Abstract
Two Neisseria gonorrhoeae isolates from Seattle and two isolates from Uruguay were resistant to erythromycin (MIC, 4 to 16 μg/ml) and had reduced susceptibility to azithromycin (MIC, 1 to 4 μg/ml) due to the presence of the self-mobile rRNA methylase gene(s) ermF or ermB and ermF. The two Seattle isolates and one isolate from Uruguay were multiresistant, carrying either the 25.2-MDa tetM-containing plasmid (Seattle) or a β-lactamase plasmid (Uruguay). Sixteen commensal Neisseria isolates (10 Neisseria perflava-N. sicca, 2 N. flava, and 4 N. mucosa) for which erythromycin MICs were 4 to 16 μg/ml were shown to carry one or more known rRNA methylase genes, including ermB, ermC, and/or ermF. Many of these isolates also were multiresistant and carried the tetM gene. This is the first time that a complete transposon or a complete conjugative transposon carrying an antibiotic resistance gene has been described for the genus Neisseria.
Neisseria gonorrhoeae isolates obtained in Denver, Colo., and Edinburgh, United Kingdom, and having high-level resistance to erythromycin (MIC, >8 μg/ml) and reduced susceptibility to azithromycin (MIC, 2 to 4 μg/ml) have been described (4, 33). The MICs for these isolates were higher than those normally associated with chromosomal mtr mutations (3, 8, 13, 15, 30). Unfortunately, these isolates were not available for examination of the mechanisms of resistance. During a gonorrhea outbreak in 1994 to 95 in Seattle, Wash., caused by strains containing the 25.2-MDa plasmid encoding tetracycline resistance of the Pro−/IA-1,2 class, two isolates resistant to both tetracycline and erythromycin (MICs, ≥16 μg/ml) were identified. Two additional gonococci, isolated in 1991 and 1995 in Uruguay and for which an erythromycin MIC (4 μg/ml) higher than that previously found in this setting was determined, were available for study. In addition to the identification of these isolates with high-level erythromycin resistance (4, 33), plasmids carrying an ermC gene (34) and conferring erythromycin resistance to both N. gonorrhoeae and N. meningitidis have been created. These findings prompted us to evaluate whether these four N. gonorrhoeae isolates had acquired one or more of the erm genes known for other urogenital species (2). Investigations further sought to define the location of these methylase genes (plasmid versus chromosome), to determine whether their location was on conjugative units, as have been found in many other species (2, 18, 19, 26, 29), and to examine the transfer of such genes to other isolates and species. The methylase gene composition of oral commensal Neisseria spp. for which the erythromycin MICs were 4 to 16 μg/ml was compared to that of the gonococcal isolates.
MATERIALS AND METHODS
Bacterial isolates.
Erythromycin-resistant (Emr) Pro−/IA-1,2 N. gonorrhoeae isolates were isolated in Seattle during the 1994-1995 gonococcal outbreak (Table 1). Both Seattle isolates (94-965 and 95-1) were also tetracycline resistant. The 1995 Uruguay isolate, 581, was Pro−/IB-3, and the 1991 Uruguay isolate, 1101, was nonrequiring Proto/IB-3. Strain 1101 carried the 3.2-MDa β-lactamase plasmid and was resistant to penicillin in addition to erythromycin, while all the other N. gonorrhoeae isolates did not carry a β-lactamase plasmid. Six other Tcr Pro−/IA-1,2 N. gonorrhoeae isolates that were from the Seattle outbreak but that were not resistant to erythromycin were available for comparison with the two Emr Tcr N. gonorrhoeae isolates. The NRL (Neisseria Reference Laboratory, University of Washington, Seattle) strains were isolated prior to 1986. The N. gonorrhoeae isolates were confirmed by biochemical methods (11). Auxotypes, protein I serovars, and plasmid contents of the gonococcal isolates were determined by established methods (5, 6, 9, 32).
TABLE 1.
MICs for and antibiotic resistance determinants of N. gonorrhoeae and commensal Neisseria spp.
| Isolate | Date | Locationa | MIC (μg/ml) ofb:
|
Resistant gene carriedc
|
|||
|---|---|---|---|---|---|---|---|
| ERY | AZM | TET | erm | tet | |||
| N. gonorrhoeaed | |||||||
| 94-965 | 1994 | Seattle | 16 | 4 | 16 | F | M |
| 95-1 | 1995 | Seattle | 16 | 4 | 16 | B, F | M |
| 1101 | 1991 | Montevideo | 4 | 1 | 1 | F | None |
| 581 | 1995 | Montevideo | 4 | 1 | 4 | F | None |
| N. perflava-N. sicca | |||||||
| CTM 1.2 | 1986 | DeKalb | 4 | ND | 1 | B, C | None |
| CTM 4.3 | 1986 | DeKalb | 8 | ND | 16 | B | Me |
| CTM 7.2 | 1986 | DeKalb | 8 | ND | 16 | B | Me |
| 10004 | 1991 | Seattle | 16 | ND | 16 | C | Me |
| 10915 | 1991 | Seattle | 16 | ND | 16 | B, C, F | Me |
| 30423 | 1991 | Seattle | 16 | ND | 8 | B, C | Me |
| 31212 | 1991 | Seattle | 16 | ND | 8 | C | Me |
| 3006 | 1995 | Seattle | 4 | ND | 8 | B, C, F | None |
| 33107 | 1995 | Seattle | 8 | ND | 1 | B | None |
| NRL 45 | Before 1986 | NRL | 4 | ND | 2 | B | None |
| N. flava | |||||||
| CTM 5.4 | 1986 | DeKalb | 16 | ND | 1 | C | None |
| NRL 69 | Before 1986 | NRL | 8 | ND | 1 | B | None |
| N. mucosa | |||||||
| CTM 2.2 | 1986 | DeKalb | 8 | ND | 16 | B, C | Me |
| CTM 8.1 | 1986 | DeKalb | 8 | ND | 16 | B, C | Me |
| 10502 | 1991 | Seattle | 8 | ND | >16 | C | None |
| NRL 76 | Before 1986 | NRL | 8 | ND | 1 | B | None |
Seattle, Wash.; Montevideo, Uruguay; DeKalb County, Ga.
