Abstract
We report on the first Polish patient diagnosed with the Aicardi-Goutières syndrome 5 (AGS5). AGS is caused by mutations in one of 9 genes (TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, ADAR, IFIH, LSM11, RNU7-1) which stimulate the type I interferon response. The diagnosis was confirmed by identifying a compound heterozygous mutation p.(Phe165Ser)/p.(Gln235*) in the SAMHD1 gene using whole-exome sequencing. The cystic lesions in the temporal lobes are an uncommon finding in the presented patient carrying a SAMHD1 mutation. Reporting new cases expands the range of phenotypes and plays the crucial role in understanding the AGS pathogenesis and creates new therapy approaches.
Keywords: Aicardi-Goutières syndrome, SAMHD1, Interferonopathy, Temporal white matter cysts, Magnetic resonance imaging
Established Facts
In Aicardi-Goutières syndrome presenting with deep white matter cysts, early-onset of the disease and the severe clinical picture are most often associated with TREX1 mutations, but the syndrome can also be associated with other mutated genes.
Novel Insights
A new variant in the SAMHD1 gene causes an uncommon neonatal onset of Aicardi- Goutières syndrome with deep white matter cysts.
Introduction
Aicardi Goutières syndrome (AGS) is a rare genetic disease with significant variability in the phenotypes, caused by mutations in any of 9 genes (TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, ADAR, IFIH, LSM11, and RNU7-1) [Crow et al., 2015; La Piana et al., 2016; Uggenti et al., 2020]. The mutations in the AGS genes cause an impairment in protein involved in nucleic acid metabolism or sensing and stimulate the type I interferon (IFN) response. As a result, an IFN signature (presence of INF-stimulated genes) is seen in the AGS patients' blood [Crow et al., 2015].
Early-onset encephalopathy, calcifications in the basal ganglia, white matter abnormalities, brain atrophy, pleocytosis, and elevated IFN alfa in the cerebrospinal fluid and in serum are characteristic features of AGS [Crow et al., 2015; La Piana et al., 2016]. AGS can have its onset prenatally, mimicking a congenital infection with a neurological dysfunction or, more often, it occurs in the first months of life as emerging encephalopathy [Crow et al., 2015]. Children with later-onset AGS usually show first symptoms after a period of a normal development and frequently have recurrent fevers without an infectious trigger. Most patients subsequently develop a progressive microcephaly and they lose acquired skills. Other observed features include seizures, chilblains, glaucoma, and hypothyroidism [Crow et al., 2015]. Vascular anomalies such as dysplastic vessels, intracranial aneurysms, or an inflammatory intracerebral large vessel disease are seen particularly in patients with SAMHD1 mutations [Ramesh et al., 2010]. The advent of new testing methods such as whole-exome sequencing (WES) and wide access to the magnetic resonance imaging (MRI) broadened AGS phenotypic diversity. The clinical history and steps of diagnostic investigations of the first patient in Poland with confirmed AGS due to an SAMHD1 mutation are described.
Case Report
The patient was born at term in good condition with Apgar score 10. He has healthy, unrelated parents. In the neonatal period, there were feeding problems, resulting in poor weight gain and growth. Since the sixth week, he started to be very irritable, crying, and he had an increased muscle tone. At that time, transfontanelle ultrasonography revealed increased periventricular echogenicity and calcifications in the right frontal lobe, confirmed by the computer tomography (CT) scan. TORCH screen was negative. At the age of 3 months, an axial hypotonia with limb hypertonia was present.
Brain MRI performed 3 months later revealed increased subcortical signal intensity mostly involving the frontal and temporal lobes.
At the age of 2.5 years, repeated brain MRI confirmed previously described changes, suggesting congenital TORCH infection or a metabolic disease with leukoencephalopathy and temporal cysts. The results of the broad metabolic analysis including urine organic acids, serum amino acids and very long chain fatty acids, lysosomal enzymes, tests for the congenital disorders of glycosylation as well as aCGH were all normal.
