Google Scholar]
- 17. Mashiyama ST, Malabanan MM, Akiva E, et al. Large‐scale determination of sequence, structure, and function relationships in cytosolic glutathione transferases across the biosphere. PLoS Biol. 2014;12:e1001843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Spence MA, Mortimer MD, Buckle AM, Minh BQ, Jackson CJ. A comprehensive phylogenetic analysis of the serpin superfamily. Mol Biol Evol. 2021;38:2915–2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vass M, Kooistra AJ, Yang D, Stevens RC, Wang MW, de Graaf C. Chemical diversity in the G protein‐coupled receptor superfamily. Trends Pharmacol Sci. 2018;39:494–512. [DOI] [PubMed] [Google Scholar]
- 20. Mullins EA, Francois JA, Kappock TJ. A specialized citric acid cycle requiring succinyl‐coenzyme A (CoA):acetate CoA‐transferase (AarC) confers acetic acid resistance on the acidophile Acetobacter aceti . J Bacteriol. 2008;190:4933–4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mullins EA, Kappock TJ. Crystal structures of Acetobacter aceti succinyl‐coenzyme A (CoA):acetate CoA‐transferase reveal specificity determinants and illustrate the mechanism used by class I CoA‐transferases. Biochemistry. 2012;51:8422–8434. [DOI] [PubMed] [Google Scholar]
- 22. Dickert S, Pierik AJ, Linder D, Buckel W. The involvement of coenzyme A esters in the dehydration of (R)‐phenyllactate to (E)‐cinnamate by Clostridium sporogenes . Eur J Biochem. 2000;267:3874–3884. [DOI] [PubMed] [Google Scholar]
- 23. Gerhardt A, Cinkaya I, Linder D, Huisman G, Buckel W. Fermentation of 4‐aminobutyrate by Clostridium aminobutyricum: cloning of two genes involved in the formation and dehydration of 4‐hydroxybutyryl‐CoA. Arch Microbiol. 2000;174:189–199. [DOI] [PubMed] [Google Scholar]
- 24. Macieira S, Zhang J, Buckel W, Messerschmidt A. Crystal structure of the complex between 4‐hydroxybutyrate CoA‐transferase from Clostridium aminobutyricum and CoA. Arch Microbiol. 2012;194:157–166. [DOI] [PubMed] [Google Scholar]
- 25. Scherf U, Buckel W. Purification and properties of 4‐hydroxybutyrate coenzyme A transferase from Clostridium aminobutyricum . Appl Environ Microbiol. 1991;57:2699–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Turroni S, Vitali B, Bendazzoli C, et al. Oxalate consumption by lactobacilli: evaluation of oxalyl‐CoA decarboxylase and formyl‐CoA transferase activity in Lactobacillus acidophilus . J Appl Microbiol. 2007;103:1600–1609. [DOI] [PubMed] [Google Scholar]
- 27. Fleck CB, Brock M. Re‐characterisation of Saccharomyces cerevisiae Ach1p: fungal CoA‐transferases are involved in acetic acid detoxification. Fungal Genet Biol. 2009;46:473–485. [DOI] [PubMed] [Google Scholar]
- 28. Berg M, Hilbi H, Dimroth P. Sequence of a gene cluster from Malonomonas rubra encoding components of the malonate decarboxylase Na + pump and evidence for their function. Eur J Biochem. 1997;245:103–115. [DOI] [PubMed] [Google Scholar]
- 29. Ricagno S, Jonsson S, Richards N, Lindqvist Y. Formyl‐CoA transferase encloses the CoA binding site at the interface of an interlocked dimer. EMBO J. 2003;22:3210–3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Toyota CG, Berthold CL, Gruez A, et al. Differential substrate specificity and kinetic behavior of Escherichia coli YfdW and Oxalobacter formigenes formyl coenzyme A transferase. J Bacteriol. 2008;190:2556–2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Berthold CL, Toyota CG, Richards NG, Lindqvist Y. Reinvestigation of the catalytic mechanism of formyl‐CoA transferase, a class III CoA‐transferase. J Biol Chem. 2008;283:6519–6529. [DOI] [PubMed] [Google Scholar]