ERM, erythromycin; AZM, azithromycin; TET, tetracycline. ND, not determined.
F, ermF; B, ermB; C, ermC; M, tetM.
Both Seattle N. gonorrhoeae isolates carried the tetM gene on the 25.2-MDa plasmid; strain 1101 carries the 3.2-MDa β-lactamase plasmid and is penicillin resistant.
The commensal species carried the tetM gene on the chromosome, where it was not mobile.
We also examined 16 isolates of commensal Neisseria spp., including 10 N. perflava-N. sicca, 2 N. flava, and 4 N. mucosa (Table 1). These isolates were confirmed by biochemical methods. The commensal Neisseria spp. were clinical isolates collected from the periodontal pockets of seven periodontitis patients seen at the Graduate Periodontics Clinic at the University of Washington, Seattle, between 1991 and 1995, isolates collected from oropharyngeal specimens from six patients attending the DeKalb County Sexually Transmitted Disease Clinic in 1986 (designated CTM before the number [7]), and type strains (NRL strains) obtained from Joan Knapp (22, 23).
Media.
GC base or GCP broth (Difco Laboratories, Detroit, Mich.) supplemented as previously described (9, 20) was used for routine culturing of N. gonorrhoeae, Neisseria spp., and Enterococcus faecalis.
Antimicrobial susceptibilities.
Mueller-Hinton medium (Difco) was used to determine the MICs for the commensal Neisseria spp., N. meningiditis, and E. faecalis transconjugants, and supplemented GC medium base (Difco) was used for N. gonorrhoeae, as recommended by the National Committee on Clinical Laboratory Standards for aerobic bacteria (12). The antibiotic concentrations tested were as follows: erythromycin, 0.06 to 32 μg/ml; azithromycin, 0.03 to 8 μg/ml; and tetracycline, 0.06 to 32 μg/ml. MIC plates were incubated at 36.5°C for 24 h with CO2 for N. gonorrhoeae and without CO2 for commensal Neisseria spp., N. meningitidis, and E. faecalis transconjugants.
Proteinase K treatment.
Isolates and transconjugants were treated with proteinase K as previously described (2) and used as templates for the PCR assays. Each proteinase K-treated sample was not used more than three times, since repeated freezing and thawing has been shown to degrade DNA samples (2).
PCR of the ermF gene.
The PCR primers used in the study were F1 (5′ CGGGTCAGCACTTTACTATTG 3′, starting at bp 1235) and F2 (5′ GGACCTACCTCATAGACAAG 3′, antisense sequence ending at bp 1700). The expected size of the PCR fragment was 466 bp (2, 14, 28). Each 100-μl reaction mixture contained 2 U of Taq polymerase (Boehringer Mannheim Indianapolis, Ind.), 200 mM deoxynucleoside triphosphate, 1× PCR buffer I (1.5 mM MgCl2), and 100 ng of each primer. Ten to 40 ng of DNA or 1 to 2 μl of proteinase K-treated bacteria were used as the DNA template. The PCR conditions were as follows: denaturing at 94°C for 30 s, annealing at 50°C for 30 s, and elongation at 72°C for 2 min. The cycle was repeated 35 times. Plasmid pBF4 (2), containing the cloned ermF gene, and water were used as positive and negative controls, respectively. The PCR products were dried on a lyophilizer, resuspended in 10 μl of sterile H2O, run on 1.5% agarose gels, and stained with ethidium bromide for visualization. Southern blots of these gels were hybridized with labeled ermF-containing plasmid probes for confirmation of PCR products as previously described (2).
PCR primers and conditions for the ermA, ermB, and ermC genes.
