Summary
SHP2 is the first oncogenic tyrosine phosphatase encoded by PTPN11, which plays a significant regulatory role in cancer and inflammation-related diseases. Although SHP2 allosteric inhibitors have been used in phase I/II clinical trials for solid tumors, whether SHP2 inhibition alleviates psoriasis remains unclear. Here we expressed and purified SHP2 related proteins, and established an enzyme activity screening system for different conformations of SHP2. We launched an iterative medicinal chemistry program and identified the lead compound, TK-453. Importantly, TK-453 possessed stronger affinity with SHP2 than SHP099, evidenced by the cocrystal structure of SHP2/TK-453, revealing that the additional aryl-S-aryl bridge in TK-453 induces a 1.8 Å shift of the dichlorophenyl ring and an approximate 20° deviation of the pyrazine ring plane relative to SHP099. Furthermore, TK-453 significantly ameliorated imiquimod-triggered skin inflammation in mice via inhibition of the IL-23/Th17 axis, proving that SHP2 is a potential therapeutic target for psoriasis.
Subject areas: Biological sciences, Biochemistry, Medical biochemistry
Graphical abstract

Highlights
-
•
We identify a SHP2 allosteric inhibitor TK-453, which has a stronger affinity with SHP2
-
•
Cocrystal structure shows that TK-453 occupies the allosteric pocket of SHP2
-
•
TK-453 alleviates psoriasis-like skin inflammation in mice
-
•
SHP2 inhibitor provides a new strategy for the treatment of psoriasis
Biological sciences; Biochemistry; Medical biochemistry
Introduction
Protein tyrosine phosphorylation plays an important role in intracellular processes such as signal transduction and modifies biological processes, including cell proliferation, differentiation, and migration (Frankson et al., 2017). The imbalance of tyrosine phosphorylation has been confirmed to be related to a variety of diseases, such as cancer, inflammation, and diabetes (Tonks, 2006; van Huijsduijnen et al., 2002). PTPN11, a member of the PTP family, is a unique proto-oncogene (Bentires-Alj et al., 2004; Cheng et al., 2013), which encodes protein tyrosine phosphatase 2 (SHP2) containing Src homology 2 and participates in a variety of intracellular signaling pathways, such as RAS-MAPK, PI3K-AKT, JAK-STAT, and PD-1/PD-L1 (Hui et al., 2017; Li et al., 2015; Neel et al., 2003; Shi et al., 2000; Xu and Qu, 2008). SHP2 is located downstream of the RTK signaling pathway, which is a common node that activates multiple RAS signaling pathways. Therefore, a suitable SHP2 inhibitor can “catch all of the mutations in different RTK genes” and has the potential to become a broad-spectrum anti-cancer drug (Liu et al., 2020a; Song et al., 2021b). Nichols et al. found that an effective and selective SHP2 allosteric inhibitor RMC-4550 can commendably inhibit the growth of tumor cells in vivo, clarifying that SHP2 inhibitors destroy SOS1-mediated RAS-GTP load to reduce oncogenic RAS/RAF/MEK/ERK signaling and cancer growth, make BRAF, NF1LOF, and KRASG12mutant tumor cells sensitive to SHP2 inhibitors (Nichols et al., 2018). Regarding the role of SHP2 in the immune system, SHP2 acts as a downstream molecule of the programmed cell death signal. SHP2 binds to the phosphotyrosine motif (ITSM) of the immune checkpoint protein PD-1 through its two tandem Src homology domains, thereby activating SHP2-mediated immune suppression (Marasco et al., 2020). Our research showed that the SHP2 inhibitor SHP099 mediates anti-tumor immunity by enhancing the cytotoxicity of CD8+T cells, and the combination of SHP099 and PD-1 antibody achieves a better therapeutic effect (Zhao et al., 2019). Therefore, SHP2 inhibitors can block the interaction between PD-1 and SHP2, and SHP2 is expected to play a better role in tumor immunotherapy, which makes SHP2 a potential drug target in tumor immunotherapy.
Psoriasis is a chronic immune skin disease that easily relapses and is difficult to cure (Boehncke and Schon, 2015). The considerable role of IL-23/Th17 axis in the pathogenesis of psoriasis has attracted attention. Researches have shown that dendritic cells and macrophages in the psoriasis dermis produce IL-23, induce the activation of Th17 cells and γδT cells, and release IL-17A, IL-17F, IL-22, IL-6 and TNFα inflammatory cytokines, promoting the progression of psoriasis (Ganguly et al., 2013; Shabgah et al., 2017; van der Fits et al., 2009). Drugs targeting the IL-23/Th17 axis have been successfully marketed to improve psoriasis and have achieved fine curative effects (Hawkes et al., 2017; Kim and Krueger, 2017). In addition, a clinician reported that patients with Noonan syndrome with activating mutations in the SHP2 encoding gene also had pustular psoriasis, suggesting the potential role of SHP2 in psoriasis (Catharino et al., 2016). Therefore, we investigated the mRNA level, protein level, and enzymatic activity of SHP2 in psoriasis patients and healthy people, and found that in psoriasis patients SHP2 was highly expressed and its enzyme activity was higher than that of healthy people, and the symptoms of mice that conditionally knock out SHP2 on macrophages in the imiquimod (IMQ)-induced psoriasis model are reduced, suggesting that SHP2 on macrophages may become a new target for the treatment of psoriasis (Zhu et al., 2021). Collectively, SHP2 is considered as an attractive target for the treatment of human diseases, and the development of highly potent and selective SHP2 inhibitors will be of great significance for drug discovery.
Previously, the development of SHP2 inhibitors mainly used traditional inhibitors that target the PTP binding site, such as PHPS1 (Hellmuth et al., 2008), NSC-87877 (Chen et al., 2006), and GS-493 (Grosskopf et al., 2015). However, traditional inhibitors usually suffer from poor cell permeability and low bioavailability. In addition, because of the high degree of homology in the catalytic sites (such as SHP1 and PTP1B), it is quite challenging to find SHP2 inhibitors with high selectivity. It is encouraging that Novartis discovered the first SHP2 allosteric inhibitor SHP099 (Chen et al., 2016), which changed the previous allosteric inhibitors and reflected progress in the discovery of structure-based allosteric modulators to guide drug development. Here, we expressed and purified SHP2 related proteins (SHP2-WT, SHP2-E76K, SHP2-PTP, SHP1, and PTP1B) through a prokaryotic expression system, and proved that they have phosphatase activity. To screen new SHP2 allosteric inhibitors, we established an enzyme activity screening system for different conformations of SHP2 based on the conformational characteristics of SHP2. We launched an iterative medicinal chemistry program and identified the lead compound TK-453. Importantly, TK-453 possessed stronger affinity with SHP2 than SHP099, as evidenced by the cocrystal structure of SHP2/TK-453, revealing that the additional aryl-S-aryl bridge in TK-453 induces a 1.8 Å shift of the dichlorophenyl ring and an approximate 20° deviation of the pyrazine ring plane relative to SHP099. Furthermore, TK-453 significantly ameliorated imiquimod-triggered skin-like inflammation in mice via inhibition of the IL-23/Th17 axis. These findings suggest that a new SHP2 allosteric inhibitor TK-453 is a promising drug candidate for the treatment of psoriasis.
Results
Screening of SHP2 inhibitors
When the SHP2 protein is in the inactive state, the amino acid residues in the N-SH2 domain invert on the catalytic surface of the PTP domain to form a self-inhibitory conformation, thereby inhibiting the activity of the SHP2 protein and restricting the substrate from entering the catalytic site. To screen and obtain new SHP2 allosteric inhibitors, first, we expressed and purified SHP2 related proteins (SHP2-WT, SHP2-E76K, SHP2-PTP) through the prokaryotic expression system, and proved that they have phosphatase activity (Figures 1A and 1B). The catalytic activity of SHP2 was detected by the rapid fluorimetric method of DiFMUP, which is an alternative substrate. In addition, in order to verify the selectivity of the compound, we also expressed and purified the SHP1 and PTP1B proteins of the SHP2 family and their high homology, and compared the basic enzyme activities of several SHP2 related proteins. Next, we established a screening method for the different conformations of SHP2 protein. SHP2-WT is in a self-inhibited state and its basic enzyme activity is low. This required a short activating peptide IRS1 (containing two phosphorylated tyrosine residues) to induce changing of the protein conformation, while SHP2-E76K and SHP2-PTP themselves are in an activated conformation. We first screened candidate compounds for their inhibitory effects on the enzyme activity of SHP2-WT protein, and eliminated compounds with no inhibitory activity. Second, the compound with an inhibitory rate of more than 50% against SHP2-WT protein was screened for its inhibitory effect on SHP2-PTP protein. When the inhibitory effect of the compound on SHP2-PTP protein was equivalent to that of SHP2-WT protein, indicating that it was a traditional enzyme activity inhibitor; if the compound had no inhibitory effect on the SHP2-PTP protein, it meant that the compound did not act on the PTP domain and might be an inhibitor of the allosteric pockets (Figures 1C and 1D).