- 32. Zhang X, Mao Y, Wang B, et al. Screening, expression, purification and characterization of CoA‐transferases for lactoyl‐CoA generation. J Ind Microbiol Biotechnol. 2019;46:899–909. [DOI] [PubMed] [Google Scholar]
- 33. Buckel W, Dorn U, Semmler R. Glutaconate CoA‐transferase from Acidaminococcus fermentans . Eur J Biochem. 1981;118:315–321. [DOI] [PubMed] [Google Scholar]
- 34. Jacob U, Mack M, Clausen T, Huber R, Buckel W, Messerschmidt A. Glutaconate CoA‐transferase from Acidaminococcus fermentans: the crystal structure reveals homology with other CoA‐transferases. Structure. 1997;5:415–426. [DOI] [PubMed] [Google Scholar]
- 35. Mack M, Bendrat K, Zelder O, Eckel E, Linder D, Buckel W. Location of the two genes encoding glutaconate coenzyme A‐transferase at the beginning of the hydroxyglutarate operon in Acidaminococcus fermentans . Eur J Biochem. 1994;226:41–51. [DOI] [PubMed] [Google Scholar]
- 36. Mack M, Buckel W. Identification of glutamate beta 54 as the covalent‐catalytic residue in the active site of glutaconate CoA‐transferase from Acidaminococcus fermentans . FEBS Lett. 1995;357:145–148. [DOI] [PubMed] [Google Scholar]
- 37. Schurmann M, Hirsch B, Wubbeler JH, Stoveken N, Steinbuchel A. Succinyl‐CoA:3‐sulfinopropionate CoA‐transferase from Variovorax paradoxus strain TBEA6, a novel member of the class III coenzyme A (CoA)‐transferase family. J Bacteriol. 2013;195:3761–3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kim J, Darley D, Selmer T, Buckel W. Characterization of (R)‐2‐hydroxyisocaproate dehydrogenase and a family III coenzyme A transferase involved in reduction of L‐leucine to isocaproate by Clostridium difficile . Appl Environ Microbiol. 2006;72:6062–6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Veit A, Rittmann D, Georgi T, Youn JW, Eikmanns BJ, Wendisch VF. Pathway identification combining metabolic flux and functional genomics analyses: acetate and propionate activation by Corynebacterium glutamicum . J Biotechnol. 2009;140:75–83. [DOI] [PubMed] [Google Scholar]
- 40. Li Y, Luxenburger E, Müller R. An alternative isovaleryl CoA biosynthetic pathway involving a previously unknown 3‐methylglutaconyl CoA decarboxylase. Angew Chem Int Ed Engl. 2013;52:1304–1308. [DOI] [PubMed] [Google Scholar]
- 41. Chen M, Huang X, Zhong C, Li J, Lu X. Identification of an itaconic acid degrading pathway in itaconic acid producing Aspergillus terreus . Appl Microbiol Biotechnol. 2016;100:7541–7548. [DOI] [PubMed] [Google Scholar]
- 42. van Grinsven KW, van Hellemond JJ, Tielens AG. Acetate:succinate CoA‐transferase in the anaerobic mitochondria of Fasciola hepatica . Mol Biochem Parasitol. 2009;164:74–79. [DOI] [PubMed] [Google Scholar]
- 43. Crowe AM, Workman SD, Watanabe N, Worrall LJ, Strynadka NCJ, Eltis LD. IpdAB, a virulence factor in Mycobacterium tuberculosis, is a cholesterol ring‐cleaving hydrolase. Proc Natl Acad Sci U S A. 2018;115:E3378–E3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jenkins LS, Nunn WD. Genetic and molecular characterization of the genes involved in short‐chain fatty acid degradation in Escherichia coli: the ato system. J Bacteriol. 1987;169:42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Korolev S, Koroleva O, Petterson K, et al. Autotracing of Escherichia coli acetate CoA‐transferase alpha‐subunit structure using 3.4 A MAD and 1.9 A native data. Acta Crystallogr D Biol Crystallogr. 2002;58:2116–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ridlon JM, Hylemon PB. Identification and characterization of two bile acid coenzyme A transferases from Clostridium scindens, a bile acid 7α‐dehydroxylating intestinal bacterium. J Lipid Res. 2012;53:66–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Savolainen K, Bhaumik P, Schmitz W, et al. Alpha‐methylacyl‐CoA racemase from Mycobacterium tuberculosis. Mutational and structural characterization of the active site and the fold. J Biol Chem. 2005;280:12611–12620. [DOI] [PubMed] [Google Scholar]