The patient was admitted to the child neurology ward in the Mother and Child Institute in Warsaw at the age of 4 years with lack of psychomotor development and suspicion of metabolic disease. Neurological examination showed he was profoundly disabled with mixed pyramidal and extrapyramidal syndrome and acquired microcephaly. Skin and oral mucosa inspection did not show chilblains or ulcers. Apart from nystagmus, the ophthalmologic examination revealed no other abnormalities, including no glaucoma. Neuropsychologically, intellectual disability was detected. He was classified in GMFCS V, MACS V and CFCS V.
According to the patient's history, clinical picture, MRI and CT scans, we first suspected nonprogressive leukoencephalopathy with temporal cysts.
Materials and Methods
WES was performed on the proband. Library was constructed using SureSelectXT Human All Exon v5 Kit (Agilent, Cedar Creek, TX, USA) according to the manufacturer's instructions and paired-end sequence (2 × 100 bp) on HiSeq 1500 (Illumina, San Diego, CA, USA) to the mean coverage of 76x (93% of targeted sequence was covered at least 20×, 97% was covered at least 10×). Bioinformatics analysis of WES data was performed as previously described [Rydzanicz et al., 2019]. Identified variants were annotated with functional information, frequency in population (including gnomAD (http://gnomad.broadinstitute.org/), and an in-house database of >3,500 Polish exomes), and known association with clinical phenotypes, based on both ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) and HGMD (http://www.hgmd.cf.ac.uk) databases. In silico pathogenicity prediction was performed based on Varsome pathogenicity and conservation scores [Kopanos et al., 2019], and by MetaSVM [Dong et al., 2015]. Variants passing a default quality were further filtered to include only those with <0.001 minor allele frequency in all tested databases and the predicted effect on the protein sequence or affecting splicing. The final list of variants was screened against known pathogenic mutations listed in ClinVar and HGMD databases, and then searched for biallelic mutations consistent with autosomal recessive inheritance and monoallelic variants potentially causative of an autosomal dominant inheritance. All prioritized variants were manually inspected with Integrative Genomics Viewer.
Variants considered as disease causing were validated in the proband and studied in proband's healthy parents by amplicon deep sequencing performed using Nextera XT Kit (Illumina) and sequenced on HiSeq 1500 (Illumina).
Results of Whole-Exome Sequencing
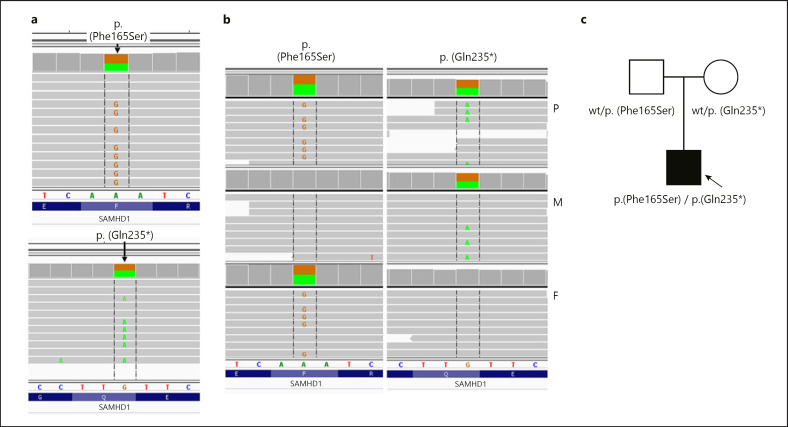
In the proband, we identified a compound heterozygous mutation (Hg19: chr20:035563447-A>G, NM_015474.3:p.(Phe165Ser)/c.494T>C/Hg19: chr20:035547916-G>A, NM_015474.3: p.(Gln235*)/c.703C>T) in the SAMHD1 gene. Missense p.(Phe165Ser) variant has a 0.000006574 frequency in the gnomAD database v 3.1.1 (http://gnomad.broadinstitute.org, access 2021-07-19), is absent from an in-house database of >3,500 Polish exomes, and was predicted as deleterious based on MetaSVM pathogenicity predictor [Dong et al., 2015]. Nonsense p.(Gln235*) variant 0.00001315 in the gnomAD database is absent from the in-house database and was predicted to be pathogenic by MetaSVM. Family study revealed that the mother is a heterozygous carrier of the p.(Gln235*) variant, while the father is a heterozygous carrier of the p.(Phe165Ser) variant (shown in Fig. 1).