Figure 1.
Establishment of SHP2 allosteric inhibitor screening system
(A) SDS-PAGE Coomassie Brilliant Blue results of SHP2-WT, SHP2-E76K, SHP2-PTP, SHP1 and PTP1B proteins separated and purified by gel filtration chromatography.
(B) Basic enzyme activity of SHP2 related protein (SHP2-WT, SHP2-E76K, SHP2-PTP, SHP1 and PTP1B). Data are represented mean ±SEM. p values are determined by two-tailed unpaired Student’s t-test. ∗∗∗p < 0.001.
(C) Screening results of candidate compounds on the enzyme activity of SHP2-WT.
(D) Screening results of candidate compounds on the enzyme activity of SHP2-PTP proteins. The initial screening concentration of the compound was 10 μM (PHPS1 is a typical PTP catalytic domain inhibitor, and SHP099 is the first reported SHP2 allosteric inhibitor).
(E) Structure of TK-453.
SHP099, a highly effective (IC50 = 71nM), selective and orally bioavailable allosteric SHP2 inhibitor, occupies the cavity formed at the interface of the N-terminal SH2, C-terminal SH2, and protein tyrosine phosphatase (PTP) domain and then stabilizes SHP2 in an auto-inhibited conformation (Chen et al., 2016). The crystal structure of the SHP2/SHP099 complex reveals key hydrogen-bond interactions formed between SHP099 and the surrounding residues of SHP2 including Arg111 (N-SH2), Glu250 (PTP), and Phe113 (C-SH2), while the 2,3-dichlorophenyl ring of SHP099 does not form strong interactions with SHP2 (PDB code: 5EHR). The binding mode highlights the importance of the 2-aminopyrazine scaffold and provides a structural basis for the design of new allosteric SHP2 inhibitors. Surface representation of SHP2 in complex with SHP099 further illustrated that the phenyl ring and 4-amino piperidine moiety in SHP099 are modifiable sites. Therefore, we initiated an iterative medicinal chemistry program, leading to the identification of the lead compound TK-453 (Figure 1E).
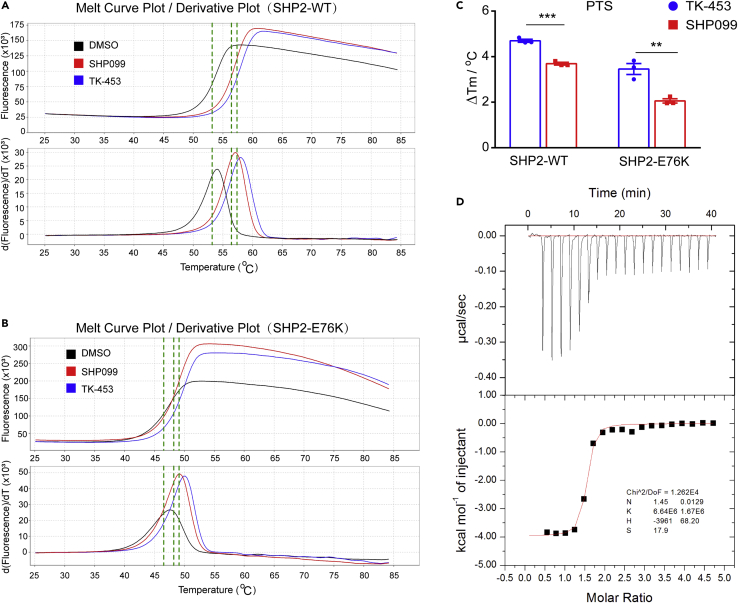
TK-453 has a higher affinity with SHP2 than SHP099 and is selective for SHP2
After preliminary screening of enzyme activity experiments, we found that the lead compound TK-453 has strong inhibitory effect on the enzyme activity of the SHP2-WT, and does not inhibit the activity of the SHP2-PTP catalytic domain (Figures 1C and 1D). Next, we further explored the affinity of TK-453 and SHP2 protein through the protein thermal shift experiment. The results showed that compound TK-453 binds SHP2-WT and SHP2-E76K very well (Figures 2A and 2C), and the affinity is higher than that of SHP099. We performed isothermal titration calorimetry (ITC) with compound TK-453 and SHP2-WT, and obtained a KD value of 150 nM (Figure 2D). The results indicate that compound TK-453 may be an inhibitor of the SHP2 allosteric pocket.
Figure 2.
The affinity of TK-453 and SHP2 is higher than that of SHP099
(A) Protein thermal shift experiment of SHP2-WT, protein thermal dissolution curve of protein incubated with different ligands.
(B) Protein thermal shift experiment of SHP2-E76K, incubated with different ligands (SHP099 and TK-453 concentrations are 20 μM).
(C) Compared with the solvent control group, SHP099 and TK-453 of ΔTm values. Data are represented mean ±SEM. p values are determined by two-tailed unpaired Student’s t-test. ∗∗∗p<0.01, ∗∗∗p<0.001.
(D) Isothermal titration calorimetry (ITC) results of TK-453 compound and SHP2-WT protein.
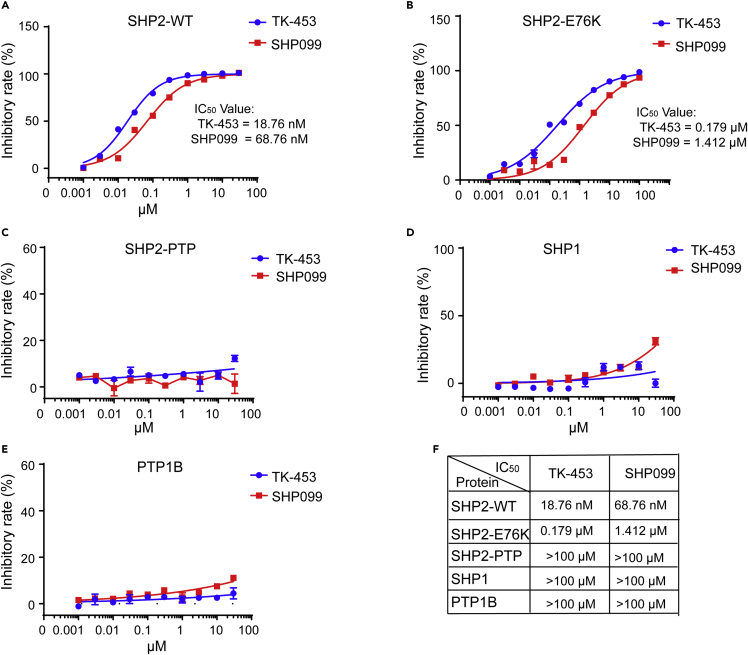
Next, we compared whether TK-453 had selective inhibitory effects on SHP2-related proteins (SHP2-WT, SHP2-E76K, SHP2-PTP) and their homologues (SHP1 and PTP1B), and compared their IC50 values. The results showed that the inhibitory effect of TK-453 on the enzyme activity of SHP2-WT and SHP2-E76K protein was better than that of SHP099. The IC50 value of SHP2-WT was 18.76 nM, and compared with the IC50 value of SHP099 (68.76 nM), the inhibitory activity of TK-453 was 3.7 times stronger (Figure 3A). TK-453 had an IC50 value of 0.179 μM for SHP2-E76K, which was 7.9 times stronger than SHP099 (IC50 = 1.412 μM) (Figure 3B). Consistently, SHP099 and TK-453 had no inhibitory effect on SHP2-PTP protein, indicating that they were allosteric inhibitors (Figure 3C), and their IC50 values for SHP2 family proteins SHP1 and PTP1B were greater than 100 μM, showing that TK-453 was selective for the inhibition of SHP2 enzyme activity (Figures 3D and 3F).
Figure 3.
TK-453 is selective for the inhibitory effect of SHP2
(A–E) Protease activity test of SHP2-WT(A), SHP2-E76K (B), SHP2-PTP (C), SHP1 (D) and PTP1B (E), gradient concentrations of SHP099 and TK-453, and calculate the IC50 values respectively.
(F) Statistical table of IC50 values of SHP099 and TK-453 for the SHP2 family and related proteins. The results of the enzyme activity experiment were repeated three times, expressed as mean ± SEM.