AF (5′ CTTCGATAGTTTATTAATATTAGT 3′) and AR (5′ TCTAAAAAGCATGTAAAAGAA 3′), BF (5′ AGTAACGGTACTTAAATTGTTTAC 3′) and BR (5′ GAAAAGGTACTCAACCAAATA 3′), and CF (5′ GCTAATATTGTTTAAATCGTCAAT 3′) and CR (5′ TCAAAACATAATATAGATAAA 3′) have been described previously (2). The PCR conditions for the ermB reaction were the same as those for the ermF reaction. The PCR assay used for ermA consisted of denaturing at 94°C for 30 s, annealing at 48°C for 1 min, and elongation at 72°C for 2 min; that used for ermC consisted of denaturing at 94°C for 30 s, annealing at 43°C for 1 min, and elongation at 72°C for 2 min.
DNA hybridization.
DNA was extracted from N. gonorrhoeae, commensal Neisseria spp., N. meningitidis, and selected transconjugants as previously described (9, 20). Uncut whole-cell DNA was visualized on a 0.7% agarose gel stained with ethidium bromide, and Southern blots were prepared. Fragment probes prepared from rRNA methylase genes from the cloned plasmids pEM9592, pJIR229, pBR328:33RV, pBF4, and pJI3, which carried the genes ermA, ermB, ermC, ermF, and tetM, respectively, or oligonucleotide probes for the appropriate genes were used (2, 16). The DNA probes were labeled with the appropriate Genius 3 chemiluminescence kit as recommended by the manufacturer (Boehringer). Hybridization under stringent conditions and detection were done according to the manufacturer’s instructions as previously described (2, 19). Positive and negative controls were included in each Southern blot.
Hybridization of PCR products.
Plasmids pEM9592, pJIR229, pBR328:33RV, and pBF4 or oligonucleotide probes for ermA, ermB, ermC, and ermF were labeled with nonradioactive Genius kits as recommended by the manufacturer (Boehringer). The labeled plasmids were used for hybridization with Southern blots of the appropriate PCR product or purified whole-cell DNA. The hybridization and wash steps were performed at stringent temperatures according to the manufacturer’s instructions. Detection was done with a CDP-Star detection kit at a reagent concentration of 1:1,000 as described by the manufacturer (Boehringer).
Sequencing.
The ermF PCR products from N. gonorrhoeae and commensal Neisseria spp. were sequenced separately with primers ermF1 and ermF2. A Taq Dye Deoxy terminator cycle sequencing kit (Applied Biosystems, Foster City, Calif.) was used for PCR amplification, and the filtered PCR products (Nuclean D50 filters; Kodak, Rochester, N.Y.) were examined on a model 373A sequencer (Applied Biosystems) (2, 19). The two sequences for each isolate were overlapped, aligned and compared with the known GenBank sequence of ermF (accession no. M14730) by use of GCG software (Genetics Computer Group, Madison, Wis.). The putative amino acid sequences were determined from the DNA sequences and also compared with the known GenBank sequence of ErmF( 19).
Mating experiments.
Recipients included N. gonorrhoeae F62, with chromosomally mediated resistance to rifampin (25 μg/ml), streptomycin (250 μg/ml), and nalidixic acid (25 μg/ml) (21, 22, 27); N. gonorrhoeae CDC36N, with chromosomally mediated resistance to nalidixic acid (25 μg/ml) and carrying the 4.4-MDa β-lactamase plasmid (20); E. faecalis JH2-2, resistant to rifampin (25 μg/ml) and fusidic acid (25 μg/ml) (2, 19, 24, 26); N. meningitidis NRL9205 (serogroup A), resistant to streptomycin (250 μg/ml) and rifampin (20 μg/ml) (23); and N. mucosa CTM 1.1, with chromosomally mediated resistance to streptomycin (250 μg/ml) and rifampin (20 μg/ml). Donors included Emr N. gonorrhoeae isolates and selected isolates from each of the commensal Neisseria species. Donors and recipients were grown separately for 24 h on agar plates. The donor and recipient isolates were each resuspended in 0.5 ml of GCP broth to form turbid suspensions (>108/ml), mixed together, and plated on a GC agar (Difco) plate without antibiotics as previously described (22, 23, 27). The mixture was incubated at 36.5°C in 5% CO2 for 24 h. N. meningitidis, N. mucosa, and N. gonorrhoeae F62 transconjugants were selected on medium containing streptomycin (150 μg/ml) and erythromycin (10 μg/ml). The transconjugants were confirmed by growth on rifampin (25 μg/ml) (21–23). N. gonorrhoeae CDC36N transconjugants were selected on medium containing penicillin (10 μg/ml) and erythromycin (10 μg/ml). The transconjugants were verified by growth on nalidixic acid (25 μg/ml) and the presence of the 4.4-MDa β-lactamase plasmid (21–23, 27). JH2-2 transconjugants were selected on medium containing rifampin (10 μg/ml) and erythromycin (10 μg/ml). The E. faecalis transconjugants were confirmed by growth on medium supplemented with streptomycin (150 μg/ml) and by use of chromosomal DNA probe specific for E. faecalis (2). N. meningitidis transconjugants were confirmed by growth on medium supplemented with rifampin (20 μg/ml).
The identity of erm genes in the transconjugants was confirmed by PCR and hybridization of the PCR products as described above (2).
PFGE.