Fig. 1.
Genetic study. a Compound heterozygosity in SAMHD1 of p.(Phe165Ser)/p.(Gln235*) identified in the proband by whole-exome sequencing (Integrative Genomic Viewer screen shot) b Verified by amplicon deep sequencing in the proband (P), proband's mother (M) and father (F). c Pedigree of examined family with phenotype/genotype information; circle represents female, squares indicate males, filled symbol shows affected individual. Arrow indicates proband. wt; wild type.
Discussion and Conclusion
We report a patient with confirmed AGS due to the mutation in the SAMHD1 gene with characteristic features of the AGS5 (MIM # 612952, https://www.omim.org) and with an uncommon clinical finding − temporal cysts (Fig. 2, 3). Both variants identified in our patient, p.(Phe165Ser) and p.(Gln235*), were not previously reported to be associated with AGS5. However, the observed low population frequency and in silico predicted pathogenicity support their role in AGS development in the proband.
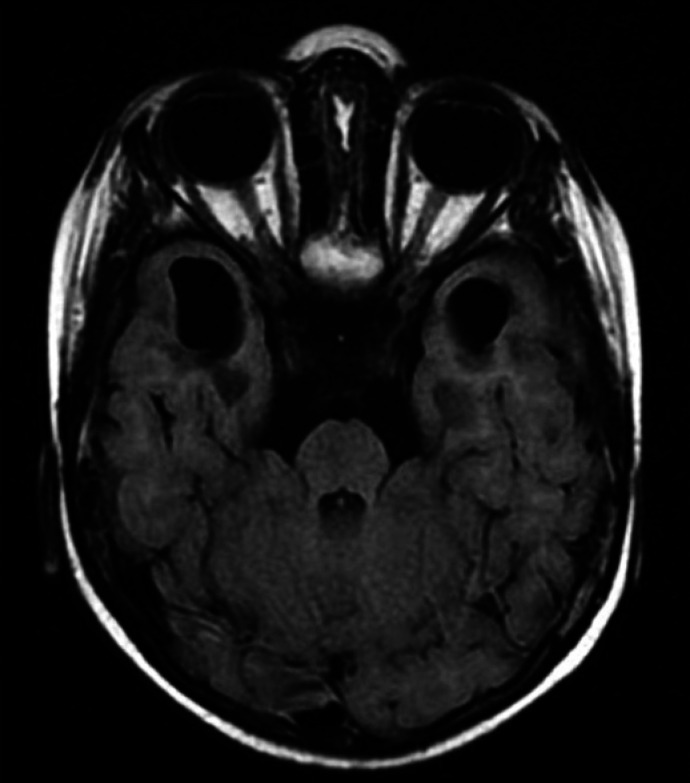
Fig. 2.
Magnetic resonance T2 flair image showing temporal cysts.