- 48. Leuthner B, Heider J. Anaerobic toluene catabolism of Thauera aromatica: the bbs operon codes for enzymes of beta oxidation of the intermediate benzylsuccinate. J Bacteriol. 2000;182:272–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Leutwein C, Heider J. Succinyl‐CoA:(R)‐benzylsuccinate CoA‐transferase: an enzyme of the anaerobic toluene catabolic pathway in denitrifying bacteria. J Bacteriol. 2001;183:4288–4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zarzycki J, Brecht V, Muller M, Fuchs G. Identifying the missing steps of the autotrophic 3‐hydroxypropionate CO2 fixation cycle in Chloroflexus aurantiacus. Proc Natl Acad Sci U S A. 2009;106:21317–21322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Oberender J, Kung JW, Seifert J, von Bergen M, Boll M. Identification and characterization of a succinyl‐coenzyme A (CoA):benzoate CoA transferase in Geobacter metallireducens . J Bacteriol. 2012;194:2501–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Borjian F, Johnsen U, Schonheit P, Berg IA. Succinyl‐CoA:mesaconate CoA‐transferase and mesaconyl‐CoA hydratase, enzymes of the methylaspartate cycle in Haloarcula hispanica . Front Microbiol. 2017;8:1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hu Y, Cronan JE. alpha‐proteobacteria synthesize biotin precursor pimeloyl‐ACP using BioZ 3‐ketoacyl‐ACP synthase and lysine catabolism. Nat Commun. 2020;11:5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Koo JH, Kim YS. Functional evaluation of the genes involved in malonate decarboxylation by Acinetobacter calcoaceticus . Eur J Biochem. 1999;266:683–690. [DOI] [PubMed] [Google Scholar]
- 55. Eichler K, Schunck WH, Kleber HP, Mandrand‐Berthelot MA. Cloning, nucleotide sequence, and expression of the Escherichia coli gene encoding carnitine dehydratase. J Bacteriol. 1994;176:2970–2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Elssner T, Engemann C, Baumgart K, Kleber HP. Involvement of coenzyme A esters and two new enzymes, an enoyl‐CoA hydratase and a CoA‐transferase, in the hydration of crotonobetaine to L‐carnitine by Escherichia coli . Biochemistry. 2001;40:11140–11148. [DOI] [PubMed] [Google Scholar]
- 57. Rangarajan ES, Li Y, Iannuzzi P, Cygler M, Matte A. Crystal structure of Escherichia coli crotonobetainyl‐CoA: carnitine CoA‐transferase (CaiB) and its complexes with CoA and carnitinyl‐CoA. Biochemistry. 2005;44:5728–5738. [DOI] [PubMed] [Google Scholar]
- 58. Hoenke S, Dimroth P. Formation of catalytically active acetyl‐S‐malonate decarboxylase requires malonyl‐coenzyme A: acyl carrier protein transacylase as auxiliary enzyme [corrected]. Eur J Biochem. 1999;259:181–187. [DOI] [PubMed] [Google Scholar]
- 59. Hoenke S, Schmid M, Dimroth P. Sequence of a gene cluster from Klebsiella pneumoniae encoding malonate decarboxylase and expression of the enzyme in Escherichia coli . Eur J Biochem. 1997;246:530–538. [DOI] [PubMed] [Google Scholar]
- 60. Engemann C, Elssner T, Pfeifer S, Krumbholz C, Maier T, Kleber HP. Identification and functional characterisation of genes and corresponding enzymes involved in carnitine metabolism of Proteus sp. Arch Microbiol. 2005;183:176–189. [DOI] [PubMed] [Google Scholar]