Pulsed-field gel electrophoresis (PFGE) was used to compare the Emr N. gonorrhoeae isolates to six Tcr Pro−/IA-1,2 Seattle N. gonorrhoeae isolates which were part of the outbreak. The isolates were digested with NheI or SpeI (Promega, Madison, Wis.) as previously described (31, 32). The PFGE patterns were compared and assumed to be genetically related if they were identical or had three or fewer band differences.
PFGE was also used to compare the N. meningitidis transconjugants with the donor and recipient N. meningitidis isolates by use of the N. gonorrhoeae protocol and one enzyme (31, 32). This procedure allowed us to verify that the N. meningitidis transconjugants were related to the recipient rather than the donor N. meningitidis.
RESULTS
Characterization of macrolide-resistant N. gonorrhoeae.
The two Seattle Pro−/IA-1,2 isolates were identified because of the high MICs of erythromycin (16 μg/ml) and azithromycin (4 μg/ml) for them (Table 1). Both Seattle isolates carried 25.2-MDa plasmids (tetracycline MIC, 16 μg/ml) which hybridized with the tetM probe (data not shown). For the two Uruguay isolates, the erythromycin MIC was 4 μg/ml and the azithromycin MIC was 1 μg/ml (Table 1). Uruguay isolate 1101 was resistant to penicillin and carried a 3.2-MDa β-lactamase plasmid (data not shown). All four N. gonorrhoeae isolates carried an ermF gene, which encodes a known rRNA methylase, and one isolate (95-1) also carried ermB (Table 1). The other isolates did not hybridize with ermA, ermB, or ermC gene probes, while 95-1 did not hybridize with ermA or ermC gene probes (data not shown). The ermF probe hybridized with the chromosomal fraction of the gel when whole-cell DNA was used for the Southern blots, suggesting a chromosomal location for the ermF gene. In addition, the 2.6- and 3.2-MDa β-lactamase plasmids or the 25.2-MDa tetM-containing plasmid common to N. gonorrhoeae was found, but no other plasmids were found in any of the four isolates.
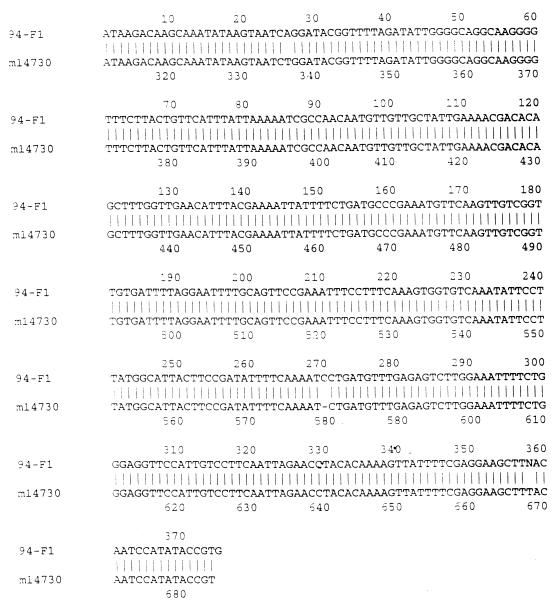
PCR fragments of the ermF genes from two strains of N. gonorrhoeae (94-965 and 1101) were sequenced. The DNA sequence and amino acid homologies between the PCR fragment from 94-965 and the ermF gene originally identified in colonic Bacteroides spp. were both 99% identical over 374 bp (Fig. 1 and 2). Results were similar (95% identity) for N. gonorrhoeae 1101 from Uruguay (data not shown). The G+C content of the PCR fragment was approximately 35%, which differs from the 50% G+C content found in the N. gonorrhoeae chromosome, suggesting a non-Neisseria origin for these genes. PCR fragments of the ermB gene from strain 95-1 were also sequenced. The DNA sequence and amino acid homologies between the PCR fragment and the ermB gene previously characterized from Clostridium perfringens (GenBank accession no. X58285) were over 99% identical over 342 bp.
FIG. 1.
DNA sequence homology between the GenBank ermF sequence (listed as m14730) and the PCR product from N. gonorrhoeae 94-965 (listed as 94-F1) (99% identity over 374 bp).
FIG. 2.
Amino acid homology between the GenBank ermF sequence (listed as m14730) and the PCR product from N. gonorrhoeae 94-965 (listed as 94.pep) (99% identity over 123 amino acids).
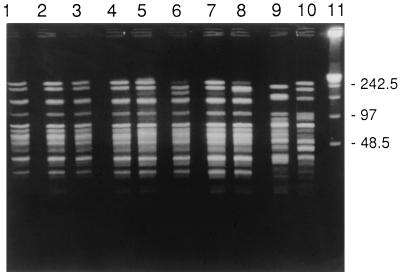
The six Tcr Pro−/IA-1,2 N. gonorrhoeae isolates obtained during the 1994-1995 Seattle outbreak had NheI PFGE patterns (Fig. 3, lanes 1 to 6) that were indistinguishable from the NheI PFGE patterns of the two Emr Tcr N. gonorrhoeae isolates (Fig. 3, lanes 7 and 8), but the Seattle Emr isolates differed from the two Emr N. gonorrhoeae isolates from Uruguay (Fig. 3, lanes 9 and 10). The two Uruguay isolates appeared unrelated to the Seattle isolates or to each other (the PFGE patterns differed by more than three fragments) (Fig. 3). Similar results were found when SpeI was used for PFGE analysis (data not shown). Both enzymes gave identical patterns for the eight Seattle isolates, strongly suggesting a very close relationship between the two Emr and the six Ems N. gonorrhoeae isolates from the outbreak.