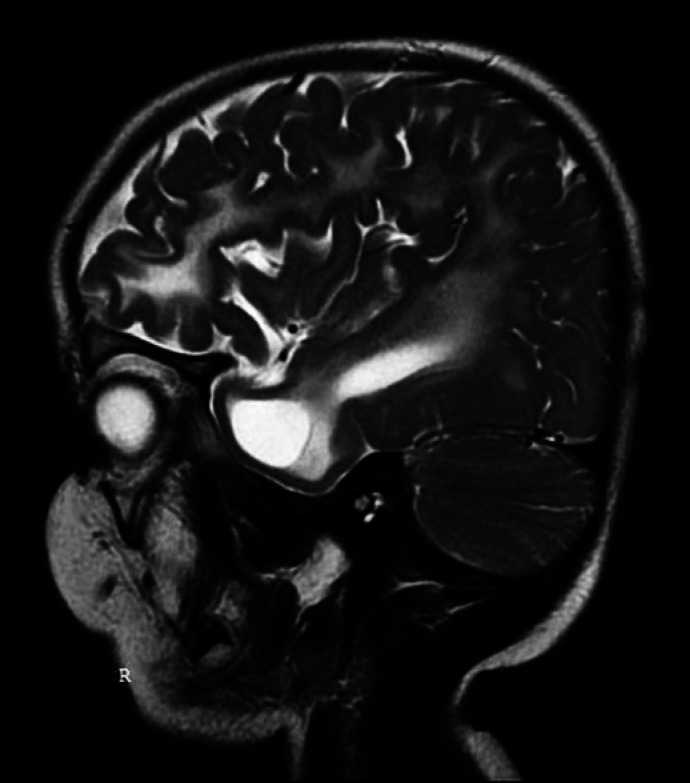
Fig. 3.
Magnetic resonance T2 image showing temporal cysts.
The p.(Phe165Ser) is present in ClinVar database being described as variant with uncertain clinical significance (https://www.ncbi.nlm.nih.gov/clinvar). We noted that in close proximity of p.(Phe165Ser), there are 2 variants with known pathogenicity − nonsense p.(Arg164*) [Dale et al., 2010] and missense p.(His167Tyr) [Ramantani et al., 2010]. All 3 variants, p.(His167Tyr), p.(Arg164*), and p.(Phe165Ser) identified in our proband, are located within a deoxyribonucleoside triphosphate triphosphohydrolase (dNTPase) catalytic core domain (encompassing amino acid residues 115–599), which has dNTPase activity, single-stranded DNA/RNA-binding activity, and exonuclease activity [Ahn, 2016]. The His167 is a part of Histidine-Aspartate residues quartet (His167-His206-Asp207-Asp311) responsible for metal ions chelating in order to correctly orient and polarize the dNTP substrate in the catalytic sites of each monomer of SMHD1 [Morris and Taylor, 2019]. The p.(His167Tyr) variant was shown to lose both oligomerization properties and the ability to block HIV-1 infection when compared to the wild-type SAMHD1 protein [White et al., 2017]. On the other hand, side chain of Arg164 residue stabilized nucleotide binding in the catalytic site [Morris and Taylor, 2019]. Variant (Arg164*) was shown to abolish nucleic acid-binding activity of SMHD1 [Dale et al., 2010]. Due to the fact that 2 functional residues located in close proximity to each other (Arg164, His167), and to Phe165 mutated in our patient, play different roles in regulation of SAMHD1 protein activity, it is hard to predict the role of Phe165 without functional studies.
The nonsense p.(Gln235*) variant in the SAMHD1 gene results in a premature termination codon (PTC). PTC-bearing transcripts may produce truncated proteins or may be degraded by an mRNA surveillance pathway termed nonsense-mediated mRNA decay [Supek et al., 2021]. p.(Gln235*) is predicted to result in premature termination of translation and cause loss of the full-length protein (627 amino acids) due to truncation after the first 235 residues. However, MutationTaster algorithm [Schwarz et al., 2014] suggests that impaired by p.(Gln235*), the SAMHD1 transcript is degraded by nonsense-mediated mRNA decay. Notably, all reported nonsense variants in the SAMHD1 gene are classified as pathogenic in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar).
Our patient, born in a good condition, demonstrated mild features early in the neonatal period resembling congenital infection. Neonatal presentation of AGS is most frequently associated with TREX1 mutations [Crow et al., 2015] and shares similarities with in-utero acquired infection.
Most AGS patients in the long-term follow-up are intellectually and physically disabled [Crow et al., 2015]; however, there are children described in the literature, carrying SAMHD1 mutations, who were mildly intellectually impaired or even retained normal intellect [Dale et al., 2010]. Our patient with neonatal-onset AGS due to a mutation in SAMHD1, is severely handicapped, has spastic quadriplegia with an extrapyramidal component, and acquired microcephaly.