- 61. Chohnan S, Akagi K, Takamura Y. Functions of malonate decarboxylase subunits from Pseudomonas putida . Biosci Biotechnol Biochem. 2003;67:214–217. [DOI] [PubMed] [Google Scholar]
- 62. Hess V, González JM, Parthasarathy A, Buckel W, Müller V. Caffeate respiration in the acetogenic bacterium Acetobacterium woodii: a coenzyme A loop saves energy for caffeate activation. Appl Environ Microbiol. 2013;79:1942–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fukao T, Shintaku H, Kusubae R, et al. Patients homozygous for the T435N mutation of succinyl‐CoA:3‐ketoacid CoA transferase (SCOT) do not show permanent ketosis. Pediatr Res. 2004;56:858–863. [DOI] [PubMed] [Google Scholar]
- 64. Shafqat N, Kavanagh KL, Sass JO, et al. A structural mapping of mutations causing succinyl‐CoA:3‐ketoacid CoA transferase (SCOT) deficiency. J Inherit Metab Dis. 2013;36:983–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kwong WK, Zheng H, Moran NA. Convergent evolution of a modified, acetate‐driven TCA cycle in bacteria. Nat Microbiol. 2017;2:17067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wang Y, Peng F, Tong W, Sun H, Xu N, Liu S. The nitrated proteome in heart mitochondria of the db/db mouse model: characterization of nitrated tyrosine residues in SCOT. J Proteome Res. 2010;9:4254–4263. [DOI] [PubMed] [Google Scholar]
- 67. Charrier C, Duncan GJ, Reid MD, et al. A novel class of CoA‐transferase involved in short‐chain fatty acid metabolism in butyrate‐producing human colonic bacteria. Microbiology. 2006;152:179–185. [DOI] [PubMed] [Google Scholar]
- 68. Hasan NM, Longacre MJ, Seed Ahmed M, et al. Lower succinyl‐CoA:3‐ketoacid‐CoA transferase (SCOT) and ATP citrate lyase in pancreatic islets of a rat model of type 2 diabetes: knockdown of SCOT inhibits insulin release in rat insulinoma cells. Arch Biochem Biophys. 2010;499:62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Fraser ME, Hayakawa K, Brown WD. Catalytic role of the conformational change in succinyl‐CoA:3‐oxoacid CoA transferase on binding CoA. Biochemistry. 2010;49:10319–10328. [DOI] [PubMed] [Google Scholar]
- 70. Lin TW, Bridger WA. Sequence of a cDNA clone encoding pig heart mitochondrial CoA transferase. J Biol Chem. 1992;267:975–978. [PubMed] [Google Scholar]
- 71. Rochet JC, Bridger WA. Identification of glutamate 344 as the catalytic residue in the active site of pig heart CoA transferase. Protein Sci. 1994;3:975–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Rochet JC, Oikawa K, Hicks LD, Kay CM, Bridger WA, Wolodko WT. Productive interactions between the two domains of pig heart CoA transferase during folding and assembly. Biochemistry. 1997;36:8807–8820. [DOI] [PubMed] [Google Scholar]
- 73. Oulton MM, Amons R, Liang P, MacRae TH. A 49 kDa microtubule cross‐linking protein from Artemia franciscana is a coenzyme A‐transferase. Eur J Biochem. 2003;270:4692–4972. [DOI] [PubMed] [Google Scholar]
- 74. Doten RC, Ngai KL, Mitchell DJ, Ornston LN. Cloning and genetic organization of the pca gene cluster from Acinetobacter calcoaceticus . J Bacteriol. 1987;169:3168–3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kowalchuk GA, Hartnett GB, Benson A, Houghton JE, Ngai KL, Ornston LN. Contrasting patterns of evolutionary divergence within the Acinetobacter calcoaceticus pca operon. Gene. 1994;146:23–30. [DOI] [PubMed] [Google Scholar]