TK-453 binds in the allosteric binding pocket at the junction of three domains and stabilizes the autoinhibition conformation of SHP2
To better understand the molecular basis of TK-453 binding to SHP2, we determined the cocrystal structure of SHP2 in complex with SHP2/TK-453 to a resolution of 2.1 Å by molecular replacement (Figure 4, Table S1). The composite omit map of TK-453 unambiguously showed that it occupies a channel comprised the N-SH2, C-SH2 and PTP domains (Figures 4A and 4B), to stabilize the inactive autoinhibition conformation of SHP2. TK-453 adopts a similar conformation as SHP099 with an overlapping piperidine motif. The extra aryl-S-aryl bridge in TK-453 induces a 1.8 Å shift of the dichlorophenyl ring and an approximate 20° deviation of the pyrazine ring plane relatively to SHP099 (Figure 4C). The structure of SHP2/TK-453 also revealed a series of protein-ligand interactions, which has primarily been observed in the structure of SHP2/SHP099 (Chen et al., 2016). The pyrazine aniline of TK-453 forms a hydrogen bond with the backbone carbonyl of Glu250 from the PTP domain and the pyrazine nitrogen serves as the hydrogen acceptor to bond with Arg111 from the N-SH2 domain. The dichlorophenyl ring of TK-453 makes hydrophobic interactions with a cluster of residues in the PTP domain including Leu254, Gln257, Arg492, Gln495 and Met496. The dichlorophenyl ring also participates in a cationic−π stacking interaction with Arg111 (Figure 4D).Additionally, the 4-(aminomethyl)-4-methylpiperidine motif of TK-453 orients toward the terminal of the ligand binding tunnel and the primary amine approaches within hydrogen bonding distance of Thr108, Glu110 and Thr253 (2.7, 2.6 and 2.7 Å respectively) (Figure 4E). The composite omit map of key residues of SHP2 which participate in the interaction with TK-453 were calculated and showed in Figure 4F. Piecing together, the binding mode of TK-453 represents a favorable shape complementarity to the allosteric pocket of SHP2, which agrees well with the high inhibitory activity of TK-453 to SHP2.
Figure 4.
Crystal structure of SHP2/TK-453
(A) Surface representation of SHP2 interacting with TK-453 shown as yellow sticks. The N-SH2 domain (residues Met1-Arg111) of SHP2 is shown in green, the C-SH2 domain (residues Trp112-Thr219) is shown in blue, and the PTP (residues Arg220-Leu525) domain is shown in beige. Inset: allosteric binding pocket at the junction of the three domains.
(B) Fo - Fc electron density (green mesh, calculated using program Phenix composite omit map) of TK453 is superimposed on the model.
(C) Conformation of TK-453 (yellow) overlaid with SHP099 (magenta).
(D) Detailed interactions between aminopyrazine ring of TK-453 and Arg111 and Glu250 of SHP2 (marked in red) and between dichlorophenyl ring of TK-453 and L54, Q257, R492, Q495 and M496 of SHP2 (marked in black).
(E) Detailed interactions between amine region of TK-453 with Thr253, Thr108, and Glu110 of SHP2 (marked in red).
(F) 2Fo - Fc electron density (blue mesh, calculated using program Phenix composite omit map) of key residues of SHP2 which participate in the interaction with TK-453 is superimposed on the model. Hydrogen bonds are shown as black dashed lines.
TK-453 alleviates imiquimod-induced inflammation in macrophages
A clinician reported that patients with Noonan syndrome with activating mutations in the gene encoding SHP2 also had psoriasis circular pustules, indicating the potential role of SHP2 in psoriasis (Catharino et al., 2016). Therefore, we investigated the mRNA level, protein level and enzyme activity of SHP2 in psoriasis patients and healthy people, and found that SHP2 is highly expressed in psoriasis patients. Mice that conditionally knock out SHP2 on macrophages have reduced disease, suggesting that SHP2 on macrophages may become a new target for the treatment of psoriasis. SHP2 inhibitors can provide a new direction for the treatment of psoriasis (Zhu et al., 2021).
We first investigated the anti-inflammatory effects of SHP2 inhibitors in mouse peritoneal macrophages (PMs) (Figure 5A) and bone marrow-derived macrophages (BMDMs) (Figure 5B), and used imiquimod to induce inflammation models to test their correlation. The gene expression levels of inflammatory factors Il23a, Il22, Il6, Tnfα, Il1b, and Ptgs2 showed that TK-453 and SHP099 significantly alleviated imiquimod-induced inflammation models in macrophages, and the anti-inflammatory activity of TK-453 was stronger than that of SHP099. Western blot experiments also showed that SHP2 inhibition inhibited the phosphorylation levels of p-IKKα/β and p-p65 (Figures 5C and 5D). Overall, these data clearly indicated that SHP2 plays an important role in IMQ-induced inflammation, and inhibiting its activity can achieve a fine anti-inflammatory effect.
Figure 5.
TK-453 alleviates imiquimod (IMQ)-induced inflammation in macrophages
(A and B) qPCR analysis of mRNA levels. After IMQ stimulation, the genes encoding the inflammatory factors Il23a, Il22, Il6, Tnfα, Il1b and Ptgs2 in mouse peritoneal macrophages PMs (A) and bone marrow-derived macrophages BMDMs (B) were detected by qPCR (the internal control is GAPDH), TK-453 and SHP099 are pretreated for 3 h at the concentration shown in the figure, and stimulated with 5 μM IMQ for 6 h. Data are represented mean ±SEM. All groups are compared with the IMQ group, one-way ANOVA with Tukey’s multiple comparisons was used to assess statistical significance, ###p<0.001, ∗p<0.05, ∗∗p<0.01, ∗∗∗p<0.001.
(C) Western blot. To detect the phosphorylation levels of inflammation-related proteins in mouse peritoneal macrophages (PMs) and bone marrow-derived macrophages (BMDMs) after IMQ stimulation, cells were pretreated with 10 μM TK-453 and SHP099 for 3 h, and then stimulated with 5 μM IMQ for 1 h.
(D) Western blot quantitation of the indicated proteins in PMs and BMDMs. Data are represented mean ±SEM. All groups are compared with the IMQ group, one-way ANOVA with Tukey’s multiple comparisons was used to assess statistical significance, ###p<0.001, ∗p<0.05, ∗∗p<0.01, ∗∗∗p<0.001.
TK-453 significantly ameliorates imiquimod-triggered skin-like inflammation in mice via inhibition of IL-23/Th17 axis
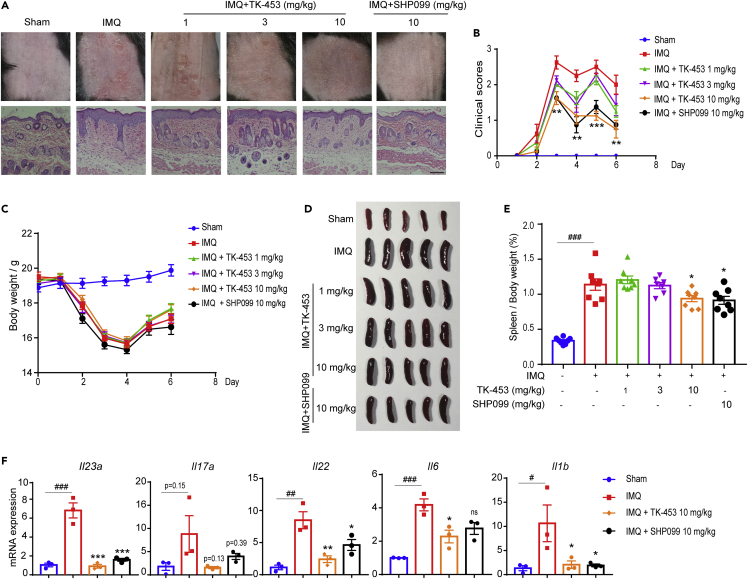
Furthermore, we verified the therapeutic effect of TK-453 on an animal model of psoriasis induced by imiquimod. After continuous administration for 5 days, the mice were anesthetized and sacrificed on the sixth day. The mice’s skin phenotype (observed and evaluated from the three indicators of erythema, scaliness, and skin thickness) showed that the TK-453 treatment group could effectively improve the redness and epidermal scales on the back skin of psoriasis model mice induced by imiquimod. The HE staining results of skin sections also showed that TK-453 and SHP099 can effectively inhibit the proliferation of epidermal cells, and the infiltration of inflammatory cells is significantly reduced (Figure 6A). Figure 6B shows the PASI score, on the third day of administration, TK-453 significantly improved the redness and scaly of the skin of mice with psoriasis, and the weight of the mice began to rise on the fourth day (Figure 6C). The imiquimod-induced psoriasis model can cause the spleen of mice to enlarge. We compared the appearance and size of the mouse spleen (Figure 6D) and organ index (spleen/body weight) between the administration group and the model group. The results proved that the organ index of mice in the administration group decreased (Figure 6E), indicating to a certain extent that TK-453 has an anti-inflammatory effect. In addition, we tested the mRNA expression levels of IL-23/IL-17A axis cytokines and other psoriasis-related cytokines in the back skin of mice, and the results showed that TK-453 significantly inhibited the expression of inflammatory factors such as Il23a (Figure 6F). In summary, our results proved that TK-453 ameliorated imiquimod-triggered skin-like inflammation in mice via inhibition of IL-23/Th17 axis, suggesting that SHP2 may be a therapeutic target for psoriasis, and SHP2 allosteric inhibitors provide a new strategy for the treatment of psoriasis.