It is worth mentioning that our patient did not experience any of the features associated particularly with an SAMHD1 mutation, such as chilblains, intracerebral large artery disease [Ramesh et al., 2010; Crow et al., 2015], glaucoma [Ramesh et al., 2010], or mouth ulcers [Dale et al., 2010]. Big vascular anomalies were not found in the MRI of our patient. Nevertheless, patients carrying SAMHD1 mutations should be regularly screened for intracranial arteriopathy [Ramesh et al., 2010]. Angio-MRI was not performed in our patient, who was discharged from the hospital before the results of the genetic test were available. Due to the patient's general condition and the risk of additional general anesthesia, we retained from performing angio-MRI afterwards.
The typical neuroradiological findings in AGS include brain calcifications, most often localized in basal ganglia, periventricular regions, and deep white matter [La Piana et al., 2016]. Moreover, 2 main patterns of leukoencephalopathy predominate, namely affecting frontotemporal regions or of a diffuse type. The third pattern resembles periventricular leukomalacia [La Piana et al., 2016]. Another radiologic hallmark of AGS is a frontotemporal white matter rarefaction. The cysts in the deep white matter were found most commonly, but not exclusively, in patients carrying a TREX1 mutation, who usually present with early-onset AGS. The cystic lesions constitute a sequela of the destructive effect of IFN on the immature brain in utero [Vanderver et al., 2015; La Piana et al., 2016]. The case of our patient conforms to this view as the imaging techniques revealed, except for calcifications and leukoencephalopathy in the frontal and temporal lobes, also the temporal cysts - the finding characteristic for neonatal-onset AGS.
There is a large diversity in AGS phenotypes. Some AGS features are a consequence of the time of disease onset and result from the impact of the pathological process on the developing organism during a vulnerable period. However, some AGS features, listed in Table 1, can be helpful to distinguish between molecular subtypes of AGS. A neonatal onset and a severe disease course, as well as a severe neuroradiological picture, should raise suspicion of a TREX1 mutation. Chilblains, glaucoma, mouth ulcers, arthropathy, and vascular lesions are particularly found in patients carrying an SAMHD1 mutation. Milder phenotypes and later disease onset are typical for mutations in ADAR and IFIH1 [Crow et al., 2015].
Table 1.
Shared and distinguishing features of AGS patients with mutations in different genes
| Shared features | Distinguishing features | Gene |
|---|---|---|
| Brain calcifications Leukoencephalopathy Cerebral atrophy White matter rarefaction Severe neurological dysfunction Hypothyroidism Chilblainsa Glaucomaa Demyelinating peripheral neuropathy |
Bilateral striatal necrosis, striatal calcification, severe dystonia | ADAR |
|
|
||
| Neonatal onset of the diseaseb, deep white matter cystsc, infantile onset hypertrophic cardiomyopathyb | TREX1 | |
|
|
||
| Vascular lesions, intracerebral large vessel disease, mouth ulcers, arthropathy | SAMHD1 | |
|
|
||
| Disease onset after 1 year of life, period of normal development prior to disease onset, systemic lupus erythematosus | ADAR, IFIH1 | |
|
|
||
| Pure spastic paraparesis, normal neuroimaging or nonspecific changes in white matter, preserved intellect | RNASEH2B, SAMHD1, IFIH1, ADAR | |
|
|
||
| Preserved limited motor or communication function | RNASEH2B, SAMHD1, ADAR, IFIH1 | |
|
|
||
| Clinically asymptomatic in late adulthood | IFIH1 | |
|
| ||
| Other rare features found in patients with AGS Gastrointestinal inflammatory disease, central diabetes insipidus, diabetes mellitus, hyperparathyroidism, growth hormone deficiency, adrenal insufficiency, antiphospholipid syndrome, panniculitis | ||
Especially SAMHD1.