- 76. Trachsel J, Bayles DO, Looft T, Levine UY, Allen HK. Function and phylogeny of bacterial butyryl coenzyme A:acetate transferases and their diversity in the proximal colon of swine. Appl Environ Microbiol. 2016;82:6788–6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Parke D. Supraoperonic clustering of pca genes for catabolism of the phenolic compound protocatechuate in Agrobacterium tumefaciens . J Bacteriol. 1995;177:3808–3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Parales RE, Harwood CS. Characterization of the genes encoding beta‐ketoadipate: succinyl‐coenzyme A transferase in Pseudomonas putida . J Bacteriol. 1992;174:4657–4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Sohling B, Gottschalk G. Molecular analysis of the anaerobic succinate degradation pathway in Clostridium kluyveri . J Bacteriol. 1996;178:871–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. MacLean AM, MacPherson G, Aneja P, Finan TM. Characterization of the beta‐ketoadipate pathway in Sinorhizobium meliloti . Appl Environ Microbiol. 2006;72:5403–5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Yang TH, Kim TW, Kang HO, et al. Biosynthesis of polylactic acid and its copolymers using evolved propionate CoA transferase and PHA synthase. Biotechnol Bioeng. 2010;105:150–160. [DOI] [PubMed] [Google Scholar]
- 82. Sato M, Yoshida Y, Nagano K, Hasegawa Y, Takebe J, Yoshimura F. Three CoA transferases involved in the production of short chain fatty acids in Porphyromonas gingivalis . Front Microbiol. 2016;7:1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Sasikaran J, Ziemski M, Zadora PK, Fleig A, Berg IA. Bacterial itaconate degradation promotes pathogenicity. Nat Chem Biol. 2014;10:371–377. [DOI] [PubMed] [Google Scholar]
- 84. Torres R, Lan B, Latif Y, Chim N, Goulding CW. Structural snapshots along the reaction pathway of Yersinia pestis RipA, a putative butyryl‐CoA transferase. Acta Crystallogr D Biol Crystallogr. 2014;70:1074–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Torres R, Swift RV, Chim N, et al. Biochemical, structural and molecular dynamics analyses of the potential virulence factor RipA from Yersinia pestis . PLoS One. 2011;6:e25084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Zhang B, Lingga C, Bowman C, Hackmann TJ. A new pathway for forming acetate and synthesizing ATP during fermentation in bacteria. Appl Environ Microbiol. 2021;87:e0295920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Stols L, Zhou M, Eschenfeldt WH, et al. New vectors for co‐expression of proteins: structure of Bacillus subtilis ScoAB obtained by high‐throughput protocols. Protein Expr Purif. 2007;53:396–403. [DOI] [PubMed] [Google Scholar]
- 88. Babnigg G, Jedrzejczak R, Nocek B, et al. Gene selection and cloning approaches for co‐expression and production of recombinant protein–protein complexes. J Struct Funct Genomics. 2015;16:113–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Corthésy‐Theulaz IE, Bergonzelli GE, Henry H, et al. Cloning and characterization of Helicobacter pylori succinyl CoA:acetoacetate CoA‐transferase, a novel prokaryotic member of the CoA‐transferase family. J Biol Chem. 1997;272:25659–25667. [DOI] [PubMed] [Google Scholar]
- 90. Zhang M, Xu HY, Wang YC, Shi ZB, Zhang NN. Structure of succinyl‐CoA:3‐ketoacid CoA transferase from Drosophila melanogaster . Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013;69:1089–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Haller T, Buckel T, Retey J, Gerlt JA. Discovering new enzymes and metabolic pathways: conversion of succinate to propionate by Escherichia coli . Biochemistry. 2000;39:4622–4629. [DOI] [PubMed] [Google Scholar]