FIG. 3.
PFGE with NheI. Lanes 1 to 6, Tcr N. gonorrhoeae, with isolates in lanes 1 to 3 being isolated before and isolates in lanes 4 to 6 being isolated after the Emr Tcr N. gonorrhoeae isolates from Seattle; lanes 7 and 8, Emr Tcr N. gonorrhoeae isolates from Seattle; lanes 9 and 10, Emr isolates from Uruguay; lane 11, λ standard. Numbers at right are molecular weight standards.
Characterization of macrolide resistance in commensal Neisseria spp.
The erythromycin MICs for the other Neisseria spp. ranged from 4 to 16 μg/ml (Table 1). Compared to the N. gonorrhoeae isolates studied, these commensal species contained a more heterologous group of known erm genes (ermB, ermC, and ermF). Among the 10 N. perflava-N. sicca isolates, 4 carried the ermB gene and had erythromycin MICs ranging from 4 to 8 μg/ml; 2 carried ermC and had an erythromycin MIC of 16 μg/ml; 2 carried both ermB and ermC and had erythromycin MICs of 4 to 16 μg/ml; and 2 carried ermB, ermC, and ermF and had erythromycin MICs of 4 to 16 μg/ml (Table 1). Among the four N. perflava-N. sicca strains isolated before 1990, three carried one erm gene, while three of six strains isolated after 1990 carried multiple erm genes. One N. flava strain carried ermC, and the other strain carried ermB (erythromycin MICs, 8 to 16 μg/ml). Among the four N. mucosa strains, two carried both ermB and ermC, one carried ermB, and one carried ermC; the erythromycin MIC for all four strains was 8 μg/ml (Table 1).
To confirm the presence of the erm genes, we used PCR sequencing. The PCR fragment of the ermF gene from N. perflava-N. sicca 10915 was sequenced; the DNA sequence homology between the PCR fragment and the ermF gene from Bacteroides spp. showed 97% identity over 374 bp, and the amino acid homology was 94% (data not shown).
Transfer of erythromycin resistance.
All four of the N. gonorrhoeae isolates and seven of the commensal Neisseria sp. isolates were examined for their ability to transfer the Emr phenotype to Neisseria and E. faecalis recipients (Table 2). N. gonorrhoeae donors transferred the ermF gene at frequencies of 10−6/recipient with the two different N. gonorrhoeae recipients, 10−7/recipient with N. meningitidis as the recipient, and 10−7 to 10−8/recipient with E. faecalis as the recipient.
TABLE 2.
Mobility of erm genes in representative transconjugants
| Donor | erm gene(s) carrieda | Recipient | Transconjugants
|
|
|---|---|---|---|---|
| No. tested | erm genes trans-ferreda | |||
| N. gonorrhoeae | ||||
| 94-965b | F | N. meningitidis 9205 | 1 | F |
| 94-965b | F | N. gonorrhoeae CDC36N | 1 | F |
| 94-965b | F | N. gonorrhoeae F62 | 1 | F |
| 94-965 | F | E. faecalis JH2-2 | 1 | F |
| 95-1 | F | E. faecalis JH2-2 | 1 | F |
| 1101 | F | N. gonorrhoeae CDC36N | 1 | F |
| E. faecalis JH2-2 | 1 | F | ||
| 581 | F | E. faecalis JH2-2 | 1 | F |
| N. perflava-N. sicca | ||||
| 10915 | B, C, F | E. faecalis JH2-2 | 2 | F |
| E. faecalis JH2-2 | 5 | C, F | ||
| 10915 | B, C, F | N. meningitidis 9205 | 1 | B, C |
| 33006 | B, C, F | E. faecalis JH2-2 | 2 | C |
| 33006 | B, C, F | N. meningitidis 9205 | 6 | C |
| 33006 | B, C, F | E. faecalis JH2-2 | 1 | B, C, F |
| 30423 | B, C | N. meningitidis 9205 | 8 | B |
| 31212 | C | N. meningitidis 9205 | 2 | C |
| N. flava NRL 69 | B | N. meningitidis 9205 | 2 | B |
| N. mucosa | ||||
| CTM 8.1 | B, C | E. faecalis JH2-2 | 1 | B |
| CTM 2.2 | B, C | E. faecalis JH2-2 | 1 | C |
| E. faecalis JH2-2 | 2 | B, C | ||
| N. mucosa CTM 1.1 | 5 | B | ||
See Table 1, footnote c, for erm gene designations.
The 25.2-MDa plasmid did transfer in these matings.