Figure 6.
TK-453 significantly ameliorates imiquimod-triggered skin-like inflammation in mice via inhibition of IL-23/Th17 axis
(A) The phenotype of the back skin of the mouse and the HE staining image of the skin section. Scale bar: 100 μm.
(B) The clinical score of the imiquimod-induced psoriasis mouse skin phenotype (evaluated from three indicators: erythema, scale, and skin thickness) (n = 8/group).
(C) The weight change of mice (n = 8/group).
(D) On the sixth day after the mice were sacrificed by anesthesia, the spleens of the mice in each test group were obtained (n = 5/group).
(E) Organ index of mice in each experimental group (n = 8/group).
(F) qPCR analysis of mRNA levels encoding IL-23/IL-17A axis cytokines and other psoriasis-related cytokines in the back skin of mice in each group, and the results were normalized to GAPDH expression (n = 3/group). Data are represented mean ±SEM. All groups are compared with the IMQ group, one-way ANOVA with Tukey’s multiple comparisons was used to assess statistical significance (B, E and F), ###p<0.001, ∗p<0.05, ∗∗p<0.01, ∗∗∗p<0.001, ns, not significant.
Discussion
Protein tyrosine phosphatases (PTPs) are widely expressed in cells and mainly play a role in signal transmission and regulation. In eukaryotic cells, protein kinases and phosphatases jointly regulate intracellular phosphorylation and maintain body balance. Abnormal function of protein kinase and phosphatase can cause disease. For a long time, precision medicine with protein tyrosine kinase (PTK) as the target has been the goal of people’s efforts. Because of the highly active sites of drugs and their essential role in cell signaling pathways, PTK has become an ideal drug target in tumor research. However, the difficulty of developing targeted inhibitors of protein tyrosine phosphatase (PTP) has led people to believe that PTP is “undruggable”, thus emphasizing the need for new methods to develop PTP drugs (Mullard, 2018). With the deepening of research, phosphatase has begun to get rid of the stigma of its “undruggable”, and SHP2 is one of the most representative. Currently, there are four main types of research strategies aimed at inhibiting SHP2 activity: (1) Traditional catalytic site inhibitors, such as PHPS1 (Hellmuth et al., 2008), NSC-87877 (Chen et al., 2006), and GS-493 (Grosskopf et al., 2015). These traditional catalytic site inhibitors due to poor cell membrane permeability, difficulty entering the blood circulation through oral administration, and nonselective selection for proteins of the same family protein, thus limiting clinical research. (2) Allosteric inhibition. Since the first allosteric inhibitor SHP099 was reported in 2016, several effective SHP2 inhibitors have been identified (Chen et al., 2016). The allosteric mechanism “molecular glue” targeting SHP2 inhibitors has aroused special interest in this long-sought goal (LaMarche et al., 2020; Nichols et al., 2018; Song et al., 2021a; Xie et al., 2017). (3) PROTAC degrader of SHP2 protein. Wang et al. found an effective small molecule SHP2 degrading agent, SHP2-D26, which can reduce SHP2 protein levels by >95% in esophageal cancer KYSE520 and acute myeloid leukemia MV4;11 cells (Wang et al., 2021). (4) Protein-protein interaction inhibitor. Fan et al. clarified that blocking the interaction between SHP2 and PD-1 denotes a novel opportunity for developing PD-1 inhibitors (Fan et al., 2020). With the development of SHP2 inhibition in recent years, SHP2 has become a moderately hot target. At present, SHP2 allosteric inhibitors, such as TNO155, RMC-4630, JAB-3068, and JAB-3312, have first entered clinical trials. These compounds are currently in different stages of clinical trials, mainly for their anti-tumor effects and their combined effects with kinase inhibitors.
Although SHP2 allosteric inhibitors have been used in phase I/II clinical trials for solid tumors, whether SHP2 inhibition alleviates psoriasis remains unclear. Many previous studies by our group focused on SHP2, and explored which signaling pathways SHP2 participates in the regulation of intracellular signaling pathways and its role in various diseases (Guo et al., 2017; Liu et al., 2021b; Shao et al., 2021). We found that SHP2 inhibition attenuates osteoarthritis in mice by promoting homeostasis of cartilage metabolism via a DOK1/AP1/UPP1/Uridine cascade (Liu et al., 2021a). In addition, in chronic low-grade inflammation type 2 diabetes, SHP2 inhibition improves high-fat diet-induced hepatic steatosis and insulin resistance (Liu et al., 2020b). Our latest research discovered that SHP2 was highly expressed in both psoriatic patients and imiquimod-induced psoriasis-like mice. Single-cell RNA sequencing of murine skin demonstrated that SHP2 inhibition impaired skin inflammation in myeloid cells, especially macrophages, and SHP2 inhibitors significantly ameliorated imiquimod-triggered skin inflammation in mice (Zhu et al., 2021). In addition, Wang et al. found that in cultured human systemic lupus erythematosus T cells, inhibition of SHP2 reduced the production of IFN-γ and IL-17A/F, indicating that inhibition of SHP2 is used to improve systemic lupus erythematosus (Wang et al., 2016). Liu et al. reviewed targeting SHP2 as a therapeutic strategy for inflammatory diseases (Liu et al., 2021c). These studies indicate that SHP2 can be used as a therapeutic target in inflammatory diseases, but SHP2 inhibitors for the treatment of these inflammatory diseases have yet to be confirmed. The development of drugs targeting phosphatases has been extremely challenging, and with the development of allosteric inhibitors, phosphatases are beginning to shed their undruggable stigma (Mullard, 2018; Stanford and Bottini, 2017). However, because SHP2 is widely expressed in cells, it may have certain side effects. It is necessary to evaluate the safety of the drug and find a disease population suitable for the use of SHP2 inhibitors. Thus, high selectivity is essential to reduce the toxicity. In conclusion, we suggest that research and development of SHP2 inhibitors for the treatment of SHP2-related diseases is of great significance.
In the past few decades, SHP2 was regarded as an “undruggable” target, but the allosteric effect of protein provides attractive prospects for the emergence of new drug targets. In conclusion, we explored the possibility of SHP2 as a target in inflammatory diseases, revealing that SHP2 inhibitors significantly ameliorate imiquimod-triggered skin-like inflammation in mice via inhibition of IL-23/Th17 axis. Our study suggests that SHP2 allosteric inhibitor TK-453 is a promising drug candidate for the treatment of psoriasis.