- 92. Friedmann S, Alber BE, Fuchs G. Properties of succinyl‐coenzyme A:D‐citramalate coenzyme A transferase and its role in the autotrophic 3‐hydroxypropionate cycle of Chloroflexus aurantiacus . J Bacteriol. 2006;188:6460–6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Friedmann S, Steindorf A, Alber BE, Fuchs G. Properties of succinyl‐coenzyme A:L‐malate coenzyme A transferase and its role in the autotrophic 3‐hydroxypropionate cycle of Chloroflexus aurantiacus . J Bacteriol. 2006;188:2646–2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Göbel M, Kassel‐Cati K, Schmidt E, Reineke W. Degradation of aromatics and chloroaromatics by Pseudomonas sp. strain B13: cloning, characterization, and analysis of sequences encoding 3‐oxoadipate:succinyl‐coenzyme A (CoA) transferase and 3‐oxoadipyl‐CoA thiolase. J Bacteriol. 2002;184:216–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Mergelsberg M, Egle V, Boll M. Evolution of a xenobiotic degradation pathway: formation and capture of the labile phthaloyl‐CoA intermediate during anaerobic phthalate degradation. Mol Microbiol. 2018;108:614–626. [DOI] [PubMed] [Google Scholar]
- 96. Nilekani S, SivaRaman C. Purification and properties of citrate lyase from Escherichia coli . Biochemistry. 1983;22:4657–4663. [DOI] [PubMed] [Google Scholar]
- 97. Schneider K, Dimroth P, Bott M. Biosynthesis of the prosthetic group of citrate lyase. Biochemistry. 2000;39:9438–9450. [DOI] [PubMed] [Google Scholar]
- 98. Marlaire S, Van Schaftingen E, Veiga‐da‐Cunha M. C7orf10 encodes succinate‐hydroxymethylglutarate CoA‐transferase, the enzyme that converts glutarate to glutaryl‐CoA. J Inherit Metab Dis. 2014;37:13–19. [DOI] [PubMed] [Google Scholar]
- 99. Bott M, Dimroth P. Klebsiella pneumoniae genes for citrate lyase and citrate lyase ligase: localization, sequencing, and expression. Mol Microbiol. 1994;14:347–356. [DOI] [PubMed] [Google Scholar]
- 100. Mochizuki K, Inaoka DK, Mazet M, et al. The ASCT/SCS cycle fuels mitochondrial ATP and acetate production in Trypanosoma brucei . Biochim Biophys Acta Bioenerg. 2020;1861:148283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Zhang A. Metabolic engineering and process development for enhanced propionic acid production by Propionibacterium acidipropionici. Columbus, OH, USA: The Ohio State University, 2009;p. 249. [Google Scholar]
- 102. van Grinsven KW, Rosnowsky S, van Weelden SW, et al. Acetate:succinate CoA‐transferase in the hydrogenosomes of Trichomonas vaginalis: identification and characterization. J Biol Chem. 2008;283:1411–1418. [DOI] [PubMed] [Google Scholar]
- 103. Fleck CB, Brock M. Characterization of an acyl‐CoA: carboxylate CoA‐transferase from Aspergillus nidulans involved in propionyl‐CoA detoxification. Mol Microbiol. 2008;68:642–656. [DOI] [PubMed] [Google Scholar]
- 104. Mullins EA, Starks CM, Francois JA, Sael L, Kihara D, Kappock TJ. Formyl‐coenzyme A (CoA):oxalate CoA‐transferase from the acidophile Acetobacter aceti has a distinctive electrostatic surface and inherent acid stability. Protein Sci. 2012;21:686–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Yang Q, Guo S, Lu Q, et al. Butyryl/caproyl‐CoA: acetate CoA‐transferase: cloning, expression and characterization of the key enzyme involved in medium‐chain fatty acid biosynthesis. bioRxiv. 2021;41:BSR20211135. 10.1101/2021.03.10.434813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Wang Z, Ammar EM, Zhang A, Wang L, Lin M, Yang ST. Engineering Propionibacterium freudenreichii subsp. shermanii for enhanced propionic acid fermentation: effects of overexpressing propionyl‐CoA:succinate CoA transferase. Metab Eng. 2015;27:46–56. [DOI] [PubMed] [Google Scholar]