The commensal species carried a variety of erm genes and were able to transfer ermF, ermC and ermF, ermB and ermC, all three ermC, or ermB (Table 2) at frequencies ranging from 10−5 to 10−9 for E. faecalis and N. meningitidis. Matings were done at least twice, and only a portion of the transconjugants were characterized and described in Table 2. The donor N. mucosa CTM 2.2 could move the ermB gene but not the ermC gene to the recipient N. mucosa CTM 1.1 (Table 2) at a frequency of 10−8/recipient. The other N. mucosa donor (CTM 8.1) and the various N. perflava-N. sicca and N. flava donors used in the matings, which transferred erm genes to E. faecalis and/or N. meningitidis recipients, could not transfer erm genes at measurable frequencies (>10−9/recipient) to the recipient N. mucosa CTM 1.1 (Table 2). Both E. faecalis and N. meningitidis recipients were able to acquire one or more erm genes. No N. meningitidis with ermF was isolated from the transconjugants with the commensal donors, but ermF was found in N. meningitidis transconjugants when N. gonorrhoeae carrying the ermF gene was used as the donor (Table 2).
The 25.2-MDa plasmid, conferring tetracycline resistance, was transferred from a Seattle N. gonorrhoeae donor to N. gonorrhoeae and N. meningitidis recipients but not to E. faecalis recipients (data not shown). No plasmids carrying the tetM gene were found in the commensal species, and we were unable to transfer tetracycline resistance from these species to either N. meningitidis or E. faecalis. However, this result was anticipated, since we have previously shown that commensal Neisseria sp. isolates carry an incomplete tetM transposon in the chromosome and were unable to transfer tetM by conjugation (16, 17, 25).
DISCUSSION
This is the first description of a known erm gene(s) in the genus Neisseria, since both the TEM β-lactamase and the tetM genes have incomplete transposons in N. gonorrhoeae, N. meningitidis, and the commensal Neisseria spp. (5, 16, 17). The data indicates that the ermF genes have been in N. gonorrhoeae since at least 1991, the ermB genes have been in N. gonorrhoeae since 1995, and various erm genes have been in three commensal species (N. perflava-N. sicca, N. flava, and N. mucosa) since at least the 1980s (Table 1). Whether erm genes are relatively new (last 20 years) in Neisseria spp. or whether they predate the identification of the β-lactamase plasmids in N. gonorrhoeae (5) is currently under investigation. Donors carrying the 25.2-MDa plasmid with the tetM gene transferred this gene into N. gonorrhoeae and N. meningitidis recipients but not into E. faecalis (data not shown), indicating that the ermF gene had a wider host range than the gonococcal 25.2-MDa plasmid (17, 22) or the gonococcal 24.5-MDa and β-lactamase plasmids (17, 23). Although 10 (63%) of the commensal Neisseria isolates carried the tetM gene (Table 1), none could move this gene, as has previously been described (16, 17).
N. gonorrhoeae with reduced susceptibility to erythromycin (MICs, 2 to 4 μg/ml) has been reported since the 1960s (1, 15). Some studies have shown a positive association between reduced susceptibility to penicillin, erythromycin, chloramphenicol, and tetracycline and mtr mutations (3). It was hypothesized that resistant N. gonorrhoeae isolates for which erythromycin MICs were 2 to 4 μg/ml were due to the presence of mtr mutations (3, 8). However, the maximum azithromycin MICs for these isolates generally were 0.25 to 0.5 μg/ml (unpublished observations). Based upon our finding with the two Uruguay isolates, for which the erythromycin MIC was 4 μg/ml and the azithromycin MIC was 1 μg/ml (Table 1), it is tempting to speculate that other N. gonorrhoeae isolates for which erythromycin MICs are 2 to 4 μg/ml also may carry erm genes with or without mtr mutations. We are currently examining isolates obtained during different decades and for which erythromycin MICs range from 0.5 to 8 μg/ml. It will be of interest to determine whether the characteristics attributed to the mtr mutations are due to the combination of mtr mutations and erm genes or whether reduced susceptibility to penicillin and tetracycline is associated with mtr mutations but reduced susceptibility to erythromycin is associated not with mtr mutations but with the presence of erm genes (3). Clinically, this information will be of interest because many infections in homosexual and bisexual men in Seattle-King County (10) are due to N. gonorrhoeae isolates with a phenotypic pattern (reduced susceptibility to erythromycin, penicillin, and tetracycline) suggesting mtr mutations. Some of these isolates have been shown to carry an mtr mutation by sequencing of PCR products. One can speculate that one or more of the four Emr N. gonorrhoeae isolates in this study may carry both erm genes and mtr mutations. However, mtr mutations cannot be transferred by conjugation, nor do they influence the transfer of coresident β-lactamase plasmids. Since the ermF-tetQ transposons in colonic Bacteroides spp. are able to transfer mobilizable plasmids between Bacteroides spp., the ability of mobile ermF to conjugally transfer gonococcal β-lactamase plasmids is under investigation (2).
Three of the N. gonorrhoeae isolates and over 60% of the commensal species isolates were multiresistant (Table 1). The ermF gene in N. gonorrhoeae and the ermB, ermC, and ermF genes in the commensal species (Table 2) were able to move themselves by conjugation to other Neisseria spp. and to E. faecalis recipients. This finding implies that these erm genes are associated with complete conjugative elements, and this is the first description of complete transposable elements in Neisseria. Previously described TEM β-lactamase genes and the tetM gene are both on incomplete elements (5, 9, 17).