Limitations of the study
There are some limitations to this study, one of which is that the psoriasis model induced by imiquimod cannot fully simulate the pathological state of patients with psoriasis. However, there is no well animal model that can fully simulate the pathological state of psoriasis at present. We only verified the anti-inflammatory activity of SHP2 allosteric inhibitors in the inflammatory environment stimulated by imiquimod, and more animal models are needed to confirm.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-p-IKKα/β | Cell Signal Technology | Cat#2697; RRID: AB_2079382 |
| Mouse monoclonal anti-IKKα | Cell Signal Technology | Cat# 11930; RRID: AB_2687618 |
| Rabbit monoclonal anti-IKKβ | Cell Signal Technology | Cat#8943; RRID: AB_11024092 |
| Rabbit monoclonal anti-p-p65 | Cell Signal Technology | Cat#3033; RRID: AB_331284 |
| Rabbit monoclonal anti-p65 | Cell Signal Technology | Cat#8242; RRID: AB_10859369 |
| Rabbit monoclonal anti-Phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) | Cell Signal Technology | Cat#4370; RRID: AB_2315112 |
| Rabbit monoclonal anti-p44/42 MAPK (Erk1/2) | Cell Signal Technology | Cat#4695; RRID: AB_390779 |
| Mouse monoclonal anti-β-actin | Cell Signal Technology | Cat#3700; RRID: AB_2242334 |
| Bacterial and virus strains | ||
| E. coli DH5α Competent Cells | WEIDI | Cat#DL1001 |
| E. coli BL21(DE3) Competent Cells | WEIDI | Cat#EC1002 |
| Chemicals, peptides, and recombinant proteins | ||
| Trypsin | OXOID | Lot#2862082 |
| Yeast Extract | OXOID | Lot#2831838-02 |
| NaCl | General-Reagent | Lot#P1780219 |
| TRIS | VETEC | Lot#WXBD0198V |
| DiFMUP | Thermo Fisher Scientific, MA, USA | Catalog number: D6567 |
| IRS1 peptide | ChinaPeptides Co., Ltd., SZ, China | N/A; designed and special ordered for enzyme activity assay |
| Critical commercial assays | ||
| Protein Thermal Shift | Thermo Fisher Scientific, MA, USA | Catalog number: 4461146 |
| Deposited data | ||
| SHP2_TK-453 complex structure | This paper | PDB: 7VXG |
| Experimental models: Cell lines | ||
| Peritoneal macrophages | C57BL/6 mice in this paper | N/A |
| Bone marrow-derived macrophages | C57BL/6 mice in this paper | N/A |
| Oligonucleotides | ||
| Gapdh forward: TGA CCT CAA CTA CAT GGT CTA CA | PrimerBank | 126012538c2 |
| Gapdh reverse: CTT CCC ATT CTC GGC CTT G | PrimerBank | 126012538c2 |
| Il23a forward: ATG CTG GAT TGC AGA GCA GTA | PrimerBank | 13752579a1 |
| Il23a reverse: ACG GGG CAC ATT ATT TTT AGT CT | PrimerBank | 13752579a1 |
| Il17a forward: TTT AAC TCC CTT GGC GCA AAA | PrimerBank | 6754324a1 |
| Il17a reverse: CTT TCC CTC CGC ATT GAC AC | PrimerBank | 6754324a1 |
| Il22 forward: ATG AGT TTT TCC CTT ATG GGG AC | PrimerBank | 21426819a1 |
| Il22 reverse: GCT GGA AGT TGG ACA CCT CAA | PrimerBank | 21426819a1 |
| Il6 forward: CTG CAA GAG ACT TCC ATC CAG | PrimerBank | 13624310c1 |
| Il6 reverse: AGT GGT ATA GAC AGG TCT GTT GG | PrimerBank | 13624310c1 |
| Tnfα forward: GTC CTG CTC TAC GTG ACG AG | PrimerBank | 30425140a1 |
| Tnfα reverse: TCT CTC CTT TTC TGC CAT CTC T | PrimerBank | 30425140a1 |
| Ptgs2 forward: TGA GCA ACT ATT CCA AAC CAG C | PrimerBank | 31981525a1 |
| Ptgs2 reverse: GCA CGT AGT CTT CGA TCA CTA TC | PrimerBank | 31981525a1 |
| Recombinant DNA | ||
| pET15b-SHP2 | This paper | N/A |
| Software and algorithms | ||
| PHENIX | (Liebschner et al., 2019) | http://www.phenix-online.org/ |
| Coot 0.9.6 | (Emsley and Cowtan, 2004) | https://www2.mrclmb.cam.ac.uk/personal/pemsley/coot/ |
| HKL2000 | (Otwinowski and Minor, 1997) | http://www.hkl-xray.com/ |
| Pymol | Schrodinger, LLC | https://pymol.org/2/ |
| Protein Thermal Shift™software v1.4 | Thermofisher | https://www.thermofisher.com/ |
| Origin software v7.0 | OriginLab | https://www.originlab.com/ |
| Graphpad Prism v7.0 | GraphPad Software | https://www.graphpad.com/ |
| Other | ||
| HiTrap Q HP 5 mL | GE Healthcare | Lot#17115401 |
| HisTrap HP 5 mL | GE Healthcare | Lot#17524802 |
| Superdex 200 Increase 10/300 column | GE Healthcare | Lot#28990944 |
| 30 kDa cutoff concentrators | Millipore | REF#UFC903096 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Yang Sun (yangsun@nju.edu.cn)
Material availability
Bacterial strains and antibodies were obtained from the commercial or academic sources described in the STAR methods key resources table. Material generated in this study will be made available upon reasonable request.
Experimental models and subject details
Protein expression in E. coli
E. coli BL21(DE3) competent cells used in the studies were from Shanghai Weidi Biotechnology Co., Ltd. Cells were grown in LB (Luria-Bertani) medium to OD600 between 0.6 and 0.8, induced at 16°C for 16 h with 0.8 mM IPTG.
Primary cell culture
Peritoneal macrophages (PMs) and Bone marrow-derived macrophages (BMDMs) from wild-type mice were extracted according to the method details. The cells were cultured in Dulbecco Modified Eagle medium (DMEM, Gibco, 11995-073) containing 10% fetal bovine serum and penicillin-streptomycin (1×), and placed in a 37°C, 5% CO2 incubator nourish. After the cells adhere to the wall and induce differentiation, they are used for experiments.
Animals
C57BL/6 mice were purchased from GemPharmatech Co. Ltd (Nanjing, China). All mice were of the C57BL/6 background strain, and we used age- and sex-matched mice 6-10-week-old female mice. Mice had unlimited access to standard laboratory chow and water under a light-dark cycle of 12 h each in a room at a temperature of 22°C. All animal experiments were carried out according to the NIH Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011) and were approved by the Experimental Animal Care and Use Committee of Nanjing University (IACUC-2006024). All efforts were made to minimize the number of animals used and their suffering.
Method details
Synthesis of compound TK-453
A suspension of 3-bromo-6-chloropyrazin-2-amine 1 (300 mg, 1.44 mmol, 1 eq.), 2,3-dichlorobenzenethiol 2 (335 mg, 1.87 mmol, 1.3 eq.), potassium carbonate (597 mg, 4.32 mmol, 3 eq.), copper iodide (27 mg, 0.1 eq., 0.144 mmol) and 1,10-phenanthroline (26 mg, 0.1 eq., 0.144 mmol) in 1,4-dioxane (degassed) was stirred for 8h at 100°C. After cooling to room temperature, the mixture was filtered through a pad of diatomite followed by wash with CH2Cl2. The filtrates were concentrated and the the resulting residue was purified by flash column chromatography (PE: EA = 8:1-5:1) to give the intermediate 6-chloro-3-((2,3-dichlorophenyl) thio) pyrazin-2-amine (3, 286 mg, yield 64.8%). 1H NMR (400 MHz, CDCl3) δ 8.02 (s, 1H), 7.60 (dd, J = 7.8, 1.8 Hz, 1H), 7.40 – 7.30 (m, 2H), 4.66 (s, 2H).
To a suspension of the 6-chloro-3-((2,3-dichlorophenyl) thio) pyrazin-2-amine 3 (286 mg, 1 eq., 0.933 mmol), N, N-Diisopropylethylamine (DIEA, 307 μL, 2 eq., 1.87 mmol) in NMP (10 mL) was added tert-butyl ((4-methylpiperidin-4-yl) methyl) carbamate (240 mg, 1.2 eq., 1.12 mmol). Then the resulting slurry was kept stirring for 4hat 140°C. After cooling to room temperature, the water (10 mL) was added to the mixture, the formed solid 4 was filtered off and used for the next step without further purification. The resulting crude product was dissolved in DCM (10 mL), followed treatment with TFA (3 mL) was added. The mixture was stirred for 2h at room temperature, and the volatiles were removed under reduced pressure, the resulting residue was purified by flash column chromatography (DCM: MeOH = 20:1-10:1) to give TK-453. White solid, m.p. 153.7-155.0°C, yield 45%. 1H NMR (400 MHz, DMSO-d6) δ 7.62 (s, 1H), 7.37 (d, J = 7.9 Hz, 1H), 7.20 (t, J = 8.0 Hz, 1H), 6.57 (d, J = 8.0 Hz, 1H), 6.14 (s, 2H), 3.85 – 3.79 (m, 2H), 3.38 – 3.26 (m, 2H), 2.58 – 2.41 (m, 2H), 1.46 (m, 2H), 1.38 – 1.25 (m, 2H), 1.02 (s, 3H). 13C NMR (100 MHz, DMSO-d6) δ 156.31, 154.24, 140.17, 132.59, 128.78, 127.78, 127.04, 124.99, 120.98, 112.66, 44.00, 43.79, 34.38, 29.11. The HPLC purity was determined to be 98.64%, the HPLC condition was MeOH/H2O (60:40, v/v), and the flow rate was 0.8 mL/min (Phenomenex column, C-18, 5.0 μm, 4.6 mm × 250 mm).