- 107. Rangarajan ES, Li Y, Ajamian E, et al. Crystallographic trapping of the glutamyl‐CoA thioester intermediate of family I CoA transferases. J Biol Chem. 2005;280:42919–42928. [DOI] [PubMed] [Google Scholar]
- 108. Cary JW, Petersen DJ, Papoutsakis ET, Bennett GN. Cloning and expression of Clostridium acetobutylicum ATCC 824 acetoacetyl‐coenzyme A:acetate/butyrate:coenzyme A‐transferase in Escherichia coli . Appl Environ Microbiol. 1990;56(1576–1):583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Petersen DJ, Cary JW, Vanderleyden J, Bennett GN. Sequence and arrangement of genes encoding enzymes of the acetone‐production pathway of Clostridium acetobutylicum ATCC 824. Gene. 1993;123:93–97. [DOI] [PubMed] [Google Scholar]
- 110. Gruez A, Roig‐Zamboni V, Valencia C, Campanacci V, Cambillau C. The crystal structure of the Escherichia coli YfdW gene product reveals a new fold of two interlaced rings identifying a wide family of CoA transferases. J Biol Chem. 2003;278:34582–34586. [DOI] [PubMed] [Google Scholar]
- 111. Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ. W‐IQ‐TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016;44:W232–W235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Sievers F, Wilm A, Dineen D, et al. Fast, scalable generation of high‐quality protein multiple sequence alignments using Clustal omega . Mol Syst Biol. 2011;7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Atkinson HJ, Morris JH, Ferrin TE, Babbitt PC. Using sequence similarity networks for visualization of relationships across diverse protein superfamilies. PLoS One. 2009;4:e4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Shannon P, Markiel A, Ozier O, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. [DOI] [PubMed] [Google Scholar]
- 116. Rost B. Twilight zone of protein sequence alignments. Protein Eng. 1999;12:85–94. [DOI] [PubMed] [Google Scholar]
- 117. Sander C, Schneider R. Database of homology‐derived protein structures and the structural meaning of sequence alignment. Proteins. 1991;9:56–68. [DOI] [PubMed] [Google Scholar]
- 118. Berg M, Hilbi H, Dimroth P. The acyl carrier protein of malonate decarboxylase of Malonomonas rubra contains 2′‐(5″‐phosphoribosyl)‐3′‐dephosphocoenzyme a as a prosthetic group. Biochemistry. 1996;35:4689–4696. [DOI] [PubMed] [Google Scholar]
- 119. Schmid M, Berg M, Hilbi H, Dimroth P. Malonate decarboxylase of Klebsiella pneumoniae catalyses the turnover of acetyl and malonyl thioester residues on a coenzyme‐A‐like prosthetic group. Eur J Biochem. 1996;237:221–228. [DOI] [PubMed] [Google Scholar]
- 120. Dimroth P. The prosthetic group of citrate‐lyase acyl‐carrier protein. Eur J Biochem. 1976;64:269–281. [DOI] [PubMed] [Google Scholar]
- 121. Maderbocus R, Fields BL, Hamilton K, et al. Crystal structure of a pseudomonas malonate decarboxylase holoenzyme hetero‐tetramer. Nat Commun. 2017;8:160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Morgat A, Lombardot T, Axelsen KB, et al. Updates in Rhea—an expert curated resource of biochemical reactions. Nucleic Acids Res. 2017;45:D415–D418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Moss GP. Recommendations of the nomenclature committee of the International Union of Biochemistry and Molecular Biology on the nomenclature and classification of enzymes by the reactions they catalyse. (2021) https://wwwqmulacuk/sbcs/iubmb/enzyme/.