Other gene mutations are also possible.
Can also be present in patients with other mutations, probably a consequence of a prenatal disease onset.
AGS is a rare and underdiagnosed disease; therefore, the phenotype characteristics might still remain incomplete. One should keep the possibility of an atypical presentation in mind, so that the occurrence of any of the listed features in any constellation should raise a suspicion of AGS.
The diagnosis of our patient could have been made in the first months of life due to the characteristic features, mimicking congenital infection.
As AGS can imitate many diseases, it should be included in the differential diagnosis of the conditions such as congenital infections, including TORCH infection, neuroinfection, autoimmune diseases, unexplained cases labelled “spastic cerebral palsy” and periventricular leukomalacia, in case of normal perinatal history. Also genetic disorders affecting the white matter share similarities with AGS, namely severe forms of Alexander disease, megalencephalic leukoencephalopathy with subcortical cysts, vanishing white matter disease, leukoencephalopathy with calcifications and cysts, RNaseT2-related disease, and mitochondrial leukoencephalopathies [Vanderver et al., 2015].
First therapy approaches with immune-modulating agents such as corticosteroids, intravenous immunoglobulins or azathioprine did not bring satisfactory results. The advances in understanding the AGS pathogenesis have opened the way to novel therapies. These include reverse transcriptase inhibitors (RTIs), Janus kinase inhibitors, anti-IFN-alpha antibodies and anti-type I IFN receptor antibodies, anti-interleukin antibodies, cGAS inhibitors such as antimalarial drugs, small molecule inhibitors, suppressive oligodeoxynucleotides, and acetylsalicylic acid. Currently Janus kinase inhibitors influencing various cytokine pathways and RTIs affecting IFN production constitute the most promising therapy options [Tonduti et al., 2020].
Determining the final diagnosis relieves the patient from the large number of diagnostic procedures searching for the cause of the condition, provides families with information about family planning and possible hereditary risks, and finally, the patients can be involved in the research of new treatment strategies.
Statement of Ethics
Written informed consent for performing genetic tests and other medical diagnostic procedures as well as publication of this case report, including any accompanying images was obtained from the patient's guardians. Ethics committee approval was not required for performing these tests, which were a part of the diagnostic investigation in search of the cause of the disease.
Conflict of Interest Statement
The authors have no conflicts of interest to declare.
Funding Sources
The authors have no funding sources to declare.
Author Contributions
Barbara Oleksy created the concept and design of the case report, took the lead in writing the manuscript.
Hanna Mierzewska and Elżbieta Szczepanik were involved in planning and supervised the work, revised the case report.
Jolanta Tryfon performed the neuropsychological assessment of the patient.
Maria Wypchło, Krystyna Wasilewska, and Małgorzata Rydzanicz carried out the genetic examination using WES.
Małgorzata Rydzanicz and Rafał Płoski performed the interpretation of the results.
Małgorzata Rydzanicz wrote the genetic part of the paper.
Zofia Zalewska-Miszkurka was the main doctor, who took care of the patient and collected the data.
Data Availability Statement
All data generated or analyzed during this study are included in this article. Further enquiries can be directed to the corresponding author.
Acknowledgement
DNA sequencing was carried out with the use of CePT infrastructure (Innovative economy 2007–13, Agreement POIG.02.02.00-14-024/08-00).