Protein expression and purification
Coding sequence for human SHP2 from residues Met1-Leu525 was cloned into the expression vector pET15b containing an N-terminal 6×His-tag followed by a tobacco etch virus (TEV) protease cleavage site. The recombinant protein SHP2 was expressed in BL21(DE3) Escherichia coli cells (Tsingke) and induced with 0.8 mM IPTG in Terrific Broth media at 18°C for 20 h. The cells were pelleted by centrifugation and resuspended in lysis buffer containing 50 mM Tris-HCl pH 8.0, 500 mM NaCl, 0.1 mM TCEP, 5% glycerol, and 1 mM phenylmethylsulfonyl fluoride (PMSF). Then the cells were lysed with a high-pressure cell disrupter, and the lysate was centrifuged for 1h at 18000 g. The supernatant was loaded onto a HisTrap HP affinity column (GE Healthcare), eluted in 50 mM Tris (pH 8.0), 500 mM NaCl, and 250 mM imidazole. The protein was further purified over a HitrapQ anion exchange column (GE Heathcare) in 50 mM Tris-HCl (pH 8.0) using a 50–300 mM NaCl gradient elution. The N-terminal His-tag was removed with an overnight incubation with TEV protease at 4°C. The protein was subsequently applied to a HisTrap HP affinity column and the flow through fractions containing SHP2 were pooled, concentrated, and loaded onto a Superdex 200 Increase 10/300 GL column (GE Healthcare) to exchange the protein into crystallization buffer (20 mM Tris-HCl pH 8.0, 100 mM NaCl, 5 mM DTT). The protein was concentrated to 15 mg/mL for crystallization.
Crystallization and structure determination
Apo-SHP2 crystals were obtained at 22°C by vapor diffusion in sitting drops using 1 μL of the protein (10 mg/mL) and 1 μL of a reservoir solution consisting of 100 mM Tris-HCl pH 8.2, 12% (vol/vol) PEG 4000, 20 mM DTT. Crystals formed within 3 days and were subsequently soaked with compound in powder. After soaking for 12 h, crystals were cryoprotected in the reservoir solution supplemented with 20% glycerol and flash frozen in liquid nitrogen. Diffraction data were collected at the Shanghai Synchrotron Radiation Facility (SSRF) beamline BL17U1. Data were processed using HKL2000 (Otwinowski and Minor, 1997).
The structure of SHP2 in complex with the compound was solved by molecular replacement (Bunkoczi and Read, 2011) using PHASER (McCoy et al., 2007) with the coordinates of apo SHP2 (Hof et al., 1998) (PDB ID 2SHP) as the search model. The complex model was refined using Phenix (Adams et al., 2010) and built using Coot (Emsley and Cowtan, 2004). Details of the data collection and refinement statistics are summarized in Table S1. The final model was validated by MolProbity (Chen et al., 2010). All structural figures were generated with PyMOL (Version 2.3.0 Schrödinger, LLC).
Enzyme activity assay of SHP2 protein
The expressed and purified SHP2-0WT protein is allosterically activated by the binding of the diacetyl phosphorylated peptide IRS1 to its Src homology 2 (SH2) domain. SHP2-E76K, SHP2-PTP, SHP1 and PTP1B do not need to add IRS1 to activate the conformation, and directly pre-incubate with the compound. Co-incubate 0.5 nM of SHP2 with 0.5 μM double phosphorylated IRS1 peptide (sequence: H2N-LN (pY) IDLDLV (dPEG8) LST (pY) ASINFQK-amide) and 0.001-100 μM compound, except for SHP2 related proteins without IRS1, other conditions are the same. After pre-incubation at 25°C for 30–60 min, the replacement substrate DiFMUP (Invitrogen) was added to the reaction and incubated at 25°C for 30 min. The reaction was then quenched by adding 20 μL of 160 μM bpV (Phen) solution (Enzo Life Sciences). The fluorescence signal was measured using a microplate reader (Envision) with excitation and emission wavelengths of 340 and 450 nm, respectively. The inhibitor dose-response curve was analyzed using standardized IC.
Protein thermal shift assay
The final reaction system is 20 μL, and the reaction is carried out on a 96-well PCR plate. First add 12.5 μL of protein diluent (the final protein content is 3 μg), and then add 5 μL of the candidate compound (concentration of 20 μM, DMSO content of 2%), finally add 2.5 μL of freshly diluted Protein Thermal Shift™ dye (8x), set 4 multiple holes, pipette up and down 10 times to mix well. Seal the plate with MicroAmp® optical film, centrifuge at 1000 rpm for 1 min, and then incubate it on ice for 30 min. Use the StepOne Plus™ instrument to set up the program, and use the Protein Thermal Shift™ software v1.4 to analyze the data to get the protein dissolution curve, Tm value, etc.
Isothermal titration calorimetry assay
SHP2 protein was dialyzed repeatedly against 50 mM phosphate buffer, pH 8.0, and 150 mM NaCl (ITC Buffer) at 4°C. Compound were dissolved in DMSO, TK-453 and SHP2-WT protein solutions were prepared at 1 mM and 50 μM in ITC buffer, respectively, with 5% DMSO. We then placed it in the titration cell, set 20 sec to titrate each drop, and calculated the heat of reaction. Titrations were performed using a VP-ITC calorimetric system (Microcal) at 25°C. Data were analyzed by Origin 7.0 (OriginLab) to obtain KD and stoichiometry.
Isolation of peritoneal macrophages
Mice were intraperitoneally injected with 1 ml starch broth per mouse. After 3 d, peritoneal cells were harvested by lavaging the peritoneal cavity with 5 ml PBS. Floating cells were removed by DMEM washing and adherent peritoneal macrophages were cultured in DMEM containing 10% FBS and 1× penicillin/streptomycin at 37°C overnight, followed by treatment with different stimuli according the experimental designs.
Generation of bone marrow-derived macrophages
Mice were sacrificed by cervical dislocation, and their femurs and tibias were removed. Bone marrow cells were isolated by flushing with DMEM. Red blood cells were lysed using NH4Cl. Bone marrow cells were cultured in DMEM containing 10% FBS supplemented with M-CSF (10 ng/ml) for 5–7 days. Two days later, non-adherent cells were removed, the media was freshly replenished, and cultured for two more days. The BMDMs were used on days 5–7.
Western blotting
Cells were lysed in radio immunoprecipitation assay (RIPA) buffer supplemented with protease and phosphatase inhibitor (MCE). Proteins were quantified by the Bradford assay (HyClone-Pierce). The proteins were then separated by SDS-polyacrylamide gel electrophoresis (PAGE) and electrophoretically transferred onto polyvinylidene difluoride membranes. The membranes were probed with antibodies overnight at 4°C, and then incubated with a horseradish peroxidase-coupled secondary antibody. Detection was performed using a LumiGLO chemiluminescent substrate system. Anti-p-IKKα/β (1:1,000; Cell Signal Technology, catalog 2697), anti-IKKα (1:1,000; Cell Signal Technology, catalog 11930), anti-IKKβ (1:1,000; Cell Signal Technology, catalog 8943), anti-p-p65 (1:1,000; Cell Signal Technology, catalog 3033), anti-p65 (1:1,000; Cell Signal Technology, catalog 8242), anti-β-actin (1:2,000, Cell Signal Technology, catalog 3700), anti-pERK (1:1,000; Cell Signal Technology, catalog 4370), anti-ERK (1:1,000; Cell Signal Technology, catalog 4695) were used.
Quantitative PCR (qPCR)
Total RNA was extracted from the skin tissues of the mice (BMDMs, PMs) using TRIzol™ (TaKaRa) as described by the manufacturer. Single-stranded cDNA was synthesized from 1 μg of total RNA by reverse transcription. Real-time PCR was performed with AceQ Universal SYBR qPCR Master Mix (Vazyme Biotech Co, Ltd, China) on a CFX 100 (Bio-Rad, Hercules, CA) cycler using the primers listed in Key resources table. The amplification program was as follows: 95°C for 2.5 min, 44 cycles at 95°C for 15 s, and 60°C for 30 s. Dissociation curves were analyzed at the end of the amplification. The level of Gapdh RNA expression was used to normalize the data. The primers were Gapdh forward (5′-TGA CCT CAA CTA CAT GGT CTA CA-3′), Gapdh reverse (5′-CTT CCC ATT CTC GGC CTT G-3′), Il23a forward (5′-ATG CTG GAT TGC AGA GCA GTA-3′), Il23a reverse (5′-ACG GGG CAC ATT ATT TTT AGT CT-3′), Il17a forward (5′-TTT AAC TCC CTT GGC GCA AAA-3′), Il17a reverse (5′-CTT TCC CTC CGC ATT GAC AC-3′), Il22 forward (5′-ATG AGT TTT TCC CTT ATG GGG AC-3′), Il22 reverse (5′-GCT GGA AGT TGG ACA CCT CAA-3′), Il6 forward (5′-CTG CAA GAG ACT TCC ATC CAG-3′), Il6 reverse (5′-AGT GGT ATA GAC AGG TCT GTT GG-3′), Tnfα forward (5′- GTC CTG CTC TAC GTG ACG AG-3′), Tnfα reverse (5′- TCT CTC CTT TTC TGC CAT CTC T-3′), Ptgs2 forward (5′-TGA GCA ACT ATT CCA AAC CAG C-3′), Ptgs2 reverse (5′-GCA CGT AGT CTT CGA TCA CTA TC-3′).