There was greater diversity among the erm genes carried by the 16 commensal Neisseria isolates than by the 4 N. gonorrhoeae isolates. Within this study, there was no association of erythromycin MIC with the number of erm genes found or with a particular gene among the commensal Neisseria spp. Further studies are needed to determine whether clinical isolates of N. gonorrhoeae carrying the ermC gene can be found or whether carriage of this gene is unique to the commensal species. Studies are needed to determine the influence of these erm genes on the treatment of gonococcal disease with the newer macrolides. It also remains to be determined if the commensal Neisseria spp. are reservoirs for these erm genes, how long these genes have actually been in the genus, whether most erm genes are on mobile conjugative elements, and whether isolates carrying erm genes are more likely to be multiresistant than the general Neisseria population.
ACKNOWLEDGMENT
This work was supported in part by National Institutes of Health grants AI-131448 and DE-10913.
REFERENCES
- 1.Amies C R. Sensitivity of N. gonorrhoeae to penicillin and other antibiotics. Br J Vener Dis. 1969;45:216–222. doi: 10.1136/sti.45.3.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chung W O, Werckenthin C, Schwarz S, Roberts M C. Host range of the ermF rRNA methylase gene in human and animal bacteria. J Antimicrob Chemother. 1999;43:5–14. doi: 10.1093/jac/43.1.5. [DOI] [PubMed] [Google Scholar]
- 3.Delahay R M, Robertson B D, Balthazar J T, Shafer W M, Ison C A. Involvement of the gonococcal MtrE protein in the resistance of Neisseria gonorrhoeae to toxic hydrophobic agents. Microbiology. 1997;143:2127–2133. doi: 10.1099/00221287-143-7-2127. [DOI] [PubMed] [Google Scholar]
- 4.Ehret J M, Nims L J, Judson F N. A clinical isolate of Neisseria gonorrhoeae with in vitro resistance to erythromycin and decreased susceptibility to azithromycin. Sex Transm Dis. 1996;23:270–272. doi: 10.1097/00007435-199607000-00004. [DOI] [PubMed] [Google Scholar]
- 5.Elwell L P, Roberts M, Mayer L W, Falkow S. Plasmid-mediated beta-lactamase production in Neisseria gonorrhoeae. Antimicrob Agents Chemother. 1977;11:528–533. doi: 10.1128/aac.11.3.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evins G M, Knapp J S. Characterization of Neisseria gonorrhoeae reference strains used in the development of a serologic classification system. J Clin Microbiol. 1988;26:358–363. doi: 10.1128/jcm.26.2.358-363.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knapp J S, Johnson S R, Zenilman J M, Roberts M C, Morse S A. High-level tetracycline resistance resulting from TetM in strains of Neisseria spp., Kingella denitrificans, and Eikenella corrodens. Antimicrob Agents Chemother. 1988;32:765–767. doi: 10.1128/aac.32.5.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maness M J, Sparling P F. Multiple antibiotic resistance due to single mutation in Neisseria gonorrhoeae. J Infect Dis. 1973;128:321–330. doi: 10.1093/infdis/128.3.321. [DOI] [PubMed] [Google Scholar]
- 9.Morse S A, Johnson S R, Biddle J W, Roberts M C. High-level tetracycline resistance in Neisseria gonorrhoeae is the result of acquisition of a streptococcal tetM determinant. Antimicrob Agents Chemother. 1986;30:664–670. doi: 10.1128/aac.30.5.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morse S A, Lysko P G, McFarland L, Knapp J S, Sandstrom E, Critchlow C, Holmes K K. Gonococcal strains from homosexual men have outer membranes with reduced permeability to hydrophobic molecules. Infect Immun. 1982;37:432–438. doi: 10.1128/iai.37.2.432-438.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C: American Society for Microbiology; 1995. [Google Scholar]
- 12.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A2. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1993. [Google Scholar]
- 13.Piot P, van Dyck E, Colaert J, Ursi J-P, Bosmans E, Meheus A. Antibiotic susceptibility of Neisseria gonorrhoeae strains from Europe and Africa. Antimicrob Agents Chemother. 1979;15:535–539. doi: 10.1128/aac.15.4.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rasmussen J L, Odelson D A, Macrina F L. Complete nucleotide sequence and transcription of ermF, a macrolide-lincosamide-streptogramin B resistance determinant from Bacteroides fragilis. J Bacteriol. 1986;168:523–533. doi: 10.1128/jb.168.2.523-533.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reyn A, Bentzon M W. Relationships between the sensitivities in vitro of Neisseria gonorrhoeae to spiramycin, penicillin, streptomycin, tetracycline and erythromycin. Br J Vener Dis. 1969;45:223–227. doi: 10.1136/sti.45.3.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roberts M C. Characterization of the Tet M determinants in urogenital and respiratory bacteria. Antimicrob Agents Chemother. 1990;34:476–478. doi: 10.1128/aac.34.3.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roberts M C. Plasmids of Neisseria gonorrhoeae and other Neisseria species. Clin Microbiol Rev. 1989;2:S18–S23. doi: 10.1128/cmr.2.suppl.s18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roberts M C, Brown M B. Macrolide-lincosamide resistance determinants in streptococcal species isolated from the bovine mammary gland. Vet Microbiol. 1994;40:253–261. doi: 10.1016/0378-1135(94)90114-7. [DOI] [PubMed] [Google Scholar]