References
- Ahn J. Functional organization of human SAMHD1 and mechanisms of HIV-1 restriction. Biol Chem. 2016;397:373–9. doi: 10.1515/hsz-2015-0260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow YJ, Chase DS, Lowenstein Schmidt J, Szynkiewicz M, Forte GM, Gornall HL, et al. Characterization of human disease phenotypes associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, ADAR, and IFIH1. Am J Med Genet A. 2015;167A:296–312. doi: 10.1002/ajmg.a.36887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale RC, Gornall H, Singh-Grewal D, Alcausin M, Rice GI, Crow YJ. Familial Aicardi-Goutières syndrome due to SAMHD1 mutations is associated with chronic arthropathy and contractures. Am J Med Genet A. 2010;152A:938–42. doi: 10.1002/ajmg.a.33359. [DOI] [PubMed] [Google Scholar]
- Dong C, Wei P, Jian X, Gibbs R, Boerwinkle E, Wang K, et al. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum Mol Genet. 2015;24:2125–37. doi: 10.1093/hmg/ddu733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopanos C, Tsiolkas V, Kouris A, Chapple CE, Albarca Aguilera M, Meyer R, et al. VarSome: the human genomic variant search engine. Bioinformatics. 2019;35:1978–80. doi: 10.1093/bioinformatics/bty897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Piana R, Uggetti C, Roncarolo F, Vanderver A, Olivieri I, Tonduti D, et al. Neuroradiologic patterns and novel imaging findings in Aicardi-Goutières syndrome. Neurology. 2016;86:28–35. doi: 10.1212/WNL.0000000000002228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris ER, Taylor IA. The missing link: allostery and catalysis in the anti-viral protein SAMHD1. Biochem Soc Trans. 2019;47:1013–27. doi: 10.1042/BST20180348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramantani G, Kohlhase J, Hertzberg C, Innes AM, Engel K, Hunger S, et al. Expanding the phenotypic spectrum of lupus erythematosus in Aicardi-Goutières syndrome. Arthritis Rheum. 2010;62:1469–77. doi: 10.1002/art.27367. [DOI] [PubMed] [Google Scholar]
- Ramesh V, Bernardi B, Stafa A, Garone C, Franzoni E, Abinun M, et al. Intracerebral large artery disease in Aicardi-Goutières syndrome implicates SAMHD1 in vascular homeostasis. Dev Med Child Neurol. 2010;52:725–32. doi: 10.1111/j.1469-8749.2010.03727.x. [DOI] [PubMed] [Google Scholar]
- Rydzanicz M, Wachowska M, Cook EC, Lisowski P, Kuźniewska B, Szymańska K, et al. Novel calcineurin A (PPP3CA) variant associated with epilepsy, constitutive enzyme activation and downregulation of protein expression. Eur J Hum Genet. 2019;27:61–9. doi: 10.1038/s41431-018-0254-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods. 2014;11:361–2. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- Supek F, Lehner B, Lindeboom RGH. To NMD or Not To NMD: Nonsense-Mediated mRNA Decay in Cancer and Other Genetic Diseases. Trends Genet. 2021;37((7)):657–68. doi: 10.1016/j.tig.2020.11.002. [DOI] [PubMed] [Google Scholar]
- Tonduti D, Fazzi E, Badolato R, Orcesi S. Novel and emerging treatments for Aicardi-Goutières syndrome. Expert Rev Clin Immunol. 2020;16:189–98. doi: 10.1080/1744666X.2019.1707663. [DOI] [PubMed] [Google Scholar]
- Uggenti C, Lepelley A, Depp M, Badrock AP, Rodero MP, El-Daher MT, et al. cGAS-mediated induction of type I interferon due to inborn errors of histone pre-mRNA processing. Nat Genet. 2020;52:1364–72. doi: 10.1038/s41588-020-00737-3. [DOI] [PubMed] [Google Scholar]
- Vanderver A, Prust M, Kadom N, Demarest S, Crow YJ, Helman G, et al. Early-Onset Aicardi-Goutières Syndrome: Magnetic Resonance Imaging (MRI) Pattern Recognition. J Child Neurol. 2015;30:1343–8. doi: 10.1177/0883073814562252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TE, Brandariz-Nuñez A, Martinez-Lopez A, Knowlton C, Lenzi G, Kim B, et al. A SAMHD1 mutation associated with Aicardi-Goutières syndrome uncouples the ability of SAMHD1 to restrict HIV-1 from its ability to downmodulate type I interferon in humans. Hum Mutat. 2017;38:658–68. doi: 10.1002/humu.23201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this article. Further enquiries can be directed to the corresponding author.