- 124. Mistry J, Chuguransky S, Williams L, et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021;49:D412–D419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Andreeva A, Kulesha E, Gough J, Murzin AG. The SCOP database in 2020: Expanded classification of representative family and superfamily domains of known protein structures. Nucleic Acids Res. 2020;48:D376–D382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Schnoes AM, Ream DC, Thorman AW, Babbitt PC, Friedberg I. Biases in the experimental annotations of protein function and their effect on our understanding of protein function space. PLoS Comput Biol. 2013;9:e1003063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Hackmann TJ. Accurate estimation of microbial sequence diversity with distanced. Bioinformatics. 2020;36:728–734. [DOI] [PubMed] [Google Scholar]
- 128. Hackmann TJ, Zhang B. Using neural networks to mine text and predict metabolic traits for thousands of microbes. PLoS Comput Biol. 2021;2:e1008757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Bodenhofer U, Bonatesta E, Horejš‐Kainrath C, Hochreiter S. Msa: An R package for multiple sequence alignment. Bioinformatics. 2015;31:3997–3999. [DOI] [PubMed] [Google Scholar]
- 130. R Core Team . R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing, 2020. [Google Scholar]
- 131. Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UFBoot2: Improving the ultrafast bootstrap approximation. Mol Biol Evol. 2018;35:518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Yu G, Smith DK, Zhu H, Guan Y, Lam TTY. Ggtree: An R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. J Method Ecol Evol. 2017;8:28–36. [Google Scholar]
- 133. Letunic I, Bork P. Interactive tree of life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:W293–W296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Pagès H, Aboyoun P, Gentleman R, DebRoy S. Efficient manipulation of biological strings. Biostrings. 2020; https://bioconductororg/packages/Biostrings. [Google Scholar]
- 135. Wickham H. ggplot2: Elegant graphics for data analysis. New York, NY: Springer‐Verlag, 2016. [Google Scholar]
- 136. Braberg H, Webb BM, Tjioe E, Pieper U, Sali A, Madhusudhan MS. SALIGN: A web server for alignment of multiple protein sequences and structures. Bioinformatics. 2012;28:2072–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Paradis E, Schliep K. Ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics. 2019;35:526–528. [DOI] [PubMed] [Google Scholar]
- 138. Zeileis A, Fisher J, Hornik K, et al. Colorspace: A toolbox for manipulating and assessing colors and palettes. J Stat Softw. 2020;96:1–49. [Google Scholar]
- 139. Dusa A. venn: Draw Venn Diagrams. (2021) https://CRANR-projectorg/package=venn.
- 140. Eddy SR. Accelerated profile HMM searches. PLoS Comput Biol. 2011;7:e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Phylogenetic tree of CoA transferases that shows subfamilies IA, IB, and IC proposed in Tielens et al. (2010). Sequences included are from Fig. 3 of Tielens et al. (2010), plus all sequences from our own analysis. Some sequences from Tielens et al. (2010) belong to putative enzymes (ones with no biochemical evidence of activity), and for this reason were not included in our own analysis.
Figure S2. Crystal structure of a Gct family protein, showing the large number of layers in domain 1 that distinguishes this family. Structure of a Cat1 family protein, with typical three layers, shown for comparison. Proteins shown are GctAB (Afe) and AarC (Aac). Structures are presented in stereo and can be seen in 3D.
Figure S3. Phylogenetic tree of CoA transferase structures after removing putative enzymes, showing positions of other families in tree are unchanged.
Figure S4. Crystal structures show the region around active site of Cat1 family differs from that of Gct and OXCT1 families. Structures were aligned together and then overlaid within family. The indel region refers to E567 to G584 in Figure 4. The catalytic residue refers to that shown in Figure 4. Only structures with an identifiable catalytic residue are shown. See Table S1 for information on these enzymes and PDB accession numbers for structures.
Figure S5. CoA transferases show different organizations of their domains. (A) Types of organization. The N‐terminus and C‐terminus of each polypeptide (subunit) is shown. (B) Enzyme by type of organization. See Table S1 for information on these enzymes.
Figure S6. Phylogenetic tree of CoA transferases with sequences split by domains, showing that sequences do not always cluster by domain. See Table S1 for information on these enzymes.
Figure S7. Phylogenetic tree of CoA transferase superfamily plus proteins from additional families, showing the additional families are not closely related. See Table S1 for information on enzymes of the CoA transferase superfamily (Cat1, OXCT1, Gct, MdcA, Frc, CitF), and see Table S7 for information on proteins from additional/other families.
Table S1. Coenzyme A transferases that have been experimentally characterized in the primary literature
Table S2. Reactions of coenzyme A transferases in the primary literature
Table S3. Coenzyme A transferases that areputative enzymes and included in this analysis only for their structures
Table S4. Accession numbers of reactions in databases
Table S5. Coenzyme A transferases that have been annotated in databases
Table S6. Reactions of coenzyme A transferases in databases
Table S7. Proteins that some authorities consider part of a CoA transferase superfamily (1 representative protein per family)
Table S8. Coenzyme A transferase sequences and proposed subfamilies from Tielens et al. (2010)