IMQ-induced psoriasis-like mouse model
Eight- to ten-week-old C57BL/6 wildtype female mice were used to establishing the IMQ-induced psoriasis-like model. The mice were treated with a daily topical dose of 62.5 mg of IMQ cream (5%, Aldara; 3M Pharmaceuticals) on the shaved dorsal skin for 4 consecutive days. Mice were sacrificed one day after the final treatment. The severity of disease was measured using the clinical Psoriasis Area and Severity Index, which was scored on a scale from 0 to 4: 0, none; 1, slight; 2, moderate; 3, marked; and 4, very marked (Shao et al., 2016). For each mouse, skin lesions were taken for hematoxylin and eosin (H&E) staining and qPCR analysis.
Chemical synthesis and characterization
Details of chemistry methods and characterization of compounds is included in supplemental information (Methods S1 and Data S1).
Quantification and statistical analysis
Whereby statistical tests are presented as asterisks in the figures, details of the statistical test are described in the figure legends. Statistical analysis was performed using GraphPad Prism 7.0. Data are presented as the mean ± SEM. We assessed data for normal distribution and similar variance between groups. One-way ANOVA with Tukey’s multiple comparisons was used to assess statistical significance, two-tailed unpaired Student’s t-test was used to detect statistically significant when only two groups were compared (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ns = not significant). Error bars depict SEM.
Acknowledgments
This work was supported by National Natural Science Foundation of China (Nos. 81872877, 91853109, 81773562, 81871639 and 81803081), Mountain-Climbing Talents Project of Nanjing University and the Program for Science & Technology Innovation Talents in Universities of Henan Province (No. 21HASTIT045).
Author contributions
Y.S., X.J., and B.Y. conceived this project and designed the study. M.W., Z.O., K.T., Y.Z., C.S., and H.S. performed the experiments. T.L. performed structural biology experiments. M.W., T.L., and K.T. analyzed the data. Y.S. and M.W. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: April 15, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104009.
Contributor Information
Bin Yu, Email: yubin@zzu.edu.cn.
Xiaoyun Ji, Email: xiaoyun.ji@nju.edu.cn.
Yang Sun, Email: yangsun@nju.edu.cn.
Supplemental information
Data and code availability
Crystallographic statistics is shown in Table S1 and the atomic coordinates of SHP2 in complex with compound TK-453 have been deposited in the Protein Data Bank under the code ID 7VXG. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentires-Alj M., Paez J.G., David F.S., Keilhack H., Halmos B., Naoki K., Maris J.M., Richardson A., Bardelli A., Sugarbaker D.J., et al. Activating mutations of the noonan syndrome-associated SHP2/PTPN11 gene in human solid tumors and adult acute myelogenous leukemia. Cancer Res. 2004;64:8816–8820. doi: 10.1158/0008-5472.CAN-04-1923. [DOI] [PubMed] [Google Scholar]
- Boehncke W.H., Schon M.P. Psoriasis. Lancet. 2015;386:983–994. doi: 10.1016/S0140-6736(14)61909-7. [DOI] [PubMed] [Google Scholar]
- Bunkoczi G., Read R.J. Improvement of molecular-replacement models with Sculptor. Acta Crystallogr. D Biol. Crystallogr. 2011;67:303–312. doi: 10.1107/S0907444910051218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catharino A., Daiha E., Carvalho C., Martinez D., Lima R.B., D'Acri A., Martins C.J., Lupi O. Possible correlations between annular pustular psoriasis and Noonan syndrome. J. Eur. Acad. Dermatol. Venereol. 2016;30:E195–E196. doi: 10.1111/jdv.13521. [DOI] [PubMed] [Google Scholar]
- Chen L., Sung S.S., Yip M.L., Lawrence H.R., Ren Y., Guida W.C., Sebti S.M., Lawrence N.J., Wu J. Discovery of a novel shp2 protein tyrosine phosphatase inhibitor. Mol. Pharmacol. 2006;70:562–570. doi: 10.1124/mol.106.025536. [DOI] [PubMed] [Google Scholar]
- Chen V.B., Arendall W.B., 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.N., LaMarche M.J., Chan H.M., Fekkes P., Garcia-Fortanet J., Acker M.G., Antonakos B., Chen C.H., Chen Z., Cooke V.G., et al. Allosteric inhibition of SHP2 phosphatase inhibits cancers driven by receptor tyrosine kinases. Nature. 2016;535:148–152. doi: 10.1038/nature18621. [DOI] [PubMed] [Google Scholar]
- Cheng Y.P., Chiu H.Y., Hsiao T.L., Hsiao C.H., Lin C.C., Liao Y.H. Scalp melanoma in a woman with LEOPARD syndrome: possible implication of PTPN11 signaling in melanoma pathogenesis. J. Am. Acad. Dermatol. 2013;69:E186–E187. doi: 10.1016/j.jaad.2013.04.033. [DOI] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fan Z.Z., Tian Y.H., Chen Z.P., Liu L., Zhou Q., He J.J., Coleman J., Dong C.J., Li N., Huang J.Q., et al. Blocking interaction between SHP2 and PD-1 denotes a novel opportunity for developing PD-1 inhibitors. EMBO Mol. Med. 2020;12:e11571. doi: 10.15252/emmm.201911571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankson R., Yu Z.H., Bai Y., Li Q., Zhang R.Y., Zhang Z.Y. Therapeutic targeting of oncogenic tyrosine phosphatases. Cancer Res. 2017;77:5701–5705. doi: 10.1158/0008-5472.CAN-17-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly D., Haak S., Sisirak V., Reizis B. The role of dendritic cells in autoimmunity. Nat. Rev. Immunol. 2013;13:566–577. doi: 10.1038/nri3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosskopf S., Eckert C., Arkona C., Radetzki S., Bohm K., Heinemann U., Wolber G., von Kries J.P., Birchmeier W., Rademann J. Selective inhibitors of the protein tyrosine phosphatase SHP2 block cellular motility and growth of cancer cells in vitro and in vivo. ChemMedChem. 2015;10:815–826. doi: 10.1002/cmdc.201500015. [DOI] [PubMed] [Google Scholar]
- Guo W., Liu W., Chen Z., Gu Y., Peng S., Shen L., Shen Y., Wang X., Feng G.S., Sun Y., et al. Tyrosine phosphatase SHP2 negatively regulates NLRP3 inflammasome activation via ANT1-dependent mitochondrial homeostasis. Nat. Commun. 2017;8:2168. doi: 10.1038/s41467-017-02351-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkes J.E., Chan T.C., Krueger J.G. Psoriasis pathogenesis and the development of novel targeted immune therapies. J. Allergy Clin. Immunol. 2017;140:645–653. doi: 10.1016/j.jaci.2017.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmuth K., Grosskopf S., Lum C.T., Wurtele M., Roder N., von Kries J.P., Rosario M., Rademann J., Birchmeier W. Specific inhibitors of the protein tyrosine phosphatase Shp2 identified by high-throughput docking. Proc. Natl. Acad. Sci. U S A. 2008;105:7275–7280. doi: 10.1073/pnas.0710468105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hof P., Pluskey S., Dhe-Paganon S., Eck M.J., Shoelson S.E. Crystal structure of the tyrosine phosphatase SHP-2. Cell. 1998;92:441–450. doi: 10.1016/s0092-8674(00)80938-1. [DOI] [PubMed] [Google Scholar]
- Hui E.F., Cheung J., Zhu J., Su X.L., Taylor M.J., Wallweber H.A., Sasmal D.K., Huang J., Kim J.M., Mellman I., et al. T cell costimulatory receptor CD28 is a primary target for PD-1-mediated inhibition. Science. 2017;355:1428–1433. doi: 10.1126/science.aaf1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Krueger J.G. Highly effective new treatments for psoriasis target the IL-23/type 17 T cell autoimmune axis. Annu. Rev. Med. 2017;68:255–269. doi: 10.1146/annurev-med-042915-103905. [DOI] [PubMed] [Google Scholar]
- LaMarche M.J., Acker M., Argintaru A., Bauer D., Boisclair J., Chan H., Chen C.H., Chen Y.N., Chen Z., Deng Z., et al. Identification of TNO155, an allosteric SHP2 inhibitor for the treatment of cancer. J. Med. Chem. 2020;63:13578–13594. doi: 10.1021/acs.jmedchem.0c01170. [DOI] [PubMed] [Google Scholar]