- 19.Roberts M C, Chung W O, Roe D E. Characterization of tetracycline and erythromycin resistance determinants in Treponema denticola. Antimicrob Agents Chemother. 1996;40:1690–1694. doi: 10.1128/aac.40.7.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roberts M C, Elwell L P, Falkow S. Molecular characterization of two beta-lactamase-specifying plasmids isolated from Neisseria gonorrhoeae. J Bacteriol. 1977;131:557–563. doi: 10.1128/jb.131.2.557-563.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roberts M C, Falkow S. Conjugal transfer of R plasmids in Neisseria gonorrhoeae. Nature. 1977;266:630–631. doi: 10.1038/266630a0. [DOI] [PubMed] [Google Scholar]
- 22.Roberts M C, Knapp J S. Host range of the conjugative 25.2-megadalton tetracycline resistance plasmid from Neisseria gonorrhoeae and related species. Antimicrob Agents Chemother. 1988;32:488–491. doi: 10.1128/aac.32.4.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roberts M C, Knapp J S. Transfer of β-lactamase plasmids from Neisseria gonorrhoeae to N. meningitidis and commensal Neisseria species by the 25.2-megadalton conjugative plasmid. Antimicrob Agents Chemother. 1988;32:1430–1432. doi: 10.1128/aac.32.9.1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roberts M C, Lansciardi J. Transferable Tet M in Fusobacterium nucleatum. Antimicrob Agents Chemother. 1990;34:1836–1838. doi: 10.1128/aac.34.9.1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roberts M C, Moncla B J. Tetracycline resistance and TetM in oral anaerobic bacteria and Neisseria perflava-N. sicca. Antimicrob Agents Chemother. 1988;32:1271–1273. doi: 10.1128/aac.32.8.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roe D E, Weinberg A, Roberts M C. Mobility of rRNA methylase genes in Campylobacter (Wolinella) rectus. J Antimicrob Chemother. 1995;36:738–740. doi: 10.1093/jac/36.4.738. [DOI] [PubMed] [Google Scholar]
- 27.Sarafian S K, Genco C A, Roberts M C, Knapp J S. Acquisition of β-lactamase and TetM-containing conjugative plasmids by phenotypically different strains of Neisseria gonorrhoeae. Sex Transm Dis. 1990;17:67–71. doi: 10.1097/00007435-199004000-00004. [DOI] [PubMed] [Google Scholar]
- 28.Smith C J. Nucleotide sequence analysis of Tn4551: use of ermFS operon fusions to detect promoter activity in Bacteroides fragilis. J Bacteriol. 1987;169:4589–4596. doi: 10.1128/jb.169.10.4589-4596.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sutcliffe J, Grebe T, Tait-Kamradt A, Wondrack L. Detection of erythromycin-resistant determinants by PCR. Antimicrob Agents Chemother. 1996;40:2562–2566. doi: 10.1128/aac.40.11.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Klingeren B, Ansink-Schipper M C, Doornbos L, Lampe A S, Wagenvoort J H T, Dessens-Kroon M, Verheuvel M. Surveillance of the antibiotic susceptibility of non-penicillinase producing Neisseria gonorrhoeae in The Netherlands from 1983 to 1986. J Antimicrob Chemother. 1998;21:737–744. doi: 10.1093/jac/21.6.737. [DOI] [PubMed] [Google Scholar]
- 31.Xia M, Whittington W L, Holmes K K, Plummer F A, Roberts M C. Pulsed-field gel electrophoresis for genomic analysis of Neisseria gonorrhoeae. J Infect Dis. 1995;171:455–458. doi: 10.1093/infdis/171.2.455. [DOI] [PubMed] [Google Scholar]
- 32.Xia M, Whittington W L, Holmes K L, Roberts M C. Genomic homogeneity of the AHU/IA-1,2 phenotype of Neisseria gonorrhoeae during its elimination from an urban population. Sex Transm Dis. 1997;24:561–566. doi: 10.1097/00007435-199711000-00002. [DOI] [PubMed] [Google Scholar]
- 33.Young H, Moyes A, McMillan A. Azithromycin and erythromycin resistant Neisseria gonorrhoeae following treatment with azithromycin. Int J Sex Transm Dis AIDS. 1997;8:299–302. doi: 10.1258/0956462971920127. [DOI] [PubMed] [Google Scholar]
- 34.Zhou D, Apicella M A. Plasmids with erythromycin resistance and catechol 2,3-dioxygenase- or β-galactosidase-encoding gene cassettes for use in Neisseria spp. Gene. 1996;171:133–134. doi: 10.1016/0378-1119(96)00103-5. [DOI] [PubMed] [Google Scholar]