- Li J., Jie H.B., Lei Y., Gildener-Leapman N., Trivedi S., Green T., Kane L.P., Ferris R.L. PD-1/SHP-2 inhibits Tc1/Th1 phenotypic responses and the activation of T cells in the tumor microenvironment. Cancer Res. 2015;75:508–518. doi: 10.1158/0008-5472.CAN-14-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebschner D., Afonine P.V., Baker M.L., Bunkoczi G., Chen V.B., Croll T.I., Hintze B., Hung L.W., Jain S., McCoy A.J., et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D. 2019;75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Zhai L., Han M., Shi D., Sun Z., Peng S., Wang M., Zhang C., Gao J., Yan W., et al. SH2 domain-containing phosphatase 2 inhibition attenuates osteoarthritis by maintaining homeostasis of cartilage metabolism via the docking protein 1/uridine phosphorylase 1/uridine cascade. Arthritis Rheumatol. 2021;74:462–474. doi: 10.1002/art.41988. [DOI] [PubMed] [Google Scholar]
- Liu Q.Q., Qu J., Zhao M.X., Xu Q., Sun Y. Targeting SHP2 as a promising strategy for cancer immunotherapy. Pharmacol. Res. 2020;152:104595. doi: 10.1016/j.phrs.2019.104595. [DOI] [PubMed] [Google Scholar]
- Liu W., Wang M., Shen L., Zhu Y., Ma H., Liu B., Ouyang L., Guo W., Xu Q., Sun Y. SHP2-mediated mitophagy boosted by lovastatin in neuronal cells alleviates parkinsonism in mice. Signal Transduct. Target. Ther. 2021;6:34. doi: 10.1038/s41392-021-00474-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Yin Y., Wang M.J., Fan T., Zhu Y.Y., Shen L.H., Peng S., Gao J., Deng G.L., Meng X.B., et al. Disrupting phosphatase SHP2 in macrophages protects mice from high-fat diet-induced hepatic steatosis and insulin resistance by elevating IL-18 levels. J. Biol. Chem. 2020;295:10842–10856. doi: 10.1074/jbc.RA119.011840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Yang X., Wang Y., Yang Y., Sun D., Li H., Chen L. Targeting SHP2 as a therapeutic strategy for inflammatory diseases. Eur. J. Med. Chem. 2021;214:113264. doi: 10.1016/j.ejmech.2021.113264. [DOI] [PubMed] [Google Scholar]
- Marasco M., Berteotti A., Weyershaeuser J., Thorausch N., Sikorska J., Krausze J., Brandt H.J., Kirkpatrick J., Rios P., Schamel W.W., et al. Molecular mechanism of SHP2 activation by PD-1 stimulation. Sci. Adv. 2020;6:eaay4458. doi: 10.1126/sciadv.aay4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullard A. Phosphatases start shedding their stigma of undruggability. Nat. Rev. Drug Discov. 2018;17:847–849. doi: 10.1038/nrd.2018.201. [DOI] [PubMed] [Google Scholar]
- Neel B.G., Gu H.H., Pao L. The 'Shp'ing news: SH2 domain-containing tyrosine phosphatases in cell signaling. Trends Biochem. Sci. 2003;28:284–293. doi: 10.1016/S0968-0004(03)00091-4. [DOI] [PubMed] [Google Scholar]
- Nichols R.J., Haderk F., Stahlhut C., Schulze C.J., Hemmati G., Wildes D., Tzitzilonis C., Mordec K., Marquez A., Romero J., et al. RAS nucleotide cycling underlies the SHP2 phosphatase dependence of mutant BRAF-, NF1-and RAS-driven cancers. Nat. Cell Biol. 2018;20:1064–1073. doi: 10.1038/s41556-018-0169-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Shabgah A.G., Navashenaq J.G., Shabgah O.G., Mohammadi H., Sahebkar A. Interleukin-22 in human inflammatory diseases and viral infections. Autoimmun. Rev. 2017;16:1209–1218. doi: 10.1016/j.autrev.2017.10.004. [DOI] [PubMed] [Google Scholar]
- Shao F., Liu Q., Zhu Y., Fan Z., Chen W., Liu S., Li X., Guo W., Feng G.S., Yu H., et al. Targeting chondrocytes for arresting bony fusion in ankylosing spondylitis. Nat. Commun. 2021;12:6540. doi: 10.1038/s41467-021-26750-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao F., Tan T., Tan Y., Sun Y., Wu X., Xu Q. Andrographolide alleviates imiquimod-induced psoriasis in mice via inducing autophagic proteolysis of MyD88. Biochem. Pharmacol. 2016;115:94–103. doi: 10.1016/j.bcp.2016.06.001. [DOI] [PubMed] [Google Scholar]
- Shi Z.Q., Yu D.H., Park M., Marshall M., Feng G.S. Molecular mechanism for the Shp-2 tyrosine phosphatase function in promoting growth factor stimulation of Erk activity. Mol. Cell Biol. 2000;20:1526–1536. doi: 10.1128/mcb.20.5.1526-1536.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y.H., Zhao M., Wu Y.H., Yu B., Liu H.M. A multifunctional cross-validation high-throughput screening protocol enabling the discovery of new SHP2 inhibitors. Acta Pharm. Sin. B. 2021;11:750–762. doi: 10.1016/j.apsb.2020.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z., Wang M., Ge Y., Chen X.P., Xu Z., Sun Y., Xiong X.F. Tyrosine phosphatase SHP2 inhibitors in tumor-targeted therapies. Acta Pharm. Sin B. 2021;11:13–29. doi: 10.1016/j.apsb.2020.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanford S.M., Bottini N. Targeting tyrosine phosphatases: Time to end the stigma. Trends Pharmacol. Sci. 2017;38:524–540. doi: 10.1016/j.tips.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonks N.K. Protein tyrosine phosphatases: from genes, to function, to disease. Nat. Rev. Mol. Cell Biol. 2006;7:833–846. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- van der Fits L., Mourits S., Voerman J.S.A., Kant M., Boon L., Laman J.D., Cornelissen F., Mus A.M., Florencia E., Prens E.P., et al. Imiquimod-induced psoriasis-like skin inflammation in mice is mediated via the IL-23/IL-17 axis. J. Immunol. 2009;182:5836–5845. doi: 10.4049/jimmunol.0802999. [DOI] [PubMed] [Google Scholar]
- van Huijsduijnen R.H., Bombrun A., Swinnen D. Selecting protein tyrosine phosphatases as drug targets. Drug Discov. Today. 2002;7:1013–1019. doi: 10.1016/s1359-6446(02)02438-8. [DOI] [PubMed] [Google Scholar]
- Wang J., Mizui M., Zeng L.F., Bronson R., Finnell M., Terhorst C., Kyttaris V.C., Tsokos G.C., Zhang Z.Y., Kontaridis M.I. Inhibition of SHP2 ameliorates the pathogenesis of systemic lupus erythematosus. J. Clin. Invest. 2016;126:2077–2092. doi: 10.1172/JCI87037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M.L., Lu J.F., Wang M., Yang C.Y., Wang S.M. Discovery of SHP2-D26 as a first, potent, and effective PROTAC degrader of SHP2 protein (vol 63, pg 7510, 2020) J. Med. Chem. 2021;64:906–908. doi: 10.1021/acs.jmedchem.0c02108. [DOI] [PubMed] [Google Scholar]
- Xie J.J., Si X.J., Gu S.L., Wang M.L., Shen J., Li H.Y., Shen J., Li D., Fang Y.J., Liu C., et al. Allosteric inhibitors of SHP2 with therapeutic potential for cancer treatment. J. Med. Chem. 2017;60:10205–10219. doi: 10.1021/acs.jmedchem.7b01520. [DOI] [PubMed] [Google Scholar]
- Xu D., Qu C.K. Protein tyrosine phosphatases in the JAK/STAT pathway. Front. Biosci. 2008;13:4925–4932. doi: 10.2741/3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M., Guo W., Wu Y., Yang C., Zhong L., Deng G., Zhu Y., Liu W., Gu Y., Lu Y., et al. SHP2 inhibition triggers anti-tumor immunity and synergizes with PD-1 blockade. Acta Pharm. Sin B. 2019;9:304–315. doi: 10.1016/j.apsb.2018.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Wu Z., Yan W., Shao F., Ke B., Jiang X., Gao J., Guo W., Lai Y., Ma H., et al. Allosteric inhibition of SHP2 uncovers aberrant TLR7 trafficking in aggravating psoriasis. EMBO Mol. Med. 2021:e14455. doi: 10.15252/emmm.202114455. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Crystallographic statistics is shown in Table S1 and the atomic coordinates of SHP2 in complex with compound TK-453 have been deposited in the Protein Data Bank under the code ID 7VXG. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.